Abstract
Community-associated acquisition of extended-spectrum beta-lactamase- (ESBL) and carbapenemase-producing Enterobacteriaceae has significantly increased in recent years, necessitating greater inquiry into potential exposure routes, including food and water sources. In high-income countries, drinking water is often neglected as a possible source of community exposure to antibiotic-resistant organisms. We screened coliform-positive tap water samples (n = 483) from public and private water systems in six states of the United States for blaCTX-M, blaSHV, blaTEM, blaKPC, blaNDM, and blaOXA-48-type genes by multiplex PCR. Positive samples were subcultured to isolate organisms harboring ESBL or carbapenemase genes. Thirty-one samples (6.4%) were positive for blaCTX-M, ESBL-type blaSHV or blaTEM, or blaOXA-48-type carbapenemase genes, including at least one positive sample from each state. ESBL and blaOXA-48-type Enterobacteriaceae isolates included E. coli, Kluyvera, Providencia, Klebsiella, and Citrobacter species. The blaOXA-48-type genes were also found in non-fermenting Gram-negative species, including Shewanella, Pseudomonas and Acinetobacter. Multiple isolates were phenotypically non-susceptible to third-generation cephalosporin or carbapenem antibiotics. These findings suggest that tap water in high income countries could serve as an important source of community exposure to ESBL and carbapenemase genes, and that these genes may be disseminated by non-Enterobacteriaceae that are not detected as part of standard microbiological water quality testing.
Introduction
Antibiotic-resistant infections are responsible for an estimated 2 million illnesses and 23,000 deaths in the United States each year1. The rising prevalence of multidrug-resistant bacteria is especially alarming, as infections with these organisms have led to increasing use of broad spectrum antibiotics such as third and fourth generation cephalosporin and carbapenem antibiotics1,2. Enzymes such as extended-spectrum beta-lactamases (ESBLs) and carbapenemases can render these antibiotics ineffective. Enterobacteriaceae harboring these enzymes are ranked among the most urgent antibiotic resistance threats according to the U.S. Centers for Disease Control and Prevention and the World Health Organization1,3. Additionally, genes encoding these enzymes are often found on mobile genetic elements that can be transferred horizontally to other bacterial species4.
ESBL- and carbapenemase-producing bacteria are commonly associated with healthcare contact1; however, community-associated infections have significantly increased in recent years5. Between 2009 and 2011, the occurrence of ESBL-producing bacteria in community-associated infections increased from 3.1% to 12.6%, while the occurrence in hospital-associated infections remained the same5. It is estimated that as many as two-thirds of all ESBL-producing Enterobacteriaceae infections are community-associated5,6. In 2013, the U.S. Centers for Disease Control warned that spread of carbapenem-resistant Enterobacteriaceae (CRE) into the community could reasonably be expected, as experienced with ESBLs1. A 2017 review on CRE in the community noted that the prevalence of CRE among U.S. community-associated study samples ranged from 5.6 to 10.8%7.
ESBL and carbapenemase genes are frequently found in Enterobacteriaceae such as Klebsiella pneumoniae, Escherichia coli, Enterobacter cloacae, and Citrobacter species4, but can also be found in non-fermenting Gram-negative species8. These bacteria are most commonly spread via fecal-oral transmission routes, including direct transmission (e.g. via hands) and indirect transmission (e.g. via the environment). Enterobacteriaceae, including ESBL- and carbapenemase-producing strains, have been reported in a number of environmental compartments including food, animals, surface waters, and drinking water9–11. Studies reporting carbapenemase or ESBL genes in drinking water have largely been performed in low-income countries9,12. In high income countries, reports of ESBL- and carbapenemase-producing bacteria in drinking water have been limited to single-cases or intrinsic genes in nonpathogenic environmental bacterial species10,13, but comprehensive studies are lacking.
In clinical infections, ESBL and carbapenemase genes are most frequently found in Enterobacteriaceae species, but these organisms are typically uncommon in chlorinated public drinking water supplies in high income countries. To potentially increase detection of ESBL and carbapenemase genes in U.S. drinking water, we targeted water samples testing positive for Enterobacteriaceae (coliform bacteria). The objectives of our study were (1) to determine whether ESBL- and carbapenemase-producing genes are present in U.S. drinking water samples that have tested positive for E. coli or total coliform bacteria, (2) to describe the sample and water system characteristics associated with samples testing positive for ESBL or carbapenemase genes, and (3) to determine if the ESBL and carbapenemase genes are present in viable bacteria isolated from the coliform-positive water samples.
Methods
Water sample collection and coliform testing
Between July 2015 and November 2016, regulatory and investigational drinking water samples testing positive for E. coli or total coliform bacteria (Enterobacteriaceae) were acquired from multiple local and state public health laboratories that perform regulatory water quality testing for water utilities in their state or county. Participating laboratories included the state public health and environmental laboratories in Wisconsin (99 samples), New York (22 samples), Pennsylvania (69 samples), Illinois (125 total samples from three laboratories), Arkansas (64 samples), and Utah. Additionally, one water utility and one county health laboratory in Utah supplied coliform-positive samples (total of 104 Utah samples). Sample acquisition was opportunistic, and may not have consistently included all coliform-positive samples from each site. Private well samples were only included in the study if positive for E. coli, to avoid testing a high proportion of private well samples, which frequently test positive for coliform bacteria. Drinking water samples collected for regulatory water quality testing are required to be 100 milliliters in volume and must be preserved with sodium thiosulfate and tested at the laboratory within 30 hours of collection, preferably held at a temperature between 0 and 10 degrees Celsius during transport14.
