Abstract
The basolateral amygdala (BLA) is a critical nucleus in mediating behavioral responses after exposure to acute social conflict. Male and female Syrian hamsters both readily establish a stable dominant-subordinate relationship among same-sex conspecifics, and the goal of the current study was to determine potential underlying genetic mechanisms in the BLA facilitating the establishment of social hierarchy. We sequenced the BLAtranscriptomes of dominant, subordinate, and socially neutral males and females, and using de novo assembly techniques and gene network analyses we compared these transcriptomes across social status within each sex. Our results revealed 499 transcripts that were differentially expressed in the BLA across both males and females and 138 distinct gene networks. Surprisingly, we found that there was virtually no overlap in the transcript changes or in gene network patterns in males and females of the same social status. These results suggest that, although males and females reliably engage in similar social behaviors to establish social dominance, the molecular mechanisms in the BLA by which these statuses are obtained and maintained are distinct.
Keywords: RNA-Seq, sex differences, social defeat, social stress, agonistic behavior, transcriptional signature
Introduction
Social stress is argued to be the most prevalent stressor experienced by humans [1] and is thought to increase the risk or to stimulate the onset of many neuropsychiatric disorders including anxiety disorders, posttraumatic stress disorder, and depression [2–6], The most commonly studied form of social stress in pre-clinical studies is social defeat. Acute and repeated exposure to social defeat stress causes many of the same outcomes in rodents (e.g., increased social avoidance, anxiety, and anhedonia) as are observed in humans after stressful experiences [7–10,5,11,12], Sex differences in response to social stress are also prevalent across taxa, including in humans wherein many stress-related neuropsychiatric disorders present with sexually dimorphic prevalence and symptoms [13–16], It can be challenging to study female responses to social stress in rodent species, however, because female rats and mice do not display spontaneous aggressive behavior, and males do not normally attack conspecific females [17]. Therefore, it is difficult to study sex differences in mechanisms and responses to social stress in ethologically meaningful ways using these animal models. In Syrian hamsters (Mesocricetus auratus), however, both sexes readily engage in agonistic behavior with a same-sex conspecific in the wild and in the laboratory [18–20], Both males and females use the same agonistic behaviors to rapidly establish stable dominant-subordinate relationships allowing for the study of “winners” (dominants) and “losers” (subordinates) in both sexes. Fortuitously, agonistic encounters in hamsters rarely result in physical injury, as contrasted with commonly used mouse models of defeat, permitting the direct study of the effects of social stress without the confounding trauma of physical injury.
Our laboratory has extensively studied the behavioral and neurochemical correlates of responses to social conflict in hamsters and have found sex differences in some of the behavioral responses to social stress. While both sexes display behavioral responses to social stress, defeated males paired with a new opponent often exhibit more overt submissive behaviors after defeat (e.g., flees and risk assessments), while defeated females exhibit less submission and more social and communicative behaviors (e.g., sniffing and flank marking)[21–24]. In both males and females, social defeat stress elicits similar levels of social avoidance of a confined opponent, however, suggesting that both sexes exhibit at least some of the same behavioral responses to defeat [21,25], We have defined a neural circuit that facilitates the behavioral responses to acute social conflict in hamsters and have identified the basolateral amygdala (BLA) as a critical nucleus mediating these changes. The BLA is necessary for the acquisition and expression of conditioned defeat, a striking change in behavior in which a defeated, subordinate animal abandons all territorial aggression and produces only submissive and defensive behavior [26,27], We have also demonstrated that de novo protein synthesis in this nucleus is required [28] and that overexpression of cyclic AMP binding protein in the BLA during social defeat enhances subsequent conditioned defeat [29]; thus, it appears clear that gene regulation in the BLA mediates, at least in part, behavioral responses to defeat. Less is known, however, about the specific genes or gene networks involved in mediating behavioral responses to acute social conflict and the subsequent formation of a dominant-subordinate relationship in males and females.
While hamsters are an ideal model for the study of acute social conflict, as outlined above, there is a dearth of genetic tools available for the investigation into genetic mechanisms underlying stress-induced behavioral changes in this species. Our lab recently published the transcriptome of male and female Syrian hamster whole brain [30], providing a starting point with which to begin genetic exploration of behavior using Syrian hamsters as a model. To further investigate the genetic mechanisms underlying the establishment of social status, we sequenced the transcripts in the BLA of dominant and subordinate males and females and compared transcript expression and gene network patterns to that of same-sex socially-neutral controls. The primary purpose of this study was to identify genes that may mediate the formation of social status and to determine whether these patterns of gene regulation are similar in males and females.
Results
Females and males exhibit similar behavioral patterns during acute social conflict
As is often true with weight-matched pairs, residence did not confer dominance. In fact, in this study intruders were dominant 65% of the time. There was no difference in the latency to first attack between males and females (Student’s t-test with Welch’s correction: t(5.942)=1.915, p=0.1045), nor was there a significant difference in the duration of agonistic behavior in males and females before a hierarchy was established (Student’s t-test: t(10)=0.8742, p=0.4025) in Trial 1 (Figure 1a). Overall, across all 3 trials, there was no main effect of sex in dominants (two-way ANOVA: F(l,40)=9.18e-013, p>0.9999; Figure 1b) or subordinates (two-way ANOVA: F(l,40)=1.652e-011, p>0.9999; Figure 1c), suggesting no significant sex differences in overall duration of each behavior within social status. There was a main effect of behavior in dominants (F(3,40)=34.91, p<0.0001) and subordinates (F(3,40)=454.1, p<0.0001), with dominant animals primarily displaying aggressive behavior and subordinate animals displaying almost exclusively submission (Figure 1b, 1c). There were subtle behavioral differences observed during Trial 1, but overall there were no substantial sex differences observed within each of the 3 behavioral trials (Supplemental Figure 1).
Fig. 1.
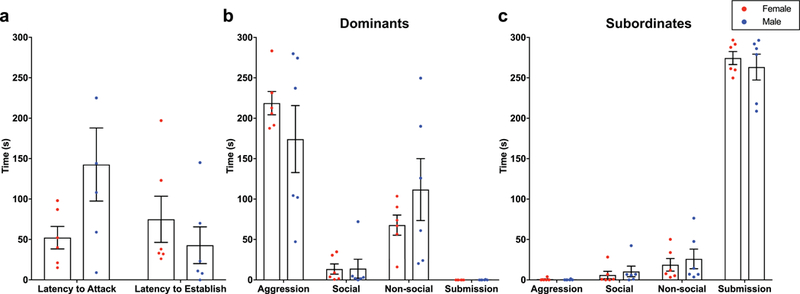
Females (red) and males (blue) show similar behavior during acute social conflict. (a) There were no differences in latency to attack in the first trial or in latency to establish a hierarchy after attack between males and females. (b-c) There were no sex differences in behavior in dominant animals (b) or subordinate animals (c).
De novo transcriptome assembly and optimization
The initial de novo assembly was created using high quality samples (Supplemental Table 1, Supplemental Figure 2) and generated 1.2M possible genes. The assembly was then optimized, as described in the Supplemental Materials and Supplemental Figure 3, resulting in 120,003 transcripts mapping to 14,493 unique BLAST identifiers.
