Abstract
Objective:
This meta-analysis is to investigate the relationship between the patatin-like phospholipase domain containing 3 (PNPLA3) rs738409 polymorphism and the susceptibility and severity of nonalcoholic fatty liver disease (NAFLD).
Methods:
Chinese Journal Full-text Database, Wanfang Database, VIP Database, and PubMed Database were subjected to case-control study retrieving, from January 2008 to December 2014. Following key words were used: fatty liver, PNPLA3, and rs738409 gene or variants or polymorphism or alleles. Meta-analysis was performed based on the retrieved articles.
Results:
In total 65 studies were first retrieved according to the key words, and finally 21 studies with 14,266 subjects were included. Meta-analysis showed that PNPLA3 rs738409 polymorphism exerted strong influence not only on fatty liver but also on the histological injury. PNPLA3 rs738409 [G] allele was a risk factor for NAFLD (GG vs CC, OR = 4.01, 95% CI 2.93–5.49; GC vs CC, OR = 1.88, 95% CI 1.58–2.24). PNPLA3 gene variant was significantly associated with the increased serum alanine aminotransferase (ALT) levels (GG vs CC, standardized mean difference = 0.47, 95% CI 0.14–0.81). In addition, nonalcoholic steatohepatitis (NASH) was more frequently observed in G allele carriers (GG vs CC, OR = 3.24, 95% CI 2.79–3.76; GC vs CC, OR = 2.14, 95% CI 1.43–3.19).
Conclusion:
PNPLA3 rs738409 polymorphism is not only a factor significantly associated with the susceptibility of NAFLD, but also related to the susceptibility of aggressive diseases.
Keywords: meta-analysis, nonalcoholic fatty liver disease (NAFLD), patatin-like phospholipase domain containing 3 (PNPLA3) polymorphism, susceptibility
1. Introduction
Patatin-like phospholipase domain containing 3 (PNPLA3) is located on chromosome 22, which belongs to the family of patatin-like phospholipases (also known as adiponutrin). It has been shown that the expression of PNPLA3 in human beings would be affected by series of factors, including diet, obesity, insulin and glucose levels, and gene mutation.[1] Moreover, a number of studies have shown that the PNPLA3 I148 M mutation on the rs738409 site is related to the genetic susceptibility, liver damage, liver fibrosis, and disease development in liver diseases, such as nonalcoholic fatty liver disease (NAFLD), alcoholic liver disease, hepatitis C virus (HCV) infection, and liver cancers.[2,3]
Studies have demonstrated that the PNPLA3 I148 M rs738409 gene polymorphism is closely associated with the elevated liver fat content in NAFLD, making the relationship between the PNPLD3 polymorphism and NAFLD pathogenesis a research hot spot. In 2008, Romeo et al[4] have first reported the association between the PNPLA3 [rs738409 C/G] gene polymorphism and NAFLD. Since then, several studies have been focusing on the relationship between gene mutation and NAFLD. However, due to the differences in the population samples, detection methods, and diagnostic standards, as well as the limitations such as subject ages and sample sizes, no consensus has been achieved in this research field. In this study, to investigate the effects of PNPLA3 rs738409 polymorphism on the genetic susceptibility and histological damages in NAFLD patients, related references were analyzed with systematic review and meta-analysis.
2. Materials and methods
2.1. Inclusion criteria
The inclusion criteria were as follows: study type: the casa-control studies based on populations or hospitals; study subjects: studies on association between the PNPLA3 rs738409 polymorphism and NAFLD; studies concerning the gene detection of the single nucleotide polymorphisms at the PNPLA3 rs738409 locus; and observation indexes: genotype distribution, clinical data of NAFLD and control subjects, and liver damage index. This is a meta-analysis and thus ethical approval is not necessary.
2.2. Exclusion criteria
The exclusion criteria were as follows: repeated publication of the same population data, or articles with unclearly described original data; review articles; and secondary causes of steatosis, including alcoholism, total parenteral nutrition, hepatitis B (HBV), and HCV infection, and drug usage.
2.3. Study retrieving and key words
Chinese Journal Full-text Database, Wanfang Database, VIP Database, and PubMed Database were subjected to study retrieving, from January 2008 to December 2014. The following key words were used for searching: fatty liver, PNPLA3, and rs738409 gene or variants or polymorphism or alleles.
2.4. Literature extraction
Following basic information was extracted from included studies: the study title, first author, publication year, study type, genetic detection method, study population, age, sex, and sample size. The retrieving and screening process was shown in Fig. 1.
Figure 1.
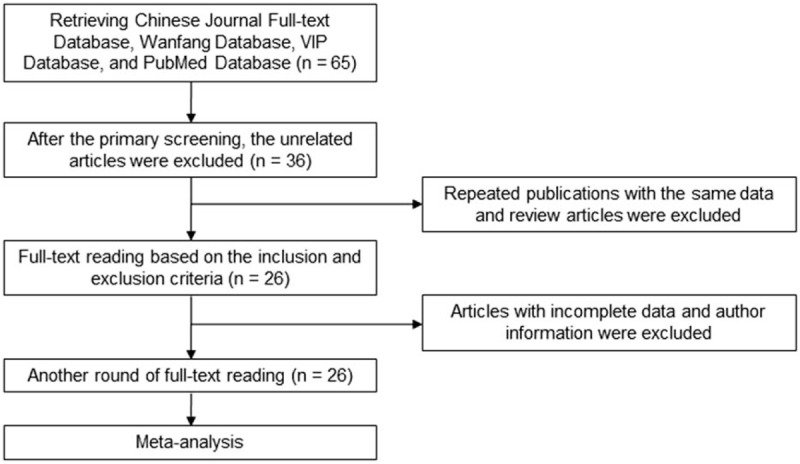
Article retrieving and screening process.
2.5. Statistical analysis
NAFLD diagnostic criteria included the application of proton magnetic resonance spectroscopy, ultrasonic detection of liver fat infiltration, and liver biopsy detection method. The genotype frequencies and the risk odds (OR and 95% CI) between different genetic models in NAFLD and control groups were recorded. The frequencies of NASH or liver fibrosis for each genotype were recorded. The biochemical indexes [alanine aminotransferase (ALT), AST, etc.] of each subject from included studies were standardized and unified. Combined statistics (OR values) were obtained for dichotomic variables. The OR and 95% CI values of genotypes between the NAFLD and control groups, and the OR and 95% CI values of genotypes between the NASH and simple steatosis groups, were analyzed and compared. All continuous variables (such as serum ALT levels and liver fibrosis scores) were expressed as mean ± SD, and the standardized mean difference (SMD) was used as the combined effect value.
Stata 12.0 software was used for statistical analysis. Heterogeneity analysis was performed using the I2 statistics: P>.05 or I2< 50% suggested satisfactory homogeneity, which was suitable for the fixed-effect model, while I2> 50% suggested greater heterogeneity, and the random-effect model was applied. OR or SMD values were calculated, and the forest plots were obtained accordingly.
