Abstract
Certain Escherichia coli strains residing in the human gut produce colibactin, a small molecule genotoxin implicated in colorectal cancer pathogenesis. However, colibactin’s chemical structure and the molecular mechanism underlying its genotoxic effects have remained unknown for over a decade. Here, we combine an untargeted DNA adductomics approach with chemical synthesis to identify and characterize a covalent DNA modification from human cell lines treated with colibactin-producing E. coli. Our data establish that colibactin alkylates DNA with an unusual electrophilic cyclopropane. We show this metabolite is formed in mice colonized by colibactin-producing E. coli and is likely derived from an initially-formed, unstable colibactin-DNA adduct. Our findings reveal a potential biomarker for colibactin exposure and provide mechanistic insights into how a gut microbe may contribute to colorectal carcinogenesis.
One Sentence Summary:
Identifying DNA adducts in cells and animals exposed to colibactin-producing gut microbes sheds light on a cancer-linked genotoxin.
The human gut harbors trillions of microorganisms capable of producing small molecules that mediate microbe-host interactions (1). For example, certain gut commensal and extraintestinal pathogenic strains of Escherichia coli and other Proteobacteria produce colibactin, a genotoxin of unknown structure implicated in colorectal cancer (CRC) pathogenesis. These organisms harbor a 54-kb biosynthetic gene cluster that encodes a nonribosomal peptide synthetase-polyketide synthase (NRPS-PKS) assembly line (pks island), which has been implicated in colibactin biosynthesis (Fig. 1A) (2). E. coli containing the pks island (pks+ E. coli) cause DNA double-strand breaks (DSBs) in human cell lines and in animals (3,4), accelerate colon tumor growth under conditions of host inflammation (5–8), and are found with increased frequency in inflammatory bowel disease, familial adenomatous polyposis, and colorectal cancer patients (4,5,8). Despite these intriguing links to human disease, our understanding of colibactin’s chemical structure and biological activity is limited, as this natural product has eluded isolation.
Fig. 1. pks+ E. coli synthesize cyclopropane-containing metabolites that may alkylate DNA.
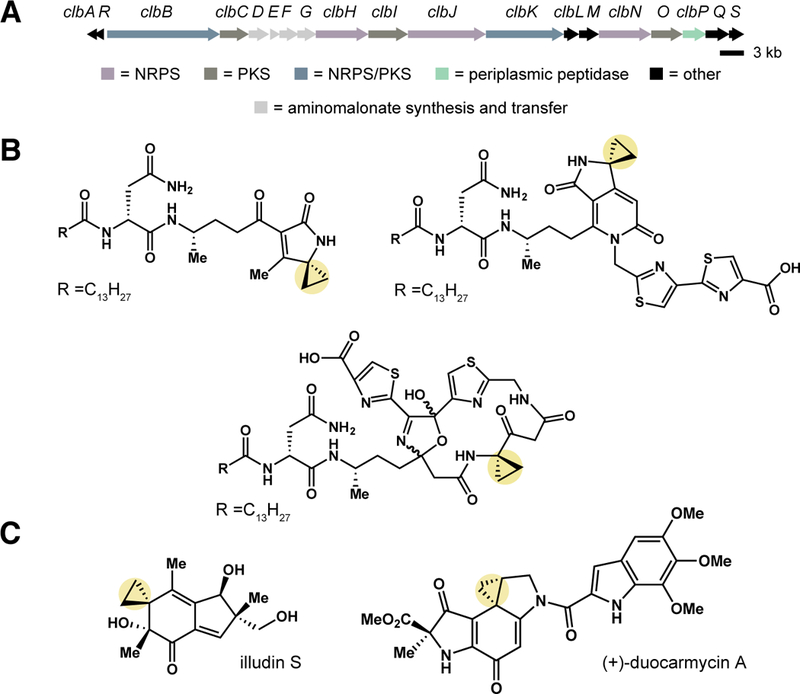
(A) pks genomic island. Open reading frames encoding nonribosomal peptide synthetase (NRPS) (purple), polyketide synthase (PKS) (brown), hybrid NRPS/PKS (blue), peptidase (ClbP) (green), aminomalonate synthesis and transfer (grey), and other (black) enzymes are highlighted. (B) Selected cyclopropane-containing candidate precolibactins isolated and structurally characterized from pks+ ΔclbP E. coli, including lactam, pyridone, and macrocyclic scaffolds. (C) Illudin S and (+)-duocarmycin A are DNA alkylating metabolites that contain a cyclopropane ring.
Colibactin has been exceptionally challenging to isolate and structurally characterize. For example, colibactin’s genotoxic activity is contact-dependent and not observed when cells are treated with pks+ E. coli culture supernatants or cell lysates (2). It is also currently unknown how colibactin is transported into mammalian cells. Attempts to directly identify colibactin using comparative metabolite analyses have been unsuccessful, indicating the active genotoxin may be unstable and/or recalcitrant to isolation. To gain information about colibactin’s structure, we and others have isolated and characterized non-genotoxic, pks-associated metabolites (9–15) from mutant strains of pks+ E. coli missing a critical peptidase enzyme (ClbP), which removes an N-myristoyl-D-asparagine ‘prodrug motif’ from a late-stage biosynthetic precursor and is required for genotoxicity (16–18) (Fig. 1B). These metabolites, termed ‘precolibactins’, are unlikely to be precursors to the mature colibactin as their synthesis requires only a subset of the biosynthetic machinery known to be essential for genotoxic activity. Notably, several ‘precolibactins’ contain a cyclopropane ring, a structural feature found in DNA alkylating natural products, such as the illudins (19) and duocarmycins (20) (Fig. 1C).