Differential expression, gene ontology, and network analyses reveal little transcriptional overlap between males and females
Expected read counts from each sample were calculated to determine which transcripts were most highly expressed. The most highly expressed transcripts in hamster amygdala (both male and female) are listed in Supplemental Table 2 and largely overlapped with those found in the whole brain of males and females [30], Differential expression analyses were then completed. Samples from dominant and subordinate hamsters were compared to samples from same-sex controls, and Supplemental Table 3 lists all the differentially expressed transcripts (DETs). There were similar numbers of DETs in males and females of different social status (Figure 2a). In total, 258 unique transcripts were differentially expressed in males (with 48 transcripts present in more than one comparison), and 263 unique DETs in females (with 47 transcripts present in more than one comparison). The majority of DETs present in more than one comparison were in both dominants and subordinates compared with controls, suggesting that these transcripts are altered by social interaction and not necessarily by social status (see Supplemental Table 3). Figure 2 shows the distribution of all DETs in males (Figure 2b,d) and females (Figure 2c,e).
Fig. 2.

The distribution of differentially expressed transcripts (DETs) in males and females follow similar patterns despite little overlap in the identity of the actual transcripts that were changed. (a) Table depicting the number of transcripts that were differentially expressed in males (left, blue) and females (right, red). Arrows indicate the number of transcripts that were higher or lower in dominants and subordinates compared with controls. (b-e) The distribution of transcripts in each comparison in males (b, d) and females (c, e). Red dots in the volcano plots (b, c) indicate the transcripts that were significantly different (FDR < 0.05). The heatmaps (d, e) show the transcript clustering dendrograms to the left. Purple indicates transcripts that had a negative log2 fold change and yellow indicates transcripts that had a positive log2 fold change. Note the difference in scale in males (−10 to 10) and females (−4 to 4).
After analyzing the DETs in males and females, we determined that very few overlapped between the sexes. Table 1 lists the transcripts that were differentially expressed in both males and females; however, it is the case that some of these changed in opposite directions. In fact, of the 521 differentially expressed transcripts, only 22 overlapped between the sexes, leaving 499 unique DETs across sex and social status (Supplemental Table 3). To determine if this lack of overlap was due to divergent expression patterns in males and females, or, alternatively, if these transcripts were regulated similarly but just failed to reach significance in both sexes, we determined the degree of overlap between expression profiles of males and females of different social status using rank rank hypergeometric overlap (RRHO). This analysis revealed no correlation between the DETs in male and female dominants (Spearman’s rho = −0.0576; Figure 3a,b) or subordinates (Spearman’s rho = 0.0256; Figure 3c,d), suggesting that there was no significant overlap in the expression patterns of transcripts after the establishment of social status. The RRHO heatmaps indicate that any overlap that exists in the DETs at all is in a set of transcripts that are regulated in opposite directions in males and females (reds in Figure 3b,d). Supplemental Figure 4 shows the degree of overlap in the expression of all transcripts in the BLA between male and female dominants and subordinates.
Table 1:
Differentially expressed transcripts that reached significance in both males and females + 2 isoforms were differentially expressed in both males and females
| Gene (Uniprot ID) | Expression Change in Females | Expression Change in Males | ||||
|---|---|---|---|---|---|---|
| Dom v. Sub | Dom v. Con | Sub v. Con | Dom v. Sub | Dom v. Con | Sub v. Con | |
| AT-hook-containing transcription factor (AKNA MOUSE) | ⬆Dom | - | - | ⬆Sub | - | - |
| ATP-dependent 6-phosphofructokinase, muscle type (PFKAM MOUSE) | - | - | ⬇ | - | - | ⬆ |
| Cullin-3 (CUL3_RAT) | - | ⬇ | - | ⬇Dom | ⬇ | - |
| DNA repair protein RAD51 homolog 4 (RA51D MOUSE) | ⬆Sub | - | - | - | ⬆ | - |
| Exportin-4 (XPO4_MOUSE) | ⬆Dom | - | - | ⬆Dom | ⬆ | - |
| Glycylpeptide N-tetradecanoyltransferase 2 (NMT2 MOUSE) | ⬆Sub | - | - | - | - | ⬇ |
| Histidine triad nucleotide-binding protein 3 (HINT3) | ⬆Sub | - | - | ⬆Sub | - | - |
| Inactive ubiquitin carboxyl-terminal hydrolase 53 (UBP53 MOUSE) | - | ⬇ | - | - | - | ⬆ |
| Integral membrane protein 2C (ITM2C_RAT)+ | - | ⬆ | ⬇ | - | ⬆ | ⬆ |
| Junctophilin-1 (JPH1_MOUSE) | - | - | ⬇ | ⬆Sub | - | - |
| Kelch repeat and BTB domain-containing protein 4 (KBTB4 MOUSE) | - | ⬆ | ⬆ | - | ⬇ | - |
| Magnesium transporter NIPA1 (NIPA1_MOUSE) | - | - | ⬇ | ⬆Dom | - | - |
| MAM domain-containing glycosylphosphatidylinositol anchor protein 2 (MDGA2 RAT) | ⬆Dom | ⬆ | - | ⬇Sub | - | ⬇ |
| Meprin A subunit alpha (MEP1A_RAT) | - | ⬇ | - | - | - | ⬆ |
| NEDD4-binding protein 2-like 1 (N42L1_MOUSE) | ⬆Dom | - | - | - | ⬆ | - |
| Protein transport protein Sec24A (SC24A_MOUSE) | - | - | ⬇ | ⬆Dom | - | - |
| Serine/threonine-protein kinase PAK 2 (PAK2_RAT) | - | - | ⬆ | ⬆Dom | - | - |
| TM2 domain-containing protein 1 (TM2D1_MOUSE) | ⬆Sub | - | ⬆ | - | ⬇ | - |
| Tubby-related protein 4 (TULP4_MOUSE) | ⬆Sub | - | - | - | - | ⬇ |
| Voltage-dependent R-type calcium channel subunit alpha-1E (CAC1E RAT) | - | ⬇ | - | - | ⬇ | ⬇ |
| Voltage-dependent T-type calcium channel subunit alpha-1H (CAC1H RAT) | ⬆Sub | - | - | - | - | ⬆ |
Fig. 3.
