Supplemental Digital Content is available in the text
Keywords: acute cellular rejection, alcoholic liver disease, chemokine, gene polymorphisms, liver transplantation
Abstract
While increased serum concentrations of CXCL9/10 are associated with acute cellular rejection (ACR) occurrence, the association between CXCL9/10 single nucleotide polymorphisms (SNPs) and ACR after liver transplantation (LT) remains unknown.
In the present case-control study, polymorphisms of CXCL9 (rs10336) and CXCL10 (rs3921) were determined by polymerase chain reaction in 215 liver transplant recipients. ACR was defined as biopsy proven within 6 months after LT. As selected SNPs were in 3’-UTR region, their possible association with protein synthesis was assessed by measuring the plasma concentration of CXCL9/10 in a cohort of 40 new transplant patients using ELISA.
There was no association between CXCL9/10 genotypes and overall incidence of ACR. However, patients with CXCL9 genotype AA developed ACR earlier than patients with GG genotype (P = .003), with similar results for CXCL10 gene (CC vs GG; P = .005). There was no statistically significant difference in plasma concentrations of CXCL9/10 between the rejectors and the non-rejectors. Of note, patients with AA CXCL9 genotype had significantly higher CXCL9 plasma concentrations than patients with AG (P = .01) or GG genotype (P = .045).
In conclusion, the SNPs of CXCL9 (rs10336) and CXCL10 (rs3921) are not associated with the incidence of ACR. However, patients with CXCL9 genotype AA developed ACR earlier and the same genotype was associated with greater plasma concentrations suggesting the involvement of CXCL9 mediated processes in ACR development.
1. Introduction
Despite the improvement and optimization of immunosuppressive protocols, acute cellular rejection (ACR) remains a common complication after liver transplantation (LT) that occurs in about 10% to 30% of transplant patients.[1,2] Contrary to previous data, a recently conducted study on a large number of recipients showed that ACR is a clinically significant event, which is associated with an increased risk of graft failure and death after LT.[1] Furthermore, clinical predisposing factors, such as younger recipient age, lack of renal impairment, higher AST levels before LT, longer cold ischemic times and older donors, do not entirely explain the risk for ACR occurrence.[2] Following the findings that immune mediators played a key role in ACR pathogenesis, it was hypothesized that single nucleotide polymorphisms (SNPs) of various cytokine and chemokine genes might be a predisposing factor for ACR occurrence or severity. Indeed, it was subsequently reported that certain SNPs of cytokine genes such as interleukin (IL) IL-4 T-33C polymorphism, IL-6 G-174C polymorphism, interferon gamma T + 874A polymorphism and transforming growth factor beta (TGFB) + 869 gene polymorphism are associated with altered susceptibility to ACR.[3–5]
Findings of recent studies suggest that cytokine CXCL9 might be a useful prognostic factor for ACR occurrence. Asaoka et al reported that intragraft expression levels of CXCL9 mRNA are increased during the ACR, while Raschzok et al found that CXCL9 serum concentrations were increased preoperatively and at the first postoperative day in patients who subsequently developed ACR.[6,7] An association between the serum levels of CXCL9 and CXCL10 and the development of early allograft dysfunction was also reported.[8] Furthermore, CXCL9 and CXCL10 polymorphisms were shown to be associated with diseases such as rheumatoid arthritis or Chagas cardiomyopathy, both conditions where T-cell response plays an important role.[9,10] However, the association between CXCL9 and CXCL10 polymorphisms and ACR after LT has not been investigated to the best of our knowledge. Therefore, the primary aim of our study was to assess possible associations between CXCL9 (rs10336) and CXCL10 (rs3921) polymorphisms and the incidence of ACR, while, as a secondary aim, we assessed associations between the polymorphisms, chemokine levels, and the time of ACR occurrence.