For the large heterogeneity, subgroup analysis was performed, based on the subject races, ages, or other related factors. If the heterogeneity persisted, sensitivity analysis was conducted, and the combined effect values and the 95% CI values were recalculated after excluding certain studies. In the sensitivity analysis, the research models (the fixed- and random-effect models) were altered. The statistics were recombined after removing the article with the largest weight, and the result stability was evaluated. Funnel plots were obtained with the OR or SMD values as the horizontal coordinates, and the standard error of OR logarithm or standard error of SMD as the vertical coordinates. Publication bias was quantitatively evaluated with the Egger regression and Begg rank correlation.
3. Results
3.1. Basic information of retrieved articles
In total 65 related articles (in which 10 articles were published in Chinese) were first retrieved based on the searching key words. After the primary screening, the unrelated articles were excluded, and 36 related articles were selected. According to the inclusion and exclusion criteria, after the full-text reading, the repeated publications with the same data and review articles were excluded, leaving 26 articles. After another round of full-text reading, based on the data extraction requirements, articles with incomplete data and author information were excluded, and in total 21 articles were finally included in this study and subjected to the meta-analysis.[5–24] Basic information of the 21 included articles was shown in Table 1.
Table 1.
Basic characteristics of the included articles.
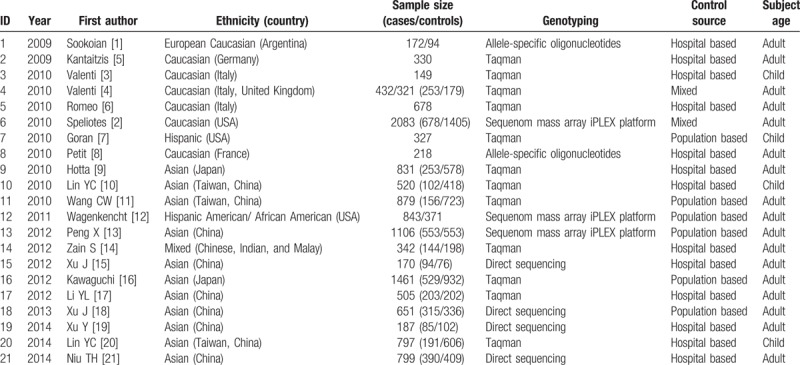
Out of these 21 articles, there were 2 published in Chinese, and 19 in English. Moreover, there were 6 case-control articles based on populations, 13 case-control articles based on hospitals, and 2 articles with mixed background. Furthermore, out of these 21 articles, 7 articles focused on the Caucasian population, 11 articles studied the Asian population, and the rest articles concerned the Hispanic, Finnish, and other races. The gene detection methods included the first generation of direct sequencing technique, Taqman probe technique, allele-specific probe technique, and single nucleotide polymorphisms genotyping detection based on time-of-flight mass spectrometry. In addition, in these articles, 20 articles reported the relationship between the NPNLA3 gene mutation and NAFLD susceptibility, and 6 articles regarded the association between the incidence of NASH and the genotypes of NAFLD patients, and 7 articles concerned the relationship between the serum ALT level and PNPLA3 gene polymorphism.
3.2. Meta-analysis on association between PNPLA3 rs738409 gene polymorphism and NAFLD
Our results showed that the PNPLA3 gene polymorphism (rs738409 [G]), that is, the 3 genotypes of CC (homozygous wild-type), GG (homozygous mutant), and CG (heterozygous mutant), were found in both the NAFLD and control groups, even though with different genotype distribution frequencies. In this meta-analysis, homozygous wild-type CC genotype was used as reference, which was compared with homozygous mutant GG and heterozygous mutant CG genotypes, respectively, in these 21 articles (involving 14,266 subjects).
Forest plots concerning the relationship between the PNPLA3 rs738409 gene mutation and NAFLD susceptibility were shown in Figs. 2 and 3. Our results indicated large heterogeneity of these 21 included articles (I2> 50%), and therefore random-effect model was applied. The combined OR value for GG versus CC was 4.01 (95% CI: 2.93–5.49), with the Z value of 8.69 (P < .05) for the statistically significance analysis. The combined OR value for GC versus CC was 1.88 (95% CI: 1.58–2.24), with the Z value of 7.01 (P < .05). Taking into account the effect of different test methods on heterogeneity, 4 articles applying the first-generation of gene detection (direct sequencing technique) were excluded,[19,21,24] and the other articles were again subjected to the meta-analysis, obtaining the combined OR values. The forest plot showed that significantly declined heterogeneity (I2 < 50%). The combined OR value for GG versus CC was 2.76 (95% CI: 2.41–3.16), with the Z value of 14.79 (P < .05). The combined OR value for GC versus CC was 1.56 (95% CI: 1.40–1.74), with the Z value of 8.08 (P < .05) (Table 2). Compared with the former combined OR values before excluding the 4 articles, the P values were less than .05. Our results showed that, compared with the CC genotype carriers, the mutant allele [G] carriers were more common in the NAFLD group. These results suggest the close association between the PNPLA3 rs738409 gene mutation and NAFLD susceptibility.
Figure 2.
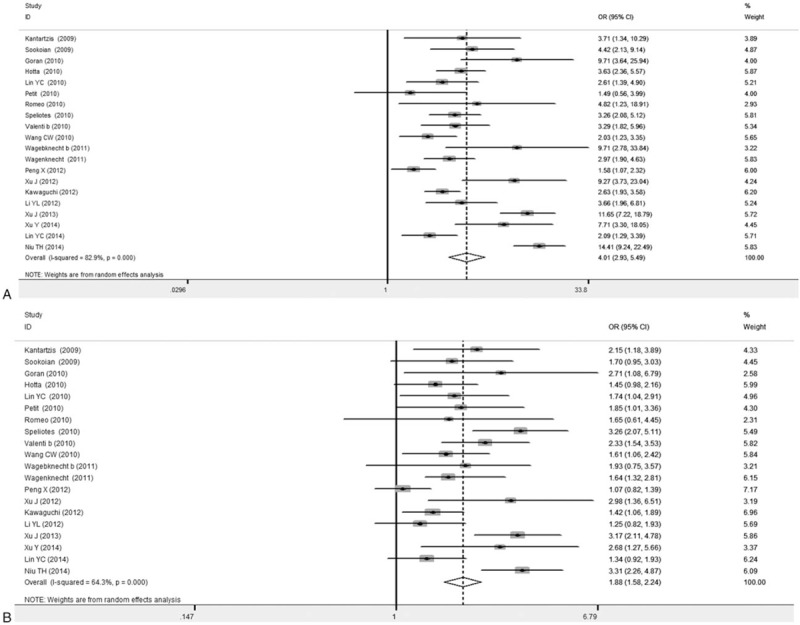
Forest plots for association between PNPLA3 rs738409 polymorphism and NAFLD susceptibility (20 articles). A, GG versus CC. B, GC versus CC. Wagebknecht b represented African-American population, and Valenti b represented adults.
Figure 3.