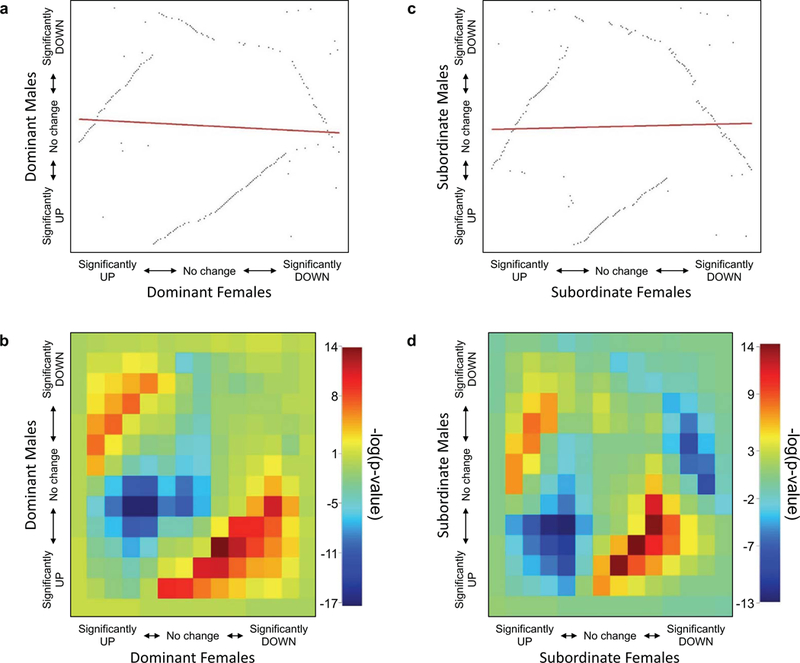
There was no correlation in dominant males and females (a, b) or in subordinate males and females (c, d) when the differentially expressed transcripts (DETs) were compared using rank rank hypergeometric overlap (RRHO). RRHO ranks the DETs with the significantly increased transcripts first (bottom left) and the significantly decreased transcripts last (top right) on each panel (a and c). The heatmaps (b, d) show the log10 transformed p-values of the overlapping transcripts in males and females according to the hypergeometric distribution. Higher (reds) values indicate transcripts that overlap more than expected by chance while lower (blues) values indicate lower than expected overlap. The reds in the lower right and upper leftcorners (b,d) indicate that the overlapping transcripts are, in fact, regulated in opposite directions in males and females.
The optimized assembly and the subsets of DETs were then analyzed using PANTHER gene ontology (GO) analysis to determine which molecular functions, biological processes, protein classes, and pathways were most represented. Catalytic activity and binding were the highest represented molecular functions, whereas metabolic and cellular processes ranked highest in biological processes (Supplemental Figure 5). The top protein classes (Supplemental Figure 5) and pathways (Supplemental Figure 6) were more evenly distributed. Analysis of the DETs within each sex revealed these GO terms were also present in the highest number of transcripts, with some sex-specific differences (Figures 4-5). GOSeq enrichment analysis further revealed that the regulation of chromatin disassembly (under Biological Processes) was significantly enriched in transcripts that were lower in subordinate males (FDR = 0.02). Furthermore, 4 GO terms, all under Molecular Function, were enriched in transcripts that were significantly higher in dominant and subordinate males (cystine: glutamate antiporter activity (FDR = 0.02), cystine secondary active transmembrane transporter activity (FDR = 0.02), sulfur-containing amino acid secondary active transmembrane transporter activity (FDR = 0.02), and neutral L-amino acid secondary active transmembrane transporter activity (FDR = 0.04)). In addition, 33 terms (24 Biological Processes, 8 Molecular Functions, and 1 Cellular Component) were enriched in transcripts that were higher in dominant and subordinate females, including dopaminergic synaptic transmission (FDR = 0.01), negative regulation of synaptic plasticity (FDR = 0.007), and receptor activator activity (FDR = 0.04) (Supplemental Table 4).
Fig. 4.
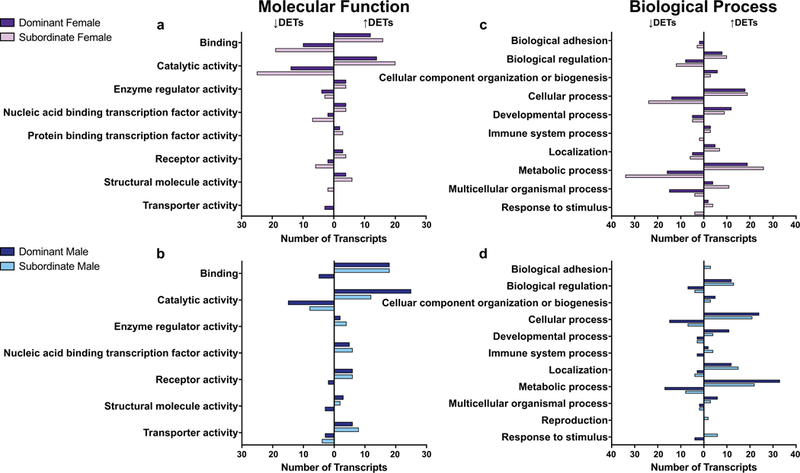
GO analysis revealed molecular functions (a, b) and biological processes (c, d) that were represented by the differentially expressed transcripts (DETs) that were higher (bars to the right) or lower (bars to the left) in dominant (dark bars) and subordinate (light bars) animals.
Fig. 5.
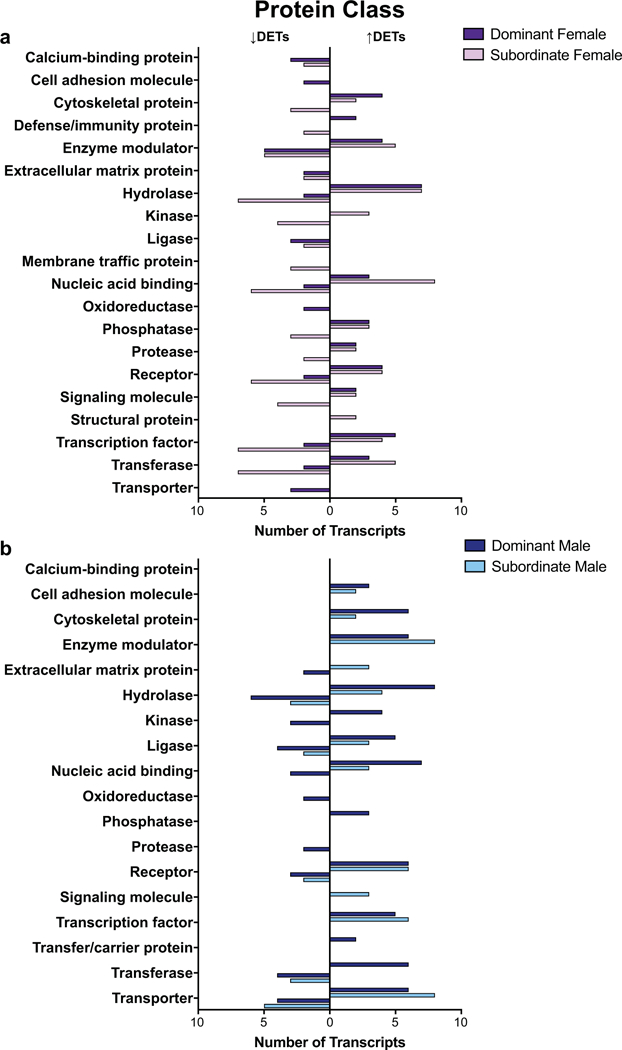
GO analysis revealed the protein classes represented by the differentially expressed transcripts (DETs) in females (a) and males (b) that were higher (bars to the right) or lower (bars to the left) in dominants (dark bars) or subordinates (light bars).
Finally, we used a weighted gene correlation network analysis (WGCNA) to determine the similarity in gene network expression patterns of the dominant, subordinate, and control samples in males and females. Analyzing transcript expression in the optimized assembly allowed us to graph the connectivity of our samples based on overall expression patterns. WGCNA grouped the 120,003 transcripts into 138 distinct modules (Supplemental Figure 7). Of these, 10 significantly correlated with subordinate status (Figure 6a), while 9 correlated with dominant status (Figure 6b). These correlations, however, were largely driven by only one sex, as can be seen in Figure 6, further suggesting minimal overlap in transcript or gene network expression patterns in females and males after social conflict and the establishment of social status.
