Abstract
Many advances in instrumentation and methodology have furthered the use of solid-state NMR as a technique for determining the structure and studying the dynamics of molecules involved in complex biological assemblies. Solid-state NMR does not require large crystals, has no inherent size limit, and with appropriate isotopic labeling schemes, supports solving one component of a complex assembly at a time. It is complementary to cryo-EM, in that it provides local, atomic-level detail that can be modeled into larger-scale structures. This review focuses on the development of high-field MAS instrumentation and methodology; including probe design, benchmarking strategies, labeling schemes, and experiments that enable the use of quadrupolar nuclei in biomolecular NMR. Current challenges facing solid-state NMR of biological assemblies and new directions in this dynamic research area are also discussed.
Keywords: Magic Angle Spinning, solid-state NMR, instrumentation, biological assemblies, MAS probe
1. Introduction
Many biological macromolecules assume their functional states only in the context of complex and heterogeneous assemblages, such as multi-protein complexes [1] or membrane proteins in their native environments [2]. Others involve a large assembly composed of many repeating units of the same simple monomer, as in amyloid fibrils [3] or the external coverings of viruses [4]. Solid-state NMR can be used either on its own or synergistically with other structural techniques, and has emerged as the method of choice for providing atomic-level detail about the structures of such non-crystalline biological solids. A few notable successes (selected from a list far too long to fully enumerate) include determination of the structures of key components of the HIV virus capsid assembly [5], a small protein in complex with a full-length human immunoglobulin [6], RNA-protein complexes [7], biomimetic photosynthetic centers [8], a bacterial chemoreceptor consisting of multiple protein components [9], and cytoskeleton-associated proteins on microtubules [10]. Several recent reviews have addressed novel spectroscopic developments [11, 12] as well as applications to biological systems [13, 14], including membrane proteins [15, 16, 17].
One prominent example of a non-crystalline biological solid where magic-angle spinning (MAS) NMR has contributed many of the known molecular-level details is the protofibril structure and assembly of amyloid fibrils, which are primarily β-sheet aggregates implicated in many protein deposition diseases. An ever more detailed picture of these aggregates has emerged in conjunction with advances in solid-state NMR instrumentation, pulse sequences and sample preparation, illustrating how technology development enables new applications [18]. Some of the earliest details of amyloid structure, beyond the basics of the common cross-β architecture first described based on EM and fiber di raction studies [19, 20, 21], came from solid-state NMR measurements using a small number of site-specific labels. Specifically, pairwise dipolar recoupling [22] and multiple quantum spin counting experiments [23] were peformed on samples labeled with 13C at particular backbone carbonyl sites. These measurements showed unambiguously that disease-relevant peptides from β-amyloid form in-register parallel β-sheets, contra the anti-parallel model that was assumed previously, and which could not be distinguished from the parallel structure based solely on the X-ray data. At the time, the structure determination protocols including full assignments and simultaneous measurement of multiple distance restraints that are taken for granted today were not feasible due to technological limitations; nevertheless, via clever experimental design, the key information needed to distinguish between competing structural models was obtained.
Later developments, enabled by advances in probes, pulse sequences, isotopic labeling schemes, and sample preparation, have focused on near-complete resonance assignments using multidimensional NMR [24]. Solid-state NMR data have been used to produce structural models for the Aβ1–40 [25, 26] and Aβ1–42 peptides [27, 28], both of which have been implicated in Alzheimer’s disease. MAS NMR and EM data were used to demonstrate that the same peptide can form parallel or anti-parallel β-sheets; and in vivo experiments show the latter structure is a toxic intermediate along the fibrillization pathway [29]. Chemical shift assignments allowed the identification of distinct fibril polymorphs associated with prion proteins from different mammalian species [30]. The steric zipper arrangement first seen in short peptides and hypothesized to be a major determinant of amyloidogenic sequences [31] was observed in fibrils of a peptide from β2-microglobulin using a large number of restraints measured from a uniformly isotopically labeled sample [32]. A similar approach was used in a WW domain, where uniform labeling was used for assignment, while specific labels helped to pinpoint long range side chain interactions [33]. Mixed labeling strategies of this type have become an important feature of solid-state NMR structure determination, where they are used to control which dipolar couplings are observed. Reducing the spectral complexity by labeling only parts of the sample or particular chemical groups is especially helpful for samples where one type of secondary structure is dominant, resulting in extensive spectral overlap, e.g. transmembrane helices in membrane proteins or β-sheets in amyloid fibrils. The compact, repeating structure of fibrils introduces the additional problem that inter- and intra-molecular distance restraints are often comparable, necessiting disambiguation via isotopic labeling [34]. Another useful labeling scheme removes background signal from natural abundance 13C and 15N by preparing mixed [15N,12C]/[14N,13C] fibrils, allowing unambiguous interstrand restraints from 13C-15N correlations [35].
Building on the intitial idea of a set of stacked, in-register sheets, subsequent structures of the repeating units have revealed much more complex arrangements, including triangular prisms built from single strands [34] or β-hairpins [26, 36], modified Greek-key folds [37], and planar sheets built from compactly folded dimers [38, 39]. Solvent exposure of different parts of the fibril structure can be measured with H-D exchange [40], by using Overhauser-effect DNP relaxometry to measure the effect of the protein on water diffusivity [41], or via the introdution of paramagnetic ions such as Cu2+, enabling measurement of longitudinal paramagnetic relaxation enhancements (PREs) from 15N nuclei in solvent-exposed locations [42]. The sensitivity enhancement a orded by dynamic nuclear polarization (DNP) has enabled MAS NMR of fibril-forming proteins that form multiple conformers or ‘strains’ [43], require segmental labeling to minimize spectral overlap [44], and interact with their cellular environments in complex ways [45].
This progression from low-resolution measurements on simple model systems to high-resolution structures of disease-relevant protein aggregates has been enabled by several contemporaneous advances in MAS NMR methodology; high-field magnets, high-resolution multi-channel MAS probes, ultrafast MAS, and sophisticated labeling and increased use of quadrupolar nuclei as spectroscopic probes; these advances are the topic of this review. Another important innovation, solid-state dynamic nuclear polarization (DNP), has greatly improved the sensitivity of solid-state NMR, and is largely complementary to these other methodological improvements. DNP is the subject of several recent reviews [46, 47], including those focused on instrumentation [48] and applications to structural biology [49, 50], and is beyond the scope of the present work. Here we discuss MAS probe design, including considerations for the choice of the sample coil, assessing probe performance, extracting biological information from quadrupolar nuclei, and some perspective on future directions.
2. High-field MAS Instrumentation
2.1. Design Strategies for Multi-channel Probes
The development of the first 800 and 900 MHz NMR magnets in the late 1990s and early 2000s held out the promise of su cient sensitivity and resolution to obtain full resonance assignments from solid-state protein samples, comparable to the explosion in solution-state NMR structures in previous years. However, these opportunities came with significant experimental challenges, many of them related to the strong 1H-1H homonuclear dipolar couplings, and to a lesser extent 1H-13C and 1H-15N heteronuclear dipolar couplings. In solution-state NMR, 1H-detected experiments anchor the entire structure determination effort, with most of the multidimensional experiments used to assign the backbone and side chain residues, as well as the two-dimensional experiments used to provide long-range distance restraints, depending on being able to fully assign proton chemical shifts. This was not feasible in the early solid-state experiments, as the proton resolution was insu cient to allow full assignments. Therefore, early methods had to rely on experiments where the detected nuclei were 13C or 15N; robust 1H decoupling was also required [51, 52].
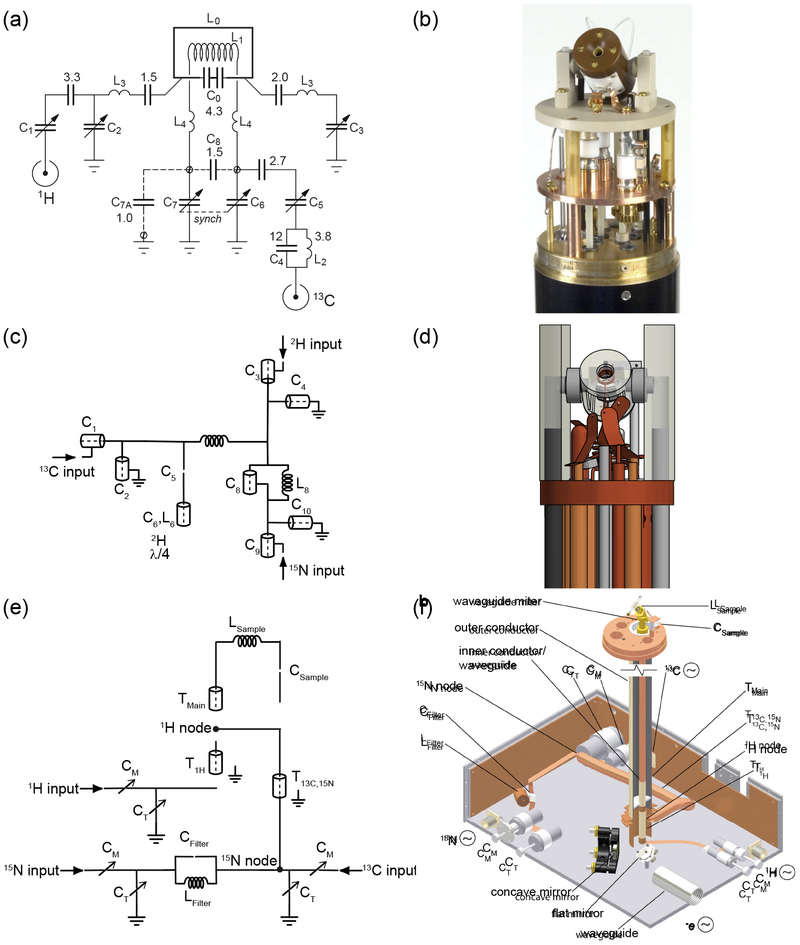
Such experiments placed stringent demands on MAS probes. They must be able to tune to three or more frequencies simultaneously (typically 1H, 13C, and 15N for biomolecular structure determination), be able to handle high radiofrequency (rf) power for cross-polarization (CP) and decoupling, and have su cient isolation among the channels to produce high sensitivity spectra, all while spinning stably at the magic angle and maintaining reliable temperature control in order to keep delicate biological samples intact. Circuit diagrams and schematics for representative examples illustrating different multiply-resonant probe designs are shown in Figure 1. Many early MAS probes provided pieces of the technology required for the first structure determination efforts, starting with early single-coil multiply-tuned circuits having either transmission line [53] or lumped-element components [54]. Isolation is a critical factor in the success of MAS experiments, particularly if high-power decoupling is needed. In these experiments, it is necessary to detect a microwatt signal at the frequency of interest while hundred-watt (or higher) decoupling pulses are applied to the very same coil. Failure to provide adequate isolation can place an undue load on the external filters, or result in the introduction of unacceptable levels of noise. Care must also be taken not to compromise effciency (defined as B1, where P is the transmitter power) too much while optimizing isolation. The excitation bandwidth in the frequency domain depends on the ability to generate short pulses, while the reciprocity principle states that the sensitivity is proportional to the probe effciency [55], and equally importantly, the extra power dissipated as heat can damage biological samples. In the traditional MAS probe design, which is characterized by its modularity and ease of fabrication, discrete circuit elements are connected by wire or ribbon leads. The current apotheosis of this design strategy is seen in recent examples built for MAS and static NMR at 1.5 GHz (Fig. 1a–b) [56]. These probes eliminate many of the leads that can cause stray reactances at high fields, instead using blocks of circuit elements attached to removable PC boards (tuning cards). These modular tuning cards enable the same probe to be used in a variety of H/X/Y combinations without sacrificing sensitivity on the detection channel. The rf coils are likewise mounted on removable inserts with additional tuning capacitors to pre-tune the detection frequency before connection to the matching network.
Figure 1:
A comparision of the different probe circuit styles discussed above, using representative examples from the literature. Shown are circuit diagrams and images of a lumped element probe (a-b) [63], a tuning tube probe (c-d) [62], and a transmission line probe (e-f) [64].
On the other end of the design spectrum are transmission line probes (Fig. 1e–f). In this type of circuit, the properties of transmission lines provide rf isolation among the channels, enabling tuning to many resonance frequencies simultaneously [57]. One elegant solution to the isolation problem is the triple-resonance probe design of Stringer and Drobny, which uses transmission lines both as isolation elements and to enable placement of the variable tuning capacitors (in this case large Polyflons) outside the magnet bore [58]. A major objective of this probe design was to provide excellent isolation between 1H and 19F, which resonate at similar rf frequencies. However, the principles and general design strategy described are useful for a wide range of MAS applications. The same transmission line elements behave very differently depending on the input frequency; for example, an open transmission line segment can simultaneously act as a low-impedance path to ground at low frequency, and essentially an open circuit at higher frequencies. This type of approach not only improves the isolation, but also simplifies analysis of the circuit. A key property of transmission line probes is that excellent isolation can be achieved by simply connecting the channels at common impedance nodes, without additional isolation elements. This was demonstrated for a fully-balanced double-resonance MAS probe where each channel consists of a tunable impedance-matching network on one side of the coil and a balun on the other [59]. The channels are connected at common impedance nodes on both sides of the sample coil, with each frequency traveling primarily though the branch tuned to that frequency, which presents the path with the lowest impedance. This techique has the advantage of eliminating trap circuit elements, which may introduce electrical losses and are diffcult to simultaneously optimize, especially as the number of channels increases. These probes are physically robust and have the advantage of placing the large tuning capacitors outside the magnet bore, which is particularly advantageous for high-field DNP, where the sample dewar, the MAS and variable temperature gas lines, the microwave waveguide, and various sensors must all be placed inside the limited bore diameter of a high-field magnet [60].
A somewhat related approach is the tuning tube design, in which nearly all circuit elements are composed of variable transmission line elements, but can easily be understood as discrete circuit elements (Fig. 1c–d). This design was used to build triple-resonance (1H, 13C, 15N) [61] and quadruple-resonance (1H, 2H, 13C, 15N) [62] MAS probes for operation at 800 MHz. In these probes, the tune and match capacitors are arranged concentrically, with all tuning and matching components associated with each rf channel contained within a grounded outer tube. In the Varian/Agilent T3 probe, which used a similar strategy, broadband operation was achieved by swapping the entire inner portion of the channel. This geometry makes use of the full length of the magnet bore, as well as minimizing the stray reactances that come with connecting discrete elements above a common ground plane, which can make tuning unreliable, particularly at high frequencies where these reactances can approach those of the principal circuit elements. As in true transmission line probes, the large physical size of the components helps to improve heat dissipation and minimize AC conduction losses.
2.2. Radiofrequency Homogeneity
Radiofrequency homogeneity is critical to biomolecular solid-state NMR, as these experiments frequently involve several polarization-transfer steps, and in many cases composite pulses or complex recoupling schemes where propagation of errors in the amplitude and phase of the rf pulses can result in substantial signal losses. Doty et al. performed a comprehensive study of several coil and stator designs, demonstrating the impact of rf homogeneity through modeling and experiments [65]. Several strategies for assessing the rf homogeneity of an NMR probe are summarized in Figures 2 & 3. CPMAS, which was first introduced by Schaefer and Stejskal [66] and is the cornerstone of many MAS experiments to this day, requires that the MAS-adapted Hartmann-Hahn condition, vH − vX = ±nvR, where n = ±1 or ±2, be met. At this condition, mutual spin flips mediated by the heteronuclear dipolar coupling conserve energy, causing the magnetization of the two nuclei to equilibrate. As CP is often used to enhance sensitivity by transferring magnetization from abundant, high γ nuclei, e.g. 1H, to rarer, low γ nuclei such as 13C and 15N, meeting this condition over as much of the sample as possible is a major objective. Many strategies exist for maximizing the overlap during the pulse sequence, the simplest being a linear ramp on one of the spin lock pulses [67]. Coil design is also important for ensuring effective polarization transfer. The rf homogeneity of an infinitely long solenoid is perfect, however in any practical implementation, the relatively short length of the coil results in a field profile that is stronger in the center but falls off toward the edges. This effect can be mitigated by winding the coil with variable pitch, such that the spacings between turns are greater in the center and smaller near the edges [68]. Alternatively, wire-wound solenoids can altogether in favor of other geometries, such as the loop-gap resonator [69, 70] or the scroll coil [71].
