Abstract
Chromodomain Y (CDY) is one of the candidate genes for male dyszoospermia related to Y chromosome microdeletion (YCM). However, the function of CDY in regulating spermatogenesis has not been completely determined. The mouse Cdyl (CDY-like) gene is the homolog of human CDY. In the present study, we generated a germline conditional knockout (cKO) model of mouse Cdyl. Significantly, the CdylcKO male mice suffered from the defects in spermatogonia maintenance and spermatozoon morphogenesis, demonstrating teratozoospermia and a progressive infertility phenotype in early adulthood. Importantly, patterns of specific histone methylation and acetylation were extensively changed, which disturbed the transcriptome in CdylcKO testis. Our findings indicated that Cdyl is crucial for spermatogenesis and male fertility, which provides novel insights into the function of CDY gene, as well as the pathogenesis of YCM-related reproductive failure.
Introduction
Since the turn of the millennium, the global incidence rate of infertility has been rising continuously. Among all infertility cases, ~50% of them can be attributed to male factors, such as idiopathic oligozoospermia or azoospermia. One of the most frequent molecular genetic causes of spermatogenic failure is deletion of the azoospermia factor region of the Y chromosome. Y chromosome microdeletion (YCM) was detected in about 2–5% of patients with severe oligozoospermia and in 5–10% of patients with azoospermia1. Studies on YCM are particularly important because of its potential for genetic transmission to the offspring. To date, genes including DAZ (deleted in azoospermia)2, RBMY (RNA binding motif protein, Y-linked)3, USP9Y (ubiquitin specific peptidase 9, Y-linked)4, TSPY (testis-specific protein, Y-linked)5, and CDY (chromodomain Y)6 have been identified as candidates for YCM. In contrast to the DAZ gene, whose critical impact upon human spermatogenesis has been fully elucidated, the functional role of the CDY gene cluster remains unknown. The deletion or low expression of CDY genes is closely correlated with male dyszoospermia7–12, however, the precise molecular mechanism involved remains to be investigated.
The human CDY gene family originated by transposition of an autosomal genomic Cdyl gene in primates6,13. In mice, there is no CDY gene on the Y chromosome; instead, the autosomal Cdyl gene is homologous to the human CDY gene family. The protein products of either human CDY/CDYL or mouse Cdyl genes share high similarity14. In the present study, we generated a Cdyl germline conditional knockout (CdylcKO) mouse model. The CdylcKO male mice demonstrated a phenotype of teratozoospermia and progressive infertility. In particular, the patterns of histone methylation and acetylation were globally altered in CdylcKO testis, which led to transcriptomic changes and various spermatogenic defects. These findings revealed an essential role of mouse Cdyl in male fertility, and provided novel insights into the mechanism of YCM-related reproductive failure.
Materials and methods
Animals
The mice used in this study were bred on the C57BL/6 × 129 background. All animal experiments were carried out in accordance with the guidelines for the Use of Animals in Research issued by the Shanghai Jiao Tong University, School of Medicine.
Whole-mount immunofluorescence staining
Embryonic day 15.5 (E15.5) embryos were dissected from euthanized pregnant females, and XY embryonic gonads (EGs) were collected according a previously published method15. EGs were washed in phosphate-buffered saline (PBS), transferred into 4% paraformaldehyde, and fixed overnight at 4 °C on a rocker. The EGs were then subjected to three 15 min washes with PBS containing 0.1% Triton-X (PBS-T). All the antibodies were diluted in PBS-T containing 1 mg/ml bovine serum albumin (BSA). The samples were incubated with primary antibodies for 2 days at 4 °C, washed with PBS-T, and then incubated with the Alexa Fluor-conjugated secondary antibodies for 1 h at room temperature. After washing with PBS-T, the EGs were viewed and photographed under a fluorescence microscope (Leica). The antibodies used in this research are listed in Table S1.
