SUMMARY
DNA methylation and H3K9me are hallmarks of heterochromatin in plants and mammals and are successfully maintained across generations. The biochemical and structural basis for this maintenance is poorly understood. The maintenance DNA methyltransferase from Zea mays, ZMET2, recognizes dimethylation of H3K9 via a chromodomain (CD) and a BAH domain, which flank the catalytic domain. Here, we show that dinucleosomes are the preferred ZMET2 substrate, with DNA methylation preferentially targeted to linker DNA. Electron microscopy shows one ZMET2 molecule bridging two nucleosomes within a dinucleosome. We find that the CD stabilizes binding, while the BAH domain enables allosteric activation by the H3K9me mark. ZMET2 further couples recognition of H3K9me to an increase in the specificity for hemimethylated versus unmethylated DNA. We propose a model in which synergistic coupling between recognition of nucleosome spacing, H3K9 methylation and DNA modification allows ZMET2 to maintain DNA methylation in heterochromatin with high fidelity.
INTRODUCTION
In mammals and plants, DNA methylation is associated with silenced regions of the genome known as heterochromatin. In mammals, disrupted DNA methylation is associated with various developmental defects and cancers (Kulis and Esteller, 2010; Messerschmidt et al., 2014; Robertson, 2005). In plants, aberrant DNA methylation patterns can lead to severe developmental phenotypes (Jacobsen and Meyerowitz, 1997; Lindroth, 2001). For DNA methylation to be effective, it must be maintained across multiple generations (Edwards et al., 2017; Law and Jacobsen, 2010; Torres and Fujimori, 2015; Zhang et al., 2018a). The biochemical and structural basis for how such DNA methylation is faithfully reproduced within the appropriate chromatin context is poorly understood.
In mammals, three DNA methyltransferase (MTase) enzymes, DNMT1, DNMT3A and DNMT3B, promote silencing of repetitive and transposable elements and methylate CG sites in the promoters of inactive genes (Smith and Meissner, 2013). DNMT1 is thought to maintain DNA methylation during DNA replication, whereas DNMT3A and DNMT3B are primarily de novo MTase enzymes. In Arabidopsis thaliana, four active DNA MTase enzymes have been identified: DRM2, CMT2, CMT3 and MET1 (Du et al., 2015). The focus of this study, ZMET2, is an ortholog of CMT3 from Zea mays (maize) and, like CMT3, is thought to primarily methylate CHG sites (where H = A, T, or C) (Bartee et al., 2001; Du et al., 2012; Lindroth, 2001; Papa et al., 2001). Both DNMT1 and CMT3/ZMET2 are classified as maintenance MTases based on their co-localization with replication machinery in vivo and, in the case of DNMT1, preference for hemimethylated DNA in vitro. While DNMT1 has been shown to have a preference for hemimethylated DNA, the relative activity of ZMET2 on hemimethylated versus unmethylated DNA substrates has not been reported (Du et al., 2012; Jeltsch and Jurkowska, 2016).
Direct coupling between DNA MTase enzymes and posttranslational modifications on histone proteins has been observed in mammals. For example, DNMT3A recognizes unmodified H3 through an ATRX-DNMT3-DNMT3L (ADD) domain, and its activity is inhibited by methylation of H3 at K4 (Li et al., 2011; Otani et al., 2009; Zhang et al., 2010). Full activity requires the formation of a complex of two DNMT3A molecules with two accessory DNMT3L molecules. The resultant tetramer allows for multivalent histone recognition, in which all four subunits make contacts with the unmodified H3 tails on nucleosomes (Jia et al., 2007). This type of multivalent histone recognition likely serves to promote fidelity and specificity for DNA MTase activity, as DNA methylation will depend not only on the successful formation of the tetrameric complex, but also successful recognition of the H3 tail by each subunit. Crystal structures of the tetramer formed by the C-terminal domains of DNMT3A and DNMT3L suggests models in which the complex bridges across nucleosomes to target DNA, but these structures do not include nucleosomes (Jia et al., 2007; Zhang et al., 2018b).
In plants, the first example of positive feedback between DNA methylation and histone methylation came from studies of CMT3 and the histone H3 MTase KRYPTONITE (KYP). Mutation of KYP was found to reduce CHG methylation, while deletion of CMT3 led to reduced levels of H3K9me (Jackson et al., 2002; Malagnac et al., 2002; Mathieu et al., 2005; Soppe et al., 2002; Tariq et al., 2003). H3K9me and CHG methylation were also shown to be highly correlated throughout the Arabidopsis genome (Bernatavichute et al., 2008). This positive reinforcement was found to be driven in part by the KYP SRA domain which recognizes methylated CHG sites and a SET domain which deposits H3K9me (Du et al., 2014). Conversely, CMT3 contains a chromodomain (CD) with a canonical aromatic cage that specifically binds H3K9me. In addition to CMT3’s H3K9me recognition via its CD, it was discovered that CMT3’s BAH domain is also competent to bind H3K9me (Du et al., 2012). Crystal structures of an N-terminal truncation of ZMET2 demonstrate recognition of methylated H3 tail peptides by either the CD or the BAH domain (Du et al., 2012). Thus CMT3 and ZMET2 appear to have bypassed the necessity for adaptor proteins or heterodimerization to engage in multivalent histone tail recognition. How CMT3 and ZMET2 harness this “all-in-one” architecture to methylate target DNA sequences with high fidelity in the context of chromatin remains an open question.
To investigate the mechanism by which ZMET2 interrogates chromatin context to ensure specificity and to understand the implications of its dual H3K9me binding capabilities, we carried out enzymatic and structural studies on ZMET2. Our results explain how a DNA MTase structurally and biochemically integrates multiple substrate cues including 3-dimensional chromatin architecture to methylate hemimethylated CHG sites and reinforce heterochromatic regions of the genome.
RESULTS
ZMET2 activity is preferentially stimulated in an H3K9me-specific manner on dinucleosomes over mononucleosomes
Previous work describing ZMET2’s dual H3K9me-binding capability via the CD and BAH raises the possibility that ZMET2 forms a 1:1 complex with a nucleosome, whereby one molecule of ZMET2 engages with both H3 tails on a single nucleosome (Du et al., 2012). Alternatively, it is possible that ZMET2 bridges across two nucleosomes, engaging one tail on each adjacent mononucleosome. Both models invite interesting hypotheses for where on chromatin ZMET2 may target DNA for methylation, and how ZMET2 may help to reinforce specific chromatin states.
To distinguish between these two possibilities, we sought to measure DNA MTase activity on various mono- and dinucleosome substrates using full-length recombinant ZMET2 (Figures 1A and S1A). Because CHG DNA methylation and H3K9me are mutually reinforcing in vivo, and ZMET2 binds specifically to H3K9me2 peptides in vitro, we tested both unmodified nucleosomes and nucleosomes modified using methyl-lysine analog (MLA) technology (denoted as H3Kc9me3) (Simon et al., 2007). To test the suitability of the MLA approach, we first compared binding of ZMET2 to native and MLA peptides. ZMET2 binds an H3Kc9me3 tail peptide ~3.5-fold more weakly than a peptide containing native H3K9me3, but the H3Kc9me3 tail affinity is still greater than that of an unmodified peptide, suggesting that the MLA is a reasonable mimic of native H3K9 methylation (Figure S1B). Further, consistent with previous work on an N-terminally truncated ZMET2, the affinity of ZMET2 for H3K9me2 and H3K9me3 tail peptides is comparable (Figure S1B, 1.4 μM and 1.9 μM, respectively), suggesting similar specificities for di- and trimethylated states of H3K9 (Du et al., 2012). Finally, ZMET2 binds H3Kc9me3 mononucleosomes at least 16-fold more strongly than WT H3K9 mononucleosomes demonstrating specificity for the H3Kc9 methyl mark in the context of a nucleosome (Figure S1C).
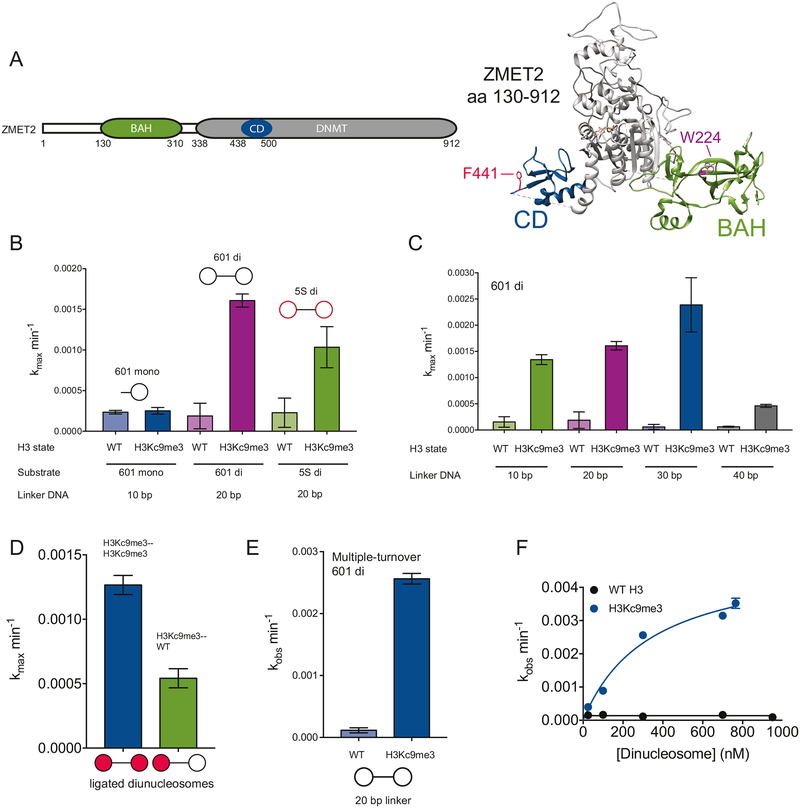
Figure 1. ZMET2 methylates H3Kc9me dinucleosomes faster than mononucleosomes.
(A) Schematic of full-length ZMET2 domain architecture and crystal structure of truncated ZMET2 (130–912) with bound SAH (PDB 4FSX). Chromodomain is in blue with aromatic cage residue F441 in red, bromo adjacent homology (BAH) domain in green with aromatic cage residue W224 in purple.
