Abstract
Reconstitution of hepatocytes by hematopoietic stem cells—a phenomenon which occurs in rodents under highly selective conditions—results from infrequent fusion between incoming myelomonocytes and host hepatocytes, with subsequent proliferation. Human hematopoietic-stem-cell-transplant recipients have been little studied, with some support for transdifferentation (direct differentiation). We studied routinely obtained autopsy liver tissue of 4 female hematopoietic cell transplant recipients with male donors, using a highly specific conjoint immunohistochemistry-in situ hybridization light microscopic technique. Hepatocyte nuclei were identified by cytokeratin (Cam5.2) staining and evaluated for X- and Y-chromosome content. Over 1.6 million hepatocytes were assessed for rare instances of donor origin, revealing a Y chromosome in 67. Mixed tetraploids (XXXY) and their nuclear truncation products (XXY, XY, Y) were directly demonstrated, with no detection of the male tetraploids (XXYY) that may result from transdifferention with susequent tetraploidization, nor their unique truncation products (XYY, YY), implicating fusion as the mechanism. To determine whether it is the sole mechanism, we modeled the chromosome distribution based on the same probability of detection of each X chromosome, deriving parameters of sensitivity and female tetraploidy by best fit. We then hypothesized that the distribution of Y-chromosome-containing cells could be predicted by a similar model. After modification to account for “clumpy” Y chromosomes, the observed results were in accord with the predicted (p=.6). These results suggest that all the Y-containing cells, including apparent XY cells, derive from mixed tetraploids, consistent with fusion as the sole mechanism.
Keywords: liver regeneration, stem cells, progenitor cells, in situ hybridization, hematopoietic stem cell transplant
INTRODUCTION
The liver can be repopulated by transplanted adult hepatocytes under selective conditions [1–3]. Fetal hepatocytes can repopulate the liver under less selective conditions [1]. Bone-marrow-derived cells have been extensively studied as a potential liver repopulation source in animal models, mainly rodents. Significant repopulation by marrow cells requires highly selective conditions [1–6]. For example, in the tyrosinemia mouse model, in which the catabolic product is toxic to host liver cells, donor-derived hepatocytes confer a strong selective growth advantage, resulting in donor derivation of greater than 50% of the liver mass [4]. Fusion between the incoming myelomonocytic cells and host hepatocytes, with subsequent proliferation, has been demonstrated as the mechanism [5–9]. The fusion-derived hepatocytes are tetraploids, but most of the progeny are aneuploids with chromosome numbers near-tetraploid, or, less often, near-diploid [10]. In less selective models, such as inducedliver injury, repopulation of donor derived hepatocytes has been extremely low or non-existent [1, 11–13].
Donor hepatocytes in human hematopoietic stem-cell-transplant (HSCT) recipients have been reported, with direct enumeration of up to 8%, with some reports suggesting transdifferentiation (direct differentiation) as the mechanism [14–16]. There has been little effort in recent times to assess the contribution of hematopoietic cells to hepatic parenchyma in humans.
Techniques to detect possible donor-derived hepatocytes in humans are limited. Experimental manipulation by necessity is restricted. Hence, the detection of donor-derived hepatocytes in humans has universally involved the demonstration of male sex chromatin in male-to-female transplant recipients. Techniques include in situ hybridization (ISH) for Y, or both X and Y, with nuclear counterstaining, and sometimes additional cell-protein staining. Difficulties result from substantial differences in the optimal treatment of tissue for ISH and immunohistochemistry (IHC). Interpretation typically involves fluorescent microscopy, sometimes standard light microscopy, with analysis of adjacent sections or comparative photomicroscopy. This has limited the certainty by which the cell identity can be determined, as the relatively thin section produces truncation artifacts, and overlying cells produce superimposition artifacts [2, 17, 18].
Here we report a conjoint chromogenic XY IHC-ISH method that allows examination of large numbers of hepatocytes in cross-sex HSCT recipients. This allows evaluation of all nuclei by light microscopy directly. This study is designed to quantitate and characterize donor-derived hepatocytes in human HSCT recipients, and infer their mechanism of production.
MATERIALS AND METHODS
Cases
Routinely formalin-fixed, paraffin-embedded liver tissue obtained at autopsy, was chosen from allogeneic HSCT patients who had died at varying times post-transplant, up to 6 months. We selected patients with a history of sinusoidal obstruction syndrome (veno-occlusive disease) (SOS/VOD) in an effort to investigate livers that might be under some regenerative stress. Four female patients with male donors were chosen as cases, and five female patients with female donors were chosen as negative controls. One male patient with a male donor was chosen as a positive control. Despite the history of SOS, it was the primary cause of death in only one patient. Patient characteristics are listed in Table 1. The tissue was sectioned at 4 μm. Hybridizations in the informative cases, female recipients with male donors, were performed at least in duplicate on different dates.
Table 1.
Patient Characteristics
| Patient No. | Recipient Sex | Donor Sex | Recipient -Donor HLA status | Preparation -Donor cell typea | Age | Diagnosis | Pregnancies (sons) | Days post-transplant | Cause of death |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Female | Male | Matched related | BM | 55 | Myeloma | 3 (3) | 21 | Diffuse alveolar damage, parainfluenza-3 pneumonia |
| 2 | Female | Male | Matched related | PBSC | 49 | Myeloma | ? (1) | 48 | Diffuse alveolar damage, hypertrophic cardiomyopathy |
| 3 | Female | Male | 1 Antigen MM related | PBSC | 56 | Myeloma | 4 (1) | 87 | Aspergillosis |
| 4 | Female | Male | 1 Antigen MM unrelated | BM | 39 | CML | ? (0) | 172 | Diffuse alveolar damage, varicella pneumonia |
| 5 | Female | Female | Matched unrelated | BM | 27 | ALL | 1 (0) | 20 | Fungal pneumonia |
| 6 | Female | Female | Matched unrelated | BM | 48 | AML | 2 (1) | 36 | Sinusoidal Obstruction Syndrome |
| 7 | Female | Female | Matched related | BM | 43 | AML | 3 (≥1) | 68 | Pseudomonas pneumonia |
| 8 | Female | Female | Matched related | BM | 39 | RAEB | 3 (≥1) | 74 | Aspergillosis, septicemia |
| 9 | Female | Female | Matched related | PBSCb | 63 | DLCL | 6 (≥1) | 83 | Aspergillus pneumonitis |
| 10 | Male | Male | Matched related | BM | 47 | RAEB | 83 | Congenital heart disease, central leukodystrophy |
All transplants were myeloablative, except where noted.
