INTRODUCTION:
Alterations of gut microbiota have been thought to be associated with irritable bowel syndrome (IBS). Many studies have reported significant alterations of gut microbiota in patients with IBS based on 16S ribosomal RNA-targeted sequencing. However, results from these studies are inconsistent or even contradictory. We performed a systematic review to explore the alterations of gut microbiota in patients with IBS compared with healthy controls (HCs).
METHODS:
The databases PubMed, Cochrane Library, Web of Science, and Embase were searched for studies published until February 28, 2018, for case–control studies detecting gut microbiota in patients with IBS. Methodological quality was assessed using the Newcastle–Ottawa Scale. The α-diversity and alterations of gut microbiota in patients with IBS compared with HCs were analyzed.
RESULTS:
Sixteen articles involving 777 patients with IBS and 461 HCs were included. Quality assessment scores of the studies ranged from 5 to 7. For most studies, patients with IBS had a lower α-diversity than HCs in both fecal and mucosal samples. Relatively consistent changes in fecal microbiota for patients with IBS included increased Firmicutes, decreased Bacteroidetes, and increased Firmicutes:Bacteroidetes ratio at the phylum level, as well as increased Clostridia and Clostridiales, decreased Bacteroidia and Bacteroidales at lower taxonomic levels. Results for mucosal microbiota were inconsistent.
CONCLUSIONS:
Alterations of gut microbiota exist in patients with IBS and have significant association with the development of IBS. Further studies are needed to draw conclusions about gut microbiota changes in patients with IBS.
TRANSLATIONAL IMPACT:
This knowledge might improve the understanding of microbial signatures in patients with IBS and would guide future therapeutic strategies.
INTRODUCTION
Irritable bowel syndrome (IBS) is a common functional gastrointestinal disorder that affects 11.2% of the world's population (1). As a chronic disease, IBS is characterized by abdominal pain or bloating associated with alterations in bowel habits and is further subtyped into four patterns: diarrhea-predominant (IBS-D), constipation-predominant (IBS-C), mixed irritable bowel syndrome (IBS-M), and un-subtyped (IBS-U) (2). Due to the lack of biological markers and organic lesions, gastroenterologists rely on clinical findings for the diagnosis of IBS, based on the Rome criteria. (3).
The human gut is a complicated living ecosystem inhabited by up to 1014 microbes (4). Metagenomic sequencing has revealed that gut microbes contain 150-fold more genes than that of the human genome (5). In a healthy person, the gut microbiota is able to absorb dietary nutrients, facilitate normal immune responses, and maintain homeostasis within the host (6). Accordingly, it is reasonable to regard gut microbiota as a virtual organ (7), and dysbiosis of this organ has been shown to exert important effects on multiple diseases (8–11). In particular, the clinical guidance regarding modulation of gut microbiota in IBS provided by the Rome Team Working Group has confirmed the concept of disturbed gut microbiota in patients with IBS (12).
Traditional culture-based methods can be used to only study approximately 0.1% of the microbes living in the human gut (13). To more completely investigate microbial ecosystems, new molecular techniques have been developed over the past decades (14). Microbiota profiling using 16S ribosomal RNA (rRNA)-based sequencing is a technique that amplifies the hypervariable regions of 16S rRNA genes and subsequently compares the sequences obtained with known sequences in databases to analyze the microbial composition (14). Due to its accuracy and high efficiency, the 16S rRNA-targeted sequencing technique has rapidly become the most widely used approach for characterization of the gut microbiota (15).
A considerable number of studies based on 16S rRNA sequencing have shown perturbed gut microbiota in patients with IBS. Nevertheless, the results of these studies are inconsistent or even contradictory. To the best of our knowledge, no systematic reviews of the alterations in gut microbiota of patients with IBS based on 16S rRNA-targeted sequencing have been conducted. This article synthesizes data from the relevant publications to evaluate the current state of knowledge and provides direction for future research.
METHODS
Literature search
Four electronic databases were searched for publications from their inception to February 28, 2018, PubMed, Cochrane Library, Web of Science, and Embase. Our search terms, which are reported in the Supplementary Materials, covered expressions for IBS, microbiota, and 16S rRNA sequencing. The reference lists of relevant articles were searched manually for other possible studies.
Study selection
The eligible inclusion criteria included (i) subjects aged 18 years or older, (ii) diagnosis of IBS based on established criteria (Manning or Rome criteria), (iii) case–control studies that compared the alterations of gut microbiota between patients with IBS and healthy controls (HCs), and (iv) studies published in English.
Quality assessment
The quality of each eligible study was carefully assessed according to the nine items of the Newcastle–Ottawa Scale, which is a valid tool for quality assessment of case–control studies (16). The selection criteria include four items: (i) is the case definition adequate? (ii) representativeness of the cases, (iii) selection of controls, and (iv) definition of controls. The comparability criteria include comparability of cases and controls on the basis of the design or analysis. The exposure criteria include 3 items: (i) ascertainment of exposure, (ii) same method of ascertainment for cases and controls, and (iii) nonresponse rate. Studies that scored ≤4 were considered to be of poor quality and were excluded.
Data extraction
Two reviewers independently extracted the following data from the eligible studies: (i) first author, (ii) publication year, (iii) country of origin, (iv) diagnostic criteria for IBS, (v) IBS subtypes, (vi) characteristics of the participants (e.g., sample size, age, and sex), (vii) sample materials, (viii) DNA extraction method, 16S rRNA variable region, sequencing platform, data analysis platform, and reference sequences database employed in the studies, and (ix) α-diversity and alterations of gut microbiota in patients with IBS compared with those in HCs. Disagreements between the reviewers regarding the data abstraction were resolved through discussion, with a consensus being reached.
