Resection of DNA double-strand break (DSB) termini is essential to create single-stranded DNA 3′ -overhangs that participate in homologous recombination (HR) DNA repair mechanisms. Early steps in DNA end processing involve a series of conserved nuclease-directed incisions followed by longer-range resection by additional exonucleases1. An extensive chromatin response at DNA breaks limits end resection by opposing nuclease action. Chromatin-directed control of end resection is spearheaded by the nucleosome-binding protein 53BP1, which directly binds to modified nucleosomes along megabase domains on either side of a DSB (ref.2). 53BP1 interaction with Rif1 occurs in response to DNA damage and is necessary to block end resection. Adding further complexity to the process, the Rev7 protein limits end resection downstream of 53BP1–Rif1, albeit by undefined mechanisms. While dispensable for much of the end-joining that occurs in response to ionizing radiation, the 53BP1–Rif1–Rev7 epistasis group is particularly important for DNA repair during class switch recombination at immunoglobulin loci, which entails long range non-homologous end joining (NHEJ) of programmed breaks. Work by Dev et al.3 in this issue of Nature Cell Biology, together with several contemporaneous studies4–8, define a previously unidentified protein complex termed Shieldin that acts in conjunction with 53BP1–Rif1 and Rev7 to limit DNA end-resection and promote NHEJ.
Limiting end-resection can also play a more sinister role in the context of HR deficiency. Mutation of the breast cancer early onset gene, BRCA1, is a major cause of hereditary breast and ovarian cancer susceptibility. BRCA1 mutant cells exhibit HR deficiency due to impaired end resection and failure to seed the Rad51 protein onto single stranded DNA. BRCA1 mutant cells that also lack 53BP1, Rif1 or Rev7 show a remarkable restoration of HR and genomic stability, while mice harbouring mutations in both BRCA1 and 53BP1 are not cancer prone2. In these scenarios, HR proceeds without fully active BRCA1, ostensibly due to removal of the chromatin block to end resection. While achieving genomic stability in a BRCA1 mutant cell is seemingly a positive outcome, paradoxically, this produces the unwanted consequence of resistance to poly(ADP)ribose polymerase (PARP) inhibitors, a recently approved therapy for BRCA1 and BRCA2 mutant ovarian and breast cancer2.
How the battle for end resection is decided is therefore a problem of considerable importance. In this light, several outstanding questions persist in relation to 53BP1-orchestrated events on damaged chromatin. Namely, how the 53BP1–Rif1 interaction is linked to Rev7; how nucleosome-bound 53BP1 communicates with single-stranded DNA regions proximal to the break; and if enzymatic activity is involved in combatting resection downstream of the 53BP1–Rif1– Rev7 axis. Work by Dev et al.3, as well as several independent studies4–9, has begun to assemble the missing pieces that link 53BP1 to end protection. CRISPR–Cas9 genome-wide screens were used to identify determinants of PARP inhibitor response in the setting of BRCA1 mutation3,4. Biochemical approaches were taken by the other groups to define 53BP1 and Rev7 interaction networks6–8. All studies complemented their respective genetic and biochemical investigations to reveal a protein complex that works in concert with 53BP1 and Rif1 to limit end resection. This, aptly named, Shieldin complex consists of C20orf196 (Shld1), Fam35A (Shld2), CTC-534A2.2 (Shl3) and Rev7 (refs3–8). Like 53BP1, Shieldin deficiency resulted in elevated end resection, while causing PARP inhibitor resistance in BRCA1 mutant cells3–8. In further agreement with a 53BP1-related role in end protection, Shieldin loss severely reduced class switch recombination at immunoglobulin loci and prevented fusion of deprotected telomeres3–7. Interestingly, BRCA1 and Shieldin double mutant cells are more sensitive to ionizing radiation and cisplatin, pointing to alternative treatment strategies in BRCA1 mutant cells that have become resistant to PARP inhibition3,4.
Several features of Shieldin address gaps in our understanding of 53BP1-mediated end protection. Notably, Shieldin components have been shown to biochemically link Rif1 to Rev7 (refs3–8). In addition, Shld2 binds single-stranded DNA in vitro, and mutations abrogating this biochemical activity disrupted end protection3,4,6. Thus, Shieldin provides a biochemical bridge between 53BP1–Rif1 and Rev7, while also connecting chromatin to single-stranded DNA at break sites. However, Shieldin itself does not contain enzymatic activity that could oppose resection. This led Mirman et al. to explore commonalities between control of single-stranded DNA end length at telomeres and resection at DSBs (ref.5). The CST complex (Ctc1, Stn1, Ten1) directs DNA polymerase-α (Polα)–primase for fill-in synthesis of the 3′ telomere overhang, which is necessary for replication-associated telomere maintenance10. Interestingly, CST localized to damaged telomeres or nontelomeric breaks in a 53BP1–Rif1–Shieldin-dependent manner5.
This data begs the question, does the resection blocking activity of CST in DSB repair share a common mechanism with its actions during telomere maintenance? Circumstantial evidence suggests that it might. Polα was recruited to site-specific breaks in a 53BP1-, Shieldin- and CST-dependent manner. Remarkably, Polα inhibition also prevented radial chromosome formation in BRCA1 mutant cells exposed to PARP inhibitors. These findings suggest that interaction between CST and Shieldin occurs at damage sites to direct Polα -driven DNA synthesis in opposition of nuclease activity5 (Fig. 1). However, quantitative detection of CST–Polα -dependent DNA synthesis at DSBs will be necessary for formal proof of this model and to understand the extent to which fill-in synthesis limits resection.