Fig. 6.
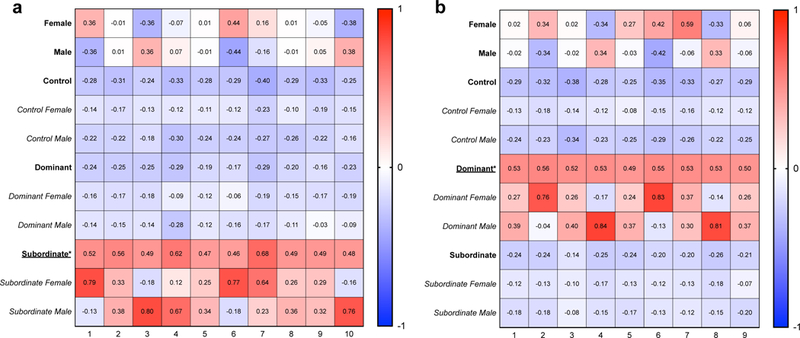
WGCNA analysis revealed several gene networks that significantly correlated with social status. 10 modules correlated with subordinate status (a) and 9 modules correlated with dominant status (b). Correlations are reported within each cell, with positive correlations shown in red and negative correlations shown in blue. All 138 modules from WGCNA, along with p-values for each module, are shown in Supplemental Figure 7. Each of the modules shown here had a p-value <0.05 for either subordinate (a) or dominant (b) status (indicated by *). These correlations were primarily driven by one sex, as can be seen in the subcategories below when separated by sex within status.
Discussion
Transcriptomic analysis of hamster amygdalae revealed striking sex differences in the transcripts and pathways that change in response to the establishment of social status in males and females. In fact, there was virtually no overlap in overall expression patterns between the sexes. In this study, we investigated the transcriptomic profiles of dominant and subordinate males and females 24hr after acute social conflict because this is the time frame when we observe marked behavioral changes in defeated hamsters. Both males and females reliably and rapidly engaged in similar agonistic behaviors upon encountering a conspecific; however, the transcripts that were regulated between the sexes after this establishment were markedly different, suggesting that while the establishment of a social hierarchy is readily observed in both sexes, the underlying genetic mechanisms are distinct. In the rare instances when the same transcripts were differentially regulated by status in males and females, it was often in opposite directions. The transcripts, pathways, and gene networks highlighted here provide a more concrete understanding of how these behaviors might be mediated in male and female hamsters. Future experiments will further test these mechanisms, focusing on the specific genes and networks that are differentially expressed between males and females of different social status.
The sex-specificity of our data supports the hypothesis that male and female brains may act via different mechanisms to produce similar behavior. These results are consistent with previous data and theories that similar behavioral outcomes may be attained via activation of very different, sex-specific pathways, or by the same pathways but in opposite directions, to reach the same ultimate goal [31,32], which in the present instance is the establishment of a stable dominance relationship within pairs of hamsters. Consistent with the data presented here, it was recently found that male and female mice have distinct gene expression patterns in hippocampal area CA3 in response to an acute stressor [33], suggesting that distinct brain regions have unique patterns of gene expression that are sexually dimorphic in response to stress. Furthermore, it was recently reported that males and females suffering from major depressive disorder have markedly different transcriptional profiles in several brain regions, including the prefrontal cortex and nucleus accumbens [34]. This study did not analyze the amygdala, but the results, in conjunction with the data presented here, further reinforce the need to include both males and females and underscore the importance of using animal models that allow the direct study of sex differences in the regulation of behavior in species wherein both sexes produce similar, ethologically-relevant behaviors.
Of note, several transcripts that were differentially expressed between animals of different social status were GABA-or glutamate-related genes that have been reported to have differential expression patterns in males and females in various animal models of neuropsychiatric disorders as well as in clinical populations. These differences are often confined to specific brain regions, and our data further support sexually dimorphic patterns of expression in the amygdala following acute social conflict. For example, Gria2, which codes for an ionotropic glutamate receptor, was significantly higher in dominant and subordinate males compared with controls, suggesting that this gene may play a role in the processing of cues during social interactions in males. Gria2 is associated with stimulus-reward learning [35] and is linked to sex differences in major depressive disorder (MDD), in that Gria2 expression is higher in the prefrontal cortex of female patients with MDD [36]. Our data suggest that Gria2 in the amygdala is not significantly altered in females and may in fact play a more significant role in males during social interactions. We also found that the alpha-4 subunit of the GABA-A receptor was more highly expressed in dominant males when compared with subordinate males. Expression of this subunit increases in the amygdala of females but also in the hippocampus of both males and females after progesterone withdrawal and is linked to increases in anxiety-like behavior [37,38]. Our data, in conjunction with these previous findings, reinforce the importance of sex-and site-specificity when interpreting differences in gene expression patterns after stress exposure.
In addition, Gad2, the gene that encodes the protein GAD65, was higher in dominant males after an acute agonistic interaction. This gene is reduced in patients with MDD [39], and low GAD65 expression is correlated with spatial learning deficits after stress in the medial amygdala of males. Conversely, high GAD65 expression in the CA1 region of the hippocampus is correlated with spatial learning deficits in females [40]. Glutamic acid decarboxylase (GAD) catalyzes the formation of GABA from glutamate, and GAD65, in particular, is involved in GABA synthesis specifically for phasic inhibition of neurotransmission [41]. GAD65 increases in several nuclei after acute and chronic stressors, including specific nuclei within the bed nucleus of the stria terminalis and hypothalamus [42], respectively, and we demonstrated here that Gad2 was higher in the amygdala of dominant males. This suggests a potential increase in GABA stores available for neurotransmission in the abundant GABAergic neurons in the amygdala. An increase in GABA neurotransmission in the amygdala during social conflict would suppress the social stress-induced behavioral phenotype, thus potentially providing a protective effect in dominant animals against the stress of the encounter. This finding is also consistent with data showing that previous dominance confers resilience to subsequent stress [43] and suggests that increased GABAergic tone in the amygdala may be one mechanism by which this resilience is obtained and maintained.
We also examined genes associated with learning and memory, mood and anxiety disorders, and social behavior. Several transcripts that had lower expression in animals that experienced an agonistic encounter have been linked to bipolar disorder (Akap5) [44], general mood disorders (Aldh1a1) [45], anxiety (Kif13a) [46], and depression (Mgat5) [47]. Other genes linked to major depressive disorder (Gad2, Gria2) [36,39], PTSD (Dicer1) [48], and anxiety (Spock3) [49] had higher expression in dominant and/or subordinate animals when compared with controls. Specifically, Dicer1, a gene directly involved in the expression of other genes by regulating the production of microRNAs, was higher in dominant males, and, consistent with this effect, increases in this gene have been linked to stress resilience [50]. Relatedly, decreases in Dicer1 are observed in patients suffering from PTSD and depression compared with healthy controls [48]. These results suggest that Dicer1 could be another gene of interest, along with Gad2, regulating the resilient behavioral phenotype obtained from achieving dominance. On the other hand, in regulating subordinate status, we found that Uba6 was lower in subordinate males. This is consistent with previously observed increases in anxiety-like behavior and social avoidance in animals with a depletion of this gene [51] and is worth future investigation into how this gene, and others up-and down-stream in its network, may correlate with social avoidance after acute social stress.
