Abstract
The pathology of type 2 diabetes mellitus is associated with the aggregation of human islet amyloid polypeptide (hIAPP) and aggregation-mediated membrane disruption. The interactions of hIAPP aggregates with lipid membrane, as well as the effects of pH and lipid composition at the atomic level, remain elusive. Herein, using molecular dynamics simulations, we investigate the interactions of hIAPP protofibrillar oligomers with lipids, and the membrane perturbation that they induce, when they are partially inserted in an anionic dipalmitoyl-phosphatidylglycerol (DPPG) membrane or a mixed dipalmitoyl-phosphatidylcholine (DPPC)/DPPG (7:3) lipid bilayer under acidic/neutral pH conditions. We observed that the tilt angles and insertion depths of the hIAPP protofibril are strongly correlated with the pH and lipid composition. At neutral pH, the tilt angle and insertion depth of hIAPP protofibrils at a DPPG bilayer reach ~52° and ~1.62 nm with respect to the membrane surface, while they become ~77° and ~1.75 nm at a mixed DPPC/DPPG membrane. The calculated tilt angle of hIAPP at DPPG membrane is consistent with a recent chiral sum frequency generation spectroscopic study. The acidic pH induces a smaller tilt angle of ~40° and a shallower insertion depth (~1.24 nm) of hIAPP at the DPPG membrane surface, mainly due to protonation of His18 near the turn region. These differences mainly result from a combination of distinct electrostatic, van der Waals, hydrogen bonding and salt-bridge interactions between hIAPP and lipid bilayers. The hIAPP-membrane interaction energy analysis reveals that besides charged residues K1, R11 and H18, aromatic residues Phe15 and Phe23 also exhibit strong interactions with lipid bilayers, revealing the crucial role of aromatic residues in stabilizing the membrane-bound hIAPP protofibrils. hIAPP-membrane interactions disturb the lipid ordering and the local bilayer thickness around the peptides. Our results provide atomic-level information of membrane interaction of hIAPP protofibrils, revealing pH-dependent and membrane-modulated hIAPP aggregation at the early stage.
Keywords: Amyloid oligomer, type 2 diabetes, hIAPP, membrane, MD simulation
1. Introduction
The formation of fibrillar amyloid deposits is implicated in many human diseases, including Alzheimer’s disease, Parkinson’s disease and type 2 diabetes (T2D) mellitus [1,2]. These deposits are observed in over 90% of T2D patients, and their cytotoxicity is closely related to the membrane damage of insulin-producing β-cells [3,4]. The primary component of islet amyloid and actual fibril-forming molecule is human islet amyloid polypeptide (hIAPP, or amylin). hIAPP is a polypeptide co-secreted with insulin by pancreatic β-cells, and it can form β-sheet-rich aggregates of varied sizes and structures [3]. Increasing evidence suggests the early intermediates of fibrillation, such as oligomers and protofibrils, contribute to trigger pathological processes [3,5,6].
Monomeric hIAPP mainly adopts a random coil structure in aqueous solutions, with residues 8–19 displaying transient α-helical conformations [7–9]. Its aggregation follows a nucleation-dependent pathway as other amyloidogenic proteins, which can be dramatically accelerated in the presence of lipid membranes [10]. It was also reported that in the presence of model membranes, the α-helical transient intermediates were observed in the early stage of membrane-hIAPP interaction [11]. Intermediate oligomers can disrupt membrane integrity and permeability, lead to ionic homeostasis and signal disturbance, and thereby cause cell death [4,6,12–14]. Earlier electron paramagnetic resonance (EPR) spectroscopy and nuclear magnetic resonance (NMR) studies showed that a membrane-bound hIAPP monomer adopts pH- and membrane composition-dependent helical structure in one (residues 9–22) or two regions (residues 7–17 and 21–28) [15,16]. Upon enrichment of membrane-adsorbed peptides and subsequent formation of toxic oligomers, hIAPPs cooperatively transform into β-sheet aggregates [17–19]. Membrane-bound hIAPP oligomers not only promote amyloid formation but also induce membrane leakage [20–22]. However, it remains challenging to experimentally detect how hIAPP oligomers interact with membranes.
hIAPP is a 37 amino acid peptide, its N-terminal 1–19 region involves membrane binding, the amyloidogenic 20–29 fragment governs fibril formation, and the C-terminal region involves peptide self-association [23]. Aromatic stacking interactions play an important role in driving the self-assembly of hIAPP [24–26]. Histidine 18 is responsible for the pH-dependence of the fibrillation and membrane interactions of hIAPP [27,28]. His18 modulates the HiAPP1–19 orientation in anionic palmitoyloleoyl-phosphatidylglycerol (POPG) liposomes, and has a reduced toxicity via membrane disruption at pH 6.0 [27]. The kinetics of fibril formation and membrane damage of full-length hIAPP was observed to be much slower at pH 5.5 than at pH 7.4, which is closely related to His18 protonation state [28]. The aggregation of membrane-bound hIAPP is also affected by lipid composition, and can be significantly accelerated in the presence of negatively charged lipids, such as phosphatidylglycerol (PG) or phosphatidylserine (PS) [3,29,30]. These studies reflect the important role of electrostatic interaction in hIAPP-membrane interaction.
All-atom molecular dynamics (MD) simulations can provide detailed information of the interactions between amyloid proteins and membranes. Increasing number of research groups have examined the membrane interaction of monomeric/oligomeric amyloidosis-related proteins, including β-amyloid (Aβ), α-synuclein, hIAPP, and huntingtin, etc [31–44]. For example, Jang et al studied the structural properties of β-sheet-rich Aβ transmembrane channels and they observed in their MD simulations that preformed Aβ channels in lipid bilayers undergo a dynamic process comprised of subunit association, dissociation, and channel rearrangements [31]. Pannuzzo et al explored Aβ oligomerization in POPC membranes using MD simulations and molecular modeling, and reported that the insertion of Aβ into lipid membranes is related to the presence of high energy “frustrated helices” which could lead to the formation of highly dynamic ion channels [32]. Tsigelny et al. reported that α-synuclein can penetrate the membrane rapidly with a helix-coil transition, and the penetration of annular α-synuclein oligomers may form pore-like structure [33]. Zhao et al explored the dynamic properties of constructed hIAPP channels and they found that annular-like hIAPP channels with different sizes and topologies lose the initial continuous β-sheet network and break into oligomeric subunits [34]. They also investigated the property of double hIAPP channels in the DOPC bilayer using MD simulations and experiments, and reported the non-selective ion channel activity of hIAPP double channels [35]. Christensen et al examined the formation process of trimers and tetramers using a highly mobile membrane mimetic model and their simulations showed that initially membrane-bound α-helical hIAPPs can self-assemble into β-sheet structures [42]. Brown et al carried out MD simulations of disordered Aβ1–42 tetramers on zwitterionic lipid bilayers and they observed that the binding of Aβ tetramers on POPC bilayers resulted in a greater membrane perturbation than that on cholesterol-rich membranes [43]. Recently, Dong et al investigated systemically the adsorption dynamics, structural stability and membrane perturbation of protofibrillar Aβ trimers constructed using three different NMR-derived fibril structures, 2BEG, 2LMN and 2M4J. Their simulations showed that regardless of the morphologies and the initial orientations of the three different protofibrillar Aβ trimers, the N-terminal β-sheet of all trimers preferentially binds to the membrane surface [44].