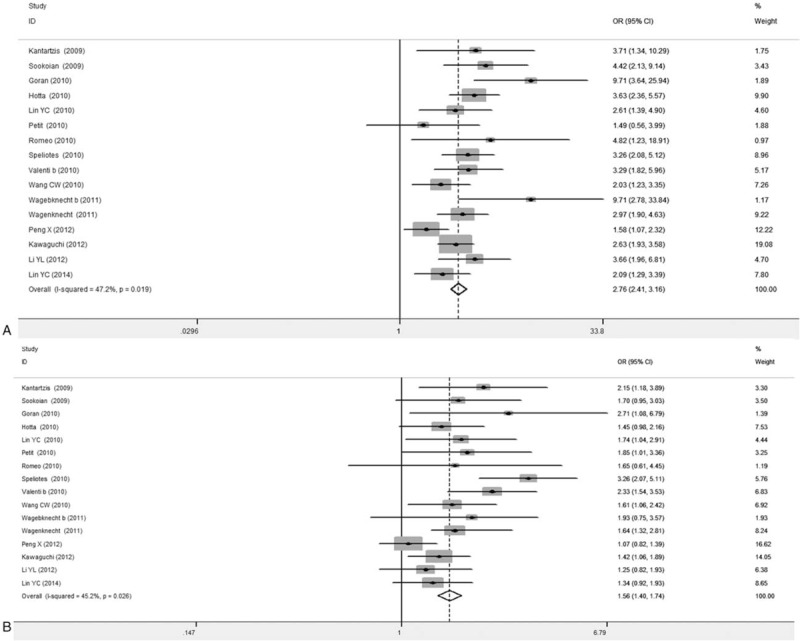
Forest plots for association between PNPLA3 rs738409 polymorphism and NAFLD susceptibility (16 articles after exclusion). A, GG versus CC. B, GC versus CC. Wagebknecht b represented African-American population, and Valenti b represented adults.
Table 2.
Association between the rs738409 gene mutation and NAFLD susceptibility before and after article exclusion.

3.3. Subgroup analyses according to subject ages and races
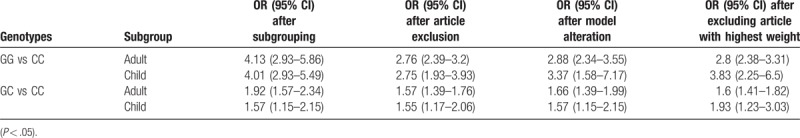
Subgroup analysis based on subject ages was first performed. Our results showed that, after excluding the 4 articles applying the first-generation of gene detection (direct sequencing technique),[19,21,24] in the remaining 16 articles, 13 articles regarded adult subjects, and the combined OR values for GG versus CC and GC versus CC were 2.76 (95% CI: 2.39–3.2) and 1.57 (95% CI: 1.39–1.76), respectively, both P < .001. In the other 3 articles concerning child subjects, the combined OR values for GG versus CC and GC versus CC were 2.75 (95% CI: 1.93–3.93) and 1.55 (95% CI: 1.17–2.06), respectively, both P < .001 (Fig. 4) (Table 3).
Figure 4.
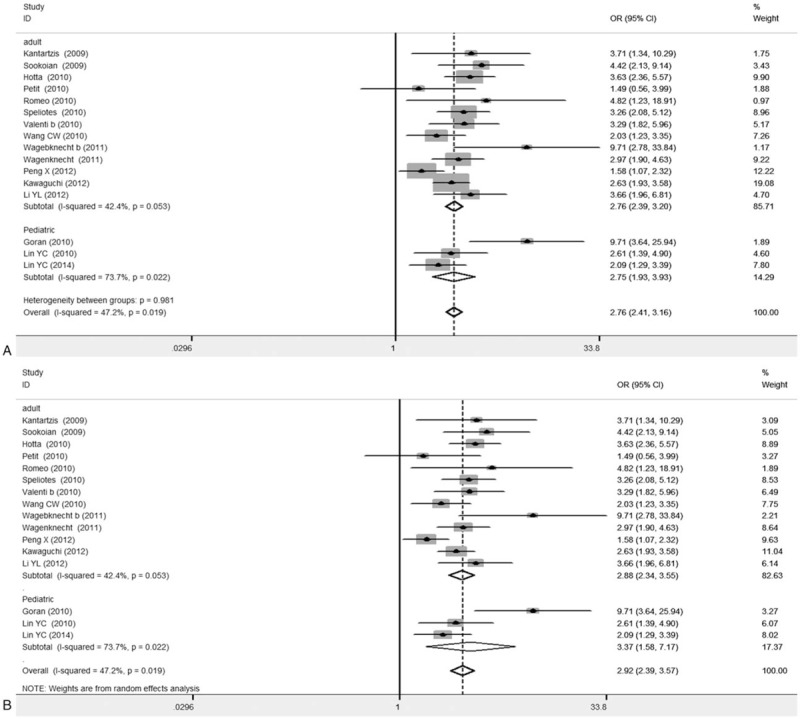
Forest plots for association between PNPLA3 rs738409 polymorphism and NAFLD susceptibility based on age sub-group analysis (16 articles). A, GG versus CC. B, GC versus CC.
Table 3.
Meta-analysis ongene polymorphism and NAFLD susceptibility based on age subgrouping.
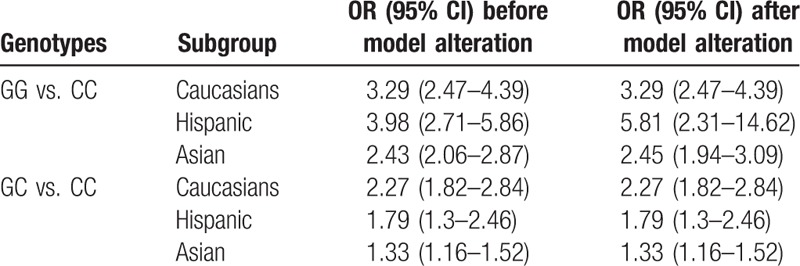
Our results from the subgroup analysis based on subject races were shown in Fig. 5. In the 16 analyzed articles, 6 articles focused on Asian populations, 7 articles regarded Caucasian people, and the remaining 3 articles concerned Hispanic population. Our results showed that, for the Asian population, the combined OR values for GG versus CC and GC versus CC were 2.43 (95% CI: 2.06–2.87) and 1.33 (95% CI: 1.16–1.52), respectively, with statistical significance. For the Caucasian people, the combined OR values for GG versus CC and GC versus CC were 3.29 (95% CI: 2.47–4.39) and 2.27 (95% CI: 1.82–2.84), respectively, both P < .05 (Table 4). These findings suggest the stability of results after model alteration.
Figure 5.