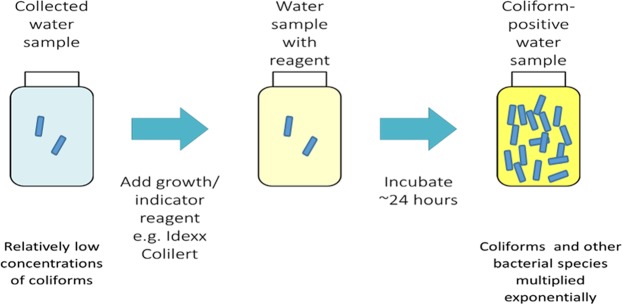
Upon arrival to each public health laboratory location, drinking water samples were tested using a conventional enzyme-substrate method that indicates the presence of E. coli and total coliform bacteria (Fig. 1)15. The substrate reagent includes a nutrient medium that creates culture conditions in the water sample and promotes bacterial growth and has chromogenic and fluorogenic indicators to detect total coliforms and E. coli, respectively. Samples were tested in either a presence/absence format using the original 100-mL collection vessel, or in a quantitative format by dispensing the 100 mL sample into a multi-well tray (Quanti-Tray, IDEXX, Westbrook, ME) for quantification by the most-probable-number technique. When E. coli and/or coliform bacteria were detected in cultured presence/absence samples, 1 mL of the positive enriched sample was placed in a sterile cryovial with 1 mL 40% glycerol (final concentration 20% glycerol). If a sealed Quanti-Tray sample was positive, the back of the tray was disinfected and a sterile syringe was used to extract the culture liquid from positive wells. When multiple wells indicated coliforms, a composite 1 mL of enriched sample was produced by extracting liquid from several wells, and the composite was mixed with glycerol as described above. All samples were cryogenically frozen, and samples from locations other than Utah were shipped on dry ice overnight to the research laboratory in Salt Lake City, Utah.
Figure 1.
Water sample coliform screening process.
Resistance gene detection and PCR amplicon sequencing
Upon arrival to the research laboratory, preserved samples were screened for the three ESBL genes most common in the U.S. (blaSHV, blaTEM, and blaCTX) by multiplex PCR, as previously described16. Samples were also screened for the three most common carbapenemase genes (blaOXA-48-type, blaNDM, and blaKPC) by a separate multiplex PCR17. Primer sequences are available in Supplemental Table 1. Four microliters of the preserved sample culture was directly used as a template for the PCR in a total reaction volume of 25 µL. Positive controls included American Type Culture Collection isolate BAA-2146 for blaSHV, blaTEM, blaCTX-M, and blaNDM detection, a clinical KPC-producing K. pneumoniae for blaKPC detection, and an OXA-48-producing Shewanella for blaOXA-48 detection. PCR products were run on a 1% agarose gel, and amplicon from samples putatively positive for any of the genes of interest were sequenced by the Sanger method using the forward and reverse primers used for PCR screening. If samples were positive for multiple genes, PCR was repeated with individual primer pairs in separate reactions, followed by amplicon sequencing. Sequences were searched against the Comprehensive Antibiotic Resistance Database (CARD) and the GenBank (BLASTn) database to confirm the presence of one of the six target ESBL or carbapenemase genes. SHV and TEM variants identified by these databases were compared against the Lahey Clinic designation of ESBL-type blaSHV and blaTEM genes18. Lack of specificity of the multiplex CTX-M primer can result in amplification of several other beta-lactamase genes, such as blaOXY16; as a result, chromatograms from all CTX-M –positive amplicon sequences were closely inspected to see if multiple genes may have been present in the sample. In the case of mixed chromatograms, samples were tested as described above using a CTX-M group-specific multiplex PCR19 (Supplemental Table 1). Samples confirmed as positive for any of the target genes by Sanger sequencing were subsequently tested for the specific gene using primers that amplified larger segments of the genes to better identify the specific alleles20–23.
Bacterial isolation and identification
Bacteria carrying the target resistance genes were isolated through selective and non-selective culture of the preserved samples as previously described24,25. Samples confirmed for the presence of blaTEM, blaSHV, or blaCTX-M genes were plated on CHROMagar OrientationTM agar plates (DRG International, Springfield, NJ) with and without a proprietary ESBL supplement. Approximately 50 µL of the preserved sample was spread onto each plate, and cefotaxime (30 µg), ceftazidime (30 µg), and aztreonam (30 µg) disks were placed on the non-selective CHROMagar plates. After overnight incubation, colonies on the ESBL CHROMagar plate and colonies falling within a distinct zone forming around the disks on the non-selective plate were tested by PCR for blaTEM, blaSHV, or blaCTX-M genes using the primer set from the ESBL multiplex PCR.
For samples confirmed as having a blaOXA-48-type gene, 50 µL of preserved sample was spread onto an mSuperCARBATM plate (DRG International, Springfield, NJ) and a non-selective CHROMagar OrientationTM plate (DRG International, Springfield, NJ) with a temocillin disk (Rosco, Taastrup, Denmark). In cases where the blaOXA-48-type producer could not be isolated by direct plating, 20 µL of the original culture sample was added to a Luria broth with 0.5, 1, 2, and 4 µg/mL imipenem. Broths testing positive by PCR were plated as described above and to sheep’s blood agar with 0.125 µg/mL ertapenem. Colonies growing on the mSuperCARBATM plate and colonies growing within the distinct zone that formed around the temocillin disk were tested for blaOXA-48-type genes using the OXA-48 primer set from the carbapenemase gene multiplex PCR and confirmed by Sanger sequencing of the amplicon. Bacterial species identification was performed by MALDI-TOF (Biotyper, Bruker, Bellerica, MA) using the full spectral library which is composed of spectra representing roughly 2750 species of microorganisms from approximately 470 genera26.
