ABSTRACT
Missing data ubiquitously occur in randomized controlled trials and may compromise the causal inference if inappropriately handled. Some problematic missing data methods such as complete case (CC) analysis and last-observation-carried-forward (LOCF) are unfortunately still common in nutrition trials. This situation is partially caused by investigator confusion on missing data assumptions for different methods. In this statistical guidance, we provide a brief introduction of missing data mechanisms and the unreasonable assumptions that underlie CC and LOCF and recommend 2 appropriate missing data methods: multiple imputation and full information maximum likelihood.
Keywords: missing data, randomized controlled trials, multiple imputation, full information maximum likelihood, missing data mechanisms
Introduction
Randomized controlled trials (RCTs), the gold standard to generate rigorous evidence on intervention effects by providing unbiased causal inference, are common in nutrition research. An ideally conducted RCT ensures that the observed difference in outcomes between control and treatment groups is only attributed to the absence or presence of the intervention (i.e., causality); if the RCT could be repeated multiple times, the estimated effects would, on average, equal the true intervention effect (i.e., unbiasedness). However, missing data may seriously compromise the credibility of causal conclusions from RCTs. Although many ideas for preventing missingness have been suggested in the design stage (1), missing data are still a ubiquitous problem in RCTs. Missing data refer to those unavailable values that are planned to be collected and are important in data analysis. We focus on missing data in RCTs here, but the missing data methods introduced in this article can also be applied to other study designs. In an RCT, missing data can happen due to various reasons, such as participants missing scheduled visits or completely dropping out from the study. A review of 121 RCTs for weight loss suggests that the drop-out rates range from 0% to 80% and increase with the duration of the trials (2). As emphasized in a recent National Research Council report, missing data must be appropriately handled using valid statistical approaches in analyzing RCTs (1, 3) in order to draw accurate inferences. Unfortunately, some proven problematic and biased methods—for example, complete case (CC) analysis and last-observation-carried-forward (LOCF)—are still commonly used in nutrition trials. In this statistical guidance, we briefly introduce the idea of the missing data mechanism, effects of missing data on statistical inference from RCTs, and why CC and single imputation methods including LOCF can lead to incorrect inferences and then recommend 2 appropriate approaches: multiple imputation (MI) and full information maximum likelihood (FIML). The goal is to encourage nutrition researchers to become fully aware of missing data issues in RCTs and some appropriate, readily available statistical tools to handle missing data, thus leading to a better practice in nutrition research.
Why Must Missing Data Be Handled Appropriately in RCTs?
An obvious reason to appropriately deal with missing outcomes is to retain the statistical power of the original RCT design. Statistical power is the probability with which a study can detect a meaningful effect given such an effect exists. RCTs are expensive in time and money, so the sample size in an RCT is usually calculated to achieve ≥80% power to detect an assumed treatment effect under a given significance level—for example, 0.05. If not handled appropriately, missing data in an RCT will shrink the effective sample size and decrease the likelihood to detect a meaningful treatment effect. Assuming a 2-armed nutrition trial with an expected medium effect size (Cohen's d = 0.6), 45 participants in each group achieves 80% power under a significance level of 0.05. If 10 participants in each group drop out and the CC analysis is conducted, the statistical power will be only 70%. In some trials, the calculated sample size is purposely inflated by a factor on the basis of the estimated drop-out rate to ensure that the final sample size is still capable of achieving required statistical power if missing data occur. However, this approach is only valid under certain (and quite restrictive) conditions, as detailed below.