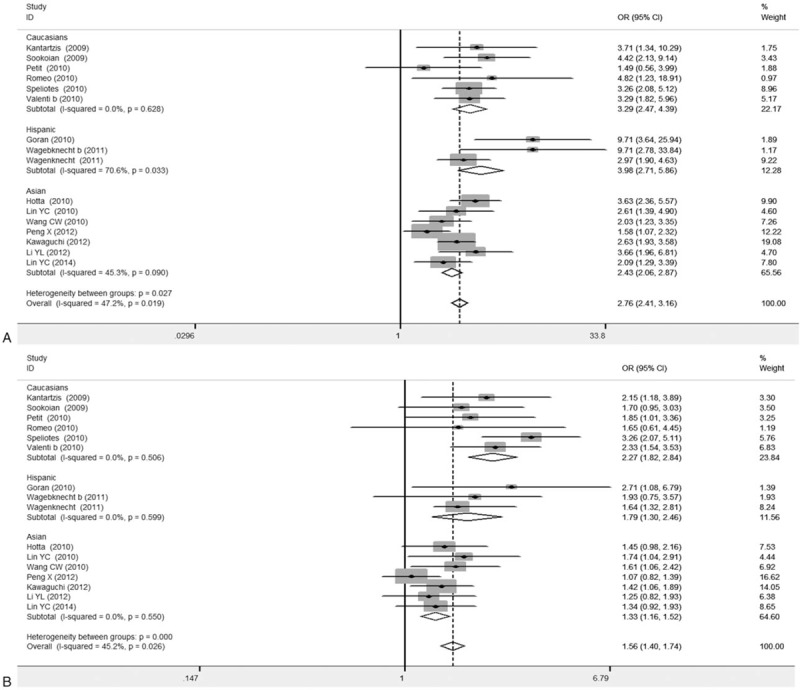
Forest plots for association between PNPLA3 rs738409 polymorphism and NAFLD susceptibility based on race subgroup analysis (16 articles). A, GG versus CC. B, GC versus CC.
Table 4.
Meta-analysis on gene polymorphism and NAFLD susceptibility based on race subgrouping.
3.4. Associations between the gene polymorphism and serum ALT level, as well as NASH incidence
The results from the association analysis between the gene polymorphism and serum ALT level were shown in Fig. 6. In the articles included in the meta-analysis, 7 articles provided data on the serum ALT level in NAFLD patients with different genotypes. Combined SMD value was used as the effect value. Corresponding effect models were applied according to the heterogeneity analysis from the forest plots. Our results showed that, huge heterogeneity was reported for GG versus CC, and therefore the random-effect model was applied, with the combined effect SMD of 0.47 (95% CI: 0.14–0.81), Z value of 2.75, and P = .006 (<.05), suggesting statistical significance. For GC versus CC, the heterogeneity test suggested P < .05, and therefore the fixed-effect model was used, with the combined SMD of 0.14 (95% CI: −0.2 to 0.29), Z value of 1.76, and P = .079 (>.05). The same results were obtained after altering the analysis models (Table 5).
Figure 6.
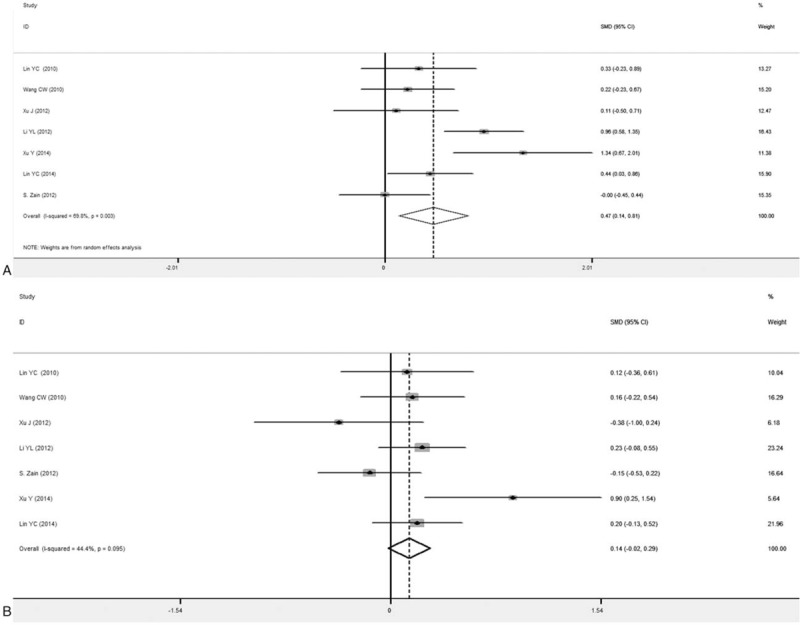
Forest plots for association between PNPLA3 rs738409 polymorphism and ALT level in NAFLD patients (7 articles). A, GG versus CC. B, GC versus CC.
Table 5.
Meta-analysis on serum ALT level and PNPLA3 re738409 gene polymorphism.

Results from the association analysis between the gene polymorphism and NASH incidence were shown in Fig. 7. In total 6 articles reported on the relationship between the risky allele G and NASH. Our results showed that, compared with CC carriers, NASH was more commonly seen in the NAFLD patients carrying GG genotype. The combined OR value was 3.24 (95% CI: 2.79–3.76) (P < .001), and consistent results were obtained for the analysis after model altering. For the comparison between the carriers of GC and CC genotypes, the combined OR value from the random-effect model was 2.14 (95% CI: 1.43–3.19), and the combined OR from the fixed-effect model was 2.71 (95% CI: 2.35–3.12) (P = .0000), with statistical significance.
Figure 7.
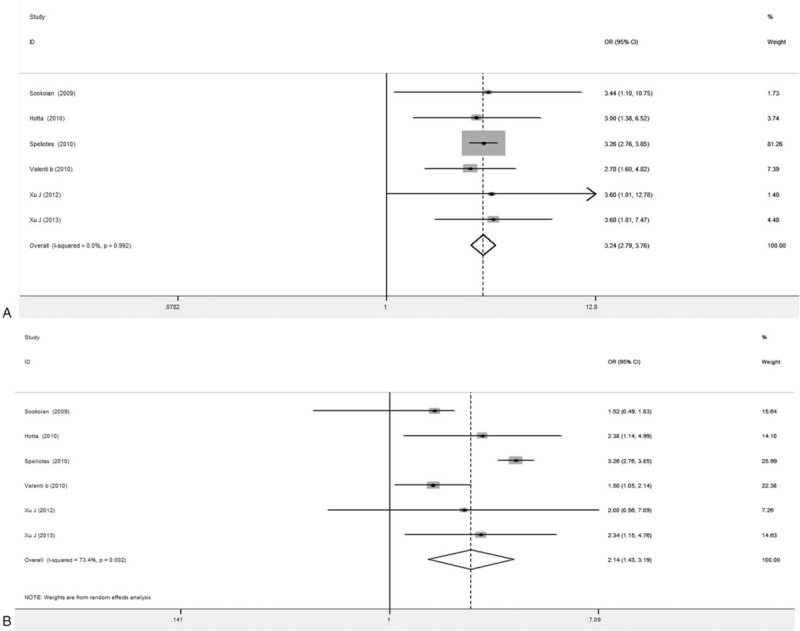
Forest plots for association between PNPLA3 rs738409 polymorphism and NASH (6 articles). A, GG versus CC. B, GC versus CC.