The adsorption of hIAPP aggregates onto anionic PG membranes was monitored by means of chiral sum frequency generation (SFG) spectroscopy, and the β-sheet aggregates were found to orient at 48° relative to the interface [45]. Inspired by the experimental study, Poojari et al. simulated the stability and orientation of membrane-embedded hIAPP trimers and tetramers [46]. Their 150-ns molecular dynamics simulations showed that these lipid bilayer-inserted hIAPP oligomers may cause water permeation and Na+ intrusion. Very recently, Zhang et al. examined the adsorption of hIAPP oligomers onto zwitterionic lipid bilayers by MD simulations [47]. They found that hIAPP has stronger interactions with mixed palmitoyloleoyl-phosphatidylcholine (POPC)/palmitoyloleoyl-phosphatidylethanolamine (POPE) lipids than with pure POPC lipids, without thinning or curving the bilayers during 200-ns simulations. Our previous studies examined the structure and dynamics of hIAPP monomer and dimer at anionic bilayers [48,49]. However, the interactions between hIAPP protofibrillar oligomers (protofibrils) and anionic membranes with different lipid compositions under different pH condition remain elusive. Here we investigated the conformational dynamics and insertion depth of hIAPP protofibrils in anionic dipalmitoyl-phosphatidylglycerol (DPPG) membrane by performing multiple independent 300-ns MD simulations. We also examined the influence of pH and lipid compositions to gain more insight into membrane interactions of hIAPP protofibrils.
2. Materials and methods
2.1. hIAPP protofibrils and lipid bilayers
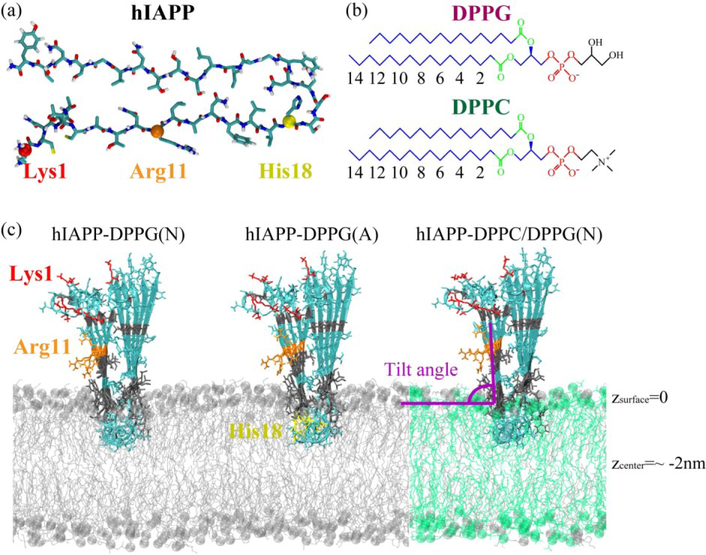
The hIAPP protofibrillar pentamer was modeled on the basis of a previous solid-state NMR study [50]. The amino acid sequence of hIAPP is 1-KCNTATCATQRLANFLVHSSNNFGAILSSTNVGSNTY-37. Each hIAPP peptide chain in the protofibril is composed of a disordered N-terminal loop with the Cys2 and Cys7 forming a disulfide bond, two β-strand regions (Ala8-Val17 and Ser28–Tyr37), and a turn region of His18-Leu27 (Fig. 1(a)). The N-terminus was charged (NH3+) and the C-terminus was amidated to mimic experimental conditions. At neutral pH, the side chains of Lys1 and Arg11 are positively charged; at acidic pH, the side chain of His18 is also positively charged.
Fig. 1.
The structures of a peptide chain in the hIAPP protofibril (a), a DPPG and a DPPC lipid molecule (b), and initial states of three simulated systems: hIAPP-DPPG(N), hIAPP-DPPG(A) and hIAPP-DPPC/DPPG(N) (c), where N and A in the parentheses denote neutral and acidic pH, respectively. Color codes: choline/glycerol group (black), phosphate group (red), ester group (green), and other carbon atoms (blue) in a lipid; residues Lys1 (red), Arg11 (orange), positively charged His18 (yellow), hydrophilic residues (cyan), and hydrophobic residues (gray) in the hIAPP protofibril; DPPG lipids (silver), DPPC lipids (green) in membrane bilayer, with phosphorus atoms in spheres. The atom index of acyl chain 1 (sn-1) in DPPG and DPPC lipids is also labeled.
The DPPG bilayer provides an ideal model to study the mechanism for the binding of hIAPP at anionic membrane and the membrane-mediated hIAPP aggregation, and it has been widely used in previous studies of hIAPP–membrane interaction [18,19,46]. To examine the influence of lipid composition, zwitterionic phosphatidylcholine (PC), the most abundant phospholipids in pancreatic islets [51], was introduced and mixed with DPPG lipids at a molar ratio of ~7:3 to mimic the membrane of pancreatic islet cells [52]. The chemical structures of a DPPG and a DPPC bilayer are shown in Fig. 1(b). A pure 2×64 DPPG bilayer and a mixed 2×64 DPPC/DPPG (90:38 in lipid number) bilayer were constructed based on a previous work [53], and they were well equilibrated in a 100-ns MD simulation at 323 K, above the gel-liquid crystal phase transition temperature of DPPG and DPPC lipid bilayers [54,55]. The molar ratio of peptide:lipid in our simulations is close to a previous experiment in which the ratio is 1:20 [28]. Na+ and Cl− ions were added to neutralize the systems and a salt concentration of 0.1 M.
The hIAPP protofibril was pre-inserted partially into the membranes by the program INFLATEGRO [56], with the backbone perpendicular to the bilayer surface. The turn region of hIAPP protofibril was located at the half depth of the upper leaflet so that hIAPP may choose to insert deeper if it favors the lipid environment or move backward to the membrane surface if it favors the water solution (Fig. 1(c)). This initial state allows the N-terminal residues to be exposed to the water environment and core hydrophobic residues 20–27 to be located in the lipid phase, as suggested in previous studies [57,58]. The systems of hIAPP protofibril with a DPPG bilayer at neutral pH, hIAPP protofibril with DPPG bilayer at acidic pH, and hIAPP protofibril with mixed DPPC/DPPG (7:3) bilayer at neutral pH, are labeled as hIAPP-DPPG(N), hIAPP-DPPG(A) and hIAPP-DPPC/DPPG(N), respectively. Figure 1 displays the structures of a peptide chain in hIAPP protofibril, a DPPG/DPPC lipid molecule, and the initial states of three systems. The average z-coordinate of the phosphorus atoms in the upper leaflet is defined as z=0 (corresponding to the membrane surface), and the z-coordinate of the bilayer center is at about −2 nm. The β-strand vector (Cα-Cα vector from T9 to V17 residues) in each peptide chain is used to define the protofibril orientation, and the initial tilt angle relative to the membrane surface is ~90° (Fig. 1(c)).