Generation of cKO mouse and genotyping
Conditional Cdyl knockout mice were generated by inserting loxP sites flanking the fifth exon of the ubiquitously expressed Cdyl transcript. The targeting vector was electroporated into embryonic stem cells (ESCs) to construct the heterozygous Cdyl+/flox ESCs, followed by injection of the Cdyl+/flox ESCs into mouse blastocysts. After the standard breeding procedures, homozygous Cdylflox/flox mice were obtained, which were phenotypically normal. The female Cdylflox/flox mice were crossed to the male Vasa-Cre transgenic mice to obtain the Vasa-Cre+, Cdylflox/flox mice (CdylcKO). Either Cdylflox/flox or Cdyl+/flox male mice were used as controls (CdylCtrl) in this study. DNA was extracted from mouse tails and subjected to PCR. The primers used to detect Vasa-Cre, Cdylflox, Cdyl Δ (Cdyl deletion) in this assay are listed in Table S2.
Mating experiment
Each CdylcKO or control male mouse was bred with two wild-type adult females continuously from 6 weeks old. The mating experiments lasted for at least 16 weeks. The date of delivery and the numbers of litters and pups were recorded.
RNA extraction and quantitative real-time reverse transcription PCR
Total RNA was extracted using the TRIzol reagent according to the manufacturer’s protocols (Invitrogen). Reverse transcription reactions were executed using PrimeScript RT reagent Kit (Takara). The quantitative real-time PCR reaction was prepared using the SYBR Premix Ex Taq Kit (Takara) and performed using an ABI 7500 System (Applied Biosystems). Gapdh was used as the internal control. The primers used in this experiment are listed in Table S2.
Protein preparation and western blotting
To prepare the proteins from testes and mouse/human sperm, the samples were lysed in Radioimmunoprecipitation assay buffer and then homogenized on ice. The lysates were then centrifuged at 12,000 × g for 10 min at 4 °C to collect the supernatant. Protein concentrations were measured using a BCA Protein Assay Kit (Beyotime). All samples were processed using 10–12% sodium dodecyl sulfate polyacrylamide gel electrophoresis and electrotransferred to polyvinylidene fluoride membranes. Membranes were blocked for 1 h at room temperature with 5% BSA and then incubated with primary antibodies overnight at 4 °C. After incubation with labeled secondary antibodies, signals were obtained using a Visualizer Western Blot Detection Kit (Millipore). The primary antibodies are listed in Table S1.
Histological and immunohistochemistry assays
Male mice were killed at 6 weeks, 14 weeks, or 5 months old. The weights of the whole body, testis, and epididymis were recorded. Tissues were fixed in Bouin’s solution overnight. Paraffin sections (5 μm) were prepared using standard procedures. Periodic acid-Schiff (PAS) staining was performed according to the manufacturer's instructions (Beyotime). Staging of mouse seminiferous tubule cross-sections was performed according to a published method16.
For the immunohistochemical assays, antigen retrieval was executed by incubation in buffered citrate (pH 6.0) for 15 min at 105 °C. The sections were blocked in counterpart IgG for 30 min at room temperature and then incubated with primary antibodies at 4 °C overnight. On the following day, the signals were visualized using a Histostain-Plus Kit (Life Technologies). Images were captured under a microscope BX53F (Olympus). The antibodies used in this assay are listed in Table S1.
CASA analysis and sperm staining
The cauda epididymis was dissected from an individual mouse, and then incubated in Tyrode’s Solution (Solarbio) at 37 °C for 15 min to release the sperms. The supernatant was collected, and sperm counts and motility were evaluated using the Computer Assisted Semen Analysis (CASA) system (Hamilton). Otherwise, the supernatant was centrifuged at 1000 g for 15 min, the sperm pellet was resuspended in 4% paraformaldehyde, and spread on precoated slides. For morphological observation, Giemsa staining was performed according to the manufacturer's instructions (Beyotime). At least 200 sperms were recorded for each sample.
Sex hormone measurements
The mouse sex hormone levels were measured at 5 months. The exact levels of testosterone (R&D Systems), luteinizing hormone (LH) (Shanghai Xinle Biotechnology), and follicle stimulating hormone (FSH) (Elabscience) in serum were detected using immunoassay kits according the manufacturer's protocols.