DNA alkylating agents act as electrophiles towards DNA bases, forming covalent modifications known as DNA adducts (21). The discovery of cyclopropane-containing ‘precolibactins’ has led to the hypothesis that colibactin’s mode of action involves DNA alkylation, but there is limited direct evidence to support this idea (10,11). Reacting a cyclopropane-containing ‘precolibactin’ with linearized plasmid DNA revealed small amounts of a putative higher molecular weight adduct by gel electrophoresis, leading to an initial proposal that colibactin cross-links DNA (10). Recent in vitro work using synthetic ‘colibactin mimics’, compounds designed based on partial biosynthetic information, showed that the cyclopropane ring in a putative ClbP cleavage product can be attacked by a thiol nucleophile and is necessary for these molecules to shear purified DNA (22). When artificially dimerized, these ‘colibactin mimics’ appear to cross-link DNA as assessed by gel electrophoresis (22). pks+ E. coli lacking both the nucleotide excision repair protein UvrB (23) and a self-resistance protein encoded in the pks island (ClbS) exhibit severe autotoxicity and impaired growth (24), providing indirect support for DNA alkylation and repair of the resulting lesions in colibactin-producing E. coli strains. ClbS can hydrolyze the cyclopropane ring of a synthetic ‘colibactin mimic’, further implicating this functional group in colibactin’s activity (25). However, experimental proof that colibactin itself alkylates DNA remains elusive, as colibactin-DNA adducts have not been structurally characterized or identified in biologically relevant settings.
Untargeted DNA adductomics can identify unknown DNA adducts
Owing to the challenges associated with isolating the active genotoxin from E. coli, we instead sought to identify the in vivo product(s) of colibactin-mediated DNA damage. We hypothesized that detecting and characterizing colibactin-DNA adducts generated in human cells treated with pks+ E. coli would yield direct information about the active genotoxin’s chemical structure and the molecular basis for its DNA damaging activity in a biologically relevant setting. While targeted liquid chromatography-mass spectrometry (LC-MS)-based methodologies exist to identify previously characterized DNA adducts in cells, detecting unknown DNA adducts represents a significant challenge because of the low abundance of these modifications and the extraneous false-positive ion signals that derive from the complex matrices of biological samples (26). Indeed, preliminary attempts to identify colibactin-DNA adducts using standard comparative metabolite profiling approaches failed to reveal differences in hydrolyzed DNA samples from HeLa cells treated with either E. coli BW25113 pBeloBAC (pks-) or BACpks (pks+) strains. To overcome this difficulty, we envisioned exploiting a newly developed, untargeted mass spectrometry-based DNA adductomics approach (26) to identify colibactin-DNA adducts in cells exposed to pks+ E. coli.
LC-MS3 DNA adductomics identifies adducts in hydrolyzed DNA samples using high-resolution/accurate mass data-dependent constant neutral loss monitoring of 2’-deoxyribose (116.0474 amu) or one of the four DNA bases (guanine, 151.0494 amu; adenine, 135.0545 amu; thymine, 126.0429 amu; and cytosine, 111.0433 amu) (Fig. 2A) (27). Accurate mass measurement of an observed DNA adduct can allow for the determination of its elemental composition, and the triggered MS2 and MS3 fragmentation spectra provide additional structural information about the modified base. We first used this DNA adductomic approach to detect characterized DNA adducts induced by illudin S, a cytotoxic agent that alkylates DNA upon cellular metabolic activation (28). LC-MS3 DNA adductomic analysis of hydrolyzed DNA obtained from HeLa cells exposed to either illudin S or DMSO identified a known illudin- derived adduct (28) with m/z 384.2030 [M+H]+ only in the illudin-treated cells, confirming the utility of this approach for adduct detection in our model (fig. S1).
Fig. 2. High-resolution accurate mass (HRAM) LC-MS3 DNA adductomic analysis identifies DNA adducts in HeLa cells and mice exposed to pks+ E. coli.