2.2. Simulation details
Atomistic MD simulations were performed in the isothermal-isobaric (NPT) ensemble using the GROMACS 4.5.3 software package [59]. The peptides and lipids were respectively described with the GROMOS [60] and modified Berger force fields [61] with an adaption of Tieleman’s lipid parameters. The systems were solvated with SPC water molecules. The integration time step is 2 fs. The pressure is kept at 1 bar using the semi-isotropic Parrinello-Rahman’s method [62,63] with a coupling constant of 1.0 ps and a compressibility of 4.5×10–5 bar−1. The temperature is maintained at 323 K using Nose-Hoover’s method [64,65] with a coupling constant of 0.1 ps. Van der Waals (vdW) interaction is described with a space cutoff of 1.4 nm. Long-range electrostatic interaction is described by the Particle Mesh Ewald (PME) method [66] with a recommended cutoff of 1.2 nm, especially for charged lipids [67]. For each system, four independent 300-ns MD runs were carried out with different initial velocities.
2.3. Analyses
MD trajectories were analyzed using in-house-developed codes and the GROMACS toolkits. The z-position of a residue is defined as the displacement of the centroid of each residue relative to the average z-coordinate of the phosphorus atoms (z=0) in the upper bilayer. An atomic contact is defined when two non-hydrogen atoms are located within 0.54 nm. The peptide-lipid interaction energy U is calculated using the GROMACS tools g_ener and mdrun-rerun (using the formula Uinter = U(peptide+lipid) - U(peptide) - U(lipid)). The number of hydrogen bonds (H-bonds) is calculated using GROMACS tool g_hbond. An H-bond is considered as formed only if the distance between donor D and acceptor A is less than 0.35 nm and the angle of D-H… A is greater than 150°. A salt bridge is considered to be formed if the minimum distance between the nitrogen atom of sidechain NH3+ group and the phosphorus atom of lipid PO4− group is less than 0.4 nm. The order parameter SCD of lipid tail is calculated using the formula SCD = 0.5<3cos2θ−1>, where θ is the angle between the bilayer normal direction and the C-H bond vector (in simulations) or the C-D bond vector (in experiments). The angular brackets denote a time and ensemble average [68]. This parameter reveals the orientation and the (dis)order degree of lipid acyl chains, correlated with the perturbation of hIAPP on the lipid tail. The larger the SCD is, the more ordered the lipid acyl chains are. The thickness of lipid bilayer is estimated by the average of the phosphorus-to-phosphorus distance [69]. The local bilayer thickness is estimated by the average z-position of the phosphorus atoms in the upper leaflet minus that in the lower leaflet within a certain cutoff from hIAPP peptides.
3. Results and discussion
3.1. Orientation and insertion depth of a membrane-bound hIAPP protofibril
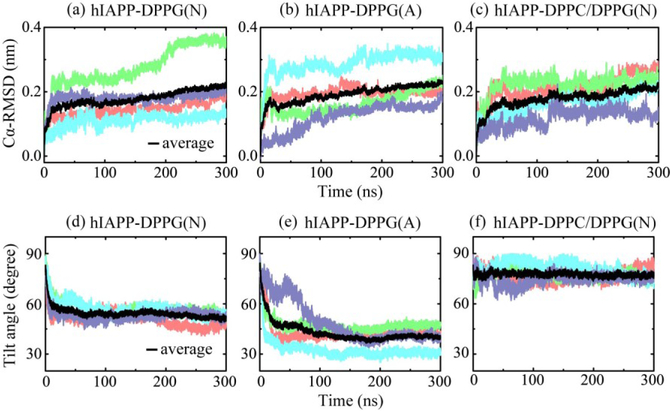
Membrane-bound hIAPP protofibrils remain inserted in the lipid bilayer, but reorient from their initial insertion direction and exhibit a stable conformational behavior. The time evolution of the root-mean-square deviation (RMSD) of Cα-atoms relative to the initial structure of hIAPP protofibril in Figs 2(a–c) shows that the Cα-RMSD in each MD run are smaller than 0.4 nm, and the average Cα-RMSD value for each of the three systems is about 0.2 nm, indicating that the hIAPP protofibril is highly stable in the presence of anionic membranes under both neutral and acidic pH conditions.
Fig. 2.
Time evolution of Cα-RMSD with respect to the initial structure of hIAPP protofibril (a-c) and the tilt angle of hIAPP protofibril relative to the bilayer surface (d-f) for different systems. Four independent MD runs for each system are denoted in different colors, and the average value is in black. The tilt angle is defined as the protofibril orientation relative to the membrane surface (see Fig. 1(c)).
The time evolution of the tilt angle of the hIAPP protofibril relative to the membrane surface in Figures 2(d) shows that the hIAPP in hIAPP-DPPG(N) system has a relaxation time of ~50 ns at the beginning, then adjusts the orientation slowly for tens of nanoseconds and finally reaches an average angle of 52°. This tilt angle is close to hIAPP orientation of 48° in DPPG monolayers, which was predicted by chiral SFG spectroscopy and quantum chemistry calculation [45]. However, at acidic pH, hIAPP reaches a smaller tilt angle of 40°, and one MD run with a long relaxation time of ~200 ns is observed (Fig. 2e). In the case of a mixed DPPC/DPPG membrane at neutral pH, the average tilt angle of hIAPP protofibril is 77° (Fig. 2f)), not far away from the initial tilt angle of 90°.