2. Patients and methods
2.1. Patients
This study included 215 adult liver transplant recipients (see Diagram 1) who underwent LT due to alcoholic liver disease (ALD) between January 2009 and June 2017 at the Merkur University Hospital, Zagreb, Croatia. All subjects were of Caucasian ethnic origin and had no other or concomitant liver etiologies (viral, autoimmune, metabolic, cryptogenic, etc.). Recipients with multi-organ transplantations or previous solid-organ transplantations were excluded. The study was performed according to the principles of the Declaration of Helsinki. The Hospital Ethics Committee approved the protocol in 2016. Patients that underwent transplantation before the start of the study were recruited during their regular checkups, while 40 new transplant patients were recruited at 3rd to 4th post-transplantation week (median 23rd (20–28) post-transplantation day). After obtaining informed, written consent to participate in the study, venous blood samples were taken and stored at −20°C until DNA isolation and gene analysis. Additionally, plasma was obtained by centrifugation from samples collected from new transplant patients and stored at −20°C until analysis. Data on age, sex, MELD score, AST, ALT, immunosuppressive therapy, date of transplantation, and date of acute rejection, where applicable, were collected from patients’ records. All liver transplant recipients received the same immunosuppressive protocol according to standard clinical practice at our institution. Induction immunosuppressive therapy consisted of steroids, which were gradually tapered in the maintenance protocol during the first 3 months and then discontinued. A combination of calcineurin inhibitors (cyclosporine or tacrolimus) and mycophenolate mofetil (3 g daily) was used from the start and/or modified in case of side effects. The dose of calcineurin inhibitor was adjusted according to the blood drug levels.
2.2. Acute cellular rejection
For the purpose of analysis, subjects were classified into 2 groups (ACR and non-ACR) according to biopsy-proven acute rejection (BPAR) episodes. Episodes of ACR were defined by typical histopathological features of Banff score ≥3 within the first 6 months after LT.[11] Liver biopsies were performed as clinically indicated based on an increase of liver enzymes and interpreted by an independent transplant pathologist. All patients responded well to an anti-rejection high-dose corticosteroid therapy and/or increase in calcineurin inhibitors.
2.3. Genotyping
DNA was extracted from the whole blood samples (200 ul) using QIAGEN QIAamp DNA Blood Mini Kit spin procedure according to manufacturer's instructions. DNA concentration and quality were determined using NanoDrop ND1000 Spectrophotometer. All samples were stored at −20°C until batch analysis. Genotypes were determined by polymerase chain reaction (PCR) analysis using commercially available TaqMan SNP assays for CXCL9 (rs10336) and CXCL10 (rs3921) with ABI 7500 instrument (Applied Biosystems).
CXCL9 and CXCL10 SNPs were selected based on the data of Hapmap (available at manufacturer's website: https://www.thermofisher.com/order/genome-database/details/genotyping/C____486222_10?CID=&ICID=&subtype, https://www.thermofisher.com/order/genome-database/details/genotyping/C____497062_10?CID=&ICID=&subtype= and NCBI (https://www.ncbi.nlm.nih.gov/projects/SNP) with minor allele frequency greater than 20% in European population.
2.4. Enzyme linked immunosorbent assay (ELISA)
Concentrations of CXCL9 and CXCL10 in plasma were determined by ELISA using commercially available kits (DCX900 cat number for CXCL9, DIP100 cat number for CXCL10, R&D Systems) in accordance with manufacturer's instructions.
2.5. Statistical analysis
Continuous variables are presented as median with interquartile range (IQR) or mean ± standard error (SEM) and compared by using Mann–Whitney/Kruskal–Wallis test or Student t test/ANOVA, as appropriate. Post-hoc multiple comparison corrections were applied to control the family-wise error rate. Categorical data were compared using Chi-Squared test, while Kendall tau (τ) test was used for correlation assessment.
Genotype frequencies of all polymorphisms were tested for Hardy–Weinberg equilibrium. For genotype analysis we tested dominant, recessive, overdominant, and codominant model. Linear regression models were built with recipient and donor age, MELD score, sex, and CXCL9 or CXCL10 genotypes as independent predictors to estimate their association with the time of ACR. For all calculations, P value < .05 was defined as statistically significant. Statistical analyses were performed using free online software SNPStats (http://bioinfo.iconcologia.net/snpstats) and R (a language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria) with figures plotted in GraphPad Prism version 6 for Windows (GraphPad Software Inc., La Jolla, CA).