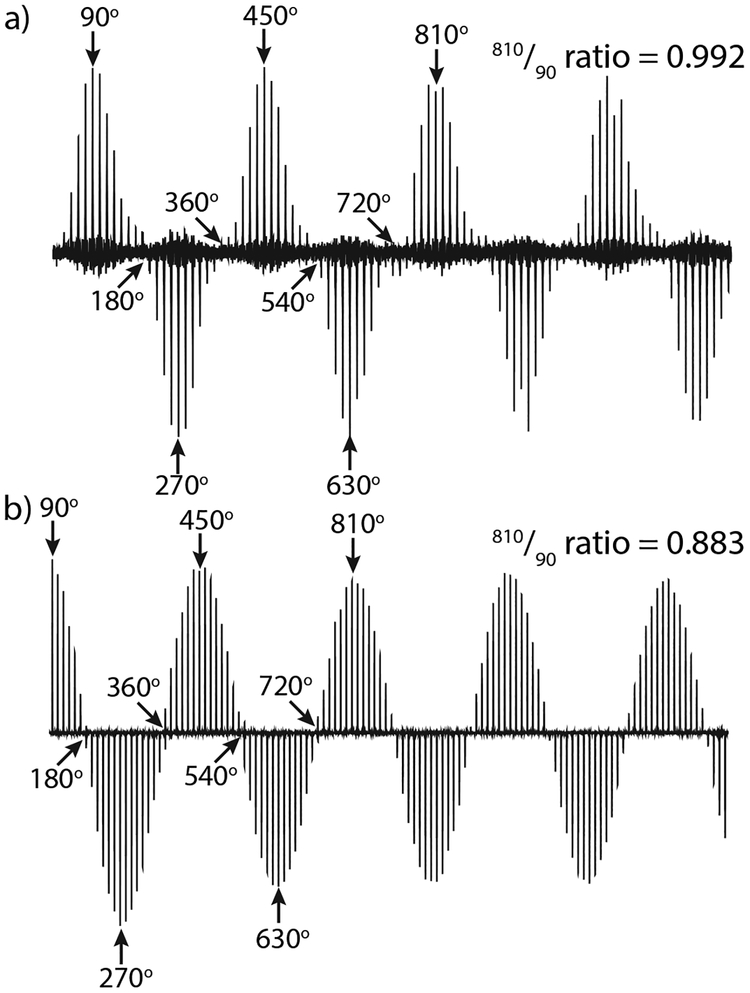
Figure 2:
A comparision of the 810°/90° ratios for the two crossed coils in a quadruple-resonance MAS probe [62]. (a) The 1H nutation on the MAG coil, and (b) the 15N nutation on the variable pitch solenoid.
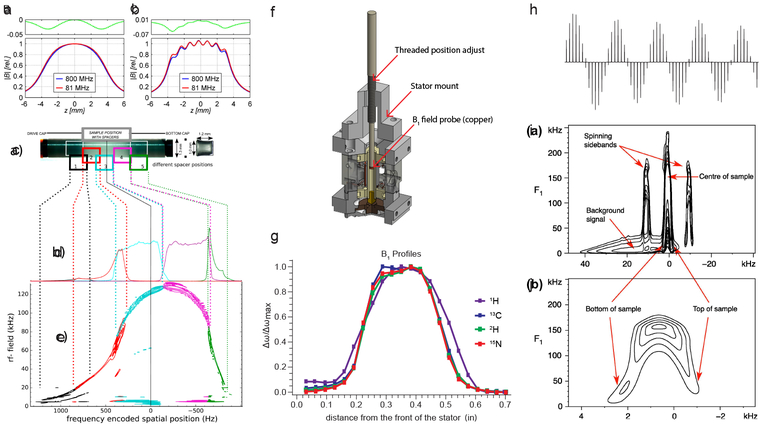
Figure 3:
Methods for assessing the rf homogeneity of an NMR probe (a-b) Magnitude profiles for the B1 field at two frequencies calculated for two positions within a 3.2 mm rotor, at the center of the rotor (a) and along the inner wall of the rotor (b) in a solenoid coil [77]; (c-e) NMR measurements of the rf field profile (c) shows the locations of the silicone spacers in the rotor, (d) shows the 1D 1H spectra of the silicone spacer at each location, and (e) is the 2D 1H nutation spectra of the same [74]; f) CAD image of a tool used to perform ball-shift measurements [90]; g) homogeneity measurements taken using the tool in (f) [62]; h) nutation curve of the 2H channel of a quadruple-resonance MAS probe [62]; i-j) 2D 1H nutation spectra of adamantane (at 400 MHz), (j) is an expansion of the center part of the spectrum in (i) [83].
For optimal results in MAS experiments, it is usually desirable to restrict the sample to the homogeneous region of the coil. This is most often done by placing spacers inside the rotor to physically restrict the sample volume. However, sample restriction can also be accomplished during the NMR experiment by using selective pulses [72] or via the application of magnetic field gradients, generated either with a specialized gradient coil in the MAS probe if one is available [73], or using the room-temperature shim coils [74]. Although at first glance these approaches seem more complicated than simply using physical spacers, they have important advantages. These include shifting the abrupt change in magnetic susceptibility at the boundary between the sample and the spacer well outside the detection volume, and the ability to calibrate it easily at the beginning of each experiment, in contrast to the practical challenges of ensuring that each sample is packed exactly within the homogeneous region of the coil before commencing NMR experiments.
Serendipitously, in many experiments Hartmann-Hahn mismatches for CPMAS due to rf inhomogeneity appear to mostly reduce the overall signal intensity rather than causing problematic artifacts [75], primarily because the majority of the signal comes from the parts of the sample experiencing the maximum rf amplitude. However, rf inhomogeneity does produce artifacts such as line-broadening and phase errors in other types of pulse sequences, notably homonuclear decoupling [76]. Rf inhomogeneity can reduce the magnetization transfer effciency to as low as 50–60% [77] as well as negatively impacting the decoupling effectiveness [78], though this can be compensated with carefully designed sequences, e.g. [79]. Because signal-to-noise is always a major concern in NMR experiments, rf homogeneity is an important design criterion for biomolecular solid-state NMR probes. A straightforward and commonly used way to measure the rf homogeneity is to collect a nutation curve and compare the intensity of the signal at 810° to that at 90° as shown in Figure 2. This measurement is often provided by probe manufacturers because it is readily interpreted and easily compared among different probes. However, it lacks spatial information, i.e. it does not indicate where along the coil a homogeneity problem exists.
The spatial information can be acquired somewhat painfully by moving a very small plug of a setup sample to different positions along the rotor [80, 81, 82], or by applying a strong magnetic field gradient (this can be achieved using the Z1 shim, or in a pinch, by pulling the probe out of the magnet until the resonance lines are su ciently broadened) and acquiring a 2D nutation signal [83] (Figure 3i–j). If the gradient is instead applied along the rotor axis at the magic angle [84], then measurement of various desirable experimental parameters is very straightforward. In this case, a nutation experiment or a spin-lock experiment performed at the rotary resonance recoupling (R3) [85] condition provides the rf amplitude distribution along the rotor for a particular rf channel, while a cross-polarization match array yields a spatial map of the correlation between the rf field profiles of the two channels [77]. In the case where there is a strong temperature dependence of the chemical shift, a similar approach can be used to map temperature gradients inside the rotor generated by MAS and/or the application of rf pulses [74]. A related method has been used to provide an elegant method of suppressing background signal: a 2D nutation experiment is performed in the presence of a linear gradient along the B0 axis [83], yielding a map of the B1 distribution that can then be used to calibrate depth pulses [86, 87]. This technique can also be used to eliminate background artifacts more directly, by identifying the sources of 1H background signals in the probe itself, allowing them to be physically eliminated. In general, this type of in situ mapping is necessary for obtaining a detailed understanding of how the rf profile behaves during NMR experiments, as the rf homogeneity is not static, but interacts with MAS to produce modulations of the pulse amplitude and phase, particularly at the ends of the rotor [77].
The experiments described above provide a detailed picture of how the rf fields inside the sample vary in space and time, which is a requirement for optimizing solid-state NMR pulse sequences that necessarily involve several interactions represented by complicated time-varying Hamiltonians. However, NMR-based testing methods are inconvenient to use during the early stages of probe development, when spectrometer testing after every minor modification is too time-consuming, not to mention a profligate use of instrument time. A more convenient and economical method is the ball-shift assay, in which the probe is tuned to the desired frequency and a small sphere or disc of conductive material is moved along the coil axis (Figure 3f). Assuming that the B field is much larger than the E field, the resulting shift in resonance frequency is proportional to (B1)2, as indicated by analytical calculations and numerical simulations [88]. The ball-shift method only measures the magnetic field, and not the electric field that is responsible for many of the sample heating effects that cause problems in NMR experiments. Dill-mann et al. have presented an analytical treatment, numerical simulations and experimental measurements of both the B and E fields of a solenoid in a double-resonance NMR probe, showing that the ball-shift measurement yields an accurate value for B1 only when the magnetic field is much larger than the electric field [88]. The homogeneity of a particular coil geometry can be predicted using a simple but broadly effective power-function model based on a Biot-Savart analysis. Here, the magnetic field along the long (z) axis of a single-turn scroll coil of length is 2L and radius r varies according to where λ is the ratio of the field in the center to that at the edges [75]. A modified version for a solenoid is based on the sample geometry rather than the solenoid windings, and takes as input parameters the rf amplitudes at the center and edges of the coil. This model allows extraction of the rf field profile from simple nutation curves measured in a homogeneous magnetic field, and has been found to agree with previously published experimental data [80, 81, 76, 82] as well as detailed numerical simulations [77] (Figure 3a–b). As the above examples demonstrate, numerical simulations of the field distribution serve as a very useful guide to optimizing multiple-coil NMR probes, as well as the microwave field in MAS probes for DNP [89].
2.3. Balancing
The above treatment assumes that the rf field profile of the coil is symmetric about the center. For a coil constructed with reasonable care, this is a good approximation at low frequencies, but it breaks down as the length of the wire making up the solenoid approaches 10% or more of the wavelength, an issue that is particularly relevant given that the wavelength is shorter in a wire-wound coil than it is in free space [91]. In this case, for an unbalanced circuit, the rf field profile is asymmetric [92]. This leads to the somewhat surprising observation that in an 800 MHz probe, the rf fields of 1H and 13C channels are not centered at the same point in the sample, even though both channels share the same sample coil [82]. The result of this is that during a CP experiment, only a small fraction of the sample at a time is experiencing the Hartman-Hahn condition. These issues can be mitigated by employing a ramped CP pulse [67], but it is also advantageous to optimize the circuit as much as possible to preserve experimental flexibility and optimize signal-to-noise. Problems stemming from failing to cross-polarize over the entire sample become much more important at high field and with increasing MAS rate, as the rotor frequency approaches or even surpasses the magnitude of the relevant heteronuclear and homonuclear dipolar couplings, making the match conditions more sensitive [93]. One can determine the correlation between the 1H and X field profiles by performing a 2D nutation experiment where the two nutation frequencies are detected in two time domains [94].
In a balanced circuit, the capacitance is split across the detection coil. This has the advantage of halving the maximum amplitude of the voltage swings and placing a voltage node in the center of the coil, allowing double the B1 field to be applied, which is highly advantageous for many experiments, notably MQMAS [95], but also any other experiment requiring a large bandwidth, robust decoupling, or very short hard pulses. Balanced circuits have long been used to reduce rf-induced sample heating in both imaging [96] and solid-state NMR experiments [65]. This approach also has the advantage of greatly increasing the rf field that can be applied before damaging the probe or the sample, a consideration that is particularly important for conductive samples such as biomolecules in hydrated crystals [97] or membrane mimetics [98].
2.4. Setting the Magic Angle
Signal quality in most MAS experiments strongly depends on accurately setting the magic angle, which is typically mechanically controlled using a worm gear or captured thread mechanism. In modern MAS probes, the angle adjustment is generally reliable once set, making it feasible to use an external standard and then switch to a rotor containing the sample of interest. Useful standards are samples that produce a spectrum that is extremely sensitive to mis-setting of the spinning angle, as in the case of quadrupolar nuclides in a highly symmetric environment, e.g. the 79Br signal of KBr [99]. The cubic crystal form of KBr would lead to cancellation of the quadrupolar interaction and the CSA in a perfect crystal, but defects and impurities create enough anisotropy to produce an easily observable sideband manifold, enabling the magic angle to be set by monitoring either the sideband intensity or equivalently, the magnitude of the rotational echoes in the time domain. Although this time-honored method is conceptually simple, implementing it is tedious when the spinning axis is far away from the magic angle.
Several innovative approaches have been proposed to improve the ease and accuracy of setting the magic angle. A Hall effect sensor can be attached to the stator with its sensitive axis perpendicular to the main magnetic field when the stator is at the magic angle, effectively measuring the angle with respect to the field itself rather than to any mechanical reference [100]. With careful initial calibration, the Hall voltage reads zero at the magic angle, enabling sensitive adjustments without resorting to the use of external standards between experiments. Another approach involves using an inexpensive optical lever to monitor the stator angle [101]. This apparatus was built by mounting a small mirror on the stator, with a laser from a laser pointer shining on it from the base of the probe. A 45° prism enables a horizontal readout at the bottom of the probe; using this apparatus, the position of the laser spot on a pre-calibrated scale indicates the stator angle. Another innovative solution is the application of a transverse field produced by an external saddle coil, which enables fine adjustment of the external magnetic field experienced by the spins [102].
2.5. Minimizing Sample Heating During MAS and Decoupling
Biological samples generally have high dielectric loss; proteins and nucleic acids require controlled solution pH to maintain their optimal structure and function, usually necessitating bu er solutions ranging in concentration from several mM to 100 mM. Even in minimally concentrated bu ers, the biomolecules themselves contain charged functional groups. This effectively acts like adding a poor capacitor with a lossy dielectric in parallel with the sample coil, which is why the full sensitivity gain promised in solution-state cryoprobes is often not realized for concentrated solutions of biomolecules. Due to dielectric loss, the water and ions in the sample dissipate electromagnetic energy as heat when subjected to MAS and rf pulses, causing problems for the many biomolecules that are subject to heat denaturation or unfolding. Sample heating can occur during sustained pulses on the lower-frequency channels or broadband high-power decoupling on the proton channel. This effect can be mitigated by using the minimum bu er concentration necessary to stabilize the sample pH, selecting bu ers with low ionic mobility [103], or simply freezing the sample. The heating caused by rf pulses is highly sample-dependent, so much so that Thompson and coworkers have advocated calibrating the expected rf heating on a sacrificial sample before performing NMR experiments on expensive labeled proteins [104]. In their scheme, a bu er solution containing natural abundance protein (or bu er alone) is doped with the compound Sm2Sn2O7, which acts as an NMR thermometer via the temperature dependence of the 119Sn chemical shift.