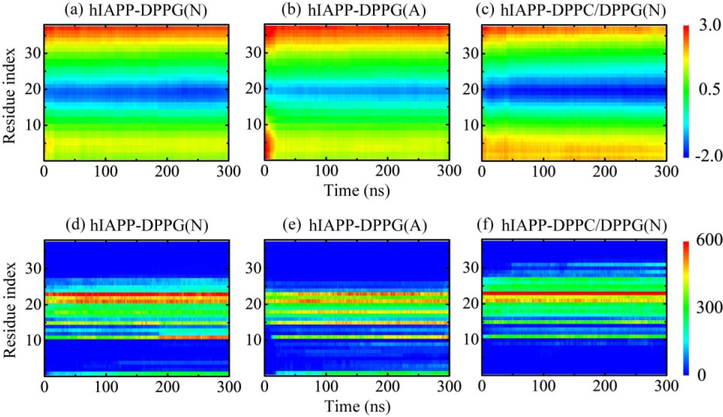
To quantify the peptide orientation relative to the membrane surface, we plot in Figs. 3(a–c) the time evolution of z-position of individual residues in a representative MD run. The low z-positions of N-terminal residues with respect to those of C-terminus residues reflect that the hIAPP protofibril leans to the N-terminal side in the DPPG bilayer at neutral pH. The z-position of each individual residue in hIAPP protofibril is quite stable during the whole 300-ns simulation time, with the turn region at 18–27 partially buried below the bilayer surface. At acidic pH, the turn region has a larger z-position in DPPG bilayer than that in hIAPP-DPPG(N) system, indicating that the insertion depth of the hIAPP protofibril at acidic pH is smaller than that at neutral pH. Clearly, the positively charged His18 at acidic pH prevents the deep membrane insertion. This change in orientation is consistent with a previous NMR experimental study showing that the hIAPP1–19 peptides tend to orient at the micelle surface at pH 6.0 [29]. The larger z-position in the N-terminal region at the very beginning of the simulation also reflects a longer relaxation time. As for the hIAPP-DPPC/DPPG(N) system, the enlarged blue region and the higher N-terminal z-position indicate a deeper insertion and a larger tilt angle with respect to those of the hIAPP-DPPG(N) system.
Fig. 3.
Time evolution of residue-based z-position (a-c) and number of contacts between each individual residue of hIAPP protofibrils and lipid membranes (d-f) in a representative MD run for each hIAPP-membrane system.
The time-dependent number of contacts between each individual residue of the hIAPP protofibril and lipid membranes in a representative MD run is given in Figs. 3(d–f). At neutral pH, the N-terminal residues Lys1, Arg11 and Phe15 and residues in the turn region of hIAPP protofibril, especially residues Asn21, Asn22 and Phe23, have a large number of contacts with the DPPG bilayer. In the hIAPP-DPPG(A) system, the contact number increased in the N-terminal region and reduced in the turn region, corresponding to a larger tilt angle and a shallower insertion depth. His18 displays a remarkably stronger interplay with DPPG, while the contacts between Arg11 and lipids get smaller. As for the hIAPP-DPPC/DPPG(N) system, the turn region 18–27 has more contacts with membranes, among which Phe23 contributes the most. Residues Arg11 and Phe15 also have impressive contacts with lipids, while Lys1 has no contact with membrane due to the large tilt angle.
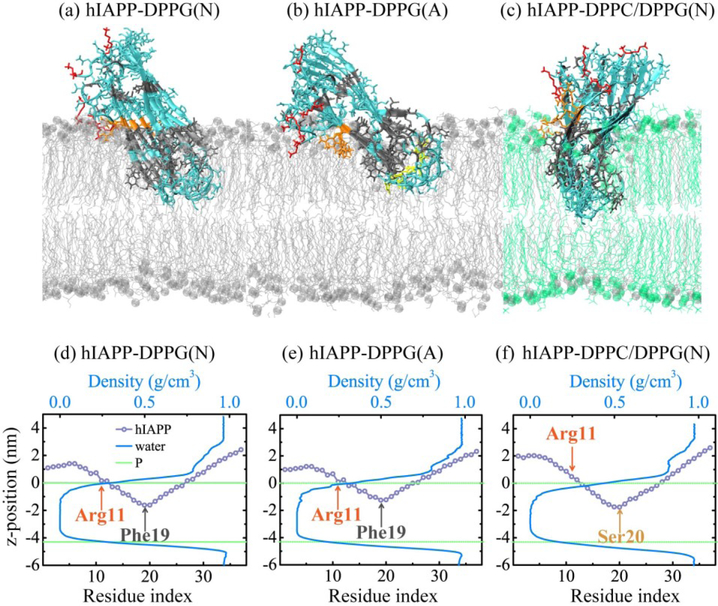
Figures 4(a–c) represent the final snapshots of the hIAPP protofibril at the membrane interface for each of the three systems. The membrane-bound hIAPP exhibits that different conformational behaviors accompany different pH and lipid composition. Arg11 always anchors at the bilayer surface and Lys1 assists to adjust the oligomer orientation, different from the binding behavior of hIAPP monomer in which two basic residues Lys1 and Arg11 anchor the peptide to charged lipid headgroups by strong electrostatic interactions [48,49]. Figures 4(d–f) display the average z-position of each residue of hIAPP protofibirl at bilayer surface and the water density profile along the membrane normal. In the presence of the DPPG bilayer, Phe19 has the lowest z-position of −1.62 nm among all the residues at neutral pH, while this value becomes −1.24 nm at acidic pH. In addition, the N-terminal residues in the hIAPP-DPPG(A) system are located closer to phosphorus atoms in the upper leaflet. These results reveal that the hIAPP protofibirl at acidic pH has a small insertion depth and a preference to interact with DPPG bilayer surface. At neutral pH, Ser20 has the lowest z-position of −1.75 nm in the mixed DPPC/DPPG membrane and is not yet able to cross the bilayer center. The N-terminal residues 1–5 are ~1 nm higher and residues 18–27 in the turn region insert more deeply into the membrane than those in pure DPPG bilayer. Arg11 is no longer the closest residue to bilayer surface. These data reveal that the reduced net charge of mixed DPPC/DPPG membrane weaken the restriction of hIAPP to the bilayer surface. The water density profile shows that no water molecules enter the hydrophobic tail region of the lower leaflets (Fig. 4(e–f)). The hIAPP trimer was reported to lose the strand-turn-strand motif and induce the diffusion of Na+ deep into the membrane in the neutralizing ion only simulation [46]. However, when additional salt was added, the peptide motif was kept and no ions diffused into the hydrophobic region of bilayer, with water flow inside the membrane being much reduced [46]. The salt concentration in our simulated systems is 0.1 M, and few water molecules are observed to penetrate into headgroup region of the upper leaflet.
Fig. 4.
(a-c) Snapshots of hIAPP protofibril at membrane interface in a representative MD run of each system. The hydrophilic, hydrophobic, Lys1, Arg11, and positively charged His18 residues are colored in cyan, gray, red, orange, and yellow, respectively; DPPG and DPPC lipids are denoted in silver and green, with phosphorus atoms in spheres. (d-f) Average z-position of each residue of hIAPP protofibirl at bilayer surface and water density profile along the membrane normal for hIAPP-DPPG(N) (d), hIAPP-DPPG(A) (e) and hIAPP-DPPC/DPPG(N) (f) systems. Average z-positions of phosphorus atoms in upper and lower leaflets are denoted by green dashed line.