3.5. Publication bias analyses of included articles
Publication bias analysis of these included articles was performed. The funnel plot of the association between the gene polymorphism and NAFLD susceptibility was shown in Fig. 8. In total 20 articles concerned the comparison of the GG and CC genotypes between the NAFLD and control subjects. Meta-analysis showed that GG carriers were more prone to suffer from NAFLD. The funnel plot showed symmetrical distribution pattern, and the Egger regression and Begg rank correlation analysis indicated P = .247 (>.05) and P = .098 (>.05), respectively, with no statistical significance, suggesting no publication bias. For the comparison of the GC versus CC model, the funnel plot showed symmetrical distribution pattern. The Egger regression and Begg rank correlation analysis based on quantitative measurement indicated P = .051 (>.05) and P = .086 (>.05), respectively, with no statistical significance, suggesting small publication bias and stable results.
Figure 8.
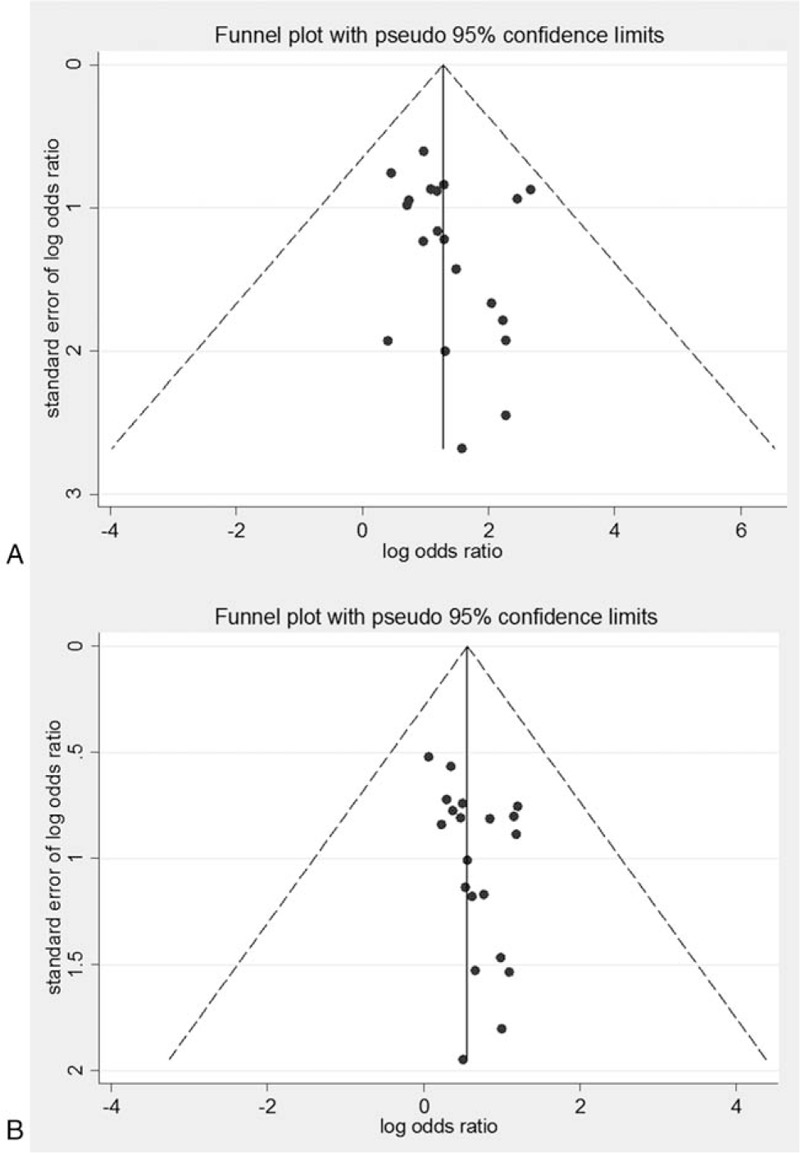
Funnel plots for association between PNPLA3 rs738409 polymorphism and NAFLD susceptibility (20 articles). A, GG versus CC. B, GC versus CC.
The funnel plot of the association between the gene polymorphism and serum ALT level was shown in Fig. 9. In total 7 articles concerned the relationship between the GG/CC genotypes and serum ALT level. Asymmetry distribution pattern was showed in the funnel plot, and the Egger regression and Begg rank correlation analysis based on quantitative measurement indicated P = .764 (>.05) and P = .951 (>.05), respectively, with no statistical significance. For the association between the GC versus CC genotypes and the serum ALT levels, the funnel plot showed symmetrical distribution pattern. The Egger regression and Begg rank correlation analysis based on quantitative measurement indicated P = .548 (>.05) and P = .917 (>.05), respectively, with no statistical significance.
Figure 9.
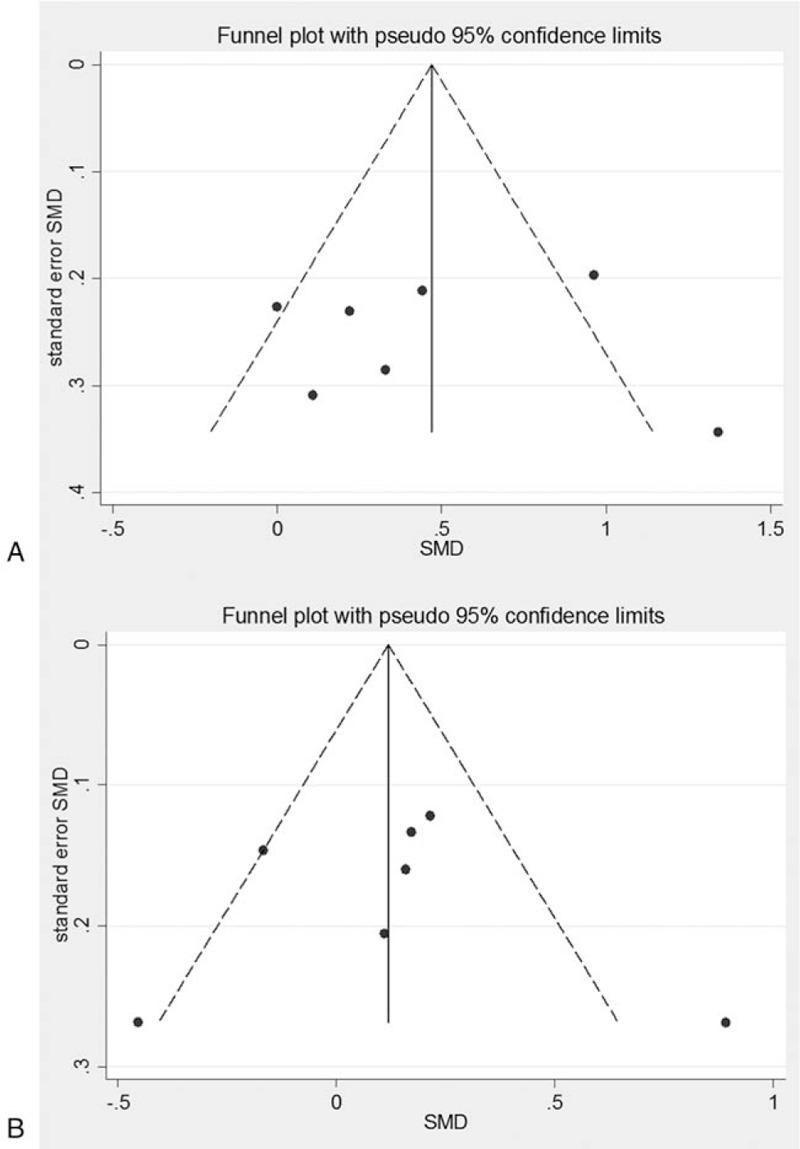
Funnel plots for association between PNPLA3 rs738409 polymorphism and ALT level in NAFLD patients (7 articles). A, GG versus CC. B, GC versus CC.