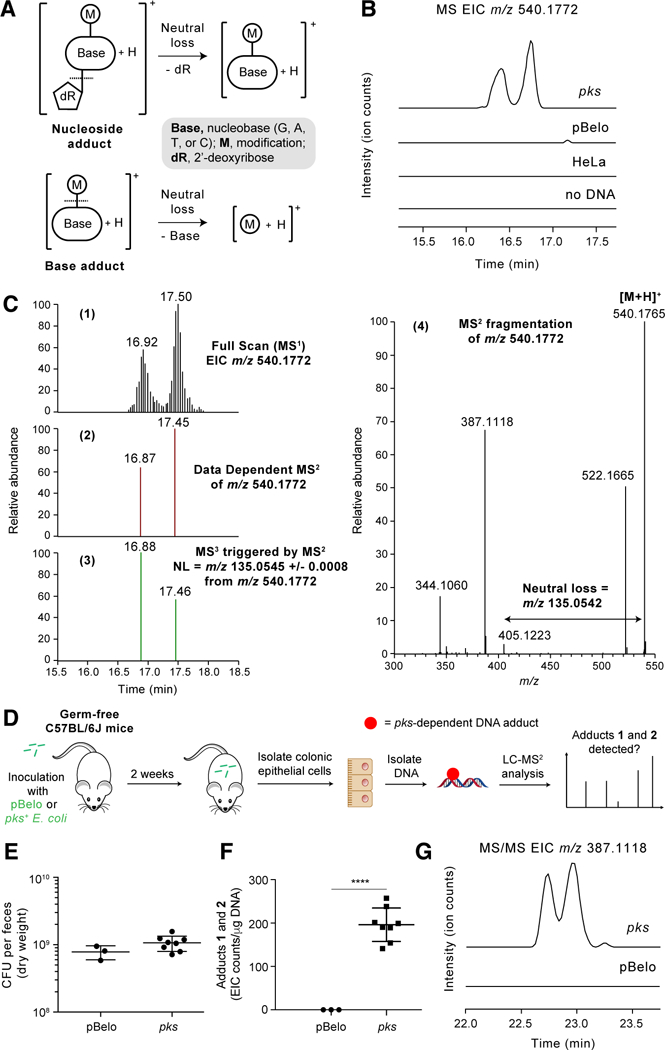
(A) Structural features of DNA adducts and detection by neutral loss monitoring. (B) Full scan extracted ion chromatogram (EIC) of DNA adducts 1 and 2 (m/z 540.1772) in HeLa cells exposed to colibactin-producing E. coli and negative controls (HeLa cells exposed to non-colibactin producing pBeloBAC E. coli, HeLa cells alone, or when no DNA was present). (C) 1. Full scan EIC of DNA adducts 1 and 2 (m/z 540.1772). 2. Signal corresponding to the data dependent MS2 events (RT = 16.87 and 17.45 min). RT = retention time. 3. Signal corresponding to MS3 events (RT = 16.88 and 17.46 min) triggered by the neutral loss of adenine. 4. MS2 mass spectrum resulting from fragmentation of m/z 540.1772 which triggered the MS3 event. (D) Flowchart of experiment detecting DNA adducts 1 and 2 in mouse colonic epithelial cells. (E) Bacterial load in the feces of mice colonized with pBelo (n = 3) or pks+ E. coli (n = 8) for 2 weeks. (F) EIC counts of DNA adducts 1 and 2 per μg of DNA in colonic epithelial cells isolated from mice colonized with pBelo (n = 3) or pks+ E. coli (n = 8) for 2 weeks. EIC counts were determined by area under the curve integrations of the most abundant MS2 fragmentation ion (m/z 387.1118 ± 0.0008) of the adducts 1 and 2 precursor ion (m/z 540.1772). (G) Representative MS/MS EIC of DNA adducts 1 and 2 (m/z 387.1118 [M+H-Ade-H2O]+), the most abundant fragment ion of m/z 540.1772. Each symbol in (E) and (G) represents an individual mouse; error bars represent mean +/− the standard error of the mean (SEM); **** P < 0.0001 (unpaired t test).
Discovery of DNA adducts in mammalian cells and mice exposed to pks+ E. coli
We next investigated whether this method could identify putative colibactin-DNA adducts. First, we isolated DNA from HeLa cells transiently infected with either pks- or pks+ E. coli. Following DNA hydrolysis, we then performed a comparative, untargeted LC-MS3 DNA adductomic screen of these samples. This analysis revealed two putative DNA adducts (1 and 2) that accumulated only in the cells treated with pks+ E. coli (Fig. 2B). We also detected these adducts in a colonic epithelial cell line exposed to pks+ E. coli and in HeLa cells exposed to native colibactin-producing strains (fig. S2 and S3). 1 and 2 eluted at 16.92 and 17.15 min, respectively, and exhibited an m/z of 540.1765 [M+H]+ (Fig. 2C). Both peaks triggered MS3 fragmentation events upon observation of the neutral loss of adenine (135.0545 amu) in the MS2 fragmentation spectra (Fig. 2C), indicating these compounds were adenine adducts. To confirm adducts 1 and 2 were pks-associated, we repeated the cell infection assays described above but included individual stable isotope labeled amino acids known to be used by the pks NRPS-PKS assembly line and integrated into pks-associated metabolites (10,12,13). These experiments revealed that the expected building blocks L-[2,3-13C2]Ala, L-[1-13C]Met, [1,2-13C2]Gly, and L-[1-13C]Cys were incorporated into the two adducts (fig. S4–8). While this work was in revision, Herzon and co-workers identified a putative DNA adduct with the same mass in plasmid DNA exposed to pks+ E. coli in vitro thus further confirming our findings (29).
Next, we sought to determine whether adducts 1 and 2 could be detected in mice exposed to colibactin-producing E. coli. Germ-free wild-type C57BL/6J mice were inoculated with either pks− orpks+ E. coli. After two weeks, colonic epithelial cells were harvested, DNA was isolated from the cells, and then adduct formation was assessed using LC-MS/MS (Fig. 2D). Both strains colonized the mice to a similar extent as assessed by fecal colony counts (Fig. 2E and table S1). We detected adducts 1 and 2 only in the mice colonized with pks+ E. coli (Fig. 2F and G, table S2, and fig. S9). These results show that the colibactin-mediated DNA damage observed in human cell lines also occurs within a genetically intact host in the absence of exogenous carcinogens or inflammatory mediators. Furthermore, these data suggest that these adducts are biomarkers for pks+ E. coli exposure. Overall, this experiment provides the first direct support for DNA alkylation playing a critical role in colibactin’s genotoxicity in vivo.