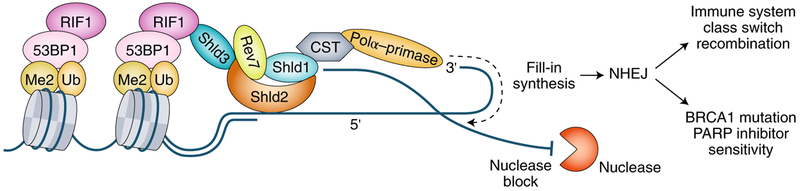
Fig. 1 |. Assembly of the Shieldin complex at DNA DSBs limits generation of single stranded overhangs to promote NHEJ.
The drawing represents a composite model derived from references3–9, although further studies will be necessary to fully understand the architecture of the Shieldin complex. The Shieldin complex links 53BP1–Rif1 recognition of nucleosomes flanking DSBs to single stranded DNA at the break site. Shieldin may limit end resection by preventing the action of nucleases that resect DNA with a 5’ to 3′ polarity. Additionally, Shieldin is required for localization of the CST complex, which interacts with DNA Polα -primase and could counteract end resecting nucleases by fill-in DNA synthesis on the resected 3′ end of single stranded DNA at break sites. This 53BP1–Rif1–Shieldin–CST pathway is necessary to promote NHEJ during class switch recombination at immunoglobulin loci and following PARP inhibitor treatment of BRCA1 mutant cells. Ub, ubiquitin on lysine 15 of histone H2A; Me2, dimethylation on lysine 20 of histone H4.
Collectively, these remarkable series of studies represent the next chapter in defining the forces that regulate end resection at DSBs. As with any provocative finding, many questions remain that will surely attract the efforts of DNA repair researchers. Factors that promote end resection are relatively well conserved across eukaryotes. This raises the unresolved issue of what evolutionary forces drove development of a complex chromatin interaction network that limits resection. Shieldin emerged in ancient vertebrates, coinciding with the advent of class switch recombination at immunoglobulin genes6,7. This process requires long-range end joining between loci that experience programmed breaks and may necessitate additional measures to reduce end resection. Selective pressure could also come from competition between BRCA1 and 53BP1 for access to chromatin along DSBs (ref.11), which is an important cell cycle determinant of whether DNA repair ensues by HR or NHEJ.
Another question emerging from these studies is whether the sequence of events, from damage recognition by 53BP1 to fill-in synthesis by CST–Polα , represents a singular pathway. Human genetics may provide clues to the answer. 53BP1 recognizes histone H2A ubiquitination at lysine 15, the substrate of the DNA-damage-responsive E3 ubiquitin ligase, RNF168 (ref.2). Mutations in RNF168 are the causative genetic etiology of RIDDLE syndrome12, a congenital condition characterized by immune deficiency due to class switch recombination failure along with hypersensitivity to ionizing radiation. CST members, CTC1 and Stn1, are mutated in Coats Plus syndrome13, which has phenotypic similarities to other telomere maintenance syndromes while also displaying features indicative of additional functional abnormalities. Surprisingly, class switch recombination defects have not been described in patients with CST mutations. Furthermore, CST-loss was reported to cause chromosomal fragility and impaired responses to replication-associated DNA damage, something not present in 53BP1-deficient cells14. These incongruities raise the suspicion that additional complexities exist. Finally, the question of whether DNA-Polα -dependent synthesis is necessary to limit end resection may also avoid simplicity. Indeed, several other polymerases have also been implicated in NHEJ (ref.15), suggesting that multiple synthesis activities may cooperate in this repair process. Further possibilities arise as to whether Polα/primase could introduce ribonucleotides or extend an RNA primer to facilitate repair by NHEJ. While the ‘Shield’ has clearly been raised, additional assembly may be required.
Footnotes
Competing interests
The author declares no competing interests.
A previously unidentified protein complex termed Shieldin acts with the nucleosome-binding protein 53BP1 to limit end resection at DNA double-strand breaks, impacting myriad biological outcomes, from immunology to cancer therapy, and highlighting the importance of chromatin responses to DNA damage in vertebrates.
References
- 1.Symington LS Crit. Rev. Biochem. Mol. Biol 51, 195–212 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hustedt N & Durocher D Nat. Cell Biol 19, 1–9 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Dev H et al. Nat. Cell Biol 10.1038/s41556-018-0140-1 (2018). [DOI] [Google Scholar]
- 4.Noordermeer SM et al. Nature 10.1038/s41586-018-0340-7 (2018). [DOI] [Google Scholar]
- 5.Mirman Z et al. Nature 10.1038/s41586-018-0324-7 (2018). [DOI] [Google Scholar]
- 6.Ghezraoui H et al. Nature 10.1038/s41586-018-0362-1 (2018). [DOI] [Google Scholar]
- 7.Gupta R et al. Cell 173, 972–988 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomida J et al. EMBO J. 37, e99543 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barazas M et al. Cell Rep. 23, 2107–2118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang F et al. Cell Rep. 2, 1096–1103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang J et al. Nat. Struct. Mol. Biol 20, 317–325 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart GS et al. Cell 136, 420–434 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Anderson BH et al. Nat. Genet 44, 338–342 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Chastain M et al. Cell Rep. 16, 2048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pryor JM et al. Proc. Natl Acad. Sci. USA 112, 4537–4545 (2015). [Google Scholar]