3. Results
3.1. General characteristic of patients with and without ACR
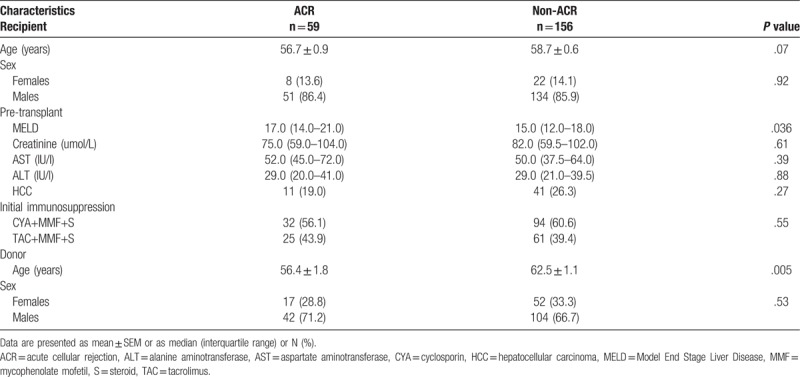
A total of 215 adult alcohol-related liver transplant recipients were divided into 2 groups; ACR 59 (27.4%) and non-ACR 156 (72.6%). There were no significant differences in pre-transplant recipients’ parameters regarding age, sex, creatinine, AST and ALT levels, or type of immunosuppression after LT, between the rejection and non-rejection group (Table 1). However, patients that developed ACR had significantly higher MELD scores (P = .036) pre-LT and received significantly younger liver grafts (56.4 ± 1.8 vs 62.5 ± 1.1, P = .005; Table 1). The median time from LT to rejection was 7 (IQR = 5–10) days.
Table 1.
General characteristic of patients with and without ACR.
3.2. CXCL9 and CXCL10 genotypes are not associated with ACR incidence
The genotype and allele frequencies of liver transplant recipients are summarized in Table 2. Genotypes were in Hardy–Weinberg equilibrium (P = .57 for CXCL9 and P = .76 for CXCL10), with strong linkage disequilibrium (D’ = .99, r = .986) between CXCL9 and CXCL10.
Table 2.
CXCL9 and 10 genotypes compared to acute cellular rejection occurrence.
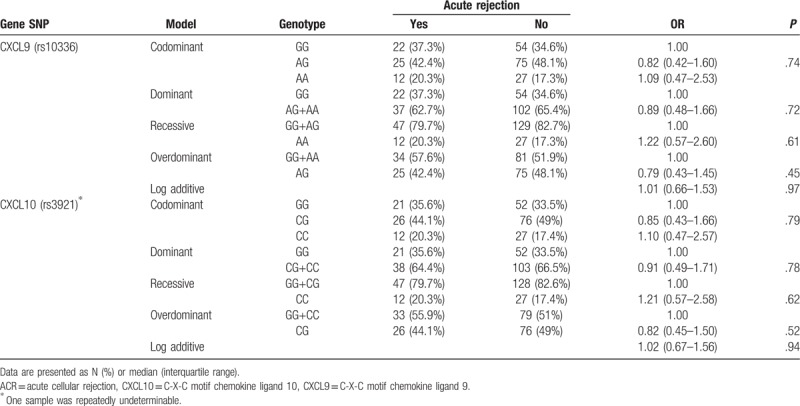
In the rejection group, 22 (37.3%) patients had GG, 25 (42.4%) had AG and 12 (20.3%) had AA genotype of the CXCL9 polymorphism, with similar genotype distribution observed in the non-rejection group, where 54 (34.6%) patients had GG, 75 (48.1%) had AG and 27 (17.3%) had AA genotype. Lack of association between CXCL9 genotypes and incidence of ACR was found in codominant, dominant, recessive, overdominant and log-additive model (P > .05) (Table 2). Similar results were obtained for CXCL10 genotypes (Table 2).
