Abstract
Cystic Fibrosis (CF) exhibits morbidity in several organs, including progressive lung disease in all patients and intestinal obstruction at birth (meconium ileus) in ~15%. Individuals with the same causal CFTR mutations show variable disease presentation which is partly attributed to modifier genes. With >6,500 participants from the International CF Gene Modifier Consortium, genome-wide association investigation identified a new modifier locus for meconium ileus encompassing ATP12A on chromosome 13 (min p = 3.83x10-10); replicated loci encompassing SLC6A14 on chromosome X and SLC26A9 on chromosome 1, (min p<2.2x10-16, 2.81x10−11, respectively); and replicated a suggestive locus on chromosome 7 near PRSS1 (min p = 2.55x10-7). PRSS1 is exclusively expressed in the exocrine pancreas and was previously associated with non-CF pancreatitis with functional characterization demonstrating impact on PRSS1 gene expression. We thus asked whether the other meconium ileus modifier loci impact gene expression and in which organ. We developed and applied a colocalization framework called the Simple Sum (SS) that integrates regulatory and genetic association information, and also contrasts colocalization evidence across tissues or genes. The associated modifier loci colocalized with expression quantitative trait loci (eQTLs) for ATP12A (p = 3.35x10-8), SLC6A14 (p = 1.12x10-10) and SLC26A9 (p = 4.48x10-5) in the pancreas, even though meconium ileus manifests in the intestine. The meconium ileus susceptibility locus on chromosome X appeared shifted in location from a previously identified locus for CF lung disease severity. Using the SS we integrated the lung disease association locus with eQTLs from nasal epithelia of 63 CF participants and demonstrated evidence of colocalization with airway-specific regulation of SLC6A14 (p = 2.3x10-4). Cystic Fibrosis is realizing the promise of personalized medicine, and identification of the contributing organ and understanding of tissue specificity for a gene modifier is essential for the next phase of personalizing therapeutic strategies.
Author summary
Cystic Fibrosis (CF) impacts the normal functioning of several organs including the pancreas, intestines and lungs. CF is caused by mutations in the CF transmembrane conductance regulator, but individuals with the same mutations have different disease severity. For example, only ~15% of individuals with CF are born with intestinal obstruction at birth (meconium ileus) while all have progressive lung disease with varying severity. Modifier genes contribute to variation across individuals with CF. In this study we identified modifiers of meconium ileus susceptibility, encompassing the previously identified SLC26A9 and SLC6A14, and two new loci ATP12A and PRSS1. To understand the mechanism by which these modifier genes impact CF severity we developed a new statistical method that assesses colocalization with regulatory information. Using this methodology we show, surprisingly, that all the meconium ileus association signals colocalize with gene expression in the pancreas rather than in intestinal tissues. A distinct and neighbouring locus near SLC6A14 also contributes to CF lung disease. Aided by transcriptomics of nasal epithelia from CF patients we found that each locus impacts variation in gene expression of SLC6A14 with tissue specificity. Understanding the contributing tissue and responsible gene are necessary to prioritize modifiers as alternative therapeutic targets.
Introduction
Greater than 348 CF transmembrane conductance regulator (CFTR) gene variants are known to cause CF, where epithelial function is impaired in several organs including the lung, pancreas, and intestine, amongst others (see CFTR2 (Web Resources in Material and Methods) and [1]). Individuals with the same causal CFTR genotype display a wide range of disease burden within and across organs and exhibit variable response to costly CFTR mutation-directed therapies. Modifier gene relationships with CFTR may involve tissue-specific facets that explain some of this variability, and need to be understood to improve therapeutic strategies [2].
Significant morbidity and mortality in CF is a consequence of progressive lung disease, for which the estimated heritability is approximately 50% [3] and correlates poorly with CFTR genotype [4]. Meconium ileus, intestinal obstruction due to failure to pass the meconium, occurs in 13–21% [5] of CF newborns, with equal frequency in males and females. Meconium ileus displays heritability estimates of ~88% [6] and can be discordant for CF siblings with identical CFTR mutations, further highlighting the importance of genetic background beyond CFTR. Meconium, or first stool of a newborn, is typically comprised of intestinal mucins, proteins, bile salts, and cellular debris that are shed from the intestinal mucosa during the fetal period [7]. The meconium exhibits increased thickness and adherent properties in CF, with blockage. Evidence for blockage or meconium ileus can be seen on ultrasound as early as the second trimester, and although there can be resolution in utero, persistence to beyond birth is fatal without enema or surgical intervention [7]. Given the early occurrence, unambiguous presentation and high heritability, genes that contribute to meconium ileus may be more amenable to gene mapping. Moreover, genetic contributors to meconium ileus have been shown to inform disease severity in other CF affected organs.
Genome-wide association studies (GWAS) have identified modifier genes that contribute to morbidity across multiple CF-affected organs [8], with the associated variants frequently annotated to putative regulatory regions of nearby genes. For example, allelic variation near the transcription start site (TSS) of SLC26A9, which encodes an anion transporter in epithelial cells that interacts with CFTR to enhance its functional expression [9–11], has been shown to associate with aspects of CF lung disease [2], meconium ileus [12], CF-related diabetes (CFRD) [13], as well as exocrine pancreatic damage at birth [14, 15]. Although the same modifier can show association with multiple CF co-morbidities, it is unclear whether these are independent effects as their timing of impact and expression levels vary. Exemplary is how SLC26A9 appears to contribute to CFRD through exocrine pancreatic damage in utero [15], an example of vertical pleiotropy [16].
Here, we are interested in understanding the relation between the intestine, where meconium develops, and the pancreas. The presence of meconium ileus is correlated with disease in other CF affected organs, in particular pancreatic disease. Direct pancreatic function studies have shown that individuals with CF have reduced digestive enzyme secretory fluid with low pH and high protein concentrations [17, 18]. Meconium ileus occurs almost exclusively in individuals with severe CFTR mutations known to confer pancreatic exocrine insufficiency [5, 19] (~90% of the CF population [20]), therefore modifier gene studies of meconium ileus have been restricted to this CF sub-population. It has been noted that those with meconium ileus on average have relatively less elevated immunoreactive trypsinogen (IRT) levels at birth compared to patients without meconium ileus ([15, 21]); elevated IRT is the newborn screening biomarker for CF and is also reflective of the extent of prenatal pancreatic injury [19, 22]. The phenotypic correlation between meconium ileus and CF pancreatic disease, including CFRD [13], leads one to consider the involvement of the pancreas in the development of meconium ileus. Evidence that gene expression in the pancreas is mediating the association between meconium ileus and the GWAS variants would support this hypothesis.
Using the largest study population of individuals with CF to date from the International CF Gene Modifier Consortium (GMC), we (1) identify genetic loci contributing to meconium ileus using a combination of whole genome genotype arrays and next generation sequencing; (2) integrate publicly available cross-tissue gene expression [23] (and epigenomic data [24–27]) with genotypes (Material and Methods and S1 Appendix) to test the hypothesis that the variants associated with meconium ileus are colocalizing with variants that impact gene expression and (3) determine the most probable originating tissue for the meconium ileus modifiers. We also (4) integrate RNA-sequencing of nasal epithelia from individuals with CF to determine cross-phenotypic mechanisms and tissue-specific GWAS signals.
Results
Participants include n = 6,770 individuals with CF from Canada, the United States and France, recruited in two phases by the GMC (Table 1), and genotyped on various Illumina platforms (Material and Methods, S1 and S2 Tables). With the incorporation of a hybrid reference [28] and after standard quality control (Material and Methods), a total of 8,763,019 (genotyped and imputed) variants were analyzed for association with meconium ileus (with clinical definitions as previously described; [12]).
Table 1. Characteristics of CF participants included in the genome-wide study of meconium ileus from the International CF Gene Modifier Consortium (GMC).
The four consortium sites are Canadian CF Gene Modifier Study (CGMS), UNC/Case Western Reserve Modifier Study (UNC/Case), Johns Hopkins University CF Twin and Sibling Study (JHU), and French Gene Modifier Study (FGMS). Additional sample details stratified by genotyping platform are provided in S1 Table.
| Consortium Site | Participantsa | Female % | CFTR genotype | meconium ileus | |||||
|---|---|---|---|---|---|---|---|---|---|
| Phe508del/ | Phe508del/ | Yes | No | %Fb with MI (p-valuec) |
|||||
| Phe508del | other | ||||||||
| Phase I-NA | Phase II-NA | Mega-all | % | % | |||||
| CGMS | 1519 | 338 | 1857 | 46.3 | 61.1 | 31.3 | 310 | 1547 | 53.2 (0.2805) |
| UNC/Case | 1303 | 602 | 1905 | 45.7 | 78.4 | 19.1 | 326 | 1579 | 46.6 (0.2448) |
| JHU | 1042 | 700 | 1742 | 47.4 | 56.2 | 37.9 | 320 | 1422 | 47.2 (0.3419) |
| FGMS | - | - | 1266 | 48.7 | 58.1 | 35.8 | 159 | 1107 | 54.7 (0.2669) |
| Total | 3864 | 1640 | 6770 | 46.8 | 64.2 | 30.4 | 1115 | 5655 | 49.8 (0.9046) |
aAll patients from the four study sites had two severe CFTR mutations associated with pancreatic insufficiency, and all patients clustered within 6 S.D. of the Hapmap (v3) [29] Utah residents with Northern and Western European ancestry from the CEPH collection (CEU) and Toscani in Italia (TSI) in a principal component analysis (PCA).
b%F indicates percentage of female participants.
cp-value tests whether the proportion of females differ from 0.5
Genome-wide association analysis
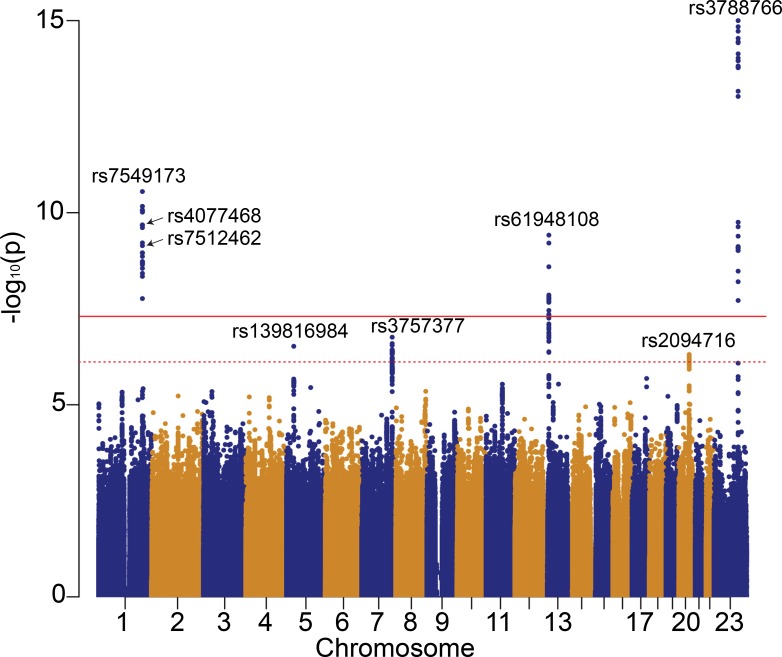
Genome-wide association analysis used generalized estimating equations with an exchangeable correlation structure (to account for siblings present in the data), and included adjustment for consortium site, genotype platform and population stratification (S1 Fig for the first three principal components). Three genome-wide significant loci with nearest genes SLC6A14, SLC26A9 and ATP12A (p-values<5x10-8 based on the conventional family-wise error control [30]), and three additional loci with nearest genes PRSS1, TARS, CEBPB (q-values<0.05 based on the false discovery rate control [31]) were evident (Fig 1).
Fig 1. Manhattan plot for the genome-wide association study with meconium ileus in the GMC sample of 6,770 individuals with CF.
All variants with MAF>1% are included in the analysis. The solid horizontal line corresponds to the genome-wide significance threshold of p = 5x10-8 [30], and the dotted horizontal line is the false discovery rate control threshold of q = 0.05 (equivalent to p<7.67x10-7) [31]. Accuracy of p-value calculation was up to 2.2x10-16 by the geeglm function [100] in R. Variants rs4077468 and rs7512462 identified in previous CF gene modifier studies also noted.
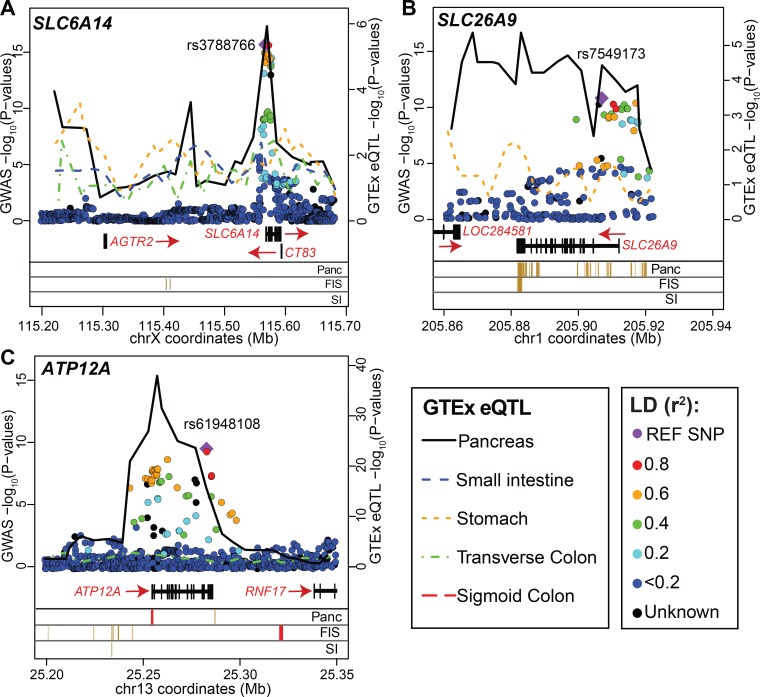
The chromosome X (SLC6A14; panel (a) of Fig 2) and chromosome 1 (SLC26A9; panel (b) of Fig 2) loci were identified as genome-wide significant in an earlier meconium ileus study by the GMC [12] using a subset of the current study sample (S3 Table); this earlier study also demonstrated that a set of 157 genes that code for constituents of the apical plasma membrane where CFTR resides (see Supplementary Table 3 in [12]) were significantly enriched for variants associated with meconium ileus, and this gene-set association was replicated in the French Gene Modifier Study (FGMS) cohort [12, 32]. The expanded sample (Table 1 and S1 Table) analyzed here provides further support for the same variants at the SLC6A14 and SLC26A9 loci (Table 2, minimum p<2.2x10-16 and p = 2.81x10-11 in column ‘Mega-all’, and S3 Table). Support for CFTR (min p = 2.34x10-5 at rs213972; gene-based permutation p = 0.0001; S2 Fig) and the association between the apical plasma membrane gene-set and meconium ileus was also observed in this study (permutation p<1x10-4 in a subset of 5,869 unrelated individuals from the full consortium data and p = 0.017 in the North American Phase II subgroup alone). The confirmation of multiple contributing genes supports the existence of genetic heterogeneity, indicating that meconium ileus is a phenotype of complex genetic etiology within a mendelian disease.
Fig 2. Overlay of the p-values of individual SNPs from the meconium ileus GWAS and GTEx eQTL association and the enhancer and promoter states from the REMC at the three genome-wide significant loci.
All the p-values are on the–log10 scale with the left y-axis representing the GWAS (with the filled diamond for the SNP with the lowest p-value and colored dots indicating the LD with this SNP) and the right y-axis representing the GTEx eQTL results (version 7; with colored lines indicated for different tissues) for SLC6A14 (A), SLC26A9 (B), and ATP12A (C). Note that eQTLs for SLC6A14 in the pancreas and ATP12A in the transverse colon were not mapped by GTEx v7, but were calculated and included here—refer to Material and Methods, GTEx Data section for details. To display the eQTL data, each physical region shown was divided into 25 equal windows and lines were drawn for each tissue by connecting the minimum GTEx eQTL p-value within each window for that gene. The absence of a line indicates insufficient gene expression for that particular tissue (see GTEx Data section for details on expression threshold). In the lower panels, the 5-mark 15-state observed chromHMM enhancer (orange) and promoter (red) states within DNase hypersensitive sites are shown (Material and Methods) from adult pancreas (Panc), fetal small intestine (FIS) and adult small intestine (SI).
Table 2. Top ranked variants by p-value from each genome-wide significant locus in the GMC sample of 6,770 individuals with CF.
