Abstract
The collective tolerance towards antimicrobial peptides (APs) is thought to occur primarily through mechanisms associated with live bacterial cells. In contrast to the focus on live cells, we discover that the LL37 antimicrobial peptide kills a subpopulation of Escherichia coli, forming dead cells that absorb the remaining LL37 from the environment. Combining mathematical modelling with population and single-cell experiments, we show that bacteria absorb LL37 at a timing that coincides with the permeabilization of their cytoplasmic membranes. Furthermore, we show that one bacterial strain can absorb LL37 and protect another strain from killing by LL37. Finally, we demonstrate that the absorption of LL37 by dead bacteria can be reduced using a peptide adjuvant. In contrast to the known collective tolerance mechanisms, we show that the absorption of APs by dead bacteria is a dynamic process that leads to emergent population behaviour.
Keywords: collective tolerance, population dynamic, antimicrobial peptide
1. Introduction
Antimicrobial peptides (APs) are short amino acid chains that counter bacterial pathogens in host innate immune systems [1,2] and that are being developed as new sources of antibacterial agents [3,4]. Unlike classical antibiotics, APs exhibit a unique mode of action towards bacteria. For instance, after the initial contact with bacterial membranes driven by the cationic property of APs, the insertion of APs into lipid bilayers leads to membrane permeabilization and cell death [5,6]. Previous studies have also shown that some APs can target DNA [7,8] and intracellular proteins [9,10]. The studies on the interactions between APs and bacteria have led to the discovery of tolerance mechanisms such as modification of bacterial surface charge [11–15] and surface shielding [16–18]. However, beyond the direct interaction between APs and bacterial targets, APs can be tolerated by certain bacterial species through collective mechanisms. The collective tolerance mechanisms are relatively well studied for classical antibiotics [19,20] when compared with antimicrobial peptides, and their implications for antibiotic treatment are well demonstrated in the literature [21–24]. For antimicrobial peptides, bacteria may exhibit collective tolerance through mechanisms such as membrane-displayed proteases that degrade APs [25–27] and secreted molecules including lipids, vesicles and proteins that sequestrate APs [16–18,28]. The sequestration mechanism occurs due to electrostatic interactions between cationic APs and negatively charged molecules or surfaces [28–32].
To understand collective tolerance caused by the sequestration mechanism, it is necessary to first track the localization and distribution of APs in a bacterial population. However, previous results have been contradictory because the minimum inhibitory concentration (MIC) of APs is at least 100 times higher than the amount necessary to kill a single bacterium, suggesting an unknown sequestration source. Through fluorescence spectroscopy of the AP PMAP-23, it was found that bacteria are killed when the AP molecules saturate the total surface area of bacterial membranes with 106–107 peptides per cell [33]. In contrast, another study has shown that the MIC of AP Pexiganan requires approximately 109 peptides per cell [34], which is much higher than the necessary amount of AP to saturate the surface area of a single bacterium [34,35]. In addition, if the membrane of live cells is the only sequestration source of APs, the MIC of bacteria must increase linearly with the inoculum size. This expectation has also been proven wrong in the literature [34]. What is the hidden factor that contributes significantly to the sequestration of APs in a bacterial population that does not exhibit any of the known tolerance mechanisms (i.e. lipid shedding and protease display)? Answers to this question may lead to a new explanation of collective tolerance dynamics during AP treatment.
Instead of focusing on live bacterial cells following current thoughts in the field, we find that dead bacterial cells can serve as a major sequestration source of an AP. We discover that a human-derived AP LL37 permeabilizes cytoplasmic membranes of a subpopulation of bacteria (Escherichia coli), which then absorbs LL37 from the environment. The sequestration of LL37 by permeabilized bacteria forms a negative feedback response to LL37 treatment, generating emergent collective tolerance dynamics that cannot be predicted without the AP-absorption mechanism. Specifically, we track the dynamics of LL37 in bacterial populations using both single-cell and population measurements based on previous work [36,37]. We first rule out known AP-tolerance mechanisms in our model system, including the modification of bacterial surface charge [11–15], the inactivation of APs by surface shielding [16–18] and the cleavage of LL37 [25–27]. Next, we show that free LL37 in the bacterial culture is absorbed by dead bacterial cells, which allows a subpopulation of E. coli to grow and repopulate the culture. We also present single-cell data and perturbation experiments that confirm the AP-absorption mechanism. Furthermore, we demonstrate that the AP-absorption leads to emergent cross-bacterial strain protection against LL37. To illustrate the importance of understanding the AP-absorption mechanism, we show that a peptide adjuvant can be supplemented to reduce the absorption of APs. Our work expands the understanding of collective bacterial tolerance towards APs, and it may be considered in a new design of AP treatment that enhances the efficacy of APs.
2. Results
2.1. Bacterial population recovers from initial killing by LL37 through a non-heritable mechanism
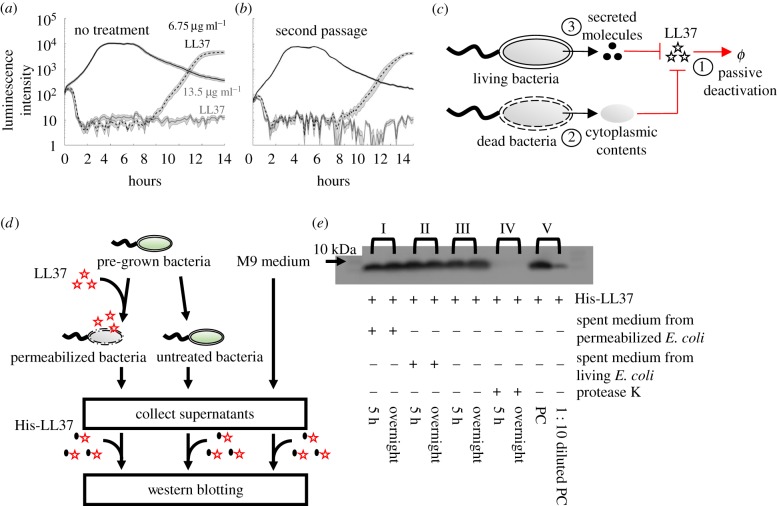
To establish the experimental conditions for our study, we first investigate real-time growth dynamics of E. coli under LL37 treatment. The E. coli BL21PRO strain expresses lux genes (BP-lux), leading to luminescence that is tracked as a surrogate of bacterial viability using a plate reader [38]. Luminescence intensity is widely used to report bacterial metabolic state under antimicrobial treatment because it exhibits higher sensitivity and larger dynamic range than optical density [38–40]. For this experiment, we initiate cultures using the M9 medium with approximately 103–104 CFU µl−1 of bacteria (see pre-growth protocol 1 in Methods (§4.1)) and measure their growth dynamics in a 96-well plate supplemented with twofold dilutions of LL37 using a plate reader for at least 14 h (see Methods (§4.2)). In typical antibiotic tests using batch cultures, bacteria will either grow or be inhibited by the antibiotic for at least 24 h before the emergence of resistant mutants [41]. Instead, for LL37, we find that bacterial populations are inhibited (decline in luminescence intensity) by LL37 at 6.75 µg ml−1 (figure 1a, black dashed line) in the first 7–8 h, after which they re-grow at the same rate as the untreated bacteria (figure 1a, black solid line). The recovery dynamic does not occur with LL37 at 13.5 µg ml−1 within the duration of the experiment (figure 1a, grey line).
Figure 1.