Structural characterization of the colibactin-derived DNA adducts
Further analysis of the LC-MS3 data revealed preliminary information about the structure(s) of adducts 1 and 2. The high-resolution accurate mass measurement of m/z 540.1765 [M+H]+ yielded a molecular formula of C23H25N9O5S (calculated, 540.1772) with 16 degrees of unsaturation. MS2 fragmentation of 1 and 2 in high-resolution mode displayed major fragment ions of m/z 522.1673 [M+H-H2O]+, 387.1123 [M+H-Ade-H2O]+, 344.1065, and 229.0973. The shared fragmentation spectra indicated these compounds were likely stereoisomeric (fig. S10). Using this information and our MS2 fragmentation data, we proposed potential structures for the in vivo-derived colibactin-DNA adducts that were analogous to a recently characterized, chemically unstable ‘model colibactin’-thiol adduct but containing an extra hydroxyl group (fig. S11) (25). However, these adducts’ low abundance in cells precluded further isolation and structural characterization efforts.
To elucidate the structures of adducts 1 and 2, we accessed authentic standards by chemically synthesizing new ‘colibactin mimics’ and reacting them with calf-thymus DNA (ctDNA) (Fig. 3A). We prepared carboxylic acid-containing cyclopropane 3 in seven steps using a route developed to access other colibactin mimics (fig. S12–29A) (22). Based on the proposed structures of the adducts identified in pks+-treated HeLa cells, 3 could react with ctDNA to give 1 and 2 directly. However, cyclopropane 3 generated only trace amounts of detectable adducts when incubated with ctDNA (fig. S30) and minimally sheared DNA at 1 mM concentration (fig. S31A). Hypothesizing that an unfavorable electrostatic interaction between the carboxylate of 3 and the negatively charged phosphate backbone of DNA greatly reduced its reactivity, we masked the carboxylic acid as an ethyl ester (fig. S12–19, S29B, and S32–35). Cyclopropane 4 was ~100-fold more potent than 3 in a DNA shearing assay and induced both G2/M cell cycle arrest and DNA DSBs in treated HeLa cells (fig. S31B, S36–38). LC-MS analysis of a ctDNA reaction with 4 revealed three major products of m/z 552.2135 [M+H]+ (5) and m/z 568.2081 [M+H]+ (6 and 7) (fig. S30 and S39). MS2 fragmentation confirmed these compounds were adenine adducts that only differed by the presence of one oxygen atom (fig. S40 and S41). The “non-oxidized” adduct 5 (m/z 552.2135) was unstable at room temperature and slowly converted to a pair of “oxidized” adducts (6 and 7) (m/z 568.2081) over the course of two days (fig. S42 and S43). Further analysis of the fragmentation data indicated that 6 and 7 contained a hydroxyl group and a fragment ion (m/z 229.0973) identical to that of the in vivo-derived adducts 1 and 2 (fig. S44). Since 6 and 7 were stable and possessed an MS2 fragmentation pattern matching those of the adducts detected in pks+ E. coli-treated HeLa cells, we targeted these compounds for isolation and structural characterization.
Fig. 3. Comparing DNA adducts generated in vivo with a synthetic standard confirms their chemical structures.
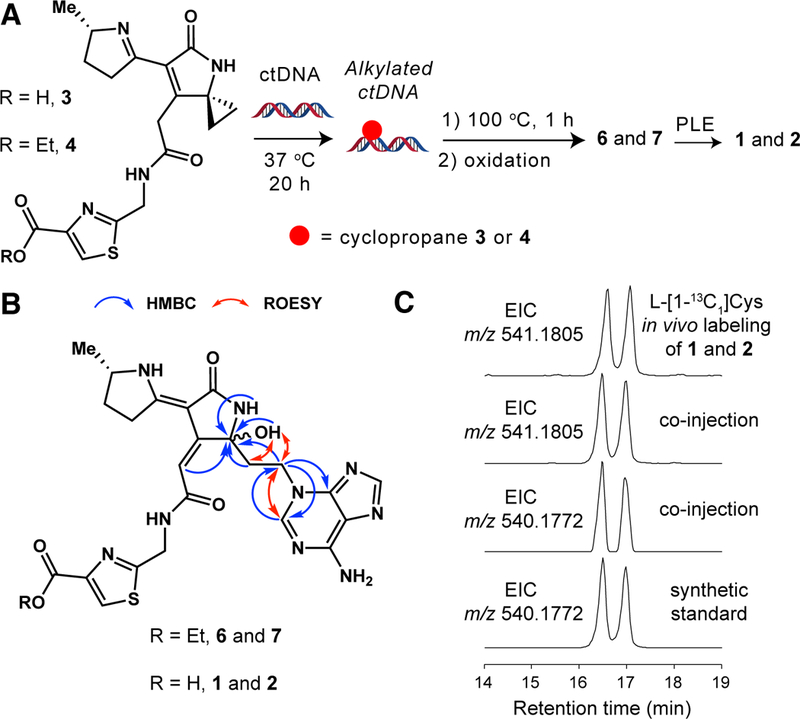
(A) In vitro DNA alkylation reaction used to generate a synthetic standard of adducts 1 and 2. ctDNA = calf-thymus DNA; PLE = pig liver esterase. (B) Chemical structures of diastereomeric DNA adducts 6 and 7 showing key 2D-NMR correlations that support the hemiaminal and N3-adenine assignments. (C) EICs of the L-[1-13C]Cys-labeled in vivo adducts 1 and 2 (m/z 541.1805), co-injection of synthetic standard (m/z 540.1772) with in vivo adducts 1 and 2 (m/z 541.1805 and residual unlabeled m/z 540.1772), and synthetic standard (m/z 540.1772).