Transmission electron microscopy
Testis tissues or sperm pellets were fixed with 2.5% glutaraldehyde in 0.2 m cacodylate buffer overnight. After washing in 0.2 m PBS, the tissues were immersed in 1% OsO4 in 0.2 m cacodylate buffer for 2 h at 4 °C. The samples were then dehydrated and embedded in resin. Ultrathin sections were cut and stained with uranyl acetate and lead citrate, and then observed using an H-7650 transmission electron microscope (HITACHI).
RNA-seq library preparation and data processing
Total RNA was extracted from 6-week old testis tissues using the TRIzol reagent (Invitrogen). Complementary DNA library construction and sequencing were performed by Beijing Genomics Institute using the BGISEQ-500 platform. High-quality reads were aligned to the mouse mm10 genome using Bowtie2. The expression levels of each of gene were normalized to fragments per kilobase of exon model per million mapped reads using RNA-seq by Expectation Maximization. The interaction network of spermatogenesis-related genes was constructed by String and Cytoscape based on the published references.
Flow cytometry
Male mice were killed at 3 weeks and their testes were harvested. A testicular cell suspension was prepared according to a previously published method17. After incubation with the appropriate antibodies, quantitative flow cytometry analysis was performed using bead-based 123count eBeads count (Invitrogen 01-1234-42)18 on CytoFlex S (Beckman). In brief, the testicular cells harvested from 3-week-old mouse were resuspended in 100 μL of PBS. Then 100 μL of beads, ~1010 beads/μL, was added to each sample. The final volume in the tube was 200 μL. Each tube was fully vortexed before collection to ensure equal dispersion of cells and beads. In each analysis, ~10,000 beads events were collected and the results were analyzed using CytExpert2.0. The absolute enumeration of Thy1 or c-Kit positive cells per mouse was calculated according to the relative ratio to beads.
Statistical analysis
Statistical analysis was performed using one-way analysis of variance, followed by Student’s t test. Values of p < 0.05 were considered statistically significant. The statistical graphs were generated using GraphPad Prism 6.
Results
Generation of Cdyl germline cKO mice
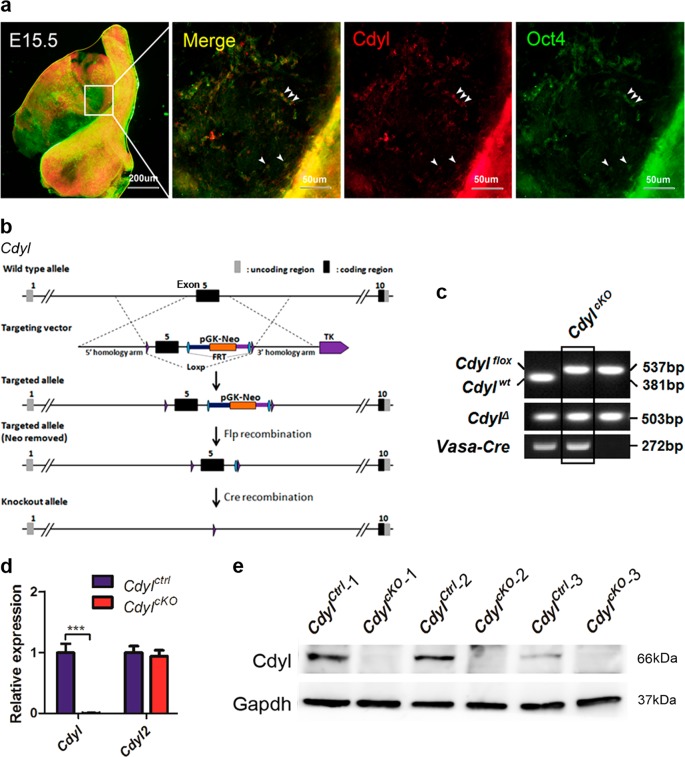
The mouse Cdyl gene produces a longer ubiquitously expressed transcript and a shorter testis-specific transcript after birth19. However, it is not clear whether Cdyl is expressed during embryonic gonadal development. By whole-mount immunofluorescence staining, we detected the co-localization of Cdyl and octamer-binding protein 4 (Oct4) in E15.5 male mouse embryonic gonads (Fig. 1a, arrowhead). As Oct4 is the acknowledged marker of primordial germ cells (PGCs)20, it was suggested that Cdyl also functions in mouse male PGCs.