The funnel plot of the association between the gene polymorphism and NASH incidence was shown in Fig. 10. For the GG versus CC model, our results showed that, GG genotype carriers were more prone to suffer from NASH. The funnel plot showed basically symmetry distribution pattern, and the Egger regression and Begg rank correlation analysis based on quantitative measurement indicated P = 1.0 (>.05) and P = .188 (>.05), respectively, with no statistical significance. For the association between the GC versus CC genotypes and NASH incidence, the funnel plot showed obviously asymmetry distribution, suggesting publication bias. The Egger regression and Begg rank correlation analysis based on quantitative measurement indicated P = 1.0 (>.05) and P = .963 (>.05), respectively, with no statistical significance.
Figure 10.
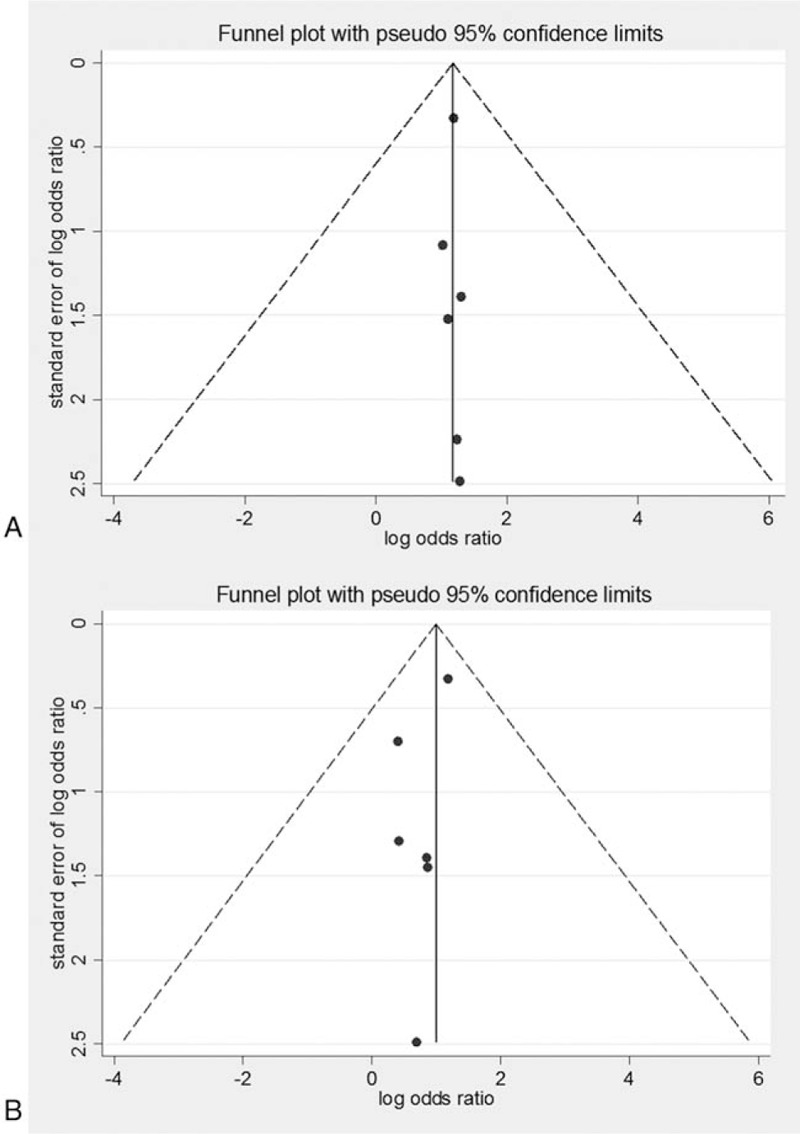
Funnel plots for association between PNPLA3 rs738409 polymorphism and NASH (6 articles). A, GG versus CC. B, GC versus CC.
4. Discussion
In this study, meta-analysis was performed on the published articles concerning the relationship between the PNPLA3 rs738409 gene polymorphism and NAFLD, to further investigate the association between gene polymorphism and disease pathogenesis, and its effects on the genetic susceptibility of NAFLD. The findings might help to understand the disease pathogenesis, and contribute to the screening of populations and individuals with high risk of NAFLD. Our results from the meta-analysis indicated the different distribution patterns of rs738409 genotype between the NAFLD and control groups. The GG and GC genotypes (carrying the mutant alleles) were more common in the NAFLD group. Using the CC genotype as reference, the GG and GC genotype carriers were compared in the NAFLD and control groups. For the GG versus CC, the combined OR value was 4.01 (95% CI: 2.93–5.49), with the Z value of 8.69 (P < .05). Due to the huge heterogeneity in the forest plot, I2 was more than 50% (P < .001). Considering the different gene detection methods applied, 4 articles using the direct sequencing technique were excluded, and meta-analysis was performed again. The results indicated significantly reduced heterogeneity, and the combined OR value was 2.76 (95% CI: 2.41–3.16) (P < .001), with statistically significant differences. For the GC versus CC model, the combined OR value was 1.88 (95% CI: 1.58–2.24) (P < .001), with statistical significance. Similarly, the heterogeneity was obviously declined after excluding the very 4 articles, and the newly calculated combined OR value was 1.56 (95% CI: 1.40–1.74) (P < .05), with statistical significance. The statistical significance before and after excluding the 4 articles with relatively poor qualities demonstrated the result stability. According to the results of this meta-analysis, the frequency of the risk gene [G] was relatively high in the NAFLD group, and the PNPLA3 rs738409 allele G polymorphism was closely linked with the susceptibility of NAFLD. This finding was in line the former reports showing that the risk gene [G] carriers are more prone to suffer from NAFLD.[19,25–30]
Numerous studies on the PNPLA3 rs738409 polymorphism have shown that, the gene polymorphism is closely related to the occurrence and development of NAFLD.[3] It has been demonstrated that PNPLA3 is an important regulating factor for triglyceride,[21] which could act as both phospholipase and acyl transfer enzyme. However, the physiological mechanism has not been fully elucidated. Although the effects of PNPLA3 on liver physiology are still unclear, it is clear that the rs738409 gene polymorphism affects not only the triglyceride deposition in liver, but also the disease progression in patients with fatty liver. At present, researches on the relationship between the PNPLA3 gene mutation and NAFLD pathogenesis have mainly concerned the effects of gene mutation on the lipid decomposition in the liver. A previous study has shown that, the triglyceride content in the liver is increased, in both the PNPLA3 mutant mice and the in vitro PNPLA3 mutant liver cells.[31] Therefore, some scholars propose that the gene mutation leads to the change from isoleucine to methionine in the expressed protein, which limits the binding of the substrate to the catalytic site and the activity of triglyceride hydrolase. In this study, our results showed that, the mutant gene G carriers were more prone to suffer from NAFLD, indicating that the gene mutation might influence the triglyceride hydrolysis in liver cells, leading to the accumulation of triglycerides in the liver and triggering the occurrence of fatty liver.