3.2. Electrostatic, van der Waals, hydrogen-bonding and salt-bridge interactions between hIAPP protofibrils and membranes
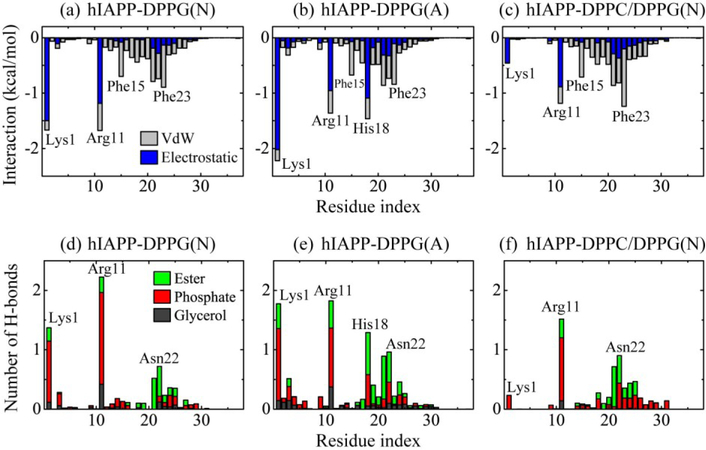
To identify the critical interactions that stabilize the orientation and structure of hIAPP protofibril in membranes, we plot in Figs. 5(a–c) the average interaction energy between each residue of hIAPP protofibril and lipid membranes (per lipid), decomposed into van der Waals (vdW) and electrostatic terms. Figure 5(a) shows that at neutral pH, residues Lys1 and Arg11 have the strongest interaction with DPPG bilayers, dominated by electrostatic components. Residues 18–27 in the turn region as well as Phe15 has a strong vdW interactions with the lipids, among which Asn21, Asn22 and Phe23 contribute the most. As Arg11 is the anchor residue, the orientation of the hIAPP protofibril results from the combination of electrostatic attraction between Lys1 and the bilayer surface and the vdW interaction between the turn region and the lipid bilayer. At acidic pH, residues Lys1, Arg11 and His18 interplay most strongly with membranes (Fig. 5b). The electrostatic attraction of Lys1/His18 with bilayers leads to a smaller tilt angle of hIAPP protofibril. This significantly enhanced attraction between His18 and the lipid bilayers reveals its key role in hIAPP-lipid interaction at acidic pH, consistent with the observation that His18 is critical in the modulation of pH-dependent hIAPP1–19 orientation in the anionic membrane. In the case of mixed DPPC/DPPG bilayer under neutral pH, the electrostatic terms of residues Lys1 with lipids are significantly reduced due to the replacement of anionic DPPG lipids by zwitterionic DPPC lipids. The turn region 18–27 has an increased interaction energy, among which Phe23 interacts strongest with the bilayer mixture. The hIAPP-membrane interaction energy analysis reveals that besides charged residues K1, R11 and H18, aromatic residues Phe15 and Phe23 also display strong interactions with lipid bilayers. This finding is consistent with a previous study of hIAPP12–18 aggregation in DMPC vesicles, showing the high affinity of Phe with lipid bilayer [24].
Fig. 5.
(a-c) The interaction energy (kcal/mol) between each individual residue of the hIAPP protofibril and lipid membranes (per lipid) for each of the three systems, decomposed into van der Waals (vdW) and electrostatic terms. (d-f) Number of H-bonds between each residue of hIAPP protofibril (per chain) and different lipid headgroups (glycerol, phosphate, and ester groups) for each simulated system. Data are averaged over the last 60 ns of four MD runs for each system.
The number of H-bonds between each residue of the hIAPP protofibril (per chain) and different lipid headgroups in Figs. 5(d–f) shows that the protofibril mostly forms H-bonds with the phosphate groups, and less with the ester groups, and least with the glycerol groups. Arg11 has the highest number of H-bonds with lipid headgroups in the three systems, which facilitate its role as the anchor point at the membrane surface. In the hIAPP-DPPG(N) system, the H-bonds are formed mostly by charged residues Lys1 and Arg11, and polar residues Asn21 and Asn22 with lipid headgroups. The number of H-bonds between His18 and the lipid heads is dramatically increased in the hIAPP-DPPG(A) system, and more DPPG ester groups are involved in H-bond formation. In the case of a mixed DPPC/DPPG bilayer at neutral pH, the total number of H-bonds between hIAPP and lipid heads is greatly reduced due to the weakened electrostatic attraction. As a result, hIAPP inserts deeper into the bilayer to maximize its interaction with the lipid bilayer. This resembles the NMR and fluorescence observation of an antimicrobial peptide MSI-78, in which the peptide inserts more deeply in PC lipid bilayers than in PG lipid bilayers [70].
The number of salt bridges formed between Lysl, Argil and His18 residues of hIAPP protofibril and phosphate groups of lipid bilayers is listed in Table 1. It shows that Argil has the most salt bridges with membranes. The Arg11-lipid salt bridges help to stabilize Arg11 at the bilayer surface. Lys1 and Arg11 form less salt bridges with DPPG lipids at acidic pH. The number of salt bridges between Lys1 and mixed DPPC/DPPG bilayer gets much smaller. The hIAPP-membrane interaction is enhanced by the additional force resulting from the formation of Lys1-PO4 and Arg11-PO4 salt bridges [47].
Table 1.
Number of salt bridges formed between Lys1, Arg11 and His18 residues and phosphate groups of lipid bilayers.
| System | Lys1 | Arg11 | His18 |
|---|---|---|---|
| hIAPP-DPPG(N) | 5.0 | 6.0 | — |
| hIAPP-DPPG(A) | 2.8 | 3.2 | 1.1 |
| hIAPP-DPPC/DPPG(N) | 1.0 | 4.1 | — |
3.3. Perturbation of the membrane-bound hIAPP protofbirl on the local ordering of lipid bilayers
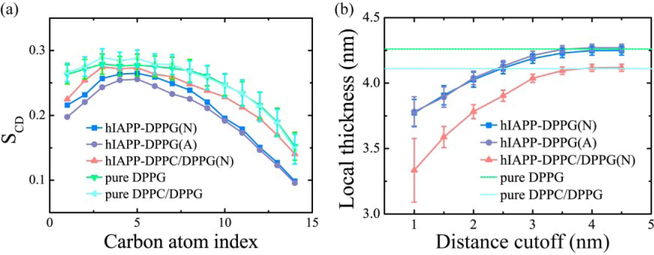
The toxicity of hIAPP oligomers is related to hIAPP-membrane interactions [12,17]. To examine the influence of membrane-bound hIAPP protofibril on the bilayers, we calculated the lipid tail order parameter SCD of acyl chain 1 (sn-1) and the local membrane thickness (Fig. 6). Figure 6(a) shows that in the presence of the DPPG bilayer, the hIAPP protofibril disturbs the membrane integrity and the impact is more obvious at acidic pH than at neutral pH. This difference is attributed to the stronger peptide-lipid electrostatic and H-bonding interactions at acidic pH. Similar perturbation effect of hIAPP on lipid order was revealed by NMR spectra at both low and neutral pH [28]. As for the hIAPP-DPPC/DPPG(N) system, the disturbance of the protofibril on the lipid integrity is much weak. The local membrane thickness within different cutoffs from peptides in Fig. 6(b) shows that hIAPP protofibril thins the neighboring lipid bilayer. Similar change of bilayer thickness was reported previously, in which the DPPG bilayer is thinned by 1–2 nm in the vicinity of hIAPP, and the bilayer farther away maintain a thickness of ~4.2 nm as without hIAPP aggregates [46]. The hIAPP protofibril has a similar impact on DPPG bilayer thickness at neutral and acidic pH, and induces a slightly thinner thickness in the mixed DPPC/DPPG bilayer with respect to the pure mixed membrane. With the increase of membrane-bound peptide concentration, the hIAPP-lipid interaction is expected to cause larger membrane disruption.