Another reason, actually the most important reason, is to protect the validity of randomization in an RCT. Among many study designs, the RCT is accepted as the “gold standard” because of its capability to obtain unbiased estimates of treatment effects by eliminating the potential confounding effects through randomized allocations. Randomization in an RCT ensures that the participants in each treatment arm have a similar distribution of baseline characteristics, which means that any difference in outcomes across study arms can be attributed to the treatments that were tested. However, if analysts simply analyze individuals with observed outcome data and do not use more principled missing data methods, the analysis samples in the treatment and control arms may no longer be comparable. For example, imagine a nutrition study in which more males drop out in the treatment group while more females drop out in the control group. In this case, the subsets of the 2 groups with observed outcomes will have different gender distributions and no longer be comparable. If the missing data are ignored and the CC analysis is conducted, we cannot rule out the possibility that the observed intervention effect is attributed to the gender difference between the groups with observed outcomes, wholly or partially. Thus, missing outcomes can jeopardize the validity of randomization unless the missing values are missing completely at random (i.e., missingness is unrelated to any observed or unobserved factors), the strictest assumption, which rarely holds in practice. This assumption is discussed further below.
To explain this further, intent-to-treat (ITT) analysis is recommended when analyzing RCTs (4); the ITT analysis is unbiased given the randomization. Under the ITT principle, randomized participants in an RCT need to be analyzed on the basis of their original treatment assignment, regardless of their adherence, actual treatment, or study withdrawal to ensure the validity of randomization. Therefore, missing data must be handled appropriately in ITT analysis for valid inferences.
Missing Data Mechanism
Missing data in RCTs can come from various sources and can be due to known or unknown reasons. Knowing the sources and reasons can greatly help in terms of both preventing the missingness and understanding how to deal with it in analyses.
As defined by Rubin (5), mechanisms under which the missing data can occur are summarized into 3 categories: missing at random (MAR), missing completely at random (MCAR), and missing not at random (MNAR). MAR indicates that whether a data point is missing is related to the observed data but not the unobserved data. An example of MAR would be if more females drop out than males in an RCT but gender is observed and thus can be adjusted for in analyses. MCAR, a special case of MAR, indicates that whether a data point is missing is completely unrelated to either observed data or unobserved data. An example of MCAR would be if some participants miss scheduled visits due to bad weather or if a random data-programming error leads to some observations being missing. MNAR indicates scenarios in which whether a data point is missing is related to some unobserved factor, such as the missing value itself. An example of MNAR would be if participants drop out from a weight-loss trial because of unsuccessful weight loss.
Under MCAR, the missingness is totally random and thus the subjects without missing data are a random subsample of the original sample, which means that the analysis with observed data only is still valid. However, analysis of the complete cases will lead to larger SE estimates because of the reduced sample size. MCAR can be partially tested by Little's MCAR test, which tests whether the multivariate distribution of observed variables differs across groups defined by whether there are missing values or not (6). The smaller the P value, the stronger the evidence that the MCAR assumption is invalid. However, tests for MCAR generally rely on fairly strong parametric assumptions. In addition, any attempt to use that P value to decide the data is or is not MCAR would be arbitrary. MCAR is the strictest assumption with regard to the missing mechanism and rarely happens in practice. MAR is the most common assumption for the modern missing data methods (e.g., MI and FIML). If the MAR assumption is met, these methods will provide unbiased estimation of the intervention effect as if no subjects have missing entries (i.e., across hypothetical repeated replications of the study these methods would, on average, give accurate parameter estimates, such as regression coefficients). Although MAR is not a testable assumption, MI and FIML methods are fairly robust to some violation of the MAR assumption especially when “auxiliary variables” (variables that are not in the primary analysis but related to missingness; discussed further below) are used in addition to the primary analysis variables (7, 8). The most difficult missing data situation is MNAR, which is also untestable. If MNAR is assumed, the missing mechanism cannot be derived from the observed data and has to be specified by researchers on the basis of the knowledge of reasons that missing data can occur (9); for example, that those with missing weight information likely had less weight loss than those with observed weight outcomes. Because this assumption is untestable and unknowable from data, sensitivity analyses are recommended, which specify a range of plausible values for missing data under an MNAR assumption to evaluate the potential departure of statistical estimates and inference from those obtained in the primary analysis under an MAR assumption (1).