Escherichia coli population tolerates LL37 through a mechanism that is not heritable or degradation. (a) Population dynamics of E. coli are tracked using their luminescence intensity. Bacteria treated with LL37 at two concentrations (black dashed line and grey line) demonstrate initial killing (before 7–8 h) compared with the one without treatment (black solid line). However, the bacterial population treated with 6.75 µg ml−1 of LL37 recovers after around 8 h. Shaded error bars are standard error of the mean (s.e.m.) from N = 11. See Methods (§4.2). (b) Recovered bacteria from LL37 treatment at 6.75 µg ml−1 (a, black dashed line) are collected and grown overnight. The second passage of recovered bacteria is treated with LL37 at two concentrations (black dashed line and grey line) following the same protocol as in (a). The second passage of bacteria exhibits similar recovery after inhibition, suggesting that genetic mutations do not cause the recovery in our system. Shaded error bars are s.e.m. from N = 12. See Methods (§4.2). (c) The schematic shows possible mechanisms that may reduce the antimicrobial activity of LL37 in bacterial culture: ①. Natural degradation or self-aggregation may alter and mask functional domains of LL37. ②. Permeabilized bacterial cells may release intracellular contents that degrade or inactivate LL37. ③. Live bacteria may secrete molecules that degrade or inactivate LL37. (d) A flow chart illustrates the experiments that investigate stability (①) of His-LL37 in medium and the degradation (② and ③) of His-LL37 in spent medium from permeabilized or live bacteria (e). Specifically, bacterial cells are permeabilized by LL37, and cytoplasmic contents in the spent medium are collected by centrifugation (left). Secreted molecules are collected by removing untreated bacteria from the medium (middle). As a control, medium without cells is included (right). His-LL37 is incubated in the spent medium or fresh medium and subjected to western blotting. See electronic supplementary material, method S2. (e) Quantification of the relative amount of His-labelled LL37 (His-LL37). We treat His-LL37 with the collected spent medium (d) for 5 h or overnight to assess its degradation. The relative amount of His-LL37 is quantified using the band intensity from western blotting. His-LL37 incubated in spent medium from permeabilized bacteria (I) or live bacteria (II) does not show a reduction in band intensity. Furthermore, the relative amount of His-LL37 does not change over time in medium without bacteria, which suggests that LL37 does not naturally degrade in the medium (III). Proteinase K retains proteolytic activity in our reaction condition (IV). Western blotting is sensitive to a 10-fold decrease in the amount of His-LL37 (V). See replicate of western blotting results in electronic supplementary material, figure S4a. See also electronic supplementary material, method S2. (Online version in colour.)
Based on the results, we define MIC in this study as the concentration of APs where no bacterial growth is observed within the entire duration of the experiment. In contrast, the concentration of APs that allows bacterial recovery is considered as sub-MIC. We validate the mode of actions of LL37 using phosphatidylserine (PS) exposure and propidium iodide (PI) staining, which have been used as markers for bactericidal antibiotics [42], and cell permeabilization and death [43] in previous studies (electronic supplementary material, figure S1). Unless otherwise noted, we use the intermediate concentrations (i.e. sub-MIC) of LL37 that allow bacterial recovery to reveal the unknown collective tolerance mechanism. The use of sub-MIC instead of MIC is well accepted in the study of tolerance mechanisms [41,44,45]. Bacterial growth dynamics are observable and sensitive to the APs at sub-MIC, increasing the feasibility of detecting any tolerance mechanisms. Despite the use of sub-MIC for our study, the revealed collective tolerance mechanisms will occur at AP concentrations above MIC and potentially reduce the efficacy of the AP.
To approach the unknown tolerance mechanism, we first consider whether some cells may escape the binding of LL37 stochastically owing to under-coverage of the AP molecules to bacterial membranes, which may contribute to the bacterial recovery. Specifically, if the amount of LL37 is not sufficient to cover the membrane areas, we expect to see an increase in the average amount of LL37 bound to bacterial membranes with a higher dose of LL37. Our flow cytometry results counteract this argument. To track LL37, we use rhodamine-labelled LL37 (Rh-LL37) that demonstrates antimicrobial activity and generates similar recovery dynamics of bacteria to unmodified LL37 (electronic supplementary material, figure S2a) to treat wild-type E. coli BL21PRO (WT-BP). Since the conjugation of rhodamine to LL37 reduces its antimicrobial activity, we use Rh-LL37 at 54 µg ml−1 (no recovery) and 27 µg ml−1 (with recovery), which lead to similar dynamics to unmodified LL37 at 13.5 µg ml−1 and 6.75 µg ml−1, respectively. Because the results from Rh-LL37 experiments are interpreted by comparing with negative controls, the specific concentrations we used do not impact the main conclusions of the experiments. We find that the initial distributions of rhodamine intensity in bacterial populations do not show any difference between Rh-LL37 treatments at the two concentrations (electronic supplementary material, figure S2c). The result implies that the average amount of Rh-LL37 bound to bacterial membranes remains the same for the Rh-LL37 treatments. The counteracting evidence between the bacterial recovery and over-coverage of LL37 molecules to the bacterial membrane (for the calculation, see the electronic supplementary material, text S2) prompts us to investigate if there are any mechanisms that significantly reduce the effective amount of LL37 and govern the population dynamics of bacteria.
We next attempt to rule out a few canonical resistance mechanisms before investigating collective tolerance mechanisms. We first determine whether the bacterial tolerance to LL37 is heritable. Specifically, we examine whether mutations may occur in our experiments and lead to the recovery of the bacterial population. We collect bacteria (BP-lux) that recovered from LL37 treatment at 6.75 µg ml−1 and passage them using fresh M9 medium supplemented with LL37. The passaged bacteria exhibit the same dynamics as the original bacterial populations (figure 1b): the passaged bacteria recover after a period of inhibition with 6.75 µg ml−1 LL37, but no recovery with 13.5 µg ml−1 LL37. In addition, real-time supplementation of LL37 during the recovery phase still inhibits the bacterial growth (electronic supplementary material, figure S3). The results indicate that the bacterial recovery is not due to random bacterial mutations or heritable resistance towards LL37.
2.2. The non-heritable mechanism is not due to degradation of LL37
Next, we investigate if the bacterial recovery is due to natural degradation of LL37 (figure 1c, ①). In subsequent experiments, we use His-tagged LL37 (His-LL37) as a surrogate for LL37 to allow quantification of the peptide. Because His-LL37 is exposed directly to the possible degrading agents in our experiments, it does not have to exhibit the same antimicrobial activity as LL37. We first add His-LL37 to medium and incubate it at 37°C for 5 h or overnight (figure 1d, right). We next compare the relative amount of His-LL37 incubated for 5 h, overnight and a positive control (fresh His-LL37 at identical concentration) using western blotting to assess its degradation. If His-LL37 is naturally degraded, we would expect a reduced intensity of the band for 5 h or overnight treatment compared with the positive control. Western blotting does not show any difference between the band intensities of the 5 h sample, overnight sample and the positive control (figure 1e-III and electronic supplementary material, figure S4a). The result implies that the amount of His-LL37 is not reduced after either 5 h or overnight incubation in the medium. We note that the western blotting is capable of distinguishing at least a 10-fold decrease in the relative amount of His-LL37 (figure 1e-V and electronic supplementary material, figure S4a). Furthermore, since the results for His-LL37 degradation tests are compared with designed controls, the main conclusions are not affected by the specific concentration of His-LL37 we choose. We further rule out the self-degradation of LL37 in our system by showing that pre-exposure of unmodified LL37 in the medium does not diminish its activity (electronic supplementary material text S3 and figure S5).
LL37 may be degraded by cytoplasmic contents released from permeabilized bacteria (figure 1c, ②). To test this hypothesis, we use western blotting to investigate if cytoplasmic contents degrade His-LL37. To collect cytoplasmic contents, we treat WT-BP with LL37 at 13.5 µg ml−1 to permeabilize bacterial membranes as previously described (figure 1a). We then extract spent medium from permeabilized bacteria by centrifugation. The cytoplasmic contents in the spent medium directly mimic the molecular concentration and composition in the extracellular environment of a bacterial culture that has undergone LL37 treatment. The spent medium is then supplemented with His-LL37 at 37°C for 5 h or overnight (figure 1d, left). We next use western blotting again to assess its degradation. We find no difference between the band intensities of the 5 h sample, overnight sample and the positive control (figure 1e-I and electronic supplementary material, figure S4a). The result implies that His-LL37 is not degraded after either 5 h or overnight incubation in the spent medium. We repeat this experiment using whole cell extract (WCE) from E. coli BL21PRO instead of spent medium from permeabilized E. coli. Again, we find no degradation of His-LL37 by the WCE (electronic supplementary material, figure S4b). To further explore the degradation of LL37 by cytoplasmic contents, we assess the antimicrobial activity of Rh-LL37 after incubation with the spent medium from permeabilized E. coli using a microscope (electronic supplementary material, S4 and figure S6a). We find that Rh-LL37 still retains its activity after pre-exposure of 5 h to spent medium from permeabilized bacteria (electronic supplementary material, figure S6c). Therefore, our results suggest that the cytoplasmic contents released from permeabilized bacteria do not degrade LL37.