3.3. CXCL9 genotype is associated with the time until rejection and CXCL9 concentration in plasma
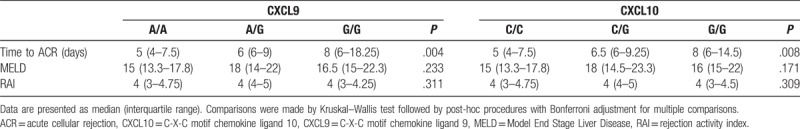
Even though there was no connection between the examined polymorphisms and ACR occurrence, we observed a significant association between both CXCL9 and CXCL10 genotypes and the time of ACR occurrence. Patients with CXCL9 genotype AA developed ACR earlier (median 5 days, IQR = 4–7.5) than patients with the GG genotype (median 8 days, IQR = 6–18.3; Bonferroni adjusted P = .003), with similar results for CXCL10 gene (CC vs GG; Bonferroni adjusted P = .005; Table 3). No significant differences were observed in MELD or RAI score across different genotypes of CXCL 9–10 in ACR patients (Table 3). In addition, there were no associations either between RAI score and the time of ACR (Kendall τ = 0.09; P = .40) or between the type of the initial immunosuppression and the time of ACR (Mann–Whitney, P = .11). To assess whether CXCL9 or CXCL10 genotype is an independent predictor of the time of ACR occurrence, we constructed a linear regression model with donor and recipient age, sex, MELD score, and genotype included as predicting variables. CXCL9 genotype remained a significant predictor of the time until ACR event, even after the addition of age, sex and MELD to analysis (β = 0.283; P = .033), while CXCL10 genotype was not significant (β = 0.222; P = .096).
Table 3.
Comparison of the grade of ACR according to Banff criteria, MELD score, and time of ACR occurrence with genotypes for CXCL 9 and 10.
Finally, we analyzed the plasma concentration of CXCL9 and CXCL10 in a cohort of 40 new transplant patients. There was no statistically significant difference between the rejectors and the non-rejectors for either of the analyzed chemokines (P = .78 and P = .68, respectively, Fig. 1A–B) with positive correlation between CXCL9 and CXCL10 concentrations (r = 0.35, P = .028, Fig. 1C). Interestingly, patients with AA CXCL9 genotype had significantly higher CXCL9 concentration (151.65 ± 22.74 pg/ml) in plasma than patients with AG (56.49 ± 16.31 pg/ml, P = .01) or GG genotype (87.1 ± 16.23 pg/ml, P = .045; Fig. 1D). There was no association between CXCL10 genotypes and CXCL10 concentration (Fig. 1E).
Figure 1.
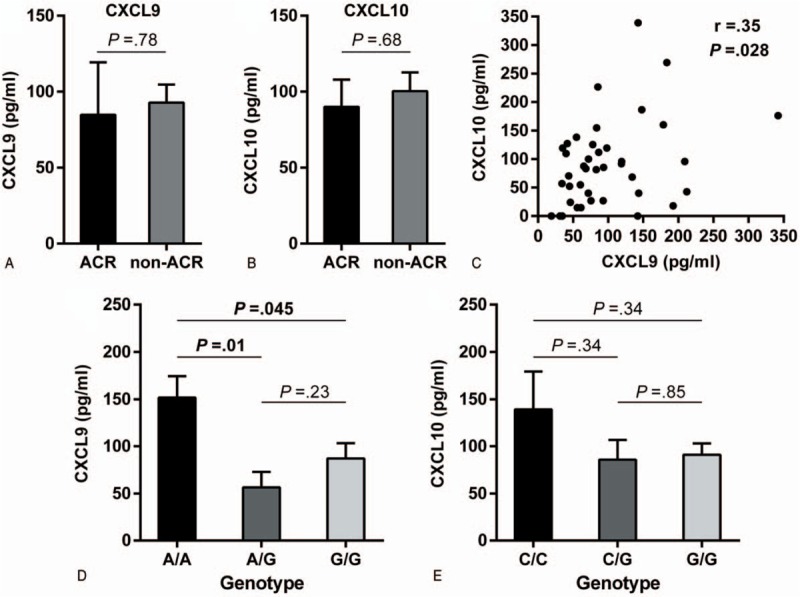
CXCL9 and CXCL10 concentration in plasma in a cohort of 40 new transplant patients. CXCL9 and CXCL10 plasma concentrations were determined by ELISA. A and B show a comparison of plasma CXCL9/10 levels between patients with and without rejection episodes. C Correlation between the CXCL9/10 levels in plasma. D and E Association between CXCL9/10 genotypes and their protein levels in plasma. Data represents mean ± standard error. ACR = acute cellular rejection, CXCL10 = C-X-C motif chemokine ligand 10, CXCL9 = C-X-C motif chemokine ligand 9.