RESULTS
Study selection
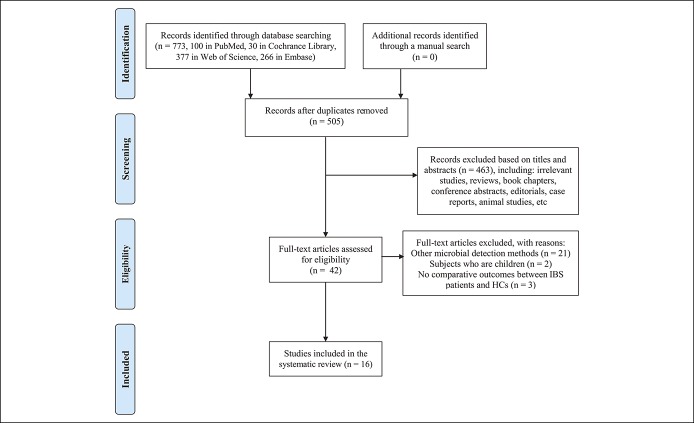
Applying the search strategy, a total of 773 records were identified from the 4 databases. The manual search yielded no additional studies. After removing duplicates, 505 records were screened by titles and abstracts, of which 42 remained for full-text review. Finally, 16 studies fulfilled the inclusion criteria for this systematic review (Figure 1) (17–32).
Figure 1.
Flow chart of study selection.
Study characteristics
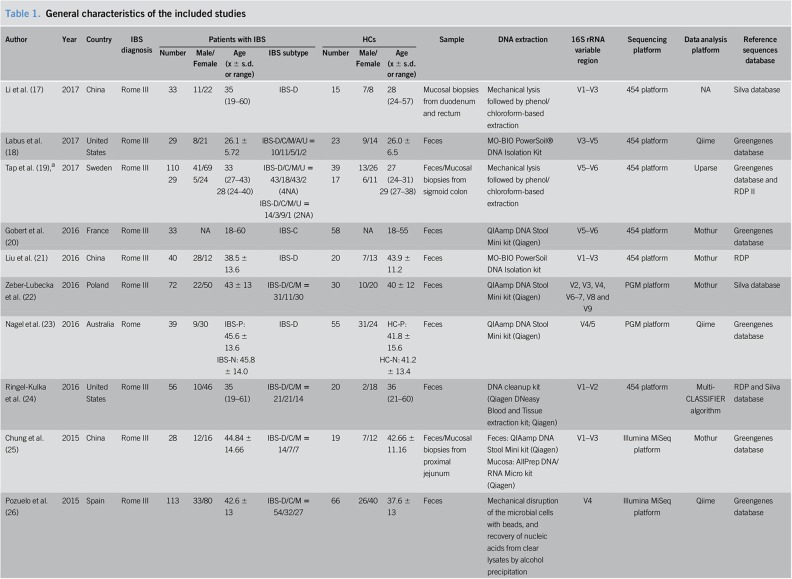
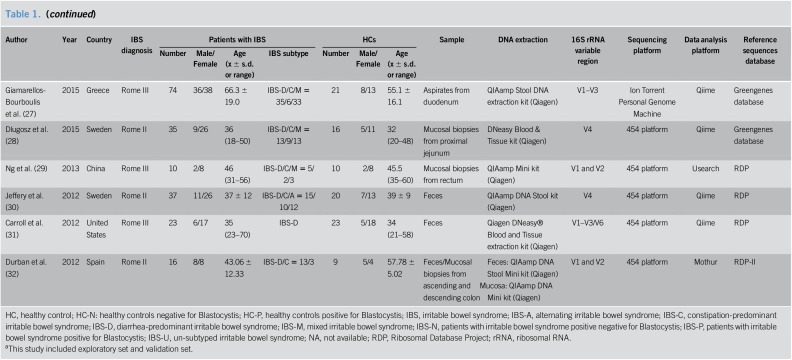
The general characteristics of the included studies are presented in Table 1. All eligible studies were published after 2010. The regions of studies involved eight countries: China, Sweden, the United States, Spain, France, Poland, Australia, and Greece. The total sample size of the 16 studies was 1,238 (777 patients with IBS and 461 HCs). Female participants made up 66.3% of the patients with IBS and 62.8% of the HCs, with one study being excluded from the gender calculation because the exact number of female participants was not reported. The average age varied from 26.1 to 66.3 in patients with IBS and 26.0 to 57.8 in HCs, with one study being excluded from the age description because only the age range was available. Most of the included studies were sex- and age-matched. As for the identification of IBS, 12 articles used Rome III criteria, 3 articles adopted Rome II criteria, and one article only mentioned Rome criteria. Eleven studies recruited various subtypes of patients with IBS, while 5 studies focused on specific subtypes (IBS-D or IBS-C). The sources of samples differed, with 9 studies using fecal samples, 3 studies using mucosal biopsies, 3 studies based on both fecal and mucosal specimens, and the remaining study adopting aspirates from the duodenum. A QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) was extensively used for DNA extraction. Different 16S rRNA variable regions were used for DNA amplification. The widely used sequencing platform was the 454 platform, and data analysis platforms were Qiime and Mothur. Reference sequence databases used in the studies included the Silva database, Greengenes database, and Ribosomal Database Project.
Table 1.
General characteristics of the included studies
Quality assessment
The results from the quality assessment are shown in Supplemental Digital Content 1 (Table 1, http://links.lww.com/CTG/A7). The quality scores of the 16 studies varied from 5 to 7 and were sufficient to conduct our systematic review.