Susceptibility testing
Minimum inhibitory concentrations of isolates were determined using Thermo ScientificTM SensititreTM Extended Spectrum Beta-lactamase plates (Trek Diagnostic Systems, Inc., Independence, OH) according to manufacturer’s directions. For isolates in which the genes were lost upon subculture, fresh colonies from the original sample were used in preparation of the antimicrobial susceptibility testing inoculum, if sufficient growth was present.
Supplementary sample data
Laboratories supplying coliform-positive samples also provided limited data on sample characteristics such as water source (e.g. groundwater, [treated] surface water, or blended), sample type (regulatory or investigational), and system type (e.g. community public water system [CPWS], non-community public water system [NCPWS], or private well, as defined by the U.S. Environmental Protection Agency)27. Sample results were described with respect to these characteristics. Statistical analyses to test specific hypotheses or make statements of inference would have been inappropriate because the sample collection was opportunistic.
Results
Sample characteristics
A total of 483 non-duplicate samples, representing 361 public and private water systems, were collected. Of the 483 samples, 27% were from private water systems, 31% were from CPWSs, and 42% were from NCPWSs. Spring water sources comprised 5% of the samples, while surface and ground water sources made up 9% and 78%, respectively. Approximately 6% of the samples were blended, meaning that they were from systems where both groundwater and treated surface water were supplied to consumers during periods of high demand.
PCR detection of ESBL and carbapenemase genes and amplicon sequencing
The 483 samples were screened for blaTEM, blaSHV, blaCTX-M, blaKPC, blaNDM, and blaOXA-48-type genes. Sixty-four samples appeared to be positive for blaCTX-M by initial PCR and agarose gel screening; however, comparison of PCR amplicon sequences to the GenBank database revealed that the majority of the putative blaCTX-M positives were attributed to extended-spectrum beta-lactamase genes that are typically chromosomally-encoded in various Enterobacteriaceae species, including blaRAHN (Rahnella aquatilis), blaFONA (Serratia fonticola), blaOXY (Klebsiella oxytoca), blaSMO (strain RUS)28, and Citrobacter amalonaticus class-A beta lactamase. Because these were not target genes in the study, we did not pursue any further analysis of these samples.
Thirty-one samples (6.4%) from twenty-six (7.2%) of the water systems were positive for the target blaCTX-M, blaOXA-48-type, or ESBL-type blaTEM or blaSHV genes. Thirteen of the samples (2.7%) from twelve different water systems (3.3%) were confirmed to have blaCTX-M genes based on amplicon sequences. Thirteen (2.7%) and ten (2.1%) samples were positive for blaSHV and blaTEM genes, respectively. One SHV-positive amplicon sequence most closely matched an ESBL-type blaSHV (blaSHV-38) and one TEM-positive amplicon sequence closely matched both ESBL- and non-ESBL-type blaTEM genes. The carbapenemase multiplex PCR screen did not reveal any NDM- or KPC-positive samples; however, blaOXA-48-type genes were detected in 19 samples (3.9%) from 15 (4.2%) of the water systems. The samples with blaOXA-48-type genes were from four states and represented between 2–7% of samples from those states. Three of the samples that tested positive for blaOXA-48-type genes were also positive for blaCTX-M genes and were each from different states. At least one ESBL or carbapenemase gene was detected in all six states comprising between 1% and 15% of the coliform-positive samples tested from each state. Of the 185 water systems with repeat samples, six had samples that tested positive for at least one ESBL or carbapenemase gene; half of these water systems had only one positive sample and one water system had all three samples positive.
Isolation of ESBL- or carbapenemase-producing bacteria and antimicrobial susceptibility testing
Bacteria carrying target ESBL or carbapenemase genes were isolated from 21 of the 31 positive samples (Table 1). The blaCTX-M gene was detected in Klebsiella oxytoca, Citrobacter freundii complex, Kluyvera ascorbata and Kluyvera georgiana, the latter two being the progenitor of blaCTX-M genes. The ESBL-type blaSHV-38 gene was present in Klebsiella pneumoniae and a blaTEM variant gene exhibiting an ESBL phenotype was present in E. coli. Species carrying blaOXA-48-type genes included E. coli, Providencia rettgeri, Acinetobacter baumannii complex, Pseudomonas putida, Pseudomonas koreensis, Pandoraea sputorum, Shewanella putrefaciens, and other Shewanella (n = 3) and Pseudomonas (n = 1) that could not be identified at the species level. The stability of ESBL and blaOXA-48-type genes were noted based on the number of subcultures where the resistance gene could still be detected in culture. The ESBL genes were very stable and were still detected after two or more subcultures. The blaOXA-48-type genes were typically lost after one subculture in non-Shewanella non-fermenting Gram-negative rods, and after two or more subcultures in Enterobacteriaceae, with or without selective pressure from imipenem concentrations ranging from 0.125 ug/mL to 4 ug/mL. Nucleic acid sequences of the PCR amplicon from isolates and from positive samples where an isolate could not be recovered are available in GenBank (Table 1). Antimicrobial susceptibility testing results for selected isolates are presented in Table 2.
Table 1.