(B) Activity of ZMET2 for different nucleosomal substrates shown as schematics above bars. All reactions were carried out under single-turnover conditions in which [ZMET2] was in excess and saturating over nucleosomes. Experiments with 601 dinucleosomes (20 bp) were performed in quadruplicate (n=4). Experiments with 601 mononucleosome and 5S dinucleosome were performed in duplicate (n=2).
(C) Dinucleosome linker length dependence for ZMET2 activity under single-turnover conditions ([ZMET2] was in excess and saturating over dinucleosomes). For ease of comparison, kcat measurements for dinucleosomes with the 20 bp linker are re-plotted from (A) next to the experiments with dinucleosomes containing 10 bp and 30 bp linkers (n=3) and 40 bp linkers (n=2).
(D) DNA methyltransferase activity for WT ZMET2 on ligated nucleosomes with symmetric (two red circles in schematic) or asymmetric (one red, one white circle in schematic) H3Kc9me3 marks (n=6). Reactions were conducted under the conditions as in (C).
(E) Activity of ZMET2 for 601 dinucleosomes with 20 bp linkers under multiple-turnover conditions in which the concentration of H3Kc9me3 dinucleosomes is in excess and saturating over the concentration of ZMET2.
(F) Dependence of ZMET2 activity on concentration of 601 dinucleosomes with 20 bp linkers. The Km for H3Kc9me3 dinucleosomes is 340 nM. The Km for WT dinucleosomes is not reliably measurable due to the low activity (n=2 for all experiments).
We next focused on the role of nucleosome architecture in ZMET2’s DNA methylation reaction post-binding. To ensure a post-binding state, DNA methylation was measured under single-turnover conditions, with saturating concentrations of ZMET2 in excess of nucleosomes (see Methods). The rate constant measured under saturating conditions was termed kmax. We found that H3Kc9me3 dinucleosomes were methylated with a kmax of 0.0016 min−1 (Figures 1B, S1D and S2B). In contrast, H3Kc9me3 mononucleosomes generated with 10 base pairs (bp) of linker DNA to recapitulate half of the dinucleosome substrate showed a kmax of 0.00025 min−1, ~ 6-fold lower than the rate constant measured for H3Kc9me3 dinucleosomes. Additionally, ZMET2 demonstrated 8-fold higher activity on H3Kc9me3 vs. H3K9 dinucleosomes, while higher activity was not observed on H3Kc9me3 vs. H3K9 mononucleosomes (Figures 1B, S1D and S2A). These results suggest that, for activity, dinucleosomes are the preferred substrate over mononucleosomes and that ZMET2 may bridge across the nucleosomes within a dinucleosome. While monononucleosomes can be methylated at a low level, the lack of H3Kc9me3 specificity suggests ZMET2 is bound non-productively to mononucleosomes, while on dinucleosomes it can engage in an optimal orientation for catalytic activity.
We next asked whether DNA sequence could influence ZMET2’s preference for dinucleosomes. We generated mono- and dinucleosomes with the 5S rRNA nucleosome positioning sequence. For the dinucleosomes, the same 20 bp linker sequence present in the 601 dinucleosomes was used. Saturation was ensured as described in the Methods (Figure S2C). We found that kmax for DNA methylation on the 5S dinucleosomes is 2-fold slower compared to the 601 dinucleosomes, suggesting a small but reproducible sequence-dependent effect (Figures 1B and S1D).
Next, we hypothesized that nucleosome bridging behavior would render ZMET2 sensitive to dinucleosome linker length. To test this hypothesis, we generated dinucleosomes with 10, 30 and 40 bp linker lengths. Dinucleosomes with a 10 bp linker were comparable to dinucleosomes with a 20 bp linker with respect to rate constant and specificity for H3Kc9me3, suggesting that ZMET2 is not sensitive to a shorter linker (Figures 1C, S1D and S2D). Dinucleosomes with a 30 bp linker and one CHG site positioned in the center of the linker were methylated modestly faster than dinucleosomes with 10 and 20 bp linkers (each also containing one center-positioned CHG site), suggesting the average nucleosome spacing in A. thaliana (30 bp) may support maximal DNA methylation in plant heterochromatin (Figures 1C and S2E) (Chodavarapu et al., 2010). We generated a variation on the 30 bp linker by positioning CHG sites at either end of the linker to test whether a center-positioned site is favored for ZMET2 activity and found significantly (15-fold) reduced activity on this substrate compared to a 30 bp linker with a central CHG site (Figure S2E). This finding supports a model in which the catalytic domain of a bridging molecule of ZMET2 is more proximal to CHG sites located in the center of a dinucleosome linker. Importantly, the dinucleosomes with a 40 bp linker were methylated ~5-fold more slowly than dinucleosomes with a 30 bp linker suggesting that ZMET2 activity is inhibited when adjacent nucleosomes are more distant from one another than 30 bp (Figure 1C and S2F). We further reasoned that if ZMET2 is maximally activated when bridging across two H3K9 methylated nucleosomes, then its activity would be sub-optimal on dinucleosomes in which only one of the nucleosomes contains the H3Kc9me3 mark. We tested this hypothesis by generating ligated symmetric (H3Kc9me3-H3Kc9me3) and asymmetric (H3Kc9me3-WT) dinucleosomes with 30 bp linkers. Using saturating concentrations of ZMET2, we observed a modest but reproducible 2.3-fold defect (kmax(asymmetric) = 0.00054 ± 7.4×10−5 min−1; kmax(symmetric) = 0.0013 ± 7.4×10−5) on the asymmetric dinucleosomes, consistent with maximal activity requiring H3K9 methylation on both octamers in the dinucleosome (Figure 1D).
ZMET2’s preference for the H3Kc9me3 dinucleosome substrate suggests a model in which the enzyme bridges across the two nucleosomes in order to engage with target CHG sites. This model is consistent with molar mass measurements of ZMET2 demonstrating that ZMET2 is a monomer at the concentrations used in the assays above (Figure S1E). However, an alternative model is that ZMET2 self-associates on dinucleosomes, with one ZMET2 molecule per mononucleosome, and that this self-assembly stimulates activity on the dinucleosome substrate. To distinguish between these models, we measured DNA MTase activity on dinucleosomes under multiple-turnover conditions such that dinucleosome concentration was in large excess over ZMET2 concentration, favoring a single molecule of ZMET2 binding to a dinucleosome. The maximal rate constants for ZMET2 DNA methylation activity under these conditions were similar to those obtained under single-turnover studies, consistent with a model in which a single molecule of ZMET2 engages both nucleosomes within the dinucleosome to achieve maximal activity (Figures 1E and S1F).
Finally, given the stimulatory effect of dinucleosomes on ZMET2 activity post-binding, we wondered if ZMET2 might also associate more tightly with dinucleosomes than mononucleosomes during the binding step of the reaction. To estimate binding to dinucleosomes, we measured the Km using a standard Michaelis-Menten approach (Figure 1F). The Km for ZMET2 on H3Kc9me3 dinucleosomes is 340 nM, which is about 2-fold tighter than the Kd measured for mononucleosomes using fluorescence polarization (Figure S1C). These results suggest that ZMET2 has a larger preference for dinucleosomes over mononucleosomes during catalysis (6-fold, Figure 1B) than during binding (2-fold). These preferences for the individual binding and catalysis steps translate to an overall preference of ~12-fold for methylating H3K9 methylated dinucleosomes over mononucleosomes.
ZMET2 uses the H3K9me mark and DNA hemimethylation for binding as well as activity
Our finding that the H3K9 methyl mark plays a role in activating ZMET2 catalysis post-binding raised the question of whether this mark plays an allosteric role or simply helps anchor ZMET2 to the dinucleosome in an activated intermediate. To test for an allosteric role we asked if the H3K9 methylated tail peptide could stimulate DNA methylation activity when added in trans. We assessed the ability of ZMET2 to methylate a 157 bp fragment of DNA in the presence and absence of the H3K9me2 (1–32) tail. Using saturating concentrations of ZMET2, we found that the H3K9me2 tail added in trans stimulates DNA MTase activity ~90-fold (Figures 2A, 2B S3A). This dramatic stimulation suggests that the H3K9me2 peptide is an allosteric activator of ZMET2 activity. The same stimulation was not observed upon addition of an unmethylated H3 tail peptide, suggesting that the stimulation requires the H3K9me2 mark (Figures 2B and S3A). All reactions were carried out under single-turnover conditions, and conditions in which [ZMET2] was in excess and saturating over [DNA]. Consequently, the results are a measure of how the tail peptide affects ZMET2 catalysis on DNA and not ZMET2 binding to DNA (Figure S3A).
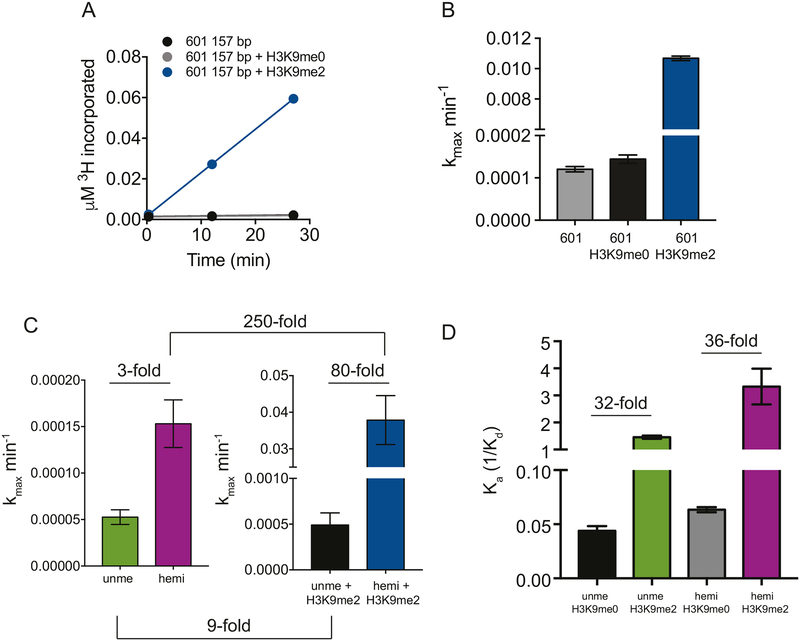
Figure 2. ZMET2 binding and activity are increased by the H3K9me mark and DNA hemimethylation.