Because MAS and rf pulses both heat the sample, the di erence between the temperature measured in the stator and that of the sample can vary dramatically, especially at fast MAS rates. It is therefore desirable to calibrate the temperature using an internal chemical shift standard, often the 207Pb signal of lead nitrate [105, 106, 107] or 79Br [99]. A combined approach using both 79Br and 13C chemical shifts has been proposed for separating the effects of temperature from those of static field variation [108]. Neat methanol, KBr, [109] or water [110] can also be used as internal or external standards.
Sample heating effects can also be mitigated by instrumentation design. For example, the effciency of a solenoid coil can be improved, and the rf heating in a conductive sample reduced, by placing a strip shield composed of thin copper strips on a Teflon backing in between the solenoid and the sample [111], or by utilizing the cross-coil probes discussed below.
2.6. Variable Temperature MAS Experiments
Variable temperature operation is required in order to preserve delicate biological samples. This can be accomplished quite simply, using either liquid nitrogen boil-off or a vortex tube cooler [112], or can be made more effective by cooling the drive and bearing gas if much lower temperatures are desired, i.e. for DNP. Cold helium can be used for the VT, drive, and bearing gas [113]. Modeling the gas flow using computational fluid dynamics enables optimization of the cooling so that temperatures as low as 6 K can be acheived [114]. A controlled mixture of nitrogen and helium can be delivered depending on the desired temperature [115], or nitrogen can be used to cool the drive and bearing, with cold helium used only as the VT gas to avoid excessive helium use and reduce operating costs. In this case, temperature gradients along the rotor due to frictional heating from the air bearings can be a major problem for low-temperature MAS. One solution is to make the rotor much longer than the rf coil, using the poor thermal conductivity of the zirconia rotor to effectively isolate the cold sample region from the MAS bearings [116, 117]. This approach can achieve temperatures in the range of 20–25 K with moderate MAS rates. Another way to reduce helium consumption is to run the cooling gas through a closed-cycle helium refrigerator, similar to those used in solution-state cryo-probes [118, 119]. In such cryo-MAS probes, the coil is cooled as well as the sample, providing sensitivity gains from reduced thermal noise as well as the Boltzmann factor associated with cooling the sample. Insulation can be improved by evacuating the region surrounding the MAS stator assembly [120]. One problem with low-temperature MAS NMR, whether it is performed for its own sake or in the context of DNP, is that changing the sample in most standard MAS probes requires removing the probe from the magnet. Under standard laboratory conditions, this is a straightforward procedure, but when it entails removing all variable temperature connections and warming up the probe components from cryogenic temepratures to room temperature and then reversing the process for the new sample, it can take hours and provide opportunities for delicate probe components to be damaged. Barnes et al. proposed a simple solution to this problem where the sample is ejected from the probe dewar through a 3D printed eject tube using dry nitrogen gas, enabling samples to be changed without disconnecting any of the VT, rf, or microwave components [64].
3. Multiple Coil Probes
3.1. Crossed-coil Circuits
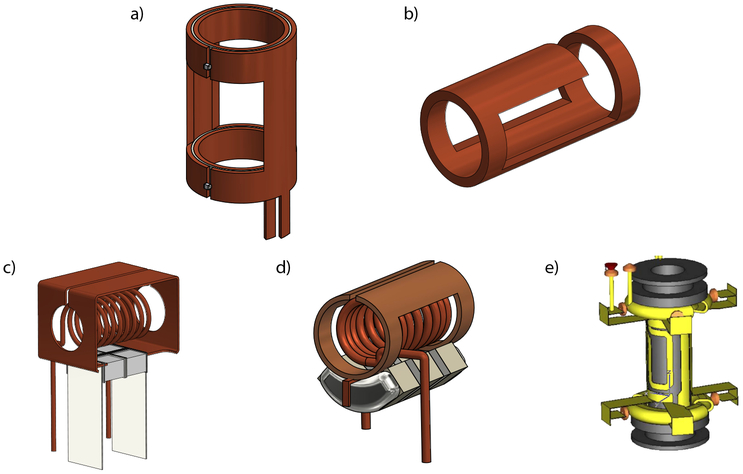
One successful solution to the problem of heating induced by broadband decoupling during detection was the development of low-inductance coils with high current and low voltage [69]. These low electric field (low E) coils are particularly advantageous at high frequency, and are often used in crossed-coil designs. A crossed-coil probe uses a low E, transverse coil (e.g., the Alderman-Grant coil [69], see Figure 4a) for the high γ nucleus (usually 1H) and a solenoid for the lower-frequency channels. This allows decoupling to be applied on the proton channel without excessive heating, while still having a higher-inductance solenoid for the detection of the heteronuclei. The sets of crossed coils can be arranged with the low E coil either inside or outside the solenoid, depending on the desired detection scheme. Examples of crossed-coil assemblies and transverse coils that can be used for the proton channel are shown in Figure 4.
Figure 4:
Crossed coil assemblies and transverse coils that can be used in such; a) the original Alderman-Grant resonator [69], b) the Zhang MAG [123], c) the Gor’kov MAS crossed coil assembly [63], d) the Martin lab MAS crossed coil assembly [62] using a version the Opella lab’s compact MAG design [122], e) the Doty XC on a ceramic support (the crossed coil assembly would have a variable-pitch solenoid wound around this) [65].
The crossed-coil assemblies designed by Gor’kov and coworkers are arranged with the low E resonator outside the solenoid. The first version of this arrangement was used for static studies of dilute membrane proteins in bilayers [121], while a later version was modified to enable MAS studies [63] (Figure 4c). These probes allow for large sample volumes and utilized a simpler coil derived from a wide single-turn solenoid. A similar, although more compact design was developed in the Opella lab [122], also for the study of membrane proteins in static samples of phospholipid bilayers. Another variation of the Opella crossed-coil design was further adapted to construct the quadruple-resonance probe designed in the Martin lab [62] (Figure 4d). This probe makes use of the low E resonator as the proton coil for decoupling, with the inner detection solenoid triple-tuned to 13C, 2H, and 15N).
More complex resonators have also been developed for cross-coil designs. The Cross Coil (XC) probe of Doty et al. [65], is an excellent example of a crossed-coil probe where the low E coil (the XC) is positioned inside the variable-pitch solenoid. This is done by creating the foil XC such that it lies flat, nearly on the rotor (see Figure 4e), enabling the solenoid to be wound as close as possible to the rotor in order to maximize the filling factor.
3.2. Microcoil MAS
Another clever application of multiple coils involves using microcoils to amplify the signal for a small sample volume. Some microcoils, such as those developed by Sakellariou et al. [124] are wound directly on a sample capillary, greatly improving the filling factor. In magic-angle coil spinning (MACS), a capacitor is soldered to the capillary-mounted microcoil to form a resonant circuit, and then the whole assembly is put into a rotor (shown in Figure 5a). The microcoil is inductively coupled to the main coil of the probe, enabling the signal enhancement of microcoils to be obtained in any commercial probe. This also allows for the whole assembly to be spun, greatly reducing any susceptibility effects from the microcoil circuit. Microfabrication of these microcoils makes it possible to manufacture them more reproducibly, with with less individual variation than normally results from hand-winding the tiny solenoids under a microscope [125]. Other types of coils have been developed for MACS assemblies, such as transmission line resonator pairs [126]. The original MACS assembly was singly resonant, allowing detection of only a single type of nucleus (usually 1H). More recently, the development of doubly-tuned MACS assemblies by Inukai and Takeda [127], has enabled the use of double-resonance methods, such as CP and proton decoupling (shown in Figure 5b).
Figure 5:
a) The Sakellariou MACS microcoil fits inside the MAS rotor [124], b) The doubly-tuned microcoil of Takeda et al enables double-resonance MAS [127], c) The Kent-gens piggyback coil enhances the signal in small samples [128].
Another technique for signal amplification in high-resolution MAS of small samples, developed by Kentgens and coworkers [128], piggy-backs a microcoil onto a conventional stator. Rather than inductively coupling the microcoil to the existing sample coil as done shown by Sakellariou et al. [124], this design uses a microcoil placed in the center of a capacitive plate that is then attached to the front of the stator. This arrangement of the coil and capacitor increases the mechanical stability of the apparatus and minimizes the lengths of the leads attaching the coil to the capacitor, decreasing the associated resistive losses of signal. The sample is loaded in a custom-machined microrotor that is mounted on the top of a conventional rotor (Figure 5c), which spins normally at MAS rates typical of that rotor size. These piggy-backed assemblies are fully tunable on multiple channels, allowing for CPMAS experiments [128, 129], and have even been triple tuned [130] making possible an even wider range of experiments.
4. Quadrupolar Nuclei in Biomolecular NMR
4.1. 17O
Oxygen-17 is an underutilized but potentially very informative nucleus in the study of biomolecules: it can be used to observe protonation states [131], the strength, distance and number of hydrogen bonds [131, 132], water intercalation [133], and the lengths [134] and orientations [132, 135] of carbonyl carbon-owygen bonds. However, in solid-state NMR it presents significant challenges due to being a quadrupolar spin 5/2 nucleus with low natural abundance (0.037%) and a sensitivity approximately 1/15th that of 13C. If these limitations can be overcome, 17O can be used to provide substantial site-specific information via analysis of the central transition (-1/2 → 1/2) and extraction of the isotropic chemical shift, quadrupolar coupling constant, and asymmetric parameters. Recent advances in proton decoupling [136, 137], high-field strength [138] and rotor synchronized sequences [137, 139] have contributed to our ability to study the central transition under MAS.
However, broadening due to the second order quadrupolar coupling (a fourth-rank spatial tensor) cannot be completely removed by MAS alone. In quadrupolar NMR, this is often addressed using MQMAS [140] and subsequently developed sequences, including satellite transition MAS (STMAS) [141] and rotor assisted population transfer (RAPT) [142]. Specialized probes that provide extra spatial degrees of freedom are also used in 17O NMR stud ies, principally those built to perform double rotation (DOR) [143] and dynamic angle spinning (DAS) [144]. DOR utilizes two rotors; the outer rotor spins at the magic angle (54.74°) while the inner rotor spins at a quadrupolar magic angle (30.6°) [145], representing the zeros of the second- and fourth-order Legendre polynomials [146]. In DAS, a single rotor changes its spinning axis during the experiment, typically between the quadrupolar magic angles (37.4° and 79.2°) [146, 147].
Specialized probes can potentially provide advantages in the form of shorter acquisition times and better sensitivity, while pulse sequences are more accessible to the majority of users. These complementary approaches have been productively used together in a number of combinations [133, 136, 137, 138], notably cross-polarized DAS [148] and double-frequency sweep DOR [149]. It has also been proposed that determination of chemical shift anisotropy (CSA) will be more reliable [133], and resolution will continue to approach that of spin 1/2 systems [138] with the advent of higher magnetic fields.
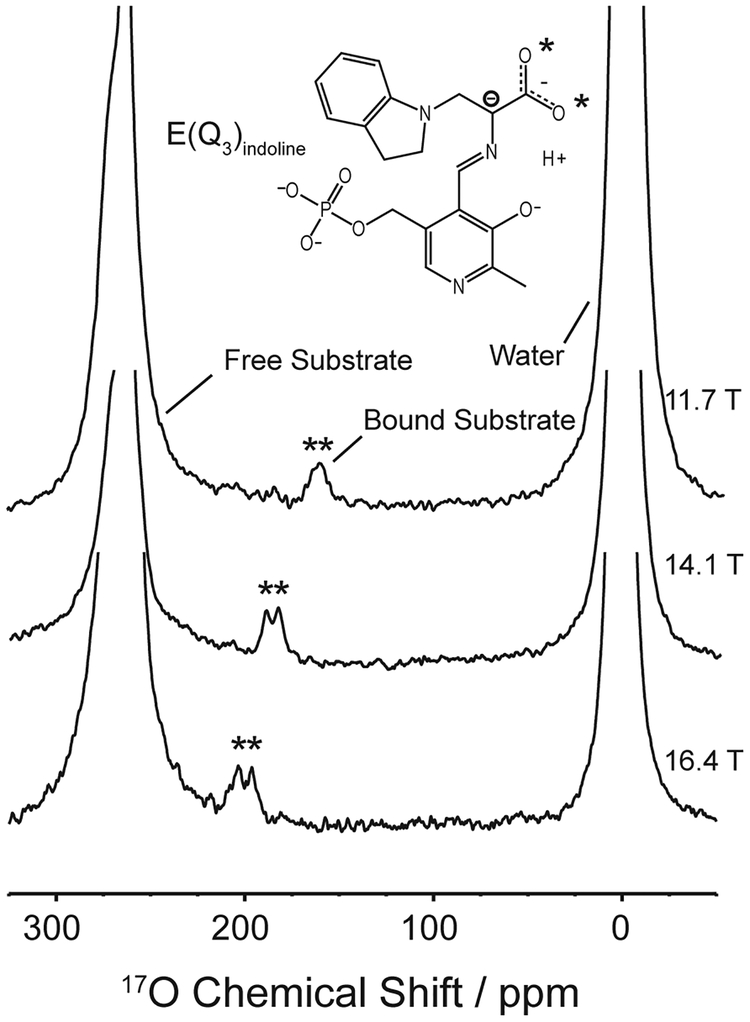
These techniques have been used in studies on important biological molecules and systems such as amino acids, nucleic acids, polypeptides, water-protein and ligand-protein interactions [150], transmembrane proteins [131], enzymatic reaction intermediates [132], ion channels [135] and with eight oxygens being the largest set of unique sites characterized to date [137]. An example of how 17O NMR was used to detect protein-ligand binding is shown in Figure 6.
Figure 6:
17O spectra demonstrating the utility of this nucleus for characterizing reaction mechanisms, particularly in acid-base chemistry. Here, the binding of a substrate to the enzyme tryptophan synthase is observed [150].
5. Deuterium
One of the biggest challenges in solid-state NMR is effectively using proton detection, as is routine for structure determination in solution. Many efforts have been directed toward taking advantage of the sensitivity associated with the high proton gyromagnetic ratio, while at the same time limiting the line broadening induced by the strong 1H-1H homonuclear dipolar coupling [151]. As is the case for many experimental methods, in practical terms, it is often possible to achieve a gain in resolution at the expense of sensitivity, or vice versa. Due to these challenges it was diffcult to achieve high-resolution 1H spectra of large biomolecules (MW <20kDa) prior to recent developments in methodology, in particular those involving fast MAS and deuterium labeling [151]. Fast and ultra-fast MAS o er improved 1H resolution, as linewidth is linearly dependent on the inverse MAS rate [152]; however these experiments can only be performed with small-diameter rotors and therefore reduced sample volumes. Similarly, extensive deuteration improves resolution by diminishing the number of homonuclear “neighbors” impacting each proton, resulting in less need for the high power rf decoupling that is one of the biggest contributors to sample heating [153, 154]. However, these gains come at the cost of sensitivity concomitant with the reduction in proton concentration. This is usually true even for experiments with heteronuclear detection, because so many sequences begin with polarization transfer from the abundant protons to the sparse 13C or 15N nuclei.