Characteristics of sources of samples positive for blaCTX-M, blaOXA-48-type, and putative blaSHV or blaTEM genes; aGene variant or cluster with greatest homology in GenBank; bCPWS: Community public water system, NCPWS: Non-community public water system; cSamples collected from the same water systems: A1 and A2; D1, D2, and D3; E2, E5, and E6; d100% nucleotide homology to uncultured bacterium clones; eGene sequence is similarly homologous with both ESBL and non-ESBL blaTEM genes, but exhibits an ESBL phenotype fBlended water is a composite of ground water and treated surface water.
| Sample ID | Gene variant or group with highest DNA sequence similaritya | Gene homology with reference sequence | Protein homology with reference sequence | GenBank Nucleotide Reference (GenBank Protein reference) | Organism | E. coli present/absent in sample | System typeb | Water source | GenBank Accession Number |
|---|---|---|---|---|---|---|---|---|---|
| A1c | CTX-M-205 | 79% | 86% | MG028655 (ATJ25945.1) | Citrobacter freundii- complex | Absent | CPWS | Ground | MG560059 |
| A2c | CTX-M-205 | 80% | 88% | MG028655 | Citrobacter freundii- complex | Present | CPWS | Ground | MG560060 |
| A3a | CTX-M-40 or CTX-M-63 | 84% | 94% | NG_048991 NG_049014 | Kluyvera georgiana | Present | Private | Ground | MG560061 |
| A3b | OXA-181 | 100% | 100% | CP023897 | Acinetobacter baumannii complex | Present | Private | Ground | MG560077 |
| A4 | CTX-M-40 or CTX-M-63 | 98% | 99% | NG_048991 NG_049014 | Kluyvera georgiana | Absent | CPWS | Ground | MG560062 |
| A5 | CTX-M-40 or CTX-M-63 | 84% | 94% | NG_048991 NG_049014 | Kluyvera georgiana | Absent | CPWS | Ground | MG560063 |
| A6 | CTX-M-9 family | 86%d | 95% | NG_049028.1 | Not Isolated | Absent | CPWS | Surface | MG560055 |
| B1 | CTX-M-10 or CTX-M-34 | 100% | 100% | NG_048898 NG_048984 | Kluyvera species | Present | Private | Ground | MG560064 |
| B2 | CTX-M-40 or CTX-M-63 | 96% | 95% | NG_048991 NG_049014 | Not Isolated | Present | Private | Unknown | MG560056 |
| B3a | CTX-M-12 | 100% | 100% | DQ821704 | Not Isolated | Present | Private | Ground | MG560057 |
| B3b | OXA-48b | 100% | 100% | KU820807 | Not Isolated | Present | Private | Ground | MG560068 |
| B4 | OXA-48b | 100% | 100% | KU820807 | E. coli | Present | Private | Ground | MG560078 |
| B5 | OXA-181 | 85%d | 93% | KJ620504 | E. coli | Present | Private | Ground | MG560079 |
| B6 | OXA-48b | 100% | 100% | KU820804 | Shewanella species | Present | Private | Ground | MG560080 |
| B7 | CTX-M-3 | 100% | 100% | KU200455 | Klebsiella oxytoca | Present | Private | Ground | MG560065 |
| B8 | OXA-181 | 85% | 94% | KJ620504 | Not Isolated | Present | Private | Ground | MG56006 |
| B9 | OXA-48b | 100% | 100% | KU820804 | Shewanella species | Present | Private | Ground | MG560081 |
| B10 | OXA-252 | 100% | 100% | CP022089 | Not Isolated | Present | Private | Ground | MG560070 |
| B11a | OXA-48b | 99% | 99% | KC902850 | Shewanella putrefaciens; | Absent | NCPWS | Ground | MG560082 |
| B11b | OXA-48b | 99% | 99% | KC902850 | Pandoraea sputorum | Absent | NCPWS | Ground | MG560083 |
| B12 | TEM-1 variante | 99% | 99% | KU664682.1 | E. coli | Present | Private | Ground | MG560088 |
| B13 | OXA-48b or OXA-252 | 100% | 100% | KU820804 NG_050608.1 | Shewanella species | Present | Private | Ground | MG560084 |
| C1 | CTX-M-3 | 99% | 99% | Y10278 | Not Isolated | Absent | CPWS | Surface | MG560058 |
| C2 | CTX-M-2-like | 89% | 96% | KX377894 | Kluyvera ascorbata | Absent | CPWS | Ground | MG560066 |
| D1c | OXA-48b | 100% | 100% | KU820807 | Providencia rettgeri | Absent | NCPWS | Ground | MG560085 |
| D2c | OXA-252 | 100% | 100% | NG_050608 | Not Isolated | Present | NCPWS | Ground | MG560071 |
| D3c | OXA-199 | 99% | 99% | NG_049495 | Not Isolated | Present | NCPWS | Ground | MG560072 |
| E1a | CTX-M-36 | 99% | 100% | NG_048986 | Klebsiella oxytoca | Absent | CPWS | Blendf | MG560067 |
| E1b | OXA-48b | 100% | 100% | KU820807 | Not Isolated | Absent | CPWS | Blendf | MG560073 |
| E2c | OXA-48b | 100% | 100% | KU820807 | Not Isolated | Absent | CPWS | Blendf | MG560074 |
| E3 | OXA-252 | 100% | 100% | NG_050608 | Pseudomonas species | Absent | CPWS | Blendf | MG560075 |
| E4 | OXA-48b | 100% | 100% | KU820802 | Not Isolated | Absent | CPWS | Blendf | MG560076 |
| E5c | OXA-48b | 100% | 100% | KU820807 | Pseudomonas koreensis | Absent | CPWS | Blendf | MG560086 |
| E6c | OXA-252 | 100% | 100% | KU820800 | Pseudomonas putida | Absent | CPWS | Blendf | MG560087 |
| F1 | SHV-38 | 100% | 100% | NG_050077 | Klebsiella pneumoniae | Present | NCPWS | Ground | MG560089 |
Table 2.