Fig. 1. Generation of germline CdylcKO mice.
a Whole-mount staining of Cdyl and Oct4 in mouse E15.5 male embryonic gonads. b Schematic overview of the gene targeting strategy to generate CdylcKO mice. c Representative result of genotyping to identify the CdylcKO mice. Cdylflox, loxP flanked allele; Cdylwt, wild-type allele; CdylΔ, Cdyl conditional deletion. d Verification of the conditional knockout efficiency by detection of the relative mRNA expression of Cdyl in CdylcKO testis (n = 4). ***p < 0.001. e Verification of the conditional knockout efficiency by detecting the protein expression of Cdyl in CdylcKO testis (n = 3)
In the present study, male Vasa-Cre transgenic mice were used to generate Cdyl germline cKO mice, considering that the recombinase would be active in Vasa-Cre+ germ cells as early as E1521. In detail, the fifth exon of the ubiquitously expressed Cdyl transcript, which was identical to exon 2 in the testis-specific transcript, was flanked by inserted loxP sites. Homozygous Cdylflox/flox mice were obtained and were physiologically normal, then the female Cdylflox/flox mice were crossed to the male Vasa-Cre transgenic mice. Eventually, we successfully established the male germline cKO Vasa-Cre+,Cdylflox/flox (CdylcKO) mice (Fig. 1b, c). The results of quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) and western blotting (Fig. 1d, e) showed that the expression level of Cdyl was dramatically decreased in the testis of 6-week-old CdylcKO mice.
Germline knockout of Cdyl caused a severe progressive infertility in male mice
We then assessed the reproductive performance of male CdylcKO mice. Six-week-old CdylcKO mice were bred with adult wild-type females for continuous 16 weeks (n = 5). The numbers of litters and pups from each tested male were recorded, as well as the exact age by which they gave the first and the last litters during the experiment (Table 1). The average litter size from the CdylcKO males was not changed (P > 0.05); however, the total number of litters produced by the CdylcKO group was only 42.86% of that from the control group (P < 0.001). The average age of CdylcKO males that delivered their first litter was apparently older than that of the control males (P < 0.05). In addition, their average age of delivery of the last litter was around 15 weeks old, whereas the control males remained fertile for at least 10 months (P < 0.01).
Table 1.
The fertility of CdylcKO male mice
| CdylCtrl (n = 5) | CdylcKO (n = 5) | P | |
|---|---|---|---|
| Age of first litter (PND) | 64.2 ± 8.70 | 76.2 ± 8.07 | <0.05 |
| Age of last litter (PND) | 142.4 ± 12.29 | 96 ± 15.95 | <0.01 |
| Total litters | 21 | 9 | |
| Average litters | 4.2 ± 0.45 | 1.8 ± 0.45 | <0.001 |