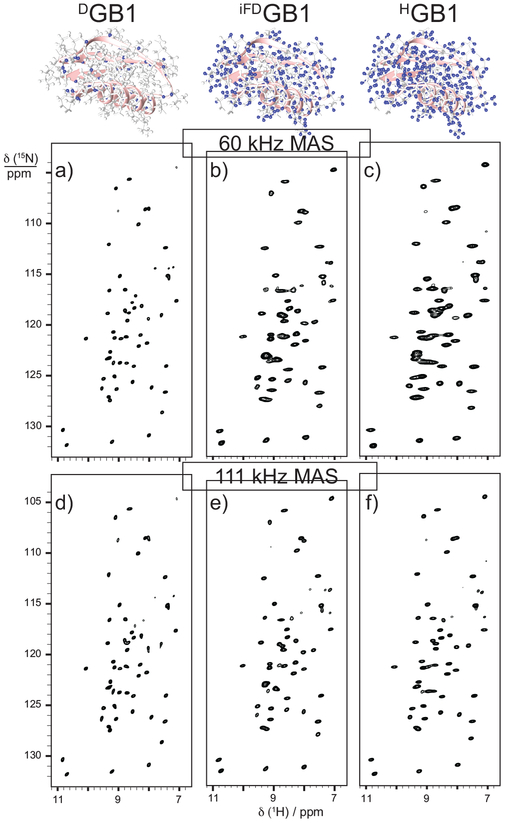
Fast MAS rates (30–100kHz) compare favorably to deuteration as a means of achieving well-resolved 1H spectra in cases where sample amount is already limited to sub-milligram quantities by sample preparation constraints and loss in sensitivity would be particularly unfavorable [155]. A comparison of the effects of fast MAS and di erent deuteration levels is shown in Figure 7 for a microscrystalline protein sample [156]. This is particularly applicable for large proteins or those that are insoluble or otherwise diffcult to express. The effectiveness of fast MAS is also dependent on the applied magnetic field strength. Proton chemical shifts can be better resolved using fast MAS at ultra-high field than using fractional deuteration on a lower-field instrument, however the extent to which this is true is dependent on the intrinsic heterogeneity of the protein sample. Perdeuterated, back-exchanged samples provide optimal resolution for the amide and alpha proton resonances, however in most other cases perdeuteration only provided a 10% increase in resolution compared to fast, high-field MAS spectra on fully protonated systems. It has been suggested that deuteration facilitates backbone resonance assignment, while full protonation is ideal for aliphatic side-chains and methyl resonances [156]. The effect of water concentration during crystalization of perdeuterated proteins has been shown to impact resolution and signal to noise ratios of exchangable amide sites as well as di erences in the effectiveness of different polarization transfer experiments such as CP versus INEPT[157]. These observations should be considered when determining the best approach for a given sample.
Figure 7:
Spectral resolution for a microcrystalline sample of the protein GB1 at 60 kHz MAS can be improved by deuteration, while at 111 kHz MAS the difference between the spectra of deuterated and fully protonated GB1 is almost negligible [156].
By taking advantage of all available techniques, it is expected that solid-state NMR will soon be able to characterize the full range of moderately sized proteins (10–100 kDa), through increased coherence lifetimes, more effcient cross-polarization and heteronuclear decoupling, site-specific relaxation and directly determined proton proximities and distance restraints [158]. This has been demonstrated successfully on five different proteins ranging from 5–30kDa with different condensed states using an adapted solution-state protocol and automated computational software, MATCH, for resonance assignment [159]. The discussion of sample deuteration above focuses on its role in simplifying the proton spectra, however 2H is also an excellent NMR nucleus in its own right. Methodological advances enabling optimal use of deuterium in biological systems include MAS probes capable of direct detection on 2H in the context of multidimensional experiments, optimized pulse sequences, extensive or perdeuteration and recrystallization from deuterated solvent [160].
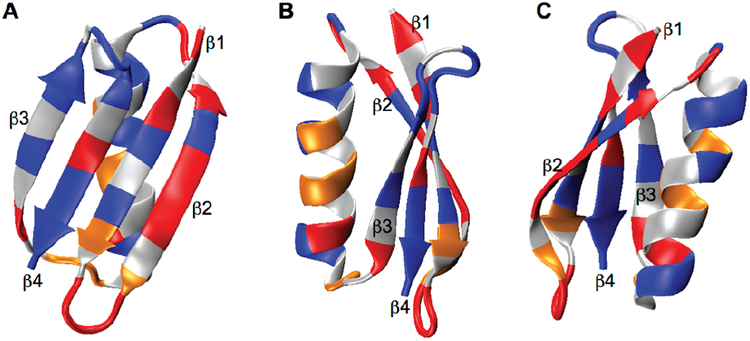
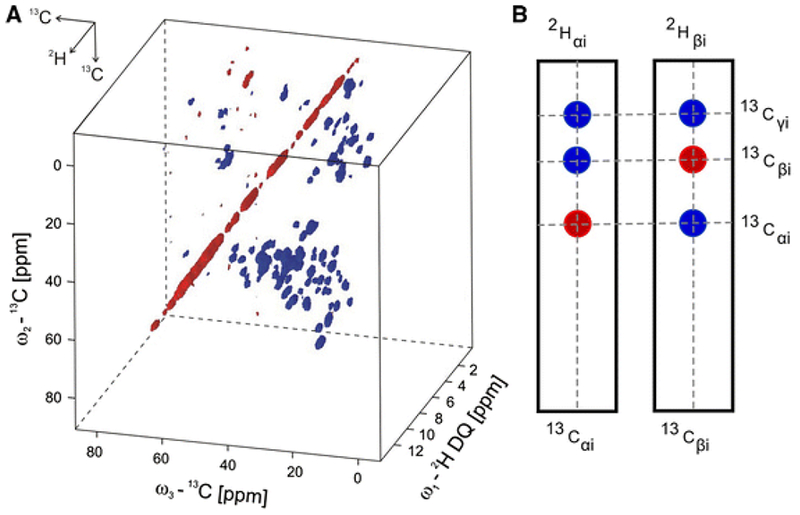
Despite being a quadrupolar (spin 1) isotope with a gyromagnetic ratio 6.5 times less sensitive than 1H, 2H can a ord other information through its anisotropic quadrupolar interaction, providing information about bond orientation, local order, and site-specific mobility [161, 162, 163, 164]. Using double quantum transitions reduces the vulnerability of this experiment to small variations in MAS rate and stability [165] and facilitates the use of 2H as a chemical shift dimension in multidimensional experiments [166]. Another useful property is that deuterium has a much faster relaxation rate than proton, making it useful over a large kinetic range [167, 168] and for dynamics studies [169, 170]. Line-shapes can be analyzed to determine quantitative rate constants [171]. This coupled with its low natural abundance of 0.015% makes it ideal for site-specific studies without interference from the natural abundance background [172]. Site-specific studies can also be extended to characterize the overall mobility of macromolecules, with further potential to elucidate function, as shown in Figure 8. 2D 2HDQ-13C and 3D 2HDQ-13C-13C correlation experiments, an example of which is shown in Figure 9, allow assignment of sidechain spin systems [173], representing a promising direction for future studies with recently developed instrumentation.
Figure 8:
Panels A, B, and C show three different orientations of GB1. The color coding indicates di erences in overall mobility based on site-specific evaluation of 140 aliphatic sidechain sites using deuterium labeling. Red, orange and blue indicate the amplitude of motion, being large, moderate and small, respectively [172].
Figure 9:
A) 3D spectrum of labeled ubiquitin from 2HDQ-13C-13 correlation experiments. B) 2D slices can be used to determine connectivities and assign resonances similar to conventional solution-state techniques. The resolution achieved was also suitable for studies on a much larger membrane protein, OmpG. [173].
An important example of a solid-state NMR probe capable of utilizing deuterium signals was built for investigating the phase behavior of lipid bilayers and conformational changes in phospholipid headgroups as a function of pressure [174]. This probe required high power for broadband excitation over the entire range of the wide-line deuterium spectrum, a necessity that can be greatly reduced with the implementation of MAS in conjunction with partial motional averaging [170, 175]. An auxillary deuterium coil for decoupling in 2H-labeled proteins was developed to provide the necessary decoupling power with a simple modification to existing triple-resonance (1H/13C/15N) biomolecular probes without the need for another instrument [176], while a quadruple-resonance probe operating at 600 MHz proton Larmor frequency was built for multidimensional experiments including 2H excitation [177]. In multidimensional experiments, the signal-to-noise was demonstrated to be comparable or slightly better when initiating excitation through deuterium, and using Rotor Echo Short Pulse IRrAdiaTION (RESPIRATION) [172] and optimal control (OC) cross polarization pulse sequences allowed for lower rf power, better tolerance of rf inhomogeneity, and improved effciency of magnetization transfer [178]. 2D experiments utilizing initial excitation on deuterium and hydrogen were compared, showing di erences in cross peaks that can be used to identify solvent-exposed structural components or membrane interiors due to their relative ability to back-exchange [177]. The Martin lab recently developed a quadruple-resonance (1H/13C/2H/15N) probe for use at 800 MHz 1H Larmor frequency. This probe includes a high-power deuterium channel for decoupling and detection of 2H in deuterated biomolecular samples [62].
Traditionally, low-γ nuclei were detected indirectly through proton in order to yield higher sensitivity, as signal to noise is proportional to γ2/3[179]. This was speculated to be compromised by extensive deuteration; however Rienstra and Zilm independently showed that this limitation could be overcome through intermediate deuteration and reverse cross polarization (RCP) [152] and CPMAS on extensively deuterated proteins [153]. At the same time, Reif and coworkers found that fully deuterating at nonexchangable sites while back-exchanging the amide protons limits spin di usion averaging of relaxation times, making it possible to extract order parameters from dipolar interactions and scaled quadrupolar tensor anisotropy parameters [154].
The demonstration of triple cross polarization (TCP) by simultaneous CP on 1H-13C and 2H-13C results in up to a fourfold gain in sensitivity compared to direct excitation on carbon. This method is particularly useful for systems where back-exchange is diffcult or impossible, such as membrane proteins, where the protein interior is not exposed to aqueous solvent [180]. Oschkinat et al. initially used the double nucleus enhanced recoupling (DONER) pulse sequence for simultaneous irradiation of protons and deuterons, providing higher sensitivity in carbon correlation spectroscopy at distances of up to 6 Å. This provided reasonable 1H-13C cross-polarization effciency even at high levels of deuteration and resulted in 3–5 times stronger peak intensities and coverage of the full aliphatic spectral range as compared to previous techniques such as PDSD and 1H-DARR [181]. The effect of the fraction of deuterated sites in conjunction with the MAS rate on resolution and sensitivity has been extensively characterized [152, 182, 183]. Recently developed pulse sequences such as frequency-selective REDOR (FSR) support experiments on deuterated samples under fast MAS enable the measurement of long-range distances along with other structural restraints [184]. Sensitivity is of particular concern in analysis of fully deuterated systems. Smaller rotor size in conjunction with fast MAS has been shown to improve sensitivity [182], especially for multidimensional experiments requiring transfer between 15N-13C and 13C-13C [183]. A high-quality review on the structure and dynamics of perdeuterated proteins using MAS provides a more detailed reference specific to this area [185].
6. Ultrafast MAS
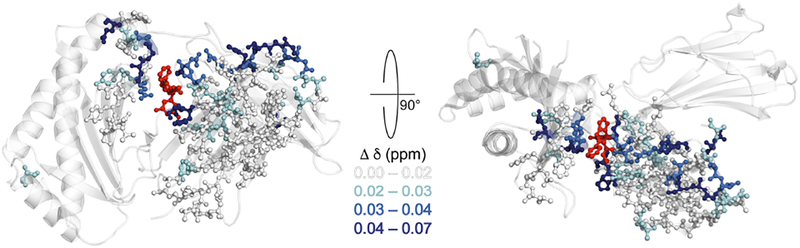
Increasing the MAS rate from on the order of 25 kHz in the late 1990s to well over 100 kHz at the present has required technical devlopments in rotors and bearings, pioneered by Samoson and co-workers [186, 187] and quickly adopted by other research groups and commercial probe manufacturers. The trend toward increasingly fast MAS rates is synergistic with other technological and methodological advances that improve the sensitivity of the NMR experiment, including high-field magnets, labeling schemes, and DNP. A recent example of work done at ultrafast MAS speeds, could set a new precedent for experiments to support drug design and testing is shown in Figure 10.
Figure 10:
Above is an illustration of observed chemical shift perturbations as they relate to the overall structure upon binding of a ligand, UCB-FcRn-303, to the extracellular domain of a 42 kDa, fully protonated neonatal Fc receptor (FcRn). This receptor was chosen because it has been established as a drug target for several autoimmune diseases. Color denotes the magnitude of the perturbation as shown by the scale [188].
Sensitivity is critically important for these experiments because faster MAS necessarily means less sample volume: the maxiumum rotor frequency is inversely dependent on the size of the rotor such that the current maxiumum of ≈ 130 kHz is achievable with 0.75 mm rotors [189]. Further experimental considerations include interference between the spin rotation by rf irradiation and mechanical rotation of the sample [190], however effcient polarization transfer has been achieved using first-order recoupling pulse sequences [158]. Experiments focused on distance restraints should be designed with longer mixing times or stronger interactions to reduce dipolar truncation at ultra-fast MAS rates, coupled with optimization in second-order pulse sequences, which is currently an active research area. Frictional heating under ultra-fast MAS can cause a 20–40 K increase in temperature, which especially problematic for biomacromolecule samples that are thermally sensitive or negatively impacted by dehydration. In modern MAS systems, these concerns are overcome using sample cooling and watertight rotor seals [155].
7. Outlook
One important category of projected improvements to MAS probes concerns technological advances that have been demonstrated, but are not yet standard equipment for most commercial probes. The routine addition of gradient coils to nearly all high-resolution solution-state NMR probes has led to the development of a plethora of complex pulse sequences that use gradients for coherence selection, eliminating the long phase cycles often used in solid-state NMR experiments. The implementation of similar sequences in MAS probes requires a slightly more complicated coil arrangement, as the gradient is most usefully applied along the magic angle. This has been demonstrated in at least three fairly early designs [191, 192, 193, 84], and commercial implementations have also become available [194]. Gradients have been used for water suppression in 1H-detected MAS experiments, both with a PFG probe [195] and using a homospoil gradient provided by the Z1 shim coil [196]. As MAS becomes more of a mature technology and throughput increases, the automation of time-consuming and diffcult optimization procedures becomes increasingly valuable. Instrumentation and procedures for automated tuning [197] and gradient shimming [198] of MAS probes have been introduced.
Another frontier of potential automation involves real-time optimization of pulse sequences and gradient waveforms for particular experiments, an approach that becomes more feasible as computational power increases, speeding up the relevant calculations. Examples include the “shim pulse” shimming method, in which a tailored gradient waveform is used to correct a mapped B0 inhomogeneity [199]. This approach would be most useful if applied in a fully or semi-automated manner, where the B0 field is first mapped and then corrected using gradient pulses, a modality that was not computationally feasible at the time of the first demonstration. In a similar vein, optimal control [200, 201] has been used to design tailored pulse shapes for a variety of NMR experiments, including dipolar recoupling sequences [202], as well as the NCO and NCA experiments that are essential for obtaining protein backbone assignments [203].
The experimental demands of solving structures of ever larger and more complex biological assemblies drives the development of ever higher-field NMR magnets. The practical limit using currently known low-temperature superconductor technology appears to be on the order of 1 GHz proton Larmor frequency [204, 205]. The impetus to increase sensitivity and resolution continues to drive the production of NMR magnets with even higher field, using a variety of different approaches, including high-temperature superconductors [206, 207, 208], powered resistive magnets used in either continuous field [209, 210] or pulsed [211, 212, 213] mode, and resistive/superconducting series-connected hybrid magnets [56].