Commonly Used but Problematic Methods
Problematic (but unfortunately common) missing data methods include CC analysis and various single imputation approaches. In a CC analysis, only subjects without missing entries are included in the analysis. Many nonstatistician researchers are inclined to conduct CC analysis because of a perception (and fear) of “making up data” in MI or the untestable missing data assumptions underlying MI or FIML. However, this point of view is specious. First, MI or FIML rests on well-established theory (8, 10). Second, CC analysis actually requires MCAR, a far more restrictive assumption than MAR, the underlying assumption for MI or FIML. In many studies, CC analysis is justified by a lack of significant differences when comparing the baseline characteristics between subjects with and without missing outcomes. This comparison is definitely helpful to understand the characteristics of subjects with missing outcomes but could only provide very weak evidence of MCAR due to the fact that the sample sizes may not be big enough to detect a significance, and no comparison can be made for unmeasured characteristics. However, the CC analysis can still be considered as part of sensitivity analyses supplementary to MI or FIML analysis when missing data occur.
In single imputation approaches, each missing value is replaced by a single value. This could be based on the sample mean, a regression-based prediction, hot-deck, LOCF, or other strategies. When single imputation is used, in the outcome analyses the imputed values will be assumed the real value, as if no data were missing. This can then lead to incorrect SEs, too small because they do not reflect the uncertainty due to the missing data. For example, single imputation based on the sample mean would assume that an individual's missing weight is 150 pounds, the mean of observed weights, with no adjustment for the fact that that is just a guess and that, in reality, that person's weight may be far from 150 pounds. The consequence is that mean imputation would make the sample appear far more homogeneous than it really was, resulting in underestimated sample variance and wrongly inflated precision. In addition, some of the single imputation approaches can lead to bias (as well as incorrect SEs) if they do not account for sufficient variables (e.g., a mean imputation essentially assumes MCAR).
LOCF is a particularly common approach in nutrition trials that have outcomes measured repeatedly over time, but it unfortunately can lead to bias in analyses. In brief, LOCF assumes that there are no changes in the outcome over time, which seems particularly strange in studies explicitly interested in longitudinal outcomes. As a specific example, LOCF would assume that someone's weight when he or she dropped out of the study would reach a plateau at that value through the end of the study period. However, this assumption is often scientifically and statistically implausible and can result in treatment effect estimates that are either too large or too small (11). The drawbacks of LOCF can be articulated in multiple ways. First, most outcomes do change over time and that is why the time of treatment and follow-up duration is important in RCTs. Second, even when there is no underlying true change over time, the repeated measures could change because of measurement error. Some argue that LOCF could be valid in situations in which outcomes only change over time in only 1 of the 2 groups because it could provide more conservative treatment effect estimates (i.e., that LOCF will give effect estimates that are smaller than the true effects) and such conservative estimates are acceptable. However, this argument can also put researchers in a difficult situation: RCTs require the analytical plan a priori, but it is almost impossible to examine assumptions about time trends without checking the data. In addition, the “true” trend can be disguised by the missing data. Because of these serious limitations, some top biomedical journals now do not accept studies using LOCF (1, 12).
Suggested Methods
There are, however, multiple appropriate procedures to handle missing outcome data in RCTs (1), including MI and FIML methods. Both methods are valid under an assumption of MAR.
MI
MI is a principled and flexible approach for handling missing data. MI involves imputing values for the missing values (as with single imputation) but doing so multiple times, creating multiple (e.g., 100) “complete” data sets. The analysis of interest is then carried out separately within each data set, with overall results obtained by combining results across data sets using standard MI “combining rules” (13, 14). In particular, the parameters of interest (e.g., regression coefficients) can be obtained using a simple average across complete data sets. Importantly, the SE is calculated by accounting for both the “within” variance (the average variance of the parameter of interest within each of the imputed data sets) and the variance across imputed data sets. In this way, the analysis accounts for the uncertainty created by the missing values. We discuss MI in more detail below. It is often an attractive approach because it can incorporate a large set of variables, can handle missingness in covariates or outcomes, and can be combined with almost any analysis. Another benefit of MI is that it can easily incorporate “auxiliary” variables that may provide useful information about the missing values but may not be of interest in the actual analyses (7).