We have previously shown that dopamine in the nucleus accumbens modulates the acquisition and expression of conditioned defeat [52]. Three transcripts representing dopamine-mediated signaling pathways (E4111, E4112, Cdk5) emerged from the gene ontology analysis as being higher in subordinate females, suggesting that these genes may be directly involved in mediating defeat-induced behavioral responses in females. In addition, the gonadotropin releasing hormone pathway was represented by four transcripts that were lower in subordinate females (Nab1, Nfyb, Bmr1a, Plcb1) and three transcripts that were higher in dominant males (Bmr1b, Pp2ba, Tba1b). It is not surprising that social stress might change these types of genes in a sexually dimorphic manner; indeed, we have previously reported that gonadal hormones modulate social stress-induced behavioral responses in both males and females [22,24]. Several additional pathways were represented in the DETs, including multiple glutamate receptor pathways, beta 1 and 2 adrenergic receptor signaling pathways, 5HT2-type receptor mediated signaling pathway, oxytocin receptor mediated signaling pathway, and GABA synthesis. Further defining and investigating these gene pathways will provide a solid framework for determining the precise molecular mechanisms underlying dominant and subordinate status in males and females and will help us to understand the sex-specific mechanisms that ultimately lead to similar behavioral outcomes.
Overall, the data presented here are the first to show that there are sex-dependent mechanisms in the amygdala underlying dominant and subordinate status after acute social conflict in hamsters. Both sexes readily establish a social dominance hierarchy, but these data indicate that the genes and gene networks by which social statuses are established and maintained are genetically distinct in the two sexes.
Methods
Animals and behavior
Adult male and female Syrian hamsters, approximately 10 weeks old and weighing between 120–140g, were obtained from Charles River Laboratories (Danvers, MA). Animals were singly housed upon arrival under a 14:10 hr light:dark cycle, as is standard in this species to maintain gonadal patency. Single housing in hamsters is standard and is not stressful for this species [53]. All animals were handled daily for at least eight days before the beginning of the study. During handling, estrous cycles of females were monitored for at least two cycles (8d) via vaginal swab to confirm estrous cycle stage and stability.
Before social defeat training, animals were weight-matched and randomly assigned as a resident, intruder, or home cage control. All females were paired on Diestrus 1, and brains were collected on Diestrus 2 because females on Diestrus 2 show the most pronounced social avoidance after defeat [21]. An equal number of males were paired each day that the females were run. Intruders were placed in the resident’s home cage three times for 5min to ensure a stable hierarchy; each pairing was separated by an inter-trial interval of 3min during which the intruder was returned to its home cage (Supplemental Figure 1). The timing for the 5min trial during the first pairing began immediately after the first agonistic behavior was displayed, as described previously [54], All behavior emitted during each trial was scored separately for each animal. Behavior was separated into the following categories: aggression (side or upright attack, chase, bite), submission (flee, side or upright submissive posture, tail lift), social (greet, sniff, allogroom), and non-social behavior (self-groom, cage exploration, general locomotion), as described in detail in Albers, Huhman, and Meisel [55], Latency to the initial agonistic interaction and time to establish a stable dominance relationship were also recorded. Home cage controls were left alone in their home cage during training and were not exposed to the behavioral suite during defeat training. All procedures and protocols were approved by the Georgia State University Institutional Animal Care and Use Committee and are in accordance with the standards outlined in the National Institutes of Health Guide for Care and Use of Laboratory Animals. Latencies to attack and to establish a hierarchy were analyzed using non-paired t-tests, with Welch’s correction for unequal variances. Behaviors scored during the agonistic encounters were analyzed using two-way ANOVAs with Sidak’s post-hoc analysis. All behavioral statistics were completed using Prism 7 for Mac OS X, version 7.0c.
Tissue collection, RNA Isolation, and sequencing
Paired animals, as well as home cage controls, were anesthetized via isoflurane exposure and decapitated 24hr after the final agonistic encounter. This is the time when we would normally test for the presence of defeat-induced social avoidance and for conditioned defeat [19,56,57], Brains were quickly extracted, frozen immediately in isopentane on dry ice, and stored at −80°C until processing. Bilateral tissue punches (1mm) aimed at the basolateral amygdala (BLA) were extracted from frozen brains and pooled for RNA isolation processing. RNA extractions followed a modified protocol using Trizol (Life Technologies, Grand Island, NY). Bilateral amygdalae from 2 animals of the same sex and social status (4 total amygdala punches per sample) were pooled together for each RNA extraction in order to minimize the effect of individual variability, as described previously [30,58–61], This resulted in 3 biological replicates for each of 6 groups (dominant males, dominant females, subordinate males, subordinate females, control males, control females). Tissue was homogenized on ice with lmL Trizol. After full homogenization, homogenate was allowed to settle at room temperature for 5min. Homogenate was then mixed with 200μΙ of chloroform, allowed to stand at room temperature for 2– 3min and then centrifuged at 12,000xg for 15min at 4°C to separate the phases. The aqueous RNA phase was removed and dispensed into a new 2mL microcentrifuge tube. The aqueous phase was washed with 200μL of chloroform, mixed well, allowed to stand 2–3min and then centrifuged at 12,000xg for lOmin at 4°C. For enhanced visualization of the pellet, 3μL of GlycoBlue (Life Technologies, Grand Island, NY) was added and mixed gently. For RNA precipitation, 500μL of 100% isopropanol was added, mixed gently and allowed to stand at room temperature for lOmin. To obtain an RNA pellet, the solution was centrifuged at 12,000xg for 20min at 4°C. The remaining liquid was carefully removed and the pellet was washed twice in lmL 75% ethanol in RNase-free water and centrifuged at 7,500xg for 5min at 4°C. The pellet was allowed to air dry for approximately 5min and was then re-suspended in 20μΙ of ultrapure water. Samples were stored at −80°C until downstream processing. RNA quality and concentration was determined using the Agilent Bioanalyzer, and samples were then sent for sequencing to Beckman Coulter Genomics (Danvers, MA). Amygdala sequencing was completed in paired-end lOObp reads, averaging 37M reads per sample.
Transcriptome assembly and optimization
The amygdala de novo transcriptome was assembled as described previously [30], All data was acquired using the computing resources at Georgia State University [62], Briefly, we used Trinity [63,64] (https://github.com/trinityrnaseq/trinityrnaseq/wiki) for the initial de novo assembly using all 18 samples from both males and females. The assembly was then optimized using TransDecoder [64] (https://transdecoder. github.io) with a minimum cut-off of 50 amino acids [65] and BLASTx [66] (http://blast.ncbi.nlm.nih.gov/Blast.cgi), using the Uniprot-rodent database from January 21, 2016 [67] (http://uniprot.org). The optimized assembly was annotated using the Trinity-recommended platform, Trinotate (https://trinotate.github.io). PANTHER (Protein Analysis Through Evolutionary Relationships, http://www.pantherdb.org) was used for functional annotation of the optimized assembly, using Mus musculus as the reference organism. All sequences are available under NCBI SRA Accession Number SRP074844.