(A) Activity of ZMET2 on 157 bp naked DNA with the 601 sequence in the absence and presence of H3 tail peptides. All peptides contain residues 1–32 of histone H3 and were used at 25 μM. Reactions were conducted under single-turnover conditions in which ZMET2 (5 μM) was in excess and saturating over DNA substrates (200 nM).
(B) Rate constants calculated from (A) by dividing initial DNA methylation rates by the concentration of substrate. The values of kmax for no peptide, H3K9me0 and H3K9me2 reactions were 0.00012 (±8.8E-6) min−1, 0.00014 (±1.4E-5) min−1, and 0.011 (±0.0002) min−1, respectively, (n=2 for each measurement).
(C) DNA methyltransferase activity of ZMET2 on unmethylated or hemimethylated 38 bp duplexed DNA in the absence (left panel) or presence (right panel) of H3K9me2 peptide. Reactions were conducted as in (D), except with 300 nM 38 bp duplexes. Rate constants for unmethylated, hemimethylated, unmethylated with H3K9me2 and hemimethylated with H3K9me2 were 5.3E-5 (±1.1E-5) min−1, 0.00015 (±3.6E-5) min−1, 0.00049 (±0.00019) min−1, and 0.038 (±0.0095) min −1, respectively, (n=2 for each measurement).
(D) Affinity of ZMET2 for 38 bp duplexed DNA with and without hemimethylation and either H3K9me0 or H3K9me2 peptides measured by fluorescence polarization. Experiments with H3K9me0 peptide were done in duplicate (n=2), unme + H3K9me2 was done in triplicate (n=3), and hemi + H3K9me2 was done in quadruplicate (n=4).
The activating potential of the H3K9me mark prompted us to investigate how hemimethylation, the other proposed feature of ZMET2’s preferred substrate, as a putative maintenance DNA MTase that is most active during DNA replication, regulates ZMET2 activity. We compared DNA MTase activity on unmethylated and hemimethylated 38 bp DNA duplexes under single-turnover conditions in which ZMET2 was in excess and saturating over DNA (Figure S3B). In the presence of the H3K9me2 (1–32) tail peptide, ZMET2 methylates the hemimethylated DNA substrate ~80-fold faster than the unmethylated DNA substrate. Surprisingly, in the absence of the H3K9me2 tail peptide, the preference for the hemimethylated DNA substrate is only ~3-fold versus the unmethylated substrate, suggesting that H3K9me2 and hemimethylation cooperate to enhance ZMET2 DNA MTase activity (Figures 2C and S3C). Analogous to the experiments in Figures 2A and 2B, the results reflect effects on catalytic activity after the binding step of the reaction. To investigate the potential for cooperative effects in the context of binding, we directly measured binding to 38mer DNA duplexes and found that ZMET2 affinity for both unmethylated and hemimethylated 38mers was dramatically enhanced in the presence of an H3K9me2 (1–32) peptide by 32-fold and 36-fold, respectively (Figures 2D and S3D).
Together, these results suggest that: (1) the H3K9me mark and hemimethylation of DNA cooperatively stimulate ZMET2 activity, and (2) the H3K9me mark increases the affinity of ZMET2 for its DNA substrate.
The ZMET2 CD recognizes H3K9me in the binding step and the BAH domain recognizes H3K9me in the catalytic step of the DNA methylation reaction
Given that both the CD and BAH domain of ZMET2 are capable of interaction with H3K9me2 peptides and our finding that H3K9me2 can allosterically activate ZMET2, we can formulate two models for CD and BAH domain function: (1) Both the CD and BAH domain are involved in H3K9me2 binding and catalytic activation or (2) one domain is a binding module, and the other serves as the allosteric regulatory domain.
To distinguish between these models, we made mutations in the aromatic cages of either the CD (F441A, CDx) or the BAH domain (W224L, BAHx) (Figure 1A, right panel). We then measured the affinity of these mutants for H3K9me0 and H3K9me2 (1–32) peptides and found that binding of the CDx mutant to H3K9me2 peptides is significantly decreased compared to wild type ZMET2 (Figure 3A). In contrast, the BAHx mutant did not show a defect, showing instead modestly tighter binding. Next, we measured ZMET2 affinity for unmodified and H3Kc9me3 mononucleosomes (Figure 3B). Mutating the CD resulted in a 4-fold decrease in affinity for H3Kc9me3 nucleosomes. In contrast, mutating the BAH domain resulted in a modestly tighter affinity for H3Kc9me3 nucleosomes. For all ZMET2 constructs, affinity for unmethylated nucleosomes was very weak, allowing us to obtain only a lower limit for the Kd. The decreased affinity for both H3K9me3 tail peptides and H3Kc9me3 nucleosomes resulting from mutation of the CD aromatic cage but not the BAH aromatic cage suggests that the CD is ZMET2’s primary H3K9me binding module.
Figure 3. The ZMET2 CD recognizes H3K9me in the binding step and the BAH domain recognizes H3K9me in the catalytic step.
(A) WT, F441A and W224L ZMET2 affinity for fluorescein-labeled H3 tail peptides measured by fluorescence polarization. (For these experiments, n=2).
(B) WT, F441A and W224L ZMET2 affinity for WT and H3Kc9me3 mononucleosomes measured by fluorescence polarization. Experiments with WT and F441A (CDx) ZMET2 were performed in duplicate (n=2). Experiments with W224L (BAHx) were performed in quadruplicate (n=4).
(C) DNA methyltransferase activity for WT, F441A and W224L ZMET2 on WT and H3Kc9me3 dinucleosomes. Reactions were conducted under single-turnover conditions in which the concentration of each ZMET2 protein was in excess and saturating over the dinucleosome concentration. (For these experiments, n=2).
We next explored the roles of both domains in catalysis. We carried out DNA MTase assays under single-turnover conditions in which the ZMET2 constructs were in excess and saturating over unmodified and H3Kc9me3 dinucleosomes (Figure S4 A-D). Remarkably, the rate constant for DNA methylation by the CDx mutant was only modestly (1.3-fold) decreased compared to wild type ZMET2, while the rate constant for DNA methylation by the BAHx mutant was decreased ~8-fold compared to wild type ZMET2 (Figure 3C). Further, the same experiment conducted with naked 157 bp DNA and H3K9me0/2 peptides in trans demonstrated an even stronger pattern whereby the rate constant for the CDx mutant was comparable to wild type ZMET2, but the BAHx mutant was impaired 17-fold (Figure S4E). Combined with the binding analysis, these data suggest that the ZMET2 CD is mainly responsible for recognizing H3K9me in the binding step, while the BAH domain recognizes H3K9me during catalysis and promotes H3K9me-induced allosteric activation of ZMET2.
ZMET2 preferentially methylates the linker DNA in H3Kc9me3 dinucleosomes
Our model that ZMET2 bridges across nucleosomes suggests that it targets linker DNA between the mononucleosomes. To test this possibility, we sought to determine ZMET2’s preferred DNA methylation target(s) on chromatin by measuring DNA methylation kinetics on dinucleosome substrates with CHG sites present in both the nucleosomal DNA and the 20 bp linker region. To control for intrinsic differences in methylation due to DNA sequence context, we compared the distribution of DNA methylation on H3Kc9me3 dinucleosomes with that on the corresponding naked dinucleosomal DNA in the presence of an H3K9me2 (1–32) tail peptide. Naked DNA with H3K9me2 peptide and H3Kc9me3 dinucleosomes were incubated with saturating concentrations of ZMET2 and time points were subjected to bisulfite conversion and Illumina sequencing (Figure S5A). The resolution of bisulfite sequencing allowed us to determine kinetic parameters for DNA methylation at each of the CHG sites on the dinucleosome (Figure S5B and S5C).
Our analysis revealed three categories of CHG sites: (1) CCG sites, which were not detectably methylated in either the naked DNA or nucleosomal context, (2) CHG sites that were methylated faster on naked DNA than on dinucleosomes and (3) a single CHG site that was methylated faster on the dinucleosome than on naked DNA (Figure 4A, CTG at position 159 and Figure 4C). The finding that CCG sites are not methylated is consistent with previous in vivo shotgun bisulfite sequencing data (Cokus et al., 2008a; Gruenbaum et al., 1981).
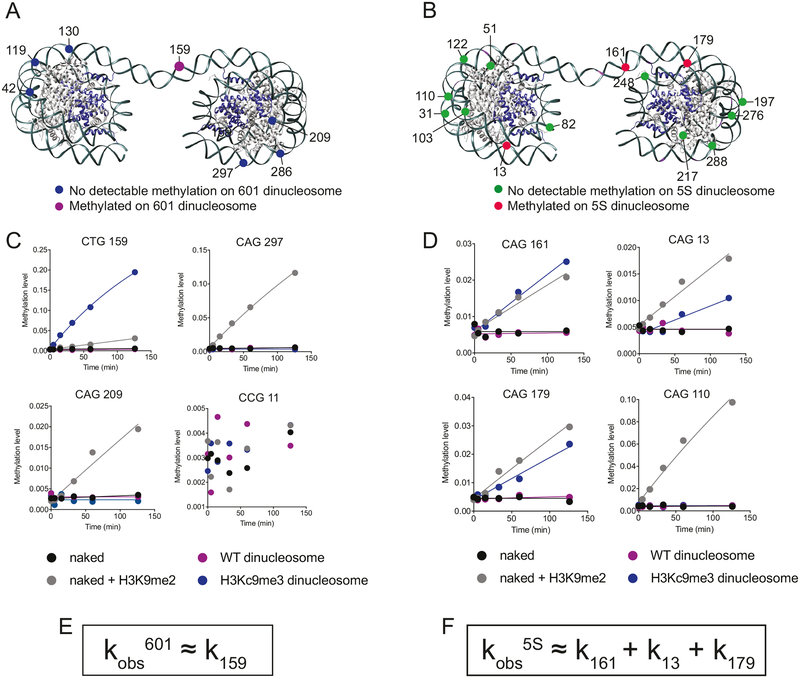
Figure 4. ZMET2 preferentially methylates the linker DNA in H3Kc9me3 dinucleosomes.