The first three rows correspond to loci with SNPs that reached genome-wide significance and the bottom three rows correspond to loci with SNPs that exceeded false discovery rate (FDR) control threshold of q = 0.05 (equivalent to p<7.67x10-7). The top SNP in the SLC6A14 region remains the same at rs3788766 as in [12], while the top SNP at the SLC26A9 locus is now rs7549173. However all significant loci reported at the SLC26A9 locus in [12] are in linkage disequilibrium with rs7549173 (S3 Table), including rs7512462 which has been used as an instrumental variable in a Mendelian Randomization analysis of CF-related diabetes and shown to associate with exocrine pancreas disease [15] and CFTR-directed therapeutics response [2]. The variants in the ATP12A and PRSS1 regions in the previous study [12] are provided in S3 Table. The chromosome 5 and 20 variants are new findings and were not previously identified in [12].
| Chr | BP | SNPs | Nearest gene | Risk Allele | 1Mega-all | 2Mega-DFDF | 3Phase I-NA | 4Phase II-NA | 5FGMS | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | P | OR | P | OR | P | OR | P | OR | P | |||||
| X | 115566839 | rs3788766 | SLC6A14 | A | 1.44 | <2.2x10-16 | 1.40 | 2.21x10−11 | 1.53 | 4.15x10−14 | 1.33 | 2.98x10−4 | 1.42 | 1.50x10−3 |
| 1 | 205906897 | rs7549173 | SLC26A9 | C | 1.37 | 2.81x10−11 | 1.41 | 3.37x10−9 | 1.40 | 8.88x10−8 | 1.37 | 5.80x10−4 | 1.24 | 0.076 |
| 13 | 25282819 | rs61948108 | ATP12A | T | 1.55 | 3.83x10-10 | 1.70 | 5.69x10−10 | 1.54 | 1.42x10−6 | 1.48 | 4.62x10−3 | 1.78 | 2.50x10−3 |
| 7 | 142455538 | rs3757377 | PRSS1 | T | 1.29 | 2.55x10-7 | 1.31 | 5.55x10−6 | 1.34 | 3.88x10−6 | 1.28 | 8.43x10−3 | 1.12 | 0.4 |
| 5 | 33471002 | rs139816984 | TARS | AAAAAAAAT | 1.35 | 3.01x10-7 | 1.30 | 1.93x10−4 | 1.33 | 1.85x10−4 | 1.32 | 0.012 | 1.58 | 6.80x10−3 |
| 20 | 48835331 | rs2094716 | CEBPB | A | 1.30 | 4.92x10-7 | 1.28 | 8.90x10−5 | 1.26 | 1.10x10−3 | 1.29 | 9.30x10−3 | 1.48 | 3.80x10−3 |
1Mega-all: all GMC CF participants (n = 6,770)
2Mega-DFDF: individuals homozygous for the Phe508del mutation (n = 4,343)
3Phase I-NA: the sample (n = 3,864) analyzed previously in [12], although the current analysis differs by inclusion of additional variants available through genome-wide imputation with the hybrid reference panel
4Phase II-NA: continued North American recruitment (n = 1,640)
5FGMS: individuals from the French CF Gene Modifier Study (n = 1,266)
The SLC6A14 association region is subject to X-inactivation based on the H3K27me3 histone methylation pattern provided by the Roadmap Epigenomics Mapping Consortium (REMC (Web Resources in Material and Methods); experiments ENCSR204NFO and ENCSR727VOB [25] in female lung tissue where strong SLC6A14 expression occurs, GTEx [23]). Therefore, the observation that the risk alleles for the genome-wide significant SNPs display lower odds ratios (OR) in females (with two versus zero copies of the risk alleles) than the ORs in males (one versus zero copies of the risk alleles), p = 0.074, 0.046, and 0.058, respectively, for rs3768766 A, rs5905177 T, and rs12710568 C risk alleles (S4 Table), suggests the possibility of sex-specific differences beyond what is expected under random X-inactivation.
A new genome-wide significant locus was identified on chromosome 13 with minimum p-value = 3.83x10-10 at rs61948108 (Table 2 and S3 Table), and located in intron 17 of the ATPase H+/K+ Transporting Non-Gastric Alpha2 Subunit (ATP12A) (panel (c) in Fig 2). Restricting the association analysis to the 4,343 individuals homozygous for Phe508del resulted in a larger effect size (OR = 1.7 vs. 1.55, Table 2, S3 Fig). Although this did not result in statistically significant GxG interaction (interaction p = 0.057), the dependence on CFTR genotype of the ATP12A-meconium ileus relationship requires further investigation. Modifier-CFTR-mutation relationships have previously been reported for CF lung disease [2, 33], and ATP12A was previously implicated as a modifier of CF lung disease using candidate gene analysis in pig models [34].
Conditional analysis at each of the three genome-wide significant loci displayed no evidence for allelic heterogeneity (S4 Fig). Top variants from the three genome-wide significant loci explain 7% of the phenotypic variability for meconium ileus. Of the three FDR significant loci (Table 2), the chromosome 7 locus (‘Mega-all’ p = 2.55x10-7 at rs3757377 located upstream of PRSS1; S5 Fig) was suggestive in the previous consortium study (Fig 1 of [12]), and is replicated in the additional North American cohort (‘Phase II-NA’ p = 8.43x10-3, Table 2). We note that the physical landscape encompassing this locus has been revised in recent genome map versions, such that it remains unclear if variation at this locus has been fully appreciated. The new chromosomes 5 and 20 loci were not identified in the previous study [12] and will need to be investigated further.
Integration of GWAS and tissue-specific gene expression data
The identified associated variants and those nearby in high linkage disequilibrium (LD) do not occur in gene coding regions; however, they include variants near the transcription start sites (TSS) of the nearest genes suggesting roles in gene regulation. PRSS1, or cationic trypsinogen, is used as a measure of early pancreatic disease in CF [35, 36]. At the PRSS1 locus, the meconium ileus associated SNP, rs3757377 (Table 2, Chr7:142455538, 1.8kb 5’ of the TSS, OR = 1.29, p = 2.55x10-7; S5 Fig) is in high LD (r2 = 0.71 and D’ = -0.99) with rs10273639, the variant that was identified in a GWAS of alcoholic-related chronic pancreatitis in Europeans [37]. A follow-up study [38] further showed that rs4726576 (chr7:142457132, 186bp upstream of PRSS1; S3 Table), a variant in high LD with both GWAS study-associated variants (rs3757377 and rs10273639), is contributing to the pancreatitis relationship by directly influencing gene expression levels based on reporter gene assays in acinar pancreatic cells. These assays and consideration that the encoded digestive enzyme function originates from the pancreas argue that variants associated with meconium ileus at the PRSS1 locus impact gene expression in this organ. The pancreatitis protective T allele of rs10273639 that is associated with less PRSS1 expression is a risk allele for meconium ileus. The meconium of individuals with CF contain high levels of protein and less trypsinogen may adversely retain consistency [39]. We did note that no eQTLs for PRSS1 are reported in the pancreas in the genotype tissue expression project (GTEx). This may be reflecting its extremely high expression (the PRSS1 transcript is the most highly expressed gene in the pancreas with median TPM of 9.91 x 104 [23]), or that eQTL determination is affected by the local genome landscape that has been altered in the most recent genome assembly (GRCh38/hg38) with added genes as well as indication of alternative assemblies.
To determine whether gene expression variation at the three genome-wide significant loci (SLC6A14, SLC26A9 and ATP12A) influence meconium ileus risk and in which tissue(s), we used data from the GTEx v7 (accessed on Oct 1, 2018) [23] and considered the enhancer and promoter states from the Regulatory Regions Map (Reg2map; see Web Resources in Material and Methods) [24, 25]. We first display the association findings (Fig 2, colored circles as in the LocusZoom plots [40]) at the three loci in the context of the available eQTLs of gastro-intestinal tissues (Fig 2, colored lines indicating different tissues; eQTLs for SLC6A14 in the pancreas and ATP12A in the transverse colon were included in the GTEx v6 but not v7. They were calculated using v7 data and included here; Material and Methods and sex-stratified analysis S6 Fig), with the locations of tissue-specific enhancers and promoters noted when available to complement the comparisons. Notably, the pattern of eQTL evidence for each of SLC6A14, SLC26A9 and ATP12A in the pancreas (black solid line) mirrors the genetic association pattern (colored circles), with the enhancer and promoter states overlapping with pancreas-specific regulatory elements for at least SLC26A9 and ATP12A. In contrast, eQTLs in the small intestine (blue dashed line) do not meet expression threshold criteria by either GTEx v6 or v7 (due to a very low level of expression) for SLC26A9 and ATP12A (panels (b) and (c) in Fig 2) or do not overlap (SLC6A14, panel (a) in Fig 2) with the GWAS findings. Based on GTEx data, the meconium ileus risk alleles are associated with decreased SLC26A9 expression, but increased ATP12A and SLC6A14 expression in the pancreas.
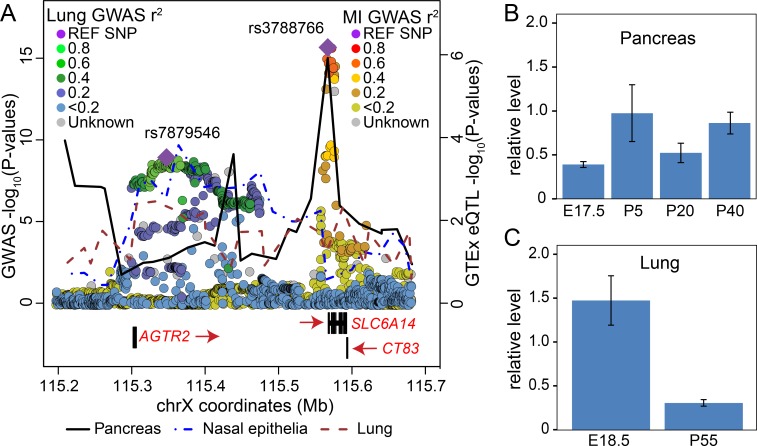
The chromosome X locus associated with meconium ileus also displayed association with CF lung function severity [41], with similar effects in males and females. However, the previously identified broad peak of association for lung function appeared shifted and was notably closer to the angiotensin II receptor type 2 (AGTR2) gene (panel (a) in Fig 3, blue/green dot palette for lung function association [41], and red/yellow dot palette for meconium ileus association). The SLC6A14 eQTLs from the GTEx lung samples (panel (a) in Fig 3, brown dash-dotted line) did not appear to colocalize with the lung GWAS association evidence. Given that the GTEx protocol for lung tissue collection largely avoids the respiratory epithelia (which include the most relevant cell types in CF), we confirmed the lung eQTLs by RNA sequencing of nasal epithelia (a surrogate airway model in CF [42]) from 63 Canadians with CF included in the GWAS (panel (a) in Fig 3, blue dash line). The SLC6A14 eQTLS from the nasal epithelia (blue dashed line) and pancreas (black solid line) tissues appeared to coincide, respectively, with lung disease and meconium ileus associated variants. eQTLs for AGTR2 in lung from GTEx also colocalize with the lung association signals in this region (S7 Fig), however AGTR2 is not expressed in the nasal epithelia.
Fig 3. SLC6A14 meconium ileus and lung association and gene expression profiles point to cis-regulation that is distinct in pancreas and lung.
(A) Overlay of p-values (on the–log10 scale) from the meconium ileus GWAS (red/yellow palette of colored dots; this study), from the previous lung function GWAS (green/blue palette of colored dots; [41]) and from the GTEx eQTL for association between SNPs and SLC6A14 expression (lines, derived as for Fig 2) for tissues of interest. (B) and (C) Slc6a14 gene expression levels measured by RT-PCR of RNA from C57BL6/J mice (n = 4) from pancreas (B) and lung (C); expression is shown relative to Gusb at gestational (E) or post-natal (P) days indicated.
To formally evaluate the colocalization patterns observed in Figs 2 and 3, we developed a frequentist regulatory integration analytic framework, referred to as the Simple Sum (SS, Material and Methods and S1 Appendix), to directly test whether eQTLs for a given gene and in a given tissue colocalize with meconium ileus-associated GWAS variants, and can also test whether one tissue (or expression for one gene) shows greater colocalization over another. Only summary statistics for the respective eQTL and association evidence are required, and the colocalization p-values are simple to calculate analytically. We developed the SS because detecting colocalization in the presence of high LD and/or allelic heterogeneity is challenging for several commonly used colocalization methods [43–48], the majority of which are Bayesian approaches, such as COLOC [43] and eCAVIAR [44], that estimate the posterior probability of co-localization under specific causal variation assumptions. COLOC and eCAVIAR can be used with summary statistics, but COLOC assumes a single causal variant in both eQTL and GWAS, and the presence of allelic heterogeneity reduces the co-localization posterior probability. eCAVIAR can accommodate allelic heterogeneity, but the posterior probability of co-localization will be averaged across several variants in high LD. In contrast, the goal of the SS approach is not to estimate which variant is causal, but rather to assess the evidence for a regulatory mechanism at the associated locus. The performance of the SS approach, including in the presence of high LD and allelic heterogeneity, was confirmed by extensive simulation studies (Material and Methods, S1 Appendix, and S5–S7 Tables), demonstrating correct type 1 error control across most settings (S8–S13 Figs, and S8 and S9 Tables) and good power performance (S11–S16 Tables), as well as agreement with permutation-based results in applications (Table 3 and S10 Table). Results of method comparison with the alternative approaches COLOC [43] and eCAVIAR[44] are also provided (Table 3, and S8, S11–S16 Tables).
Table 3. Results of Simple Sum colocalization and contrasting colocalization analyses for the three meconium ileus genome-wide significant loci, and colocalization posterior probabilities from COLOC and eCAVIAR.
The eQTL evidence is the GTEx p-value based on the -log10(eQTL p) scale for a specified gene and tissue. The meconium ileus-SNP association evidence is the -distributed Wald test statistic obtained in this study. Analytical (and permutation-based) Simple Sum (SS) colocalization p-values correspond to each individual tissue and evaluates if the eQTLs and meconium ileus-associated variants colocalize. Simple Sum contrasting (SSC) colocalization p-value evaluates if the eQTLs in the pancreas colocalize with meconium ileus-associated variants more than eQTLs in another tissue; NAs are listed for the pancreas since we do not contrast pancreas with itself. All p-values are one-sided because only positive association implies eQTL-association colocalization (i.e. eQTL peaks coincide with association peaks). COLOC [43] PP represents the posterior probability of eQTL-association colocalization calculated for each individual tissue. eCAVIAR [44] calculates SNP-level colocalization posterior probability (CLPP) for each variant in a locus for a given gene and in a given tissue. The regional colocalization probability (RCP) can be defined by summation of the SNP-level CLPP in a given LD block; RCP is recommended in [49] and the results are provided in the ‘eCAVIAR RCP’ column. eCAVIAR maximum regional CLPP provides similar qualitative results as eCAVIAR RCP thus are not shown here.
| Gene | Tissue | SS Analytical colocalization p-value |
SS Permutation colocalization p-value |
SSC colocalization p-value (Pancreas vs. other tissue) |
COLOC PP |
eCAVIAR RCP |
|---|---|---|---|---|---|---|
| SLC6A14 | Pancreas | 1.12x10-10 | 0 | NA | 0.9966 | 0.4916 |
| Esophagus | 1 | 1 | 9.97x10-9 | 0.0191 | 0.0035 | |
| Transverse Colon | 9.49x10-9 | 0 | 7.65x10-9 | 0.3755 | 0.0213 | |
| Stomach | 0.983 | 0.983 | 7.64x10-8 | 0.0265 | 0.0035 | |
| Lung | 1 | 1 | 7.68x10-9 | 0.014 | 0.0039 | |
| SLC26A9 | Pancreas | 4.48x10-5 | 2.00x10-5 | NA | 0.6926 | 0.0172 |
| Esophagus | 0.0301 | 0.0303 | 5.47x10-5 | 0.0064 | 0.002 | |
| Stomach | 1 | 1 | 5.73x10-6 | 0.0275 | 0.0046 | |
| Lung | 0.641 | 0.642 | 4.05x10-7 | 0.0241 | 0.006 | |
| ATP12A | Pancreas | 3.35x10-8 | < 10−5 | NA | 0.285 | 0.0188 |
| Transverse Colon | 1 | 1 | 1.67x10-9 | 0.06 | 0.0017 | |
| Esophagus | 1 | 1 | 8.85x10-9 | 0.0474 | 0.0026 | |
| Lung | 1 | 1 | 1.86x10-8 | 0.0372 | 0.0028 |
Consistent with the patterns seen in Fig 2, the eQTL evidence for SLC6A14, SLC26A9 and ATP12A in the pancreas (as measured by −log10 (eQTL p) are significantly correlated with the GWAS evidence (as measured by meconium ileus-SNP Wald association statistic) at each of these three loci, with SS p-values = 1.12x10-10, 4.48x10-5 and 3.35x10-8, respectively for the three loci (Table 3, analytical results, confirmed by the permutation-based results also provided in Table 3). Posterior probabilities of colocalization using the alternative COLOC [43] and eCAVIAR [44] methods provide supportive although less conclusive findings (Table 3), particularly for the ATP12A locus. In addition, eCAVIAR, which calculates SNP-level colocalization statistics and can be challenged by substantial LD in regions of interest, did not provide a regional colocalization probability that exceeded 0.50 across any of the three regions or tissues; the eCAVIAR maximum occurred at rs3788766 at chromosome X for the pancreas.