Alpha-diversity of gut microbiota in patients with IBS
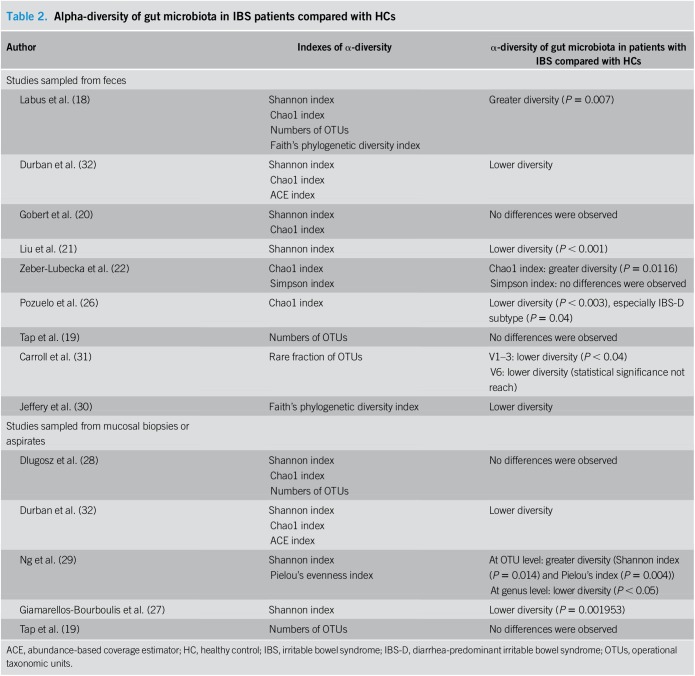
Table 2 displays the results of the α-diversity of gut microbiota in patients with IBS compared with that in HCs, and the indexes are used to describe the diversity in the included studies. The most frequently adopted α-diversity estimator was the Shannon index, followed by the Chao1 index. As for alterations of microbial α-diversity in patients with IBS, the studies were discrepant in their results, presenting a greater diversity (18,22,29), a lower diversity (21,26,27,29–32), or no differences (19,20,22,28) compared with that of HCs. However, we were still able to capture a relatively consistent trend with patients with IBS having a lower microbial α-diversity than HCs in both fecal and mucosal samples for most of the studies. Furthermore, the majority of the studies showed that the differences were statistically significant.
Table 2.
Alpha-diversity of gut microbiota in IBS patients compared with HCs
Alterations in the fecal microbiota composition of patients with IBS
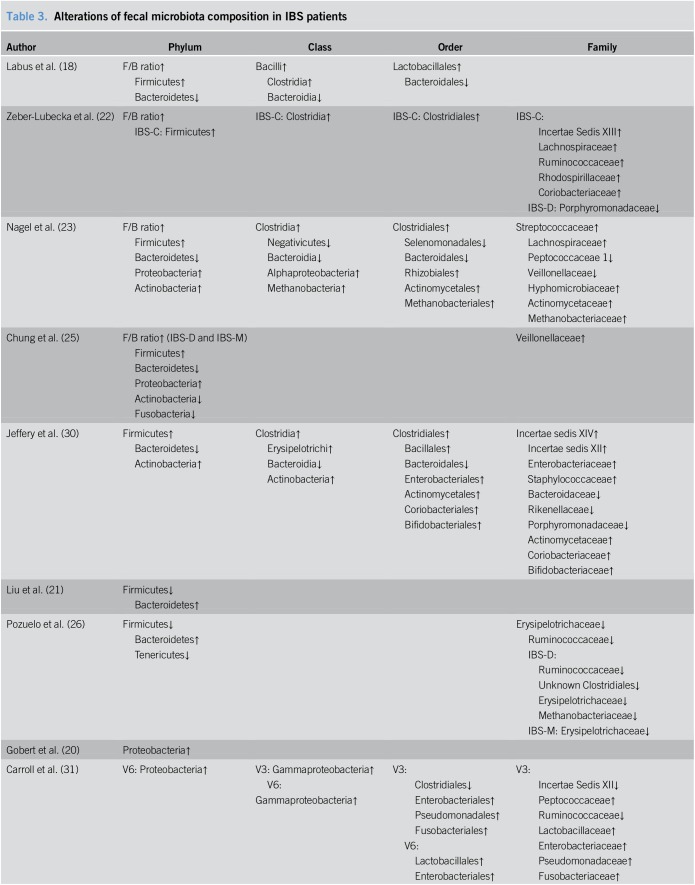
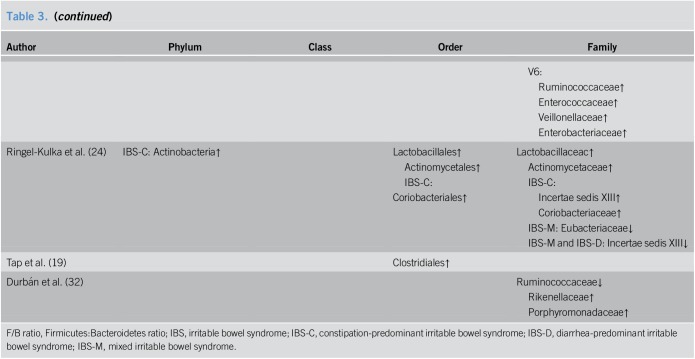
Differences in the fecal microbiota composition between patients with IBS and HCs are listed in Table 3. On the whole, there were some microorganisms that showed relatively consistent tendencies for variation, whereas others presented inconsistent or even contradictory differences. In general, the detected bacterial phyla changes in those who suffered from IBS in the eligible studies included Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, Tenericutes, and Fusobacteria, with the majority of species attributed to the 2 largest phyla colonizing the human gut, Firmicutes and Bacteroidetes. In all but two of the included studies (21,26), we observed that there were trends at the phylum level for patients with IBS to harbor a higher abundance of Firmicutes and a lower abundance of Bacteroidetes compared to those for HCs and thus an increased Firmicutes:Bacteroidetes ratio (F/B ratio, approximately 1.2–3.5-fold in patients with IBS than in HCs from the available data) was identified (18,22,23,25,30). The mean relative abundances of Firmicutes and Bacteroidetes, and F/B ratio in the relevant studies are presented in Supplemental Digital Content 1 (Table 2, http://links.lww.com/CTG/A7).