(A) Schematic of 601 dinucleosome with CHG sites that are either methylated (purple) or not methylated (blue) on the dinucleosome. Only non-CCG sites are colored. Histone H3 is in dark blue (PDB 1ZBB).
(B) Schematic of 601 dinucleosome, used to represent approximate structure of a dinucleosome assembled with the 5S positioning sequence. CHG sites that are either methylated (red) or not methylated (green) are labeled. Only non-CCG sites are colored. Histone H3 is in dark blue (PDB 1ZBB).
(C) Upper left, bisulfite sequencing time course for ZMET2 activity at 601 CTG 159, the CHG site in the linker region of the dinucleosome, a site where H3Kc9me3 dinucleosomes are methylated faster than naked DNA plus H3K9me2 peptide; upper right, bisulfite sequencing time course for ZMET2 activity at 601 CAG 297, a site where naked DNA plus H3K9me2 peptide is methylated much faster than H3Kc9me3 dinucleosomes; lower left, bisulfite sequencing time course for ZMET2 activity at 601 CAG 209, a site resembling the pattern seen at CAG 297; lower right, bisulfite sequencing time course for ZMET2 activity at 601 CCG 11. This site is representative of all CCG sites in the DNA sequence, in which no DNA methyltransferase activity was detectable.
(D) Upper left panel, bisulfite sequencing time course for ZMET2 activity at 5S CAG 161, the CHG site in the linker region of the dinucleosome; upper right panel, bisulfite sequencing time course for ZMET2 activity at 5S CAG 13, a CHG site located near the entry/exit site of the nucleosome; lower left panel, bisulfite sequencing time course for ZMET2 activity at 5S CAG 179, a CHG site located near the entry/exit site of the nucleosome; lower right panel, bisulfite sequencing time course for ZMET2 activity at 5S CAG 110, a site in which methyltransferase activity was much faster on naked DNA plus H3K9me2 peptide than on the dinucleosome.
(E) The observed rate constant (kobs) for DNA methylation on 601 dinucleosomes as measured by the radioactive assay (Figure 1B) is mostly dominated by the rate constant for methylating CHG 159 (k159) as measured using bisulfite sequencing.
(F) The observed rate constant (kobs) for DNA methylation on 5S dinucleosomes as measured by the radioactive assay can be explained by contributions from the rate constants of the three most methylated sites as measured by bisulfite sequencing (k161, k13, and k179).
Earlier comparison of bulk DNA MTase activity on dinucleosomes generated with the 5S rRNA positioning sequence versus the 601 positioning sequence suggested a small reduction in activity on the 5S dinucleosomes (Figure 1A). However, nucleosomes assembled using the 5S DNA sequence have also been shown to exhibit higher DNA unpeeling at the entry-exit site compared to nucleosomes assembled on the 601 sequence (Anderson et al., 2002). We therefore wondered whether the 5S dinucleosomes might allow internal nucleosomal CHG sites to be methylated compared to the 601 dinucleosomes. To address this question, we repeated bisulfite sequencing using 5S dinucleosomes or the corresponding naked DNA with H3K9me2 (1–32) tail peptides. Similar to the 601 bisulfite sequencing results, (1) a site within the linker DNA (161) was methylated faster in the context of dinucleosomes compared to the naked DNA and, (2) several sites were methylated rapidly on naked DNA but not detectably methylated when they were nucleosomal (Figures 4B, 4D and S5C). However, unlike with the 601 dinucleosomes, nucleosomal sites located near the DNA entry/exit sites of the 5S sequence (13 and 179) were detectably methylated. For comparison, sites 13 and 179 were methylated 165-fold and 56-fold faster than the nearest corresponding positions in the 601 dinucleosome (sites 297 and 130), respectively (Table S1). While the position of these targets sites on the 5S and 601 dinucleosomes do not exactly correspond with respect to the nucleosome structure, the results suggest that the more breathable DNA in the 5S dinucleosome promotes DNA methylation by ZMET2 at nucleosomal sites that are proximal to the entry/exit site. Nonetheless, the rate of methylation for the linker DNA site on the 5S dinucleosome was substantially slower than for the linker site in the 601 dinucleosome (Table S1). As a result, the overall rate constant of methylation on the 5S dinucleosomes, which is the sum of all the individual rate constants, is still slower (~5-fold) than that with 601 dinucleosomes as measured by bisulfite sequencing, consistent with the slower 5S methylation seen in bulk. (Figures 1A, 4E and 4F).
Visualization of ZMET2 bridging the dinucleosome by negative stain electron microscopy
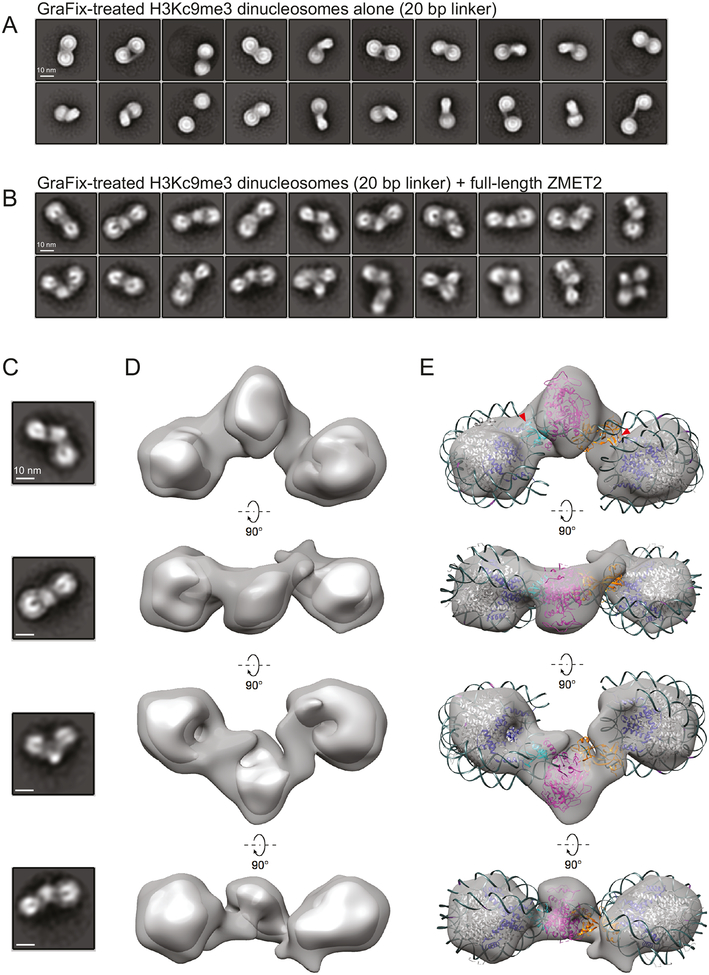
To further test the nucleosome bridging model, we generated ~512 ZMET2/H3Kc9me3 dinucleosome complexes using a glutaraldehyde crosslinking approach (GraFix, see Methods) and subjected these complexes to single particle electron microscopy (Figure S6A). Two-dimensional classification rendered classes with clearly visible nucleosomal features, where the nucleosomal DNA and histone octamer are apparent (Figures 5B and S6B). The majority of the 2D classes, (> 80% of particles), show a single molecule of ZMET2 bound to the dinucleosome, while a small subset of the classes (~12% of the particles) show two ZMET2 molecules bound. heterogeneity observed in the 2D classes is likely attributable to different ZMET2/dinucleosome binding modes or potential conformational flexibility. comparison we also generated GraFix-treated dinucleosomes in the absence of ZMET2 and produced 2D class averages that suggest a more significant range of flexibility between the two constituent nucleosomes than we observed in the ZMET2-bound complexes (Figures 5A and 5B). These 2D class averages suggest ZMET2 may stabilize specific dinucleosome conformations when bound. We proceeded by selecting all particles corresponding to classes with singly-bound ZMET2/dinucleosome complexes and determined a 3D reconstruction with RELION using an ab initio 3D map generated from a smaller data set as an initial model (Figure S6C).
Figure 5. Visualization of ZMET2 bridging the dinucleosome by negative stain electron microscopy.
(A) Two-dimensional class averages of GraFix-treated H3Kc9me3 dinucleosomes (2N_20) alone. The dinucleosomes contain a 20 bp linker with one CHG site positioned 11 bp from one nucleosome and 8 bp from the other.
(B) Two-dimensional class averages of GraFix-treated complex formed between H3Kc9me3 dinucleosomes (2N_20) and ZMET2.
(C) Two-dimensional class averages depicting different views of the ZMET2/H3Kc9me3 dinucleosome complex. Scale bar is 10 nm.
(D) Four different 90° rotational views of a 3D reconstruction of the ZMET2/H3Kc9me3 complex. The complex is represented at two different threshold levels.
(E) The same views from (B), with the dinucleosome and ZMET2 (130–912) crystal structures manually fitted into the map. Red arrowheads represent location of emergence of the H3 tail from the globular portion of histone H3 (blue). Colors highlight different domains of ZMET2 (130–912): magenta, catalytic domain; blue, CD; orange, BAH.
The map, at a resolution of ~28 Å, clearly shows a single molecule of ZMET2 bridging across the two nucleosomes of the dinucleosome (Figure 5D). Different rotational views of the map closely correspond with 2D class averages of the ZMET2/dinucleosome complex (Figure 5C). The shape of the density corresponding to ZMET2 is consistent with the triangular architecture seen in the unbound ZMET2 (130–912) crystal structure, where there is a large central density and two extensions on either side. The crystal structure of a 601 dinucleosome (PDB 1KX5) fits well into the map, highlighting the twisted orientation of the constituent mononucleosomes in the dinucleosome and the positioning of the central ZMET2 density over the linker DNA between the mononucleosomes (Schalch et al., 2005). The two extensions make contacts with regions of the mononucleosomes that are proximal to the location of the H3 tails. We were also able to manually fit the ZMET2 (130–912) crystal structure (PDB 4FSX) into the central density (Du et al., 2012). Based on our biochemical data suggesting that the CD makes the most ground state contacts with the H3K9me modification, we fit the ZMET2 (130–912) crystal structure so that the CD occupies the interface with more ZMET2-nucleosome contacts in the 3D reconstruction (Figure 5E).