At the chromosome X lung GWAS locus (Fig 3 and S7 Fig), application of the SS colocalization analysis indicates that eQTLs from GTEx for AGTR2 in the lungs show evidence of colocalization with the lung-associated variants but not for SLC6A14 (colocalization p-values = 1 and 2.82x10-7 for SLC6A14 and AGTR2, respectively; S17 Table). Given sub-optimal content of epithelia in the GTEx lung tissue samples by design, we calculated SLC6A14 eQTLs from the CF nasal epithelia which provided colocalization evidence with the lung disease associated variants(p = 2.4x10-4); AGTR2 was not expressed in the nasal epithelia. Individually, the lung and meconium ileus GWAS association findings could be interpreted as simply indicating distinct genes for the two CF phenotypes. However, the integration of the tissue-specific eQTL patterns for SLC6A14 (Fig 3) support a role for SLC6A14 in both CF lung and pancreas. Further, as regulatory elements can exert effects over long distances, the differential positioning of the GWAS peaks with the co-alignment of eQTL peaks could accordingly reflect alternate modes of cis-regulation of SLC6A14.
Significant colocalization can occur in more than one tissue (Table 3, S10 Table and S14 Fig) or for expression of more than one gene. Therefore, it is also of interest to directly contrast the colocalization evidence between tissues or genes (Material and Methods, Table 3, S10 and S17 Tables), suggesting for example, the most probable tissue(s) of origin or target gene for an association locus identified via GWAS. Contrast colocalization analysis for the meconium ileus GWAS loci showed that eQTLs for SLC6A14, SLC26A9 and ATP12A in the pancreas colocalize with meconium ileus-associated variants significantly more than eQTLs for these genes in the other meconium ileus-relevant tissues tested including the intestine (Table 3, SSC colocalization p-value results). The maximum posterior probability of colocalization from COLOC and eCAVIAR also occurred with the pancreas eQTLs versus the other tissues evaluated (Table 3). These findings are consistent with the pancreas-specific PRSS1 locus in that variation in SLC6A14, SLC26A9 and ATP12A gene expression in the pancreas appears to mediate the association between meconium ileus and the GWAS variants.
Is exocrine pancreatic damage a causal contributor to meconium ileus?
We previously used the SLC26A9 variant rs7512462 (S3 Table) as an instrumental variable in a Mendelian Randomization (MR) study to assess whether damage to the exocrine pancreas is a causal contributor to CF-related diabetes [15]. To determine whether variation in exocrine pancreatic injury in utero (measured by immunoreactive trypsinogen (IRT) at birth) is a causal contributor to meconium ileus, we now likewise applied a two-sample MR procedure [50, 51] using the same instrumental variable (Material and Methods). The two-sample method first assessed the effect of rs7512462 on meconium ileus in the Canadian Gene Modifier Study (CGMS) Phase I sample (n = 1,661, OR = 1.39, 95% CI = [1.14, 1.70]). Then a sub-sample of n = 126 was used, for whom IRT measurements were available, to estimate the effect of rs7512462 on log(IRT) levels estimated at birth (mean change in log(IRT) per additional rs7512462 risk allele (T) = -0.60, 95% CI = [-1.06, -0.14]). These lead to the estimate of the causal effect of pancreatic damage on meconium ileus risk (OR = e[log(1.39)/-0.60] = 0.58, 95% CI = [0.34, 0.99]). This significant causal estimate is similar to that obtained from a logistic regression of estimated log(IRT) on meconium ileus status (OR = 0.54, 95% CI = [0.40, 0.70]), suggesting this direct association is largely free of confounding. Together these data contribute to evidence that variation in gene expression in the exocrine pancreas may be a causal contributor to meconium ileus, although the MR assumption of no horizontal pleiotropy [16] is difficult to confirm for single-instrument analyses. In support of this assumption, we note that expression of SLC26A9 is reported in the pancreas (median TPM 1.60), with essentially none in the relevant terminal ileum tissue (i.e. small intestine; median TPM 0.02) in human (GTEx, [23]). Similarly, Slc26a9 was noted in the pancreas but was undetectable in the distal segments of the small intestine by RT-PCR analysis of murine tissues at adult stages (see below and [52]).
Discussion
Together with the identification of PRSS1 as a meconium ileus modifier, the integration of GTEx eQTL evidence and meconium ileus susceptibility associations through colocalization analysis support that variation at the identified modifier loci may reflect SLC6A14, SLC26A9 and ATP12A gene expression, and in the CF pancreas rather than in the intestine or any other meconium ileus-relevant tissues tested. The underlying mechanism of this complex intestinal obstruction phenotype that requires loss of CFTR in the intestine [53] must then include the critical roles played by the transporters in early development and/or the early tissue damage that arises with loss of CFTR in the pancreas.
SLC6A14 encodes a neutral and cationic amino acid transporter [54] that based on GTEx information is most notably expressed in human lung. Various cancers of epithelial origin, including breast [55], colon [56] and pancreas [57] also exhibit robust expression that is relatively elevated from their source tissues. The SLC6A14 rs3788766 was the most significant SNP in this (Table 2, panel (a) in Fig 2) and our previous meconium ileus GWAS [12] (S3 Table), and its risk allele T corresponds to increased transcript level based on the GTEx eQTL data of adult pancreas tissue (panel (a) in Fig 2). Although human data from early stages of development would be preferable, we did examine pancreatic and lung tissue from the mouse spanning late embryonic to adult stages of development (panel (b) and (c) in Fig 3) to find maintained, albeit relatively low, Slc6a14 transcript levels in the pancreas, with decreasing levels in lung over time. The rs3788766 SNP is in perfect LD (r2 = 0.8 and D’ = 1) with rs12710568, which is just 714bp 5’ of the SLC6A14 TSS. ChIP-seq evidence [58] demonstrates the binding of primed AR (androgen receptor), which has been shown in seven independent studies (GSE58428, GSE84432, GSE56288, GSE28950, GSE56086, GSE65478, GSE32892) and this SNP falls within the androgen receptor binding motif (5'-GGA/TACA[N]NNTGTTCT-3'—SNP position marked in brackets [59]) and could potentially contribute to the sex-specific risk differences observed (S4 Table, S6 Fig).
In contrast to the chromosome 13 and X loci, there are additional genes and tissues that show evidence of colocalization with the GWAS variants (S14 Fig) within 1Mbp of the chromosome 1 locus. As a consequence it is more challenging to pinpoint the responsible gene and tissue from the colocalization evidence alone. However, published experimental evidence for SLC26A9 complements the colocalization results. SLC26A9 has been characterized as an anion channel in epithelial cells [60, 61]. Slc26a9-/- /Cftr-/- double knock out models have a lower rate of survival post-birth than either Cftr-/- or Slc26a9-/- mice [52]. SLC26A9 is expressed in several tissues including the lung, stomach, upper intestinal tract, kidney, and pancreas [25, 52, 62, 63]. Several studies indicate that CFTR and SLC26A9 interact in lung tissue, with biochemical and electrophysiological studies indicating that interaction enhances functional expression of CFTR [9–11]. Interestingly, this genome-wide study, as well as previous studies of early-onset pancreatic phenotypes that identified SLC26A9 as a CF modifier [12–14], highlight that SLC26A9 and CFTR may not require interaction in the pancreas in the same way apparent for the lung phenotype. The meconium ileus association is seen in patient cohorts composed of many different CFTR genotypes, and the effect size is similar in individuals who are Phe508del/Phe508del (OR = 1.41, se = 0.058) versus other CFTR genotypes (OR = 1.32, se = 0.083). Indeed, notable differences in murine mRNA levels between late gestation and adult stages for Slc26a9 in the pancreas and the lung, in opposing directions, suggests the function of SLC26A9 may be different across time and tissue (panel (c) in Fig 4 of [2]). Detailed analysis of murine Slc26a9 expression in the intestine by others have also indicated changes in expression with young versus older ages [52]. Taken together with previous reports of SLC26A9-associated pancreatic phenotypes [13–15] we purport that SLC26A9 may provide alternative chloride transport at early developmental stages, and that its contribution to meconium ileus, is at least partly, from the pancreas.
ATP12A encodes the α-subunit of the non-gastric H+/K+ transporter. In a recent study, its expression in human and pig versus the negligible expression in the murine airway was argued to explain differences in airway surface liquid pH, viscosity and host defense in human and pig versus murine CF airways [34]. Although we were not able to detect Atp12a mRNA in total mouse pancreas using RT-PCR, ATP12A expression has been seen in human pancreatic ductal lines [64] and in isolated rat pancreatic ducts with localization toward the lumen [65]. The essential contributions of H+/HCO3- movements and pH maintenance for fluid secretion in the pancreatic duct has been studied extensively, and any disturbance with decrease in pH would increase concern for effective digestive enzyme transport and risk of auto-activation. With the eQTL findings pointing to the pancreas, the common allele variants in ATP12A that associated with meconium ileus risk are also associated with increased ATP12A expression, where even modest increases in proton secretion, may be critical in the absence of CFTR [66].
Meconium ileus has complex etiology. The pig and murine models have highlighted the engagement of the gastrointestinal tract, where confined restoration of CFTR is sufficient to ameliorate both meconium ileus [53] and the intestinal obstruction phenotypes to substantial degrees [67, 68]. Although the genes identified in this GWAS are candidates for contributions to CF given their roles in epithelial transport and function, a working model in which their contribution to intestinal obstruction at birth is through common variation of their expression in the CF pancreas was not immediately obvious, beyond PRSS1. Understanding the organ of origin for any modifier gene is highly relevant as we embark on personalized medicine, first, toward designing the most optimal functional studies to test therapies and second, toward development of the complex models and signatures of prediction for both prognosis and therapeutic responses. Access to tissue-specific expression data for colocalization analysis here, beyond the nasal epithelia data, was limited to that available in the GTEx consortium project, and therefore our conclusions must be viewed in that context. Consideration of expression of modifier genes in early pancreatic stages rather than just adult tissue would be helpful for confirmation.
There is substantial variability in response to the recently developed CFTR-modulatory therapies between [69, 70] and within [71] individuals, and modifier studies highlight that individual level genetic background beyond CFTR genotype contributes to these differences. For example, we have shown that variation in lung response to modulator therapy can be explained in part by the SLC26A9 modifier gene, possibly via interaction with CFTR [2]. Consideration of temporal and spatial expression of CFTR and modifier genes is needed when assessing CFTR function and therapeutic response for the purpose of choosing therapeutic strategies. This is specifically relevant where current testing paradigms use primary CF-tissue models such as nasal and lung epithelia, or rectal and lung organoid models [72–74]. Maximum benefit will be achieved with consideration of genetic background, as these models are being considered to decipher how to implement therapeutic decision-making and achieve personalized medicine [73].
Predictive genetic signatures that include organ specific aspects hold great promise to direct and complement therapeutic decision making in CF and other inherited diseases. They have additional potential in assisting clinical monitoring regimes, and toward stratification of patients for clinical trials to optimize outcome.
Material and methods
Ethics statement
Studies were approved by institutional review boards at participating sites and include Johns Hopkins School of Medicine eIRB2 (Committee: IRB-3); Research Ethics Board of the Hospital for Sick Children; Biomedical Institutional Review Board, Office of Human Research, University of North Carolina at Chapel Hill; University Hospitals Case Medical Center, Institutional Review Board for Human Investigation; and the French ethical committee (CPP n°2004/15) and the information collection was approved by CNIL (n°04.404). Written informed consent was obtained from adults, and for patients<18 years old consent was obtained from parents or guardians for participation in the study. The research ethics approval number for this cystic fibrosis gene modifier study was obtained from the Hospital for Sick Children Research Ethics Board, number 1000016662.
Study sample
The International CF GMC consists of individuals from four consortium sites, Johns Hopkins University (JHU), the University of North Carolina/Case Western Reserve study (UNC/Case), the Canadian Gene Modifier Study (CGMS) and the French Gene Modifier Study (FGMS). The North American (NA) data were genotyped in two phases, Phase I-NA with 5,132 individuals and Phase II-NA with 2,830 individuals (before quality control; see below). Data from the FGMS were genotyped separately and include 1,300 individuals with CF. The Phase I-NA data were the basis of the first published meconium ileus genome-wide association study (GWAS) from the International CF GMC [12].
Genotyping and quality control
Genotyping used four different Illumina platforms: the 610Quad, CNV370, 660W and Omni5, where 660W genotyping was performed in three batches (660W-NA, 660W-JHU and 660W-FR). Genotype calling was performed using GenomeStudio V2011.1. Quality control (QC) of genotypes and phenotypes was performed separately for each of the six genotyping platforms and batches: 610Quad, 660W-NA, 660W-JHU, Omni5, CNV370 and 660W-FR. Sample details after data quality control (QC) are listed in S1 Table, and QC procedures for SNP exclusion are presented in S2 Table.
SNP position and annotation information were based on Genome Reference Consortium 37 (GRCh37). The number of probes for each platform before QC was 570,572 for the 610Quad, 655,214 for 660W-NA and 660W-JHU, 4,301,332 for the Omni5, 309,012 for CNV370 and 554,649 for 660W-FR (S2 Table). SNPs were removed if they were not annotated to chromosomes 1–22 or X, with no rs number, or had a call rate <90%. Duplicated variants (occurred on the Omni5 chip after mapping kgp SNPs into dbSNPs) were also removed. Additionally, SNPs on chromosome X with heterozygosity rate more than 10% in the male sample were removed [75]. To reduce potential false negatives, the SNP QC criteria used here were relatively lenient, but any apparent association signals were subject to additional QC scrutiny to minimize potential false positives.
Detailed QC procedure for sample exclusion is as follows: individuals were removed if the initial call rates were <95%, or had extreme heterozygosity rates where the threshold was defined as in [12]: that is, 10 S.D. away from the mean autosomal heterozygous rate calculated from a set of pruned common SNPs (r2<0.2, MAF>0.05). Plink (v1.07, Web Resources in Material and Methods and [76]) inferred sex for each individual was compared to the reported gender and individuals with mismatched sex were then removed. After the aforementioned QC was performed separately for each platform and genotyping batch, all six subsets were then merged together and KING (Web Resources in Material and Methods and [77]) was used to identify any cryptic familial relationships among all individuals. Duplicated individuals/identical twins were identified and for each pair the ones with more genotype data were kept. Individuals without phenotypic information were excluded. All analyses were restricted to individuals with two severe CFTR mutations associated with pancreatic insufficiency (PI) since individuals with more mild mutations are in general at lower risk of meconium ileus [5]. Lastly, KING was used for population stratification using principal component analysis (PCA), and individuals identified as non-Europeans were excluded. Specifically, genotyped SNPs common across all platforms with MAF>0.05 and in low linkage disequilibrium (LD) with each other (r2<0.2) were included for PCA on the combined sample. Following the PCA procedure used in [12], non-European descendants were first excluded, defined as 6 S.D. away from the center of the HapMap3 [29] European (CEU/TSI) cluster. The Tracy-Widom statistic, computed using Eigensoft [78–81], was then used to determine the statistical significance of each principal component; six principal components were determined to be significant (p<0.05) and were included in subsequent association analyses as covariates. S1 Fig displays the pairwise comparison of the first three principal components.
Imputation
A subset (n = 101) of the Canadian CF Gene Modifier cohort that mirrored the Canadian severe CF population also underwent whole genome sequencing by Complete Genomics (CG) at an average of 30X coverage. These participants were of European ancestry and had two CFTR mutations associated with pancreatic insufficiency [5, 12, 19], with 64.2% being homozygous for the most common CFTR mutation, Phe508del. The genome sequences were integrated with the 1000 Genomes (IKG) Phase 3 data [82] to generate a hybrid reference panel [83, 84] that was then used for genome-wide imputation of the full GMC sample using Beagle (Web Resources in Material and Methods) [85]. Details on the methodology can be found in [28]. The hybrid reference with in-sample whole genome sequence ensured the presence of CFTR disease-causing haplotypes that were not well represented in publicly available reference panels such as the 1000 Genomes Phase 3 reference [82] or the Haplotype reference consortium panel [86]. The importance for constructing this hybrid reference that includes individuals representative of the study population (CF in our case) is best illustrated by S2 Fig, which demonstrates the gain in fine-mapping achieved by augmenting the 1KG reference with WGS from 101 patients with CF, suggesting better coverage of study population-specific variants [28].
After obtaining this CF-specific hybrid reference panel, Beagle V4.1 [87] was used for phasing and imputing missing genotype data for our samples. We applied the same method as in [28] for whole genome pre-imputation phasing. Samples were phased and imputed separately for the six genotyping platforms. Approximately 9% of our samples displayed evidence of non-European ancestry by principal component analysis, thus all individuals in the reference panel were used for phasing and imputation.
Beagle used a sliding window of 250,000 markers from our genotype data with 25,000 markers overlapping with the next window for the phasing and then imputation steps. The phasing step included all individuals from our samples and from the reference panel to conduct 10 burn-in phasing iterations, followed by additional 5 phasing iterations to ensure accuracy, and the imputation step followed to fill in the markers not genotyped. Imputed SNPs with Beagle quality score AR2<0.3 were set to missing and SNPs with more than one alternative allele or with a minor allele frequency <1% were excluded from the association analysis.