LL37 may also be degraded or sequestered by secreted molecules from live bacteria (figure 1c, ③). Here, we collect spent medium from WT-BP without LL37 treatment, which contains secreted molecules from bacteria. We supplement the spent medium with His-LL37 and compare its relative concentration after either 5 h or overnight incubation with a positive control using western blotting. Again, we observe no difference between the band intensities of the 5 h sample, overnight sample and the positive control (figure 1e-II; electronic supplementary material, figure S4a), which implies that the amount of His-LL37 is not decreased by the spent medium. Next, we explore the antimicrobial activity of Rh-LL37 after incubation with the spent medium from untreated E. coli using a microscope (electronic supplementary material, figure S6a). We find that the incubated Rh-LL37 co-localizes with fresh bacterial cells, indicating that the spent medium from untreated bacterial culture does not diminish the antimicrobial activity of Rh-LL37 (electronic supplementary material, figure S6b). Our results suggest that degradation of LL37 by secreted molecules of bacteria does not occur in our experiments.
2.3. LL37 is absorbed by permeabilized bacteria
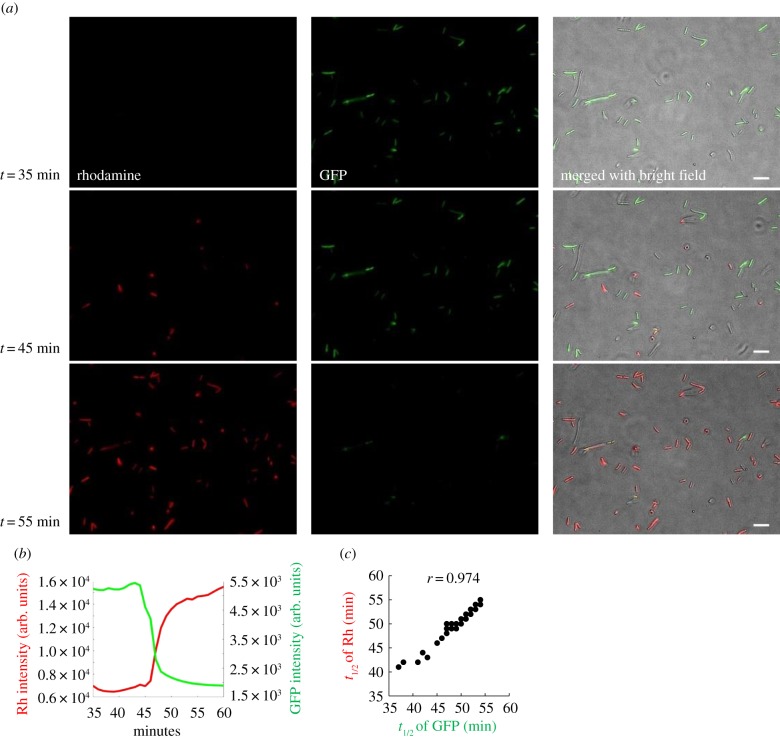
The above results have ruled out the loss of LL37 activity by either active or passive degradation. To shed light on the unknown mechanism that leads to bacterial recovery, we track the dynamics of Rh-LL37 at the single bacterium level. E. coli BL21AI expresses green fluorescent proteins (BA-GFP), which are leaked outside of bacteria when their cytoplasmic membranes are permeabilized [36] (figure 2a). The leakage of intracellular GFP has been used as an indicator of bacterial cell death [46]. The bacteria are incubated in a culture chamber at room temperature for 30 min to allow their adhesion to the bottom surface of the chamber, and then supplemented with Rh-LL37 at 54 µg ml−1. Thirty-five minutes after supplementation, all bacterial cells show strong GFP signals, suggesting bacterial cells are not permeabilized yet. At 45 min, some bacterial cells exhibit strong Rh-LL37 signals, indicating co-localization of the AP with bacterial cells. Ten minutes later, most bacteria exhibit strong Rh-LL37 signals (figure 2a). Quantification of the GFP and Rh-LL37 dynamics shows that accumulation of Rh-LL37 co-localized with bacteria coincides with the loss of cytoplasmic GFP (figure 2b and electronic supplementary material, figure S7). We observe that the rhodamine intensity does not show measurable fluctuations before the drop of GFP intensity, suggesting that any binding events of Rh-LL37 (e.g. binding to the outer membrane and periplasmic space [36]) before cytoplasmic membrane leakage may be below the detectable limit of our wide-field microscopy. Furthermore, the accumulation of Rh-LL37 in bacterial cells is not due to the intercalation of rhodamine dye into DNA (electronic supplementary material, text S1 and figure S8) [47]. Moreover, the half-time of fluorescence signal fluctuations from bacterial cells shows a positive correlation between GFP and Rh-LL37 (with a Pearson correlation coefficient (r) of 0.974; figure 2c). Next, we separate dead bacteria treated with Rh-LL37 and culture medium through centrifugation. We find that the activity of Rh-LL37 remaining in the medium is significantly reduced (electronic supplementary material, text S5 and figure S9). Altogether, the observed LL37 accumulation inside bacteria and loss of activity in spent medium suggest that LL37 permeabilizes and kills a subpopulation of bacteria, which then absorbs LL37 from the environment, leading to the regrowth of the living bacteria.
Figure 2.
The permeabilization of bacteria under Rh-LL37 treatment correlates with the accumulation of the peptide in dead bacterial cells. (a) Single bacterium microscopy shows the accumulation of Rh-LL37 in bacteria. E. coli constitutively expressing green fluorescent proteins (GFP) is treated by Rh-LL37. Scale bar represents 10 µm. See electronic supplementary material, method S15. (b) The representative dynamics of rhodamine and GFP intensity of one bacterium show that the leakage of GFP from bacterial cytoplasm correlates with the accumulation of Rh-LL37 in bacterial cells. See additional single-cell dynamics in electronic supplementary material, figure S7. See also electronic supplementary material, method S15. (c) Single-cell GFP and rhodamine dynamics are analysed using Matlab to identify a correlation between the dynamics. Times to reach half of the maximum changes in GFP and rhodamine intensities (t1/2) are positively correlated. N = 54. See electronic supplementary material, method S15. (Online version in colour.)
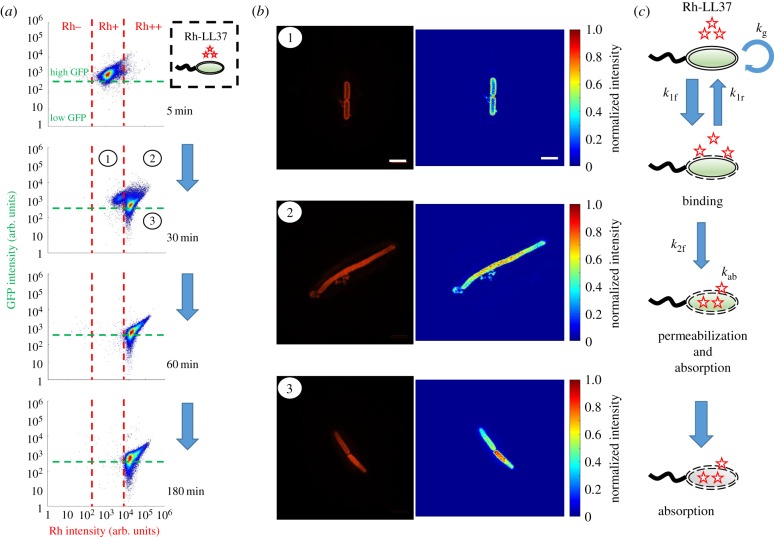
To link our observations from single-cell measurements to population dynamics, we perform flow cytometry to track the fates of E. coli BL21PRO expressing GFP (BP-GFP) under Rh-LL37 treatment. Specifically, approximately 103 CFU µl−1 of BP-GFP (see pre-growth protocol 2 in Methods (§4.1)) is treated with Rh-LL37 at 27 µg ml−1 for several durations and subjected to flow cytometry. We set one threshold for GFP intensity based on the negative controls (WT-BP) that do not express GFP (figure 3a, and electronic supplementary material, figure S10, for negative control). For rhodamine intensity, we first set one threshold to separate the negative control (Rh−; electronic supplementary material, figure S10, for negative control) and Rh-LL37 associated subpopulations (Rh+). We find that the majority (approx. 97%) of the bacterial cells have high GFP and Rh+ after 5 min of treatment. After 30 min of treatment, another subpopulation emerges at higher rhodamine intensity (figure 3a). We set another threshold for rhodamine intensity based on the emergent subpopulation (Rh++). At 60 and 180 min, the majority of bacterial cells (approx. 99% for both time points) have shifted to Rh++. The results suggest a dynamic transition of bacterial cell states during Rh-LL37 treatment (i.e. Rh negative → ① → ② and ③ from figure 3a). The majority of cells (Rh++) are dead after 180 min because, after Rh-LL37 accumulates in the bacterial cells, we do not observe any cell division or growth using single-cell microscopy. We notice that high rhodamine intensity may cause unspecific GFP signals (high GFP signals in figure 3a at late time points) in our flow cytometry measurements (electronic supplementary material, figure S11a).