A major obstacle to the construction of high-field NMR magnets is field stabilization. The highest field NMR magnets often drift significantly on the timescale of the type of multidimensional experiment needed for protein structure determination. One approach that is often successful in super-conducting magnets is to measure the drift and increment the Z0 room-temperature shim accordingly, although it is tedious to calibrate and does not account for sudden perturbations. An external lock probe located outside the center of the field [214] allows field regulation during sample changes, and is an attractive approach for very high field magnets that are subject to large field variations. With their dramatic increase in sensitivity, high-field magnets promise to reduce the sample volume and experiment time needed for NMR experiments, enabling experiments on samples that are diffcult to prepare in large quantities, surfaces, and materials with limited stability that may not be stable over the several days currently needed for protein structure determination.
Hybrid techniques, where solid-state NMR synergizes with other methods are another important future direction. A combined MAS NMR and SAXS study provided an early look at the large functional oligomers of the holdase chaperone αB-crystallin, with SAXS providing the overall size and shape of the complex and NMR data filling in the molecular details [215]. Solid-state NMR has been used with CS-Rosetta [216] to solve the structure of a filamentous phage capsid [217]. Cryo-EM can be used in conjunction with molecular modeling software [218, 219] to provide a framework for the overall assembly and relative orientation of the subunits, with atomic-level detail as well as dynamics information provided by MAS NMR [220]. Alternatively, the overall model can be built and refined from solid-state NMR and EM restraints simultaneously [221]. Combinations of NMR, Cryo-EM, and molecular modeling approaches are likely to play an increasingly important role in future structure determination efforts [222].
8. Acknowledgments
The Martin group’s work on the development of solid-state NMR probes is supported by NSF grant CHE-1308231 and NIH grant 1R01EY021514; our work on amyloid fibrils is supported by NSF grant DMS-1361425. JIK acknowledges support from NIH training grant T32 GM108561 and the NSF GRFP. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1321846. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. We thank L. Mueller, H. Oschkinat, D. Piehl, G. Pintacuda, C. Rienstra, and D. Stöppler for providing figures illustrating recent advances.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Fricke P, Chevelkov V, Shi C, Lange A, Strategies for solid-state NMR investigations of supramolecular assemblies with large subunit sizes, Journal of Magnetic Resonance 253 (2015) 2–9. [DOI] [PubMed] [Google Scholar]
- [2].Ding Y, Yao Y, Marassi FM, Membrane protein structure determination in membrana, Accounts of Chemical Research 46 (9) (2013) 2182–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tycko R, Amyloid polymorphism: Structural basis and neurobiological relevance, Neuron 86 (3) (2015) 632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Marvin D, Symmons M, Straus S, Structure and assembly of filamentous bacteriophages, Progress in Biophysics and Molecular Biology 114 (2014) 80–122. [DOI] [PubMed] [Google Scholar]
- [5].Han Y, Hou G, Suiter CL, Ahn J, Byeon I-JL, Lipton AS, Burton S, Hung I, Gorkov PL, Gan Z, Brey W, Rice D, Gronenborn AM, Polenova T, Magic angle spinning NMR reveals sequence-dependent structural plasticity, dynamics, and the spacer peptide 1 conformation in HIV1 capsid protein assemblies, Journal of the American Chemical Society 135 (2013) 17793–17803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lamley JM, Iuga D, Oster C, Sass H, Rogowski M, Oss A, Past J, Reinhold A, Grzesiek S, Samoson A, Lewandowski JR, Solid-state NMR of a protein in a precipitated complex with a full-length antibody, Journal of the American Chemical Society 136 (2014) 16800–16806. [DOI] [PubMed] [Google Scholar]
- [7].Marchanka A, Simon B, Altho-Ospelt G, Carlomagno T, RNA structure determination by solid-state NMR spectroscopy, Nature Communications 6 (2015) 7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Pandit A, Ocakoglu K, Buda F, van Marle T, Holzwarth AR, de Groot HJM, Structure determination of a bio-inspired self-assembled light-harvesting antenna by solid-state NMR and molecular modeling, The Journal of Physical Chemistry B 117 (2013) 11292–11298. [DOI] [PubMed] [Google Scholar]
- [9].Harris MJ, Struppe JO, Wylie BJ, McDermott AE, Thompson LK, Multidimensional solid-state nuclear magnetic resonance of a functional multiprotein chemoreceptor array, Biochemistry 55 (2016) 3616–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yana S, Guo C, Hou G, Zhang H, Lua X, Williams JC, Polenova T, Atomic-resolution structure of the CAP-Gly domain of dynactin on polymeric microtubules determined by magic angle spinning NMR spectroscopy, Proceedings of the National Academy of Sciences of the United States of America 112 (47) (2015) 14611–14616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Goldbourt A, Biomolecular magic-angle spinning solid-state NMR: recent methods and applications, Current Opinion in Biotechnology 24 (2013) 705–715. [DOI] [PubMed] [Google Scholar]
- [12].Linser R, Solid-state NMR spectroscopic trends for supramolecular assemblies and protein aggregates, Solid State Nuclear Magnetic Resonance 87 (2017) 45–53. [DOI] [PubMed] [Google Scholar]
- [13].Habenstein B, Loquet A, Solid-state NMR: An emerging technique in structural biology of self-assemblies, Biophysical Chemistry 210 (2016) 14–26. [DOI] [PubMed] [Google Scholar]
- [14].Quinn CM, Polenova T, Structural biology of supramolecular assemblies by magic-angle spinning NMR spectroscopy, Quarterly Reviews of Biophysics 50 (2017) e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Brown LS, Ladizhansky V, Membrane proteins in their native habitat as seen by solid-state NMR spectroscopy: Membrane Proteins in their Native Habitat, Protein Science 24 (9) (2015) 1333–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wylie BJ, Do HQ, Borcik CG, Hardy EP, Advances in solid-state NMR of membrane proteins, Molecular Physics 114 (24) (2016) 3598–3609. [Google Scholar]
- [17].Molugu TR, Lee S, Brown MF, Concepts and Methods of Solid-State NMR Spectroscopy Applied to Biomembranes, Chemical Reviews 117 (19) (2017) 12087–12132. [DOI] [PubMed] [Google Scholar]
- [18].Meier BH, Riek R, Böckmann A, Emerging structural understanding of amyloid fibrils by solid-state NMR, Trends in Biochemical Sciences 42 (10) (2017) 777–787. [DOI] [PubMed] [Google Scholar]
- [19].Sunde M, Serpell LC, Bartlam M, Fraser PE, Pepys MB, Blake CCF, Common core structure of amyloid fibrils by synchrotron x-ray di raction, Journal of Molecular Biology 273 (3) (1997) 729–739. [DOI] [PubMed] [Google Scholar]
- [20].Serpell LC, Sunde M, Benson MD, Tennent GA, Pepys MB, Fraser PE, The protofilament substructure of amyloid fibrils, Journal of Molecular Biology 300 (5) (2000) 1033–1039. [DOI] [PubMed] [Google Scholar]
- [21].Serpell LC, Smith JM, Direct visualisation of the beta-sheet structure of synthetic alzheimer’s amyloid, Journal of Molecular Biology 299 (1) (2000) 225–231, 318NC J MOL BIOL. [DOI] [PubMed] [Google Scholar]
- [22].Benzinger TLS, Gregory DM, Burkoth TS, Miller-Auer H, Lynn DG, Botto RE, Meredith SC, Propagating structure of Alzheimer’s β-amyloid (10–35) is parallel β-sheet with residues in exact register, Proceedings of the National Academy of Sciences of the United States of America 95 (1998) 13407–13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Antzutkin ON, Balbach JJ, Leapman RD, Rizzo NW, Reed J, Tycko R, Multiple quantum solid-state NMR indicates a parallel, not antiparallel, organization of β-sheets in Alzheimer’s β-amyloid fibrils, Proceedings of the National Academy of Sciences of the United States of America 97 (24) (2000) 13045–13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tang M, Comellas G, Rienstra CM, Advanced solid-state NMR approaches for structure determination of membrane proteins and amyloid fibrils, Account of Chemical Research 46 (9) (2013) 2080–2088. [DOI] [PubMed] [Google Scholar]
- [25].Petkova AT, Leapman RD, Guo ZH, Yau WM, Mattson MP, Tycko R, Self-propagating, molecular-level polymorphism in alzheimer’s beta-amyloid fibrils, Science 307 (5707) (2005) 262–265. [DOI] [PubMed] [Google Scholar]
- [26].Paravastu AK, Leapman RD, Yau W-M, Tycko R, Molecular structural basis for polymorphism in Alzheimer’s β-amyloid fibrils, Proceedings of the National Academy of Sciences 105 (47) (2008) 18349–18354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Döbeli H, Schubert D, Riek R, 3D structure of Alzheimer’s amyloid-β(1–42) fibrils, Proceedings of the National Academy of Sciences of the United States of America 102 (48) (2005) 17342–17347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Xiao Y, Ma B, McElheny D, Parthasarathy S, Long F, Hoshi M, Nussinov R, Ishii Y, Aβ(1–42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease, Nature Structural & Molecular Biology 22 (6) (2015) 499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Qiang W, Yau W-M, Luo Y, Mattson MP, Tycko R, Antiparallel β-sheet architecture in Iowa-mutant β-amyloid fibrils, Proceedings of the National Academy of Sciences 109 (12) (2012) 4443–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Theint T, Nadaud PS, Aucoin D, Helmus JJ, Pondaven SP, Surewicz K, Surewicz WK, Jaroniec CP, Species-dependent structural polymorphism of Y145Stop prion protein amyloid revealed by solid-state NMR spectroscopy, Nature Communications 8 (2017) 753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sawaya M, Sambashivan S, Nelson R, Ivanova M, Sievers SA, Astopol M, Thompson M, Balbirnie M, Wiltzius J, Macfarlane H, Madsen AO, Riekel C, Eisenberg D, Atomic structures of amyloid cross-beta spine reveal varied steric zippers, Nature 447 (2007) 453–457. [DOI] [PubMed] [Google Scholar]
- [32].Iwata K, Fujiwara T, Matsuki Y, Akutsu H, Takahashi S, Naiki H, Goto Y, 3D structure of amyloid protofilaments of β2-microglobulin fragment probed by solid-state NMR, Proceedings of the National Academy of Sciences of the United States of America 102 (48) (2006) 18119–18124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ferguson N, Becker J, Tidow H, Tremmel S, Sharpe TD, Krause G, Flinders J, Petrovich M, Berriman J, Oschkinat H, et al. , General structural motifs of amyloid protofilaments, Proceedings of the National Academy of Sciences of the United States of America 103 (44) (2006) 16248–16253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Van Melckebeke H, Wasmer C, Lange A, Ab E, Loquet A, Böckmann A, Meier B, Atomic-resolution three-dimensional structure of HET-s(218–289) amyloid fibrils by solid-state NMR spectroscopy, Journal of the American Chemical Society 132 (39) (2010) 13765–13775. [DOI] [PubMed] [Google Scholar]
- [35].Bayro MJ, Debelouchina GT, Eddy MT, Birkett NR, MacPhee CE, Rosay M, Maas WE, Dobson CM, Gri n RG, Inter-molecular structure determination of amyloid fibrils with magic-angle spinning and dynamic nuclear polarization NMR, Journal of the American Chemical Society 133 (2011) 13967–13974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lu J-X, Qiang W, Yau W-M, Schwieters CD, Meredith SC, Tycko R, Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue, Cell 154 (6) (2013) 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Tuttle MD, Comellas G, Nieuwkoop AJ, Covell DJ, Berthold DA, Kloepper KD, Courtney JM, Kim JK, Barclay AM, Kendall A, Wan W, Stubbs G, Schwieters CD, Lee VM, George JM, Rienstra CM, Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein, Nature Structural and Molecular Biology 23 (2016) 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Schütz AK, Vagt T, Huber M, Ovchinnikova OY, Cadal-bert R, Wall J, Güntert P, Böckmann A, Glockshuber R, Meier BH, Atomic-resolution three-dimensional structure of amyloid β fibrils bearing the Osaka mutation, Angewandte Chemie International Edition 54 (1) (2015) 331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wälti MA, Ravotti F, Arai H, Glabe CG, Wall JS, Böckmann A, Güntert P, Meier BH, Riek R, Atomic-resolution structure of a disease-relevant Aβ (1–42) amyloid fibril, Proceedings of the National Academy of Sciences 113 (34) (2016) E4976–E4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Damo S, Phillips A, Young A, Li S, Woods V Jr.., Wemmer D, Probing the conformation of a prion protein fibril with hydrogen exchange, Journal of Biological Chemistry 285 (2010) 32303–32311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Pavlova A, Cheng C-Y, Kinnebrew M, Lew J, Dahlquist FW, Han S, Protein structural and surface water rearrangement constitute major events in the earliest aggregation stages of tau, Proceedings of the National Academy of Sciences of the United States of America 113 (2) (2016) E127–E136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Aucoin D, Xia Y, Theint T, Nadaud PS, Surewicz K, Surewicz WK, Jaroniec CP, Protein-solvent interfaces in human Y145Stop prion protein amyloid fibrils probed by paramagnetic solid-state NMR spectroscopy, Journal of Structural Biology in press (2018) 10.1016/j.jsb.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Frederick KK, Debelouchina GT, Kayatekin C, Dorminy T, Jacavone AC, Gri n RG, Lindquist S, Distinct prion strains are defined by amyloid core structure and chaperone binding site dynamics, Chemistry & Biology 21 (2014) 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Frederick KK, Michaelis VK, Caporini MA, Andreas LB, Debelouchina GT, Gri n RG, Lindquist S, Combining DNP NMR with segmental and specific labeling to study a yeast prion protein strain that is not parallel in-register, Proceedings of the National Academy of Sciences of the United States of America 114 (14) (2017) 3642–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Frederick KK, Michaelis VK, Corzilius B, Ong T-C, Jacavone AC, Gri n RG, Lindquist S, Sensitivity-enhanced NMR reveals alterations in protein structure by cellular milieus, Cell 163 (2015) 620–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lee D, Hediger S, DePaëpe G, Is solid-state NMR enhanced by dynamic nuclear polarization?, Solid State Nuclear Magnetic Resonance 66–67 (2015) 6–20. [DOI] [PubMed] [Google Scholar]
- [47].Thankamony ASL, Wittmann M, Kaushik Johannes J., Corzilius B Dynamic nuclear polarization for sensitivity enhancement in modern solid-state NMR, Progress in Nuclear Magnetic Resonance Spectroscopy 102–103 (2017) 120–195. [DOI] [PubMed] [Google Scholar]
- [48].Rosay M, Blank M, Engelke F, Instrumentation for solid-state dynamic nuclear polarization with magic angle spinning NMR, Journal of Magnetic Resonance 264 (2016) 88–98. [DOI] [PubMed] [Google Scholar]