Association analysis
Genome-wide association analysis was carried out using Generalized Estimating Equations (GEE) with an exchangeable covariance structure to account for sibling relationships in the cohorts, as implemented previously [12]. Covariates included the six significant principal components, as well as three indicators for the four consortium sites (CGMS as baseline, JHU, UNC/Case and FGMS) and three indicators for the four genotyping platforms (namely, 610Quad as baseline, Omni5, CNV370 and 660W as defined in S1 and S2 Tables) to account for potential batch effects. Genotypes were coded additively for all the autosomal SNPs. For X chromosome SNPs females were coded additively, and males were coded as 0 and 2. Genome-wide significance was defined as p≤5x10-8 [30] and the accuracy of p-value calculation was up to 2.2×10−16 by the geeglm function [100] in R.
Gene-based analysis of CFTR
In total, 241 (genotyped and imputed) SNPs with MAF ≥1% and within 10kb of the boundaries of CFTR were included in a gene-based permutation test as in [12] using the 5,869 unrelated individuals. For the observed data, the association analysis was performed using logistic regression adjusting for site and platform effects, for each of the 241 SNPs. The SNP-level Wald test statistics were aggregated, and the sum represented the gene-level association evidence. The observed meconium ileus phenotype values were then permuted within each of the site and platform groups, independently 10,000 times, and the corresponding sum of the Wald association statistics were obtained. The empirical p-value was the proportion of the permutation samples whose sum was larger than that calculated from the observed sample.
Replication of the apical plasma membrane hypothesis
Previous studies demonstrated that multiple constituents of the apical plasma membrane that reside alongside CFTR also contribute to meconium ileus susceptibility [12]. To confirm this gene-set association, all 157 apical plasma membrane genes were extracted from the Refseq table in August, 2016 and the grand union of the boundaries in hg19 from multiple transcripts were used to extract corresponding SNPs. To be consistent with the original analysis in [12], the analysis here was limited to SNPs genotyped on the Illumina 610 Quad in the updated gene region with +/-10kb of the gene boundaries; corresponding SNPs for samples genotyped using other platforms were imputed. In total, 3,951 SNPs were included in the apical gene set analysis. A permutation test (using 10,000 replicates), similar to that for gene-based analysis above, was then carried out to obtain the empirical p-value for the gene-set.
Chromosome X analysis
To investigate sex-specific effects of variants from the SLC6A14 X chromosome locus, besides the association test above, GEE analyses were also conducted separately for males and females using either additive or genotypic coding. A contrast test comparing males with the risk allele (e.g. D) to females homozygous for the risk allele (DD) was also implemented to test for sex-SNP interaction effects. This test assumed a common baseline risk of meconium ileus between males and females without the risk alleles (e.g. d and dd), and the statistical model was constructed as
where IFemale,1, IFemale,2 and IMale,1 are indicator variables representing, respectively, the female dD and DD genotype groups and male D genotype group, and g(μ) is the logit link function. Under the X-inactivation assumption supported by evidence from the Roadmap Epigenomics Mapping Consortium [25], the DD genotype is expected to express one allele. Thus, testing whether β2 = β3 provides evidence for sex-specific differences in effect size or an interaction effect. This contrast test was performed using the doBy package (version 4.5–15) in R (Web Resources in Material and Methods).
Estimation of phenotypic variance explained
Pseudo R2 was used as a measure of the phenotypic variance explained by the associated SNPs. The estimate was calculated based on multivariate logistic regression (covariates include the top significant SNP from each of the three genome-wide significant loci in addition to the six PCs and six site and platform indicators), applied to 5,869 unrelated individuals (a subset of the original sample of 6,770 that included siblings), using the rms package (version 4.5–0) in R.
Mendelian randomization
Immunoreactive trypsinogen (IRT) is a biomarker of exocrine pancreatic damage (PD) [22]. In the CGMS subset, n = 126 individuals had longitudinal measures of IRT. A linear mixed effects (LME) regression model, accounting for the limits of detection of the IRT assay (<3ng/ml) by the statistical model [88], was used to estimate patient-specific IRT measurements at birth. IRT and age (days) were natural log transformed. The patient-specific birth IRT estimates from this model were then used as measures of prenatal pancreatic damage as in earlier biomarker studies [15].
To determine if variation in the exocrine pancreas contributes to the cause of meconium ileus, we estimated the causal effect using an application of instrumental variable analysis known as Mendelian Randomization (MR) [51]. We compared the estimate of causal effect to the (possibly confounded) direct effect estimate obtained by regressing meconium ileus status on estimated values of log(IRT) at birth. The causal effect on meconium ileus was estimated using the two-sample MR method with SLC26A9 rs7512462 genotype (coded additively) as the instrumental variable [50, 51], applied to the CGMS Phase I genotyped sample (i.e. sample one, n = 1,661) and the sub-sample for whom IRT measurements were available (i.e. sample two, n = 126). Dividing log(OR), the association estimate obtained from the logistic regression of meconium ileus status on rs7512462 using sample one, by the average increase in log(IRT) at birth per additional rs7512462 risk allele, obtained from linear regression of log(IRT) on rs7512462 using sample two, gives the log(OR) that represents the association of genetically determined log(IRT) levels at birth with meconium ileus risk [51]. We obtained a cluster robust standard error for the estimated OR in sample one, and used the delta method to estimate the standard error for the two-sample causal OR estimate [89].
The application of MR required that the SLC26A9 instrument be robustly associated with the exposure (exocrine PD). Although we obtained a modest F statistic of 6.7 from the regression of log(IRT) at birth on rs7512462 genotype, the log(IRT)-rs7512462 association has now been demonstrated in two independent samples from Canada [15] and the United States [14]. In addition, our use of the delta method to estimate the confidence interval surrounding the causal estimate is conservative [89]. MR assumes that the relationship between the instrument (SLC26A9) and meconium ileus was mediated by variation in prenatal exocrine PD, and not due to any pathway independent of PD. This critical assumption of ‘no horizontal pleiotropy’ [16], although difficult to prove in practice [90], is supported here by a lack of SLC26A9 gene expression in intestinal tissue consistent with the ‘sites’ of meconium ileus in the ileum and proximal colon [52].
Enhancer and promoter regions
The enhancer and promoter region data are obtained from the Broad Institute website [24, 25]. The enhancer regions included were defined as those with DNaseI signal with -log10P>2 and falling in enhancer states 6,7,12 in the 5-mark 15-state chromHMM observed data across 127 epigenomes from the Roadmap [25] and ENCODE [24] tissues. The promoter regions were defined as those falling within a DNaseI signal with–log10P>2 and 15-state chromHMM states 1,2,10.
Genotype tissue expression consortium (GTEx) data
We investigated all genes in the relevant tissues that satisfied the GTEx v6 threshold for expression, which is a gene expression threshold of >0.1 RPKM in at least 10 individuals and > = 6 reads in at least 10 individuals. This criteria was slightly more liberal than in v7 which calculated eQTLs when there was >0.1 TPM in at least 20% of samples and > = 6 reads in at least 20% of samples. All eQTLs were obtained from the GTEx v7 portal with the exception of SLC6A14 in the pancreas and ATP12A in the transverse colon, which satisfied v6 but were just shy of satisfying the v7 criteria. We calculated these eQTLs following the GTEx protocol. Briefly, normalization of the publicly available GTEx expression matrix (version 7, 15-Jan-2016 data freeze, phe000020.v1) was subsetted for the samples for the specific tissue of interest and expression values were normalized between samples [91], and each gene was then inverse quantile normalized across samples using normalize_expression.py script on GTEx’s Broad Institute’s GitHub page (Web Resources in Material and Methods). Probabilistic Estimation of Expression Residuals (PEER) [92] were calculated using run_PEER.R script from GTEx, while principal components were calculated using the PC-AiR package in R [93], with the kinship matrix input calculated using KING version 2.0 [77]. Sex and sequencing platform covariates were obtained from phs000424.v7. Genotypes were obtained from variants called and QC’d from WGS (phg000830.v1). The FastQTL version 2.0 program [94] was used for mapping cis-eQTLs for each variant-gene pair. Nominal p-values with a mapping window of 1 Mbp were used. The eQTL p-values obtained closely matched those provided on the GTEx v6 portal, although the v7 results provided more significant p-values for SLC6A14 in the pancreas.
RNA-sequencing
We conducted eQTL analysis with RNA sequencing of human nasal epithelial (HNE) cells from 63 CF Canadians enrolled in the GMC. Samples that were included had HNE cells collected that were drug-naïve, and their RNA had an RNA integrity number (RIN) > = 7. Sample processing and RNA sequencing was conducted in two batches following two protocols, n = 32 as part of the CF-Canada SickKids Program for Individualized Therapy (CFIT; [95]) and n = 31 from a UNC-led initiative [96]. In brief, HNE cells for the CFIT were collected by cytology brushes, and RNA sequencing was conducted by the Illumina HiSeq 2500 with the High Throughput Run Mode, producing 35 million paired-end reads with a length of 124 base pairs per sample. The HNE cells from the UNC-led initiative were collected by a Rhino-probe curette, and the Illumina HiSeq 2000 platform was used for RNA sequencing, producing 25 million paired-end reads with 49 base pair length per sample.
The sequencing reads were aligned to the hg19 genome reference using STAR v2.4.2a [97], aided by the GENCODE 19 gene annotation. Gene expression read counts and RPKM were calculated by RNA-SeQC [98] with the strictMode option, based on a collapsed GENCODE 19 gene model as describe by the GTEx consortium [23]. The eQTL analysis followed the GTEx v6p pipeline [23]. Genes with RPKM >0.1 and read counts > = 6 in at least 10 individuals were included. RPKM of selected genes at the chromosome X locus were first quantile normalized to the average empirical distribution across samples, then inverse quantile normalized to the standard normal across samples for each gene. The cis-eQTL mapping was conducted by FastQTL [94] with its default settings. The Beagle imputed SNPs with MAF>5% and within a one mega-base window up and downstream of the transcription start sites were included. Covariates included the first three genotype principal components calculated by KING, 15 hidden variables estimated by PEER [99], normalized RPKM of CD45 (an adjustment for immune cell composition) and sex.
Colocalization Simple Sum testing
At each of the genome-wide significant loci, to statistically determine whether the GWAS associations were mediated through gene expression, and in which tissue, formal colocalization analysis was required to integrate the association evidence with available tissue-specific gene expression data. We implemented a frequentist colocalization framework, the Simple Sum (SS). We outline the method and evaluation strategies below, and we provide details in the Supporting Information including analytic derivations of the method, simulation design and parameter settings, and results of method evaluation and comparisons. The Simple Sum application and simulation results were compared to two other frequently implemented Bayesian colocalization methods designed for summary level data, COLOC [43] and eCAVIAR [44].
To carry out the Simple Sum approach, the meconium ileus-SNP association statistics are the Wald statistics obtained from this GWAS study. To facilitate the permutation-based method that was used to confirm the analytic results, we used the association results obtained from 5,869 unrelated CF participants. The tissue-specific gene expression-SNP association summary statistics (the eQTL p-values) were obtained from the GTEx version 7 (15-Jan-2016 data freeze) [23], except in the case of SLC6A14 in the pancreas and ATP12A in the transverse colon; we calculated these using v7 data as explained above in the GTEx data section. eQTLs from the nasal epithelia of the 63 CF Canadians were used for colocalization analysis at the chromosome X locus to complement the lung eQTLs obtained from GTEx.
The Simple Sum analytical framework
GWAS association-eQTL SS colocalization test for a single-tissue (or gene)
At each genome-wide significant locus, we extract all m genotyped or imputed SNPs that are within 0.1 Mbp on either side of the lead (most significant) SNP at the locus. Using the pancreas as an example, an intuitive approach is to assign each SNP to one of the two groups according to whether it is an eQTL or not based on the GTEx data. The definition of an eQTL can be subjective, e.g. GTEx p<0.05 or <0.005, but for the moment let us first consider using a p-value threshold for eQTL. In that case, let tjk be the gene-expression indicator variable for SNP j and tissue k, then a Simple Sum (SS) colocalization test statistic at a given locus can be defined as
| (1) |
where Sj, j = 1,…,m is the phenotype-SNP association statistic (e.g. the Wald statistic, ) for the jth SNP. This SS statistic represents the difference in average association test statistics for eQTL and non-eQTL variants, and a significant positive value intuitively suggests colocalization of association and eQTL evidence, implying the test must be one-sided which we elaborate on later.
The p-value for the SS test can be evaluated by a permutation procedure similar to that used in the gene-based and gene-set analyses above. That is, we permute the phenotype to preserve the LD pattern between SNPs within each cohort and platform, independently, say 105 times. For each permutation sample, the corresponding phenotype-SNP association Wald statistics and the SS colocalization quantity shown in Eq (1) are re-calculated. The empirical colocalization (one-sided) p-value is the proportion of the permutation samples whose SS test statistics are larger than that in the observed sample. However, this permutation-based approach can be computationally challenging.
Alternatively, we derived the exact distribution of the SS statistic by expressing SS as a quadratic form Z′AZ, where Z = (Z1,Z2, …,Zm)’ is a vector containing the phenotype-SNP association statistics for the m SNPs in the region, and A = diag(a1,a2,…am) with . Z′AZ is distributed as a mixture of chi-squared distributions under the simple null hypothesis of no association and no-colocalization (S1 Appendix), thus the SS p-value can be obtained analytically without the need for permutation. Importantly, this analytic form allows for the use of a continuous measure of gene expression evidence (e.g. tjk = −log10(GTEx p)) in the SS statistic, making the subjective definition of eQTL (e.g. eQTL p<0.05 or 0.005) unnecessary. Moreover, the intuitive interpretation when using the p-values rather than a cut-off is that the colocalization test assesses the similarity in pattern of eQTL and GWAS association, as is visualized in Fig 2. The validity of the proposed analytic method was confirmed by both the application (Table 3 and S10 Table, analytic vs. permutation p-value) and simulation studies (S8–S13 Figs and S8 and S9 Tables for empirical type 1 error control).
We first note that although the proposed SS method is motivated by a statistic that uses the difference in the sum of association test statistics for eQTLs and non-eQTLs, it inherently adjusts for LD between SNPs through properly calibrating the variance of the SS test statistic based on the LD pattern in the region of interest; this is similar to a phenotype-permutation-based empirical approach that preserves the LD pattern between SNPs.
We also note that the test must be one-sided. Besides the intuitive argument above, consider the case where both association and eQTL are significant but at distinct, uncorrelated SNPs (i.e. the H3 scenario of COLOC [43]). In that case, a large tjk corresponds to a small Sj, and it is easy to see that the SS test statistic would be negative, resulting in a large p-value based on the proposed one-sided test. Thus, although the complement to the alternative of interest includes association without colocalization, the one-sided test ensures that this scenario is ruled out. We conducted simulation studies to provide corroborating empirical evidence (Case 4 simulation design in S6 Table with corresponding type 1 error results in S8 Table).
GWAS association-eQTL colocalization contrasting SSC test, contrasting two tissues (or genes)
To determine if there is better colocalization with tissue k (or gene) than with tissue h (or gene), intuitively, we can define a SS contrasting colocalization statistic that assesses the difference in average phenotype-SNP association test statistics for SNPs that are eQTLs for tissue k versus for tissue h,
where tjk and tjh are the gene-expression indicator variables for SNP j in tissue k and h, respectively (e.g. eQTL p<0.05 or 0.005). Again, we were able to reformulate this SSC test statistic in a quadratic form that is also distributed as a mixture of chi-squared distributions under the null hypothesis (S1 Appendix). Thus, we can obtain p-values for SSC efficiently without the need for permutations; p-values can be calculated by using the R package “CompQuadForm”. Similar to the single-tissue colocalization case, we can also use the original continuous measure of gene expression evidence without arbitrarily defining an eQTL (e.g. eQTL p<0.05 or 0.005). S15–S20 Figs provide simulation results for all three loci analyzed and demonstrate that the proposed analytic method controls the type 1 error.
The proposed single-tissue and tissue contrasting colocalization SS framework is quite general, and it requires only summary statistics for the respective eQTL and phenotype-SNP association evidence. The tjk can be −log10(eQTL p) as presented or a Wald eQTL test statistic as was used for the phenotype-SNP association component. The developed analytical approach to obtain p-values for SS makes the implementation easier when applied genome-wide or across several tissues and genes at a locus (S14 Fig). In addition, the proposed method implicitly allows for allelic heterogeneity (S7 and S11–S16 Tables). If there were two (or more) causal variants in a locus of interest and both contributed through regulating gene expression of the same gene (e.g. Alter 6 scenario in S7 Table), then the phenotype-SNP association and eQTL evidence would be statistically correlated, thus be identified by the SS method given sufficient sample size (e.g. Power in S16 Table for the Alter 6 scenario).
Method evaluation and comparisons
We outline the evaluation strategies here and provide additional details of the simulation designs and results in the Supporting Information.