Table 3.
Alterations of fecal microbiota composition in IBS patients
The change in Firmicutes was relatively consistent through the downstream taxonomic hierarchy. As a predominant class within the phylum Firmicutes, Clostridia was present at increased levels in patients with IBS (18,22,23,30), and at the order level, a coincidently higher abundance of Clostridiales was observed (19,22,23,30). Within the order Clostridiales, we noticed that conflicting results were reported for one family (Ruminococcaceae) being present at a high frequency in the included studies, with both increases (22,31) and decreases (26,31,32) being observed in patients with IBS. As for microbial alterations at the genus level, enormous numbers of genera were detected that differed between patients with IBS and HCs and the results among the different studies varied dramatically (Supplemental Digital Content 1, Table 3, http://links.lww.com/CTG/A7). Unlike the class Clostridia, the class Bacilli, which is another member of Firmicutes, was demonstrated to be increased in patients with IBS in only one study (18), but its lower taxon Lactobacillales was consistently shown to be at elevated levels in patients (18,24,31). Studies also presented an increase in the abundance of Lactobacillaceac at the family level (24,31) and Lactobacillus at the genus level (24), which is one of the most commonly known probiotics.
As mentioned above, the second most abundant phylum in the human gut, Bacteroidetes, was found at a lower proportion in patients with IBS. Consistent with this finding, decreases in Bacteroidia at the class level and Bacteroidales at the order level were also detected in patients (18,23,30). Prevotella, a genus within the order Bacteroidales, which was identified as one of the three enterotypes in the human gut microbiome by Arumugam et al. and Liu et al. (21,33), was shown to be both increased (20,21,25) and decreased (19) in patients with IBS.
A higher proportion of Proteobacteria, another main phylum detected in the human gut, was found in several studies to be associated with IBS (20,23,25,31). Specifically, the order Enterobacteriales and family Enterobacteriaceae within the phylum showed increased levels in patients with IBS (30,31).
However, unlike the above 3 phyla, there was no consistent alteration for the phylum Actinobacteria, which showed both increased (23,24,30) and decreased (25) levels in patients with IBS. This phylum consists of many genera, including Bifidobacterium, another bacteria commonly used as a probiotic. However, reports on the changes for this probiotic were limited and conflicting with both increased (30) and decreased (21) levels found in patients with IBS.
Furthermore, we did a subtype analysis based on fecal microbiota. Alterations of fecal microbiota composition in different subtypes of patients with IBS are listed in Supplemental Digital Content 1 (Table 4, http://links.lww.com/CTG/A7). There were limited studies regarding each subtype, and we did not find consistent change trends in subtype analyses.
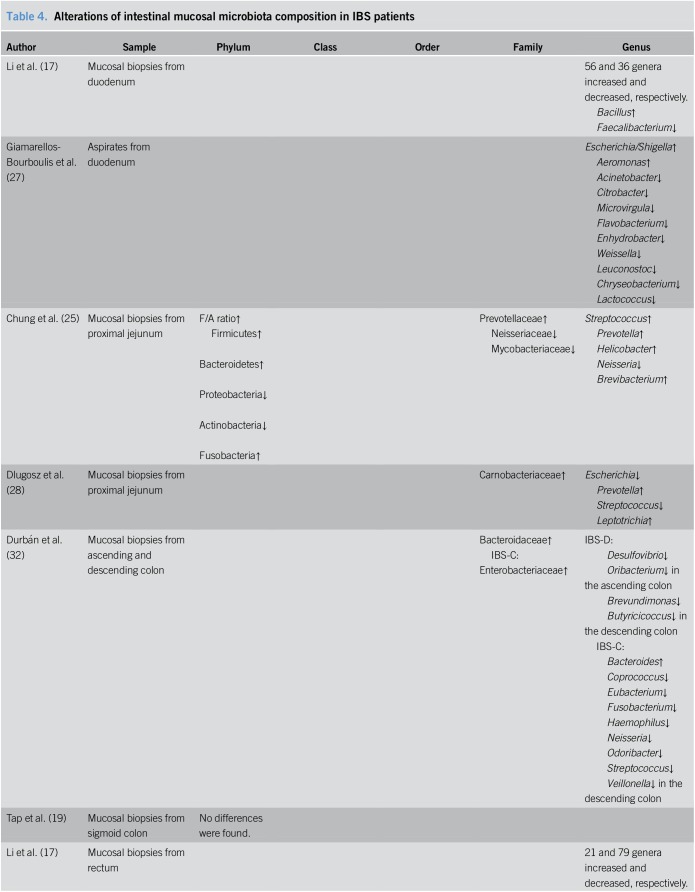
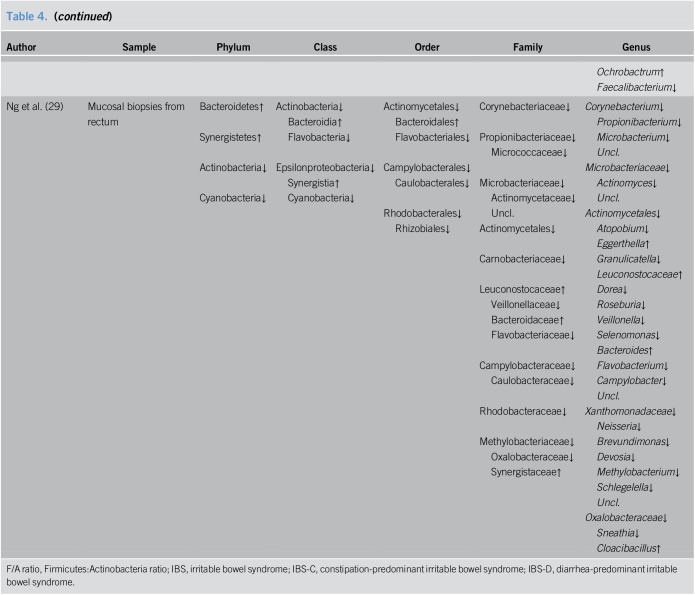
Alterations in intestinal mucosal microbiota composition of patients with IBS
Six of the studies focused on alterations in intestinal mucosal microbiota of patients with IBS. In addition, one study evaluated duodenal aspirates, which we include here for the purpose of convenient discussion. The results from these studies are summarized in Table 4.