There are 2 main approaches for generating the (multiple) imputations: a joint modeling approach and “fully conditional specification” (FCS; also sometimes referred to as multiple imputation by chained equations). Joint modeling involves imposing a joint distribution [usually multivariate normal (MVN)] on all of the variables in the analysis and imputing values from that model. Joint modeling under MVN assumption does not require all the variables be continuous, and the categorical variables can be jointly modeled with other continuous variables using a latent variable approach (8). However, MVN might not be a reasonable assumption when most of the variables are categorical. FCS involves specifying a series of conditional models, with each variable modeled as a function of the others (e.g., Y ∼ X1 + X2, X1 ∼ Y + X2, X2 ∼ Y + X1). FCS allows for more flexible imputation models (e.g., logistic regression for binary variables, Poisson for counts), but a drawback is that fitting a series of conditional models does not necessarily imply a coherent joint distribution. However, this does not seem to be a large problem in practice (15, 16). With either approach, the distributions or associations seen in the observed data are used to estimate plausible values for the missing data, with some randomness induced to reflect the uncertainty. For example, if an FCS approach is used, a model of weight as a function of other characteristics may be fit using a linear regression. Imputations for each person are generated by generating a series of predicted values from that regression model; the variability across those imputations will reflect the uncertainty in the regression model.
A crucial point is that all variables (including interactions) to be included in the analyses of interest should also be included in the imputations (17, 18). Not doing so would assume no association between that factor and other variables, which can lead to bias. An implication of this is that, even when imputing covariates, the outcome variable should be included in the imputation process to respect associations between covariates and outcome (19). Similarly, in a trial context, it often makes sense to impute separately for treatment and control arms to allow for different associations between covariates and outcomes in each group (e.g., to allow for treatment effect heterogeneity).
Current advice is to generate a larger number of imputations than was originally recommended, with White et al. (19) noting that often ≥100 imputations are needed to get maximum power and full reproducibility. Imputation diagnostics should include checking the distributions of observed and imputed values [although discrepancies may be totally reasonable and result because of differences between the cases with observed and missing values (20)].
After creating the imputations, the analysis of interest is run within each complete data set. Suppose there is interest in estimating the effect of a dietary intervention on sodium intake. The outcome analysis might involve a regression model of sodium intake as a function of treatment status and baseline covariates, with primary interest in the coefficient on the treatment indicator. This model would be run within each imputed data set. The overall estimate of effect would be obtained by averaging the coefficients from each imputed data set. The variance of that estimated effect is calculated as a function of the average variance of the coefficient within each imputed data set and the variability of the coefficients across data sets. Full details and formulas on the combining rules can be found in the literature (13, 14, 19); note that some quantities (such as P values) cannot be easily combined (19). Providentially, the combining rules have been implemented in most MI software that can provide the overall point estimate, interval estimate, and test statistics directly. Again, a benefit of MI is that researchers can run any analysis they want in each of the imputed data sets, treating it as if no data were missing. Nevertheless, the combining rules ensure that the final inferences reflect the uncertainty induced by the missing values.
In-depth descriptions of MI, choices in implementation, and a summary of recent developments can be found in White et al. (19) and Murray (21). Many statistical software programs have easily available packages for creating the imputations and analyzing the multiply imputed data, such as packages “mi” and “mice” in R, the multiple imputation suite in Stata, and proc MI and proc mianalyze in SAS.
FIML
FIML, also called “raw maximum likelihood” or just “maximum likelihood,” is another commonly used missing data approach that is valid assuming MAR. Unlike MI, missing data are not imputed in this method but handled in the analysis by utilizing a case-wise likelihood function using only the variables that are observed for each case. If the MAR assumption is met, FIML and MI will produce similar estimates and statistical inference (7).