Minimum inhibitory concentrations (MIC) (µg/mL) of isolates to selected antibiotics by broth microdilution.
| Sample ID | Organism (gene type) | ESBL pheno-typea | TAZ | FOT | FAZ | FEP | FOX | CEP | POD | AXO | IMI | MEM | GEN | AMP | CIP | P/T4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | C. freundii-complex (CTX) | No | ≤0.25 | ≤0.25 | 16* | ≤1 | 8* | >16* | 1 | ≤1 | 1 | ≤1 | ≤4 | ≤8* | ≤1 | 32/4 |
| A2 | C. freundii-complex (CTX) | No | ≤0.25 | 0.5 | >16* | ≤1 | 8* | >16* | 1 | ≤1 | ≤0.5 | ≤1 | ≤4 | 16* | ≤1 | ≤4/4 |
| A3a | K. georgianab (CTX) | No | ≤0.25 | ≤0.25 | >16 | ≤1 | ≤4 | >16 | 2 | ≤1 | ≤0.5 | ≤1 | ≤4 | >16 | ≤1 | ≤4/4 |
| A3b | A. baumannii complex (OXA-48) | Yes | 8 | 8 | >16 | 2 | 64 | >16 | 8 | 8 | ≤0.5 | ≤1 | ≤4 | 16* | ≤1 | 64/4 |
| A4 | K. georgianab (CTX) | No | ≤0.25 | ≤0.25 | >16 | ≤1 | ≤4 | >16 | 1 | ≤1 | 4 | ≤1 | ≤4 | >16 | ≤1 | 64/4 |
| A5 | K. georgianab (CTX) | No | 0.5 | 0.5 | >16 | ≤1 | ≤4 | >16 | 2 | 2 | ≤0.5 | ≤1 | ≤4 | >16 | ≤1 | ≤4/4 |
| B1 | Kluyvera speciesb (CTX) | Yes | 32 | 2 | >16 | 8 | 8 | >16 | 16 | 4 | ≤0.5 | ≤1 | ≤4 | >16 | ≤1 | ≤4/4 |
| B4 | E. coli (OXA-48) | Yes | 8 | 8 | 16 | 2 | 64 | >16 | 0.5 | ≤1 | ≤0.5 | ≤1 | 8 | ≤8 | ≤1 | >64/4 |
| B5 | E. coli (OXA-48) | Yes | 8 | 4 | ≤8 | >16 | ≤4 | >16 | 1 | ≤1 | 1 | ≤1 | ≤4 | ≤8 | ≤1 | >64/4 |
| B6 | Shewanella speciesb (OXA-48) | No | ≤0.25 | 2 | >16 | ≤1 | ≤4 | >16 | ≤0.25 | ≤1 | ≤0.5 | >8 | ≤4 | 16 | ≤1 | ≤4/4 |
| B7 | K. oxytoca (CTX) | No | 1 | ≤0.25 | ≤8 | ≤1 | ≤4 | ≤8 | ≤0.25 | ≤1 | ≤0.5 | ≤1 | ≤4 | >16* | ≤1 | ≤4/4 |
| B11 | S. putrefaciensb (OXA-48) | Yes | 0.5 | 2 | >16 | ≤1 | 8 | ≤8 | ≤0.25 | 64 | ≤0.5 | 4 | ≤4 | 16 | ≤1 | ≤4/4 |
| B12 | E. coli (TEM) | Yes | 8 | 8 | ≤8 | >16 | 8 | >16 | 16 | 4 | ≤0.5 | ≤1 | ≤4 | >16 | ≤1 | >64/4 |
| B13 | Shewanella speciesb (OXA-48) | No | ≤0.25 | ≤0.25 | >16 | ≤1 | ≤4 | >16 | ≤1 | ≤1 | ≤0.5 | 2 | 16 | ≤8 | ≤1 | ≤4/4 |
| C2 | K. ascorbatab (CTX) | No | ≤0.25 | ≤0.25 | >16 | ≤1 | ≤4 | >16 | 1 | ≤1 | ≤0.5 | ≤1 | ≤4 | ≤8 | ≤1 | ≤4/4 |
| D1 | P. rettgeri (OXA-48) | No | 0.5 | ≤0.25 | >16* | 2 | 64 | >16* | ≤0.25 | ≤1 | ≤0.5 | ≤1 | ≤4 | ≤8* | ≤1 | ≤4/4 |
| E1a | K. oxytoca (CTX) | No | 0.5 | 1 | ≤8 | ≤1 | ≤4 | >16 | 4 | 16 | ≤0.5 | ≤1 | ≤4 | ≤8* | ≤1 | ≤4/4 |
| E3 | Pseudomonas speciesb (OXA) | No | 1 | 2 | >16 | ≤1 | >64 | >16 | 2 | 8 | ≤0.5 | ≤1 | ≤4 | >16 | ≤1 | ≤4/4 |
| F1 | K. pneumoniae (SHV) | Yes | 64 | 4 | ≤8 | ≤1 | ≤4 | >16 | 4 | ≤1 | ≤0.5 | ≤1 | 16 | >16* | ≤1 | >64/4 |
TAZ: ceftazidime, FOT: cefotaxime, FAZ: cefazolin, FEP: cefepime, FOX: cefoxitin, CEP: cephalothin, POD: cefpodoxime, AXO: ceftriaxone, IMI: imipenem, MEM: meropenem, GEN: gentamicin, AMP: ampicillin, CIP: ciprofloxacin, P/T4: piperacillin/tazobactam. Cells marked with a * indicate antibiotics to which the organism is considered intrinsically resistant according to the 2019 Clinical and Laboratory Standards Institute (CLSI) M100 ED29:2019 document34; aAn ESBL phenotype is defined as an bacterial phenotype that exhibits a ≥3 two-fold concentration decrease in a MIC for ceftazidime or cefotaxime tested in combination with clavulanate vs the MIC of ceftazidime or cefotaxime when tested alone. Although the result is listed for all organisms in Table 2, this test is intended for Klebsiella pneumoniae, Klebsiella oxytoca, E. coli, and Proteus mirabilis per CLSI m10034; bIndicates organisms for which an intrinsic resistance profile is not available in the CLSI m100 document.