Evaluation through data application
We applied the proposed Simple Sum colocalization and contrasting colocalization methods to the three meconium ileus genome-wide significant loci, SLC6A14, SLC26A9 and ATP12A (Table 3, using -log10(eQTL p) as a continuous measure of the gene expression evidence); a heatmap of the colocalization test is shown in S14 Fig. To validate the analytical p-values, we also provide permutation-based p-values in Table 3. To be comprehensive, S10 Table also provides the corresponding results when we dichotomize the gene expression evidence using various thresholds (e.g. eQTL p< 0.05, <0.005 or <0.0005). For the SCL6A14 locus on Chromosome X, S17 Table provides the results when the gene expression evidence for lung was obtained from GTEx or HNE, and when the contrasting colocalization test was performed comparing expression of SLC6A14 with other genes in the lung tissue models. This example also illustrates the flexibility of the proposed contrasting colocalization method: it can be used to contrast tissues (as in Table 3 and S10 Table) or genes (as in S17 Table).
For method comparisons with alternatives existing in the literature, we focused on COLOC [43] and eCAVIAR [44], two frequently implemented Bayesian approaches that can be used with summary statistics. There are several other colocalization methods (the majority being Bayesian) including ENLOC [49] and SHERLOCK [45]. In a recent study [49], it has been shown that eCAVIAR is extremely conservative with low false positive rate and low power, which we also observe here in our simulation studies (S8 and S11–S16 Tables) with the exceptions of S13 and S16 Tables. In these two scenarios (Alter 3 and Alter 6 in S7 Table), the allelic heterogeneity is such that colocalization exists at the second peak, which was simulated in a region of low LD; the parameter setting for which eCAVIAR is reported to perform well. COLOC is a special case of ENLOC, is easy to implement for simulation studies, and agrees with ENLOC results in the presence of strong signals. Results in Table 3 include the corresponding posterior probability (PP) of eQTL-association colocalization based on COLOC, and the regional posterior probability (RCP) of eCAVIAR as defined and recommended in [49].
Evaluation through simulation: Type 1 error of SS and false positive rate of COLOC and eCAVIAR
For type 1 error evaluation, we considered a comprehensive set of null cases where Case 1 represents the simple null of no association and no eQTL, while Cases 2–4 correspond to composite null scenarios where for example both association and eQTL may be significant but distinct. Detailed descriptions and illustrations of the scenarios and their corresponding parameter values considered are provided in S5 and S6 Tables.
For Case 1, S8, S10 and S12 Figs demonstrate type 1 error control of the SS test when data were simulated based on the different LD patterns at the SLC6A14, SLC26A9 and ATP12A loci, respectively, and when the eQTL evidence was measured as −log10(eQTL p). To be comprehensive, S9, S11 and S13 Figs provide the corresponding type 1 error control results when eQTL p-values were dichotomized using a liberal threshold of p<0.05 when calculating the SS test statistic. Similarly, S15–S20 Figs provide evidence of type 1 error rate control of the SSC contrasting test, for the three loci and using either continuous or dichotomized measures of the eQTL evidence.
For the other cases, without loss of generality we focus on using the LD pattern at the SLC6A14 locus. S8 and S9 Tables provide the empirical type 1 error rates of the SS method, as well as the false positive rates of COLOC and eCAVIAR using three different cut-off values (0.5, 0.75 and 0.9) for the posterior probability (S8 Table).
Evaluation through simulation: Power of SS and true positive rate of COLOC and eCAVIAR
For power evaluation, we considered a variety of alternatives including allelic heterogeneity in addition to the simple case where one single GWAS signal colocalizes with one single eQTL signal. For example, there might be two eQTLs but only one of the two colocalizes with a GWAS signal, and vice versa. Detailed descriptions and illustrations of the scenarios considered are given in S7 Table, and the corresponding results are provided in S11–S16 Tables including the power of SS and the true positive rate of COLOC and eCAVIAR.
Web resources
CFTR2, https://cftr2.org; Plink, http://zzz.bwh.harvard.edu/plink/; KING, http://people.virginia.edu/~wc9c/KING/kingpopulation.html; Beagle, https://faculty.washington.edu/browning/beagle/b4_1.html; R, https://cran.r-project.org/; Roadmap Epigenomics Mapping Consortium (REMC), http://www.roadmapepigenomics.org/; The Encyclopedia of DNA Elements (ENCODE), https://www.encodeproject.org; Haploreg, http://archive.broadinstitute.org/mammals/haploreg/haploreg.php; Genotype-Tissue Expression Project (GTEx) Portal, https://www.gtexportal.org/home/; Regulatory Regions Map (Reg2Map), https://personal.broadinstitute.org/meuleman/reg2map/HoneyBadger2-intersect_release/; The International Hapmap Project, https://www.genome.gov/10001688/international-hapmap-project/; UCSC browser, https://genome.ucsc.edu/; Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo/; JASPAR, http://jaspar.genereg.net/. GTEx’s Broad Institute’s GitHub page, https://github.com/broadinstitute/gtex-pipeline
Supporting information
(DOCX)
Different solid dots show the clusters of the samples from the International Hapmap consortium [7]. The red circles correspond to our samples with outliers highlighted as black dots; outliers defined as 6 S.D. away from the center of the HapMap3 European (CEU/TSI) cluster. See the detailed description of the Hapmap samples at http://www.sanger.ac.uk/resources/downloads/human/hapmap3.html.
(TIF)
The Locus zoom plot [8] of CFTR association imputed using (A) the 1000 Genome Project Phase 3 [3], and (B) the hybrid reference (augmenting the 1000 genome reference with the whole genome sequencing from 101 patients with CF). Imputed variants with MAF>1% were analyzed.
(TIF)
(TIF)
All subfigures are plotted in 200kb regions surrounding (A) SLC6A14, (B) SLC26A9, and (C) ATP12A. The GWAS signals are obliterated after conditioning on the top SNP in each region. The color of each dot represents the amount of LD of the SNP to the purple diamond point, which is the top SNP in each region after the conditional analysis.
(TIF)
(TIF)
GTEx expression data was analyzed separately by sex for eQTL association with SLC6A14 in the pancreas using the same linear regression model described in Materials and Methods, GTEx Data without the sex covariate. Dots in the figure represent the association with MI, while the lines depict the association pattern of eQTLs for SLC6A14 in the pancreas when analyzed in males (black solid line) and in females (dashed blue line) separately.
(TIF)
Overlay of p-values (on the −log10 scale) from the meconium ileus GWAS (red/yellow palette of colored dots; this study), lung function GWAS (green/blue palette of colored dots; [9]) and GTEx (v7, [4]) eQTLs association for AGTR2 expression (colored lines, derived the same as for Fig 2) for the different tissues of interest. AGTR2 is not expressed in the CF nasal epithelia.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the SS colocalization p-value on the original scale, (B) QQ-plot of the SS colocalization p-value on the −log10 scale, and (C) the histogram of the SS colocalization p-value that is expected to follow a Unif(0,1) distribution under the null. The empirical Type 1 error is 0.0501 at the 0.05 nominal level, and 0.0053 at the 0.005 level.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the SS colocalization p-value on the original scale, (B) QQ-plot of the SS colocalization p-value on the −log10 scale, and (C) the histogram of the SS colocalization p-value that is expected to follow a Unif(0,1) distribution under the null. The empirical Type 1 Error is 0.048 at the 0.05 nominal level, and 0.0049 at the 0.005 level.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the SS colocalization p-value on the original scale, (B) QQ-plot of the SS colocalization p-value on the −log10 scale, and (C) the histogram of the SS colocalization p-value that is expected to follow a Unif(0,1) distribution under the null. The empirical Type 1 error is 0.052 at the 0.05 nominal level, and 0.0064 at the 0.005 level.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the SS colocalization p-value on the original scale, (B) QQ-plot of the SS colocalization p-value on the −log10 scale, and (C) the histogram of the SS colocalization p-value that is expected to follow a Unif(0,1) distribution under the null. The empirical Type 1 error is 0.0509 at the 0.05 nominal level, and 0.0051 at the 0.005 level.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the SS colocalization p-value on the original scale, (B) QQ-plot of the SS colocalization p-value on the −log10 scale, and (C) the histogram of the SS colocalization p-value that is expected to follow a Unif(0,1) distribution under the null. The empirical Type 1 error is 0.0487 at the 0.05 nominal level, and 0.0034 at the 0.005 level.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the SS colocalization p-value on the original scale, (B) QQ-plot of the SS colocalization p-value on the −log10 scale, and (C) the histogram of the SS colocalization p-value that is expected to follow a Unif(0,1) distribution under the null. The empirical Type 1 error is 0.0513 at the 0.05 nominal level, and 0.0046 at the 0.005 level.
(TIF)
The SS colocalization test evaluates if the eQTLs for a given gene and in a given tissue colocalize with meconium ileus-associated variants in the regions of (A) chromosome X, (B) chromosome 1, and (C) chromosome 13. In each panel, each row shows the SS colocalization evidence for the specified tissue across all genes within 1Mbp of the peak GWAS variant. SS colocalization evidence for each gene is calculated for SNPs within 0.1Mbp of the peak GWAS variants; the genes on the x-axis are ordered by their chromosomal positions. Each column shows the SS colocalization evidence for the specified gene across each tissue tested. The color intensity corresponds to the SS colocalization evidence as measured by −log10(SS p-value), with red representing −log10(p) = 6 and white representing −log10(p) = 0. Grey indicates either insufficient expression levels attained for the gene in the tissue under study, or that there were no significant eQTLs for the gene in that tissue. The eQTL analyses used for all gene/tissue pairs are those conducted by GTEx version 7 release, except the boxes indicated on the margins. eQTL analysis for the boxes on the margins were calculated in version 6 but were not calculated in GTEx version 7 due to a more stringent expression threshold criteria set in GTEx v7 versus v6 (see Material and Methods for specifics); these analyses were conducted using the publicly available expression matrix (phe000020.v1) and genotypes from WGS (phg000830.v1) following GTEx’s protocol for eQTL analysis.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the simple sum contrasting (SSC) colocalization p-value on the original scale, (B) QQ-plot of the SSC colocalization p-value on the −log10 scale, and (C) the histogram of the SSC colocalization p-value that is expected to follow a Unif(0,1) distribution under the null hypothesis. The empirical Type 1 Error is 0.0536 at the 0.05 nominal level, and 0.0048 at the 0.005 level.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the simple sum contrasting (SSC) colocalization p-value on the original scale, (B) QQ-plot of the SSC colocalization p-value on the −log10 scale, and (C) the histogram of the SSC colocalization p-value that is expected to follow a Unif(0,1) distribution under the null hypothesis. The empirical Type 1 error is 0.0454 at the 0.05 nominal level, and 0.0043 at the 0.005 level.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the simple sum contrasting (SSC) colocalization p-value on the original scale, (B) QQ-plot of the SSC colocalization p-value on the −log10 scale, and (C) the histogram of the SSC colocalization p-value that is expected to follow a Unif(0,1) distribution under the null hypothesis. The empirical Type 1 error is 0.0457 at the 0.05 nominal level, and 0.0039 at the 0.005 level.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the simple sum contrasting (SSC) colocalization p-value on the original scale, (B) QQ-plot of the SSC colocalization p-value on the −log10 scale, and (C) the histogram of the SSC colocalization p-value that is expected to follow a Unif(0,1) distribution under the null hypothesis. The empirical Type 1 error is 0.0512 at the 0.05 nominal level, and 0.0052 at the 0.005 level.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the simple sum contrasting (SSC) colocalization p-value on the original scale, (B) QQ-plot of the SSC colocalization p-value on the −log10 scale, and (C) the histogram of the SSC colocalization p-value that is expected to follow a Unif(0,1) distribution under the null hypothesis. The empirical Type 1 error is 0.052 at the 0.05 nominal level, and 0.0048 at the 0.005 level.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the simple sum contrasting (SSC) colocalization p-value on the original scale, (B) QQ-plot of the SSC colocalization p-value on the −log10 scale, and (C) the histogram of the SSC colocalization p-value that is expected to follow a Unif(0,1) distribution under the null hypothesis. The empirical Type 1 error is 0.0503 at the 0.05 nominal level, and 0.0051 at the 0.005 level.
(TIF)
The total number of individuals used for the meconium ileus association analysis after quality control of both genotypes and phenotypes, stratified by consortium site and Illumina genotyping platform.
(DOCX)
The number of SNPs before quality control (QC) and the number of SNPs excluded by each stated QC criterion, stratified by consortium sites and Illumina genotyping platform.
(DOCX)
SNPs significant in either study (this study with n = 6770 and previous consortium GWAS published in Sun et al [10]) and of particular functional relevance are provided in this table.
(DOCX)
The three variants here include the top two ranked SNPs, rs3788766 and rs5905177 as in S3 Table, and putative functional variant rs12710568.
(DOCX)
Values of standardized true effect size of the associated SNP for GWAS or an eQTL used in the various simulation settings, with the corresponding GWAS or eQTL power of individually detecting the SNP at the 10−8 significance level, and the expected -log10(p-value) of the GWAS association or the eQTL analysis if the observed signal strength is the true effect size. See S1 Appendix for other simulation details.
(DOCX)
Plots (a)-(d) provide the general visualization of GWAS (red line) and eQTL (blue) patterns (on the -log10 p scale) in a region of interest (e.g. the SLC6A14 locus). The value of represents the standardized true effect size of a GWAS associated variant, and represents the true standardized effect size of an eQTL variant. The corresponding power of detecting the GWAS SNP or finding the eQTL are provided in S5 Table. For illustration purposes but without loss generality, if there was a GWAS association, was set to be 5.73 such that 0.5 power could be achieved to detect the signal. If there was an eQTL from the gene-expression study, was set to be 7.01 such that 0.9 power could be achieved to detect the signal. See S1 Appendix for other simulation details.
(DOCX)
Plots (a)-(f) provide the general visualization of GWAS (red line) and eQTL (blue) patterns (on the -log10 p scale) in a region of interest (e.g. the SLC6A14 locus). The parameter values are varied in the power study, thus they are provided in the corresponding referred tables.
(DOCX)
The null cases considered are detailed in S6 Table. The LD pattern at the simulated region follows that at the SLC6A14 locus. For the SS method, the nominal type 1 error was set at alpha = 0.05 or alpha = 0.005. The eQTL evidence was measured continuously as -log10 (eQTL p-value), or dichotomized using the eQTL p<0.05 or <0.005 threshold. For COLOC and eCAVIAR, the false positive rates were calculated by applying the 0.5, 0.75 or 0.9 threshold (as in [1]) to the colocalization posterior probability. In total, 104 replications were simulated to obtain each cell of the table. See S1 Appendix for other simulation details.
(DOCX)
All the null cases considered are detailed in S6 Table. Case 3 is the situation when there is a GWAS associated SNP but there is no eQTL (plot (c) in S6 Table). In that case, colocalization results should be interpreted with caution if the observed eQTL signal is weak with examples demonstrated below (i.e. the null hypothesis is not rejected if the maximum of -log10 (eQTL p-value) is below a threshold (column 1)). Among the remaining replicates, the null is rejected if the Simple Sum colocalization p-value is smaller than the nominal type 1 error level (alpha = 0.05 or alpha = 0.005). The LD pattern at the simulated region follows that at the SLC6A14 locus. In total, 104 replications were simulated to obtain each cell of the table. See S1 Appendix for other simulation details.
(DOCX)
The eQTL evidence was dichotomized by using thresholds of eQTL p<0.05, <0.005 or <0.0005 instead of based on the -log10(p-value) as in Table 3. Analytical and permutation-based (# of replicates = 105) Simple Sum (SS) colocalization p-values evaluate if the eQTLs for a given gene and tissue colocalize with meconium ileus-associated variants. All colocalization p-values were one-sided because only positive association implies eQTL-association colocalization (i.e. eQTL peaks coincide with association peaks). Simple Sum Contrasting (SSC) colocalization p-value evaluates if the eQTLs in the pancreas colocalize with meconium ileus-associated variants more than eQTLs in another tissue; NAs are listed for the pancreas since we do not contrast pancreas with itself. Other NAs are used when there are no SNPs with eQTLs p less than the thresholds considered (0.05, 0.005 or 0.0005) for that gene and tissue.
(DOCX)
The LD pattern at the simulated region follows that at the SLC6A14 locus. For the SS method, the nominal type 1 error was set at alpha = 0.05 or alpha = 0.005. The eQTL evidence was measured continuously as -log10 (eQTL p-value), or dichotomized using the eQTL p<0.05 or <0.005 threshold. For COLOC and eCAVIAR, the false positive rates were calculated by applying the 0.5, 0.75 or 0.9 threshold to the colocalization posterior probability. The value of represents the standardized true effect size of the GWAS associated variant, and represents the standardized true effect size of the eQTL variant, as detailed in S5 Table. Here, is set to be 5.73 such that 0.5 power is achieved to detect the GWAS association at significance level of 10−8, while is set to be 3.4, 4.09, 4.45, 5.21 or 5.73 for each row of the table such that 0.01, 0.05, 0.1, 0.3, or 0.5 power is achieved to detect the eQTL association at significance level 10−8. In total, 104 replications were simulated to obtain each cell of the table. See S1 Appendix for other simulation details.