Nonmyeloablative.
BM bone marrow, PBSC peripheral blood stem cells, MM mismatched
Conjoint IHC-ISH
The conjoint IHC-ISH procedure is similar to Myerson et al [19] and presented in detail in Online Resource 1. Briefly, endogenous peroxidase was inhibited with hydrogen peroxide in methanol, and the slides were successively treated with Dako pH 6 target retrieval solution at 100°C, Dako serum-free protein block, anti-cytokeratin Cam5.2, biotinylated horse anti-mouse, and ABC-alkaline phosphatase. Cam5.2 was reapplied, and steps through ABC-AP were repeated to intensify staining. Cam5.2 was detected with Vector Red. The slides were further treated with 0.1N HCl, 0.01 M sodium citrate pH 4 at 100°C, and pepsin 0.02–0.5mg/ml (titrated for optimal results on each block). Mixed digoxigenin-X probe DXZ1 [20] and FITC-Y probe DYZI [21] was applied. Slides were denatured at 99°C for 15 minutes and hybridized overnight at 40°C. The slides were subsequently stringently washed, followed by alkaline phosphatase-coupled sheep anti-digoxigenin and rabbit anti-FITC mixed together, and peroxidase-labeled goat anti-rabbit. The chromogens, Vector Blue and diaminobenzidine (DAB), were sequentially applied. Slides were lightly counterstained with Gill’s hematoxylin.
Microscopy
Sections were optically examined and cells enumerated. Staining for hepatocyte cytokeratin (Cam5.2) and subsequent X and Y ISH were interpreted on a single slide by light microscopy. Digital photographs were color corrected globally in Adobe Photoshop Elements to a white background, with no further corrections performed.
Enumeration
Hepatocytes were easily recognized as large cells, with nuclei about 10 μm in diameter. Cam5.2 stained the hepatocyte cytoplasm, with more intense staining at the cell membrane, making the delineation of hepatocytes more certain. To reduce the enumeration of cells that were not hepatocytes, and to reduce superimposition artifacts, only hepatocyte nuclei completely surrounded by Cam5.2 positive (red) cytoplasm were evaluated. Portal tract cells, including bile duct cells, were not evaluated. A chromosome was considered stained if the nucleus showed a clear signal of blue (X) or brown (Y). Usually the sex chromosome signals appeared widely separated. The two chromatids of a single replicated chromosome may produce two very closely spaced signals of similar appearance; these were considered only one chromosome.
Y-containing hepatocytes in female livers were enumerated and categorized by examining entire slides, with the sex chromatin recorded for each nucleus. Hepatocytes without Y chromosomes and the male control were evaluated in five random fields on each slide, with the sex-chromatin content recorded for each nucleus, including those containing no visible sex chromatin. Hybridized slides chosen for analysis were all those that showed greater than 65% of hepatocytes with at least one X signal, a cutoff that was empirically determined as evidence of good hybridization. Three slides were thus excluded.
Statistical Modeling
Statistics were performed on the numbers of actual cells observed. For modeling, a uniform sensitivity of X chromosome detection was assumed. Let pX = chance of detecting X (qX = not detecting X). Let pTET = frequency of tetraploid hepatocytes (qTET = diploid). The predicted distribution of X chromosomes in nuclei (XXXX, XXX, XX, X, null) were the coefficients of the binomial expansion qTET*(pX+qX)2+pTET*(pX+qX)4. For each slide, pX and pTET were optimized together by comparing the observed enumerations to the predicted (expected) enumerations with a chi-square test, maximizing the chi-square p value, using the Microsoft Excel “Solver” tool with the GRG Nonlinear method (Online Resource 2). A similar model was constructed for the XY male control [i.e., qTET*(pX+qX)(pY+qY)+pTET*(pX+qX)2(pY+qY)2]. Here, pX, pY, and pTET were optimized all together.