Figure 3.
LL37 is absorbed by permeabilized bacterial cells, and the absorption causes depletion of LL37 in bacterial cultures. (a) Flow cytometry results demonstrate the transition of bacterial states over time during Rh-LL37 treatment. E. coli constitutively expressing green fluorescent proteins (GFP) is treated by Rh-LL37. Green dashed line separates populations with intact (high GFP) and permeabilized (low GFP) membrane. Rh negative (Rh−) represents bacterial population without Rh-LL37 association (see electronic supplementary material, figure S10, for negative controls). Bacterial cells can transit from ‘high GFP, Rh positive (Rh+)’ to ‘high GFP, Rh double positive (Rh++)’ over time, and the permeabilization (transition from high GFP to low GFP) only occurs in Rh++ bacteria. See electronic supplementary material, method S16. (b) Structured illumination microscopy (SIM) reveals co-localization of Rh-LL37, bacterial membrane and intracellular space. Bacterial populations are sorted to collect subpopulations under Rh-LL37 treatment (①, ②, and ③ from figure 3a). SIM images (left) and intensity heat maps (right) show that the Rh-LL37+ population (①) has Rh-LL37 co-localized at the perimeter of the cell membrane, whereas the Rh-LL37++ population (② and ③) has Rh-LL37 co-localized in the intracellular space of the bacteria. See electronic supplementary material, figure S20, for more SIM images. Scale bar represents 2 µm. See also electronic supplementary material, method S17. (c) Proposed model for the transition of bacterial states under Rh-LL37 treatment. We propose three bacterial states to explain the population dynamics: growing population, binding population (① from figure 3a) and absorbing population (② and ③ from figure 3a). Kinetics of the transitions between states are governed by three reaction rate constants (k1f, k1r and k2f). The depletion rate of Rh-LL37 from the medium by the absorbing population is governed by kab. Intrinsic bacterial growth has a rate constant of kg. See equation (4.1) and Methods (§4.3). (Online version in colour.)
To better measure the subcellular localization of LL37, we treat E. coli BP-GFP with Rh-LL37 for 30 min as described above (figure 3a) and sort three subpopulations: high GFP, Rh+ (① in figure 3a), high GFP, Rh++ (② in figure 3a) and low GFP, Rh++ (③ in figure 3a). The sorted samples are subjected to high-resolution structured illumination microscopy (SIM) to identify the localization of Rh-LL37 molecules. We find that Rh-LL37 molecules accumulate at the perimeter of bacterial cells for the Rh+ subpopulation (① and figure 3b, top), indicated by the high Rh intensities on bacterial membranes. When the bacterial cells progress to Rh++ subpopulations (② and ③), Rh-LL37 molecules co-localize within the intracellular space of the bacteria (figure 3b, middle and bottom), indicated by the high Rh intensities in the cytoplasm. Furthermore, we note that the bacterial population treated with Rh-LL37 at the MIC demonstrates a similar bacterial state transition (i.e. the transition from Rh−, Rh+ to Rh++) to the one treated at sub-MIC, implying that the AP absorption occurs at concentrations of LL37 above MIC (electronic supplementary material, figure S11b).
According to the observations, we propose a phenomenological model to describe the sequence of events during Rh-LL37 treatment (figure 3c). Specifically, we define three states of bacteria in our model: growing (Rh−), binding (Rh+, ①) and absorbing (Rh++, ②, and ③) states. First, free Rh-LL37 molecules bind to bacterial cells and transfer them from the ‘growing’ to the ‘binding’ state. We assume that the cells can recover at a certain rate from the ‘binding’ to the ‘growing’ state owing to dissociation of bound Rh-LL37. The assumption is used to formulate our mathematical model (see Methods (§4.3)). Meanwhile, bound Rh-LL37 can further progress towards permeabilizing bacterial membranes (transition from ‘binding’ to ‘absorbing’ state). This event corresponds to the leakage of intracellular contents (reported by GFP; figure 2b), as well as the absorption of free Rh-LL37 molecules (figures 2b and 3a). Next, we build a mathematical model to quantitatively explore the proposed model (see Methods (§4.3)). Specifically, the progression of sequential events is governed by several reaction rate constants: kg for growth rate, k1f and k1r for forward and reverse transitions, respectively, between ‘growing’ and ‘binding’ states, k2f for transition from the ‘binding’ to the ‘absorbing’ state, and kab for the AP absorbing rate (figure 3c). We estimate the parameters in our model by fitting to three biological replicates of the flow cytometry experiments (electronic supplementary material, figure S12, and Methods (§4.3) for details). The mathematical model is then extended to provide insights for population and collective tolerance dynamics of bacteria by assuming bacteria will undergo a similar transition of states during treatment by unmodified LL37. The model successfully captures the recovery of the bacterial population under AP treatments (electronic supplementary material, figure S21).
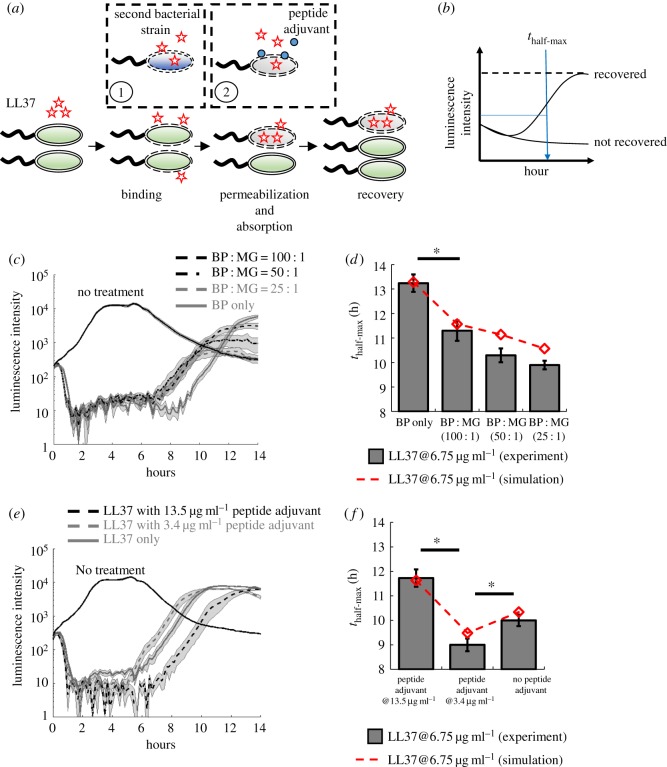
2.4. AP absorption by dead bacteria is perturbed by the presence of another bacterial strain and reduced by a peptide adjuvant
Depending on the membrane surface charge, lipid composition, intracellular composition and other factors [48,49], bacterial strains and species may display different kinetics of state transitions (k1f, k1r, k2f from figure 3c) and AP absorption (kab, from figure 3c, and Kab, which is the half-maximal constant for absorption). Therefore, the growth dynamics of one bacterial strain may be perturbed by the presence of the second strain during LL37 treatment (① in figure 4a). To start, we first estimate the kinetic parameters of bacterial state transitions and AP absorption for the E. coli MG1655 strain that expresses GFP (MG-GFP) following the same protocol as BP-GFP (see electronic supplementary material, S16, and Methods (§4.3) for details). We estimate that MG-GFP demonstrates faster growth rate (larger kg), as well as faster recovery from the ‘binding’ to the ‘growing’ state (larger k1r), than BL21PRO (electronic supplementary material, figure S12b). Furthermore, MG-GFP also has a faster permeabilization rate and absorption rate for Rh-LL37 (larger k2f and kab; electronic supplementary material, figure S12b). The faster permeabilization and absorption rates of MG-GFP should lead to an earlier emergence of the recovered subpopulation than BP-GFP, which is demonstrated in our flow cytometry results (electronic supplementary material, figure S13). The E. coli MG1655 strain has different cell wall and outer membrane compositions than the BL21PRO strain, which may lead to the difference in the kinetic parameters. Furthermore, we show that cytoplasmic or secreted contents from MG1655 do not degrade Rh-LL37 (electronic supplementary material, text S4 and figure S14).