Maximum likelihood (ML) estimation is the most widely used method to estimate the parameters in a statistical model, by maximizing the likelihood function (L). Suppose that a data set contains n independent cases (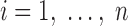 ) and k variables (
) and k variables (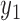 ,
, 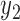 ,…,
,…, 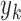 ); for each case i there exists a joint probability function,
); for each case i there exists a joint probability function, 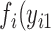 ,
, 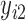 ,…,
,…, 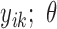 ), where
), where 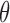 is a set of parameters (e.g., the coefficients in a regression model) to be estimated. Without missing data, the likelihood function (L) is built by multiplying the joint probability functions for all cases (i.e.,
is a set of parameters (e.g., the coefficients in a regression model) to be estimated. Without missing data, the likelihood function (L) is built by multiplying the joint probability functions for all cases (i.e., 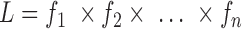 ). The ML estimates of
). The ML estimates of 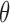 are the values of
are the values of 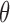 that maximize L. Now suppose that missing data exist and the missing mechanism is MAR. For simplicity, but without loss of generality, we assume cases 1 to m have no missing data, whereas cases
that maximize L. Now suppose that missing data exist and the missing mechanism is MAR. For simplicity, but without loss of generality, we assume cases 1 to m have no missing data, whereas cases 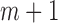 to n have missing data for variable
to n have missing data for variable 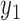 . Under the regular ML method, the joint probability function
. Under the regular ML method, the joint probability function 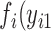 ,
, 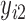 ,…,
,…, 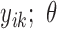 ) will be specified for the cases without missing data (cases 1 to m), whereas no joint probability function will be specified for the cases with missing data (cases
) will be specified for the cases without missing data (cases 1 to m), whereas no joint probability function will be specified for the cases with missing data (cases 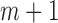 to n). The likelihood function is then calculated by multiplying the joint probability functions for cases without missing values only (i.e.,
to n). The likelihood function is then calculated by multiplying the joint probability functions for cases without missing values only (i.e., 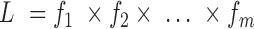 ). Therefore, the regular ML estimation discards the cases with missing data, becoming a CC analysis. When FIML is used, the joint probability function
). Therefore, the regular ML estimation discards the cases with missing data, becoming a CC analysis. When FIML is used, the joint probability function 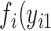 ,
, 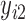 , …,
, …, 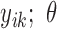 ) is specified for the cases without missing data (cases 1 to m), whereas a different joint probability function,
) is specified for the cases without missing data (cases 1 to m), whereas a different joint probability function, 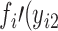 ,
, 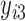 ,…,
,…, 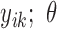 ), is specified for the cases with missing data (cases
), is specified for the cases with missing data (cases 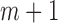 to n). Note that the joint probability function for cases with missing data only contain the variables without missing values (
to n). Note that the joint probability function for cases with missing data only contain the variables without missing values (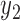 ,
, 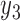 ,…,
,…, 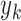 ). The L is then calculated by multiplying the joint probability functions for all cases (i.e.,
). The L is then calculated by multiplying the joint probability functions for all cases (i.e., 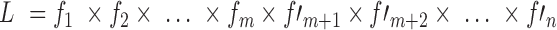 ). Hence, the FIML method allows all cases to be included in the analysis and contribute to the likelihood function, regardless of the missingness and using the data available for each case. The details about FIML can be found in the literature (7, 22).
). Hence, the FIML method allows all cases to be included in the analysis and contribute to the likelihood function, regardless of the missingness and using the data available for each case. The details about FIML can be found in the literature (7, 22).
Although FIML and MI will produce similar results under MAR, FIML should be used with more caution. Because this method is used in the analysis model, it is not easy to use “auxiliary variables” and conduct sensitivity analysis under MNAR assumptions. FIML has been implemented in many software packages, such as in SAS, R, Stata, etc. However, users may need to know how it works in different software or packages because some can only handle missing data on dependent variables but not covariates, or assume a normal distribution for all of the variables with missing values. Currently, the commercial statistical software Mplus (Muthén & Muthén) provides the most comprehensive missing data handling using FIML, for a range of data types, and is able to incorporate missingness in both dependent and independent variables in a broad range of regression models.