We isolated and purified 6 and 7 from multiple small-scale ctDNA alkylation reactions to give approximately 1 mg of pure material. Analysis by one-dimensional (1H) and two-dimensional (gCOSY, gHSQC, gHMBC, and ROESY) NMR (fig. S45–52 and table S3 and S4), as well as DP4 computational analysis (30) (table S5–7 and fig. S53), revealed a 1:1 diastereomeric mixture of a single adduct that contains a 5-hydroxypyrrolidin-2-one ring system with an attached N3-substituted adenine ring (Fig. 3B). The N3-adenine and hemiaminal substitution assignments were supported by key 1H-13C heteronuclear multiple bond (HMBC) and through-space 1H-1H correlations. Interestingly, the observed preference for N3-adenine alkylation resembles that of other cyclopropane-containing DNA alkylating agents (28,31).
We hypothesized that the structures of the monoadducts obtained in vitro (6 and 7) and the adducts identified from pks+ E. coli-treated cells (1 and 2) differed only in the presence of an ester versus carboxylic acid functional group based on their shared MS2 fragmentation patterns. To confirm the structure of the adducts generated in vivo (1 and 2), ester-containing adducts 6 and 7 were hydrolyzed using pig liver esterase to give an authentic standard of the corresponding carboxylic acids (fig. S54). LC-MS1 and MS2 analysis revealed that this standard possessed the same m/z, retention time, and MS2 fragmentation pattern as the adducts (1 and 2) identified in pks+-treated mammalian cells (Fig. 3C and fig. S55), thereby confirming their chemical structures. Based on the observed reactivity of the ‘non-oxidized’ adduct 5 in vitro and the reactivity of related synthetic compounds (22,25), we propose that the 5-hydroxypyrrolidin-2-one ring found in adducts 1 and 2 likely arises from oxidation of an initial, chemically unstable enamide-containing adduct (fig. S56).
Successful characterization of monoadducts 1 and 2 confirms that exposure of host cells to pks+ E. coli results in DNA alkylation. This finding provides direct information about colibactin’s structure, resolves questions surrounding the active genotoxin’s electrophilic cyclopropane and mode of activation, and allows us to propose a mechanism for how exposure to colibactin leads to DNA damage. The ‘precolibactins’, isolated to date, possess a variety of cyclopropane-containing scaffolds, including linear structures (13), an unsaturated lactam (10–12), a pyridone (13,14), and a macrocycle (15). The structures of 1 and 2 strongly suggest that the cyclopropane in colibactin is conjugated to an α,β-unsaturated imine and is not embedded within a linear framework or pyridone. Furthermore, our findings provide in vivo evidence that cleavage of ‘precolibactin’ by peptidase ClbP generates the active colibactin genotoxin by triggering an intramolecular cyclodehydration to form an α,β-unsaturated imine, enhancing the reactivity of the cyclopropane toward DNA (Fig. 4A) (10,11,22).
Fig. 4. The characterized DNA adducts may derive from a colibactin-DNA interstrand cross-link.
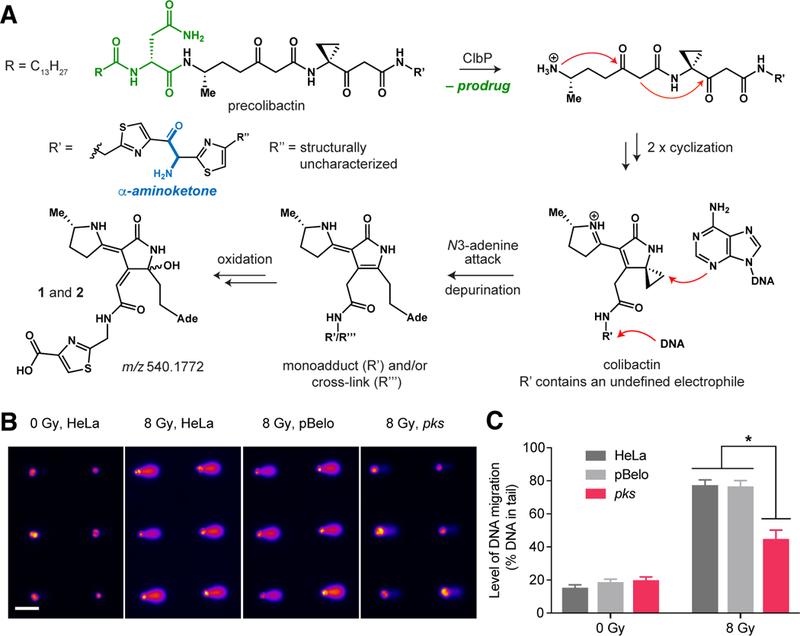
(A) Proposed model for colibactin DNA alkylation and formation of DNA adducts 1 and 2 (DNA = deoxyribonucleic acid; Ade = adenine). (B) Arrayed microwell comets from untreated HeLa cells and HeLa cells treated with pBelo or pks+ E. coli for 1 h at 37 °C [multiplicity of infection (MOI) = 1,000]. Scale bar indicates 100 μm. (C) Quantification of % DNA in tail from untreated HeLa cells and HeLa cells treated with pBelo or pks+ E. coli for 1 h at 37 °C (MOI = 1,000). Data points and error bars represent mean +/− SEM, respectively, of three independent experiments. Student’s t-test was performed to compare each treated dose to the corresponding negative controls (* P < 0.05).