| Total pups | 150 | 58 | |
| Average litter size | 7.14 ± 2.52 | 6.44 ± 2.69 | >0.05 |
CdylcKO mice were bred to adult wild-type females for continuous 16 weeks, the numbers of litters and pups from each tested male were recorded (n = 5). Values represent the mean ± SEM; statistical analyses were conducted using Student’s t test. PND, post-natal day
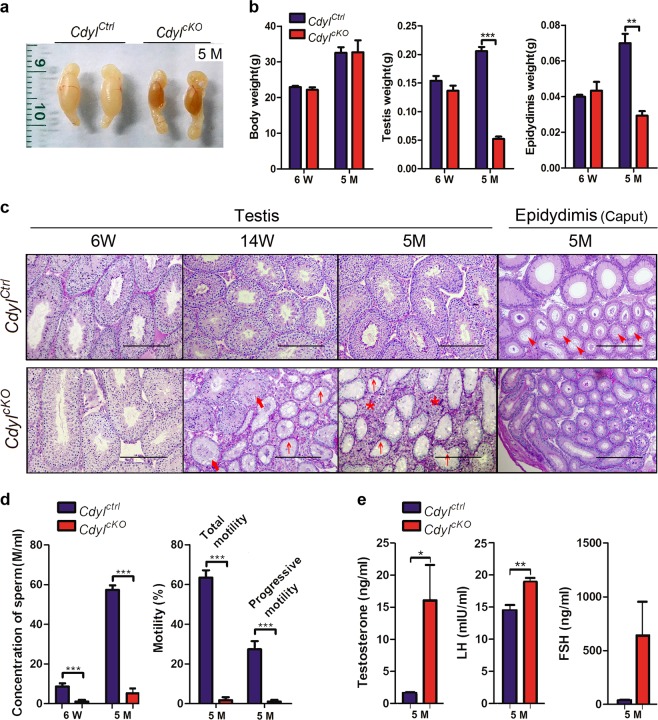
By 5 months after birth, the testis from CdylcKO mice had a shrunken and sclerotic appearance (Fig. 2a). Although there was no difference in body weight, the weight of the testis or epididymis was significantly reduced compared with those of the wild-type (n = 6) (Fig. 2b). Histopathological examination revealed that a progressive degeneration of spermatogenesis happened in CdylcKO testis from 6 weeks until 5 months (Fig. 2c); the germ cells were gradually lost in most of the seminiferous tubules (bold arrow). We observed a typical phenotype of Sertoli-cell-only syndrome (SCOS)22 in 5-month-old CdylcKO testis (arrow), that only Sertoli cells were lining the seminiferous tubules. Meanwhile, hyperplasia of the interstitial tissues was noticed (asterisk). Usually, the lumen of epididymis is full of the seminal fluid in adult mice (arrowhead), however, only a few spermatozoa were found in epididymal sections of 5-month-old CdylcKO males.
Fig. 2. CdylcKO male mice exhibited the progressive infertility.
a Testes harvested from CdylCtrl and CdylcKO mice at 5 months. b Weight of body, testis, and epidydimis in CdylCtrl and CdylcKO group (n = 6). c Pathology of CdylcKO testis at 6 weeks, 14 weeks, and 5 months; Pathology of CdylcKO epidydimis at 5 months. Bar = 200 μm. Bold arrow, loss of spermatogenic cells; arrow, Sertoli-cell-only syndrome phenotype; asterisk, hyperplasia of the interstitial tissues; arrowhead, seminal fluid in the lumen of epididymis. d Computer Assisted Semen Analysis of CdylcKO males at 6 weeks and 5 months (n = 6). e Serum sex hormone levels of CdylcKO males at 5 months (n = 5). *p < 0.05; **p < 0.01; ***p < 0.001
Along with these findings, the CASA analysis disclosed a decrease in the sperm concentration in CdylcKO male mice. Compared with that in the control group, it was only about 18.88% (1.62 ± 0.50 m/ml vs. 8.58 ± 3.18 m/ml) in 6-week-old CdylcKO males, and then declined to 8.99% (5.16 ± 6.15 m/ml vs. 57.34 ± 5.19 m/ml) by 5 months (n = 6) (Fig. 2d). We next measured the serum sex hormone levels in 5-month-old CdylcKO male mice (n = 5). We observed upregulation of testosterone (P < 0.05) and LH (P < 0.01), and a non-significant increase in FSH (n = 5) (Fig. 2e). In conclusion, the CdylcKO male mice suffered from the progressive infertility and displayed a severe oligozoospermia phenotype by 5 months.