RESULTS
Y-containing hepatocytes in female recipients with male donors
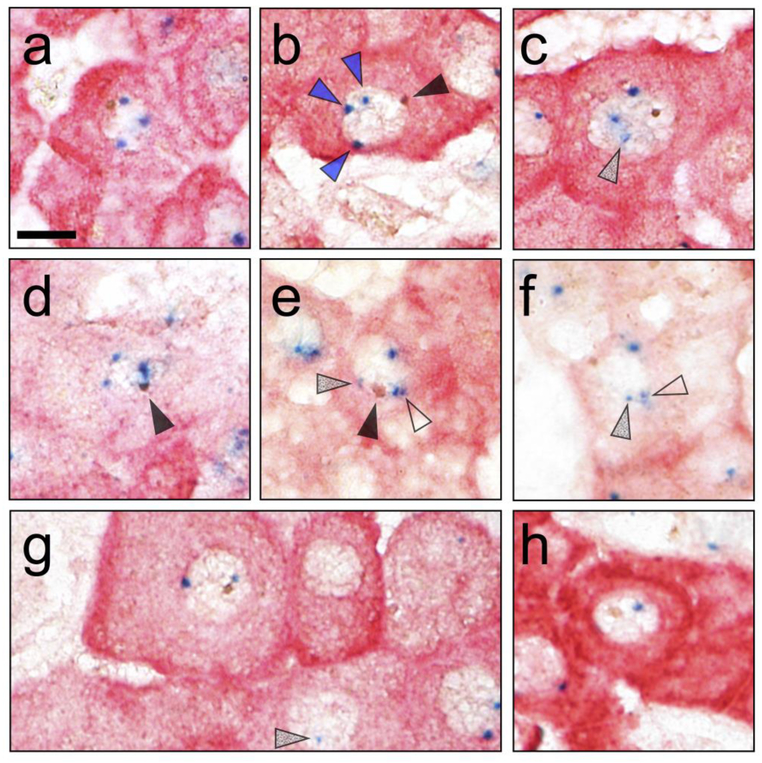
Over 1,600,000 nuclei were evaluated (4 cases, 9 slides). The results were striking, finding 6 mixed tetraploids (XXXY), and no male tetraploids (XXYY). Photomicrographs of all the mixed tetraploids, unselected for quality, and a representative XXY and XY are shown in Fig. 1. Mixed tetraploids were directly detected in 3 of the 4 cases studied. The XXY truncation product of mixed tetraploids was observed in all of the cases studied. The unique truncation products of male tetraploids (XYY and YY) were absent. All Y-chromosome–containing hepatocytes constituted 0.0041% (41 per 106) of the hepatocytes. Complete enumeration is in Table 2; the fraction calculated in Table 3. Fusion was immediately suggested as the mechanism. However, the question then arose as to whether fusion could lead to all the non-unique truncation products observed (Y, XY, XXY). These products could arise from fusion, but could also arise from transdifferentiation directly (XY) or transdifferentiation with tetraploidization-producing male tetraploids. Further studies explored whether all the Y-containing hepatocytes could be the result of initial fusion events.
Fig.1.
Y-chromosome containing hepatocytes in cross-sex hematopoietic stem cell transplants. Examples of sex chromosomes are highlighted with arrows. Closed arrows show brown Y chromosomes. Blue arrows show welldefined blue X chromosomes. Open arrows show X chromosomes as a pair of chromatids separated by a narrow space. Grey arrows show examples of lightly staining X chromosomes. (a-f) Each of the 6 XXXY mixed tetraploid hepatocytes. (g) Representative XXY hepatocyte, with typical diploid XX hepatocyte on right edge. (h) Representative XY hepatocyte. Size bar = 10 μm.
Table 2.
Enumeration of X and Y chromosomes in nuclei of Cam5.2-positive hepatocytes in females
| Patient | Hepatocytes without Y signal | Hepatocytes with Y signal | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| (slide) | Cells enumerated per area (by sampling) | Cells per slide (extrapolated)a | Cells enumerated per slide (by complete examination) | ||||||||
| M to F | Total | null | X | XX | XXX | XXXX | Total | Y | XY | XXY | XXXY |
| la | 1219 | 246 | 655 | 293 | 22 | 3 | 360700 | 1 | 4 | 3 | 1 |
| lb | 1255 | 288 | 627 | 320 | 18 | 2 | 371300 | 0 | 5 | 4 | 0 |
| 2a | 1171 | 279 | 496 | 327 | 62 | 7 | 71400 | 0 | 0 | 0 | 1 |
| 2b | 1106 | 291 | 492 | 288 | 27 | 8 | 67400 | 0 | 0 | 2 | 0 |
| 3a | 1426 | 269 | 650 | 455 | 48 | 4 | 139700 | 0 | 4 | 5 | 0 |
| 3b | 1329 | 177 | 689 | 410 | 35 | 18 | 275800 | 0 | 7 | 5 | 0 |
| 4a | 1062 | 307 | 453 | 268 | 30 | 4 | 117800 | 0 | 6 | 0 | 2 |
| 4b | 1097 | 231 | 524 | 308 | 25 | 9 | 121700 | 0 | 3 | 1 | 0 |
| 4c | 1029 | 319 | 458 | 226 | 22 | 4 | 114200 | 0 | 5 | 6 | 2 |
| Total M to F | 10694 | 2407 | 5044 | 2895 | 289 | 59 | 1640100 | 1 | 34 | 26 | 6 |
| F to F | |||||||||||
| 5 | 1169 | 205 | 602 | 338 | 19 | 5 | 152900 | 0 | 1 | 0 | 0 |
| 6 | 1193 | 248 | 634 | 276 | 22 | 13 | 141400 | 0 | 0 | 0 | 0 |
| 7 | 908 | 266 | 418 | 188 | 27 | 9 | 170400 | 0 | 0 | 0 | 0 |
| 8 | 832 | 143 | 483 | 185 | 18 | 3 | 86600 | 0 | 0 | 0 | 0 |
| 9 | 1321 | 322 | 673 | 296 | 28 | 2 | 205400 | 0 | 1 | 0 | 0 |
| Total F to F | 5423 | 1184 | 2810 | 1283 | 114 | 32 | 756700 | 0 | 2 | 0 | 0 |
| Total All | 16117 | 3591 | 7854 | 4178 | 403 | 91 | 2396800 | ||||
The area of each section is utilized to compute the extrapolated cells per slide. Block 3a was partially cut through, with a relatively lower area.
Table 3.