Resistance gene presence and tap water source characteristics
The blaCTX-M, blaOXA-48-type, or ESBL-type blaTEM or blaSHV genes were found in 5.6% of samples from ground water sources and 7.0% of samples from treated surface water sources, and from 10.0% of private and 5.1% of public water systems: 8.7% of CPWS and 2.5% of NCPWS samples. Overall, 6.8% of the coliform-positive samples from public water systems also tested positive for E. coli; however, when considering those samples that tested positive for an ESBL or blaOXA-48-type gene, 22.2% were positive for E. coli.
Discussion
Community-associated antibiotic-resistant infections caused by ESBL- and carbapenemase-producing bacteria have increased significantly in the U.S. over the past decade. Many possible transmission routes have been studied; but in high-income countries, drinking water has not been adequately assessed as a potential source. We detected ESBL genes (blaCTX-M, blaTEM, or blaSHV) or blaOXA-48-type carbapenemase genes in more than 6% of the coliform-positive U.S. drinking water samples screened. Coliform-positive drinking water samples were targeted for testing because ESBL and carbapenemase genes are most commonly found in Enterobacteriaceae in clinical settings.
Non-E. coli coliforms, such as Klebsiella, Citrobacter, Enterobacter, and Serratia species are considered to be non-pathogenic by regulatory agencies and are primarily used as indicators of potential fecal contamination or water distribution system breaches. However, it is important to consider that these species are also some of the most common carriers of ESBL and carbapenemase genes, regardless of pathogenicity24. In 2015, the presence of coliform bacteria was reported in 1909 U.S. water systems serving over 10 million in total people, although audits and other compliance reports have found that underreporting of drinking water contaminants in public water systems is likely a widespread issue29. Coliform bacteria are even more common in private wells, which supply approximately 15% of the U.S. population with drinking water30.
This study utilized a convenience sample of coliform-positive water samples from state and local public health and environmental laboratories to increase the potential for finding ESBL- or carbapenemase-producing Enterobacteriaceae, not to estimate the overall frequency of occurrence of these genes in U.S. water supplies. The study design limits the conclusions that can be drawn regarding the overall prevalence of these genes in U.S. water systems and limits access to some information associated with individual samples, such as the type of water treatment used by the water systems. There is also a slight possibility that some coliform-positive samples resulted from accidental contamination during original sample collection; however, regulatory samples are typically collected by public water system personnel trained in proper sample collection technique.
The true prevalence of the target ESBL and carbapenemase genes in these samples may be underestimated due to testing limitations. A very small proportion of the preserved water sample culture was tested, potentially missing ESBL and carbapenemase genes present in low copy numbers in the samples. Additionally, DNA was not extracted from the preserved samples prior to PCR testing, and PCR inhibitors may have been present, affecting gene detection. We were also only able to determine presence or absence of the target resistance genes and could not assess the abundance in the original water sample due to the culture step in the coliform screening process.
ESBL and carbapenemase genes other than the target genes were also clearly present in these water samples. Some of these genes may have been carried by the bacterial isolates in addition to the target ESBL or carbapenemase genes that were detected, potentially contributing to any observed phenotype. The majority of these non-target genes are known to be chromosomally-located and carried by nonpathogenic bacterial species; however, some, such as blaOXY, can be plasmid-mediated, and may still be a cause for clinical concern31. The blaCTX-M and blaOXA-48-type genes also arise from relatively nonpathogenic bacteria that are commonly found in water sources (Kluyvera and Shewanella species, respectively), but have been widely disseminated to other bacterial species via mobile genetic elements. In this study, blaOXA-48-type genes in non-Shewanella species were lost in subsequent subcultures, a phenomenon also observed with non- fermenting Gram-negative rods carrying the blaNDM-1 carbapenemase gene isolated from New Delhi drinking water9.
Bacterial isolates were identified using the full spectral Bruker MALDI-TOF library used by diverse laboratories. Despite the broad organism coverage, it is possible that some environmental species may have limited representation in this system. In our study, all of the blaCTX-M carriers were identified as Enterobacteriaceae, with approximately half being classified as Kluyvera species. Enterobacteriaceae carrying blaOXA-48-type genes were also isolated from some water samples. The blaOXA-48-like genes were also found in non-Enterobacteriaceae species, such as Shewanella, Acinetobacter, and Pseudomonas. The coliform species harboring resistance genes would trigger a positive coliform water test, which would theoretically be followed by attempts to remediate the contamination issue; however non-Enterobacteriaceae species harboring ESBL and carbapenemase genes would evade detection by the most commonly used coliform screening methods resulting in “silent” dissemination of these genes via tap water that meets all current regulatory requirements.