Figure 4.
LL37 absorption by dead cells leads to cross-bacterial strain protection and can be reduced by a peptide adjuvant. (a) The perturbations of AP absorption by another bacterial strain (①) or a competitive molecule for absorption (②). Specifically, one bacterial strain may have different LL37 absorbing kinetics (e.g. faster absorption rate) that modulate recovery of another strain during AP treatment. In addition, a peptide adjuvant that can compete with LL37 for unintended AP absorption may increase the antibacterial efficacy of LL37. (b) We evaluate the recovery of the bacterial population under AP treatment using the time when the population recovers to half of its maximum growth capacity within 14 h. (c,d) Two strains of E. coli (BP-lux and MG1655) are mixed at different ratios, where the total CFU of each mixture is kept constant. (c) Dynamic curves show the cross-bacterial strain protection. In the presence of MG1655 (black dashed and grey dashed lines), the recovery times of BP-lux (which is tracked through luminescence intensity) are shifted to an earlier time than the one with only BP-lux (grey line). Shaded error bars are s.e.m. from N = 8. (d) Quantified recovery times from experiment (grey bars) and simulation (dashed line) showing cross-bacterial strain protection (see electronic supplementary material, method S14, for the model, and electronic supplementary material, table S1, for estimated parameters). Asterisk indicates significant difference (p < 0.01), and error bars are s.e.m. from N = 8. See electronic supplementary material, method S18. (e,f) A peptide adjuvant is added to the culture of BP-lux during LL37 treatment. (e) Dynamic curves show perturbation by the peptide adjuvant. The recovery is delayed in the presence of the peptide adjuvant at 13.5 µg ml−1 (black dashed line) but accelerated in the presence of the peptide adjuvant at 3.4 µg ml−1 (grey dashed line) compared with the one with only BP-lux (grey line). Shaded error bars are s.e.m. from N = 8. (f) Quantified recovery times from experiment (grey bars) and simulation (dashed line) (see electronic supplementary material, method S14, for the model, and electronic supplementary material, table S1, for estimated parameters). Asterisk indicates significant difference (p < 0.01), and error bars are s.e.m. from N = 8. See electronic supplementary material, method S19. (Online version in colour.)
Based on the difference in the kinetics, we hypothesize that the recovery of BL21PRO can be accelerated to an early time in the presence of MG1655, which has a faster absorption rate (① in figure 4a). We first expand the mathematical model (equation (4.1)) to include MG1655 (electronic supplementary material, equation (S1)) with the estimated kinetic parameters (electronic supplementary material, table S1). In the model, two strains compete for common resources (see electronic supplementary material, method S14, for model expansion). Indeed, with the same total bacterial densities, our simulation shows that the recovery time of BL21PRO during LL37 treatment is accelerated by the presence of MG1655 (dashed line in figure 4d and electronic supplementary material, figure S22a) due to fast depletion of free LL37 molecules (electronic supplementary material, figure S22b). To test our hypothesis, we mix BP-lux and wild-type MG1655 (WT-MG) with various CFU ratios and track the recovery of BP-lux under LL37 treatment at 6.75 µg ml−1 using a plate reader. To better quantify the recovery time of BP-lux, we define a metric named thalf-max, where the population is recovered to half of its growth capacity after initial inhibition by an AP (figure 4b). We find that, for all ratios of the two strains (BP : MG = 100 : 1, 50 : 1, 25 : 1), the recovery of BP-lux is accelerated by approximately 2–3 h (figure 4c,d). Furthermore, the total initial cell density of all mixtures is tightly controlled to be identical (approx. 103 CFU µl−1; see pre-growth protocol 2 in Methods (§4.1)), so that our observations are not affected by initial bacterial densities. The results corroborate that the AP absorption forms a feedback response to the AP, which generates emergent collective tolerance dynamics.
For an AP and a bacterial strain that exhibit AP absorption, we speculate that a peptide can compete with the absorption of the AP and delay the depletion of free AP molecules. We choose a peptide adjuvant that exhibits amphipathicity based on previous work [50]. Consistent with our expectation, we find that the peptide adjuvant at 13.5 µg ml−1 delays recovery time of BP-lux (see pre-growth protocol 1 in Methods (§4.1)) by approximately 2 h compared with the culture under only LL37 treatment (figure 4e). We note that the peptide adjuvant alone does not inhibit bacterial growth at the tested concentrations (electronic supplementary material, figure S16a). However, the peptide adjuvant displays a concentration-dependent effect. That is, it delays bacterial recovery at high concentration (13.5 µg ml−1) but accelerates it at low concentration (3.4 µg ml−1) (figure 4e,f). Flow cytometry results show that the peptide adjuvant at 13.5 µg ml−1 delays the transition from the ‘binding’ to the ‘absorbing’ state (electronic supplementary material, figure S16b). However, we find that the supplementation of the peptide adjuvant barely affects the distribution of the ‘binding’ population after 5 min of AP treatment, implying that the initial binding of AP to bacterial membranes dominates over the binding of the peptide adjuvant to bacterial membranes at early time points (electronic supplementary material, figure S16b).
We next expand the mathematical model (equation (4.1)) to provide some insights into the effects of the peptide adjuvant (electronic supplementary material, equation (S2)). Based on the experimental results, we assume that the peptide adjuvant can competitively bind to both bacterial membranes and intracellular absorbing sites against the AP. Since the initial binding of the AP to the membranes is a faster event than that of the peptide adjuvant, we assume that the ‘growing’ to ‘binding’ transition is not significantly affected by the peptide adjuvant (electronic supplementary material, figure S23a). Furthermore, we assume that the peptide adjuvant delays the ‘binding’ to ‘absorbing’ transition (electronic supplementary material, figure S23a). We also assume that the boost of the ‘binding’ to ‘growing’ transition is possibly due to the strong affinity of the peptide adjuvant to bacterial membrane. Our model demonstrates that the concentration dependency of the peptide adjuvant may arise from its stronger modulation of the AP absorption rate at higher peptide concentrations (electronic supplementary material, figure S23b). Indeed, the simulation results agree with our experiments (dashed line in figure 4f, and electronic supplementary material, method S14, for model expansion). The results suggest that peptide adjuvants may be supplemented to APs to reduce or abolish AP absorption.
3. Discussion
Through a series of deductive experiments, we discover a bacterial collective tolerance mechanism towards an AP, in which LL37 permeabilizes and kills a subpopulation of E. coli, which then absorbs the LL37 from the environment, leading to regrowth of living bacteria. The collective tolerance can only occur at the population level because the permeabilized and dead bacteria, in turn, absorb LL37, enhancing the escape of other bacteria in the same population. We rule out classical resistance mechanisms of bacteria, including bacterial mutation and proteolytic cleavage of LL37 (figure 1). Both flow cytometry and single-cell microscopy corroborate the role of AP absorption, as well as suggest a phenomenological model for bacterial population dynamics during treatment (figures 2 and 3). Facilitated by the mathematical models, we demonstrate cross-bacterial strain protection of bacteria against LL37 owing to AP absorption, as well as a potential peptide adjuvant to tackle the tolerance mechanism (figure 4). Furthermore, permeabilized bacteria absorb LL37 at both sub-MIC and MIC concentrations. Therefore, for a broad concentration range of an AP where bacteria are permeabilized by the AP, our results suggest that the absorption of LL37 by dead bacteria can decrease the efficacy of the AP and generate emergent population dynamics in heterogeneous environments.