- [49].Akbey Ü, Franks WT, Linden A, Lange S, Gri n RG, van Rossum B, Oschkinat H, Dynamic nuclear polarization of deuterated proteins, Angewandte Chemie International Edition 49 (42) (2010) 7803–7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Akbey Ü, Oschkinat H, Structural biology applications of solid state MAS DNP NMR, Journal of Magnetic Resonance 269 (2016) 213–224. [DOI] [PubMed] [Google Scholar]
- [51].McDermott AE, Polenova T, Bockmann A, Zilm KW, Paulson EK, Martin RW, Montelione GT, Partial nmr assignments for uniformly (13c-15n)-enriched bpti in the solid state, Journal of Biomolecular NMR 16 (2000) 209–219. [DOI] [PubMed] [Google Scholar]
- [52].Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H, Structure of a protein determined by solid-state magic-angle-spinning nmr spectroscopy, Nature 420 (6911) (2002) 98–102. [DOI] [PubMed] [Google Scholar]
- [53].Cross VR, Hester RK, Waugh JS, Single coil probe with transmission-line tuning for nuclear magnetic double-resonance, Review of Scientific Instruments 47 (12) (1976) 1486–1488. [Google Scholar]
- [54].Kan S, Fan M, Courtieu J, A single-coil triple resonance probe for NMR experiments, Review of Scientific Instruments 51 (7) (1980) 887–890. [Google Scholar]
- [55].Hoult D, Richards R, The signal-to-noise ratio of the nuclear magnetic resonance experiment, Journal of Magnetic Resonance 24 (1976) 71–85. [DOI] [PubMed] [Google Scholar]
- [56].Gan Z, Hung I, Wang X, Paulino J, Wu G, Litvak IM, Gor’kov PL, Brey WW, Lendi P, Schiano JL, Bird MD, Dixon IR, Toth J, Boebinger GS, Cross TA, NMR spectroscopy up to 35.2 T using a series-connected hybrid magnet, Journal of Magnetic Resonance 284 (2017) 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].McKay RA, Transmission Line Probes, Vol. 6 of Encyclopedia of Nuclear Magnetic Resonance, Wiley, Chichester, 1996, pp. 3768–3771. [Google Scholar]
- [58].Stringer JA, Drobny GP, Methods for the analysis and design of a solid state nuclear magnetic resonance probe, Review of Scientific Instuments 69 (9) (1998) 3384–3391. [Google Scholar]
- [59].Markhasin E, Hu J, Su Y, Herzfeld J, Grin RG, Effcient, balanced, transmission line rf circuits by back propagation of common impedance nodes, Journal of Magnetic Resonance 231 (2013) 32–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Barnes AB, Markhasin E, Daviso E, Michaelis VK, Nanni EA, Jawla SK, Mena EL, DeRocher R, Thakkar A, Woskov PP, Herzfeld J, Temkin c. R. J., Grin RG, Dynamic nuclear polarization at 700 MHz/460 GHz, Journal of Magnetic Resonance 224 (2012) 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Martin RW, Paulson E, Zilm KW, Design of a triple resonance magic angle sample spinning probe for high field solid state nuclear magnetic resonance, Review of Scientific Instruments 74 (6) (2003) 3045–3061. [Google Scholar]
- [62].Collier KA, Sengupta S, Espinosa CA, Kelly JE, Kelz JI, Martin RW, Design and construction of a quadruple-resonance MAS NMR probe for investigation of extensively deuterated biomolecules, Journal of Magnetic Resonance 285 (2017) 8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].McNeill SA, Gor’kov P, Shetty K, Brey WW, Long JR, A low-E Magic Angle Spinning probe for biological solid-state NMR at 750 MHz, Journal of Magnetic Resonance 197 (2009) 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Barnes AB, Mak-Jurkauskas ML, Matsuki Y, Bajaj VS, van der Wel PC, DeRocher R, Bryant J, Sirigiri JR, Temkin RJ, Lugtenburg J, Herzfeld J, Gri n RG, Cryogenic sample exchange NMR probe for magic angle spinning dynamic nuclear polarization, Journal of Magnetic Resonance 198 (2009) 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Doty F, Kulkarni J, Turner C, Entzminger G, Bielecki A, Using a cross-coil to reduce RF heating by an order of magnitude in triple-resonance multinuclear MAS at high fields, Journal of Magnetic Resonance 182 (2006) 239–253. [DOI] [PubMed] [Google Scholar]
- [66].Schaefer J, Stejskal EO, C-13 nuclear magnetic resonance of polymers spinning at magic angle, Journal of the American Chemical Society 98 (4) (1976) 1031–1032. [Google Scholar]
- [67].Metz G, Wu XL, Smith SO, Ramped-amplitude cross-polarization in magic angle spinning NMR, Journal of Magnetic Resonance Series A 110 (2) (1994) 219–227. [Google Scholar]
- [68].Idziak S, Haeberlen U, Design and construction of a high homogeneity rf coil for solid-state multiple-pulse NMR, Journal of Magnetic Resonance 50 (2) (1982) 281–288. [Google Scholar]
- [69].Alderman D, Grant D, Effcient decoupler coil design which reduces heating in conductive samples in superconducting spectrometers, Journal of Magnetic Resonance 36 (3) (1979) 447–451. [Google Scholar]
- [70].Larsen F, Daugaard P, Jakobsen H, Nielsen N, Improving rf field homogeneity in solid-state MAS NMR using a loopgap resonator, Journal of Magnetic Resonance Series A 115 (1995) 283–286. [Google Scholar]
- [71].Stringer JA, Bronniman C, Mullen C, Zhou D, Stellfox S, Li Y, Williams E, Rienstra C, Reduction of rf-induced sample heating with a scroll coil resonator structure for solid-state nmr probes, Journal of Magnetic Resonance 173 (2005) 40–48. [DOI] [PubMed] [Google Scholar]
- [72].Charmont P, Sakellariou D, Emsley L, Sample restriction using radiofrequency field selective pulses in high-resolution solid-state NMR, Journal of Magnetic Resonance 154 (1) (2002) 136–141. [DOI] [PubMed] [Google Scholar]
- [73].Charmont P, Lesage A, Steuernagel S, Engelke F, Emsley L, Sample restriction using magnetic field gradients in high-resolution solid-state nmr, Journal of Magnetic Resonance 145 (2) (2000) 334–339. [DOI] [PubMed] [Google Scholar]
- [74].Nagashima H, Trebosc J, Lafon O, Pourpoint F, Paluch P, Potrzebowski MJ, Amoureux J-P, Imaging the spatial distribution of radiofrequency field, sample and temperature in MAS NMR rotor, Solid State Nuclear Magnetic Resonance 87 (2017) 137–142. [DOI] [PubMed] [Google Scholar]
- [75].Gupta R, Hou G, Polenova T, Vega A, RF inhomogeneity and how it controls CPMAS, Solid State Nuclear Magnetic Resonance 72 (2015) 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Vega AJ, Controlling the effects of pulse transients and rf inhomogeneity in phase-modulated multiple-pulse sequences for homonuclear decoupling in solid-state proton NMR, Journal of Magnetic Resonance 170 (1) (2004) 22–41. [DOI] [PubMed] [Google Scholar]
- [77].Tošner Z, Purea A, Struppe JO, Wegner S, Engelke F, Glaser SJ, Reif B, Radiofrequency fields in MAS solid state NMR probes, Journal of Magnetic Resonance 284 (2017) 20–32. [DOI] [PubMed] [Google Scholar]
- [78].Purusottam RN, Bodenhausen G, Tekely P, Sensitivity improvement during heteronuclear spin decoupling in solid-state nuclear magnetic resonance experiments at high spinning frequencies and moderate radio-frequency amplitudes, Chemical Physics Letters 614 (2014) 220–225. [Google Scholar]
- [79].Purusottam RN, Bodenhausen G, Tekely P, Effects of inherent rf field inhomogeneity on heteronuclear decoupling in solid-state NMR, Chemical Physics Letters 635 (2015) 157–162. [Google Scholar]
- [80].Campbell G, Galya L, Beeler A, English A, Effect of rf inhomogeneity upon quantitative solid-state NMR measurements, Journal of Magnetic Resonance Series A 112 (2) (1995) 225–228. [Google Scholar]
- [81].Nishimura K, Fu R, Cross T, The effect of rf inhomogeneity on heteronuclear dipolar recoupling in solid state NMR: practical performance of SFAM and REDOR, Journal of Magnetic Resonance 152 (2) (2001) 227–233. [Google Scholar]
- [82].Paulson E, Martin RW, Zilm KW, Cross polarization, radio frequency field homogeneity, and circuit balancing in high field solid state nmr probes, Journal of Magnetic Resonance 171 (2004) 314–323. [DOI] [PubMed] [Google Scholar]
- [83].Odedra S, Wimperis S, Imaging of the B1 distribution and background signal in a MAS NMR probehead using inhomogeneous B0 and B1 fields, Journal of Magnetic Resonance 231 (2013) 95–99. [DOI] [PubMed] [Google Scholar]
- [84].Maas W, Bielecki A, Ziliox M, Laukien F, Cory D, Magnetic field gradients in solid state magic angle spinning NMR, Journal of Magnetic Resonance 141 (1999) 29–33. [DOI] [PubMed] [Google Scholar]
- [85].Oas TG, Gri n RG, Levitt MH, Rotary resonance recoupling of dipolar interactions in solid-state nuclear magnetic resonance spectroscopy, The Journal of Chemical Physics 89 (2) (1988) 692–695. [Google Scholar]
- [86].Cory D, Ritchey W, Suppression of signals from the probe in Bloch decay spectra, Journal of Magnetic Resonance 80 (1988) 128–132. [Google Scholar]
- [87].Odedra S, Wimperis S, Improved background suppression in 1HMAS NMR using composite pulses, Journal of Magnetic Resonance 221 (2012) 41–50. [DOI] [PubMed] [Google Scholar]
- [88].Dillmann B, Elbayed K, Zeiger H, Weingertner M-C, Piotto M, Engelke F, A novel low-E field coil to minimize heating of biological samples in solid-state multinuclear NMR experiments, Journal of Magnetic Resonance 187 (2007) 10–18. [DOI] [PubMed] [Google Scholar]
- [89].Nanni EA, Barnes AB, Matsuki Y, Woskov PP, Corzilius B, Gri n RG, Temkin RJ, Microwave field distribution in a magic angle spinning dynamic nuclear polarization NMR probe, Journal of Magnetic Resonance 210 (2011) 16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Litvak IM, Pneumatically switched variable angle spinning NMR probe with capacitively coupled RF coil for solid state NMR of oriented systems, Doctor of Philosophy, University of California, Irvine: (2010). [Google Scholar]
- [91].Stuhlman O Jr., Githens S Jr., The magnetic field of a solenoid oscillating at radio frequencies, Review of Scientific Instruments 3 (1932) 561–571. [Google Scholar]
- [92].Engelke F, Electromagnetic wave compression and radio frequency homogeneity in NMR solenoidal coils: Computational approach, Concepts in Magnetic Resonance, Part A 15 (2) (2002) 129–155. [Google Scholar]
- [93].Marks D, Vega S, A theory for cross-polarization NMR of nonspinning and spinning samples, Journal of Magnetic Resonance Series A 118 (2) (1996) 157–172. [Google Scholar]
- [94].Paluch P, Trébosc J, Nishiyama Y, Potrzebowski M, Malon M, Amoureux J, Theoretical study of CP-VC: A simple, robust and accurate MAS NMR method for analysis of dipolar C–H interactions under rotation speeds faster than ca. 60 kHz, Journal of Magnetic Resonance 252 (2015) 67–77. [DOI] [PubMed] [Google Scholar]
- [95].Gan Z, Gor’kov PL, Brey WW, Sideris PJ, Grey CP, Enhancing MQMAS of low-γ nuclei by using a high B1 field balanced probe circuit, Journal of Magnetic Resonance 200 (2009) 2–5. [DOI] [PubMed] [Google Scholar]
- [96].Murphy-Boesch J, Koretsky A, An in vivo NMR probe circuit for improved sensitivity, Journal of Magnetic Resonance 54 (1983) 526–532. [Google Scholar]
- [97].Martin RW, Zilm KW, Preparation of protein nanocrystals and their characterization by solid state NMR, Journal of Magnetic Resonance 165 (1) (2003) 162–174. [DOI] [PubMed] [Google Scholar]
- [98].Gor’kov P, Chekmenev E, Fu R, Hu J, Cross T, Cotten M, Brey W, A large volume flat coil probe for oriented membrane proteins, Journal of Magnetic Resonance 181 (2006) 9–20. [DOI] [PubMed] [Google Scholar]
- [99].Frye JS, Maciel GS, Setting the magic angle using a quadrupolar nuclide, Journal of Magnetic Resonance 48 (1982) 125–131. [Google Scholar]
- [100].Mamone S, Dorsch A, Johannessen OG, Naik MV, Madhu PK, Levitt M, A Hall effect angle detector for solid-state NMR, Journal of Magnetic Resonance 190 (1) (2008) 135–141. [DOI] [PubMed] [Google Scholar]
- [101].Mihaliuk ER, Gullion T, Optical lever for monitoring of the magic angle, Journal of Magnetic Resonance 223 (2012) 46–50. [DOI] [PubMed] [Google Scholar]
- [102].Matsunaga T, Mizuno T, Takegoshi K, An X0 shim coil for precise magic-angle adjustment, Journal of Magnetic Resonance 256 (2015) 1–8. [DOI] [PubMed] [Google Scholar]
- [103].Kelly AE, Ou HD, Withers R, Dötsch V, Low-conductivity bu ers for high-sensitivity NMR measurements, Journal of The American Chemical Society 124 (2002) 12013–12019. [DOI] [PubMed] [Google Scholar]
- [104].Fowler DJ, Harris MJ, Thompson LK, Heat management strategies for solid-state NMR of functional proteins, Journal of Magnetic Resonance 222 (2012) 112–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Bielecki A, Burum D, Temperature dependence of 207Pb MAS spectra of solid lead nitrate: An accurate, sensitive thermometer for variabletemperature MAS, Journal of Magnetic Resonance A 116 (1995) 215–220. [Google Scholar]
- [106].Neue G, Dybowski C, Determining temperature in a magic-angle spinning probe using the temperature dependence of the isotropic chemical shift of lead nitrate, Solid State Nuclear Magnetic Resonance 7 (1997) 333–336. [DOI] [PubMed] [Google Scholar]
- [107].Guan X, Stark RE, A general protocol for temperature calibration of MAS NMR probes at arbitrary spinning speeds, Solid State Nuclear Magnetic Resonance 38 (2010) 74–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Purusottam RN, Bodenhausen G, Tekely P, Determination of sample temperature in unstable static fields by combining solid-state 79Br and 13C NMR, Journal of Magnetic Resonance 246 (2014) 69–71. [DOI] [PubMed] [Google Scholar]
- [109].Thurber KR, Tycko R, Measurement of sample temperatures under magic-angle spinning from the chemical shift and spin-lattice relaxation rate of 79Br in KBr powder, Journal of Magnetic Resonance 196 (1) (2009) 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Lakomek N-A, Frey L, Bibow S, Böckmann A, Riek R, Meier BH, Proton-Detected NMR Spectroscopy of Nanodisc-Embedded Membrane Proteins: MAS Solid-State vs Solution-State Methods, The Journal of Physical Chemistry B 121 (32) (2017) 7671–7680. [DOI] [PubMed] [Google Scholar]
- [111].Wu CH, Grant CV, Cook G, Park SH, Opella S, A strip-shield improves the effciency of a solenoid coil in probes for high-field solid-state nmr of lossy biological samples, Journal of Magnetic Resonance 200 (2009) 74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Martin RW, Zilm KW, Variable temperature system using vortex tube cooling and fiber optic temperature sensor for low temperature magic angle spinning NMR, Journal of Magnetic Resonance 168 (2004) 202–209. [DOI] [PubMed] [Google Scholar]
- [113].Matsuki Y, Ueda K, Idehara T, Ikeda R, Ogawa I, Nakamura S, Toda M, Anai T, Fujiwara T, Helium-cooling and -spinning dynamic nuclear polarization for sensitivity-enhanced solid-state NMR at 14 T and 30 K, Journal of Magnetic Resonance 225 (2012) 1–9. [DOI] [PubMed] [Google Scholar]
- [114].Sesti EL, Alaniva N, Rand PW, Choi EJ, Albert BJ, Saliba EP, Scott FJ, Barnes AB, Magic angle spinning NMR below 6 K with a computational fluid dynamics analysis of fluid flow and temperature gradients, Journal of Magnetic Resonance in press (2017) doi: 10.1016/j.jmr.2017.11.002. [DOI] [PubMed] [Google Scholar]
- [115].Doty F, Stabilizing control of a saturated cold gas stream, uS Patent 9,062,904 (Jun. 23 2015).