Evaluation of X and Y content of nuclei in Cam5.2-positive hepatocytes in females
| Patient | Hepatocyte nuclei without Y signal | Hepatocyte nuclei with Y signal | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| (slide) | Efficiency evaluation | Female tetraploids XXXX | Mixed tetraploids XXXY | ||||||||
| Null | Diploids XX visualizeda | Sensitivity of X detection | Directly observed | Various adjustments to correct for insensitivity | Directly Observed | Adjustments | |||||
| Directly observed including XXX | Adjusted for efficiency of diploids visualized | Best-fit model | Directly observed including XXY | Adjusted for efficiency of diploids visualized | |||||||
| pTET= | |||||||||||
| PX= | |||||||||||
| Null | Diploids | Best-fit | XXXX | XXXX+XXX | Adj. XXXX+XXX | XXXX | X2 | XXXY | XXXY+XXY | Adj. XXXY+XXY | |
| M to F | (%) | (%) | (%) | (%) | (%) | (%) | p= | (%) | (%) | (%) | |
| la | 20.2 | 24.5% | 0.511 | 0.2% | 2.1% | 8.4% | 6.0% | .004 | 0.0003% | 0.0011% | 0.0045% |
| lb | 22.9 | 25.9 | 0.505 | 0.2 | 1.6 | 6.2 | 5.1 | .720 | 0 | 0.0011 | 0.0042 |
| 2a | 23.8 | 29.7 | 0.474 | 0.6 | 5.9 | 19.9 | 23.3 | .141 | 0.0014 | 0.0014 | 0.0047 |
| 2b | 26.3 | 26.9 | 0.478 | 0.7 | 3.2 | 11.8 | 11.9 | .0001 | 0 | 0.0030 | 0.0110 |
| 3a | 18.9 | 33.1 | 0.551 | 0.3 | 3.6 | 11.0 | 10.0 | .103 | 0 | 0.0036 | 0.0108 |
| 3b | 13.3 | 32.1 | 0.583 | 1.4 | 4.0 | 12.4 | 8.7 | .0001 | 0 | 0.0018 | 0.0056 |
| 4a | 28.9 | 26.1 | 0.450 | 0.4 | 3.2 | 12.3 | 14.7 | .066 | 0 | 0.0020 | 0.0075 |
| 4b | 21.1 | 29.0 | 0.524 | 0.8 | 3.1 | 10.7 | 8.9 | .971 | 0 | 0.0008 | 0.0028 |
| 4c | 31.0 | 22.5 | 0.428 | 0.4 | 2.5 | 11.2 | 12.6 | .613 | 0.0018 | 0.0070 | 0.0311 |
| Average M to Fb | 22.5% | 28.0% | 0.507 | 0.6% | 3.3% | 11.6% | 10.2% | 0.0004% | 0.0020% | 0.0070% | |
| F to F | |||||||||||
| 5 | 17.5% | 29.5% | 0.551 | 0.4% | 2.1% | 7.0% | 5.1% | .178 | 0 | 0 | 0 |
| 6 | 20.8 | 23.8 | 0.504 | 1.1 | 2.9 | 12.3 | 9.2 | .0003 | 0 | 0 | 0 |
| 7 | 29.3 | 21.6 | 0.423 | 1.0 | 4.0 | 18.4 | 19.2 | .611 | 0 | 0 | 0 |
| 8 | 17.2 | 22.8 | 0.516 | 0.4 | 2.5 | 11.1 | 6.8 | .0000004 | 0 | 0 | 0 |
| 9 | 24.4 | 22.9 | 0.476 | 0.2 | 2.3 | 9.9 | 8.2 | .119 | 0 | 0 | 0 |
| Average F to Fb | 21.8% | 24.3% | 0.498 | 0.6% | 2.7% | 11.1% | 8.3% | 0 | 0 | 0 | |
| Average Allb | 22.3% | 26.7% | 0.504 | 0.6% | 3.1% | 11.5% | 9.5% | ||||
Diploids (XX) were expressed per hepatocyte with 2 or fewer sex chromosomes (XX,X, or null), to provide the most conservative adjustment for diploids visualized.
Averages for nuclei without Y are the mean of an equal sample from all slides. Averages for nuclei with Y are the mean of all cells on all slides. Note that in the cases of nuclei with Y, the average of all the slides is not necessarily equal to the average of the averages from each slide.
Y-containing hepatocytes in female recipients with female donors
As a control, Y-containing hepatocyte nuclei in recipients with female donors were examined (5 cases, 5 slides). Over 700,000 nuclei were examined, finding two Y-containing hepatocytes (Table 2). They were both XY cells, constituting 3 per 106 hepatocytes. Neither triploid nor tetraploid Y-containing hepatocytes were identified.
Tetraploidy in female livers
Tetraploid hepatocytes (XXXX) are present at unknown fractions in each liver [22, 23], confounding a straight forward attempt to assess insensitivity or truncation that may result in tetraploids appearing as apparent diploids (XX). We attempted to determine the true frequency of tetraploidy in each recipient. In this study, we present adjustments of decreasing stringency, determining the tetraploid fraction for each slide, by 1) direct enumeration, 2) adjusted for triploids, 3) adjusted for triploids with insensitivity, and a 4) binomial model assuming all X hybridization signals are equally likely to be observed. Validation is attempted by comparing the results of the adjustment methods.
Direct enumeration
The X chromatin content of each hepatocytes nucleus is evaluated on each slide in cases of female HSCT recipients with either female or male donors (Tables 2 and 3). We endeavor to determine the true fraction of female diploid hepatocytes (XX) and female tetraploid hepatocytes (XXXX). Heterokaryons (2 nuclei in one hepatocyte) also exist [24] but for this purpose are considered nuclear diploids. Without any experimental limitations, we would expect all the nuclei would appear as XX diploids or XXXX tetraploids, neglecting possible octoploids. As observed, however, the hepatocytes were null (no X chromosome), X, XX, XXX, or XXXX. Directly observed female tetraploids averaged 0.6% (range 0.2–1.4%) of the total hepatocytes (Table 3). These represent the minimum number of these cells actually present, excluding the low-frequency possibility that the tetraploids represent truncated cells of higher ploidy. Note there is no expectation that the fraction of tetraploids are equal in each case.