Both MI and FIML are advanced statistical tools to appropriately handle missing data and can provide unbiased estimates and valid statistical inferences if the MAR assumption holds. However, pitfalls exist in practice. Hence, researchers are always suggested to consult an experienced statistician when missing data arise in their research.
Sensitivity Analysis
As noted above, most standard MI and FIML approaches assume the data are MAR, but of course, this may not hold in practice. It can be made more plausible by including a large set of factors in the imputation model, which is straightforward to implement. Alternatively, in cases with strong concern about potential MNAR missingness, sensitivity analyses can be done to assess sensitivity to violations of MAR. Two broad classes of sensitivity analysis methods are pattern mixture methods and selection models [see Linero and Daniels (23) for an overview]. Pattern mixture models structure models by assuming some differences in the variables of interest between people with missing and observed values [beyond that accounted for by the observed variables; e.g., that people with missing weight were actually 10% heavier than those with observed weights (24)]. Selection models instead use a sensitivity parameter that relates more directly to the probability of having an observed compared with unobserved value (25, 26). For all of these sensitivity analysis approaches it is important to consider a range of values for the (inherently unobservable) sensitivity parameter. A broad framework for doing so is called a “global” sensitivity analysis, which uses sensitivity parameters and determines at what point results from an RCT would change due to MNAR missingness on outcomes (27).
Conclusions
Missing data are ubiquitous in RCTs. Data analysis that ignores or inappropriately handles missing data may lead to biased and invalid statistical inference and result in compromised scientific conclusions. Some statistical approaches are available to treat missing data issues, providing unbiased estimates and valid statistical inference under the assumption of MAR. Hence, it is always recommended to consult a statistician when missing data occur.
ACKNOWLEDGEMENTS
Both authors declared no conflict of interest. Both authors: read and approved the final manuscript.
Notes
Supported by the National Center for Advancing Translational Sciences of the NIH (award no. UL1TR001417, to PL; principal investigator: R Kimberly; award no. R01HL127491, to EAS; principal investigator: J Siddique). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Abbreviations used: CC, complete case; FCS, fully conditional specification; FIML, full information maximum likelihood; ITT, intent-to-treat; LOCF, last-observation-carried-forward; MAR, missing at random; MCAR, missing completely at random; MI, multiple imputation; ML, maximum likelihood; MNAR, missing not at random; MVN, multivariate normal; RCT, randomized controlled trial.
References
- 1. Little RJ, D'Agostino R, Cohen ML, Dickersin K, Emerson SS, Farrar JT, Frangakis C, Hogan JW, Molenberghs G, Murphy SA et al.. The prevention and treatment of missing data in clinical trials. N Engl J Med. 2012;367(14):1355–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Elobeid MA, Padilla MA, McVie T, Thomas O, Brock DW, Musser B, Lu K, Coffey CS, Desmond RA, St-Onge MP et al.. Missing data in randomized clinical trials for weight loss: scope of the problem, state of the field, and performance of statistical methods. PLoS One. 2009;4(8):e6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Panel on Handling Missing Data in Clinical Trials, Committee on National Statistics, Division of Behavioral and Social Sciences and Education. The prevention and treatment of missing data in clinical trials. Washington (DC): National Academies Press; 2010. [PubMed] [Google Scholar]