- [116].Thurber KR, Potapov A, Yau W-M, Tycko R, Solid state nuclear magnetic resonance with magic-angle spinning and dynamic nuclear polarization below 25 K, Journal of Magnetic Resonance 226 (2013) 100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Thurber K, Tycko R, Low-temperature dynamic nuclear polarization with helium-cooled samples and nitrogen-driven magic-angle spinning, Journal of Magnetic Resonance 264 (2016) 99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Matsuki Y, Nakamura S, Fukui S, Suematsu H, Fujiwara T, Closed-cycle cold helium magic-angle spinning for sensitivity-enhanced multidimensional solid-state NMR, Journal of Magnetic Resonance 259 (2015) 76–81. [DOI] [PubMed] [Google Scholar]
- [119].Matsuki Y, Idehara T, Fukazawa J, Fujiwara T, Advanced instrumentation for DNP-enhanced MAS NMR for higher magnetic fields and lower temperatures, Journal of Magnetic Resonance 264 (2016) 107–115. [DOI] [PubMed] [Google Scholar]
- [120].Doty F, Shevgoor S, Improved NMR cryomas probe for high-field wide-bore magnets, cA Patent App. CA 2,672,798 (Jun. 12 2008).
- [121].Gor’kov PL, Chekmenev EY, Li CG, Cotten M, Bu y JJ, Traaseth NJ, Veglia G, Brey WW, Using low-E resonators to reduce RF heating in biological samples for static solid-state NMR up to 900 MHz, Journal of Magnetic Resonance 185 (1) (2007) 77–93. [DOI] [PubMed] [Google Scholar]
- [122].Grant CV, Yang Y, Glibowicka M, Wu CH, Park SH, Deber C, Opella S, A modified Alderman-Grant coil makes possible an effcient cross-coil probe for high field solid-state NMR of lossy biological samples, Journal of Magnetic Resonance 201 (2009) 87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Zhang Z, Hammel PC, Moore GJ, Application of a novel rf coil design to the magnetic resonance force microscope, Review of Scientific Instruments 67 (9) (1996) 3307–3309. [Google Scholar]
- [124].Sakellariou D, Le Go G, Jacquinot JF, High-resolution, high-sensitivity NMR of nanolitre anisotropic samples by coil spinning, Nature 447 (2007) 694–697. [DOI] [PubMed] [Google Scholar]
- [125].Badilita V, Kratt K, Burger T, Korvink JG, Wallrabe U, 3D high aspect ratio, MEMS integrated micro-solenoids and Helmholtz micro-coils, in: TRANSDUCERS 2009 – 2009 International Solid-State Sensors, Actuators and Microsystems Conference, 2009, pp. 1106–1109. [Google Scholar]
- [126].Lehmann-Horn JA, Jacquinot JF, Ginefri JC, Bonhomme C, Sakellariou D, Monolithic MACS micro resonators, Journal of Magnetic Resonance 271 (Supplement C) (2016) 46–51. [DOI] [PubMed] [Google Scholar]
- [127].Takeda K, Inukai M, Double-resonance magic angle coil spinning, Journal of Magnetic Resonance 202 (2) (2010) 274–278. [DOI] [PubMed] [Google Scholar]
- [128].Kentgens APM, Janssen H, Brinkmann A, van Eck ERH, van Bentum JM, Microcoil high-resolution magic angle spinning NMR spectroscopy, Journal of the American Chemical Society 128 (27) (2006) 8722–8723. [DOI] [PubMed] [Google Scholar]
- [129].Kentgens APM, Bart J, van Bentum PJM, Brinkmann A, van Eck ERH, Gardeniers JGE, Janssen JWG, Knijn P, Vasa S, Verkuijlen HW, High-resolution liquid- and solid-state nuclear magnetic resonance of nanoliter sample volumes using microcoil detectors, Journal of Chemical Physics 128 (5) (2008) 052202. [DOI] [PubMed] [Google Scholar]
- [130].Brauckmann JO, Janssen JWGH, Kentgens APM, High resolution triple resonance micro magic angle spinning NMR spectroscopy of nanoliter sample volumes, Physical Chemistry Chemical Physics 18 (6) (2016) 4902–4910. [DOI] [PubMed] [Google Scholar]
- [131].Caulkins BG, Young RP, Kudla RA, Yang C, Bittbauer TJ, Bastin B, Hilario E, Fan L, Marsella MJ, Dunn MF, Mueller LJ, NMR crystallography of a carbanionic intermediate in tryptophan synthase: chemical structure, tautomerization, and reaction specificity, Journal of the American Chemical Society 138 (46) (2016) 15214–15226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Tang AW, Kong X, Terskikh V, Wu G, Solid-state 17O NMR of unstable acyl-enzyme intermediates: a direct probe of hydrogen bonding interactions in the oxyanion hole of serine proteases, The Journal of Physical Chemistry B 120 (43) (2016) 11142–11150. [DOI] [PubMed] [Google Scholar]
- [133].Michaelis VK, Keeler EG, Ong T-C, Craigen KN, Penzel S, Wren JEC, Kroeker S, Gri n RG, Structural insights into bound water in crystalline amino acids: experimental and theoretical 17O NMR, The Journal of Physical Chemistry B 119 (25) (2015) 8024–8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Lemaître V, de Planque MRR, Howes AP, Smith ME, Dupree R, Watts A, Solid-state 17O NMR as a probe for structural studies of proteins in biomembranes, Journal of the American Chemical Society 126 (47) (2004) 15320–15321. [DOI] [PubMed] [Google Scholar]
- [135].Hu J, Chekmenev EY, Gan Z, Gor’kov PL, Saha S, Brey WW, Cross TA, Ion solvation by channel carbonyls characterized by 17O solid-state NMR at 21 T, Journal of the American Chemical Society 127 (34) (2005) 11922–11923. [DOI] [PubMed] [Google Scholar]
- [136].Howes AP, Anupõld T, Lemaitre V, Kukol A, Watts A, Samoson A, Smith ME, Dupree R, Enhancing resolution and sensitivity of 17O solid-state NMR through combining double rotation, 1H decoupling and satellite modulation for biomolecular applications, Chemical Physics Letters 421 (2006) 42. [Google Scholar]
- [137].Wong A, Howes AP, Yates JR, Watts A, Anupôld T, Past J, Samoson A, Dupree R, Smith ME, Ultra-high resolution 17O solid-state NMR spectroscopy of biomolecules: A comprehensive spectral analysis of monosodium L-glutamate·monohydrate, Physical Chemistry Chemical Physics 13 (26) (2011) 12213. [DOI] [PubMed] [Google Scholar]
- [138].Wong A, Howes AP, Pike KJ, Lemaître V, Watts A, Anupôld T, Past J, Samoson A, Dupree R, Smith ME, New limits for solid-state 17O NMR spectroscopy: complete resolution of multiple oxygen sites in a simple biomolecule, Journal of the American Chemical Society 128 (24) (2006) 7744–7745. [DOI] [PubMed] [Google Scholar]
- [139].Wu G, Dong S, Two-dimensional 17O multiple quantum magic-angle spinning NMR of organic solids, Journal of the American Chemical Society 123 (37) (2001) 9119–9125. [DOI] [PubMed] [Google Scholar]
- [140].Frydman L, Harwood JS, Isotropic spectra of half-integer quadrupolar spins from bidimensional magic angle spinning NMR, Journal of the American Chemical Society 117 (19) (1995) 5367–5368. [Google Scholar]
- [141].Ashbrook SE, Wimperis S, Satellite-transition MAS NMR of spin I = 3/2, 5/2, 7/2, and 9/2 nuclei: Sensitivity, resolution, and practical implementation, Journal of Magnetic Resonance 156 (2002) 269–281. [DOI] [PubMed] [Google Scholar]
- [142].Prasad S, Kwak HT, Clark T, Grandinetti PJ, A simple technique for determining nuclear quadrupole coupling constants with RAPT solid-state NMR spectroscopy, Journal of the American Chemical Society 124 (2002) 4964. [DOI] [PubMed] [Google Scholar]
- [143].Wu Y, Sun BQ, Pines A, Samoson A, Lipmaa E, NMR experiments with a new double rotor, Journal of Magnetic Resonance 89 (1990) 297–309. [Google Scholar]
- [144].Eastman MA, Grandinetti PJ, Lee YK, Pines A, Double-tuned hopping-coil probe for dynamic-angle-spinning NMR, Journal of Magnetic Resonance 98 (2) (1992) 333–341. [Google Scholar]
- [145].Hung I, Wong A, Howes AP, Anupöld T, Past J, Samoson A, Mo X, Wu G, Smith ME, Brown SP, Dupree R, Determination of NMR interaction parameters from double rotation NMR, Journal of Magnetic Resonance 188 (2) (2007) 246–259. [DOI] [PubMed] [Google Scholar]
- [146].Chmelka BF, Mueller KT, Pines A, Stebbins J, Wu Y, Zwanziger JW, 17O NMR in solids by dynamic-angle spinning and double rotation, Nature 339 (6219) (1989) 42–43. [Google Scholar]
- [147].Mueller KT, Sun BQ, Chingas GC, Zwanziger JW, Terao T, Pines A, Dynamic-angle spinning of quadrupolar nuclei, Journal of magnetic resonance 86 (1990) 470–487. [DOI] [PubMed] [Google Scholar]
- [148].Gann S, Baltisberger J, Wooten E, Zimmermann H, Pines A, Cross polarization and dynamic-angle spinning of 17O in L-alanine. Bulletin of Magnetic Resonance 16 (1) (1994) 68–72. [Google Scholar]
- [149].Kentgens APM, van Eck ERH, Ajithkumar TG, Anupold T, Past J, Reinhold A, Samoson A, New opportunities for double rotation NMR of half-integer quadrupolar nuclei, Journal of Magnetic Resonance 178 (2006) 212. [DOI] [PubMed] [Google Scholar]
- [150].Young RP, Caulkins BG, Borchardt D, Bulloch DN, Larive CK, Dunn MF, Mueller LJ, Solution-state 17 O quadrupole central-transition NMR spectroscopy in the active site of tryptophan synthase, Angewandte Chemie International Edition 55 (4) (2016) 1350–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Sattler M, Fesik SW, Use of deuterium labeling in nmr: Overcoming a sizeable problem, Structure 4 (11) (1996) 1245–1249. [DOI] [PubMed] [Google Scholar]
- [152].Zhou DH, Graesser DT, Franks WT, Rienstra CM, Sensitivity and resolution in proton solid-state NMR at intermediate deuteration levels: Quantitative linewidth characterization and applications to correlation spectroscopy, Journal of Magnetic Resonance 178 (2) (2006) 297–307. [DOI] [PubMed] [Google Scholar]
- [153].Morcombe CR, Gaponenko V, Byrd RA, Zilm KW, C-13 CPMAS spectroscopy of deuterated proteins: CP dynamics, line shapes, and T-1 relaxation, Journal of the American Chemical Society 127 (1) (2005) 397–404. [DOI] [PubMed] [Google Scholar]
- [154].Hologne M, Faelber K, Diehl A, Reif B, Characterization of dynamics of perdeuterated proteins by MAS solid-state NMR, Journal of the American Chemical Society 127 (32) (2005) 11208–11209. [DOI] [PubMed] [Google Scholar]
- [155].Böckmann A, Ernst M, Meier BH, Spinning proteins, the faster, the better?, Journal of Magnetic Resonance 253 (2015) 71–79. [DOI] [PubMed] [Google Scholar]
- [156].Cala-De Paepe D, Stanek J, Jaudzems K, Tars K, Andreas LB, Pintacuda G, Is protein deuteration beneficial for proton detected solid-state NMR at and above 100 kHz magic-angle spinning?, Solid State Nuclear Magnetic Resonance 87 (2017) 126–136. [DOI] [PubMed] [Google Scholar]
- [157].Akbey Ü, Lange S, Franks WT, Linser R, Rehbein K, Diehl A, van Rossum B-J, Reif B, Oschkinat H, Optimum levels of exchangeable protons in perdeuterated proteins for proton detection in MAS solid-state NMR spectroscopy, Journal of Biomolecular NMR 46 (2010) 67–73. [DOI] [PubMed] [Google Scholar]
- [158].Andreas LB, Le Marchand T, Jaudzems K, Pintacuda G, High-resolution proton-detected NMR of proteins at very fast MAS, Journal of Magnetic Resonance 253 (2015) 36–49. [DOI] [PubMed] [Google Scholar]
- [159].Barbet-Massin E, Pell AJ, Retel JS, Andreas LB, Jaudzems K, Franks WT, Nieuwkoop AJ, Hiller M, Higman V, Guerry P, Bertarello A, Knight MJ, Felletti M, Le Marchand T, Kotelovica S, Akopjana I, Tars K, Stoppini M, Bellotti V, Bolognesi M, Ricagno S, Chou JJ, Gri n RG, Oschkinat H, Lesage A, Emsley L, Herrmann T, Pintacuda G, Rapid proton-detected NMR assignment for proteins with fast magic angle spinning, Journal of the American Chemical Society 136 (35) (2014) 12489–12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Reif B, Deuterated peptides and proteins: structure and dynamics studies by MAS solid-state NMR, in: Shekhtman A, Burz DS (Eds.), Protein NMR Techniques, Vol. 831, Humana Press, 2012, pp. 279–301, 10.1007/978-1-61779-480-3_16. [DOI] [PubMed] [Google Scholar]
- [161].Gall CM, DiVerdi JA, Opella SJ, Phenylalanine ring dynamics by solid-state deuterium NMR, Journal of the American Chemical Society 103 (17) (1981) 5039–5043. [Google Scholar]
- [162].Ketchem RR, Roux B, Cross TA, High-resolution polypeptide structure in a lamellar phase lipid environment from solid state NMR derived orientational constraints, STRUCTURE 5 (12) (1997) 1655–1669. [DOI] [PubMed] [Google Scholar]
- [163].Gröbner G, Choi G, Burnett IJ, Glaubitz C, Verdegem PJ, Lugtenburg J, Watts A, Photoreceptor rhodopsin: structural and conformational study of its chromophore 11cis retinal in oriented membranes by deuterium solid state NMR, FEBS Letters 422 (2) (1998) 201–204. [DOI] [PubMed] [Google Scholar]
- [164].Bechinger B, Weik M, Deuterium solid-state NMR investigations of exchange labeled oriented purple membranes at different hydration levels, Biophysical Journal 85 (1) (2003) 361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Eckman R, Müller L, Pines A, Deuterium double quantum NMR with magic angle spinning, Chemical Physics Letters 74 (2) (1980) 376–378. [Google Scholar]
- [166].Agarwal V, Diehl A, Skrynnikov N, Reif B, High resolution 1Hdetected 1H, 13C correlation spectra in MAS solid-state NMR using deuterated proteins with selective 1H, 2H isotopic labeling of methyl groups, Journal of the American Chemical Society 128 (39) (2006) 12620–12621. [DOI] [PubMed] [Google Scholar]
- [167].Hoatson G, Vold R, 2H-NMR Spectroscopy of Solids and Liquid Crystals., in: Solid-State NMR III Organic Matter, Vol. 32 of NMR Basic Principles and Progress, 1994, pp. 1–67. [Google Scholar]
- [168].Schmidt-Rohr K, Spiess HW, Multidimensional Solid-State NMR and Polymers, Elsevier Science, Burlington, 2012. [Google Scholar]