Adjusted-direct enumeration
Apparent-triploid cells (XXX) may reasonably be considered to be the result of technical insensitivity or truncation artifacts of female tetraploid (XXXX) cells, as although autosomal aneuploidy may exist, triploid sex chromosomes in hepatocytes have not been described [25, 26]. Apparent triploids occur about 4 times as often as tetraploids. “Adjusted-direct” enumeration of tetraploids, which includes enumerated tetraploids and apparent triploids, averages 3.1% (range 1.6–5.9%) of the hepatocytes (Table 3).
Diploid correction for insensitivity
A further, less well supported, “insensitivity” adjustment may be made. Since diploid XX cells are not detected with perfect efficiency, a reasonable approximation may be that tetraploids are detected about as efficiently. The “directly observed” female diploids (XX) averaged 26.7% (range 21.6–33.1%), considering only hepatocytes with no more than two sex chromosomes (yielding a slightly more conservative correction than total hepatocytes). Adjustment for “diploid correction” assumes this efficiency and corrects by increasing the frequency of presumed tetraploids (XXXX + XXX) by 1/0.267, suggesting an average tetraploid frequency averaging 11.5% (range 7.0–18.4%) (Table 3).
Binomial model
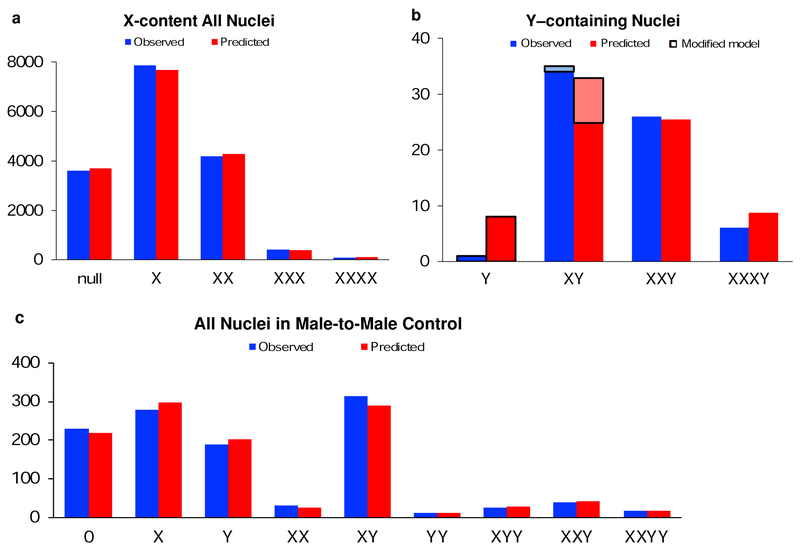
Lastly, because we have directly enumerated over 16,000 nuclei, sufficient data are available for a statistical approach to correct for insensitivity. On each slide, we assume the probability of detection for each X chromosome (pX) is the same, and derive it, along with the fraction of tetraploids (pTET). The binomial model predicts the distribution, and pX and pTET are optimized for best-fit with the observed data, per Methods. In the aggregate, the average sensitivity (pX) is 0.504, (range 0.423–0.583), and the average tetraploid frequency (pTET) is 9.5% (range 5.1–23.3%) (Table 3). Fig 2a depicts reasonable accord in the comparison between the observed and predicted results for the distribution on the slides in aggregate. Note that in this example we aggregate the cases, but there is not necessarily an expectation that the distribution would be in accord with the model, as the true tetraploid frequency for each case may be different. However, in the case by case comparison the results are comparable to the preceding diploid adjustment (Table 3).
Fig.2.
a Observed distribution of X-chromosome nuclear content of the hepatocytes in the aggregated slides of female recipients, regardless of the sex of the donor. The predicted distribution is the best-fit model assuming equal probability of detecting any given X signal, optimizing the probability of detection (pX) and the tetraploid frequency (pTET) to maximize the chi-square statistic. The best fit result was pX=0.507. b The hypothesis that the Ycontaining nuclei in female recipients of male donors also followed an equal probability model was tested. With the best fit pX (as above) as the input to the binomial best-fit model, the chromosomal content was predicted. The observed and the predicted were compared (the outlined XY not included), with chi-squared p=.015 A modified model was developed, assuming X is “clumpy” in tetraploids. The outlined Y–only data was added to the original XY data (indicated as outlined XY data) and the original Y–only data was not included in the modified model. The observed and the predicted were compared, with chi-squared p=.6 2c The male recipient of a male transplant compared the observed and predicted sex chromatin, with chi-squared p=.6.
The binomial model is expected to explain results on a slide by slide basis. While most slides showed the observed distribution of nuclear X content not significantly differing from the predicted distribution with the best fit by a chisquare, there were a few slides at wide variance (p<<.001) (Table 3). The duplicate slides (assessed in cases of male donors) offer some insight, as pTET should be the same in these cases. Tetraploids are essentially the same in two pairs/triplets of cases—5.1 vs.6.0%, 8.7 vs. 10.0%—with one case some showing small differences (triplicate) — 8.9 vs. 12.6 vs. 14.7%—and one case showing a larger, almost 2-fold difference —11.9 vs. 23.3%.
Hepatocytes in male recipient with male donor
To further confirm the validity of the technology, a positive control liver of a male-to-male HSCT recipient was examined. Hepatocyte nuclei were categorized and enumerated (Fig. 2c observed, Online Resource 3). We readily observed the expected male diploids (XY), male tetraploids (XXYY), and their truncation products, including those unique to male tetraploids (XYY, YY). Neither mixed tetraploids (XXXY) nor their unique truncation products (XXX) were identified. Male tetraploids (XXYY) were directly demonstrated as 1.5% of the hepatocytes (17/1133).
Male tetraploidy was analyzed similarly to the female livers (Fig 2c, Online Resource 3). The “adjusted-direct” male tetraploids, which include the apparent triploids XXY and XYY as likely truncated male tetraploids, were 7.2%. The diploid XY was observed in 31% of the null, X, Y, or XY hepatocytes. This “diploid correction” (1/0.31) suggests a true male tetraploid frequency of 23%. The best-fit binomial model, in concordance, yielded pTET=24% tetraploids in this liver, with chi-square p=.6, consistent with a good modeling of distribution. The results indicate efficient detection of male tetraploids and their unique truncation products.