Abnormal spermiogenesis in CdylcKO male mice
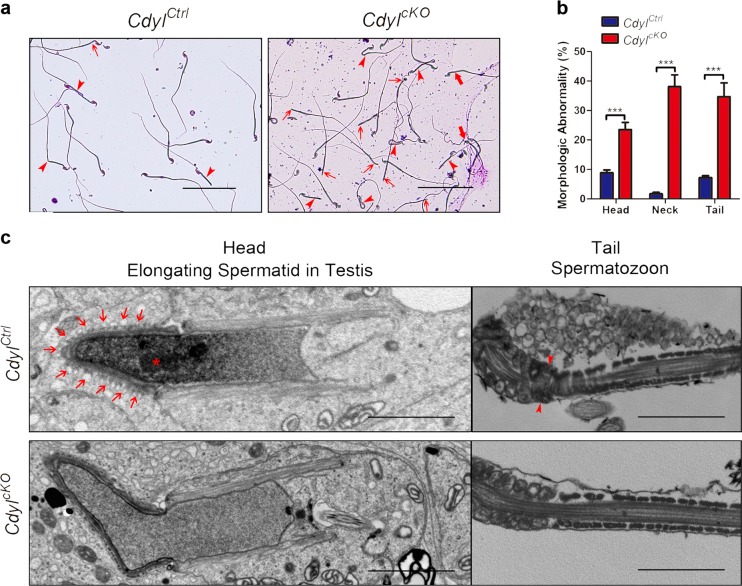
In addition to the quantitative loss of sperm, the total and progressive sperm motilities were also badly damaged in 5-month-old CdylcKO males (P < 0.001) (Fig. 2d). We also observed malformation of spermatozoon in CdylcKO mice (Fig. 3a, b), for example, the abnormal shape of head (bold arrow), detached head caused by a faulty neck structure (arrow), and bending or looping of flagellum (arrowhead) (n = 5, P < 0.001). Many sperms carried head and tail defects at the same time.
Fig. 3. Abnormal morphogenesis of spermatozoon in CdylcKO mice.
a Morphology of spermatozoa from 14-week-old CdylcKO male mice under light microscopy. Bar = 200 μm. Bold arrow, abnormal shape of head; arrow, detached head due to the faulted neck structure; arrowhead, bending or looping of flagellum. b Percentage of sperm malformation in 14-week-old CdylcKO male mice (n = 5). ***p < 0.001. c Morphology of spermatid and spermatozoa from 14-week-old CdylcKO male mice under transmission electron microscopy. Bar = 2 μm. Asterisk, organization center of nuclear concentration; arrow, ectoplasmic specialization; arrowhead, annulus structure
Using transmission electron microscopy, we inspected the ultrastructure of the spermatids in testis sections or the spermatozoon extracted from the cauda epididymis (Fig. 3c). Compared with the control samples, many of the elongating spermatids from CdylcKO male lacked the organization center of nuclear concentration (asterisk), which was replaced by homogenous diffused nucleoplasm. Ectoplasmic specialization (arrows), the particular cell junctions that connect the spermatids and Sertoli cells23, were also disrupted in CdylcKO testis. In another aspect, the annulus structure (arrowhead) partitions the middle piece and the principle piece along the sperm tail24. However, this structure was frequently missing in the CdylcKO spermatozoa, contributing to the bending of the flagellum at that exact spot. In summary, the conditional deletion of Cdyl might lead to the incorrect morphogenesis of spermatozoon and a teratozoospermia phenotype.