Differential expression analysis and statistics
Differential expression analysis was completed using expected read counts from RNA-Seq by Expectation-Maximization (RSEM) [68] (http://deweylab.github.io/RSEM ) in an exact test using the Bioconductor package edgeR [69] (https://bioconductor.org/packages/release/bioc/html/edgeR.html), as described previously [30], Transcripts were considered to significantly differ if the false discovery rate (FDR) was < 0.05. FDR is adjusted for multiple comparisons in RNA-Seq data, and is thus a more stringent threshold to use when determining significance when compared with unadjusted p-values.
For further analysis of our differential expression data, we entered the data into RRHO (Rank Rank Hypergeometric Overlap; https://bioconductor.org/packages/release/bioc/html/RRHO.html) [70], using the recommended settings, to determine the degree of overlap in the expression patterns between males and females. We also used PANTHER (pantherdb.org) [71–73] and GOSeq (https://bioconductor.org/packages/release/bioc/html/goseq.html) [74] for gene ontology analyses. Lastly, we used a signed, weighted gene co-expression network analysis (WGCNA, https://labs.genetics.ucla.edu/horvath/CoexpressionNetwork/Rpackages/WGCNA/) to cluster our individual samples by gene expression patterns in the amygdala following the recommended settings [75,76],
Supplementary Material
Acknowledgements:
The authors gratefully acknowledge Dr. James C. Walton, Alisa Norvelle, M.S., and Georgia State University’s Research Solutions and High Performance Computing Group for their expert assistance. The authors would also like to thank Reed A. Gilbert, Atit A. Patel, and Dr. Brittany M. Thompson for their technical assistance. This project was supported by NIH R01MH062044 to KLH, Georgia State University Brains and Behavior Seed Grant to KLH, Georgia State University Dissertation Grant to KEM, Brains and Behavior Fellowships to KEM, DMS, and AMR, Honeycutt Fellowships to KEM and AMR, and a Next Generation Postdoctoral Fellowship from the Center for Behavioral Neuroscience to LQB. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or Georgia State University.
Footnotes
Conflict of Interest: The authors declare they have no conflict of interest.
References
- 1.Bjorkqvist K (2001) Social defeat as a stressor in humans. Physiology & behavior 73 (3):435–442 [DOI] [PubMed] [Google Scholar]
- 2.Agid O, Kohn Y, Lerer B (2000) Environmental stress and psychiatric illness. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie 54 (3):135–141. doi: 10.1016/s0753-3322(00)89046-0 [DOI] [PubMed] [Google Scholar]
- 3.Ehlers A, Maercker A, Boos A (2000) Posttraumatic stress disorder following political imprisonment: the role of mental defeat, alienation, and perceived permanent change. Journal of abnormal psychology 109 (1):45–55 [PubMed] [Google Scholar]
- 4.Kelleher I, Harley M, Lynch F, Arseneault L, Fitzpatrick C, Cannon M (2008) Associations between childhood trauma, bullying and psychotic symptoms among a school-based adolescent sample. The British journal of psychiatry : the journal of mental science 193 (5):378–382. doi: 10.1192/bjp.bp.108.049536 [DOI] [PubMed] [Google Scholar]
- 5.Borghans B, Homberg JR (2015) Animal models for posttraumatic stress disorder: An overview of what is used in research. World journal of psychiatry 5 (4):387–396. doi: 10.5498/wjp.v5.i4.387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamashiro KL, Nguyen MM, Sakai RR (2005) Social stress: from rodents to primates. Frontiers in neuroendocrinology 26 (1):27–40. doi: 10.1016/j.yfrne.2005.03.001 [DOI] [PubMed] [Google Scholar]
- 7.Huhman KL (2006) Social conflict models: can they inform us about human psychopathology? Hormones and behavior 50 (4):640–646. doi: 10.1016/j.yhbeh.2006.06.022 [DOI] [PubMed] [Google Scholar]
- 8.Krishnan V (2014) Defeating the fear: new insights into the neurobiology of stress susceptibility. Experimental neurology 261:412–416. doi: 10.1016/j.expneurol.2014.05.012 [DOI] [PubMed] [Google Scholar]
- 9.Kudryavtseva NN, Bakshtanovskaya IV, Koryakina LA (1991) Social model of depression in mice of C57BL/6J strain. Pharmacology, biochemistry, and behavior 38 (2):315–320 [DOI] [PubMed] [Google Scholar]
- 10.Toth I, Neumann ID (2013) Animal models of social avoidance and social fear. Cell and tissue research 354 (1):107–118. doi: 10.1007/s00441-013-1636-4 [DOI] [PubMed] [Google Scholar]
- 11.Chaouloff F (2013) Social stress models in depression research: what do they tell us? Cell and tissue research 354 (1):179–190. doi: 10.1007/s00441-013-1606-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hollis F, Kabbaj M (2014) Social defeat as an animal model for depression. ILAR journal 55 (2):221–232. doi: 10.1093/ilar/ilu002 [DOI] [PubMed] [Google Scholar]
- 13.Altemus M (2006) Sex differences in depression and anxiety disorders: potential biological determinants. Hormones and behavior 50 (4):534–538 [DOI] [PubMed] [Google Scholar]
- 14.Breslau N, Davis GC, Andreski P, Peterson EL, Schultz LR (1997) Sex differences in posttraumatic stress disorder. Archives of general psychiatry 54 (11):1044–1048 [DOI] [PubMed] [Google Scholar]
- 15.Nolen-Hoeksema S (1987) Sex differences in unipolar depression: evidence and theory. Psychological bulletin 101 (2):259. [PubMed] [Google Scholar]
- 16.Weissman MM, Klerman GL (1977) Sex differences and the epidemiology of depression. Archives of general psychiatry 34 (1):98. [DOI] [PubMed] [Google Scholar]
- 17.St John RD, Corning PA (1973) Maternal aggression in mice. Behavioral biology 9 (5):635–639 [DOI] [PubMed] [Google Scholar]
- 18.Hennessey AC, Huhman KL, Albers HE (1994) Vasopressin and sex differences in hamster flank marking. Physiology & behavior 55 (5):905–911 [DOI] [PubMed] [Google Scholar]
- 19.Huhman KL, Solomon MB, Janicki M, Harmon AC, Lin SM, Israel JE, Jasnow AM (2003) Conditioned defeat in male and female Syrian hamsters. Hormones and behavior 44 (3):293–299 [DOI] [PubMed] [Google Scholar]
- 20.Taravosh-Lahn K, Delville Y (2004) Aggressive behavior in female golden hamsters: development and the effect of repeated social stress. Hormones and behavior 46 (4):428–435. doi: 10.1016/j.yhbeh.2004.03.007 [DOI] [PubMed] [Google Scholar]
- 21.Rosenhauer AM, McCann KE, Norvelle A, Huhman KL (2017) An acute social defeat stressor in early puberty increases susceptibility to social defeat in adulthood. Hormones and behavior 93:31–38. doi: 10.1016/j.yhbeh.2017.04.002 [DOI] [PubMed] [Google Scholar]
- 22.Faruzzi AN, Solomon MB, Demas GE, Huhman KL (2005) Gonadal hormones modulate the display of submissive behavior in socially defeated female Syrian hamsters. Hormones and behavior 47 (5):569–575. doi: 10.1016/j.yhbeh.2004.11.023 [DOI] [PubMed] [Google Scholar]