Modeling of Y-containing hepatocytes in female recipients with male donors
We next investigated whether the data is explained by fusion as the sole mechanism of producing mixed tetraploid truncation products. Some could also potentially derive from male transdifferentiation, and subsequent tetraploidization (XY, XXY). Totals of directly enumerated mixed tetraploids (XXXY) (N=6) were 0.0004% (4 per 106) hepatocytes. The “adjusted-direct” enumeration, which also includes XXY apparent-triploids (N=26), suggested mixed tetraploids represent 20 per 106 hepatocytes. Applying the “diploid correction”, with the XX diploids detected at a sensitivity of 28%, to the “adjusted-direct”, there are 70 per 106 mixed tetraploid hepatocytes (Tables 2 and 3).
Fit to binomial distribution
We hypothesized that fusion could account for all the Y-containing hepatocytes. A binomial model was constructed for cells containing a single Y with the distribution of the remaining X chromosomes reflecting an equal sensitivity of detection. The predicted (expected) frequencies were the coefficients of the expansion of pY*(pX +qX)3. The input parameter was the average efficiency of X hybridization (pX=0.507), which was previously determined from examination of X chromosomes (Table 3). pY was arbitrarily assigned the same efficiency, but affects only the null (no Y signal)-cell prediction, with any reasonable estimate acceptable. This predicted (expected) distribution was compared to the observed (Fig. 2b, Online Resource 4). The model predicts 8.7 XXXY (vs. 6 observed), 25.5 XXY (vs. 26 observed), 24.8 XY (vs. 34 observed), and 8.4 Y (vs. 1 observed). Chi-square was p=.015, indicating a significant difference between the observed and predicted. The null hypothesis (prediction reflects observed results) is thus rejected.
Fit with “clumpy” Y modification
There was a small deficit of observed XY, balanced by a similar excess of Y. A modified model was developed, informed by this discrepant result. If the Y were closely adjacent to one of the 3 X chromosomes in a mixed tetraploid nucleus (a “clumpy” distribution), it would rarely be expected to appear alone in a truncated nucleus. Modeling this was accomplished by replacing the XY enumeration with the sum of Y-only and XY, both for the observed and the predicted, and not including Y-only in the model (Fig. 2b with outline, Online Resource 4). Chisquare was p=.6, not rejecting the null hypothesis.
The frequency of mixed tetraploids was estimated from the binomial model. Mixed tetraploid truncation products may contain no Y (null) and thus not be differentiated from the host female liver. Our enumeration documented 67 Y-containing hepatocytes, 41 per 106. With a presumed pY of 0.507, there are 65.2 similar Y-containing cells in the liver, but without an observed Y chromosome [67 *(0.493/0.507)], appearing null for a Y signal. Therefore, there were about 132 Y-containing hepatocytes, or 81 per 106 hepatocytes, suggesting the average prevalence of mixed tetraploids.
DISCUSSION
Our conjoint chromogenic IHC-ISH procedure enables the identification of hepatocytes and X and Y chromosomes on the same tissue section, obviating some causes of error and enabling the reliable evaluation of large numbers of cells, with staining in three colors and an additional nuclear counterstain delineating the hepatocytes. Background staining was very low. This proved to be a strong technique for specifically detecting donor-associated hepatocytes at extremely low frequency.
However, the complexity of the procedure led to considerable variability in results—typical for ISH procedures with poorly controlled formalin-fixed, paraffin-embedded autopsy tissues. Inevitable gross variation in fixation times, tissue thickness, necrosis, autolysis, and other known or unknown factors, necessitated different conditions for each tissue block to attain acceptable results, as measured by the sensitivity of detecting X–containing hepatocytes. Assessing the cross-sex hybridizations performed in duplicate or triplicate, the best fit female tetraploid frequency was generally consistent. The one outlier (#2) showed an almost 2-fold difference between the duplicates (11.9 vs. 23.3% female tetraploids), which was attributable to an absolute difference in apparent triploids (XXY), (27 vs. 62). However, the delineated adjustments and best-fit model provided generally equivalent tetraploid and mixed tetraploid frequencies over a 5-log range. The frequency of normal female or male tetraploidy as calculated by bestfit analysis exhibited a large range, but is consistent with findings of others [22, 23].
X and Y signals may be compromised for many reasons, falling into 2 general classes. First, “technical insensitivity”, which results in a less than 100% efficiency of hybridization, leading to undercounting sex chromosomes in a single nucleus. Second, because hepatocyte nuclei average 10 μm in diameter and the tissue sections are cut at 4 μm, all of the nuclei are transected, exhibiting “truncation artifact”, leading to a reduced number of chromosomes observed depending on the adjacency of the sex chromosomes. We attempted to correct for these insensitivities without regard to cause with a series of decreasingly stringent adjustments, culminating in the binomial modelling. The final adjustment to directly enumerated sex chromosomes, the “diploid correction”, yielded an estimate of 70 mixed tetraploids per 106 hepatocytes, tending to validate the finding of the binomial model of 81 per 106 hepatocytes. Female tetraploid estimates were reinforced similarly.
Of the possible artifacts that could confound interpretation of these results, the only significant problem is the possibility that small-size donor male leukocytes could be superimposed onto female hepatocytes and falsely interpreted as mixed tetraploids. This possibility is reduced by the rigorous determination of a hepatocyte, but is not definitively eliminated. Artifacts might also result from the superimposition of diploid hepatocytes or superimposition of nuclei from binucleate hepatocytes [17]. If this artifact were responsible for mixed tetraploids, however, there should be a much larger number of XY hepatocytes than was observed. Lastly, possible cross-staining artifacts are not supported—if an X-stained chromosome were to artifactually appear as a Y-stained chromosome, one would expect to observe it in the female controls.