Cdyl cKO influenced the patterns of histone modifications in testis
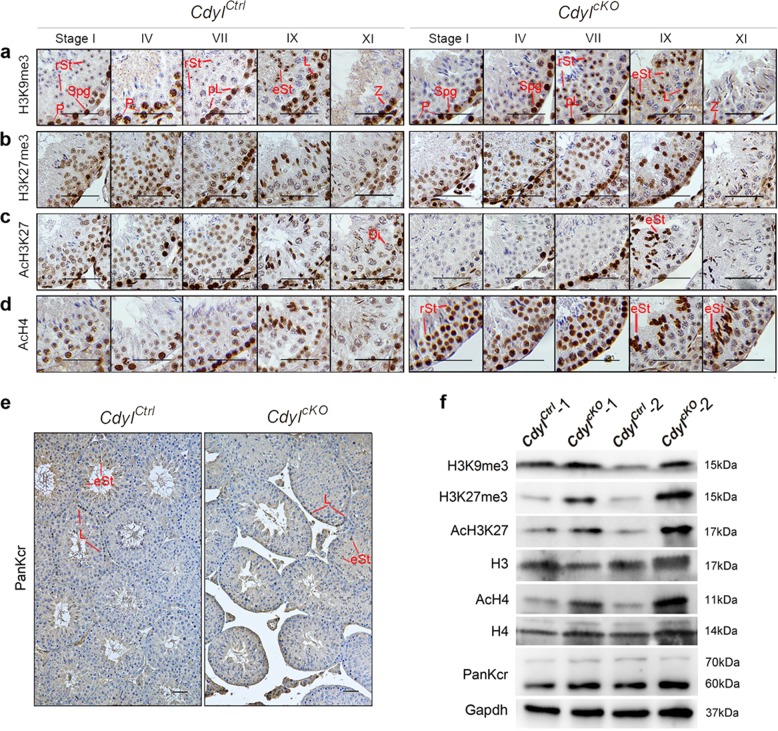
As shown above, the Cdyl cKO had complicated consequences for mouse spermatogenesis. The results implied that Cdyl has important roles in multiple ways and stages during this process. To date, our knowledge of CDYL/Cdyl proteins has focused on their functions in epigenetic regulation, especially histone methylation25–27 and crotonylation28. According to these reports, we evaluated the histone H3 lysine 9 trimethylation (H3K9me3) expression in 6-week-old testicular sections (Fig. 4a). In the normal control group, a strong nuclear signal of H3K9me3 was present in spermatogonia and middle-pachytene spermatocytes, and then disappeared in the late-pachytene phase. After meiosis, H3K9me3 formed a condensed spot in round spermatid nuclei, persisting until step 9–10 elongating spermatids. In comparison, the H3K9me3 signal almost vanished in the pachytene spermatocytes of the CdylcKO testis; however, it became much stronger than that in the control group through step 6–11 of spermiogenesis.
Fig. 4. Histone modification patterns were disturbed in CdylcKO testis by 6 weeks.
Immunostaining of a H3K9me3; b H3K27me3; c AcH3K27; d AcH4; e PanKcr in CdylcKO testis. Bar = 50 μm. Classification of stage I, IV, VII, IX, XI seminiferous tubules was performed according to a published method16. Spg, spermatogonia; pL, preleptotene spermatocytes; L, leptotene spermatocytes; Z, zygotene spermatocytes; P, pachytene spermatocytes; Di, diplotene spermatocytes; rSt, round spermatids; eSt, elongating spermatids. f Expression level of given histone modifications in CdylcKO testis by western blotting detection
We then examined the expression of histone H3 lysine 27 trimethylation (H3K27me3). In control tissues, a modest H3K27me3 signal was detected in all stages of spermatogenic cells, reaching a peak in leptotene spermatocytes and elongating spermatids (Fig. 4b). However, the H3K27me3 signal in the CdylcKO testis specifically increased in intermediate/type B spermatogonia, as well as in step 7–11 spermatids undergoing transformation. In contrast, we detected a fluctuating pattern of the acetylated histone H3 lysine 27 (AcH3K27) signal in the CdylcKO testis. Compared with that in the control group, the AcH3K27 signal accumulated in leptotene spermatocytes, but gradually weakened in later spermatocytes and round spermatids, and increased again in elongating spermatids (Fig. 4c). We next studied the expression of acetylated histone H4 (AcH4). In spermatogonia and spermatocytes, the AcH4 signal was indistinctive to that in control samples. Surprisingly, in the majority of the spermatids, including the early round and later elongating ones, the AcH4 signal was markedly elevated (Fig. 4d). Recently, human CDYL was reported to be a negative regulator of histone lysine crotonylation, and overexpressing Cdyl caused male subfertility in mice28. Therefore, we tested the global histone crotonylation (PanKcr) level in 6-week-old CdylcKO testis. Unexpectedly, the signal of PanKcr was not affected (Fig. 4e).