4. Discussion
In this study, we analyzed the associations between CXCL9 and CX10 polymorphisms and ACR after LT. There were no significant associations between CXCL9 and CXCL10 gene polymorphisms and ACR development. However, patients with CXCL9 genotype AA developed ACR earlier and the same genotype was associated with greater CXCL9 plasma levels, suggesting the involvement of CXCL9 mediated processes in the ACR development.
Our analysis of clinical risk factors for ACR showed an association between ACR and younger donor age. This is in agreement with the study by Onaca et al, which showed that patients age >60 had a higher incidence of ACR when receiving organs from donors <50 years of age.[12] The most likely explanation relates to the fact that the immune response gradually decreases with age, and that grafts from younger donors are more immunogenic. However, a more recent study investigating the impact of using elderly donors (>60 years) did not find a significant association between donor age and incidence of ACR.[13] These discrepancies regarding donor age and ACR are probably due to possible interactions between donor age and other risk factor affecting ACR such as recipient age, etiology of liver disease or induction with IL-2 receptor antagonist.[14] Our results also show an association between higher MELD score and ACR. However, lack of models and data regarding this connection, as well as its questionable biological relevance and plausibility, demands cautious interpretation, and warrants further research into this topic.
To our knowledge, this is the first study that investigated the association between CXCL9 (rs10336) and CXCL10 (rs3921) SNPs and solid-organ graft rejection. So far, CXCL9 and CXC10 gene polymorphisms were investigated in hepatic fibrosis, several autoimmune diseases, infectious diseases, and bone marrow transplantation (BMT). Bruck et al studied SNPs of CXCL9 (rs10336, rs3733236) and CXCL10 (rs3921, rs35795399, and rs8878) genes in patients with diabetes type I and found no associations in German population.[15] Kotrych et al examined the associations of CXCL9 (rs3733236) and CXCL10 (rs8878) polymorphisms with rheumatoid arthritis, showing that CXCL10 GG genotype carriers had significantly higher frequency of extra-articular manifestations vs AA+AG carriers.[9] Association between the CXCL9–11 (rs10336 for CXCL9, rs3921 for CXCL10, and rs4619915 for CXCL11) polymorphisms and hepatic fibrosis was first reported in HIV/HCV-coinfected patients by Pineda et al.[16] Recently, Jimenez-Sousa et al analyzed the association between CXCL9–11 (rs10336 for CXCL9, rs3921 for CXCL10, and rs4619915 for CXCL11) polymorphisms and liver fibrosis in HCV-infected patients, further supporting their association with the likelihood of hepatic fibrosis.[17] In transplantation medicine, the impact of an SNP (rs3921) in the CXCL10 gene was studied regarding the outcomes in patients who underwent BMT for hematologic malignancies. The carriers of C/G or G/G genotypes had a significantly better 5-year survival rate and lower transplant-related mortality than the C/C genotype carriers.[18] In agreement with the findings of Pineda-Tenor et al, we found strong linkage disequilibrium between the CXCL9 and CXCL10 genotypes.[16]
Raschzok et al reported that patients who developed ACR had elevated plasma levels of CXCL9 before the transplantation and at first post-operative day.[7] Aiming to investigate if there were any long-term CXCL9/10 elevations, we measured CXCL9/10 levels at a later point in time—in the third to fourth post-transplantation week and found no difference in CXCL9/10 levels between the rejectors and non-rejectors. This can probably be attributed to a good response to the ACR therapy. Unfortunately, we have not taken samples at earlier points in time to confirm this hypothesis.