- 4. Johnston BC, Guyatt GH. Best (but oft-forgotten) practices: intention-to-treat, treatment adherence, and missing participant outcome data in the nutrition literature. Am J Clin Nutr. 2016;104(5):1197–201. [DOI] [PubMed] [Google Scholar]
- 5. Rubin DB. Inference and missing data. Biometrika. 1976;63(3):581–92. [Google Scholar]
- 6. Roderick JAL. A test of missing completely at random for multivariate data with missing values. J Am Stat Assoc. 1988;83(404):1198–202. [Google Scholar]
- 7. Collins LM, Schafer JL, Kam CM. A comparison of inclusive and restrictive strategies in modern missing data procedures. Psychol Methods. 2001;6(4):330–51. [PubMed] [Google Scholar]
- 8. Schafer JL. Analysis of incomplete multivariate data. New York: Chapman & Hall; 1997. [Google Scholar]
- 9. Carpenter JR, Kenward MG. Multiple imputation and its application. New York: John Wiley & Sons; 2013. [Google Scholar]
- 10. Hartley HO, Hocking RR. The analysis of incomplete data. Biometrics. 1971;27(4):783–823. [Google Scholar]
- 11. Lachin JM. Fallacies of last observation carried forward analyses. Clin Trials. 2016;13(2):161–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Newgard CD, Lewis RJ. Missing data: how to best account for what is not known. JAMA. 2015;314(9):940–1. [DOI] [PubMed] [Google Scholar]
- 13. Rubin DB. Multiple imputation for nonresponse in surveys. New York: Wiley; 1987. [Google Scholar]
- 14. Schafer JL, Olsen MK. Multiple imputation for multivariate missing-data problems: a data analyst's perspective. Multivariate Behav Res. 1998;33(4):545–71. [DOI] [PubMed] [Google Scholar]
- 15. Schafer JL, Graham JW. Missing data: our view of the state of the art. Psychol Methods. 2002;7(2):147–77. [PubMed] [Google Scholar]
- 16. Van Buuren S, Brand JPL, Groothuis-Oudshoorn CGM, Rubin DB. Fully conditional specification in multivariate imputation. J Stat Comput Sim. 2006;76(12):1049–64. [Google Scholar]
- 17. Graham JW. Missing data analysis: making it work in the real world. Annu Rev Psychol. 2009;60:549–76. [DOI] [PubMed] [Google Scholar]
- 18. Barnard J, Meng XL. Applications of multiple imputation in medical studies: from AIDS to NHANES. Stat Methods Med Res. 1999;8(1):17–36. [DOI] [PubMed] [Google Scholar]
- 19. White IR, Royston P, Wood AM. Multiple imputation using chained equations: issues and guidance for practice. Stat Med. 2011;30(4):377–99. [DOI] [PubMed] [Google Scholar]
- 20. Stuart EA, Azur M, Frangakis C, Leaf P. Multiple imputation with large data sets: a case study of the Children's Mental Health Initiative. Am J Epidemiol. 2009;169(9):1133–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Murray JS. Multiple imputation: a review of practical and theoretical findings. Statist Sci. 2018;33(2):142–59. [Google Scholar]
- 22. Graham JW, Hofer SM, MacKinnon DP. Maximizing the usefulness of data obtained with planned missing value patterns: an application of maximum likelihood procedures. Multivariate Behav Res. 1996;31(2):197–218. [DOI] [PubMed] [Google Scholar]
- 23. Linero AR, Daniels MJ. Bayesian approaches for missing not at random outcome data: the role of identifying restrictions. Statist Sci. 2018;33(2):198–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Little RJA. Pattern-mixture models for multivariate incomplete data. J Am Stat Assoc. 1993;88(421):125–34. [Google Scholar]
- 25. Little RJA, Rubin DB. Statistical analysis with missing data. New York: John Wiley & Sons; 2002. [Google Scholar]
- 26. Enders CK. Applied missing data analysis. New York: Guilford Press; 2010. [Google Scholar]
- 27. Scharfstein D, McDermott A, Díaz I, Carone M, Lunardon N, Turkoz I. Global sensitivity analysis for repeated measures studies with informative drop-out: a semi-parametric approach. Biometrics. 2018;74(1):207–19. [DOI] [PubMed] [Google Scholar]