Table 4.
Alterations of intestinal mucosal microbiota composition in IBS patients
Among the selected studies, 2 publications analyzed the microbiota from the duodenum, with duodenal biopsies used in one study and duodenal aspirates used in the other. Li et al. (17) identified 56 genera in duodenal biopsies from patients with IBS-D that increased and 36 that decreased, with the most prevalent genera having increased and decreased frequencies being Bacillus and Faecalibacterium, respectively. Meanwhile, analysis of the duodenal aspirates by Giamarellos-Bourboulis et al. (27) detected significant changes for 12 genera, including the overrepresentation of Escherichia/Shigella and Aeromonas and the underrepresentation of Acinetobacter, Citrobacter, Microvirgula, Flavobacterium, Enhydrobacter, Weissella, Leuconostoc, Chryseobacterium, and Lactococcus.
Two studies focused on mucosal microbiota from the proximal jejunum, with one study using single-balloon enteroscopy and the other using sterile Watson capsules to biopsy the jejunum. Chung et al. found a higher Firmicutes:Actinobacteria ratio in patients with IBS compared to that in HCs. Increased Prevotellaceae and decreased Mycobacteriaceae and Neisseriaceae at the family level, and higher proportions of Streptococcus, Prevotella, Helicobacter, Brevibacterium and a lower proportion of Neisseria at the genus level were also observed in patients with IBS (25). In contrast, Dlugosz et al. (28) found no statistical differences among major phyla or genera between patients with IBS and HCs. Regardless, some operational taxonomic units exhibited a trend toward differential expression with increased levels of Carnobacteriaceae, Prevotella, and Leptotrichia and decreased levels of Escherichia and Streptococcus.
Other studies searched for alterations of mucosal microbiota in the colon or rectum. Durbán et al. (32) reported that mucosal biopsies from the ascending and descending colon in patients with IBS presented with higher counts of Bacteroidaceae. Tap et al. (19) failed to report significant differences between patients and controls regarding the mucosal microbiota from the sigmoid colon. As for the mucosal microbiota in the rectum, Li et al. (17) showed 21 genera in patients with IBS-D with increased levels and 79 genera with decreased levels, with the most common genera having increased and decreased frequencies being Ochrobactrum and Faecalibacterium, respectively. Ng et al. (29) detected some members that differed significantly between the 2 groups with increased levels of Bacteroidetes and Synegitestes and reduced levels of Actinobacteria and Cyanobacteria in patients with IBS. And the differences in abundance at the lower taxonomic levels are shown in detail in Table 4.
DISCUSSION
Numerous studies have focused on gut microbiota alterations in patients with IBS. A better understanding of microbial signatures is an important prerequisite if intervention is to be used in managing the disease (34). In the current study, we choose 16S rRNA-targeted sequencing as the single method in the inclusion criteria to avoid methodology-based heterogeneity. Results from the eligible studies showed both inconsistencies and several common trends pertaining to microbial alterations of patients with IBS.
Microbial diversity is a crucial property of communities because it is a primary descriptor of the community structure and a major determinant of community function (35). In this review, we found that most of the studies observed a lower microbial α-diversity in both fecal and mucosal samples from patients with IBS compared to those from HCs. Similar findings have also been confirmed by other microbial detection methods, including terminal-restriction fragment length polymorphism (36), DNA microarrays (37) and metagenomic gene-targeted approach (38). Given these findings, it can be easily speculated that microbial dysbiosis may be a result of increases or decreases of specific microbial groups and the disappearance of global homeostasis (39), which are thought to be markers for negative conditions in patients with IBS.
As noted above, the reports from different studies on fecal microbial profiles in patients with IBS are mixed but include several consistent trends. For instance, increased levels of Firmicutes, decreased levels of Bacteroidetes, and increased F/B ratio were found. The phylum Firmicutes makes up the largest portion of the human gut microbiota and has been shown to be associated with energy extraction and potentially related to obesity and diabetes (9). In contrast, the phylum Bacteroidetes generally produces butyrate, which is suggested to reduce inflammation and plays a role in the normal development of the gut (6,9). In general terms, the F/B ratio is regarded to be of significant relevance in signaling human gut microbiota status (40). Altered F/B ratio has been reported in connection with diet and some chronic diseases such as obesity (41,42). A 2-fold higher F/B ratio was also found in patients with IBS compared to that in HCs in the study of Rajilić-Stojanović et al. (43), in which fecal samples were analyzed using a human intestinal tract chip. However, even with these findings, at this time there remains no clear consensus on how the changes are associated with IBS. Clemente et al. (44) proposed that the shift in abundances of Firmicutes and Bacteroidetes in obese individuals resulted in an increased capacity for harvesting energy from food and producing low-grade inflammation. The fatty acids in high-fat diets have been shown to increase the F/B ratio, which is associated with increased gut epithelial permeability and low-grade inflammation (18). Therefore, we can speculate that the alterations in bacterial phyla of patients with IBS may be related to alterations in epithelial permeability and low-grade inflammation, which have been implicated as possible components of the pathogenesis of IBS (45). In contrast, 2 of the studies detected increases in Bacteroidetes and decreases in Firmicutes in patients with IBS. The divergence in the results may be attributed to different dietary habits, geographical environments, or even methodological heterogeneity.