Fig. 6.
Influence of membrane-bound hIAPP protofibril on the lipid tail order parameter SCD of acyl chain 1 (sn-1) (a) and the local membrane thickness (b). In the SCD calculation, only the lipids within a minimum distance of 1.0 nm from hIAPP peptides are considered. The SCD of a neat DPPG or a mixed DPPC/DPPG (7:3) lipid bilayer is also given, averaged over the last 20 ns data of a respective 100-ns MD run. The average thicknesses of a neat DPPG lipid bilayer and a mixed DPPC/DPPG (7:3) lipid bilayer are 42.6±0.3 Å (green dashed line) and 41.1±0.4 Å (cyan dashed line) respectively, consistent with previous reports [46,71].
4. Conclusions
In this study, we have investigated the conformational behavior and lipid interaction of hIAPP protofibril at neutral pH in the anionic DPPG lipid membrane by carrying out multiple MD simulations. We have also simulated hIAPP at acidic pH and in the presence of a mixed DPPC/DPPG (7:3) lipid membrane to examine the influence of pH and lipid compositions. We found that the hIAPP protofibril reaches a tilt angle of 52° and an insertion depth of −1.62 nm relative to the bilayer surface. Arg11 anchors hIAPP protofibril to anionic lipid headgroups, and the peptide orientation results from a combination of Lys1-lipid electrostatic attraction and vdW interaction between the turn region and lipid bilayer. In addition to charged residues K1, R11 and H18, aromatic residues Phe15 and Phe23 also display strong interactions with lipid bilayers, indicating the important role of aromatic residues in stabilizing the membrane-bound hIAPP protofibrils. The H-bonds and salt bridges formed between hIAPP protofibril and membrane provide additional forces to stabilize peptide-lipid interaction. The acidic pH induces a smaller tilt angle of 40° and a shallower depth of −1.24 nm, in which the protonated His18 plays a key role in the modulation of hIAPP-membrane interaction. The presence of zwitterionic DPPC lipids induces a larger tilt angle of 77° and a deeper depth of −1.75 nm, due to the reduced electrostatic restriction for hIAPP to the bilayer surface. The hIAPP-membrane interaction leads to the disturbance of lipid integrity and a thinned bilayer thickness near hIAPP. Our results provide atomic-level information of membrane-hIAPP-protofibril interactions, which may help illuminate pH-dependent and membrane lipid composition-modulated hIAPP aggregation.
Acknowledgements
This work has been supported by the National Key R&D Program of China (Grant No. 2016YFA0501702), the NSF of China (Grant No. 11674065) and the China Postdoctoral Science Foundation (grant no.: 2016M601634). This project has also been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Simulations were performed at the National High Performance Computing Center of Fudan University and the High Performance Computing Server of Shanghai University of Sport. We thank Dr. Robert Tycko for kindly providing the coordinates of hIAPP protofibril.
References
- [1].Selkoe DJ, Folding proteins in fatal ways, Nature 426 (2003) 900–904. [DOI] [PubMed] [Google Scholar]
- [2].Chiti F, Dobson CM, Protein misfolding, functional amyloid, and human disease, Annu. Rev. Biochem 75 (2006) 333–366. [DOI] [PubMed] [Google Scholar]
- [3].Khemtémourian L, Killian JA, Höppener JW, Engel MF, Recent insights in islet amyloid polypeptide-induced membrane disruption and its role in beta-cell death in type 2 diabetes mellitus, Exp. Diabetes Res 2008 (2008) 421287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Höppener JWM, Ahrén B, Lips CJM, Islet amyloid and type 2 diabetes mellitus, N. Engl. J. Med 343 (2000) 411–419. [DOI] [PubMed] [Google Scholar]
- [5].Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG, Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis, Science 300 (2003) 486–489. [DOI] [PubMed] [Google Scholar]
- [6].Engel MFM, Membrane permeabilization by islet amyloid polypeptide, Chem. Phys. Lipids 160 (2009) 1–10. [DOI] [PubMed] [Google Scholar]
- [7].Williamson JA, Miranker AD, Direct detection of transient alpha-helical states in islet amyloid polypeptide, Protein Sci 16 (2007) 110–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yonemoto IT, Kroon GJA, Dyson HJ, Balch WE, Kelly JW, Amylin proprotein processing generates progressively more amyloidogenic peptides that initially sample the helical state, Biochemistry 47 (2008) 9900–9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Vaiana SM, Best RB, Yau W-M, Eaton WA, Hofrichter J, Evidence for a partially structured state of the amylin monomer, Biophys. J 97 (2009) 2948–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Butterfield SM, Lashuel HA, Amyloidogenic protein-membrane interactions: mechanistic insight from model systems, Angew. Chem. Int. Ed 49 (2010) 5628–5654. [DOI] [PubMed] [Google Scholar]
- [11].Pannuzzo M, Raudino A, Milardi D, La Rosa C, Karttunen M, α-Helical structures drive early stages of self-assembly of amyloidogenic amyloid polypeptide aggregate formation in membranes, Sci. Rep 3 (2013) 2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Brender JR, Salamekh S, Ramamoorthy A, Membrane disruption and early events in the aggregation of the diabetes related peptide IAPP from a molecular perspective, Acc. Chem. Res 45 (2012) 454–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jayasinghe SA, Langen R, Lipid membranes modulate the structure of islet amyloid polypeptide, Biochemistry 44 (2005) 12113–12119. [DOI] [PubMed] [Google Scholar]
- [14].Sciacca Michele F.M., Lolicato F, Di Mauro G, Milardi D, D’Urso L, Satriano C, Ramamoorthy A, La Rosa C, The role of cholesterol in driving IAPP-membrane interactions, Biophys. J 111 (2016) 140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Apostolidou M, Jayasinghe SA, Langen R, Structure of alpha-helical membrane-bound human islet amyloid polypeptide and its implications for membrane-mediated misfolding, J. Biol. Chem 283 (2008) 17205–17210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nanga RPR, Brender JR, Vivekanandan S, Ramamoorthy A, Structure and membrane orientation of IAPP in its natively amidated form at physiological pH in a membrane environment, Biochim. Biophys. Acta, Biomembr 1808 (2011) 2337–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Knight JD, Hebda JA, Miranker AD, Conserved and cooperative assembly of membrane-bound alpha-helical states of islet amyloid polypeptide, Biochemistry 45 (2006) 9496–9508. [DOI] [PubMed] [Google Scholar]