We found a significant association between CXCL9/10 genotypes and the time of ACR occurrence. The median time until first ACR episode was 7 days, which is consistent with the data in previous studies.[2] However, patients with CXCL9 genotype AA and CXCL10 genotype CC developed ACR earlier than patients with the CXCL9 and 10 GG genotypes, respectively. In the regression analysis only CXCL9 genotype AA has been identified as an independent predictor of time to ACR. In line with this, we found greater levels of CXCL9 in the plasma of patients with AA genotype. To the best of our knowledge, this is the first study that analyzed the association between these CXCL9/10 gene polymorphisms and their concentration in plasma. Since studies on animal models showed that CXCL9 accelerates acute transplant rejection our finding of greater CXCL9 concentrations in genotype (AA) associated with earlier ACR occurrence seems plausible.[19]
This study has several limitations that should be considered carefully. As one of the main conclusions of the study suggests that there is no association between the ACR incidence and chemokine genotypes, the question of power should be addressed. The number of patients is equal or moderately lower than the number of patients in similar studies. It should be taken into account that minor allele frequency (MAF) for both selected genotypes is high (41.4% and 42.1% for CXCL9 and CXCL10 genotype, respectively), which considerably reduces the number of patients needed to reach specific power. Similar studies with positive findings report relative MAF difference between rejectors and non-rejectors between 40% and 50%.[5,20] Taking these limits into account, the calculated power of our study was 65% to 85%. Furthermore, as the P values for different models range between .42 and .97 and are far from the significance threshold of .05, it is highly unlikely that inclusion of an additional number of patients would change the conclusion that there is a lack of association between the ACR and studied genotypes. However, it should be noted that we tested only one polymorphism of each gene and no conclusions can be made regarding the other variants of CXCL9 and 10 genes. Furthermore, we have not analyzed the expression of CXCL9/10 ligand CXCR3, which might have an influence on the interpretation of data. As only patients transplanted due to end stage of alcoholic disease were included in the study, the result cannot be generalized to other indications. Finally, protocolled biopsies were not performed at our center, thus this study did not include subclinical rejection episodes.
In conclusion, despite previous evidence regarding the association of serum levels CXCL 9 and 10 with ACR, here, we report no connection between the CXCL9 rs10336 and CXCL10 rs3921 polymorphisms and ACR in the later course. However, CXCL9 rs10336 AA genotype is associated with earlier ACR occurrence and greater CXCL9 concentrations in plasma.
Author contributions
Conceptualization: Tomislav Kelava, Anna Mrzljak.
Data curation: Ana Ostojic, Antonio Markotic.
Formal analysis: Antonio Markotic, Tomislav Kelava.
Funding acquisition: Tomislav Kelava.
Investigation: Ana Ostojic, Antonio Markotic, Tomislav Kelava, Anna Mrzljak.
Methodology: Ana Ostojic, Antonio Markotic.
Resources: Anna Mrzljak.
Software: Antonio Markotic.
Supervision: Tomislav Kelava, Anna Mrzljak.
Validation: Anna Mrzljak.
Visualization: Antonio Markotic.
Writing – original draft: Ana Ostojic.
Writing – review & editing: Tomislav Kelava, Anna Mrzljak.
Supplementary Material
Footnotes
Abbreviations: ACR = acute cellular rejection, ALD = alcoholic liver disease, ALT = alanine aminotransferase, AST = aspartate aminotransferase, BMT = bone marrow transplantation, BPAR = biopsy-proven acute rejection, CXCL10 = C-X-C motif chemokine ligand 10, CXCL9 = C-X-C motif chemokine ligand 9, CXCR3 = C-X-C motif chemokine receptor 3, DNA = deoxyribonucleic acid, ELISA = enzyme linked immunosorbent assay, IL = interleukin, IQR = interquartile range, LT = liver transplantation, MAF = minor allele frequency, MELD = model end-stage liver disease, mRNA = messenger ribonucleic acid, PCR = polymerase chain reaction, SNP = single nucleotide polymorphism, TGFB = transforming growth factor beta.
Ana Ostojic and Antonio Markotic These authors equally contributed to the manuscript.
This Study was supported by Hrvatska Zaklada za znanost grant number: UIP-2017-05-1965 and the University of Zagreb grants.
The authors have no conflicts of interest to disclose.