The tendencies for the change of Firmicutes and Bacteroidetes in patients with IBS persisted at the lower taxonomic levels with higher abundances of Clostridia and Clostridiales and lower abundances of Bacteroidia and Bacteroidales. Clostridia is a wide and heterogenic class that includes species degrading various organic compounds and are devoted to acid production (46). Disturbed metabolism of intestinal short-chain fatty acids has been associated with IBS (11), so it is intriguing to speculate that bacteria of the class Clostridia may play a role in the development of IBS as a result of its capacity to produce acids. As for decreased levels of Bacteroidia and Bacteroidales in patients with IBS, it remains unclear about their relationship with IBS. Nevertheless, using an animal model of experimental colitis, a previous study showed that Bacteroides fragilis, which is one of the most important species within the order Bacteroidales, protected its host from inflammation and that this beneficial activity required polysaccharide A, a natural antiinflammatory molecule of Bacteroides fragilis (47).
Increased Proteobacteria, as well as the order Enterobacteriales and family Enterobacteriaceae within the phylum were found in patients with IBS, which encompass many known pathogenic species with potential inflammation causing mechanisms such as Escherichia coli (6). Thus, it is likely that these bacteria are among the potential pathogens that contribute to the progression of IBS (14).
Thus far, there have been 2 meta-analyses focusing on gut microbiota alterations in patients with IBS, and all of the studies included in these 2 meta-analyses were based on culture or real-time quantitative polymerase chain reaction methods (48,49). The first meta-analysis found increased levels of Escherichia coli and Enterobacter and decreased levels of Bifidobacteria and Lactobacillus in patients with IBS from China compared with those in HCs. In addition, Bacteroides and Bifidobacteria were shown to be increased and decreased, respectively in patients with IBS from other regions of the world. (48) The second meta-analysis pooled seven studies that were based on quantitative polymerase chain reaction and showed significant decreases in the levels of Lactobacillus, Bifidobacterium, and Faecalibacterium prausnitzii in patients with IBS compared with those in HCs (49). In these 2 studies, there was a trend with patients with IBS showing decreased levels of Lactobacillus and Bifidobacterium, the 2 most widely used probiotics. Most of the previous clinical trials demonstrated that the probiotics Lactobacillus spp. and Bifidobacterium spp. have positive effects on the overall symptoms of IBS by altering the composition of gut microbiota (50,51), suggesting a preventive role for these microorganisms in IBS. However, this finding is not universal with some studies recording no significant improvement or even unfavorable effects on patient symptoms (52,53). Our current review indicated that the previous reports on the 2 probiotics are limited and inconsistent. Thus, it remains unclear whether there are IBS-specific effects related to Lactobacillus and Bifidobacterium.
Most studies on gut microbiota of patients with IBS carried out to date have drawn conclusions by analyzing fecal samples since they are easily collected in a noninvasive manner. In contrast, mucosal samples must be obtained through an endoscopic procedure (54). However, there is increasing evidence that the mucosa-associated microbiota significantly differs from the feces-associated microbiota (55). The microbial composition in feces does not necessarily reflect the organisms directly related to disorders (54), which may be partly explained by the fact that amplification of DNA from feces also identifies nonviable and transient microbes that may not be biologically active (27). The mucosa-associated microbiota may play a more prominent pathogenic role due to their closer contact with host epithelial, immune, and enteroendocrine cells (32,56). However, studies analyzing mucosal microbiota composition are limited, and their results are not comparable because of the difference in sampling and the heterogeneity of methodologies. Hence, more studies concerning mucosa-associated microbiota in patients with IBS should be included in future research.
Considering that IBS is a multifactorial disorder with many putative causes and a wide range of symptoms, it can be reasonably stated that results from these studies might not represent patients with IBS as a whole, but rather as only a portion of patients. In addition, a variety of factors may affect the microbial identification, such as sample size, geographical environment, dietary habits, diagnostic criteria, and sample sources (11). Furthermore, the specific processes of the 16S rRNA sequencing method may also drastically affect the quality of results, including the DNA extraction techniques, primers used for amplicon generation, bioinformatic pipelines, data transformations, and statistical approaches (14). Thus, it is difficult to compare the results from various studies and make a general inference on microbial profiles in patients with IBS. From this point of view, our work depicting alterations of gut microbiota in patients with IBS based on 16S rRNA sequencing may be viewed as a preliminary study.
We acknowledge several limitations regarding this systematic review. First, we only listed alterations in fecal microbiota according to limited studies involving subtypes, because most of the studies were based on the assessment of various subtypes of patients with IBS, making subgroup analysis difficult. Therefore, additional studies considering IBS subtypes are needed in the future. Second, because this was a qualitative review and we only included peer-reviewed published studies, it is impossible to comment on the likelihood of publication bias.
CONCLUSION
In conclusion, our systematic review found that based on 16S rRNA-targeted sequencing, there are alterations in the gut microbiota of patients with IBS compared with that in HCs. Patients with IBS had a lower α-diversity than HCs in both fecal and mucosal samples. Relatively consistent changes in fecal microbiota for patients with IBS included increased Firmicutes, decreased Bacteroidetes, and increased F/B ratio at the phylum level, as well as increased Clostridia and Clostridiales and decreased Bacteroidia and Bacteroidales at lower taxonomic levels. Results for mucosal microbiota were inconsistent. Further studies, especially studies regarding mucosa-associated microbiota and studies based on IBS subtype analyses, are needed to draw conclusions about the alterations of gut microbiota in patients with IBS.