It is certainly possible that differentiated myelomonocytes have fused to hepatocytes as has been observed in rodents. There were no technical problems in detecting such donor cells, as CD68(PG-M1)-positive macrophages were found to occur in the livers of the 4 cases, averaging 93% XY, using a similar procedure (data not shown). Two observations support fusion events. First, we found 41 in 106 hepatocytes directly observed to contain a Y chromosome (as Y, XY, XXY, and XXXY), in male-to-female HSCT recipients while only 3 in 106 hepatocytes contain a Y chromosome (as XY), in our female-to-female HSCT recipients—a 13-fold increase. Both the higher frequency and difference in ploidy suggest fusion is the likely mechanism generating these cells. Second, the demonstration of 6 XXXY hepatocytes also strongly supports fusion as the mechanism, as it is the only plausible mechanism that could produce XXXY mixed tetraploids. The modified binomial model of fusion accounts for all the visualized results, including the frequency of XY.
Fetal-derived cells may arise as a normal aspect of pregnancy and can persist for decades [27–31]. These fetal cells are detected as XY cells in mothers, and do not exhibit signs of fusion. In hepatocytes, such microchimerism is increased in autoimmune diseases [32], and areas of tissue damage [33]. Fetal-maternal microchimerism in hepatocytes occurs in frequencies similar to our controls [27], and would play no major role in confounding our results.
In human HSCT recipients, donor cells of nonhematopoietic type have been demonstrated in the Purkinje cells of the cerebellum with a very rare apparent mixed triploid [34]. There are experimental results in rodent models showing binucleate Purkinje cells occurring as stable heterokaryons [35, 36]. If stable mixed heterokaryons occurred in our hepatocytes, they would be assessed as a male XY nucleus, not explaining our results.
We utilized patients with livers damaged by SOS/VOD, presumably under some regenerative stress, to maximize the chances of finding donor hepatocytes. The stress, however, is far weaker than the strong selective pressure necessary in rodent models to produce hematopoietic stem-cell-derived hepatocytes. In contrast to original reports of high frequencies of donor-derived hepatocytes in humans, we find rare events, similar to frequencies found in rodents under no or low selective pressure [1–3, 5, 11–13, 18]. Fusion of hematopoietic cells with hepatocytes may be a rare normal event in humans. Our result suggests that fusion is the sole mechanism for producing hematopoietic donor–derived hepatocytes in humans.
Supplementary Material
Acknowledgments:
We thank Dr. Ted A. Gooley for statistics advice.
Funding: This work was supported in part by grants from the National Institutes of Health CA18029, CA15074, and HL36444.
Footnotes
Compliance with ethical standards: The study was performed under a protocol approved by the Institutional Review Board of the Fred Hutchinson Cancer Research Center.
Conflict of Interest: The authors declare that they have no conflict of interest.
REFERENCES
- 1.Shafritz DA, Oertel M (2011) Model systems and experimental conditions that lead to effective repopulation of the liver by transplanted cells. The International Journal of Biochemistry & Cell Biology 43:198–213. doi: 10.1016/j.biocel.2010.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fausto N (2004) Liver regeneration and repair: Hepatocytes, progenitor cells, and stem cells. Hepatology 39:1477–1487. doi: 10.1002/hep.20214 [DOI] [PubMed] [Google Scholar]
- 3.Kallis YN, Alison MR, Forbes SJ (2007) Bone marrow stem cells and liver disease. Gut 56:716–724. doi: 10.1136/gut.2006.098442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagasse E, Connors H, Al-Dhalimy M, et al. (2000) Purified hematopoietic stem cells can differentiate into hepatocytes in vivo. Nat Med 6:1229–1234. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Willenbring H, Akkari Y, et al. (2003) Cell fusion is the principal source of bone-marrow-derived hepatocytes. Nature 422:897–901. [DOI] [PubMed] [Google Scholar]
- 6.Vassilopoulos G, Wang PR, Russell DW (2003) Transplanted bone marrow regenerates liver by cell fusion. Nature 422:901–904. [DOI] [PubMed] [Google Scholar]
- 7.Resaz R, Emionite L, Vanni C, et al. (2011) Treatment of newborn G6pc−/− mice with bone marrow-derived myelomonocytes induces liver repair. Journal of Hepatology 55:1263–1271. doi: 10.1016/j.jhep.2011.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willenbring H, Bailey AS, Foster M, et al. (2004) Myelomonocytic cells are sufficient for therapeutic cell fusion in liver. Nat Med 10:744–748. doi: 10.1038/nm1062 [DOI] [PubMed] [Google Scholar]
- 9.Camargo FD, Finegold M, Goodell MA (2004) Hematopoietic myelomonocytic cells are the major source of hepatocyte fusion partners. J Clin Invest 113:1266–1270. doi: 10.1172/JCI21301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan AW, Hickey RD, Paulk NK, et al. (2009) Ploidy reductions in murine fusion-derived hepatocytes. PLoS Genet 5:e1000385. doi: 10.1371/journal.pgen.1000385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thorgeirsson SS, Grisham JW (2006) Hematopoietic cells as hepatocyte stem cells: A critical review of the evidence. Hepatology 43:2–8. doi: 10.1002/hep.21015 [DOI] [PubMed] [Google Scholar]