DISCUSSION
In this study, we sought to understand the mechanistic roles of ZMET2/H3K9me interactions that were uncovered previously using genetics and structural biology (Du et al., 2012). Our biochemical and structural results suggest that the two H3K9me recognition domains in ZMET2 play binding and allosteric roles allowing ZMET2 to couple maximal DNA methylation to recognition of the appropriate DNA methylation status and chromatin architecture. Below we discuss the mechanistic implications of our findings.
A biophysical explanation of feedback between H3K9 methylation and DNA methylation
In both mammals and plants, positive feedback between DNA methylation and histone methylation has been proposed as a mechanism to ensure high specificity. This feedback is thought to be enabled in part by the presence of specific protein domains that recognize the methylation state of histones. In the case of the Arabidopsis thaliana DNA methyltransferase CMT3 and its Zea mays ortholog ZMET2, the chromo- and BAH domains are implicated in recognition of the H3K9me mark on nucleosomes (Du et al., 2012). The simplest explanation would be that both the chromo- and BAH domains aid in the binding of ZMET2 to H3K9me-decorated chromatin. However, we find that the CD functions primarily in the binding step of ZMET2’s DNA MTase reaction, and the BAH domain functions primarily in the allosteric activation step.
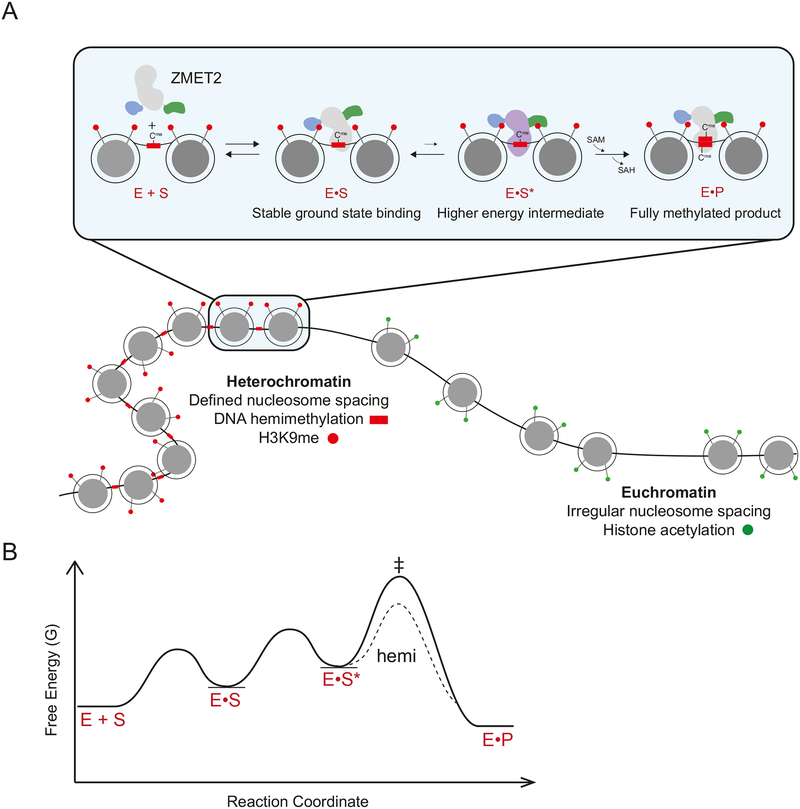
Allosteric activation is also suggested by our observation that an H3K9me2 peptide can stimulate DNA MTase activity when added in trans. We further find that ZMET2 affinity for H3Kc9me3-modified mononucleosomes is tighter than the affinity for the corresponding H3Kc9me3 peptide, suggesting that ZMET2 makes additional interactions with the nucleosome, such as with nucleosomal DNA, in the ground state (defined as the most stable bound state) (Kd(mono) = 0.89 μM versus Kd(peptide) = 5.1 μM; Figures 3B and S1B). Based on these results we suggest a model in which the CD stabilizes ground state binding to the nucleosomal template whereas the BAH domain promotes a productive catalytic orientation of ZMET2 (Figure 6A). In this way, division of labor between the CD and BAH domain promotes the most favorable action of ZMET2 on nucleosome arrays, and helps explain the strong preference for ZMET2 to methylate CHG sites contained within linker DNA between nucleosomes. This model also provides a mechanism to reduce spurious DNA methylation by ZMET2 molecules that may bind to appropriately spaced dinucleosomes in euchromatin.
Figure 6. Model for ZMET2 assembly and activity on chromatin.
(A) ZMET2 interrogates chromatin context by scanning for the correct architecture, and the presence of H3K9me and DNA hemimethylation. In heterochromatic regions of the genome, the ZMET2 CD recognizes the H3K9me mark in the ground state. The ZMET2 BAH domain recognizes the H3K9me mark post-binding in the context of a high-energy intermediate, activating the enzyme for DNA methylation. SAM is utilized for methyl transfer during product formation.
(B) Free energy profile for the model depicted in (A). The crosshatch represents the transition state for the chemical step of the reaction. The dotted line represents a lower energy transition state in the presence of a hemimethylated DNA substrate.
In vitro evidence for H3K9me-driven maintenance methyltransferase activity
Maintenance MTases are typically characterized by their interaction with components of the replication machinery and their preference for hemimethylated DNA in vitro. A direct analysis of DNA MTase activity by CMT3 or ZMET2 on hemimethylated substrates has been lacking. Furthermore, it remains to be seen how ZMET2 coordinates other substrate cues such as H3K9me with hemimethylation. We find that ZMET2 exhibits a substantially larger preference for hemimethylated DNA in the presence of the H3K9me2 peptide than in the absence of the peptide (Figure 2C, ~250-fold vs. 3-fold, respectively). We propose that the combinatorial effect of these two substrate cues serves to increase fidelity for target loci during replication. Additionally, we speculate that optimal ZMET2 activity takes place once modified histones are deposited on recently replicated chromatin, or once the action of histone MTases on newly deposited histones is complete.
A nucleosome-bridging model provides a physical explanation for linker DNA specificity
DNA methylation at CHG sites in plants is correlated with nucleosome positioning in vivo, suggesting that nucleosomes may contribute to DNA methylation patterning by directing the activity of DNA MTases (Chodavarapu et al., 2010; Cokus et al., 2008b). Furthermore, DNA methylation by de novo MTases in mammals is primarily restricted to linker DNA when mapped in vitro (Felle et al., 2011). Interestingly, asymmetric CHH DNA methylation in plant heterochromatin is dependent on the chromatin remodeling enzyme DDM1. It is proposed that DDM1 antagonizes linker histone H1, suggesting that CHH DNA methylation may also preferentially occur in linker DNA (Zemach et al., 2013). Our finding that ZMET2 has a strong preference to methylate dinucleosomes over mononucleosomes, suggests that a single ZMET2 molecule is able to sense higher-order chromatin architecture. The ZMET2 preference to methylate linker DNA is further consistent with a model in which the active site is positioned between two nucleosomes. Negative stain EM provides direct visualization of the 3D architecture of the complex. While the limited resolution of the structure precludes analysis of detailed conformational changes or residue-level interactions, the overall architecture is consistent with our biochemical data, which suggests that ZMET2 bridges across the dinucleosome. Along with H3K9me and DNA hemimethylation, chromatin architecture is thus an additional substrate cue that ZMET2 may use during target recognition and methylation within heterochromatin.
The structural data, paired with our biochemical analyses allow us to form a model for ZMET2 assembly and activity on chromatin in which the CD recognizes dinucleosomes via H3K9me in the binding step of the reaction (Figure 6A). In subsequent steps, ZMET2 forms an activated intermediate upon interaction with H3K9me on the adjoining nucleosome via the BAH domain. The higher energy intermediate formed by these two interactions is then poised to methylate linker DNA between nucleosomes in heterochromatic regions of the genome. (Figure 6A and 6B). We hypothesize that interactions with hemi-methylated DNA in ZMET2’s active site may lower the activation energy for the chemical step required for product formation (Figure 6B).
This study demonstrates one way in which eukaryotic DNA methyltransferases have evolved to read a specific chromatin context, namely via the presence of accessory domains that tightly regulate binding and catalytic activity in response to chromatin marks and nucleosome architecture. Further, this study challenges the pervasive model in which nucleosomes serve as a barrier to DNA methylation. While octamer-bound DNA may indeed be less accessible to methylation, it is the presence of tandem nucleosomes that is activating for ZMET2. The mechanism proposed here implies that nucleosome spacing in heterochromatin regions participates in a feedback loop with DNA methylating enzymes that recognize, methylate, and thus preserve the existing nucleosome spacing by promoting recruitment of histone H3 K9 MTases that contain DNA methyl binding domains (Du et al., 2014). We anticipate that future detailed biochemical studies on mammalian DNA methyltransferases will uncover how these enzymes are regulated by their specific chromatin contexts. Comparative studies between plant and mammalian DNA methyltransferase systems would then reveal how different evolutionary constraints impact specific mechanistic solutions in the context of genome regulation.