Exposure to pks+ E. coli generates interstrand cross-links in cells
Multiple lines of evidence suggest adducts 1 and 2 are unlikely to represent the immediate product of DNA alkylation with the mature colibactin genotoxin and instead arise from degradation of a larger mono- and/or cross-linked adduct. First, the carboxylic acid-containing ‘model colibactin’ 3 is a poor DNA alkylating agent. Second, while 3 could be generated using a subset of the colibactin biosynthetic enzymes, the entire NRPS-PKS assembly line is essential for genotoxicity (2). Finally, recent work reported the accumulation of a putative interstrand cross-link in DNA incubated with pks+ E. coli as assessed by gel electrophoresis and found cell lines deficient in interstrand cross-link repair were more sensitive to colibactin-producing E. coli (32).
To explore the correlation between DNA cross-linking and the formation of 1 and 2, we used a modified alkaline single cell gel electrophoresis assay (comet assay) (33) to assess the presence of interstrand cross-links in cells exposed to pks+ E. coli. This assay utilizes the high degree of strand breaks induced by γ-radiation to measure interstrand cross-link formation. Unlike monoadducts, interstrand cross-links inhibit the denaturation of DNA under alkaline conditions and therefore decrease the level of DNA migration, reducing the ability to detect radiation-induced strand breaks. We first tested whether CometChip, a high-throughput platform and more robust version of the comet assay (34,35), could detect interstrand cross-links generated by cisplatin, a bifunctional cross-linking agent. Cells were exposed to varying concentrations (0–200 μg/mL) of cisplatin and then analyzed for cross-links 6 h after drug treatment. As expected, cisplatin caused a significant decrease in DNA migration in treated cells thus confirming the utility of this assay (fig. S57).
Next, we applied CometChip to investigate whether interstrand cross-links are formed in HeLa cells exposed to colibactin at the same time point at which we detected adducts 1 and 2. HeLa cells were infected with pks+ E. coli for 1 h and cross-link formation was measured immediately afterwards. In pks+ E. coli-treated HeLa cells exposed to γ-irradiation (8 Gy), we detected a significant level of cross-links as indicated by the 32% decrease in DNA tail moment compared to both controls (Fig. 4B and C). In contrast, we observed minimal strand breaks in non-γ-irradiated pks+ E. coli-treated cells. Thus, these results indicate that interstrand cross-links are present in the pks+ E. coli-treated HeLa cells from which we isolate 1 and 2. However, we could not identify any masses corresponding to putative interstrand cross-links in our untargeted DNA adductomics datasets, suggesting that they may be unstable to our isolation, purification, or MS conditions.
Possible origins of colibactin-derived DNA adducts
Knowledge of colibactin biosynthesis suggests a mechanism for how DNA adducts or cross-links degrade to form 1 and 2. Bioinformatic analyses of the NRPS-PKS assembly line and isolation of ‘precolibactins’ have indicated that colibactin likely contains a bithiazole (13,14) and/or a related ring system with an α-aminoketone inserted between two thiazole rings (Fig. 1B) (15). However, searching our untargeted DNA adductomics datasets did not reveal masses corresponding to putative bithiazole or α-aminoketone-containing colibactin-DNA adducts. Because adducts 1 and 2 contain only one thiazole heterocycle, we propose that they derive from oxidative C–C cleavage of a larger, α-aminoketone-containing colibactin-DNA monoadduct or cross-link (Fig. 4A). While bithiazole rings are stable, α-aminoketones undergo oxidative carbon-carbon bond cleavage in the presence of reactive oxygen species to give carboxylic acids (36). Therefore, the structures of 1 and 2 strongly suggests that the active genotoxin contains an α-aminoketone, a positively charged functional group that may enhance colibactin’s affinity for DNA and thus increase its potency (22). Further experiments will be needed to clarify the nature of the interstrand cross-link, the origin and timing of the proposed oxidative degradation event, and how the specific lesions(s) generated by colibactin lead to the formation of DBSs.