CONFLICTS OF INTEREST
Guarantor of the article: Liping Duan, MD.
Specific author contributions: R.D. carried out the study selection and drafted the manuscript. R.D., S.Z., and B.W. contributed to extraction and analysis of the data. L.D. designed and supervised the study. All authors commented on drafts of the paper and approved the final manuscript.
Financial support: The study was supported by a grant from the National Natural Science Foundation of China (81670491) and the Capital Health Research and Development of Special (2016-2-4093).
Potential competing interests: None.
Supplementary Material
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A7
REFERENCES
- 1.Lovell RM, Ford AC. Global prevalence of and risk factors for irritable bowel syndrome: A meta-analysis. Clin Gastroenterol Hepatol 2012;10:712–21.e4. [DOI] [PubMed] [Google Scholar]
- 2.Chey WD, Kurlander J, Eswaran S. Irritable bowel syndrome: a clinical review. JAMA 2015;313:949–58. [DOI] [PubMed] [Google Scholar]
- 3.Lacy BE, Patel NK. Rome criteria and a diagnostic approach to irritable bowel syndrome. J Clin Med 2017;6:E99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006;124:837–48. [DOI] [PubMed] [Google Scholar]
- 5.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennet SM, Ohman L, Simren M. Gut microbiota as potential orchestrators of irritable bowel syndrome. Gut Liver 2015;9:318–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Distrutti E, Monaldi L, Ricci P, et al. Gut microbiota role in irritable bowel syndrome: New therapeutic strategies. World J Gastroenterol 2016;22:2219–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong SH, Kwong TNY, Wu CY, et al. Clinical applications of gut microbiota in cancer biology. Semin Cancer Biol 2018. [Epub ahead of print May 18, 2018.] [DOI] [PubMed] [Google Scholar]
- 9.Pascale A, Marchesi N, Marelli C, et al. Microbiota and metabolic diseases. Endocrine 2018;61:357–71. [DOI] [PubMed] [Google Scholar]
- 10.Bruce-Keller AJ, Salbaum JM, Berthoud HR. Harnessing gut microbes for mental health: Getting from here to there. Biol Psychiatry 2018;83:214–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan WT, Ding C, Xu NN, et al. Close association between intestinal microbiota and irritable bowel syndrome. Eur J Clin Microbiol Infect Dis 2017;36:2303–17. [DOI] [PubMed] [Google Scholar]
- 12.Simren M, Barbara G, Flint HJ, et al. Intestinal microbiota in functional bowel disorders: A Rome foundation report. Gut 2013;62:159–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiergeist A, Glasner J, Reischl U, et al. Analyses of intestinal microbiota: Culture versus sequencing. ILAR J 2015;56:228–40. [DOI] [PubMed] [Google Scholar]
- 14.Taverniti V, Guglielmetti S. Methodological issues in the study of intestinal microbiota in irritable bowel syndrome. World J Gastroenterol 2014;20:8821–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mancabelli L, Milani C, Lugli GA, et al. Identification of universal gut microbial biomarkers of common human intestinal diseases by meta-analysis. FEMS Microbiol Ecol 2017;93. [DOI] [PubMed] [Google Scholar]
- 16.Wells G, Shea B, O'Connell D, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomized studies in meta-analyses. (http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp). Accessed on April 11, 2018.
- 17.Li G, Yang M, Jin Y, et al. Involvement of shared mucosal-associated microbiota in the duodenum and rectum in diarrhea-predominant irritable bowel syndrome. J Gastroenterol Hepatol 2018;33:1220–6. [DOI] [PubMed] [Google Scholar]
- 18.Labus JS, Hollister EB, Jacobs J, et al. Differences in gut microbial composition correlate with regional brain volumes in irritable bowel syndrome. Microbiome 2017;5:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tap J, Derrien M, Tornblom H, et al. Identification of an Intestinal Microbiota Signature Associated With Severity of Irritable Bowel Syndrome. Gastroenterology 2017;152:111–23.e8. [DOI] [PubMed] [Google Scholar]
- 20.Gobert AP, Sagrestani G, Delmas E, et al. The human intestinal microbiota of constipated-predominant irritable bowel syndrome patients exhibits anti-inflammatory properties. Sci Rep 2016;6:39399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Zhang L, Wang X, et al. Similar fecal microbiota signatures in patients with diarrhea-predominant irritable bowel syndrome and patients with depression. Clin Gastroenterol Hepatol 2016;14:1602–11.e5. [DOI] [PubMed] [Google Scholar]
- 22.Zeber-Lubecka N, Kulecka M, Ambrozkiewicz F, et al. Limited prolonged effects of rifaximin treatment on irritable bowel syndrome-related differences in the fecal microbiome and metabolome. Gut Microbes 2016;7:397–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagel R, Traub RJ, Allcock RJ, et al. Comparison of faecal microbiota in Blastocystis-positive and Blastocystis-negative irritable bowel syndrome patients. Microbiome 2016;4:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ringel-Kulka T, Benson AK, Carroll IM, et al. Molecular characterization of the intestinal microbiota in patients with and without abdominal bloating. Am J Physiol Gastrointest Liver Physiol 2016;310:G417–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung CS, Chang PF, Liao CH, et al. Differences of microbiota in small bowel and faeces between irritable bowel syndrome patients and healthy subjects. Scand J Gastroenterol 2016;51:410–9. [DOI] [PubMed] [Google Scholar]
- 26.Pozuelo M, Panda S, Santiago A, et al. Reduction of butyrate- and methane-producing microorganisms in patients with irritable bowel syndrome. Sci Rep 2015;5:12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giamarellos-Bourboulis E, Tang J, Pyleris E, et al. Molecular assessment of differences in the duodenal microbiome in subjects with irritable bowel syndrome. Scand J Gastroenterol 2015;50:1076–87. [DOI] [PubMed] [Google Scholar]