METHOD DETAIL
Protein expression and purification
All ZMET2 constructs were generated from a codon-optimized commercially synthesized version of the coding gene and cloned into a pSUMO-based bacterial expression vector with a TEV-cleavable N-terminal 6xHis tag and N-terminal SUMO solubility tag (DNA2.0). The expression vector was a kind gift from Dr. Jiamu Du and Dr. Dinshaw Patel at Memorial Sloan Kettering Center. All ZMET2 constructs were expressed in Rosetta (DE3) E. coli (Novagen). ZMET2 expression and purification was performed using a protocol provided by Jiamu Du with some modifications. Starter cultures were inoculated from a glycerol stock of transformed Rosetta cells and allowed to grow overnight at 37°C. Large cultures (1 L each, 20 g Tryptone, 10 g Yeast Extract, 10 g NaCl) were inoculated with 12–15 mL of starter culture and allowed to reach an OD600 of ~0.9–1.2 at 37°C. Cultures were then transferred to 20°C for 30 minutes, followed by induction with 0.2 mM IPTG and subsequent expression for 16–18 h. Cells were then harvested by centrifugation and resuspended in Lysis Buffer (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 20 mM imidazole, 1 μg/mL pepstatin A, 3 μg/mL leupeptin, 2 μg/mL aprotinin, 1 mM PMSF) and lysed under high pressure with an Emulsiflex-C3 homogenizer (Avestin). DNase1 was added during high-pressure lysis to digest DNA in the lysate (Roche, ~1 mg/6 L culture). Lysates were pelleted by spinning at 30,000 × g for 25 minutes and the cleared supernatant was incubated with TALON cobalt affinity resin (Clontech, 1.5 mL slurry for each 1 L of culture) for 1–3 hours with gentle rotation. Bound resin was pelleted by spinning at 1,000 × g for 3 minutes and was washed three times with Lysis Buffer. Bound resin was applied to a disposable column (1 column per 3 L of culture; GE Lifesciences) pre-equilibrated with 20 mL H2O and 20 mL Lysis Buffer, and any remaining wash buffer was allowed to flow through. Purified ZMET2 was eluted with Elution Buffer (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 500 mM imidazole; 20 mL per 3 L culture) and concentrated to < 2 mL using a spin concentrator (EMD Millipore). TEV protease was added to a final concentration of ~0.1 mg/mL and the mixture was dialyzed overnight into Low Salt Buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 5 mM DTT). The dialyzed sample was then subjected to centrifugation to clear any precipitation (10,000 × g, 5 min.) and injected onto a Q HP anion exchange column (5 mL, GE Lifesciences) pre-equilibrated with two column volumes of Low Salt Buffer. Bound ZMET2 was eluted using an increasing gradient of NaCl (150 mM to 1 M over 30 column volumes). Pure fractions were determined by gel and then pooled and concentrated to < 2 mL. The concentrated sample was then further purified by size exclusion chromatography on a HiLoad Superdex 200 26/60 column (320 mL bed volume, GE Lifesciences) equilibrated with 360 mL Low Salt Buffer. Pure fractions were pooled and concentrated. In preparation for storage at −80°C, 10% v/v glycerol was added and samples were flash frozen in liquid N2.
ZMET2 point mutants (W224L, F441A) were generated by site-directed mutagenesis of the full-length ZMET2 construct and were prepared as described above.
Preparation of substrate and nucleosomal DNA
Unmethylated and hemimethylated short duplexed substrates were generated from 38 bp single stranded DNA oligos (IDT). Oligos were annealed by heating complementary strands to 100°C and cooling slowly to room temperature. Annealing was checked by agarose gel and DNA concentration was measured by Nanodrop.
For all mononucleosomal DNA, PCR was used to use amplify 601 or 5S sequences. Amplified DNA was ethanol precipitated and then separated from template and excess nucleotide over a 5% acrylamide gel and the band corresponding to the nucleosomal DNA was excised using UV shadowing. DNA was extracted from the gel matrix by first passing the gel through a syringe, then rocking the gel in 1X TE overnight at room temperature. Samples were filter-purified, and DNA extracted using ethanol precipitation. Plasmids containing dinucleosomal 601 sequences were a generous gift from Dr. Karim Armache (NYU). Plasmids containing dinucleosomal 5S sequences were custom synthesized, using the 5S positioning sequence for both nucleosomal portions and the same linker sequence from the 601 construct (DNA2.0). To prepare large quantities of dinucleosomal DNA, plasmids containing the appropriate sequence were transformed into dam-/dcm- E. coli (NEB) and Giga preps were performed following the manufacturers instructions (Qiagen). Typically, a 2–3 L Giga prep would yield ~10–14 mg of plasmid, and after all purification procedures, ~1–2 mg of usable dinucleosomal DNA.
Assembly of mononucleosomes and dinucleosomes
Mononucleosomes and dinucleosomes were assembled from purified nucleosomal DNA and purified histone octamer prepared from bacterially expressed Xenopus laevis histones. Histone H3 was either wild type or a K9C mutant to allow for conjugation of a methyl lysine analog (MLA) to generate H3Kc9me3 nucleosomes (Simon, 2010). Optimal DNA:octamer:dimer ratios for nucleosome assembly were determined empirically. Nucleosomes were reconstituted by salt dialysis over 36–72 hours and purified over 10–30% glycerol gradients using ultracentrifugation.
Assembly of ligated symmetric and asymmetric dinucleosomes
Ligated dinucleosomes were assembled using an adapted version of an established protocol using DraIII to generate digested fragments for ligation after mononucleosome assembly (Poepsel et al., 2018). Mononucleosomal DNA fragments were generated using PCR from existing 601 dinucleosome plasmids. Primers included extensions of at least 9 base pairs beyond the DraIII target site in order to accommodate DraIII for optimal digestion of fragment ends. PCR products resulting from 10–30 mL PCR reactions were ethanol precipitated and purified over 75 mL 5% acrylamide gels, as described above. Fragments were then digested with DraIII enzyme (NEB) for 24 hours at 37°C, ethanol precipitated, and gel-purified again. Test ligation reactions were performed using the naked DNA fragments to optimize T4 ligase concentration and to check for non-specific ligation products. Addition of too much T4 ligase causes massive non-specific over-ligation, so a titration of ligase was necessary to determine ideal concentration. Mononucleosomes were assembled using DraIII-digested and purified fragment DNA as described above. Ligation reactions containing ~300–500 nM of each mononucleosome (with complimentary digested ends for ligation), 1 U/uL T4 ligase, and 1X T4 ligase buffer were mixed to a final volume of 500 uL and incubated at room temperature for 45 minutes. Reactions were split equally over two 5 mL 10–30% glycerol gradients and centrifuged at 31,000 rcf for 17 hours at 4°C. Gradients were analyzed by gel and fractions containing pure ligated dinucleosomes were pooled and concentrated for DNA methyltransferase assays.
Fluorescence polarization
For all peptide binding assays, fluorescein-labeled peptides were synthesized commercially (CPC Scientific). H3Kc9me3 peptides were generated using the standard MLA installation procedure, followed by mass spectrometry and HPLC purification. For all nucleosome binding assays, fluorescein-labeled DNA was generated by polymerase chain reaction using fluorescein-conjugated primers. Fluorescent nucleosomes were prepared as described above. Wild type or mutated ZMET2 was either dialyzed overnight into FP buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM DTT), or was buffer exchanged a minimum of three times into FP buffer using spin concentrators (Amicon). Serial dilutions of ZMET2 were prepared in FP buffer. 10X stock solutions of either fluorescein-labeled peptides or nucleosomes were prepared in the presence of 0.01% NP-40 and pre-loaded into a 384-well plate. Using a multi-channel pipette, serially diluted ZMET2 was added and mixed with peptide or nucleosome samples. Final peptide or nucleosome concentration was between 5–10 nM. Following brief centrifugation, binding reactions were allowed to equilibrate at room temperature for 30 minutes. Plates were then scanned in fluorescence polarization mode (Analyst HD).
Size-exclusion chromatography coupled to multi angle light scattering
A Wyatt S 050 column was attached to an HPLC system with downstream multi-angle light scattering detection and refractive index detection (Wyatt). Filtered ZMET2 samples were injected onto the system after overnight equilibration of the column in MALS buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM DTT). All data were analyzed using ASTRA software.
DNA methyltransferase assay
For all single-turnover WT and F441A ZMET2 DNA methyltransferase assays, 5 μM ZMET2 was incubated with 50–800 nM substrate (nucleosomes or naked DNA) in 1X DNA MTase buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 10% glycerol, 1 mM DTT) in a 96-well plate (Corning). Naked DNA substrates sequences are as follows:
Unmethylated 38 bp DNA sequence: AACGCAGCATGCGCTGCTAGC GCAGCTAGCGCTGCATG
Methylated 38 bp DNA sequence (annealed to unmethylated complement): AACG(meC)AGCATGCG(meC)TGCTAGCG(meC)AGCTAGCG(meCTG)CATG
601 mononucleosomal DNA sequence: CTGGAGAATCCCGGTGCCGAGGCCGCTCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCCTGT
For single-turnover reactions using W224L ZMET2, 20 μM protein was incubated with substrate and the salt concentration in 1X MTase buffer was adjusted to account for the additional salt provided by the protein storage buffer. For reactions containing H3 tail peptides, at least a 4-fold excess of peptide over [ZMET2] was used. Peptide sequences were commercially synthesized by CPC Scientific and sequences are as follows:
H3K9me peptide (aa 1–32): ARTKQTARKSTGGKAPRKQLATKAARKSAPAT
H3K9me2 peptide (aa 1–32): ARTKQTAR(Kme2)STGGKAPRKQLATKAARKSAPAT
H3K9me3 peptide (aa 1–32): ARTKQTAR(Kme3)STGGKAPRKQLATKAARKSAPAT
H3Kc9me3 peptide (aa 1–32): ARTKQTARCSTGGKAPRKQLATKAARKSAPAT
All reactions were initiated by transferring protein/substrate mixtures to wells pre-loaded with S-Adenosyl-L-Methionine (3H-SAM) containing a methyl group labeled with tritium for a final 3H-SAM concentration of 4 μM (Perkin Elmer). For each time point, 10 μL of the reaction was removed and mixed in a well pre-loaded with 10 μL of 10 mM cold SAM iodide to quench the reaction (Sigma). Quenched time points were applied to circles of DE81 anion exchanging filter paper and the paper was allowed to dry (Whatman). The circles were washed twice for 5 minutes in 200 mM ammonium bicarbonate, once for 5 minutes in water and once for 5 minutes in ethanol. After drying, each individual circle was combined with scintillation cocktail and the samples were counted (Grainger, Beckman).
To ensure a post-binding state for DNA methyltransferase assays, DNA methylation was measured with saturating concentrations of ZMET2 in excess of nucleosomes for single-turnover reactions. For the majority of experiments, we ensured saturation conditions by using ZMET2 at concentrations in which a 5-fold increase caused less than a 2-fold increase in the rate constant (kobs). The rate constant measured under these saturating conditions was termed kmax. For experiments with 5S dinucleosomes, we found ZMET2 binding to H3Kc9me3 5S mononucleosomes to be about 1.5-fold tighter than to 601 mononucleosomes, so we used concentrations of ZMET2 that were 10-fold above the Kd for 5S mononucleosomes (see Figure S2C).