References
- [1].Levitsky J, Goldberg D, Smith AR, et al. Acute rejection increases risk of graft failure and death in recent liver transplant recipients. Clin Gastroenterol Hepatol 2017;15:584–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wiesner RH, Demetris AJ, Belle SH, et al. Acute hepatic allograft rejection: incidence, risk factors, and impact on outcome. Hepatology 1998;28:638–45. [DOI] [PubMed] [Google Scholar]
- [3].Kamei H, Masuda S, Ishigami M, et al. Association of interleukin4 gene polymorphisms of recipients and donors with acute rejection following living donor liver transplantation. Clin Res Hepatol Gastroenterol 2016;40:179–85. [DOI] [PubMed] [Google Scholar]
- [4].Karimi MH, Daneshmandi S, Pourfathollah AA, et al. Association of IL-6 promoter and IFN-( gene polymorphisms with acute rejection of liver transplantation. Mol Biol Rep 2011;38:4437–43. [DOI] [PubMed] [Google Scholar]
- [5].Zhang XX, Bian RJ, Wang J, et al. Relationship between cytokine gene polymorphisms and acute rejection following liver transplantation. Genet Mol Res 2016;15:1–7. [DOI] [PubMed] [Google Scholar]
- [6].Asaoka T, Marubashi S, Kobayashi S, et al. Intragraft transcriptome level of CXCL9 as biomarker of acute cellular rejection after liver transplantation. J Surg Res 2012;178:1003–14. [DOI] [PubMed] [Google Scholar]
- [7].Raschzok N, Reutzel-Selke A, Schmuck RB, et al. CD44 and CXCL9 serum protein levels predict the risk of clinically significant allograft rejection after liver transplantation. Liver Transpl 2015;21:1195–207. [DOI] [PubMed] [Google Scholar]
- [8].Friedman BH, Wolf JH, Wang L, et al. Serum cytokine profiles associated with early allograft dysfunction in patients undergoing liver transplantation. Liver Transpl 2012;18:166–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kotrych D, Dziedziejko V, Safranow K, et al. CXCL9 and CXCL10 gene polymorphisms in patients with rheumatoid arthritis. Rheumatol Int 2015;35:1319–23. [DOI] [PubMed] [Google Scholar]
- [10].Nogueira LG, Santos RHB, Ianni BM, et al. Myocardial chemokine expression and intensity of myocarditis in chagas cardiomyopathy are controlled by polymorphisms in CXCL9 and CXCL10. PLoS Negl Trop Dis 2012;6:e1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Demetris AJ, Batts KP, Dhillon AP, et al. Banff schema for grading liver allograft rejection: an international consensus document. Hepatology 1997;25:658–63. [DOI] [PubMed] [Google Scholar]
- [12].Onaca N, Mckenna G, Testa G, et al. Lower incidence of acute cellular rejection in liver transplant recipients more than 60 years of age allows minimization of immunosuppression. Transplantation 2012;94:132.22766769 [Google Scholar]
- [13].Kamo N, Kaido T, Hammad A, et al. Impact of elderly donors for liver transplantation: a single-center experience Naoko. Liver Transpl 2015;21:591–8. [DOI] [PubMed] [Google Scholar]
- [14].Au KP, Chan SC, Chok KSH, et al. Clinical factors affecting rejection rates in liver transplantation. Hepatobiliary Pancreat Dis Int 2015;14:367–73. [DOI] [PubMed] [Google Scholar]
- [15].Brück P, Bartsch W, Penna-Martinez M, et al. Polymorphisms of CXCR3-binding chemokines in type 1 diabetes. Hum Immunol 2009;70:552–5. [DOI] [PubMed] [Google Scholar]
- [16].Pineda-Tenor D, Berenguer J, García-álvarez M, et al. Single nucleotide polymorphisms of CXCL9-11 chemokines are associated with liver fibrosis in HIV /HCV-coinfected patients. J Acquir Immune Defic Syndr 2015;68:386–95. [DOI] [PubMed] [Google Scholar]
- [17].Jiménez-Sousa MÁ, Gómez-Moreno AZ, Pineda-Tenor D, et al. CXCL9-11 polymorphisms are associated with liver fibrosis in patients with chronic hepatitis C: a cross-sectional study. Clin Transl Med 2017;6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nakata K, Takami A, Espinoza JL, et al. The recipient CXCL10 +1642C>G variation predicts survival outcomes after HLA fully matched unrelated bone marrow transplantation. Clin Immunol 2013;146:104–11. [DOI] [PubMed] [Google Scholar]
- [19].Ma T, Xu J, Zhuang J, et al. Combination of C-X-C motif chemokine 9 and C-X-C motif chemokine 10 antibodies with FTY720 prolongs the survival of cardiac retransplantation allografts in a mouse model. Exp Ther Med 2015;9:1006–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yu X, Wei B, Dai Y, et al. Genetic polymorphism of Interferon Regulatory Factor 5 (IRF5) correlates with allograft acute rejection of liver transplantation. PLoS One 2014;9:2–7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