Furthermore, our findings show that the AP absorption is a major process that influences bacterial population dynamics during AP treatment. For example, if the bacterial recovery (figure 1a) is merely due to the growth of some cells that are not bound by AP, the recovery time should be independent of the presence of a second bacterial strain in the population with a constant initial bacterial density (figure 4). Instead, the observed cross-bacterial strain protection against LL37 suggests the critical role of AP absorption in regulating population dynamics during AP treatment. Moreover, if the binding of AP to live bacterial membrane is the only factor that controls population dynamics, the supplementation of the peptide adjuvant should always accelerate bacterial recovery because of competitive binding between the peptide adjuvant and LL37 to the bacterial membrane. Instead, we find that the peptide adjuvant at 13.5 µg ml−1 delays the bacterial recovery.
The AP molecules may be absorbed by intracellular contents of permeabilized bacteria that exhibit negative charges, such as DNA molecules and proteins. We find that the externally supplemented DNA molecules reduce the efficacy of LL37 (see electronic supplementary material, method S11 and figure S15). In addition, bacterial cells permeabilized by the AP remain positive for propidium iodide staining (electronic supplementary material, figure S1). Since propidium iodide is a fluorescent intercalating agent of DNA molecules, the finding suggests that the intracellular contents can absorb the AP without leakage to the extracellular environment. We speculate that some cellular extract may enter the extracellular environment and sequester the AP molecules. However, the sequestration by the cellular extract should have minor contributions to the observed dynamics in this study for two reasons. First, we do not find complete lysis of bacterial cells from single cell microscopy under the tested conditions (figure 2a). Thus, the amount of cellular extract released should be limited. Second, our models are trained using flow cytometry results, which do not detect cellular extract. Since the models successfully capture the emergent population dynamics, the released cellular extract in the environment should have a limited impact on our conclusions.
The AP-absorption tolerance mechanism may be generalizable to other bacterial species and APs for a few reasons. First, the absorption of APs likely relies on generic electrostatic interactions between APs and bacterial components, which are ubiquitous across bacterial species. However, the kinetics of events leading to AP absorption likely depends on the composition of the membranes and cytoplasm that control the insertion and transport of antimicrobial peptides. Second, bacterial permeabilization is a common mechanism of action of a major class of APs. Upon the initial permeabilization, cationic APs may diffuse into cells and bind to negatively charged cellular components. Since the electrostatic interaction is not unique to LL37, other cationic APs are likely to be tolerated by bacteria through the same absorption mechanism. Indeed, we may investigate the AP-absorption mechanism using recovery dynamics similar to those of E. coli under the treatment of indolicidin and bac2A (APs originated and derived from bovine neutrophils) (electronic supplementary material, figure S17).
The discovery of a novel collective tolerance mechanism based on AP absorption by dead bacteria spawns a new research area with several open questions. From a qualitative point of view, the intrinsic heterogeneity of bacterial populations may cause the stochastic bifurcation of cell states during treatment (e.g. some bacterial cells are permeabilized faster than others owing to the intrinsic heterogeneity). It is unclear if any genes or proteins are associated with the heterogeneous behaviour during AP treatment, which may be investigated through cell sorting and mRNA profiling. In addition, we have used a peptide adjuvant to reduce the AP absorption and improve the efficacy of LL37. APs may be sequestered by multiple negatively charged bacterial components including DNA, peptidoglycan [36], lipopolysaccharide and f-actin [30]. Designing adjuvant molecules that compete for AP absorption may provide a new way to improve AP efficacy. From a quantitative point of view, AP absorption is a highly dynamic process that has the potential to generate emergent dynamics. The kinetics of bacterial death, AP binding and bacterial recovery from bound AP affect the dynamics of AP absorption, which in turn affect population dynamics under AP treatment. Our study also reveals a negative feedback loop between an AP and bacteria. Specifically, an AP permeabilizes bacteria and induces bacterial cell death, but the dead bacterial cells, in turn, absorb AP and diminish the efficacy of the AP. Different from previous studies on collective AP tolerance, the feedback loop highlights the role of bacterial death on population survival during AP treatment, which may suggest a new direction towards improving AP efficacy by perturbing the feedback loop. Furthermore, we have shown that even the same species (E. coli) but different strains (BL21PRO and MG1655) exhibit different kinetics for AP absorption, which are sufficient to generate the cross-bacterial strain protection against LL37. It remains unclear how APs could dynamically shape the composition of a multi-species/strains environment under treatment due to AP absorption, especially when interactions between species/strains are involved.
4. Material and methods
4.1. Bacterial strains and chemicals (M1)
Escherichia coli BL21PRO strain carrying plasmid that constitutively expresses lux genes (BP-lux) was used for measurement in a plate reader. Wild-type E. coli BL21PRO (WT-BP) and E. coli BL21AI constitutively expressing GFP (BA-GFP) were used for tracking Rh-LL37 dynamics with wide-field microscopy. E. coli BL21PRO and MG1655 expressing GFP (BP-GFP and MG-GFP) were used for flow cytometry and SIM. All strains were maintained as glycerol stocks at −80°C for long-term storage or on Luria broth (LB) agar plates at 4°C for short-term storage. Bacteria were grown overnight at 37°C in LB (VWR) with no antibiotic (WT-BP), with kanamycin sulfate at 30 µg ml−1 (BP-lux), with carbenicillin at 100 µg ml−1 (BA-GFP) or with chloramphenicol at 100 µg ml−1 (BP-GFP and MG-GFP) working concentration before experiments. Two pre-growth protocols were used before any treatments to ensure bacteria cells enter the exponential growth phase.
Pre-growth protocol 1: Fresh overnight cultures were diluted 1 : 1000 into the M9 minimal medium (VWR) supplemented with 0.2% glucose and 0.2% casamino acids without any antibiotic selection and grown at 37°C on a shaker for 2 h.
Pre-growth protocol 2: Fresh overnight cultures were diluted 1 : 1000 into the M9 medium without any antibiotic selection. The cultures were grown at 37°C on a shaker for 3 h. To better control bacterial cell density, OD600, luminescence or GFP intensity of the pre-grown cultures was measured, and its CFU µl−1 was back-calculated according to calibration curves. The pre-grown cultures were then diluted with M9 to have approximately 103 CFU µl−1 for later experiments. The number of CFU was also calculated for diluted cultures to check the quality of the density control.
Unmodified LL37 was purchased from AnaSpec. Rhodamine-conjugated LL37 (Rh-LL37) was purchased from Rockland. The APs were reconstituted in nanopore water (Thermo Scientific) before use and stored at −20°C. Six histidine residues were fused to the N-terminus of LL37 (see electronic supplementary material, method S5 and figure S18, for plasmid construction, and electronic supplementary material, method S6, for peptide expression and purification). Carbenicillin and chloramphenicol were purchased from Sigma, and kanamycin sulfate was purchased from Amresco.
The peptide adjuvant was synthesized from Biomatik according to the amino acid sequence obtained from the previous literature [50]: RVQGRWKVRKSFFK with FITC linked to the N-terminus. The peptide adjuvant was reconstituted in nanopore water before use and stored at −20°C.
4.2. Measurement of bacterial growth dynamics using a plate reader (M2)
BP-lux was grown with pre-growth protocol 1. A 100 µl aliquot of bacterial culture was added to each well of a black, flat-bottomed 96-well microplate (Corning Costar). LL37 was supplemented to the wells at 6.75 or 13.5 µg ml−1 working concentrations (figure 1a). The time series of luminescence was measured using a Tecan M1000Pro plate reader at 37°C with shaking (orbital, 20 s every min). The parameters for luminescence measurement were automatic attenuation and 1000 ms integration time.
To test for bacterial mutation (figure 1b), BP-lux was treated with LL37 at 6.75 µg ml−1, and luminescence was tracked in the plate reader as described above. When a bacterial population started to recover (approx. 7–8 h after treatment), 50 µl of the bacterial culture was extracted from the wells and added into 3 ml of LB medium with kanamycin sulfate at 30 µg ml−1 working concentration to start a new overnight culture. The new overnight culture was then treated with LL37 again following the same protocol as stated above.