Preparation of samples for bisulfite sequencing
DNA methylation reactions for subsequent bisulfite conversion and sequencing were initiated as described above, using single-turnover conditions, with SAM iodide in place of 3H-SAM. Time points were removed and samples were flash frozen in liquid nitrogen to stop the methylation reaction and stored at −80°C. Each time point was rapidly thawed at room temperature and 1 μL Proteinase K (Roche) was added to digest ZMET2 and histone proteins for 30 minutes at 37°C. Subsequently, 10 μL 1X SDS/TE was added to each sample. DNA was extracted using Phenol:Chloroform:Isoamyl alcohol (25:24:1, v/v, ThermoFisher) and back-extracted one time with chloroform. The aqueous layer was transferred to a fresh tube and ethanol precipitated overnight at −20°C. Samples were centrifuged at 4°C for 15 minutes at 10,000 rpm. The supernatant was pipetted away and pellets were allowed to air dry for 1 hour. Precipitated DNA was then resuspended in 25 μL 1X TE. DNA samples were checked by native polyacrylamide gel.
Library generation, bisulfite conversion and sequencing
Libraries for Illumina sequencing were generated using the KAPA Hyper Prep Kit (Kapa Biosystems) and Illumina library adaptors. After adaptor ligation, bisulfite conversion was carried out on the DNA using the EZ DNA Methylation Kit (Zymo). Finally, adaptor-ligated, bisulfite-treated DNA was amplified for 12 cycles using MyTaq DNA Polymerase (Bioline) and Illumina PCR Primer Mix.
Bisulfite PCR products were sequenced on a MiSeq (Illumina) in 250 bp paired end mode. Read quality was checked using FastQC (v0.11.3, http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Since our reference sequence used for these experiments was only 314 bp, overlapped 250 bp paired end reads were merged using PANDAseq (v2.10) (Masella et al., 2012). Merged reads with 314 bp in length were aligned using BSseeker2 (v2.0.9) with –s 1 –e 314 options (to specify the start and end of reads) (Guo et al., 2013). After aligning, methylation over cytosines was calculated using customized R scripts. The reference sequences used are listed below:
601 dinucleosome: ATCGAGAATCCCGGTGCCGAGGCCGCTCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGGATTCTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCCGATGGTTAACGGATCTGGCCGCCATCGAGAATCCCGGTGCCGAGGCCGCTCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGGATTCTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCCGAT
5S dinucleosome: ATCCCCGACCCTGCTTGGCTTCCGAGATCAGACGATTTCGGGCACTTTCAGGGTGGTATGGCCGTAGGCGAGCACAAGGCTGACTTTTCCTCCCCTTGTGCTGCCTTCTGGGGGGGGCCCAGCCGGATCCCCGGGCGAGCTCGAATTGGTTAACGGATCTGGCCGCCGGCCCGACCCTGCTTGGCTTCCGAGATCAGACGATTTCGGGCACTTTCAGGGTGGTATGGCCGTAGGCGAGCACAAGGCTGACTTTTCCTCCCCTTGTGCTGCCTTCTGGGGGGGGCCCAGCCGGATCCCCGGGCGAGCTCGAAGAT
Preparation of H3Kc9me3 dinucleosome and ZMET2 complexes
Dinucleosomes and FL-ZMET2 were separately buffer exchanged into EM buffer (50 mM NaCl, 10 mM HEPES pH 7.5, 1 mM DTT) using spin concentrators (Amicon). Dinucleosomes and FL-ZMET2 were combined at final concentrations of 5.1 μM and 15 μM, respectively, in a final volume of 60 μL and allowed to equilibrate at room temperature for 15 minutes. Complexes were stabilized using the GraFix procedure (Kastner et al., 2008). Briefly, glycerol gradients were generated with 10% and 30% glycerol buffers (Upper: 50 mM NaCl, 10 mM HEPES pH 7.5, 10% glycerol, 1 mM DTT and Lower: 50 mM NaCl, 10 mM HEPES pH 7.5, 30% glycerol, 1 mM DTT and 0.15% glutaraldehyde). Samples were centrifuged at 4°C for 14.5–15.5 hours at a speed of 35,000 rpm and fractionated. 5–10 μL of each fraction was loaded onto a 5% polyacrylamide/0.5X TBE gel. Similar fractions were pooled and dialyzed overnight into a quenching buffer (50 mM NaCl, 10 mM Tris-HCl pH 8.0, 1 mM DTT). Samples were then concentrated and estimated nucleosome concentration in each sample was calculated by measuring DNA absorbance at 260 nm.
Negative stain electron microscopy and image processing
Pooled fractions from the GraFix procedure were screened using negative stain EM and the fraction with the least non-specifically cross-linked material was used for further data collection. To prepare grids for negative stain EM, 3 μL of 140–180 nM complex was applied to 400 mesh carbon-coated copper grids (Ted Pella), which were glow discharged for 30 seconds using the EasiGlow system (Pelco). Grids were blotted and stained with 3 μL of 0.75% uranyl formate solution three times with blotting in between each application.
A preliminary dataset was collected using a Tecnai T12 microscope (FEI Company) equipped with a LaB6 filament and operated at 120 kV. Images were collected with an Ultrascan 4096 × 4096 pixel CCD camera (Gatan) at a nominal magnification of 52,000× which corresponds to a pixel size of 2.12. A total of 40 micrographs were collected, binned by a factor of two, and 4,656 particles were selected manually and extracted using RELION. After removing non-particles from the stack using 2D alignment and classification, an ab initio 3D reconstruction was calculated using cryoSPARC (Punjani et al., 2017). Two classes were requested during 3D ab initio reconstruction and the best model, corresponding to 98.6% of the particles was selected.
An additional, larger dataset of 392 micrographs was collected using a Tecnai T20 microscope (FEI Company) equipped with a LaB6 filament and operated at 200 kV. Images were collected using SerialEM (Mastronarde, 2005) and a TemCam F816 8k × 8k (TVIPS, Germany) camera at a nominal magnification of 50,000x, which corresponds to a pixel size of 1.57. 21,526 particles were selected manually and extracted using RELION (Scheres, 2012). Micrographs were binned by a factor of two, and GCTF was used for CTF estimation (Zhang, 2016). All subsequent image processing was performed in RELION. 2D classification was performed by sorting particles into 200 2D classes and from those, a dataset of ~5000 particles with only one bound ZMET2 molecule were used for 3D classification. 3D classification into four classes yielded structures that were very similar in conformation. The final structure presented corresponded to 24.6% of the data and had the least noise.
QUANTIFICATION AND STATISTICAL ANALYSIS
All binding data were plotted using Prism software (Graphpad) and fit to the following equation to determine Kd:
For DNA methyltransferase assays, most time courses were fit to a straight line to determine initial rates. For single-turnover reactions fit to a straight line, initial rates were divided by [substrate] to determine rate constants. For experiments comparing dinucleosomes to mononucleosomes, [substrate] was normalized for the number of “mononucleosomes” contained in the substrate. For example, 100 nM dinucleosome was considered equivalent to 200 nM mononucleosomes. For multiple-turnover reactions, initial rates were divided by [ZMET2] to determine rate constants. For single-turnover time courses that reached near-completion, the data were fit to a single exponential and rate constants were extracted directly. To test background levels of 3H-methyl in the methyltransferase assay, reactions were allowed to proceed without enzyme. All plotting and fitting was done using Prism software (Graphpad).
All binding equilibrium constants and DNA methyltransferase activity rate constants were plotted as mean ± SEM. Replicate values for each experiment are listed in the corresponding figure legend.