Conclusions
In summary, we have presented direct evidence that the gut bacterial genotoxin colibactin alkylates DNA in vivo. The ability of pks+ E. coli to generate DNA adducts in mammalian cells and in mice strengthens support for the involvement of colibactin in cancer development or progression since misrepaired mono- and cross-linked adducts may generate mutations in oncogenes or tumor suppressor genes, contributing to tumorigenesis (37,38). Our findings will enable efforts to decipher the molecular details of this process. Importantly, this work has also uncovered a candidate metabolite biomarker of colibactin exposure and cancer risk. The ability to directly assess whether exposure to colibactin has occurred in animal models and human patients will help address the critical question of whether pks+ E. coli contribute to colorectal carcinogenesis in patient cohorts (39). Finally, this study represents the first use of untargeted DNA adductomics for the identification and elucidation of unknown gut microbial-derived DNA modifications, highlighting the power of emerging analytical techniques in studying human microbiota metabolites and host-microbiota interactions.
Supplementary Material
ACKNOWLEDGMENTS
We thank G. Heffron and C. Sheehan (Harvard Medical School, Boston, MA) for help with NMR experiments, M. Volpe and N. Braffman (Balskus lab, Harvard University) for assistance with DP4 computational analysis, L. Zhang (Balskus lab, Harvard University) for helping with Sephadex LH-20 chromatography, and L. Zha, V. M. Rekdal, A. Waldman, T. Ng, and N. Koppel (Harvard University) for helpful discussions. We thank W. G. Tilly (MIT) for providing the TK6 cell line and C. Woo (Harvard University) for providing the HT29 cell line. Financial support was provided by the Packard Fellowship for Science and Engineering (E.P.B.), the Damon Runyon-Rachleff Innovation Award (E.P.B), and National Institutes of Health Grants R01 CA208834 (E.P.B.), R01 CA154426 (W.S.G), and R01 ES022872 (L.D.S.). This work was also supported by National Institute of Environmental Health Sciences grant R44 ES024698 (B.P.E.) and Center for Environmental Health Sciences grant P30 ES002109 (B.P.E.). Salary support for P.W.V. was provided by the U.S. National Institutes of Health and National Cancer Institute [Grant R50-CA211256]. Mass spectrometry was carried out in the Analytical Biochemistry Shared Resource of the Masonic Cancer Center, supported in part by the U.S. National Institutes of Health and National Cancer Institute [Cancer Center Support Grant CA-77598]. M.R.W. acknowledges support from the American Cancer Society-New England Division Postdoctoral Fellowship PF-16–122-01-CDD. C.A.B. is the Dennis and Marsha Dammerman fellow of the Damon Runyon Cancer Research Foundation (DRG-2205–14). Mention of commercial products does not constitute endorsement. The experimental data obtained in this study are available as supplementary materials.
REFERENCES AND NOTES
- 1.Lynch SV, Pedersen O, The human intestinal microbiome in health and disease. N. Engl. J. Med. 375, 2369–2379 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Nougayrède J-P, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E, Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313, 848–851 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède J-P, Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 107, 11537–11542 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arthur JC, Peréz-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, Jobin C, Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buc E, Dubois D, Sauvanet P, Raisch J, Delmas J, Darfeuille-Michaud A, Pezet D, Bonnet R, High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS ONE 8, e56964 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arthur JC, Gharaibeh RZ, Mühlbauer M, Perez-Chanona E, Uronis JM, McCafferty J, Fodor AA, Jobin C, Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat. Commun. 5, 4724 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eklof V, Löfgren-Burström A, Zingmark C, Edin S, Larsson P, Karling P, Alexeyev O, Rutegård J, Wikberg ML, Palmqvist R, Cancer-associated fecal microbial markers in colorectal cancer detection. Int. J. Cancer. 141, 2528–2536 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, Wu X, DeStefano Shields CE, Hechenbleikner EM, Huso DL, Anders RA, Giardiello FM, Wick EC, Wang H, Wu S, Pardoll DM, Housseau F, Sears CL, Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359, 592–597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vizcaino MI, Engel P, Trautman E, Crawford JM, Comparative metabolomics and structural characterizations illuminate colibactin pathway-dependent small molecules. J. Am. Chem. Soc. 136, 9244–9247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vizcaino MI, Crawford JM, The colibactin warhead crosslinks DNA. Nat. Chem. 7, 411–417 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.C. A. Brotherton, M. R. Wilson, G. Byrd, E. P. Balskus, Isolation of a metabolite from the pks island provides insights into colibactin biosynthesis and activity. Org. Lett. 17, 1545–1548 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Bian XY, Plaza A, Zhang YM, Müller R, Two more pieces of the colibactin genotoxin puzzle from Escherichia coli show incorporation of an unusual 1-aminocyclopropanecarboxylic acid moiety. Chem. Sci. 6, 3154–3160 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li ZR, Li Y, Lai JY, Tang J, Wang B, Lu L, Zhu G, Wu X, Xu Y, Qian PY, Critical intermediates reveal new biosynthetic events in the enigmatic colibactin pathway. ChemBioChem 16, 1715–1719 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Zha L, Wilson MR, Brotherton CA, Balskus EP, Characterization of polyketide synthase machinery from the pks island facilitates isolation of a candidate