(DOCX)
The LD pattern at the simulated region follows that at the SLC6A14 locus. For the SS method, the nominal type 1 error was set at alpha = 0.05 or alpha = 0.005. The eQTL evidence was measured continuously as -log10 (eQTL p-value) or dichotomized using the eQTL p<0.05 or <0.005 threshold. For COLOC and eCAVIAR, the false positive rates were calculated by applying the 0.5, 0.75 or 0.9 threshold to the colocalization posterior probability. The values of and represent the standardized true effect sizes of two GWAS associated variants, while and represent the standardized true effect sizes of two eQTL variants. Here, is set to be 6.57 and is set to be 5.73 such that, respectively, 0.8 and 0.5 power are achieved to detect the two GWAS signals at significance level 10−8. is set to be 3.4, 4.09, 4.45, 5.21 or 5.73 for each row of the table such that 0.01, 0.05, 0.1, 0.3, or 0.5 power is achieved to detect the eQTL association at significance level of 10−8 and is set to be 0. In total, 104 replications were simulated to obtain each cell of the table. See S1 Appendix for other simulation details.
(DOCX)
The LD pattern at the simulated region follows that at the SLC6A14 locus. For the SS method, the nominal type 1 error was set at alpha = 0.05 or alpha = 0.005. The eQTL evidence was measured continuously as -log10 (eQTL p-value) or dichotomized using the eQTL p<0.05 or <0.005 threshold. For COLOC and eCAVIAR, the false positive rates were calculated by applying the 0.5, 0.75 or 0.9 threshold to the colocalization posterior probability. The values of and represent the standardized true effect sizes of two GWAS associated variants, while and represent the standardized true effect sizes of two eQTL variants. Here, is set to be 6.57 and is set to be 5.73 such that 0.8 and 0.5 power are achieved to detect GWAS signals at significance level of 10−8. is set to be 0, and is set to be 3.4, 4.09, 4.45, 5.21 or 5.73 for each row of the table such that 0.01, 0.05, 0.1, 0.3, or 0.5 power is achieved to detect the eQTL association at significance level 10−8. In total, 104 replications were simulated to obtain each cell of the table. See S1 Appendix for other simulation details.
(DOCX)
The LD pattern at the simulated region follows that at the SLC6A14 locus. For the SS method, the nominal type 1 error was set at alpha = 0.05 or alpha = 0.005. The eQTL evidence was measured continuously as -log10 (eQTL p-value) or dichotomized using the eQTL p<0.05 or <0.005 threshold. For COLOC and eCAVIAR, the false positive rates were calculated by applying the 0.5, 0.75 or 0.9 threshold to the colocalization posterior probability. The values of and represent the standardized true effect sizes of two GWAS associated variants, while and represent standardized true effect sizes of two eQTL variants. Here, is set to be 6.57 such that 0.8 power is achieved to detect that GWAS signal at significance level 10−8 and is set to be 0. is set to be 3.4, 4.09, 4.45, 5.21 or 5.73 for each row of the table such that 0.01, 0.05,0.1,0.3, or 0.5 power are achieved to detect the eQTL association at significance level of 10−8, and is set to be 5.73 such that 0.5 power is achieved to detect that eQTL signal at significance level of 10−8. In total, 104 replications were simulated to obtain each cell of the table. See S1 Appendix for other simulation details.
(DOCX)
The LD pattern at the simulated region follows that at the SLC6A14 locus. For the SS method, the nominal type 1 error was set at alpha = 0.05 or alpha = 0.005. The eQTL evidence was measured continuously as -log10 (eQTL p-value) or dichotomized using the eQTL p<0.05 or <0.005 threshold. For COLOC and eCAVIAR, the false positive rates were calculated by applying the 0.5, 0.75 or 0.9 threshold to the colocalization posterior probability. The values of and represent the standardized true effect sizes of two GWAS associated variants, while and represent the standardized true effect sizes of eQTL variants. Here, is set to be 6.57 such that 0.8 power is achieved to detect that GWAS signal at significance level of 10−8 and is set to be 0. is set to be 3.4, 4.09, 4.45, 5.21 or 5.73 for each row of the table such that 0.01, 0.05,0.1,0.3, or 0.5 power are achieved to detect the eQTL association at significance level of 10−8 and is set to be 7.01 such that 0.9 power is achieved to detect that eQTL signal at significance level of 10−8. In total, 104 replications were simulated to obtain each cell of the table. See S1 Appendix for other simulation details.
(DOCX)
The LD pattern at the simulated region follows that at the SLC6A14 locus. For the SS method, the nominal type 1 error was set at alpha = 0.05 or alpha = 0.005. The eQTL evidence was measured continuously as -log10 (eQTL p-value) or dichotomized using the eQTL p<0.05 or <0.005 threshold. For COLOC and eCAVIAR, the false positive rates were calculated by applying the 0.5, 0.75 or 0.9 threshold to the colocalization posterior probability. The values of and represent the standardized true effect sizes of two GWAS associated variants, while and represent the standardized true effect sizes of eQTL variants. Here, is set to be 6.57 and is set to be 5.73 such that, respectively, 0.8 and 0.5 power are achieved to detect the two GWAS signals at significance level of 10−8. is set to be 3.4, 4.09, 4.45, 5.21 or 5.73 for each row of the table such that 0.01, 0.05,0.1,0.3, or 0.5 power are achieved to detect the eQTL association at significance level of 10−8 and is set to be 7.01 such that 0.9 power is achieved to detect that eQTL signal at significance level of 10−8. In total, 104 replications were simulated to obtain each cell of the table. See S1 Appendix for other simulation details.
(DOCX)
The exact region is 115248275–115448275 bp as in human genome reference assembly GRCh37. The eQTL evidence used include the -log10 transform of eQTL p value, and dichotomized eQTL p-value indicator by thresholds of eQTL p<0.05 or <0.005 for each specified gene and tissue. We focus on the analysis evaluating whether the eQTLs for SLC6A14 in lung (or human nasal epithelial; HNE) colocalize with lung-associated variants more than eQTLs for AGTR2, PLS3 and CXorf61 in lung (or HNE). The contrasting colocalization test for SLC6A14 is listed as NA since we do not contrast SLC6A14 with itself; NA in other cells means no eQTL SNP with p<0.05 or p<0.005 for that gene. The column ‘No. of eQTL SNPs’ shows the number of SNPs with eQTL p-values< 0.05 or 0.005 in the 0.1Mbp region; it refers to the number of SNPs for which eQTL p-values were available at the locus when -log10(eQTLp) is used (first 4 rows of the table). All p-values are one-sided to ensure colocalization rather than negative correlation.
(DOCX)
Acknowledgments
We thank the US CF Foundation for the use of CF Foundation Patient Registry data. We thank the Cystic Fibrosis Canada–SickKids Program for Individualized Therapy for access to the RNA-sequencing of the nasal epithelia. We thank Drs. Rhonda Pace and Jackie Stonebraker for their work collecting and curating source documents to confirm meconium ileus status in the UNC/Case cohort. We thank the patients, care providers and clinic coordinators at CF Centers throughout the US and Canada for their contributions to the CF Foundation Patient Registry and Canadian Gene Modifier Study. Some of the datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/gap through dbGaP accession number phs000424.GTEx.v7.p2.c1. We would like to acknowledge the major contributions of the late Dr. Peter Durie to the development of the Canadian and International CF Gene Modifier Consortia resources.
Data Availability
Summary statistics from the GWAS are available in the NHGRI-EBI catalog. R scripts for the simple sum method are available at http://lab.research.sickkids.ca/strug/publications-software/. Access to the RNA-sequencing data from the nasal epithelia are available through the CF Canada – SickKids Program for Individualized Therapy Biobank http://lab.research.sickkids.ca/cfit/). Genotype data cannot be shared publicly because the consents restrict access to Cystic Fibrosis researchers. Genotype data are available by application to the Cystic Fibrosis Canada National data registry for researchers who meet the criteria for access to confidential clinical data for the purpose of CF research. Application to the Cystic Fibrosis Canada National data registry does not include a requirement to collaborate and/or publish with the authors of the current manuscript.
Funding Statement
National Heart, Lung and Blood Institute 1DP2OD007031, R01HL068890, R01HL095396, R01HL68927; National Institute of Diabetes and Digestive and Kidney Diseases 1R01DK61886-01, K23DK083551, P30DK079637; National Human Genome Research Institute R21HG007840; Cystic Fibrosis Foundation KNOWLE00A0, DRUMM0A00, CUTT00AO, CUTT06PO, BLACKM16G0, STRUG17PO; Canadian Institutes of Health Research (CIHR MOP 258916, MOP 117978), Cystic Fibrosis Canada (CFC; #2626) and the CFIT Program funded by the SickKids Foundation and CF Canada; Natural Sciences and Engineering Research Council of Canada (NSERC RGPIN-2015-03742, 250053-2013); Genome Canada through the Ontario Genomics Institute (2004-OGI-3-05; 2018-OGI-148); Institut National de la Santé et de la Recherche Médicale, Assistance Publique-Hôpitaux de Paris, Université Pierre et Marie Curie Paris, Agence Nationale de la Recherche (R09186DS), DGS, Association Vaincre La Mucoviscidose, Chancellerie des Universités (Legs Poix), Association Agir Informer Contre la Mucoviscidose, GIS-Institut des Maladies Rares. Genome-wide genotyping of subjects in North America provided by the US and CFC Foundations. The funders of the study play no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Sosnay PR, Siklosi KR, Van Goor F, Kaniecki K, Yu H, Sharma N, et al. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nature genetics. 2013;45(10):1160–7. 10.1038/ng.2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strug LJ, Gonska T, He G, Keenan K, Ip W, Boelle PY, et al. Cystic fibrosis gene modifier SLC26A9 modulates airway response to CFTR-directed therapeutics. Hum Mol Genet. 2016;25(20):4590–600. 10.1093/hmg/ddw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vanscoy LL, Blackman SM, Collaco JM, Bowers A, Lai T, Naughton K, et al. Heritability of Lung Disease Severity in Cystic Fibrosis. Am J Respir Crit Care Med. 2007;175(10):1036–43. 10.1164/rccm.200608-1164OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corey M, Edwards L, Levison H, Knowles M. Longitudinal analysis of pulmonary function decline in patients with cystic fibrosis. J Pediatr. 1997;131(6):809–14. [DOI] [PubMed] [Google Scholar]
- 5.Dupuis A, Keenan K, Ooi CY, Dorfman R, Sontag MK, Naehrlich L, et al. Prevalence of meconium ileus marks the severity of mutations of the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) gene. Genet Med. 2016;18(4):333–40. 10.1038/gim.2015.79 [DOI] [PubMed] [Google Scholar]
- 6.Blackman SM, Deering–Brose R, McWilliams R, Naughton K, Coleman B, Lai T, et al. Relative Contribution of Genetic and Nongenetic Modifiers to Intestinal Obstruction in Cystic Fibrosis. Gastroenterology. 2006;131(4):1030–9. 10.1053/j.gastro.2006.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlyle BE, Borowitz DS, Glick PL. A review of pathophysiology and management of fetuses and neonates with meconium ileus for the pediatric surgeon. J Pediatr Surg. 2012;47(4):772–81. 10.1016/j.jpedsurg.2012.02.019 [DOI] [PubMed] [Google Scholar]
- 8.Li W, Soave D, Miller MR, Keenan K, Lin F, Gong J, et al. Unraveling the complex genetic model for cystic fibrosis: pleiotropic effects of modifier genes on early cystic fibrosis-related morbidities. Hum Genet. 2014;133(2):151–61. 10.1007/s00439-013-1363-7 [DOI] [PubMed] [Google Scholar]
- 9.Avella M, Loriol C, Boulukos K, Borgese F, Ehrenfeld J. SLC26A9 stimulates CFTR expression and function in human bronchial cell lines. J Cell Physiol. 2011;226(1):212–23. 10.1002/jcp.22328 [DOI] [PubMed] [Google Scholar]
- 10.Bertrand CA, Zhang R, Pilewski JM, Frizzell RA. SLC26A9 is a constitutively active, CFTR-regulated anion conductance in human bronchial epithelia. J Gen Physiol. 2009;133(4):421–38. 10.1085/jgp.200810097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ousingsawat J, Schreiber R, Kunzelmann K. Differential Contribution of SLC26A9 to Cl- conductance in polarized and non-polarized epithelial cells. Cell Physiol Biochem. 2011;227(6):2323–29. [DOI] [PubMed] [Google Scholar]
- 12.Sun L, Rommens JM, Corvol H, Li W, Li X, Chiang TA, et al. Multiple apical plasma membrane constituents are associated with susceptibility to meconium ileus in individuals with cystic fibrosis. Nat Genet. 2012;44(5):562–9. 10.1038/ng.2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blackman S, Commander C, Watson C, Arcara K, Strug L, Stonebraker J, et al. Genetic modifiers of cystic fibrosis-related diabetes. Diabetes. 2013;62:3627–35. 10.2337/db13-0510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller MR, Soave D, Li W, Gong J, Pace RG, Boëlle PY, et al. Variants in Solute Carrier SLC26A9 Modify Prenatal Exocrine Pancreatic Damage in Cystic Fibrosis. J Pediatr. 2015;166(5):1152–7 e6. 10.1016/j.jpeds.2015.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soave D, Miller MR, Keenan K, Li W, Gong J, Ip W, et al. Evidence for a causal relationship between early exocrine pancreatic disease and cystic fibrosis-related diabetes: a Mendelian randomization study. Diabetes. 2014;63(6):2114–9. 10.2337/db13-1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nature genetics. 2018;50(5):693–8. 10.1038/s41588-018-0099-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kopelman H, Corey M, Gaskin K, Durie P, Weizman Z, Forstner G. Impaired chloride secretion, as well as bicarbonate secretion, underlies the fluid secretory defect in the cystic fibrosis pancreas. Gastroenterology. 1988;95(2):349–55. [DOI] [PubMed] [Google Scholar]
- 18.Kopelman H, Durie P, Gaskin K, Weizman Z, Forstner G. Pancreatic fluid secretion and protein hyperconcentration in cystic fibrosis. N Engl J Med. 1985;312(6):329–34. 10.1056/NEJM198502073120601 [DOI] [PubMed] [Google Scholar]
- 19.Ooi CY, Durie PR. Cystic fibrosis transmembrane conductance regulator (CFTR) gene mutations in pancreatitis. J Cyst Fibros. 2012;11(5):355–62. 10.1016/j.jcf.2012.05.001 [DOI] [PubMed] [Google Scholar]
- 20.Kristidis P, Bozon D, Corey M, Markiewicz D, Rommens J, Tsui LC, et al. Genetic determination of exocrine pancreatic function in cystic fibrosis. Am J Hum Genet. 1992;50(6):1178–84. [PMC free article] [PubMed] [Google Scholar]
- 21.Sontag MK, Corey M, Hokanson JE, Marshall JA, Sommer SS, Zerbe GO, et al. Genetic and physiologic correlates of longitudinal immunoreactive trypsinogen decline in infants with cystic fibrosis identified through newborn screening. The Journal of pediatrics. 2006;149(5):650–7.e2. 10.1016/j.jpeds.2006.07.026 [DOI] [PubMed] [Google Scholar]
- 22.Durie PR, Forstner GG, Gaskin KJ, Moore DJ, Cleghorn GJ, Wong SS, et al. Age-related alterations of immunoreactive pancreatic cationic trypsinogen in sera from cystic fibrosis patients with and without pancreatic insufficiency. Pediatr Res. 1986;20(3):209–13. 10.1203/00006450-198603000-00002 [DOI] [PubMed] [Google Scholar]
- 23.Consortium GTEx. The Genotype-Tissue Expression (GTEx) project. Nature genetics. 2013;45(6):580–5. 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Encode Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–30. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–81. 10.1093/nar/gkv1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kheradpour P, Kellis M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 2014;42(5):2976–87. 10.1093/nar/gkt1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panjwani N, Xiao B, Xu L, Gong J, Keenan K, Lin F, et al. Improving imputation in disease-relevant regions: lessons from cystic fibrosis. NPJ Genom Med. 2018;3:8 10.1038/s41525-018-0047-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.International HapMap C, Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–8. 10.1038/nature09298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 2008;32(3):227–34. 10.1002/gepi.20297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Storey JD. A direct approach to false discovery rates. J Roy Stat Soc B. 2002;64:479–98. [Google Scholar]
- 32.Soave D, Corvol H, Panjwani N, Gong J, Li W, Boëlle PY, et al. A Joint Location-Scale Test Improves Power to Detect Associated SNPs, Gene Sets, and Pathways. Am J Hum Genet. 2015;97(1):125–38. 10.1016/j.ajhg.2015.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wright FA, Strug LJ, Doshi VK, Commander CW, Blackman SM, Sun L, et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat Genet. 2011;43(6):539–46. 10.1038/ng.838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah VS, Meyerholz DK, Tang XX, Reznikov L, Abou Alaiwa M, Ernst SE, et al. Airway acidification initiates host defense abnormalities in cystic fibrosis mice. Science. 2016;351(6272):503–7. 10.1126/science.aad5589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ratjen F, Bell SC, Rowe SM, Goss CH, Quittner AL, Bush A. Cystic fibrosis. Nature Reviews Disease Primers. 2015;1:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solomon S, Gelrud A, Whitcomb DC. Low penetrance pancreatitis phenotype in a Venezuelan kindred with a PRSS1 R122H mutation. JOP. 2013;14(2):187–9. 10.6092/1590-8577/1276 [DOI] [PubMed] [Google Scholar]