Key Resource Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Rosetta (DE3) | Novagen | 70954 |
| BL21 (DE3) pLysS | Promega | L1195 |
| Dam-/dcm- | NEB | C2925I |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DNase I | Roche | 10104159001 |
| (2-Bromoethyl)trimethylammonium bromide | Aldrich | 117196 |
| (2-Bromoethyl)trimethylammonium bromide | Aldrich | 117196 |
| S-[methyl-3H]-Adenosyl-L-Methionine | Perkin Elmer | NET155H250UC |
| S-(5’-Adenosyl)-L-Methionine Iodide | Sigma | A4377 |
| Proteinase K | Roche | 3115887001 |
| EZ DNA Methylation Kit | Zymo | D5001 |
| H3K9me peptide (aa 1–32) ARTKQTARKSTGGKAPRKQLATKAARKSAPAT |
CPC Scientific | N/A |
| H3K9me2 peptide (aa 1–32) ARTKQTAR(Kme2)STGGKAPRKQLATKAARKSAPAT |
CPC Scientific | N/A |
| H3K9me3 peptide (aa 1–32) ARTKQTAR(Kme3)STGGKAPRKQLATKAARKSAPAT |
CPC Scientific | N/A |
| H3Kc9me3 peptide (aa 1–32) ARTKQTARCSTGGKAPRKQLATKAARKSAPAT |
CPC Scientific | N/A |
| DraNI | NEB | R3510L |
| Deposited Data | ||
| Negative stain 3D reconstruction of full-length ZMET2 bound to H3Kc9me3 601 dinucleosome with 20 bp DNA linker | This paper | EMD-9127 |
| Oligonucleotides | ||
| unmethylated 38 bp oligo AACGCAGCATGCGCTGCTAGC GCAGCTAGCGCTGCATG |
This paper | N/A |
| hemimethylated 38 bp oligo (annealed with unmethylated
complement) AACG(meC)AGCATGCG(meC)TGCTAGCG(meC)AGCTAGCG(meCTG)CATG |
This paper | N/A |
| 601 mononucleosome DNA
sequence CTGGAGAATCCCGGTGCCGAGGCCGCTCAATTGGTCGTAGACAGCTCTAGCA CCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGAT TACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCCTGT |
This paper | N/A |
| 601 dinucleosome DNA
sequence ATCGAGAATCCCGGTGCCGAGGCCGCTCAATTGGTCGTAGACAGCTCTAGCA CCGCTTAAACGCACGTACGGATTCTCCCCCGCGTTTTAACCGCCAAGGGGATT ACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCCGATGGTTAACGGAT CTGGCCGCCATCGAGAATCCCGGTGCCGAGGCCGCTCAATTGGTCGTAGACA GCTCTAGCACCGCTTAAACGCACGTACGGATTCTCCCCCGCGTTTTAACCGCC AAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCCGAT |
This paper | N/A |
| 5S dinucleosome DNA
sequence ATCCCCGACCCTGCTTGGCTTCCGAGATCAGACGATTTCGGGCACTTTCAGGG TGGTATGGCCGTAGGCGAGCACAAGGCTGACTTTTCCTCCCCTTGTGCTGCCT TCTGGGGGGGGCCCAGCCGGATCCCCGGGCGAGCTCGAATTGGTTAACGGA TCTGGCCGCCGGCCCGACCCTGCTTGGCTTCCGAGATCAGACGATTTCGGGC ACTTTCAGGGTGGTATGGCCGTAGGCGAGCACAAGGCTGACTTTTCCTCCCCT TGTGCTGCCTTCTGGGGGGGGCCCAGCCGGATCCCCGGGCGAGCTCGAAGA T |
This paper | N/A |
| Recombinant DNA | ||
| Pet3a_H2A (Xenopus laevis) | Canzio et al., 2011 | N/A |
| Pet3a_H2B (Xenopus laevis) | Canzio et al., 2011 | N/A |
| Pet3a_H3 (Xenopus laevis) | Canzio et al., 2011 | N/A |
| Pet3a_H4 (Xenopus laevis) | Canzio et al., 2011 | N/A |
| pUC157 2_167 601 dinucleosome (20 bp linker) | Dr. Karim Armache | N/A |
| pUC157 2_157 601 dinucleosome (10 bp linker) | This paper | N/A |
| pUC157 2_177 601 dinucleosome (30 bp linker, one CHG site) | This paper | N/A |
| pUC157 2_177 601 dinucleosome (30 bp linker, two CHG sites) | This paper | N/A |
| pUC157 2_187 601 dinucleosome (40 bp linker) | This paper | N/A |
| pSUMO full-length ZMET2 (codon optimized for bacterial expression) | This paper | N/A |
| Software and Algorithms | ||
| Prism | Graphpad | N/A |
| ASTRA | Wyatt | N/A |
| FastQC (v0.11.3) | http://www.bioinformatics.babraham.ac.uk/projects/fastqc | N/A |
| PANDAseq (v2.10) | Masella et al., 2012 | N/A |
| BSseeker2 (v2.0.9) | W. Guo et al., 2013 | N/A |
| cryoSPARC | Punjani et al., 2017 | |
| SerialEM | Mastronarde, 2005 | |
| RELION | Scheres, 2012 | |
| GCTF | K. Zhang, 2016 | |
Supplementary Material
ACKNOWLEDGMENTS
We thank D. Patel for ZMET2 expression plasmids and initial protein purification advice. We thank K. Armache for initial 601 dinucleosomal DNA plasmids, the Kruglyak lab for use of the Illumina MiSeq sequencer, S. Poepsel for guidance on generating ligated dinucleosomes, and E. Palovcak, A. Lyon, M. Ravalin and D. Elnatan for technical assistance. We thank R.S. Isaac and P.A. Dumesic for critical reading of the manuscript and members of the Narlikar, Cheng, and Jacobsen labs for useful discussion. This work was funded by an NSF grant (1517081) to G.J.N.; an NSF Graduate Research Fellowship (1144247) awarded to C.I.S.; a NSF-CAREER (MCB-1552455) and NIH-MIRA (R35GM124806) to X.Z.; a NIH grant (GM60398) to S.E.J.; and NIH grants (S10OD020054, R01GM098672 and R01GM082893) awarded to Y.C. S.E.J. and Y.C. are investigators of the Howard Hughes Medical Institute. We thank B. Carragher and Z. Zhang for preliminary EM experiments conducted at the National Resource for Automated Molecular Microscopy (NRAMM) located at the New York Structural Biology Center, supported by the NIH (GM103310) and the Simons Foundation (Grant 349247).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Anderson JD, Thåström A, and Widom J (2002). Spontaneous access of proteins to buried nucleosomal DNA target sites occurs via a mechanism that is distinct from nucleosome translocation. Molecular and Cellular Biology 22, 7147–7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee L, Malagnac F, and Bender J (2001). Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes & Development 15, 1753–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, and Jacobsen SE (2008). Genome-Wide Association of Histone H3 Lysine Nine Methylation with CHG DNA Methylation in Arabidopsis thaliana. PLoS ONE 3, e3156–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen P-Y, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, et al. (2010). Relationship between nucleosome positioning and DNA methylation. Nature 466, 388–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, and Jacobsen SE (2008a). Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, and Jacobsen SE (2008b). Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Johnson LM, Groth M, Feng S, Hale CJ, Li S, Vashisht AA, Gallego-Bartolome J, Wohlschlegel JA, Patel DJ, et al. (2014). Mechanism of DNA methylation-directed histone methylation by KRYPTONITE. Mol. Cell 55, 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Johnson LM, Jacobsen SE, and Patel DJ (2015). DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol 16, 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Zhong X, Bernatavichute YV, Stroud H, Feng S, Caro E, Vashisht AA, Terragni J, Chin HG, Tu A, et al. (2012). Dual Binding of Chromomethylase Domains to H3K9me2-Containing Nucleosomes Directs DNA Methylation in Plants. Cell 151, 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JR, Yarychkivska O, Boulard M, and Bestor TH (2017). DNA methylation and DNA methyltransferases. Epigenetics Chromatin 10, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felle M, Hoffmeister H, Rothammer J, Fuchs A, Exler JH, and Langst G (2011). Nucleosomes protect DNA from DNA methylation in vivo and in vitro. Nucleic Acids Research 39, 6956–6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Naveh-Many T, Cedar H, and Razin A (1981). Sequence specificity of methylation in higher plant DNA. Nature 292, 860–862. [DOI] [PubMed] [Google Scholar]
- Guo W, Fiziev P, Yan W, Cokus S, Sun X, Zhang MQ, Chen P-Y, and Pellegrini M (2013). BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics 14, 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, and Jacobsen SE (2002). Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416, 556–560. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, and Meyerowitz EM (1997). Hypermethylated SUPERMAN epigenetic alleles in arabidopsis. Science 277, 1100–1103. [DOI] [PubMed] [Google Scholar]
- Jeltsch A, and Jurkowska RZ (2016). Allosteric control of mammalian DNA methyltransferases - a new regulatory paradigm. Nucleic Acids Research 44, 8556–8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Jurkowska RZ, Zhang X, Jeltsch A, and Cheng X (2007). Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 449, 248–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner B, Fischer N, Golas MM, Sander B, Dube P, Boehringer D, Hartmuth K, Deckert J, Hauer F, Wolf E, et al. (2008). GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods 5, 53–55. [DOI] [PubMed] [Google Scholar]
- Kulis M, and Esteller M (2010). DNA methylation and cancer. Adv. Genet 70, 27–56. [DOI] [PubMed] [Google Scholar]
- Law JA, and Jacobsen SE (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature Reviews Genetics 11, 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B-Z, Huang Z, Cui Q-Y, Song X-H, Du L, Jeltsch A, Chen P, Li G, Li E, and Xu G-L (2011). Histone tails regulate DNA methylation by allosterically activating de novo methyltransferase. Nature Publishing Group 21, 1172–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM (2001). Requirement of CHROMOMETHYLASE3 for Maintenance of CpXpG Methylation. Science 292, 2077–2080. [DOI] [PubMed] [Google Scholar]
- Malagnac F, Bartee L, and Bender J (2002). An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. Embo J. 21, 6842–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masella AP, Bartram AK, Truszkowski JM, Brown DG, and Neufeld JD (2012). PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics 13, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51. [DOI] [PubMed] [Google Scholar]
- Mathieu O, Probst AV, and Paszkowski J (2005). Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. Embo J. 24, 2783–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt DM, Knowles BB, and Solter D (2014). DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes & Development 28, 812–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani J, Nankumo T, Arita K, Inamoto S, Ariyoshi M, and Shirakawa M (2009). Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 10, 1235–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa CM, Springer NM, Muszynski MG, Meeley R, and Kaeppler SM (2001). Maize chromomethylase Zea methyltransferase2 is required for CpNpG methylation. The Plant Cell Online 13, 1919–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poepsel S, Kasinath V, and Nogales E (2018). Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nature Publishing Group 25, 154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Robertson KD (2005). DNA methylation and human disease. Nature Reviews Genetics 6, 597–610. [DOI] [PubMed] [Google Scholar]
- Schalch T, Duda S, Sargent DF, and Richmond TJ (2005). X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 436, 138–141. [DOI] [PubMed] [Google Scholar]
- Scheres SHW (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD (2010). Installation of site-specific methylation into histones using methyl lysine analogs. Curr Protoc Mol Biol Chapter 21, Unit21.18.1–Unit21.18.10. [DOI] [PubMed] [Google Scholar]
- Simon MD, Chu F, Racki LR, la Cruz, de CC, Burlingame AL, Panning B, Narlikar GJ, and Shokat KM (2007). The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128, 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, and Meissner A (2013). DNA methylation: roles in mammalian development. Nature Reviews Genetics 14, 204–220. [DOI] [PubMed] [Google Scholar]
- Soppe WJJ, Jasencakova Z, Houben A, Kakutani T, Meister A, Huang MS, Jacobsen SE, Schubert I, and Fransz PF (2002). DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. Embo J. 21, 6549–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq M, Saze H, Probst AV, Lichota J, Habu Y, and Paszkowski J (2003). Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc. Natl. Acad. Sci. U.S.a. 100, 8823–8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres IO, and Fujimori DG (2015). Functional coupling between writers, erasers and readers of histone and DNA methylation. Curr. Opin. Struct. Biol 35, 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh P-H, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, and Zilberman D (2013). The Arabidopsis Nucleosome Remodeler DDM1 Allows DNA Methyltransferases to Access H1-Containing Heterochromatin. Cell 153, 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lang Z, and Zhu J-K (2018a). Dynamics and function of DNA methylation in plants. Nature Publishing Group 6, 597. [DOI] [PubMed] [Google Scholar]
- Zhang K (2016). Gctf: Real-time CTF determination and correction. J. Struct. Biol 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jurkowska R, Soeroes S, Rajavelu A, Dhayalan A, Bock I, Rathert P, Brandt O, Reinhardt R, Fischle W, et al. (2010). Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Research 38, 4246–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z-M, Lu R, Wang P, Yu Y, Chen D, Gao L, Liu S, Ji D, Rothbart SB, Wang Y, et al. (2018b). Structural basis for DNMT3A-mediated de novo DNA methylation. Nature 554, 387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.