precolibactin. ACS Chem. Biol. 11, 1287–1295 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Li ZR, Li J, Gu JP, Lai JY, Duggan BM, Zhang WP, Li ZL, Li YX, Tong RB, Xu Y, Lin DH, Moore BS, Qian PY, Divergent biosynthesis yields a cytotoxic aminomalonate-containing precolibactin. Nat. Chem. Biol. 12, 773–775 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubois D, Baron O, Cougnoux A, Delmas J, Pradel N, Boury M, Bouchon B, Bringer MA, Nougayrède JP, Oswald E, Bonnet R, ClbP is a prototype of a peptidase subgroup involved in biosynthesis of nonribosomal peptides. J. Biol. Chem. 286, 35562–35570 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brotherton CA, Balskus EP , A prodrug resistance mechanism is involved in colibactin biosynthesis and cytotoxicity. J. Am. Chem. Soc. 135, 3359–3362 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Bian X, Fu J, Plaza A, Herrmann J, Pistorius D, Stewart AF, Zhang Y, Müller R, In vivo evidence for a prodrug activation mechanism during colibactin maturation. ChemBioChem 14, 1194–1197 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Tanasova M, Sturla SJ, Chemistry and biology of acylfulvenes: sesquiterpene-derived antitumor agents. Chem. Rev. 112, 3578–3610 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Ghosh N, Sheldrake HM, Searcey M, Pors K, Chemical and biological explorations of the family of CC-1065 and the duocarmycin natural products. Curr. Top. Med. Chem. 9, 1494–1524 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Fu D, Calvo JA, Samson LD, Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer 12, 104–120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Healy R, Nikolayevskiy H, Patel JR, Crawford JM, Herzon SB, A mechanistic model for colibactin-induced genotoxicity. J. Am. Chem. Soc. 138, 15563–15570 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Truglio JJ, Croteau DL, Van Houten B, Kisker C, Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 106, 233–252 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Bossuet-Greif N, Dubois D, Petit C, Tronnet S, Martin P, Bonnet R, Oswald E, Nougayrède J-P, Escherichia coli ClbS is a colibactin resistance protein. Mol. Microbiol. 99, 897–908 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Tripathi P, Shine EE, Healy AR, Kim CS, Herzon SB, Bruner SD, Crawford JM, ClbS is a cyclopropane hydrolase that confers colibactin resistance. J. Am. Chem. Soc. 139, 17719–17722 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balbo S, Hecht SS, Upadhyaya P, Villalta PW, Application of a high-resolution mass-spectrometry-based DNA adductomics approach for identification of DNA adducts in complex mixtures. Anal. Chem. 86, 1744–1752 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stornetta A, Villalta PW, Hecht SS, Sturla SJ, Balbo S, Screening for DNA alkylation mono and cross-linked adducts with a comprehensive LC-MS3 adductomic approach. Anal. Chem. 87, 11706–11713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pietsch KE, van Midwoud PM, Villalta PW, Sturla SJ, Quantification of acylfulvene-and Illudin S-DNA adducts in cells with variable bioactivation capacities. Chem. Res. Toxicol. 26, 146–155 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue M, Shine E, Wang W, Crawford JM, Herzon SB, Characterization of natural colibactin-nucleobase adducts by tandem mass spectrometry and isotopic labeling. Support for DNA alkylation by cyclopropane ring opening. Biochemistry. 57, 6391–6394 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith SG, Goodman JM, Assigning stereochemistry to single diastereomers by GIAO NMR calculation: the DP4 probability. J. Am. Chem. Soc. 132, 12946–12959 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Boger DL, Ishizaki T, Zarrinmayeh H, Isolation and characterization of the duocarmycin-adenine DNA adduct. J. Am. Chem. Soc. 113, 6645–6649 (1991). [Google Scholar]
- 32.Bossuet-Greif N, Vignard J, Taieb F, Mirey G, Dubois D, Petit C, Oswald E, Nougayrede J-P, The colibactin genotoxin generates DNA interstrand cross-links in infected cells. mBio 9, e02393–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olive PL, Banáth JP, Kinetics of H2AX phosphorylation after exposure to cisplatin. Cytometry B Clin. Cytom. 76, 79–90 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Wood DK, Weingeist DM, Bhatia SN, Engelward BP, Single cell trapping and DNA damage analysis using microwell arrays. Proc. Natl. Acad. Sci. U.S.A. 107, 10008–10013 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ge J, Chow DN, Fessler JL, Weingeist DM, Wood DK, Engelward BP, Micropatterned comet assay enables high throughput and sensitive DNA damage quantification. Mutagenesis 30, 11–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wenkert D, Eliasson KM, Rudisill D, Hydrogen peroxide oxidation of α-(N,N-dialkyl)aminoketones. J. Chem. Soc., Chem Commun. 7, 392–393 (1983). [Google Scholar]
- 37.Grady WM, Carethers JM, Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology 135, 1079–1099 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poirier MC, Chemical-induced DNA damage and human cancer risk. Nat. Rev. Cancer 4, 630–637 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Wassenaar TM, coli E and colorectal cancer: a complex relationship that deserves a critical mindset. Crit. Rev. Microbiol. 44, 619–632 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Healy AR, Vizcaino MI, Crawford JM, Herzon SB, Convergent and modular synthesis of candidate precolibactins. Structural revision of precolibactin A. J. Am. Chem. Soc. 138, 5426–5432 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pierens GK, 1H and 13C NMR scaling factors for the calculation of chemical shifts in commonly used solvents using density functional theory. J. Comput. Chem. 35, 1388–1394 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