4.3. Mathematical model and parameter estimation using flow cytometry data (M3)
We constructed a deterministic model using a system of ordinary differential equations (ODEs) to explore the population dynamics of single bacterial strain under LL37 treatment (equation (4.1)),
| 4.1 |
Here, A represents bacteria that were not affected by LL37 (growing population); Abind represents bacteria that had LL37 bound to their perimeters (binding population); Aabsorb represents bacteria that had been permeabilized and were absorbing free LL37 molecules (absorbing population). APfree is free LL37 molecules in the medium. The five reaction rate constants were kg, k1f, k1r, k2f with a unit of [min]−1, and kab with a unit of [µg/ml][min]−1[CFU/nl]−1. K1f and Kab were half-maximum constants with a unit of [µg/ml]. r was the proportional coefficient between Abind and bound LL37 molecules with a unit of [µg/ml][CFU/nl]−1. We set r = 0.05, which corresponded to roughly 6.7 × 106 LL37 molecules to one bacterium, based on the saturation level of the AP PMAP-23 on the bacterial membrane [33]. K1f was approximated to be 45 µg ml−1 (roughly equal to 10 µM) using the apparent dissociation constant of LL37 on a bio-membrane [51]. The growth rate of A was governed by kg and had a capacity of cap = 100 (corresponding to approximately 100-fold changes from the initial density to growth capacity from figure 1a). kg was estimated to be 0.022 for BL21PRO or 0.04 for MG1655 so that the culture took approximately 6–7 h for BL21PRO or approximately 3–4 h for MG1655 to reach its capacity, which was similar to our experimental results with pre-growth protocol 2 (electronic supplementary material, figure S19). Forward and reverse transitions between A and Abind were represented using and , respectively. Permeabilization of Abind was governed by k2f. APfree could be depleted from medium through either absorption or binding to bacterial membrane . The depletion of free LL37 molecules through binding was directly proportional to Abind. We assume that the transition from the ‘binding’ to the ‘absorbing’ population would release some LL37 molecules from the bacterial membrane owing to the loss of membrane integrity (a positive term in ). This assumption does not affect our conclusions (electronic supplementary material, figure S24).
To estimate kinetic parameters, we first extracted the ratio of each subpopulation from the flow cytometry data of BP-GFP and MG-GFP under Rh-LL37 treatment. Specifically, two thresholds for rhodamine intensity were set as described for samples collected at different time points. Ratios of Rh−, Rh+ and Rh++ to the entire population were recorded and used as experimental data for parameter estimation. We used Matlab function fmincon to obtain the first estimation based on our mathematical model (equation (4.1)) using three replicates of flow cytometry data (electronic supplementary material, figure S12a). The loss function for fmincon calculated the summation of the square difference between simulated and three replicates of experimental data. Next, parameters were further refined to fit both the flow cytometry and recovery time measurements (see electronic supplementary material, table S1, for simulation parameters).
4.4. Statistical test
All statistical tests were performed using at least six replicates. To compare the means of two groups, a one-tail t-test was used with p < 0.01. The Pearson correlation coefficient was calculated to estimate the linear correlation between two variables.
Supplementary Material
Acknowledgements
We appreciate the discussion of the manuscript with members of the Tan laboratory.
Data accessibility
All data supporting this study are provided in the results section and as the electronic supplementary material accompanying this paper.
Authors' contributions
F.W. and C.T. designed the experiments. F.W. performed the experiments and analysed the results. Both authors wrote the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work is supported by Society-in-Science: Branco-Weiss Fellowship to C.T. Cell sorting is supported by the National Institutes of Health under award no. S10OD018223.
References
- 1.Cole JN, Nizet V. 2016. Bacterial evasion of host antimicrobial peptide defenses. Microbiol Spectr. 4 ( 10.1128/microbiolspec.VMBF-0006-2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nizet V, et al. 2001. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 414, 454–457. ( 10.1038/35106587) [DOI] [PubMed] [Google Scholar]
- 3.Hancock RE, Sahl HG. 2006. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24, 1551–1557. ( 10.1038/nbt1267) [DOI] [PubMed] [Google Scholar]
- 4.Gordon YJ, Romanowski EG, McDermott AM. 2005. A review of antimicrobial peptides and their therapeutic potential as anti-infective drugs. Curr. Eye Res. 30, 505–515. ( 10.1080/02713680590968637) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teixeira V, Feio MJ, Bastos M. 2012. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog. Lipid Res. 51, 149–177. ( 10.1016/j.plipres.2011.12.005) [DOI] [PubMed] [Google Scholar]
- 6.Nguyen LT, Haney EF, Vogel HJ. 2011. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 29, 464–472. ( 10.1016/j.tibtech.2011.05.001) [DOI] [PubMed] [Google Scholar]
- 7.Podda E, Benincasa M, Pacor S, Micali F, Mattiuzzo M, Gennaro R, Scocchi M. 2006. Dual mode of action of Bac7, a proline-rich antibacterial peptide. Biochim. Biophys. Acta Gen. Subj. 1760, 1732–1740. ( 10.1016/j.bbagen.2006.09.006) [DOI] [PubMed] [Google Scholar]
- 8.Hsu C-H, et al. 2005. Structural and DNA-binding studies on the bovine antimicrobial peptide, indolicidin: evidence for multiple conformations involved in binding to membranes and DNA. Nucleic Acids Res. 33, 4053–4064. ( 10.1093/nar/gki725) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kragol G, Lovas S, Varadi G, Condie BA, Hoffmann R, Otvos L. 2001. The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry 40, 3016–3026. ( 10.1021/bi002656a) [DOI] [PubMed] [Google Scholar]
- 10.Otvos L, Snyder C, Condie B, Bulet P, Wade JD. 2005. Chimeric antimicrobial peptides exhibit multiple modes of action. Int. J. Pept. Res. Ther. 11, 29–42. ( 10.1007/s10989-004-1719-x) [DOI] [Google Scholar]
- 11.Poyart C, Pellegrini E, Marceau M, Baptista M, Jaubert F, Lamy MC, Trieu-Cuot P. 2003. Attenuated virulence of Streptococcus agalactiae deficient in D-alanyl-lipoteichoic acid is due to an increased susceptibility to defensins and phagocytic cells. Mol. Microbiol. 49, 1615–1625. ( 10.1046/j.1365-2958.2003.03655.x) [DOI] [PubMed] [Google Scholar]
- 12.Fabretti F, Theilacker C, Baldassarri L, Kaczynski Z, Kropec A, Holst O, Huebner J. 2006. Alanine esters of enterococcal lipoteichoic acid play a role in biofilm formation and resistance to antimicrobial peptides. Infect. Immun. 74, 4164–4171. ( 10.1128/IAI.00111-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovacs M, Halfmann A, Fedtke I, Heintz M, Peschel A, Vollmer W, Hakenbeck R, Bruckner R. 2006. A functional dlt operon, encoding proteins required for incorporation of d-alanine in teichoic acids in gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J. Bacteriol. 188, 5797–5805. ( 10.1128/JB.00336-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Starner TD, Swords WE, Apicella MA, McCray PB Jr. 2002. Susceptibility of nontypeable Haemophilus influenzae to human beta-defensins is influenced by lipooligosaccharide acylation. Infect. Immun. 70, 5287–5289. ( 10.1128/IAI.70.9.5287-5289.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, Miller SI. 1998. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95, 189–198. ( 10.1016/S0092-8674(00)81750-X) [DOI] [PubMed] [Google Scholar]