- 12.Wagers AJ, Sherwood RI, Christensen JL, Weissman IL (2002) Little evidence for developmental plasticity of adult hematopoietic stem cells. Science 297:2256. [DOI] [PubMed] [Google Scholar]
- 13.Grompe M (2005) Bone marrow-derived hepatocytes. Novartis Found Symp 265:20–7– discussion 28–34–92–7. [PubMed] [Google Scholar]
- 14.Theise ND, Badve S, Saxena R, et al. (2000) Derivation of hepatocytes from bone marrow cells in mice after radiation-induced myeloablation. Hepatology 31:235–240. doi: 10.1002/hep.510310135 [DOI] [PubMed] [Google Scholar]
- 15.Körbling M, Katz RL, Khanna A, et al. (2002) Hepatocytes and epithelial cells of donor origin in recipients of peripheral-blood stem cells. N Engl J Med 346:738–746. [DOI] [PubMed] [Google Scholar]
- 16.Alison MR, Poulsom R, Jeffery R, et al. (2000) Hepatocytes from non- hepatic adult stem cells. Nature 406:257. [DOI] [PubMed] [Google Scholar]
- 17.Wu T, Cieply K, Nalesnik MA, et al. (2003) Minimal evidence of transdifferentiation from recipient bone marrow to parenchymal cells in regenerating and long-surviving human allografts. Am J Transplant 3:1173–1181. [DOI] [PubMed] [Google Scholar]
- 18.Lizier M, Castelli A, Montagna C, et al. (2018) Cell fusion in the liver, revisited. WJH 10:213–221. doi: 10.4254/wjh.v10.i2.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myerson D, Parkin RK, Benirschke K, et al. (2006) The pathogenesis of villitis of unknown etiology: analysis with a new conjoint immunohistochemistry-in situ hybridization procedure to identify specific maternal and fetal cells. Pediatr Dev Pathol 9:257–265. doi: 10.2350/08-05-0103.1 [DOI] [PubMed] [Google Scholar]
- 20.Willard HF, Smith KD, Sutherland J (1983) Isolation and characterization of a major tandem repeat family from the human X chromosome. Nucleic Acids Research 11:2017–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lau YF, Dozy AM, Huang JC, Kan YW (1984) A rapid screening test for antenatal sex determination. The Lancet 323:14–16. doi: 10.1016/s0140-6736(84)90182-x [DOI] [PubMed] [Google Scholar]
- 22.Kudryavtsev BN, Kudryavtseva MV, Sakuta GA, Stein GI (1993) Human hepatocyte polyploidization kinetics in the course of life cycle. Virchows Arch, B, Cell Pathol 64:387–393. [DOI] [PubMed] [Google Scholar]
- 23.Toyoda H, Bregerie O, Vallet A, et al. (2005) Changes to hepatocyte ploidy and binuclearity profiles during human chronic viral hepatitis. Gut 54:297–302. doi: 10.1136/gut.2004.043893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gentric G, Desdouets C (2014) Polyploidization in liver tissue. Am J Pathol 184:322–331. doi: 10.1016/j.ajpath.2013.06.035 [DOI] [PubMed] [Google Scholar]
- 25.Duncan AW, Hanlon Newell AE, Smith L, et al. (2012) Frequent Aneuploidy Among Normal Human Hepatocytes. Gastroenterology 142:25–28. doi: 10.1053/j.gastro.2011.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knouse KA, Wu J, Whittaker CA, Amon A (2014) Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proc Natl Acad Sci USA 111:13409–13414. doi: 10.1073/pnas.1415287111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stevens AM, Michael McDonnell W, Mullarkey ME, et al. (2004) Liver biopsies from human females contain male hepatocytes in the absence of transplantation. Lab Invest 84:1603–1609. doi: 10.1038/labinvest.3700193 [DOI] [PubMed] [Google Scholar]
- 28.Bianchi DW (2007) Fetomaternal cell trafficking: a story that begins with prenatal diagnosis and may end with stem cell therapy. Journal of Pediatric Surgery 42:12–18. doi: 10.1016/j.jpedsurg.2006.09.047 [DOI] [PubMed] [Google Scholar]
- 29.Rijnink EC, Penning ME, Wolterbeek R, et al. (2015) Tissue microchimerism is increased during pregnancy: a human autopsy study. Molecular Human Reproduction 21:857–864. doi: 10.1093/molehr/gav047 [DOI] [PubMed] [Google Scholar]
- 30.Boddy AM, Fortunato A, Wilson Sayres M, Aktipis A (2015) Fetal microchimerism and maternal health: A review and evolutionary analysis of cooperation and conflict beyond the womb. Bioessays 37:1106–1118. doi: 10.1002/bies.201500059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guettier C, Sebagh M, Buard J, et al. (2005) Male cell microchimerism in normal and diseased female livers from fetal life to adulthood. Hepatology 42:35–43. doi: 10.1002/hep.20761 [DOI] [PubMed] [Google Scholar]
- 32.Khosrotehrani K, Bianchi DW (2005) Multi-lineage potential of fetal cells in maternal tissue: a legacy in reverse. Journal of Cell Science 118:1559–1563. doi: 10.1242/jcs.02332 [DOI] [PubMed] [Google Scholar]
- 33.O’Donoghue K, Sultan HA, Al-Allaf FA (2008) Microchimeric fetal cells cluster at sites of tissue injury in lung decades after pregnancy. Reproductive BioMedicine Online 16:382–390. doi: 10.1016/s14726483(10)60600-1 [DOI] [PubMed] [Google Scholar]
- 34.Weimann JM, Charlton CA, Brazelton TR, et al. (2003) Contribution of transplanted bone marrow cells to Purkinje neurons in human adult brains. Proc Natl Acad Sci USA 100:2088–2093. doi: 10.1073/pnas.0337659100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weimann JM, Johansson CB, Trejo A, Blau HM (2003) Stable reprogrammed heterokaryons form spontaneously in Purkinje neurons after bone marrow transplant. Nat Cell Biol 5:959–966. doi: 10.1038/ncb1053 [DOI] [PubMed] [Google Scholar]
- 36.Johansson CB, Youssef S, Koleckar K, et al. (2008) Extensive fusion of haematopoietic cells with Purkinje neurons in response to chronic inflammation. Nat Cell Biol 10:575–583. doi: 10.1038/ncb1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.