- [169].Gall C, DiVerdi J, Opella S, Phenylalanine Ring Dynamics by Solid-state 2H NMR. Journal of the American Chemical Society 103 (1981) 5039–5043. [Google Scholar]
- [170].Hologne M, Chen Z, Reif B, Characterization of dynamic processes using deuterium in uniformly 2H,13C,15N enriched peptides by MAS solid-state NMR, Journal of Magnetic Resonance 179 (1) (2006) 20–28. [DOI] [PubMed] [Google Scholar]
- [171].Keniry MA, Solid-state Deuterium nuclear magnetic resonance spectroscopy of proteins. Methods in Enzymology 176 (1989) 376–386. [DOI] [PubMed] [Google Scholar]
- [172].Shi X, Rienstra CM, Site-Specific Internal Motions in GB1 Protein Microcrystals Revealed by 3D 2H-13C-13C Solid-State NMR Spectroscopy, Journal of the American Chemical Society 138 (12) (2016) 4105–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Lalli D, Schanda P, Chowdhury A, Retel J, Hiller M, Higman V, Handel L, Agarwal V, Reif B, van Rossum B, Akbey Ü, Oschkinat H, Three-dimensional deuterium-carbon correlation experiments for high-resolution solid-state MAS NMR spectroscopy of large proteins, Journal of Biomolecular NMR 5 (4) (2011) 477–485. [DOI] [PubMed] [Google Scholar]
- [174].Bonev BB, Morrow MR, Simple probe for variable pressure deuterium nuclear magnetic resonance studies of soft materials, Review of Scientific Instruments 68 (4) (1997) 1827–1830. [Google Scholar]
- [175].McDermott AE, Creuzet FJ, Kolbert AC, Gri n RG, High-resolution magic-angle-spinning NMR spectra of protons in deuterated solids, Journal of Magnetic Resonance (1969) 98 (2) (1992) 408–413. [Google Scholar]
- [176].Huber M, With O, Schanda P, Verel R, Ernst M, Meier BH, A supplementary coil for 2H decoupling with commercial HCN MAS probes, Journal of Magnetic Resonance 214 (2012) 76–80. [DOI] [PubMed] [Google Scholar]
- [177].Akbey Ü, Nieuwkoop AJ, Wegner S, Voreck A, Kunert B, Bandara P, Engelke F, Nielsen NC, Oschkinat H, Quadruple-Resonance Magic-Angle Spinning NMR Spectroscopy of Deuterated Solid Proteins, Angewandte Chemie International Edition 53 (9) (2014) 2438–2442. [DOI] [PubMed] [Google Scholar]
- [178].Wei D, Akbey Ü, Paaske B, Oschkinat H, Reif B, Bjerring M, Nielsen NC, Optimal 2H rf pulses and 2H–13C cross-polarization methods for solid-state 2H MAS NMR of perdeuterated proteins, The Journal of Physical Chemistry Letters 2 (11) (2011) 1289–1294. [DOI] [PubMed] [Google Scholar]
- [179].Cavanagh J (Ed.), Protein NMR spectroscopy: Principles and Practice, 2nd Edition, Academic Press, 2007. [Google Scholar]
- [180].Akbey Ü, Camponeschi F, van Rossum B-J, Oschkinat H, Triple Resonance Cross-Polarization for More Sensitive 13C MAS NMR Spectroscopy of Deuterated Proteins, ChemPhysChem 12 (11) (2011) 2092–2096. [DOI] [PubMed] [Google Scholar]
- [181].Akbey Ü, Rossum B.-J. v., Oschkinat H, Practical aspects of high-sensitivity multidimensional 13C MAS NMR spectroscopy of perdeuterated proteins, Journal of Magnetic Resonance 217 (2012) 77–85. [DOI] [PubMed] [Google Scholar]
- [182].Asami S, Szekely K, Schanda P, Meier BH, Reif B, Optimal degree of protonation for 1H detection of aliphatic sites in randomly deuterated proteins as a function of the MAS frequency, Journal of Biomolecular NMR 54 (2) (2012) 155–168. [DOI] [PubMed] [Google Scholar]
- [183].Nieuwkoop AJ, Franks WT, Rehbein K, Diehl A, Akbey Ü, Engelke F, Emsley L, Pintacuda G, Oschkinat H, Sensitivity and resolution of proton detected spectra of a deuterated protein at 40 and 60 kHz magic-angle-spinning, Journal of Biomolecular NMR 61 (2) (2015) 161–171. [DOI] [PubMed] [Google Scholar]
- [184].Ghosh M, Rienstra CM, 1H-detected REDOR with fast magic-angle spinning of a deuterated protein, The Journal of Physical Chemistry B 121 (36) (2017) 8503–8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Reif B, Ultra-high resolution in MAS solid-state NMR of perdeuterated proteins: implications for structure and dynamics, Journal of Magnetic Resonance 216 (2012) 1–12. [DOI] [PubMed] [Google Scholar]
- [186].Samoson A, Introducing new variables to MAS NMR, Joint EUROMAR 2010 and 17th ISMAR Conference, Florence, Italy (July 4–9 2010). [Google Scholar]
- [187].Samoson A, Tuherm T, Past J, Reinhold A, Heinmaa I, Anupôld T, Smith M, Pike K, Fast magic-angle spinning: Implications, in: Harris R, Wasylishen R (Eds.), Encyclopedia of Magnetic Resonance, John Wiley & Sons Ltd., 2010, p. 120. [Google Scholar]
- [188].Stöppler D, Macpherson A, Smith-Penzel S, Basse N, Lecomte F, Deboves H, Taylor RD, Norman T, Porter J, Waters LC, Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR, PLoS biology 16 (5) (2018) e2006192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Nishiyama Y, Fast magic-angle sample spinning solid-state NMR at 60100 kHz for natural abundance samples, Solid State Nuclear Magnetic Resonance 78 (2016) 2436. [DOI] [PubMed] [Google Scholar]
- [190].Ernst M, Samoson A, Meier BH, Low-power decoupling in fast magic-angle spining NMR, Chemical Physics Letters 348 (2001) 293–302. [Google Scholar]
- [191].Maas WE, Laukien FH, Cory D, Gradient, high resolution, magic angle sample spinning NMR, Journal of the American Chemical Society 118 (51) (1996) 13085–13086. [Google Scholar]
- [192].Zhang W, Cory D, Pulsed gradient NMR probes for solid-state studies, Journal of Magnetic Resonance 132 (1998) 144–149. [DOI] [PubMed] [Google Scholar]
- [193].Barbara TM, Bronnimann CE, Target field design for magic angle gradient coils, Journal of Magnetic Resonance 140 (1999) 285–288. [DOI] [PubMed] [Google Scholar]
- [194].Doty FD, The Encyclopedia of NMR, Vol. 7, Wiley, 2007, 10.1002/9780470034590.emrstm1001. [DOI] [Google Scholar]
- [195].Chevelkov V, van Rossum BJ, Castellani F, Rehbein K, Diehl A, Hohwy M, Steuernagel S, Engelke F, Oschkinat H, Reif B, 1H detection in MAS solid-state NMR spectroscopy of biomacromolecules employing pulsed field gradients for residual solvent suppression, Journal of the American Chemical Society 125 (26) (2003) 7788–7789. [DOI] [PubMed] [Google Scholar]
- [196].Zhou DH, Rienstra CM, High-performance solvent suppression for proton detected solid-state NMR, Journal of Magnetic Resonance 192 (2008) 167–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Koczor B, Sedyó I, Rohonczy J, An alternative solution for computer controlled tuning and matching of existing NMR probes, Journal of Magnetic Resonance 259 (2015) 179–185. [DOI] [PubMed] [Google Scholar]
- [198].Nishiyama Y, Tsutsumi Y, Utsumi H, MAGIC SHIMMING: Gradient shimming with magic angle sample spinning, Journal of Magnetic Resonance 216 (2012) 197–200. [DOI] [PubMed] [Google Scholar]
- [199].Topgaard D, Martin RW, Sakellariou D, Meriles CA, Pines A, “Shim pulses” for NMR spectroscopy and imaging, Proceedings of the National Academy of Sciences of the United States of America 101 (51) (2004) 17576–17581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Kehlet CT, Sivertsen AC, Bjerring M, Reiss TO, Khaneja N, Glaser SJ, Nielsen NC, Improving solid-state nmr dipolar recoupling by optimal control, Journal of the American Chemical Society 126 (33) (2004) 10202–10203. [DOI] [PubMed] [Google Scholar]
- [201].Khaneja N, Reiss T, Kehlet C, Schulte-Herbruggen T, Glaser SJ, Optimal control of coupled spin dynamics: design of nmr pulse sequences by gradient ascent algorithms, Journal of Magnetic Resonance 172 (2) (2005) 296–305. [DOI] [PubMed] [Google Scholar]
- [202].Hansen JO, Kehlet C, Bjerring M, Vosegaard T, Glaser SJ, Khaneja N, Nielsen NC, Optimal control based design of composite dipolar recoupling experiments by analogy to single-spin inversion pulses, Chemical Physics Letters 447 (1–3) (2007) 154–161. [Google Scholar]
- [203].Kehlet C, Bjerring M, Sivertsen AC, Kristensen T, Enghild JJ, Glaser SJ, Khaneja N, Nielsen NC, Optimal control based NCO and NCA experiments for spectral assignment in biological solid-state nmr spectroscopy, Journal of Magnetic Resonance 188 (2) (2007) 216–230, kehlet, Cindie Bjerring, Morten Sivertsen, Astrid C. Kristensen, Torsten Enghild, Jan J. Glaser, Ste en J. Khaneja, Navin Nielsen, Niels Chr. [DOI] [PubMed] [Google Scholar]
- [204].Bhattacharya A, Chemistry: breaking the billion-hertz barrier, Nature 463 (2010) 605–606. [DOI] [PubMed] [Google Scholar]
- [205].Polenova T, Budinger T, Ultra high field NMR and MRI: science at a crossroads. report on a jointly-funded NSF, NIH and DOE workshop, held on November 12–13, 2015 in Bethesda, Maryland, USA, Journal of Magnetic Resonance 266 (2016) 81–86. [DOI] [PubMed] [Google Scholar]
- [206].Yanagisawa Y, Piao R, Iguchi S, Nakagome H, Takao T, Komi-nato K, Hamada M, Matsumoto S, Suematsu H, Jin X, Takahashi M, Yamazaki T, Maeda H, Operation of a 400 MHz NMR magnet using a (RE:Rare Earth)Ba2Cu3O7x high-temperature superconducting coil: Towards an ultra-compact super-high field NMR spectrometer operated beyond 1 GHz, Journal of Magnetic Resonance 249 (2014) 38–48. [DOI] [PubMed] [Google Scholar]
- [207].Hashi K, Ohki S, Matsumoto S, Nishijima G, Goto A, Deguchi K, Yamada K, Noguchi T, Sakai S, Takahashi M, Yanagisawa Y, Iguchi S, Yamazaki T, Maeda H, Tanaka R, Nemoto T, Suematsu H, Miki T, Saito K, Shimizu T, Achievement of 1020 MHz NMR, Journal of Magnetic Resonance 256 (2015) 30–33. [DOI] [PubMed] [Google Scholar]
- [208].Maeda H, Yamazaki T, Nishiyama Y, Hamada M, Hashi K, Shimizu T, Suematsu H, Yanagisawa Y, Development of super-high-field NMR operated beyond 1 GHz using high-temperature superconducting coils, Vol. 5 of eMagRes, John Wiley & Sons, Ltd., 2016, Ch. 2. [Google Scholar]
- [209].Lin Y, Ahn S, Murali N, Brey W, Bowers W, and Warren CR, High-resolution, >1 GHz NMR in unstable magnetic fields, Physical Review Letters 85 (2000) 3732–3735. [DOI] [PubMed] [Google Scholar]
- [210].Shapira B, Shetty K, Brey W, Gan Z, Frydman L, Single-scan 2D NMR spectroscopy on a 25 T Bitter magnet, Chemical Physics Letters 442 (2007) 478–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Abou-Hamad E, Bontemps P, Rikken G, NMR in pulsed magnetic field, Solid State Nuclear Magnetic Resonance 40 (2011) 42–44. [DOI] [PubMed] [Google Scholar]
- [212].Kohlrautz J, Reichardt S, Green E, Kühne H, Wosnitza J, Haase J, NMR shift and relaxation measurements in pulsed high-field magnets up to 58 T, Journal of Magnetic Resonance 263 (2016) 1–6. [DOI] [PubMed] [Google Scholar]
- [213].Orlova A, Frings P, Suleiman G, and Rikken M, New high homogeneity 55 T pulsed magnet for high field NMR, Journal of Magnetic Resonance 268 (2016) 82–87. [DOI] [PubMed] [Google Scholar]
- [214].Paulson EK, Zilm KW, External field-frequency lock probe for high resolution solid state NMR, Review of Scientific Instruments 76 (2). [Google Scholar]
- [215].Jehle S, Rajagopal P, Bardiaux B, Markovic S, Kühne R, Stout JR, Higman VA, Klevit RE, Barth-Jan v., H. Oschkinat, Solid-state NMR and SAXS studies provide a structural basis for the activation of αB-crystallin oligomers, Nature Structural & Molecular Biology 17 (9) (2010) 1037–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Das R, André I, Shen Y, Wu Y, Lemak A, Bansal S, Arrowsmith C, Szyperski T, B. D, Simultaneous prediction of protein folding and docking at high resolution, Proceedings of the National Academy of Sciences of the United States of America 106 (45) (2009) 18978–18983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Moraga O, Sgourakis NG, Baker D, Goldbourt A, The NMR–Rosetta capsid model of M13 bacteriophage reveals a quadrupled hydrophobic packing epitope, Proceedings of the National Academy of Sciences of the United States of America 112 (4) (2015) 971–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].DiMaio F, Tyka MD, Baker ML, Chiu W, Baker D, Refinement of protein structures into low-resolution density maps using Rosetta, Journal of Molecular Biology 392 (2009) 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Vernon R, Shen Y, Baker D, Lange OF, Improved chemical shift based fragment selection for CS-Rosetta using Rosetta3 fragment picker, Journal of Biomolecular NMR 57 (2013) 117–127. [DOI] [PubMed] [Google Scholar]
- [220].Demers J, Habenstein B, Loquet A, Vasa SK, Giller K, Becker S, Baker D, Lange A, Sgourakis NG, High-resolution structure of the Shigella type-III secretion needle by solid-state NMR and cryo-electron microscopy, Nature Communications 5 (4976) (2014) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Sborgia L, Ravotti F, Dandey VP, Dick MS, Mazur A, Reckel S, Chami M, Scherer S, Huber M, Böckmann A, Egelman EH, Stahlberg H, Broz P, Meier BH, Hiller S, Structure and assembly of the mouse ASC inflammasome by combined NMR spectroscopy and cryo-electron microscopy, Proceedings of the National Academy of Sciences of the United States of America 112 (43) (2015) 13237–13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Cuniasse P, Tavares P, Orlova EV, Zinn-Justin S, Structures of biomolecular complexes by combination of NMR and cryoEM methods, Current Opinion in Structural Biology 43 (2017) 104–113. [DOI] [PubMed] [Google Scholar]