Studies reporting ESBL- and carbapenemase-producing bacteria in drinking water in high-income countries are extremely rare. CTX-M-producing E. coli was discovered in drinking water in France in a single water sample10, and carbapenemase-producing Serratia fonticola has previously been reported in drinking water from Portugal32, as have non-fermenting intrinsic carbapenemase-producers13. To our knowledge, this is the first extensive study of drinking water from multiple regions in a high income country that has revealed the geographically widespread distribution of ESBL- and carbapenemase-producing isolates of serious clinical concern. Our findings suggest that community exposure to these organisms may be more common than currently realized, and consequently, their prevalence in the general population may be underestimated.
In this era of increasing antimicrobial resistance, it is critical to determine the public health significance of antibiotic resistance genes in community drinking water regardless of country income-level classification. Public water systems provide an effective means by which pathogens and antibiotic-resistant organisms can be transmitted to large segments of the population. In high-income countries, approaches such as increased consumption of bottled water are not adequate solutions, as bottled water is often derived from the same sources as tap water, and may also contain trace levels of total coliform bacteria33. More research is needed to better characterize the problem, understand the associated risk, and devise solutions to combat antimicrobial-resistant bacteria in tap water2.
Supplementary information
Acknowledgements
A portion of this work was performed in the laboratory of Catherine Loc-Carrillo, PhD at the Veterans Affairs Medical Center campus in Salt Lake City, Utah and through resources provided by the Salt Lake City VA IDEAS center (VA Center of Innovation Award #I50HX001240 from the Health Services Research and Development of the Office of Research and Development of the US Department of Veterans Affairs). Funding for this project was provided through the Health Studies Fund of the University of Utah Department of Family and Preventive Medicine and through internal University Seed Funding. We are grateful for the outreach efforts of Sarah Wright and the Association of Public Health Laboratories (APHL) in finding participating APHL member laboratories. We also appreciate the assistance of the Arkansas Department of Health Public Health Laboratory, Illinois Department of Health laboratories in Chicago, Springfield, and Carbondale, Pennsylvania Department of Environmental Protection Laboratories, Biggs Laboratory at the New York State Department of Health Wadsworth Center, Wisconsin State Laboratory of Hygiene, Utah Public Health Laboratory, Davis, Utah County Health Department Laboratory, and the Weber Basin Water Conservancy District laboratory (Utah) in providing and preserving samples for this project. We are also grateful to the Derek Warner and the University of Utah Core DNA Sequencing Laboratory for consulting on and assistance in preparations for the PCR amplicon sequencing.
Author Contributions
W.T. performed all PCR testing, bacterial isolation, and susceptibility testing. J.V., R.G. and A.G. participated in study design and manuscript preparation. M.L. performed all descriptive statistical analysis. M.F. performed MALDI-TOF testing. Authors from the Arkansas, Illinois, New York, Pennsylvania, Utah, Wisconsin, and Davis County public health laboratories and the Weber Basin Water Conservancy District assisted in preserving coliform-positive samples and compiling associated sample data. All authors were involved in compiling and reviewing data for the report and approved the final version. Authors from the Arkansas, Illinois, New York, Pennsylvania, Utah, Wisconsin, and Davis County public health laboratories and the Weber Basin Water Conservancy District all contributed equally.
Data Availability
ESBL or carbapenemase gene sequence data that support the findings of this study have been deposited into GenBank with the accession numbers listed in Table 1. The data that support the descriptive statistical analyses of sample characteristics and resistance gene presence are available from the corresponding author upon reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-40420-0.
References
- 1.Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2013, https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf (2013).
- 2.World Health Organization. Antimicrobial Resistance: Global Report on Surveillance, https://www.who.int/drugresistance/documents/surveillancereport/en/ (2014).
- 3.World Health Organization. WHO publishes list of bacteria for which new antibiotics are urgently needed, http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/ (2017).
- 4.Bush K. Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr Opin Microbiol. 2010;13:558–564. doi: 10.1016/j.mib.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Bouchillon SK, Badal RE, Hoban DJ, Hawser SP. Antimicrobial susceptibility of inpatient urinary tract isolates of gram-negative bacilli in the United States: results from the study for monitoring antimicrobial resistance trends (SMART) program: 2009–2011. Clin Ther. 2013;35:872–877. doi: 10.1016/j.clinthera.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 6.Doi Y, et al. Community-associated extended-spectrum beta-lactamase-producing Escherichia coli infection in the United States. Clin Infect Dis. 2013;56:641–648. doi: 10.1093/cid/cis942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly AM, Mathema B, Larson EL. Carbapenem-resistant Enterobacteriaceae in the community: a scoping review. Int J Antimicrob Agents. 2017;50:127–134. doi: 10.1016/j.ijantimicag.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Potron A, Poirel L, Nordmann P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: Mechanisms and epidemiology. Int J Antimicrob Agents. 2015;45:568–585. doi: 10.1016/j.ijantimicag.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Walsh TR, Weeks J, Livermore DM, Toleman MA. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: An environmental point prevalence study. Lancet Infect Dis. 2011;11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 10.Madec JY, et al. ST48 Escherichia coli carrying the blaCTX-M-1 IncI1/ST3 plasmid in drinking water, France. Antimicrob Agents Chemother. 2016;60:6430–6432. doi: 10.1128/AAC.01135-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Randall LP, et al. Evaluation of meat, fruit and vegetables from retail stores in five United Kingdom regions as sources of extended-spectrum beta-lactamase (ESBL)-producing and carbapenem-resistant Escherichia coli. Int J Food Microbiol. 2016;241:283–290. doi: 10.1016/j.ijfoodmicro.2016.10.036. [DOI] [PubMed] [Google Scholar]
- 12.Talukdar PK, et al. Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PLoS One. 2013;8:e61090. doi: 10.1371/journal.pone.0061090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henriques IS, et al. Prevalence and diversity of carbapenem-resistant bacteria in untreated drinking water in Portugal. Microbial Drug Resist. 2012;18:531–537. doi: 10.1089/mdr.2012.0029. [DOI] [PubMed] [Google Scholar]
- 14.Revisions to the Total Coliform Rule, Final Rule. 40 CFR Parts 141 and 142. United States (2013).