In this meta-analysis, 7 articles reported on the relationship between the elevated ALT levels and the PNPLA3 rs738409 gene polymorphism. The serum ALT levels were analyzed and compared between the GG and CC genotype carriers, as well as between the GC and CC genotype carriers, and the combined effect SMD value was calculated. Our results showed that, the homozygous GG mutant genotype was associated with the elevated serum ALT levels. The combined SMD for GG versus CC was 0.47 (95% CI: 0.14–0.81), with the Z value of 2.75 and P = .006 <.05, suggesting statistical significance. Consistent results before and after model alteration proved satisfactory stability. For GC versus CC, the combined effect values indicated P > .05, and the unchanged results after model alteration suggested that there was no difference in the ALT level between the NAFLD patients carrying GC and CC genotypes. These results were consistent with the previous findings that the PNPLA3 rs738409 [G] mutation is linked with the elevated serum ALT level,[32,33] which might be caused by the liver inflammation or liver cell damages. Because ALT is an indicator of liver function, with high sensitivity and specificity, the elevated ALT levels in patients with fatty liver might be due to the mild liver damages. However, it is till unclear whether the PNPLA3 mutation leads to the fat accumulation in the liver and the consequent enhanced liver enzymes, or the PNPLA3 variation results in the ALT elevation via inducing liver cell damages and affecting liver function. Further in-depth studies are still needed to address the issues.
Related researches have reported that the rs738409 gene polymorphism not only affects the deposition of liver lipids, but also is closely related to the progress of NAFLD disease.[3] A previous study based on gene polymorphism detection has shown that PNPLA3 rs738409 [G] gene could increase the risk of NASH.[8] This meta-analysis included 6 articles concerning the relationship between the NASH and PNPLA3 rs738409 gene polymorphism. Our results showed that, in the patients with NAFLD, NASH is more common in the GG carriers compared with CC carriers, for both fixed- and random-effects models (PR: 3.24; 95% CI: 2.79–3.76), without obvious publication bias (P > .05). For the comparison of GC and CC genotypes, the meta-analysis showed that, the NAFLD patients carrying GC genotype were more prone to suffer from NASH, with the combined effect OR of 2.14 (95% CI: 1.43–3.19). Consistent results before and after model alteration suggest the result stability.
In recent years, several studies have reported the association between the PNPLA3 rs738409 gene polymorphism and the NAFLD in children.[34] In this meta-analysis, parts of the involved subjects in the included studies were children. According to the subgroup meta-analysis based on ages, in both the child and adult groups, the mutant gene [G] was significantly related to the occurrence of NAFLD. For GG versus CC, the combined OR values in adult and child subjects were 2.76 (95% CI: 2.39–3.2) and 2.75 (95% CI: 1.93–3.93), respectively, both P < .001. For the GC versus CC, the combined OR values in adult and child subjects were 1.57 (95% CI: 1.39–1.76) and 1.55 (95% CI: 1.17–2.06), respectively, both P < .001. After altering the effect models and subsequently excluding the references with heavy weights, the same results were obtained, suggesting the satisfactory stability. These findings indicate that the effects of PNPLA3 gene polymorphism on NAFLD susceptibility could not be influenced by the age factor, which would start since the childhood. Due to the limited sample sizes of child subjects, further in-depth studies with large sample sizes are sill needed.
In a previous study on the association between genetic polymorphism and NAFLD, European population was involved and the genetic polymorphisms at 7 loci were investigated 66. The results have shown that, the PNPLA3 s738409 G allele would increase the incidence of NAFLD (OR: 3.26; 95% CI: 2.11–7.21). These findings suggest that, the association between the PNPLA3 rs738409 and NAFLD susceptibility did not differ between different races. In this meta-analysis, 4 articles with high heterogeneities were excluded, and the remaining 16 articles were subjected to the subgroup analysis based on races. Our results showed that, for the comparison between GG and CC genotypes, the combined OR and 95% CI values for the Caucasian, Asian, and Hispanic populations were 3.29 (2.47–4.39), 2.43 (2.06–2.87), and 3.98 (2.71–5.86), respectively, all P < .001. For GC versus CC, the OR and 95% CI values were 2.27 (1.82–2.84), 1.33 (1.16–1.52), and 1.79 (1.3–2.46), respectively, all P < .001, with statistical significance. The findings were not changed before and after model alteration. These results suggest that the effects of PNPLA3 re738409 gene polymorphism on NAFLD exist over different races.
This meta-analysis has some limitations. First, general observational meta-analyses often involve dichotomous variables, however, herein, the PNPLA3 re738409 gene polymorphism included 3 genotypes, that is, the homozygous wild-type CC, homozygous mutant GG, and heterozygote mutant GC genotypes. The meta-analysis software (including the stata software) is designed only for pair-wise comparison, and no direct comparison among these 3 genotypes could be performed. Therefore, in this study, the homozygous wild-type CC was used as reference, and the comparisons between the GG and CC genotypes, as well as between the GC and CC genotypes, were performed. The multiple comparisons would decline the credibility of statistical analysis and increase the Type I error. Second, because of the lack of a unified evaluation standard for literature quality, no quantitative assessment was conducted herein. All the articles were included based on subjective selection and evaluation, leading to attenuated objectivity and enhanced heterogeneity, which might affect the stability of the results of this meta-analysis. Moreover, since meta-analysis per se is an observational and retrospective study, bias with certain degree exists. Due to these limitations, further in-depth and systematical evaluation of the case-control studies with enlarged samples sizes and multiethnic populations with high qualities is still needed.
In summary, this meta-analysis provided evidence for the association between the PNPLA3 re738409 gene polymorphism and NAFLD pathogenesis. PNPLA3 rs738409 polymorphism is not only a factor significantly associated with the susceptibility of NAFLD, but also related to the susceptibility of aggressive diseases. These finding would contribute to the screening of high risky populations and susceptible individuals to NAFLD in clinic.
Author contributions
Data curation: Xiaomei Li.
Formal analysis: Pengfei Liu.
Investigation: Guangrong Dai.
Methodology: Shuixiang He.
Supervision: Xiaoyan Zhou.
Writing – original draft: Guangrong Dai.
Writing – review & editing: Shuixiang He.
Footnotes
Abbreviations: ALT = alanine aminotransferase, HCV = hepatitis C virus, NAFLD = non-alcoholic fatty liver disease, NASH = nonalcoholic steatohepatitis, PNPLA3 = patatin-like phospholipase domain containing 3.
This work was supported by the Project of Science and Technology of Social Development in Shaanxi Province grant (No. 2016SF-120).
The authors have no conflicts of interest to disclose.