- 23.Solomon MB, Karom MC, Huhman KL (2007) Sex and estrous cycle differences in the display of conditioned defeat in Syrian hamsters. Hormones and behavior 52 (2):211–219. doi: 10.1016/j.yhbeh.2007.04.007 [DOI] [PubMed] [Google Scholar]
- 24.Solomon MB, Karom MC, Norvelle A, Markham CA, Erwin WD, Huhman KL (2009) Gonadal hormones modulate the display of conditioned defeat in male Syrian hamsters. Hormones and behavior 56 (4):423–428. doi: 10.1016/j.yhbeh.2009.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCann KE, Rosenhauer AM, Jones GMF, Norvelle A, Choi DC, Huhman KL (2017) Histone deacetylase and acetyltransferase inhibitors modulate behavioral responses to social stress. Psychoneuroendocrinology 75:100–109. doi: 10.1016/j.psyneuen.2016.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jasnow AM, Huhman KL (2001) Activation of GABA(A) receptors in the amygdala blocks the acquisition and expression of conditioned defeat in Syrian hamsters. Brain research 920 (1–2):142–150 [DOI] [PubMed] [Google Scholar]
- 27.Markham CM, Taylor SL, Huhman KL (2010) Role of amygdala and hippocampus in the neural circuit subserving conditioned defeat in Syrian hamsters. Learning & memory (Cold Spring Harbor, NY) 17 (2):109–116. doi: 10.1101/lm.1633710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markham CM, Huhman KL (2008) Is the medial amygdala part of the neural circuit modulating conditioned defeat in Syrian hamsters? Learning & memory (Cold Spring Harbor, NY) 15 (1):6–12. doi: 10.1101/lm.768208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jasnow AM, Shi C, Israel JE, Davis M, Huhman KL (2005) Memory of social defeat is facilitated by cAMP response element-binding protein overexpression in the amygdala. Behavioral neuroscience 119 (4):1125–1130. doi: 10.1037/0735-7044.119.4.1125 [DOI] [PubMed] [Google Scholar]
- 30.McCann KE, Sinkiewicz DM, Norvelle A, Huhman KL (2017) De novo assembly, annotation, and characterization of the whole brain transcriptome of male and female Syrian hamsters. Scientific reports 7:40472. doi: 10.1038/srep40472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Vries GJ (2004) Minireview: Sex differences in adult and developing brains: compensation, compensation, compensation. Endocrinology 145 (3):1063–1068. doi: 10.1210/en.2003-1504 [DOI] [PubMed] [Google Scholar]
- 32.de Vries GJ, Forger NG (2015) Sex differences in the brain: a whole body perspective. Biology of sex differences 6:15. doi: 10.1186/s13293-015-0032-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marrocco J, Petty GH, Rios MB, Gray JD, Kogan JF, Waters EM, Schmidt EF, Lee FS, McEwen BS (2017) A sexually dimorphic pre-stressed translational signature in CA3 pyramidal neurons of BDNF Val66Met mice. Nature communications 8 (1):808. doi: 10.1038/s41467-017-01014-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Labonte B, Engmann O, Purushothaman I, Menard C, Wang J, Tan C, Scarpa JR, Moy G, Loh YE, Cahill M, Lorsch ZS, Hamilton PJ, Calipari ES, Hodes GE, Issler O, Kronman H, Pfau M, Obradovic ALJ, Dong Y, Neve RL, Russo S, Kazarskis A, Tamminga C, Mechawar N, Turecki G, Zhang B, Shen L, Nestler EJ (2017) Sex-specific transcriptional signatures in human depression. Nature medicine 23 (9):1102–1111. doi: 10.1038/nm.4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mead AN, Stephens DN (2003) Involvement of AMPA receptor GluR2 subunits in stimulus-reward learning: evidence from glutamate receptor gria2 knock-out mice. The Journal of neuroscience 23 (29):9500–9507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gray AL, Hyde TM, Deep-Soboslay A, Kleinman JE, Sodhi MS (2015) Sex differences in glutamate receptor gene expression in major depression and suicide. Molecular psychiatry 20 (9):1057–1068. doi: 10.1038/mp.2015.91 [DOI] [PubMed] [Google Scholar]
- 37.Gulinello M, Gong QH, Smith SS (2002) Progesterone withdrawal increases the alpha4 subunit of the GABA(A) receptor in male rats in association with anxiety and altered pharmacology - a comparison with female rats. Neuropharmacology 43 (4):701–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gulinello M, Orman R, Smith SS (2003) Sex differences in anxiety, sensorimotor gating and expression of the alpha4 subunit of the GABAA receptor in the amygdala after progesterone withdrawal. The European journal of neuroscience 17 (3):641–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tripp A, Oh H, Guilloux JP, Martinowich K, Lewis DA, Sibille E (2012) Brain-derived neurotrophic factor signaling and subgenual anterior cingulate cortex dysfunction in major depressive disorder. The American journal of psychiatry 169 (11):1194–1202. doi: 10.1176/appi.ajp.2012.12020248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ortiz JB, Taylor SB, Hoffman AN, Campbell AN, Lucas LR, Conrad CD (2015) Sex-specific impairment and recovery of spatial learning following the end of chronic unpredictable restraint stress: potential relevance of limbic GAD. Behavioural brain research 282:176–184. doi: 10.1016/j.bbr.2014.12.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feldblum S, Erlander MG, Tobin AJ (1993) Different distributions of GAD65 and GAD67 mRNAS suggest that the two glutamate decarboxylases play distinctive functional roles. Journal of Neuroscience Research 34 (6):689–706. doi: 10.1002/jnr.490340612 [DOI] [PubMed] [Google Scholar]
- 42.Bowers G, Cullinan WE, Herman JP (1998) Region-specific regulation of glutamic acid decarboxylase (GAD) mRNA expression in central stress circuits. The Journal of neuroscience 18 (15):5938–5947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morrison KE, Swallows CL, Cooper MA (2011) Effects of dominance status on conditioned defeat and expression of 5-HT1A and 5-HT2A receptors. Physiology & behavior 104 (2):283–290. doi: 10.1016/j.physbeh.2011.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernstein HG, Dobrowolny H, Schott BH, Gorny X, Becker V, Steiner J, Seidenbecher CI, Bogerts B (2013) Increased density of AKAP5-expressing neurons in the anterior cingulate cortex of subjects with bipolar disorder. Journal of psychiatric research 47 (6):699–705. doi: 10.1016/j.jpsychires.2012.12.020 [DOI] [PubMed] [Google Scholar]
- 45.Qi XR, Zhao J, Liu J, Fang H, Swaab DF, Zhou JN (2015) Abnormal retinoid and TrkB signaling in the prefrontal cortex in mood disorders. Cerebral cortex (New York, NY : 1991) 25 (1):75–83. doi: 10.1093/cercor/bht203 [DOI] [PubMed] [Google Scholar]
- 46.Zhou R, Niwa S, Guillaud L, Tong Y, Hirokawa N (2013) A molecular motor, KIF13A, controls anxiety by transporting the serotonin type 1A receptor. Cell reports 3 (2):509–519. doi: 10.1016/j.celrep.2013.01.014 [DOI] [PubMed] [Google Scholar]
- 47.Soleimani L, Roder JC, Dennis JW, Lipina T (2008) Beta N-acetylglucosaminyltransferase V (Mgat5) deficiency reduces the depression-like phenotype in mice. Genes, brain, and behavior 7 (3):334–343. doi: 10.1111/j.1601-183X.2007.00358.x [DOI] [PubMed] [Google Scholar]