- [18].Fu L, Ma G, Yan ECY, In situ misfolding of human islet amyloid polypeptide at interfaces probed by vibrational sum frequency generation, J. Am. Chem. Soc 132 (2010) 5405–5412. [DOI] [PubMed] [Google Scholar]
- [19].Fu L, Liu J, Yan ECY, Chiral sum frequency generation spectroscopy for characterizing protein secondary structures at interfaces, J. Am. Chem. Soc 133 (2011) 8094–8097. [DOI] [PubMed] [Google Scholar]
- [20].Cao P, Abedini A, Wang H, Tu L-H, Zhang X, Schmidt AM, Raleigh DP, Islet amyloid polypeptide toxicity and membrane interactions, Proc. Natl. Acad. Sci. U. S. A 110 (2013) 19279–19284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Scalisi S, Sciacca MFM, Zhavnerko G, Grasso DM, Marletta G, La Rosa C, Self-assembling pathway of hIAPP fibrils within lipid bilayers, ChemBioChem 11 (2010) 1856–1859. [DOI] [PubMed] [Google Scholar]
- [22].Zhang M, Ren B, Chen H, Sun Y, Ma J, Jiang B, Zheng J, Molecular simulations of amyloid structures, toxicity, and inhibition, Isr. J. Chem 57 (2017) 586–601. [Google Scholar]
- [23].Brender JR, Lee EL, Cavitt MA, Gafni A, Steel DG, Ramamoorthy A, Amyloid fiber formation and membrane disruption are separate processes localized in two distinct regions of IAPP, the type-2-diabetes-related peptide, J. Am. Chem. Soc 130 (2008) 6424–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Milardi D, Sciacca MFM, Pappalardo M, Grasso DM, La Rosa C, The role of aromatic side-chains in amyloid growth and membrane interaction of the islet amyloid polypeptide fragment LANFLVH, Eur. Biophys. J 40 (2011) 1–12. [DOI] [PubMed] [Google Scholar]
- [25].Mo Y, Lei J, Sun Y, Zhang Q, Wei G, Conformational ensemble of hIAPP dimer: Insight into the molecular mechanism by which a green tea extract inhibits hIAPP aggregation, Sci. Rep 6 (2016) 33076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wiltzius JJW, Sievers SA, Sawaya MR, Eisenberg D, Atomic structures of IAPP (amylin) fusions suggest a mechanism for fibrillation and the role of insulin in the process, Protein Sci. 18 (2009) 1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Brender JR, Hartman K, Reid KR, Kennedy RT, Ramamoorthy A, A single mutation in the nonamyloidogenic region of islet amyloid polypeptide greatly reduces toxicity, Biochemistry 47 (2008) 12680–12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Khemtémourian L, Doménech E, Doux JPF, Koorengevel MC, Killian JA, Low pH acts as inhibitor of membrane damage induced by human islet amyloid polypeptide, J. Am. Chem. Soc 133 (2011) 15598–15604. [DOI] [PubMed] [Google Scholar]
- [29].Nanga RPR, Brender JR, Xu J, Veglia G, Ramamoorthy A, Structures of rat and human islet amyloid polypeptide IAPP1–19 in micelles by NMR spectroscopy, Biochemistry 47 (2008) 12689–12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Caillon L, Lequin O, Khemtémourian L, Evaluation of membrane models and their composition for islet amyloid polypeptide-membrane aggregation, Biochim. Biophys. Acta, Biomembr 1828 (2013) 2091–2098. [DOI] [PubMed] [Google Scholar]
- [31].Jang H, Zheng J, Nussinov R, Models of β-amyloid ion channels in the membrane suggest that channel formation in the bilayer is a dynamic process, Biophys. J 93 (2007) 1938–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Pannuzzo M, Milardi D, Raudino A, Karttunen M, La Rosa C, Analytical model and multiscale simulations of A[small beta] peptide aggregation in lipid membranes: towards a unifying description of conformational transitions, oligomerization and membrane damage, Phys. Chem. Chem. Phys 15 (2013) 8940–8951. [DOI] [PubMed] [Google Scholar]
- [33].Tsigelny IF, Sharikov Y, Wrasidlo W, Gonzalez T, Desplats PA, Crews L, Spencer B, Masliah E, Role of α-synuclein penetration into the membrane in the mechanisms of oligomer pore formation, FEBS J. 279 (2012) 1000–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhao J, Luo Y, Jang H, Yu X, Wei G, Nussinov R, Zheng J, Probing ion channel activity of human islet amyloid polypeptide (amylin), Biochim. Biophys. Acta, Biomembr 1818 (2012) 3121–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhao J, Hu R, Sciacca MFM, Brender JR, Chen H, Ramamoorthy A, Zheng J, Non-selective ion channel activity of polymorphic human islet amyloid polypeptide (amylin) double channels, Phys. Chem. Chem. Phys 16 (2014) 2368–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhao LN, Chiu S-W, Benoit J, Chew LY, Mu Y, Amyloid β peptides aggregation in a mixed membrane bilayer: A molecular dynamics study, J. Phys. Chem. B 115 (2011) 12247–12256. [DOI] [PubMed] [Google Scholar]
- [37].Jang H, Arce FT, Ramachandran S, Capone R, Lal R, Nussinov R, β-Barrel topology of Alzheimer’s β-amyloid ion channels, J. Mol. Biol 404 (2010) 917–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Côté S, Wei G, Mousseau N, Atomistic mechanisms of huntingtin N-terminal fragment insertion on a phospholipid bilayer revealed by molecular dynamics simulations, Proteins 82 (2014) 1409–1427. [DOI] [PubMed] [Google Scholar]
- [39].Chang Z, Luo Y, Zhang Y, Wei G, Interactions of Aβ25–35 β-barrel-like oligomers with anionic lipid bilayer and resulting membrane leakage: An all-atom molecular dynamics study, J. Phys. Chem. B 115 (2011) 1165–1174. [DOI] [PubMed] [Google Scholar]
- [40].Jang H, Connelly L, Teran Arce F, Ramachandran S, Kagan BL, Lal R, Nussinov R, Mechanisms for the insertion of toxic, fibril-like β-amyloid oligomers into the membrane, J. Chem. Theory. Comput 9 (2013) 822–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhang M, Hu R, Ren B, Chen H, Jiang B, Ma J, Zheng J, Molecular understanding of Aβ-hIAPP cross-seeding assemblies on lipid membranes, ACS Chem. Neurosci 8 (2017) 524–537. [DOI] [PubMed] [Google Scholar]
- [42].Christensen M, Skeby KK, Schiøtt B, Identification of key interactions in the initial self-assembly of amylin in a membrane environment, Biochemistry 56 (2017) 4884–4894. [DOI] [PubMed] [Google Scholar]