References
- [1].Aragones G, Auguet T, Armengol S, et al. PNPLA3 expression is related to liver steatosis in morbidly obese women with non-alcoholic fatty liver disease. Int J Mol Sci 2016;17: E630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Tai CM, Huang CK, Tu HP, et al. Interactions of a PPARGC1A variant and a PNPLA3 variant affect nonalcoholic steatohepatitis in severely obese Taiwanese patients. Medicine (Baltimore) 2016;95:e3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Fan JH, Xiang MQ, Li QL, et al. PNPLA3 rs738409 polymorphism associated with hepatic steatosis and advanced fibrosis in patients with chronic hepatitis c virus: a meta-analysis. Gut Liver 2016;10:456–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sookoian S, Castano GO, Burgueno AL, et al. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J Lipid Res 2009;50:2111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Speliotes EK, Butler JL, Palmer CD, et al. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology 2010;52:904–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Valenti L, Alisi A, Galmozzi E, et al. I148 M patatin-like phospholipase domain-containing 3 gene variant and severity of pediatric nonalcoholic fatty liver disease. Hepatology 2010;52:1274–80. [DOI] [PubMed] [Google Scholar]
- [7].Valenti L, Al-Serri A, Daly AK, et al. Homozygosity for the patatin-like phospholipase-3/adiponutrin I148 M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology 2010;51:1209–17. [DOI] [PubMed] [Google Scholar]
- [8].Kantartzis K, Peter A, Machicao F, et al. Dissociation between fatty liver and insulin resistance in humans carrying a variant of the patatin-like phospholipase 3 gene. Diabetes 2009;58:2616–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Romeo S, Sentinelli F, Dash S, et al. Morbid obesity exposes the association between PNPLA3 I148 M (rs738409) and indices of hepatic injury in individuals of European descent. Int J Obes (Lond) 2010;34:190–4. [DOI] [PubMed] [Google Scholar]
- [10].Goran MI, Walker R, Le KA, et al. Effects of PNPLA3 on liver fat and metabolic profile in Hispanic children and adolescents. Diabetes 2010;59:3127–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Petit JM, Guiu B, Masson D, et al. Specifically PNPLA3-mediated accumulation of liver fat in obese patients with type 2 diabetes. J Clin Endocrinol Metab 2010;95:E430–6. [DOI] [PubMed] [Google Scholar]
- [12].Hotta K, Yoneda M, Hyogo H, et al. Association of the rs738409 polymorphism in PNPLA3 with liver damage and the development of nonalcoholic fatty liver disease. BMC Med Genet 2010;11:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lin YC, Chang PF, Hu FC, et al. A common variant in the PNPLA3 gene is a risk factor for non-alcoholic fatty liver disease in obese Taiwanese children. J Pediatr 2011;158:740–4. [DOI] [PubMed] [Google Scholar]
- [14].Wang CW, Lin HY, Shin SJ, et al. The PNPLA3 I148 M polymorphism is associated with insulin resistance and nonalcoholic fatty liver disease in a normoglycaemic population. Liver Int 2011;31:1326–31. [DOI] [PubMed] [Google Scholar]
- [15].Wagenknecht LE, Palmer ND, Bowden DW, et al. Association of PNPLA3 with non-alcoholic fatty liver disease in a minority cohort: the Insulin Resistance Atherosclerosis Family Study. Liver Int 2011;31:412–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Peng XE, Wu YL, Lin SW, et al. Genetic variants in PNPLA3 and risk of non-alcoholic fatty liver disease in a Han Chinese population. PLoS One 2012;7:e50256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zain SM, Mohamed R, Mahadeva S, et al. A multi-ethnic study of a PNPLA3 gene variant and its association with disease severity in non-alcoholic fatty liver disease. Hum Genet 2012;131:1145–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Xu J, Xin YN, Zhang DD. Research based on effects of PNPLA3 I148 M polymorphisms on adiponectin levels and genetic susceptibility of nonalcoholic fatty liver disease in Chinese Han population. Prog Mod Biomed 2012;12:2451–6. [Google Scholar]
- [19].Kawaguchi T, Sumida Y, Umemura A, et al. Genetic polymorphisms of the human PNPLA3 gene are strongly associated with severity of non-alcoholic fatty liver disease in Japanese. PLoS One 2012;7:e38322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li Y, Xing C, Cohen JC, et al. Genetic variant in PNPLA3 is associated with nonalcoholic fatty liver disease in China. Hepatology 2012;55:327–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Xu J, Xin YN, Lv WH. Relationship between PNPLA3 rs738409 polymorphism and genetic susceptibility to nonalcoholic fatty liver disease. Chin J Hepatol 2013;21:619–23. [DOI] [PubMed] [Google Scholar]
- [22].Lin YC, Chang PF, Chang MH, et al. Genetic variants in GCKR and PNPLA3 confer susceptibility to nonalcoholic fatty liver disease in obese individuals. Am J Clin Nutr 2014;99:869–74. [DOI] [PubMed] [Google Scholar]
- [23].Niu TH, Jiang M, Xin YN, et al. Lack of association between apolipoprotein C3 gene polymorphisms and risk of nonalcoholic fatty liver disease in a Chinese Han population. World J Gastroenterol 2014;20:3655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2008;40:1461–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pan Q, Zhang RN, Wang YQ, et al. Linked PNPLA3 polymorphisms confer susceptibility to nonalcoholic steatohepatitis and decreased viral load in chronic hepatitis B. World J Gastroenterol 2015;21:8605–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Marzuillo P, Miraglia del Giudice E, Santoro N. Pediatric fatty liver disease: role of ethnicity and genetics. World J Gastroenterol 2014;20:7347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Marzuillo P, Del Giudice EM, Santoro N. Pediatric non-alcoholic fatty liver disease: new insights and future directions. World J Hepatol 2014;6:217–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liu ZT, Chen TC, Lu XX, et al. PNPLA3 I148 M variant affects non-alcoholic fatty liver disease in liver transplant recipients. World J Gastroenterol 2015;21:10054–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Petta S, Grimaudo S, Camma C, et al. IL28B and PNPLA3 polymorphisms affect histological liver damage in patients with non-alcoholic fatty liver disease. J Hepatol 2012;56:1356–62. [DOI] [PubMed] [Google Scholar]
- [30].Gallego-Duran R, Romero-Gomez M. Epigenetic mechanisms in non-alcoholic fatty liver disease: an emerging field. World J Hepatol 2015;7:2497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Marzuillo P, Grandone A, Perrone L, et al. Understanding the pathophysiological mechanisms in the pediatric non-alcoholic fatty liver disease: the role of genetics. World J Hepatol 2015;7:1439–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].He S, McPhaul C, Li JZ, et al. A sequence variation (I148 M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem 2010;285:6706–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Marzuillo P, Grandone A, Conte M, et al. Novel association between a nonsynonymous variant (R270H) of the G-protein-coupled receptor 120 and liver injury in children and adolescents with obesity. J Pediatr Gastroenterol Nutr 2014;59:472–5. [DOI] [PubMed] [Google Scholar]
- [34].Clemente MG, Mandato C, Poeta M, et al. Pediatric non-alcoholic fatty liver disease: recent solutions, unresolved issues, and future research directions. World J Gastroenterol 2016;22:8078–93. [DOI] [PMC free article] [PubMed] [Google Scholar]