- 48.Wingo AP, Almli LM, Stevens JJ, Klengel T, Uddin M, Li Y, Bustamante AC, Lori A, Koen N, Stein DJ, Smith AK, Aiello AE, Koenen KC, Wildman DE, Galea S, Bradley B, Binder EB, Jin P, Gibson G, Ressler KJ (2015) DICER1 and microRNA regulation in post-traumatic stress disorder with comorbid depression. Nat Commun 6:10106. doi: 10.1038/ncomms10106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamamoto A, Uchiyama K, Nara T, Nishimura N, Hayasaka M, Hanaoka K, Yamamoto T (2014) Structural abnormalities of corpus callosum and cortical axonal tracts accompanied by decreased anxiety-like behavior and lowered sociability in spock3-mutant mice. Developmental neuroscience 36 (5):381–395. doi: 10.1159/000363101 [DOI] [PubMed] [Google Scholar]
- 50.Dias C, Feng J, Sun H, Shao NY, Mazei-Robison MS, Damez-Werno D, Scobie K, Bagot R, LaBonte B, Ribeiro E, Liu X, Kennedy P, Vialou V, Ferguson D, Pena C, Calipari ES, Koo JW, Mouzon E, Ghose S, Tamminga C, Neve R, Shen L, Nestler EJ (2014) beta-catenin mediates stress resilience through Dicer1/microRNA regulation. Nature 516 (7529):51–55. doi: 10.1038/nature13976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee JY, Kwak M, Lee PC (2015) Impairment of social behavior and communication in mice lacking the Uba6-dependent ubiquitin activation system. Behavioural brain research 281:78–85. doi: 10.1016/j.bbr.2014.12.019 [DOI] [PubMed] [Google Scholar]
- 52.Gray CL, Norvelle A, Larkin T, Huhman KL (2015) Dopamine in the nucleus accumbens modulates the memory of social defeat in Syrian hamsters (Mesocricetus auratus). Behavioural brain research 286:22–28. doi: 10.1016/j.bbr.2015.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ross AP, Norvelle A, Choi DC, Walton JC, Albers HE, Huhman KL (2017) Social housing and social isolation: Impact on stress indices and energy balance in male and female Syrian hamsters (Mesocricetus auratus). Physiology & behavior 177:264–269. doi: 10.1016/j.physbeh.2017.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Potegal M, Huhman K, Moore T, Meyerhoff J (1993) Conditioned defeat in the Syrian golden hamster (Mesocricetus auratus). Behavioral and neural biology 60 (2):93–102 [DOI] [PubMed] [Google Scholar]
- 55.Albers HE, Huhman KL, Meisel RL (2002) Hormonal basis of social conflict and communication. Hormones, brain and behavior 1:393–433 [Google Scholar]
- 56.McCann KE, Bicknese CN, Norvelle A, Huhman KL (2014) Effects of inescapable versus escapable social stress in Syrian hamsters: the importance of stressor duration versus escapability. Physiology & behavior 129:25–29. doi: 10.1016/j.physbeh.2014.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCann KE, Huhman KL (2012) The effect of escapable versus inescapable social defeat on conditioned defeat and social recognition in Syrian hamsters. Physiology & behavior 105 (2):493–497. doi: 10.1016/j.physbeh.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hodes GE, Pfau ML, Purushothaman I, Ahn HF, Golden SA, Christoffel DJ, Magida J, Brancato A, Takahashi A, Flanigan ME, Menard C, Aleyasin H, Koo JW, Lorsch ZS, Feng J, Heshmati M, Wang M, Turecki G, Neve R, Zhang B, Shen L, Nestler EJ, Russo SJ (2015) Sex Differences in Nucleus Accumbens Transcriptome Profiles Associated with Susceptibility versus Resilience to Subchronic Variable Stress. The Journal of neuroscience : the official journal of the Society for Neuroscience 35 (50):16362–16376. doi: 10.1523/jneurosci.1392-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bagot RC, Cates HM, Purushothaman I, Vialou V, Heller EA, Yieh L, LaBonte B, Pena CJ, Shen L, Wittenberg GM, Nestler EJ (2017) Ketamine and Imipramine Reverse Transcriptional Signatures of Susceptibility and Induce Resilience-Specific Gene Expression Profiles. Biological psychiatry 81 (4):285–295. doi: 10.1016/j.biopsych.2016.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feng J, Wilkinson M, Liu X, Purushothaman I, Ferguson D, Vialou V, Maze I, Shao N, Kennedy P, Koo J, Dias C, Laitman B, Stockman V, LaPlant Q, Cahill ME, Nestler EJ, Shen L (2014) Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome biology 15 (4):R65. doi: 10.1186/gb-2014-15-4-r65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Konczal M, Koteja P, Stuglik MT, Radwan J, Babik W (2014) Accuracy of allele frequency estimation using pooled RNA-Seq. Molecular ecology resources 14 (2):381–392. doi: 10.1111/1755-0998.12186 [DOI] [PubMed] [Google Scholar]
- 62.Sarajlic S, Edirisinghe N, Lukinov Y, Walters M, Davis B, Faroux G (2016) Orion: Discovery Environment for HPC Research and Bridging XSEDE Resources. Paper presented at the Proceedings of the XSEDE16 Conference on Diversity, Big Data, and Science at Scale, Miami, USA, [Google Scholar]
- 63.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature biotechnology 29 (7):644–652. doi: 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature protocols 8 (8):1494–1512. doi: 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feng NY, Fergus DJ, Bass AH (2015) Neural transcriptome reveals molecular mechanisms for temporal control of vocalization across multiple timescales. BMC genomics 16:408. doi: 10.1186/s12864-015-1577-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. Journal of molecular biology 215 (3):403–410. doi: 10.1016/s0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 67.UniProt C (2015) UniProt: a hub for protein information. Nucleic Acids Res 43 (Database issue):D204–212. doi: 10.1093/nar/gku989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12 (1):1–16. doi: 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England) 26 (1):139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Plaisier SB, Taschereau R, Wong JA, Graeber TG (2010) Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic acids research 38 (17):e169. doi: 10.1093/nar/gkq636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mi H, Muruganujan A, Casagrande JT, Thomas PD (2013) Large-scale gene function analysis with the PANTHER classification system. Nature protocols 8 (8):1551–1566. doi: 10.1038/nprot.2013.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD (2016) PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic acids research 44 (D1):D336–342. doi: 10.1093/nar/gkv1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mi H, Thomas P (2009) PANTHER pathway: an ontology-based pathway database coupled with data analysis tools. Methods in molecular biology (Clifton, NJ) 563:123–140. doi: 10.1007/978-1-60761-175-2_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome biology 11 (2):R14. doi: 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Langfelder P, Zhang B, Horvath S (2008) Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 24. doi: 10.1093/bioinformatics/btm563 [DOI] [PubMed] [Google Scholar]
- 76.Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9 (1):1–13. doi: 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