- 16.Cole JN, Pence MA, von Köckritz-Blickwede M, Hollands A, Gallo RL, Walker MJ, Nizet V. 2010. M protein and hyaluronic acid capsule are essential for in vivo selection of covRS mutations characteristic of invasive serotype M1T1 group A Streptococcus. MBio 1, e00191–10. ( 10.1128/mBio.00191-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spinosa MR, Progida C, Tala A, Cogli L, Alifano P, Bucci C. 2007. The Neisseria meningitidis capsule is important for intracellular survival in human cells. Infect. Immun. 75, 3594–3603. ( 10.1128/IAI.01945-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campos MA, Vargas MA, Regueiro V, Llompart CM, Alberti S, Bengoechea JA. 2004. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect. Immun. 72, 7107–7114. ( 10.1128/IAI.72.12.7107-7114.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meredith HR, Srimani JK, Lee AJ, Lopatkin AJ, You L. 2015. Collective antibiotic tolerance: mechanisms, dynamics and intervention. Nat. Chem. Biol. 11, 182 ( 10.1038/nchembio.1754) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vega NM, Gore J. 2014. Collective antibiotic resistance: mechanisms and implications. Curr. Opin. Microbiol. 21, 28–34. ( 10.1016/j.mib.2014.09.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chait R, Palmer AC, Yelin I, Kishony R. 2016. Pervasive selection for and against antibiotic resistance in inhomogeneous multistress environments. Nat. Commun. 7, 10333 ( 10.1038/ncomms10333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hol FJ, Hubert B, Dekker C, Keymer JE. 2016. Density-dependent adaptive resistance allows swimming bacteria to colonize an antibiotic gradient. ISME J. 10, 30 ( 10.1038/ismej.2015.107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yurtsev EA, Chao HX, Datta MS, Artemova T, Gore J. 2013. Bacterial cheating drives the population dynamics of cooperative antibiotic resistance plasmids. Mol. Syst. Biol. 9, 683 ( 10.1038/msb.2013.39) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vega NM, Allison KR, Samuels AN, Klempner MS, Collins JJ. 2013. Salmonella typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc. Natl Acad. Sci. USA. 110, 14420–14425. ( 10.1073/pnas.1308085110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidtchen A, Frick IM, Andersson E, Tapper H, Bjorck L. 2002. Proteinases of common pathogenic bacteria degrade and inactivate the antibacterial peptide LL-37. Mol. Microbiol. 46, 157–168. ( 10.1046/j.1365-2958.2002.03146.x) [DOI] [PubMed] [Google Scholar]
- 26.Johansson L, et al. 2008. Cathelicidin LL-37 in severe Streptococcus pyogenes soft tissue infections in humans. Infect. Immun. 76, 3399–3404. ( 10.1128/IAI.01392-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sieprawska-Lupa M, et al. 2004. Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob. Agents Chemother. 48, 4673–4679. ( 10.1128/AAC.48.12.4673-4679.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frick I-M, Åkesson P, Rasmussen M, Schmidtchen A, Björck L. 2003. SIC, a secreted protein of Streptococcus pyogenes that inactivates antibacterial peptides. J. Biol. Chem. 278, 16 561–16 566. ( 10.1074/jbc.M301995200) [DOI] [PubMed] [Google Scholar]
- 29.Weiner DJ, Bucki R, Janmey PA. 2003. The antimicrobial activity of the cathelicidin LL37 is inhibited by F-actin bundles and restored by gelsolin. Am. J. Respir. Cell Mol. Biol. 28, 738–745. ( 10.1165/rcmb.2002-0191OC) [DOI] [PubMed] [Google Scholar]
- 30.Bucki R, Byfield FJ, Janmey PA. 2007. Release of the antimicrobial peptide LL-37 from DNA/F-actin bundles in cystic fibrosis sputum. Eur. Respir. J. 29, 624–632. ( 10.1183/09031936.00080806) [DOI] [PubMed] [Google Scholar]
- 31.Llobet E, Tomas JM, Bengoechea JA. 2008. Capsule polysaccharide is a bacterial decoy for antimicrobial peptides. Microbiology 154, 3877–3886. ( 10.1099/mic.0.2008/022301-0) [DOI] [PubMed] [Google Scholar]
- 32.Starr CG, He J, Wimley WC. 2016. Host cell interactions are a significant barrier to the clinical utility of peptide antibiotics. ACS Chem. Biol. 11, 3391–3399. ( 10.1021/acschembio.6b00843) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roversi D, Luca V, Aureli S, Park Y, Mangoni ML, Stella L. 2014. How many antimicrobial peptide molecules kill a bacterium? The case of PMAP-23. ACS Chem. Biol. 9, 2003–2007. ( 10.1021/cb500426r) [DOI] [PubMed] [Google Scholar]
- 34.Jepson AK, Schwarz-Linek J, Ryan L, Ryadnov MG, Poon WCK. 2016. What is the ‘Minimum Inhibitory Concentration’ (MIC) of Pexiganan acting on Escherichia coli?—a cautionary case study. In Biophysics of infection (ed. Leake MC.), pp. 33–48. Cham, Switzerland: Springer International Publishing. [DOI] [PubMed] [Google Scholar]
- 35.Wimley WC. 2010. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem. Biol. 5, 905–917. ( 10.1021/cb1001558) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sochacki KA, Barns KJ, Bucki R, Weisshaar JC. 2011. Real-time attack on single Escherichia coli cells by the human antimicrobial peptide LL-37. Proc. Natl Acad. Sci. USA 108, E77–E81. ( 10.1073/pnas.1101130108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fantner GE, Barbero RJ, Gray DS, Belcher AM. 2010. Kinetics of antimicrobial peptide activity measured on individual bacterial cells using high-speed atomic force microscopy. Nat. Nanotechnol. 5, 280–285. ( 10.1038/nnano.2010.29) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeh P, Tschumi AI, Kishony R. 2006. Functional classification of drugs by properties of their pairwise interactions. Nat. Genet. 38, 489–494. ( 10.1038/ng1755) [DOI] [PubMed] [Google Scholar]
- 39.Kishony R, Leibler S. 2003. Environmental stresses can alleviate the average deleterious effect of mutations. J. Biol. 2, 14 ( 10.1186/1475-4924-2-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bjarnason J, Southward CM, Surette MG. 2003. Genomic profiling of iron-responsive genes in Salmonella enterica serovar typhimurium by high-throughput screening of a random promoter library. J. Bacteriol. 185, 4973–4982. ( 10.1128/JB.185.16.4973-4982.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan C, et al. 2012. The inoculum effect and band-pass bacterial response to periodic antibiotic treatment. Mol. Syst. Biol. 8, 617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dwyer DJ, Camacho DM, Kohanski MA, Callura JM, Collins JJ. 2012. Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis. Mol. Cell 46, 561–572. ( 10.1016/j.molcel.2012.04.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davey HM, Hexley P. 2011. Red but not dead? Membranes of stressed Saccharomyces cerevisiae are permeable to propidium iodide. Environ. Microbiol. 13, 163–171. ( 10.1111/j.1462-2920.2010.02317.x) [DOI] [PubMed] [Google Scholar]
- 44.Müller A, et al. 2016. Daptomycin inhibits cell envelope synthesis by interfering with fluid membrane microdomains. Proc. Natl Acad. Sci. USA 113, E7077–E7086. ( 10.1073/pnas.1611173113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pader V, Hakim S, Painter KL, Wigneshweraraj S, Clarke TB, Edwards AM. 2017. Staphylococcus aureus inactivates daptomycin by releasing membrane phospholipids. Nat. Microbiol. 2, 16194 ( 10.1038/nmicrobiol.2016.194) [DOI] [PubMed] [Google Scholar]
- 46.Lowder M, Unge A, Maraha N, Jansson JK, Swiggett J, Oliver J. 2000. Effect of starvation and the viable-but-nonculturable state on green fluorescent protein (GFP) fluorescence in GFP-tagged Pseudomonas fluorescens A506. Appl. Environ. Microbiol. 66, 3160–3165. ( 10.1128/AEM.66.8.3160-3165.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Islam MM, Chakraborty M, Pandya P, Al Masum A, Gupta N, Mukhopadhyay S. 2013. Binding of DNA with Rhodamine B: spectroscopic and molecular modeling studies. Dyes Pigm. 99, 412–422. ( 10.1016/j.dyepig.2013.05.028) [DOI] [Google Scholar]
- 48.Matsuzaki K, Sugishita K, Fujii N, Miyajima K. 1995. Molecular basis for membrane selectivity of an antimicrobial peptide, magainin 2. Biochemistry 34, 3423–3429. ( 10.1021/bi00010a034) [DOI] [PubMed] [Google Scholar]
- 49.Henzler WKA, Lee D-K, Ramamoorthy A. 2003. Mechanism of lipid bilayer disruption by the human antimicrobial peptide, LL-37. Biochemistry 42, 6545–6558. ( 10.1021/bi0273563) [DOI] [PubMed] [Google Scholar]
- 50.Araña MDJ, et al. 2003. Inhibition of LPS-responses by synthetic peptides derived from LBP associates with the ability of the peptides to block LBP-LPS interaction. J. Endotoxin Res. 9, 281–291. [DOI] [PubMed] [Google Scholar]
- 51.Sood R, Domanov Y, Pietiainen M, Kontinen VP, Kinnunen PK. 2008. Binding of LL-37 to model biomembranes: insight into target vs host cell recognition. Biochim. Biophys. Acta 1778, 983–996. ( 10.1016/j.bbamem.2007.11.016) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting this study are provided in the results section and as the electronic supplementary material accompanying this paper.