- 15.American Public Health Association, American Water Works Association & Water Environment Federation. 9223 Enzyme-substtrate coliform test, 21st edition. Standard Methods for the Examination of Water and Wastewater (American Public Health Association, 2005).
- 16.Monstein HJ, et al. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in Enterobacteriaceae. APMIS. 2007;115:1400–1408. doi: 10.1111/j.1600-0463.2007.00722.x. [DOI] [PubMed] [Google Scholar]
- 17.Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Bush, K., Palzkill, T. & Jacoby, G. ß-Lactamase Classification and Amino Acid Sequences for TEM, SHV and OXA Extended-Spectrum and Inhibitor Resistant Enzymes, www.lahey.org/studies (2018).
- 19.Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum (beta)-lactamases. J Antimicrobial Chemother. 2006;57:154–155. doi: 10.1093/jac/dki412. [DOI] [PubMed] [Google Scholar]
- 20.Nuesch-Inderbinen MT, Hachler H, Kayser FH. Detection of genes coding for extended-spectrum SHV beta-lactamases in clinical isolates by a molecular genetic method, and comparison with the E test. Eur J Clin Microbiol Infect Dis. 1996;15:398–402. doi: 10.1007/BF01690097. [DOI] [PubMed] [Google Scholar]
- 21.Mena A, et al. Characterization of a large outbreak by CTX-M-1-producing Klebsiella pneumoniae and mechanisms leading to in vivo carbapenem resistance development. J Clin Microbiol. 2006;44:2831–2837. doi: 10.1128/JCM.00418-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brinas L, Zarazaga M, Saenz Y, Ruiz-Larrea F, Torres C. Beta-lactamases in ampicillin-resistant Escherichia coli isolates from foods, humans, and healthy animals. Antimicrob Agents Chemother. 2002;46:3156–3163. doi: 10.1128/AAC.46.10.3156-3163.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poirel L, Heritier C, Tolun V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Payne SJ, et al. Molecular techniques and data integration: Investigating distribution system coliform events. J Water Supply Res Technol. 2010;59:298–311. doi: 10.2166/aqua.2010.026. [DOI] [Google Scholar]
- 25.Tanner WD, et al. Development and field evaluation of a method for detecting carbapenem-resistant bacteria in drinking water. Syst Appl Microbiol. 2015;38:351–357. doi: 10.1016/j.syapm.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Khot PD, et al. Optimization of Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry Analysis for Bacterial Identification. J Clin Microbiol. 2012;50:3845–3852. doi: 10.1128/JCM.00626-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.U.S. Environmental Protection Agency. Causes of Total Coliform-Positive Occurances in Distribution Systems, http://www.epa.gov/ogwdw/disinfection/tcr/pdfs/issuepaper_tcr_causes.pdf (2006).
- 28.Lartigue MF, et al. Characterization of an extended-spectrum class A beta-lactamase from a novel enterobacterial species taxonomically related to Rahnella spp./Ewingella spp. J Antimicrobial Chemother. 2013;68:1733–1736. doi: 10.1093/jac/dkt122. [DOI] [PubMed] [Google Scholar]
- 29.Natural Resources Defense Council. Threats on tap: widespread violations highlight need for investment in water infrastructure and protections, https://www.nrdc.org/sites/default/files/threats-on-tap-water-infrastructure-protections-report.pdf (2017).
- 30.U.S. Environmental Protection Agency. About Private Water Wells, https://www.epa.gov/privatewells/about-private-water-wells (2017).
- 31.Gonzalez-Lopez JJ, et al. First detection of plasmid-encoded blaOXY beta-lactamase. Antimicrob Agents Chemother. 2009;53:3143–3146. doi: 10.1128/AAC.01473-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henriques I, et al. Draft Genome Sequence of Serratia fonticola UTAD54, a Carbapenem-Resistant Strain Isolated from Drinking Water. Genome Announc. 2013;1:e00970–13. doi: 10.1128/genomeA.00970-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.U.S. Food and Drug Administration. CFR - Code of Federal Regulations Title 21, https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/CFRSearch.cfm?fr=165.110 (2017).
- 34.Clinical and Laboratory Standards Institute (2019). Performance Standards for Antimicrobial SusceptibilityTesting, 29th Edition. CLSI M100:ED29:2019, http://em100.edaptivedocs.net/dashboard.aspx (2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ESBL or carbapenemase gene sequence data that support the findings of this study have been deposited into GenBank with the accession numbers listed in Table 1. The data that support the descriptive statistical analyses of sample characteristics and resistance gene presence are available from the corresponding author upon reasonable request.