- 37.Derikx MH, Kovacs P, Scholz M, Masson E, Chen JM, Ruffert C, et al. Polymorphisms at PRSS1-PRSS2 and CLDN2-MORC4 loci associate with alcoholic and non-alcoholic chronic pancreatitis in a European replication study. Gut. 2015;64(9):1426–33. 10.1136/gutjnl-2014-307453 [DOI] [PubMed] [Google Scholar]
- 38.Boulling A, Sato M, Masson E, Genin E, Chen JM, Ferec C. Identification of a functional PRSS1 promoter variant in linkage disequilibrium with the chronic pancreatitis-protecting rs10273639. Gut. 2015;64(11):1837–8. 10.1136/gutjnl-2015-310254 [DOI] [PubMed] [Google Scholar]
- 39.Brock DJ, Barron L. Biochemical analysis of meconium in fetuses presumed to have cystic fibrosis. Prenat Diagn. 1986;6(4):291–8. [DOI] [PubMed] [Google Scholar]
- 40.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–7. 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corvol H, Blackman SM, Boëlle PY, Gallins PJ, Pace RG, Stonebraker JR, et al. Genome-wide association meta-analysis identifies five modifier loci of lung disease severity in cystic fibrosis. Nat Commun. 2015;6:8382 10.1038/ncomms9382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moraes T. Bioelectric Properties & Drug-Response Measured in Nasal Cells Reflect Those Measured in Bronchial Cells (Abstract 161). NACFC; October 18–20; Denver2018. [Google Scholar]
- 43.Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10(5):e1004383 10.1371/journal.pgen.1004383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hormozdiari F, van de Bunt M, Segre AV, Li X, Joo JW, Bilow M, et al. Colocalization of GWAS and eQTL Signals Detects Target Genes. Am J Hum Genet. 2016;99(6):1245–60. 10.1016/j.ajhg.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He X, Fuller CK, Song Y, Meng Q, Zhang B, Yang X, et al. Sherlock: detecting gene-disease associations by matching patterns of expression QTL and GWAS. Am J Hum Genet. 2013;92(5):667–80. 10.1016/j.ajhg.2013.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nica AC, Montgomery SB, Dimas AS, Stranger BE, Beazley C, Barroso I, et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6(4):e1000895 10.1371/journal.pgen.1000895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plagnol V, Smyth DJ, Todd JA, Clayton DG. Statistical independence of the colocalized association signals for type 1 diabetes and RPS26 gene expression on chromosome 12q13. Biostatistics. 2009;10(2):327–34. 10.1093/biostatistics/kxn039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wallace C, Rotival M, Cooper JD, Rice CM, Yang JH, McNeill M, et al. Statistical colocalization of monocyte gene expression and genetic risk variants for type 1 diabetes. Hum Mol Genet. 2012;21(12):2815–24. 10.1093/hmg/dds098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wen X, Pique-Regi R, Luca F. Integrating molecular QTL data into genome-wide genetic association analysis: Probabilistic assessment of enrichment and colocalization. PLoS Genet. 2017;13(3):e1006646 10.1371/journal.pgen.1006646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Angrist JD, Krueger AB. The Effect of Age at School Entry on Educational Attainment: An Application of Instrumental Variables with Moments from Two Samples. J Am Stat Asssoc. 1992;87:328–36. [Google Scholar]
- 51.Didelez V, Meng S, Sheehan NA. Assumptions of IV Methods for Observational Epidemiology. Stat Sci. 2010;25:22–40. [Google Scholar]
- 52.Liu X, Li T, Riederer B, Lenzen H, Ludolph L, Yeruva S, et al. Loss of Slc26a9 anion transporter alters intestinal electrolyte and HCO3(-) transport and reduces survival in CFTR-deficient mice. Pflugers Arch. 2015;467(6):1261–75. 10.1007/s00424-014-1543-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoltz DA, Rokhlina T, Ernst SE, Pezzulo AA, Ostedgaard LS, Karp PH, et al. Intestinal CFTR expression alleviates meconium ileus in cystic fibrosis pigs. J Clin Invest. 2013;123(6):2685–93. 10.1172/JCI68867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sloan JL, Mager S. Cloning and functional expression of a human Na(+) and Cl(-)-dependent neutral and cationic amino acid transporter B(0+). J Biol Chem. 1999;274(34):23740–5. [DOI] [PubMed] [Google Scholar]
- 55.Karunakaran S, Ramachandran S, Coothankandaswamy V, Elangovan S, Babu E, Periyasamy-Thandavan S, et al. SLC6A14 (ATB0,+) protein, a highly concentrative and broad specific amino acid transporter, is a novel and effective drug target for treatment of estrogen receptor-positive breast cancer. J Biol Chem. 2011;286(36):31830–8. 10.1074/jbc.M111.229518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gupta N, Miyauchi S, Martindale RG, Herdman AV, Podolsky R, Miyake K, et al. Upregulation of the amino acid transporter ATB0,+ (SLC6A14) in colorectal cancer and metastasis in humans. Biochim Biophys Acta. 2005;1741(1–2):215–23. 10.1016/j.bbadis.2005.04.002 [DOI] [PubMed] [Google Scholar]
- 57.Coothankandaswamy V, Cao S, Xu Y, Prasad PD, Singh PK, Reynolds CP, et al. Amino acid transporter SLC6A14 is a novel and effective drug target for pancreatic cancer. Br J Pharmacol. 2016;173(23):3292–306. 10.1111/bph.13616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Griffon A, Barbier Q, Dalino J, van Helden J, Spicuglia S, Ballester B. Integrative analysis of public ChIP-seq experiments reveals a complex multi-cell regulatory landscape. Nucleic Acids Res. 2015;43(4):e27 10.1093/nar/gku1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5(4):276–87. 10.1038/nrg1315 [DOI] [PubMed] [Google Scholar]
- 60.Loriol C, Dulong S, Avella M, Gabillat N, Boulukos K, Borgese F, et al. Characterization of SLC26A9, facilitation of Cl(-) transport by bicarbonate. Cell Physiol Biochem. 2008;22(1–4):15–30. 10.1159/000149780 [DOI] [PubMed] [Google Scholar]
- 61.Ohana E, Yang D, Shcheynikov N, Muallem S. Diverse transport modes by the solute carrier 26 family of anion transporters. J Physiol. 2009;587(Pt 10):2179–85. 10.1113/jphysiol.2008.164863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lonsdale J, Thomas J. et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5. 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amlal H, Xu J, Barone S, Zahedi K, Soleimani M. The chloride channel/transporter Slc26a9 regulates the systemic arterial pressure and renal chloride excretion. J Mol Med (Berl). 2013;91(5):561–72. [DOI] [PubMed] [Google Scholar]
- 64.Wang J, Barbuskaite D, Tozzi M, Giannuzzo A, Sorensen CE, Novak I. Proton Pump Inhibitors Inhibit Pancreatic Secretion: Role of Gastric and Non-Gastric H+/K+-ATPases. PLoS One. 2015;10(5):e0126432 10.1371/journal.pone.0126432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Novak I, Wang J, Henriksen KL, Haanes KA, Krabbe S, Nitschke R, et al. Pancreatic bicarbonate secretion involves two proton pumps. J Biol Chem. 2011;286(1):280–9. 10.1074/jbc.M110.136382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saint-Criq V, Gray MA. Role of CFTR in epithelial physiology. Cell Mol Life Sci. 2017;74(1):93–115. 10.1007/s00018-016-2391-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hodges CA, Grady BR, Mishra K, Cotton CU, Drumm ML. Cystic fibrosis growth retardation is not correlated with loss of Cftr in the intestinal epithelium. Am J Physiol Gastrointest Liver Physiol. 2011;301(3):G528–36. 10.1152/ajpgi.00052.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou L, Dey CR, Wert SE, DuVall MD, Frizzell RA, Whitsett JA. Correction of lethal intestinal defect in a mouse model of cystic fibrosis by human CFTR. Science. 1994;266(5191):1705–8. [DOI] [PubMed] [Google Scholar]
- 69.Ramsey BW, Davies J, McElvaney NG, Tullis E, Bell SC, Drevinek P, et al. A CFTR potentiator in patients with cystic fibrosis and the G551D mutation. N Engl J Med. 2011;365(18):1663–72. 10.1056/NEJMoa1105185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wainwright CE, Elborn JS, Ramsey BW, Marigowda G, Huang X, Cipolli M, et al. Lumacaftor-Ivacaftor in Patients with Cystic Fibrosis Homozygous for Phe508del CFTR. N Engl J Med. 2015;373(3):220–31. 10.1056/NEJMoa1409547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hayes D Jr., McCoy KS, Sheikh SI. Resolution of cystic fibrosis-related diabetes with ivacaftor therapy. Am J Respir Crit Care Med. 2014;190(5):590–1. 10.1164/rccm.201405-0882LE [DOI] [PubMed] [Google Scholar]
- 72.Wong AP, Chin S, Xia S, Garner J, Bear CE, Rossant J. Efficient generation of functional CFTR-expressing airway epithelial cells from human pluripotent stem cells. Nat Protoc. 2015;10(3):363–81. 10.1038/nprot.2015.021 [DOI] [PubMed] [Google Scholar]
- 73.Dekkers JF, Berkers G, Kruisselbrink E, Vonk A, de Jonge HR, Janssens HM, et al. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci Transl Med. 2016;8(344):344ra84 10.1126/scitranslmed.aad8278 [DOI] [PubMed] [Google Scholar]
- 74.Gentzsch M, Boyles SE, Cheluvaraju C, Chaudhry IG, Quinney NL, Cho C, et al. Pharmacological Rescue of Conditionally Reprogrammed Cystic Fibrosis Bronchial Epithelial Cells. Am J Respir Cell Mol Biol. 2017;56(5):568–74. 10.1165/rcmb.2016-0276MA [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc. 2010;5(9):1564–73. 10.1038/nprot.2010.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Purcell S NB, Todd-Brown K, Thomas L, Ferreira MAR, Bender D MJ, Sklar P, de Bakker PIW, Daly MJ & Sham PC. PLINK: a toolset for whole-genome association and population-based linkage analysis. American Journal of Human Genetics. 2007;81(3):559–75. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–73. 10.1093/bioinformatics/btq559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Galinsky KJ, Bhatia G, Loh PR, Georgiev S, Mukherjee S, Patterson NJ, et al. Fast Principal-Component Analysis Reveals Convergent Evolution of ADH1B in Europe and East Asia. Am J Hum Genet. 2016;98(3):456–72. 10.1016/j.ajhg.2015.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Galinsky KJ, Loh PR, Mallick S, Patterson NJ, Price AL. Population Structure of UK Biobank and Ancient Eurasians Reveals Adaptation at Genes Influencing Blood Pressure. Am J Hum Genet. 2016;99(5):1130–9. 10.1016/j.ajhg.2016.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190 10.1371/journal.pgen.0020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics. 2006;38(8):904–9. 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- 82.Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hoffmann TJ, Witte JS. Strategies for Imputing and Analyzing Rare Variants in Association Studies. Trends Genet. 2015;31(10):556–63. 10.1016/j.tig.2015.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tachmazidou I, Suveges D, Min JL, Ritchie GRS, Steinberg J, Walter K, et al. Whole-Genome Sequencing Coupled to Imputation Discovers Genetic Signals for Anthropometric Traits. Am J Hum Genet. 2017;100(6):865–84. 10.1016/j.ajhg.2017.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81(5):1084–97. 10.1086/521987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nature genetics. 2016;48(10):1279–83. 10.1038/ng.3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Browning BL, Browning SR. Genotype Imputation with Millions of Reference Samples. Am J Hum Genet. 2016;98(1):116–26. 10.1016/j.ajhg.2015.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vaida F, Liu L. Fast Implementation for Normal Mixed Effects Models With Censored Response. J Comput Graph Stat. 2009;18(4):797–817. 10.1198/jcgs.2009.07130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thomas DC, Lawlor DA, Thompson JR. Re: Estimation of bias in nongenetic observational studies using "Mendelian triangulation" by Bautista et al. Annals of epidemiology. 2007;17(7):511–3. 10.1016/j.annepidem.2006.12.005 [DOI] [PubMed] [Google Scholar]
- 90.Hemani G, Bowden J, Davey Smith G. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet. 2018;27(R2):R195–R208. 10.1093/hmg/ddy163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–93. [DOI] [PubMed] [Google Scholar]
- 92.Stegle O, Parts L, Durbin R, Winn J. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput Biol. 2010;6(5):e1000770 10.1371/journal.pcbi.1000770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Conomos MP, Miller MB, Thornton TA. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet Epidemiol. 2015;39(4):276–93. 10.1002/gepi.21896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ongen H, Buil A, Brown AA, Dermitzakis ET, Delaneau O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics. 2016;32(10):1479–85. 10.1093/bioinformatics/btv722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eckford PDW, McCormack J, Munsie L, He G, Stanojevic S, Pereira SL, et al. The CF Canada-Sick Kids Program in individual CF therapy: A resource for the advancement of personalized medicine in CF. J Cyst Fibros. 2018. [DOI] [PubMed] [Google Scholar]
- 96.Polineni D, Dang H, Gallins PJ, Jones LC, Pace RG, Stonebraker JR, et al. Airway Mucosal Host Defense Is Key to Genomic Regulation of Cystic Fibrosis Lung Disease Severity. Am J Respir Crit Care Med. 2018;197(1):79–93. 10.1164/rccm.201701-0134OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire MD, Williams C, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28(11):1530–2. 10.1093/bioinformatics/bts196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stegle O, Parts L, Piipari M, Winn J, Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc. 2012;7(3):500–7. 10.1038/nprot.2011.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Halekoh U, Hojsgaard S, Yan J. The R Package geepack for Generalized Estimating Equations. J Stat Softw. 2006;15(2):1–11. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Different solid dots show the clusters of the samples from the International Hapmap consortium [7]. The red circles correspond to our samples with outliers highlighted as black dots; outliers defined as 6 S.D. away from the center of the HapMap3 European (CEU/TSI) cluster. See the detailed description of the Hapmap samples at http://www.sanger.ac.uk/resources/downloads/human/hapmap3.html.
(TIF)
The Locus zoom plot [8] of CFTR association imputed using (A) the 1000 Genome Project Phase 3 [3], and (B) the hybrid reference (augmenting the 1000 genome reference with the whole genome sequencing from 101 patients with CF). Imputed variants with MAF>1% were analyzed.
(TIF)
(TIF)
All subfigures are plotted in 200kb regions surrounding (A) SLC6A14, (B) SLC26A9, and (C) ATP12A. The GWAS signals are obliterated after conditioning on the top SNP in each region. The color of each dot represents the amount of LD of the SNP to the purple diamond point, which is the top SNP in each region after the conditional analysis.
(TIF)
(TIF)
GTEx expression data was analyzed separately by sex for eQTL association with SLC6A14 in the pancreas using the same linear regression model described in Materials and Methods, GTEx Data without the sex covariate. Dots in the figure represent the association with MI, while the lines depict the association pattern of eQTLs for SLC6A14 in the pancreas when analyzed in males (black solid line) and in females (dashed blue line) separately.
(TIF)
Overlay of p-values (on the −log10 scale) from the meconium ileus GWAS (red/yellow palette of colored dots; this study), lung function GWAS (green/blue palette of colored dots; [9]) and GTEx (v7, [4]) eQTLs association for AGTR2 expression (colored lines, derived the same as for Fig 2) for the different tissues of interest. AGTR2 is not expressed in the CF nasal epithelia.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the SS colocalization p-value on the original scale, (B) QQ-plot of the SS colocalization p-value on the −log10 scale, and (C) the histogram of the SS colocalization p-value that is expected to follow a Unif(0,1) distribution under the null. The empirical Type 1 error is 0.0501 at the 0.05 nominal level, and 0.0053 at the 0.005 level.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the SS colocalization p-value on the original scale, (B) QQ-plot of the SS colocalization p-value on the −log10 scale, and (C) the histogram of the SS colocalization p-value that is expected to follow a Unif(0,1) distribution under the null. The empirical Type 1 Error is 0.048 at the 0.05 nominal level, and 0.0049 at the 0.005 level.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the SS colocalization p-value on the original scale, (B) QQ-plot of the SS colocalization p-value on the −log10 scale, and (C) the histogram of the SS colocalization p-value that is expected to follow a Unif(0,1) distribution under the null. The empirical Type 1 error is 0.052 at the 0.05 nominal level, and 0.0064 at the 0.005 level.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the SS colocalization p-value on the original scale, (B) QQ-plot of the SS colocalization p-value on the −log10 scale, and (C) the histogram of the SS colocalization p-value that is expected to follow a Unif(0,1) distribution under the null. The empirical Type 1 error is 0.0509 at the 0.05 nominal level, and 0.0051 at the 0.005 level.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the SS colocalization p-value on the original scale, (B) QQ-plot of the SS colocalization p-value on the −log10 scale, and (C) the histogram of the SS colocalization p-value that is expected to follow a Unif(0,1) distribution under the null. The empirical Type 1 error is 0.0487 at the 0.05 nominal level, and 0.0034 at the 0.005 level.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the SS colocalization p-value on the original scale, (B) QQ-plot of the SS colocalization p-value on the −log10 scale, and (C) the histogram of the SS colocalization p-value that is expected to follow a Unif(0,1) distribution under the null. The empirical Type 1 error is 0.0513 at the 0.05 nominal level, and 0.0046 at the 0.005 level.