- [43].Brown Anne M., Bevan David R., Molecular dynamics simulations of amyloid β-peptide (1–42): Tetramer formation and membrane interactions, Biophys. J 111 (2016) 937–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Dong X, Sun Y, Wei G, Nussinov R, Ma B, Binding of protofibrillar Aβ trimers to lipidbilayer surface enhances Aβ structural stability and causes membrane thinning, Phys. Chem. Chem. Phys 19 (2017) 27556–27569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Xiao D, Fu L, Liu J, Batista VS, Yan ECY, Amphiphilic adsorption of human islet amyloid polypeptide aggregates to lipid/aqueous interfaces, J. Mol. Biol 421 (2012) 537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Poojari C, Xiao D, Victor S Batista, B. Strodel, Membrane permeation induced by aggregates of human islet amyloid polypeptides, Biophys. J 105 (2013) 2323–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zhang M, Ren B, Liu Y, Liang G, Sun Y, Xu L, Zheng J, Membrane interactions of hIAPP monomer and oligomer with lipid membranes by molecular dynamics simulations, ACS Chem. Neurosci 8 (2017) 1789–1800. [DOI] [PubMed] [Google Scholar]
- [48].Zhang Y, Luo Y, Deng Y, Mu Y, Wei G, Lipid interaction and membrane perturbation of human islet amyloid polypeptide monomer and dimer by molecular dynamics simulations, PLoS ONE 7 (2012) e38191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Jia Y, Qian Z, Zhang Y, Wei G, Adsorption and orientation of human islet amyloid polypeptide (hIAPP) monomer at anionic lipid bilayers: implications for membrane-mediated aggregation, Int. J. Mol. Sci 14 (2013) 6241–6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Luca S, Yau W-M, Leapman R, Tycko R, Peptide conformation and supramolecular organization in amylin fibrils: constraints from solid-state NMR, Biochemistry 46 (2007) 13505–13522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hallberg A, Effects of starvation and different culture conditions on the phospholipid content of isolated pancreatic islets, Biochim. Biophys. Acta, Lipid. Lipid Met 796 (1984) 328–335. [DOI] [PubMed] [Google Scholar]
- [52].Rustenbeck I, Matthies A, Lenzen S, Lipid composition of glucose-stimulated pancreatic islets and insulin-secreting tumor cells, Lipids 29 (1994) 685–692. [DOI] [PubMed] [Google Scholar]
- [53].Chandrasekhar I, Kastenholz M, Lins RD, Oostenbrink C, Schuler LD, Tieleman DP, van Gunsteren WF, A consistent potential energy parameter set for lipids: dipalmitoylphosphatidylcholine as a benchmark of the GROMOS96 45A3 force field, Eur. Biophys. J 32 (2003) 67–77. [DOI] [PubMed] [Google Scholar]
- [54].Morrow MR, Temple S, Stewart J, Keough KMW, Comparison of DPPC and DPPG environments in pulmonary surfactant models, Biophys. J 93 (2007) 164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lehnert R, Eibl H-J, Müller K, Order and dynamics in lipid bilayers from 1,2-dipalmitoyl-sn-glycero-phospho-diglycerol as studied by NMR spectroscopy, J. Phys. Chem. B 108 (2004) 12141–12150. [Google Scholar]
- [56].Kandt C, Ash WL, Peter Tieleman D, Setting up and running molecular dynamics simulations of membrane proteins, Methods 41 (2007) 475–488. [DOI] [PubMed] [Google Scholar]
- [57].Goldsbury C, Goldie K, Pellaud J, Seelig J, Frey P, Müller SA, Kistler J, Cooper GJS, Aebi U, Amyloid fibril formation from full-length and fragments of amylin, J. Struct. Biol 130 (2000) 352–362. [DOI] [PubMed] [Google Scholar]
- [58].Madine J, Jack E, Stockley PG, Radford SE, Serpell LC, Middleton DA, Structural insights into the polymorphism of amyloid-like fibrils formed by region 20–29 of amylin revealed by solid-state NMR and X-ray fiber diffraction, J. Am. Chem. Soc 130 (2008) 14990–15001. [DOI] [PubMed] [Google Scholar]
- [59].Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC, GROMACS: Fast, flexible, and free, J. Comput. Chem 26 (2005) 1701–1718. [DOI] [PubMed] [Google Scholar]
- [60].Van Gunsteren W, Berendsen H, Gromos-87 Manual, Biomos BV Nijenborgh 4, 9747 AG Groningen, The Netherlands, 1987. [Google Scholar]
- [61].Berger O, Edholm O, Jähnig F, Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature, Biophys. J 72 (1997) 2002–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Parrinello M, Rahman A, Polymorphic transitions in single crystals: A new molecular dynamics method, J. Appl. Phys 52 (1981) 7182–7190. [Google Scholar]
- [63].Nosé S, Klein ML, Constant pressure molecular dynamics for molecular systems, Mol. Phys 50 (1983) 1055–1076. [Google Scholar]
- [64].Hoover WG, Canonical dynamics: Equilibrium phase-space distributions, Phys. Rev. A 31 (1985) 1695–1697. [DOI] [PubMed] [Google Scholar]
- [65].Nosé S, A molecular dynamics method for simulations in the canonical ensemble, Mol. Phys 52 (1984) 255–268. [Google Scholar]
- [66].Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG, A smooth particle mesh Ewald method, J. Chem. Phys 103 (1995) 8577–8593. [Google Scholar]
- [67].Patra M, Karttunen M, Hyvönen MT, Falck E, Lindqvist P, Vattulainen I, Molecular dynamics simulations of lipid bilayers: major artifacts due to truncating electrostatic interactions, Biophys. J 84 (2003) 3636–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Vermeer L, Groot B, Réat V, Milon A, Czaplicki J, Acyl chain order parameter profiles in phospholipid bilayers: computation from molecular dynamics simulations and comparison with 2H NMR experiments, Eur. Biophys. J 36 (2007) 919–931. [DOI] [PubMed] [Google Scholar]
- [69].Elmore DE, Molecular dynamics simulation of a phosphatidylglycerol membrane, FEBS Lett. 580 (2006) 144–148. [DOI] [PubMed] [Google Scholar]
- [70].Lee D-K, Brender JR, Sciacca MFM, Krishnamoorthy J, Yu C, Ramamoorthy A, Lipid composition-dependent membrane fragmentation and pore-forming mechanisms of membrane disruption by pexiganan (MSI-78), Biochemistry 52 (2013) 3254–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Muddana HS, Gullapalli RR, Manias E, Butler PJ, Atomistic simulation of lipid and DiI dynamics in membrane bilayers under tension, Phys. Chem. Chem. Phys 13 (2011) 1368–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]