- 28.Dlugosz A, Winckler B, Lundin E, et al. No difference in small bowel microbiota between patients with irritable bowel syndrome and healthy controls. Sci Rep 2015;5:8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng SC, Lam EF, Lam TT, et al. Effect of probiotic bacteria on the intestinal microbiota in irritable bowel syndrome. J Gastroenterol Hepatol 2013;28:1624–31. [DOI] [PubMed] [Google Scholar]
- 30.Jeffery IB, O'Toole PW, Ohman L, et al. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut 2012;61:997–1006. [DOI] [PubMed] [Google Scholar]
- 31.Carroll IM, Ringel-Kulka T, Siddle JP, et al. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil 2012;24:521–30, e248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durban A, Abellan JJ, Jimenez-Hernandez N, et al. Structural alterations of faecal and mucosa-associated bacterial communities in irritable bowel syndrome. Environ Microbiol Rep 2012;4:242–7. [DOI] [PubMed] [Google Scholar]
- 33.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature 2011;473:174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fischbach MA. Microbiome: Focus on causation and mechanism. Cell 2018;174:785–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haegeman B, Hamelin J, Moriarty J, et al. Robust estimation of microbial diversity in theory and in practice. ISME J 2013;7:1092–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carroll IM, Ringel-Kulka T, Keku TO, et al. Molecular analysis of the luminal- and mucosal-associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol 2011;301:G799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sundin J, Rangel I, Fuentes S, et al. Altered faecal and mucosal microbial composition in post-infectious irritable bowel syndrome patients correlates with mucosal lymphocyte phenotypes and psychological distress. Aliment Pharmacol Ther 2015;41:342–51. [DOI] [PubMed] [Google Scholar]
- 38.Lopetuso LR, Petito V, Graziani C, et al. Gut microbiota in health, diverticular disease, irritable bowel syndrome, and inflammatory bowel diseases: Time for microbial marker of gastrointestinal disorders. Dig Dis 2018;36:56–65. [DOI] [PubMed] [Google Scholar]
- 39.Lozupone CA, Hamady M, Kelley ST, et al. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 2007;73:1576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mariat D, Firmesse O, Levenez F, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol 2009;9:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA 2010;107:14691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ley RE, Turnbaugh PJ, Klein S, et al. Microbial ecology: human gut microbes associated with obesity. Nature 2006;444:1022–3. [DOI] [PubMed] [Google Scholar]
- 43.Rajilić-Stojanović M, Biagi E, Heilig HG, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology 2011;141:1792–801. [DOI] [PubMed] [Google Scholar]
- 44.Clemente JC, Ursell LK, Parfrey LW, et al. The impact of the gut microbiota on human health: An integrative view. Cell 2012;148:1258–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Defrees DN, Bailey J. Irritable bowel syndrome: Epidemiology, pathophysiology, diagnosis, and treatment. Prim Care 2017;44:655–71. [DOI] [PubMed] [Google Scholar]
- 46.Durre P. Physiology and sporulation in clostridium. Microbiol Spectr 2014;2:TBS-0010-2012. [DOI] [PubMed] [Google Scholar]
- 47.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008;453:620–5. [DOI] [PubMed] [Google Scholar]
- 48.Zhuang X, Xiong L, Li L, et al. Alterations of gut microbiota in patients with irritable bowel syndrome: A systematic review and meta-analysis. J Gastroenterol Hepatol 2017;32:28–38. [DOI] [PubMed] [Google Scholar]
- 49.Liu HN, Wu H, Chen YZ, et al. Altered molecular signature of intestinal microbiota in irritable bowel syndrome patients compared with healthy controls: A systematic review and meta-analysis. Dig Liver Dis 2017;49:331–7. [DOI] [PubMed] [Google Scholar]
- 50.Tiequn B, Guanqun C, Shuo Z. Therapeutic effects of Lactobacillus in treating irritable bowel syndrome: A meta-analysis. Intern Med 2015;54:243–9. [DOI] [PubMed] [Google Scholar]
- 51.Ringel-Kulka T, Palsson OS, Maier D, et al. Probiotic bacteria Lactobacillus acidophilus NCFM and Bifidobacterium lactis Bi-07 versus placebo for the symptoms of bloating in patients with functional bowel disorders: A double-blind study. J Clin Gastroenterol 2011;45:518–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ligaarden SC, Axelsson L, Naterstad K, et al. A candidate probiotic with unfavourable effects in subjects with irritable bowel syndrome: A randomised controlled trial. BMC Gastroenterol 2010;10:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sondergaard B, Olsson J, Ohlson K, et al. Effects of probiotic fermented milk on symptoms and intestinal flora in patients with irritable bowel syndrome: A randomized, placebo-controlled trial. Scand J Gastroenterol 2011;46:663–72. [DOI] [PubMed] [Google Scholar]
- 54.Rangel I, Sundin J, Fuentes S, et al. The relationship between faecal-associated and mucosal-associated microbiota in irritable bowel syndrome patients and healthy subjects. Aliment Pharmacol Ther 2015;42:1211–21. [DOI] [PubMed] [Google Scholar]
- 55.Li G, Yang M, Zhou K, et al. Diversity of duodenal and rectal microbiota in biopsy tissues and luminal contents in healthy volunteers. J Microbiol Biotechnol 2015;25:1136–45. [DOI] [PubMed] [Google Scholar]
- 56.Ohman L, Tornblom H, Simren M. Crosstalk at the mucosal border: Importance of the gut microenvironment in IBS. Nat Rev Gastroenterol Hepatol 2015;12:36–49. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.