(TIF)
The SS colocalization test evaluates if the eQTLs for a given gene and in a given tissue colocalize with meconium ileus-associated variants in the regions of (A) chromosome X, (B) chromosome 1, and (C) chromosome 13. In each panel, each row shows the SS colocalization evidence for the specified tissue across all genes within 1Mbp of the peak GWAS variant. SS colocalization evidence for each gene is calculated for SNPs within 0.1Mbp of the peak GWAS variants; the genes on the x-axis are ordered by their chromosomal positions. Each column shows the SS colocalization evidence for the specified gene across each tissue tested. The color intensity corresponds to the SS colocalization evidence as measured by −log10(SS p-value), with red representing −log10(p) = 6 and white representing −log10(p) = 0. Grey indicates either insufficient expression levels attained for the gene in the tissue under study, or that there were no significant eQTLs for the gene in that tissue. The eQTL analyses used for all gene/tissue pairs are those conducted by GTEx version 7 release, except the boxes indicated on the margins. eQTL analysis for the boxes on the margins were calculated in version 6 but were not calculated in GTEx version 7 due to a more stringent expression threshold criteria set in GTEx v7 versus v6 (see Material and Methods for specifics); these analyses were conducted using the publicly available expression matrix (phe000020.v1) and genotypes from WGS (phg000830.v1) following GTEx’s protocol for eQTL analysis.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the simple sum contrasting (SSC) colocalization p-value on the original scale, (B) QQ-plot of the SSC colocalization p-value on the −log10 scale, and (C) the histogram of the SSC colocalization p-value that is expected to follow a Unif(0,1) distribution under the null hypothesis. The empirical Type 1 Error is 0.0536 at the 0.05 nominal level, and 0.0048 at the 0.005 level.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the simple sum contrasting (SSC) colocalization p-value on the original scale, (B) QQ-plot of the SSC colocalization p-value on the −log10 scale, and (C) the histogram of the SSC colocalization p-value that is expected to follow a Unif(0,1) distribution under the null hypothesis. The empirical Type 1 error is 0.0454 at the 0.05 nominal level, and 0.0043 at the 0.005 level.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the simple sum contrasting (SSC) colocalization p-value on the original scale, (B) QQ-plot of the SSC colocalization p-value on the −log10 scale, and (C) the histogram of the SSC colocalization p-value that is expected to follow a Unif(0,1) distribution under the null hypothesis. The empirical Type 1 error is 0.0457 at the 0.05 nominal level, and 0.0039 at the 0.005 level.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the simple sum contrasting (SSC) colocalization p-value on the original scale, (B) QQ-plot of the SSC colocalization p-value on the −log10 scale, and (C) the histogram of the SSC colocalization p-value that is expected to follow a Unif(0,1) distribution under the null hypothesis. The empirical Type 1 error is 0.0512 at the 0.05 nominal level, and 0.0052 at the 0.005 level.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the simple sum contrasting (SSC) colocalization p-value on the original scale, (B) QQ-plot of the SSC colocalization p-value on the −log10 scale, and (C) the histogram of the SSC colocalization p-value that is expected to follow a Unif(0,1) distribution under the null hypothesis. The empirical Type 1 error is 0.052 at the 0.05 nominal level, and 0.0048 at the 0.005 level.
(TIF)
Simulation method is outlined in S1 Appendix, and the null case considered is Case 1 described in S6 Table where there is no signal for either GWAS or eQTL. In total, 104 replications were simulated to obtain (A) QQ-plot of the simple sum contrasting (SSC) colocalization p-value on the original scale, (B) QQ-plot of the SSC colocalization p-value on the −log10 scale, and (C) the histogram of the SSC colocalization p-value that is expected to follow a Unif(0,1) distribution under the null hypothesis. The empirical Type 1 error is 0.0503 at the 0.05 nominal level, and 0.0051 at the 0.005 level.
(TIF)
The total number of individuals used for the meconium ileus association analysis after quality control of both genotypes and phenotypes, stratified by consortium site and Illumina genotyping platform.
(DOCX)
The number of SNPs before quality control (QC) and the number of SNPs excluded by each stated QC criterion, stratified by consortium sites and Illumina genotyping platform.
(DOCX)
SNPs significant in either study (this study with n = 6770 and previous consortium GWAS published in Sun et al [10]) and of particular functional relevance are provided in this table.
(DOCX)
The three variants here include the top two ranked SNPs, rs3788766 and rs5905177 as in S3 Table, and putative functional variant rs12710568.
(DOCX)
Values of standardized true effect size of the associated SNP for GWAS or an eQTL used in the various simulation settings, with the corresponding GWAS or eQTL power of individually detecting the SNP at the 10−8 significance level, and the expected -log10(p-value) of the GWAS association or the eQTL analysis if the observed signal strength is the true effect size. See S1 Appendix for other simulation details.
(DOCX)
Plots (a)-(d) provide the general visualization of GWAS (red line) and eQTL (blue) patterns (on the -log10 p scale) in a region of interest (e.g. the SLC6A14 locus). The value of represents the standardized true effect size of a GWAS associated variant, and represents the true standardized effect size of an eQTL variant. The corresponding power of detecting the GWAS SNP or finding the eQTL are provided in S5 Table. For illustration purposes but without loss generality, if there was a GWAS association, was set to be 5.73 such that 0.5 power could be achieved to detect the signal. If there was an eQTL from the gene-expression study, was set to be 7.01 such that 0.9 power could be achieved to detect the signal. See S1 Appendix for other simulation details.
(DOCX)
Plots (a)-(f) provide the general visualization of GWAS (red line) and eQTL (blue) patterns (on the -log10 p scale) in a region of interest (e.g. the SLC6A14 locus). The parameter values are varied in the power study, thus they are provided in the corresponding referred tables.
(DOCX)
The null cases considered are detailed in S6 Table. The LD pattern at the simulated region follows that at the SLC6A14 locus. For the SS method, the nominal type 1 error was set at alpha = 0.05 or alpha = 0.005. The eQTL evidence was measured continuously as -log10 (eQTL p-value), or dichotomized using the eQTL p<0.05 or <0.005 threshold. For COLOC and eCAVIAR, the false positive rates were calculated by applying the 0.5, 0.75 or 0.9 threshold (as in [1]) to the colocalization posterior probability. In total, 104 replications were simulated to obtain each cell of the table. See S1 Appendix for other simulation details.
(DOCX)
All the null cases considered are detailed in S6 Table. Case 3 is the situation when there is a GWAS associated SNP but there is no eQTL (plot (c) in S6 Table). In that case, colocalization results should be interpreted with caution if the observed eQTL signal is weak with examples demonstrated below (i.e. the null hypothesis is not rejected if the maximum of -log10 (eQTL p-value) is below a threshold (column 1)). Among the remaining replicates, the null is rejected if the Simple Sum colocalization p-value is smaller than the nominal type 1 error level (alpha = 0.05 or alpha = 0.005). The LD pattern at the simulated region follows that at the SLC6A14 locus. In total, 104 replications were simulated to obtain each cell of the table. See S1 Appendix for other simulation details.
(DOCX)
The eQTL evidence was dichotomized by using thresholds of eQTL p<0.05, <0.005 or <0.0005 instead of based on the -log10(p-value) as in Table 3. Analytical and permutation-based (# of replicates = 105) Simple Sum (SS) colocalization p-values evaluate if the eQTLs for a given gene and tissue colocalize with meconium ileus-associated variants. All colocalization p-values were one-sided because only positive association implies eQTL-association colocalization (i.e. eQTL peaks coincide with association peaks). Simple Sum Contrasting (SSC) colocalization p-value evaluates if the eQTLs in the pancreas colocalize with meconium ileus-associated variants more than eQTLs in another tissue; NAs are listed for the pancreas since we do not contrast pancreas with itself. Other NAs are used when there are no SNPs with eQTLs p less than the thresholds considered (0.05, 0.005 or 0.0005) for that gene and tissue.
(DOCX)
The LD pattern at the simulated region follows that at the SLC6A14 locus. For the SS method, the nominal type 1 error was set at alpha = 0.05 or alpha = 0.005. The eQTL evidence was measured continuously as -log10 (eQTL p-value), or dichotomized using the eQTL p<0.05 or <0.005 threshold. For COLOC and eCAVIAR, the false positive rates were calculated by applying the 0.5, 0.75 or 0.9 threshold to the colocalization posterior probability. The value of represents the standardized true effect size of the GWAS associated variant, and represents the standardized true effect size of the eQTL variant, as detailed in S5 Table. Here, is set to be 5.73 such that 0.5 power is achieved to detect the GWAS association at significance level of 10−8, while is set to be 3.4, 4.09, 4.45, 5.21 or 5.73 for each row of the table such that 0.01, 0.05, 0.1, 0.3, or 0.5 power is achieved to detect the eQTL association at significance level 10−8. In total, 104 replications were simulated to obtain each cell of the table. See S1 Appendix for other simulation details.
(DOCX)
The LD pattern at the simulated region follows that at the SLC6A14 locus. For the SS method, the nominal type 1 error was set at alpha = 0.05 or alpha = 0.005. The eQTL evidence was measured continuously as -log10 (eQTL p-value) or dichotomized using the eQTL p<0.05 or <0.005 threshold. For COLOC and eCAVIAR, the false positive rates were calculated by applying the 0.5, 0.75 or 0.9 threshold to the colocalization posterior probability. The values of and represent the standardized true effect sizes of two GWAS associated variants, while and represent the standardized true effect sizes of two eQTL variants. Here, is set to be 6.57 and is set to be 5.73 such that, respectively, 0.8 and 0.5 power are achieved to detect the two GWAS signals at significance level 10−8. is set to be 3.4, 4.09, 4.45, 5.21 or 5.73 for each row of the table such that 0.01, 0.05, 0.1, 0.3, or 0.5 power is achieved to detect the eQTL association at significance level of 10−8 and is set to be 0. In total, 104 replications were simulated to obtain each cell of the table. See S1 Appendix for other simulation details.
(DOCX)
The LD pattern at the simulated region follows that at the SLC6A14 locus. For the SS method, the nominal type 1 error was set at alpha = 0.05 or alpha = 0.005. The eQTL evidence was measured continuously as -log10 (eQTL p-value) or dichotomized using the eQTL p<0.05 or <0.005 threshold. For COLOC and eCAVIAR, the false positive rates were calculated by applying the 0.5, 0.75 or 0.9 threshold to the colocalization posterior probability. The values of and represent the standardized true effect sizes of two GWAS associated variants, while and represent the standardized true effect sizes of two eQTL variants. Here, is set to be 6.57 and is set to be 5.73 such that 0.8 and 0.5 power are achieved to detect GWAS signals at significance level of 10−8. is set to be 0, and is set to be 3.4, 4.09, 4.45, 5.21 or 5.73 for each row of the table such that 0.01, 0.05, 0.1, 0.3, or 0.5 power is achieved to detect the eQTL association at significance level 10−8. In total, 104 replications were simulated to obtain each cell of the table. See S1 Appendix for other simulation details.
(DOCX)
The LD pattern at the simulated region follows that at the SLC6A14 locus. For the SS method, the nominal type 1 error was set at alpha = 0.05 or alpha = 0.005. The eQTL evidence was measured continuously as -log10 (eQTL p-value) or dichotomized using the eQTL p<0.05 or <0.005 threshold. For COLOC and eCAVIAR, the false positive rates were calculated by applying the 0.5, 0.75 or 0.9 threshold to the colocalization posterior probability. The values of and represent the standardized true effect sizes of two GWAS associated variants, while and represent standardized true effect sizes of two eQTL variants. Here, is set to be 6.57 such that 0.8 power is achieved to detect that GWAS signal at significance level 10−8 and is set to be 0. is set to be 3.4, 4.09, 4.45, 5.21 or 5.73 for each row of the table such that 0.01, 0.05,0.1,0.3, or 0.5 power are achieved to detect the eQTL association at significance level of 10−8, and is set to be 5.73 such that 0.5 power is achieved to detect that eQTL signal at significance level of 10−8. In total, 104 replications were simulated to obtain each cell of the table. See S1 Appendix for other simulation details.
(DOCX)
The LD pattern at the simulated region follows that at the SLC6A14 locus. For the SS method, the nominal type 1 error was set at alpha = 0.05 or alpha = 0.005. The eQTL evidence was measured continuously as -log10 (eQTL p-value) or dichotomized using the eQTL p<0.05 or <0.005 threshold. For COLOC and eCAVIAR, the false positive rates were calculated by applying the 0.5, 0.75 or 0.9 threshold to the colocalization posterior probability. The values of and represent the standardized true effect sizes of two GWAS associated variants, while and represent the standardized true effect sizes of eQTL variants. Here, is set to be 6.57 such that 0.8 power is achieved to detect that GWAS signal at significance level of 10−8 and is set to be 0. is set to be 3.4, 4.09, 4.45, 5.21 or 5.73 for each row of the table such that 0.01, 0.05,0.1,0.3, or 0.5 power are achieved to detect the eQTL association at significance level of 10−8 and is set to be 7.01 such that 0.9 power is achieved to detect that eQTL signal at significance level of 10−8. In total, 104 replications were simulated to obtain each cell of the table. See S1 Appendix for other simulation details.
(DOCX)
The LD pattern at the simulated region follows that at the SLC6A14 locus. For the SS method, the nominal type 1 error was set at alpha = 0.05 or alpha = 0.005. The eQTL evidence was measured continuously as -log10 (eQTL p-value) or dichotomized using the eQTL p<0.05 or <0.005 threshold. For COLOC and eCAVIAR, the false positive rates were calculated by applying the 0.5, 0.75 or 0.9 threshold to the colocalization posterior probability. The values of and represent the standardized true effect sizes of two GWAS associated variants, while and represent the standardized true effect sizes of eQTL variants. Here, is set to be 6.57 and is set to be 5.73 such that, respectively, 0.8 and 0.5 power are achieved to detect the two GWAS signals at significance level of 10−8. is set to be 3.4, 4.09, 4.45, 5.21 or 5.73 for each row of the table such that 0.01, 0.05,0.1,0.3, or 0.5 power are achieved to detect the eQTL association at significance level of 10−8 and is set to be 7.01 such that 0.9 power is achieved to detect that eQTL signal at significance level of 10−8. In total, 104 replications were simulated to obtain each cell of the table. See S1 Appendix for other simulation details.
(DOCX)
The exact region is 115248275–115448275 bp as in human genome reference assembly GRCh37. The eQTL evidence used include the -log10 transform of eQTL p value, and dichotomized eQTL p-value indicator by thresholds of eQTL p<0.05 or <0.005 for each specified gene and tissue. We focus on the analysis evaluating whether the eQTLs for SLC6A14 in lung (or human nasal epithelial; HNE) colocalize with lung-associated variants more than eQTLs for AGTR2, PLS3 and CXorf61 in lung (or HNE). The contrasting colocalization test for SLC6A14 is listed as NA since we do not contrast SLC6A14 with itself; NA in other cells means no eQTL SNP with p<0.05 or p<0.005 for that gene. The column ‘No. of eQTL SNPs’ shows the number of SNPs with eQTL p-values< 0.05 or 0.005 in the 0.1Mbp region; it refers to the number of SNPs for which eQTL p-values were available at the locus when -log10(eQTLp) is used (first 4 rows of the table). All p-values are one-sided to ensure colocalization rather than negative correlation.
(DOCX)
Data Availability Statement
Summary statistics from the GWAS are available in the NHGRI-EBI catalog. R scripts for the simple sum method are available at http://lab.research.sickkids.ca/strug/publications-software/. Access to the RNA-sequencing data from the nasal epithelia are available through the CF Canada – SickKids Program for Individualized Therapy Biobank http://lab.research.sickkids.ca/cfit/). Genotype data cannot be shared publicly because the consents restrict access to Cystic Fibrosis researchers. Genotype data are available by application to the Cystic Fibrosis Canada National data registry for researchers who meet the criteria for access to confidential clinical data for the purpose of CF research. Application to the Cystic Fibrosis Canada National data registry does not include a requirement to collaborate and/or publish with the authors of the current manuscript.