Finally, we checked the overall protein levels of the above histone modifications using western blotting (Fig. 4f). Except for PanKcr, all the other histone modifications detected were upregulated in 6-week-old CdylcKO testis. Taken together, we revealed that Cdyl conditional deletion resulted in disrupted patterns of histone modification.
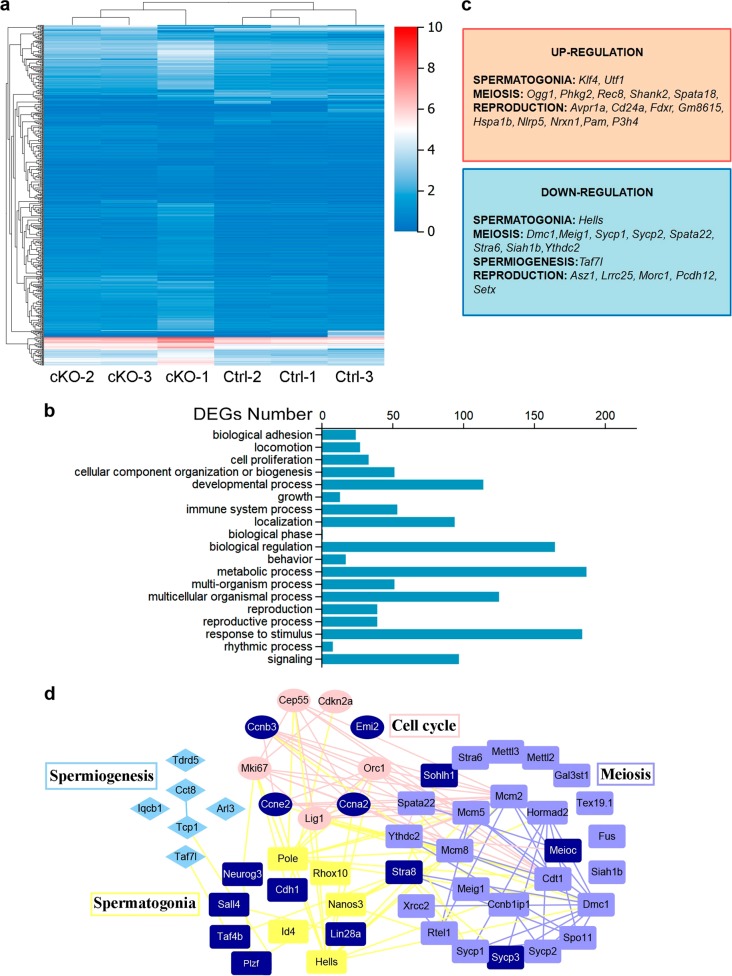
Cdyl conditional deletion disturbed the transcriptome in the testis
Transcriptional activity is dynamically regulated by epigenetic modifications, including histone methylation and acetylation. Therefore, we compared the transcriptome between the 6-week-old CdylcKO testis and their control counterparts using high-throughput RNA sequencing (RNA-seq) (n = 3 each). We identified 675 differentially expressed genes (DEGs) in the CdylcKO group (Fig. 5a). These DEGs were enriched in gene clusters of the cell proliferation, developmental process, and reproduction (Fig. 5b, c). Genes with functions in spermatogenesis29,30 were mapped into an interaction network using String and Cytoscape (Fig. 5d). In particular, we checked the sex chromosome-linked genes that reported highly expressed in round spermatids and are regulated by histone crotonylation modification28. However, no significant change was found in the expression of these genes from the RNA-seq analysis (Supplementary Table S3).
Fig. 5. Cdyl conditional deletion altered the transcriptome in CdylcKO testis by 6 weeks.
a Heatmap showing the differentially expressed genes between CdylCtrl and CdylcKO testes (n = 3). Twofold expression difference and adjust p ≤ 0.01 were used as the cutoff. b The most affected gene ontology (GO) Terms in CdylcKO testis. c Representatives of the dysregulated spermatogenesis-related genes in CdylcKO testis. d Interaction network showing the dysregulated genes in CdylcKO testis that involved in spermatogenesis regulation. The genes validated by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) are marked in dark blue
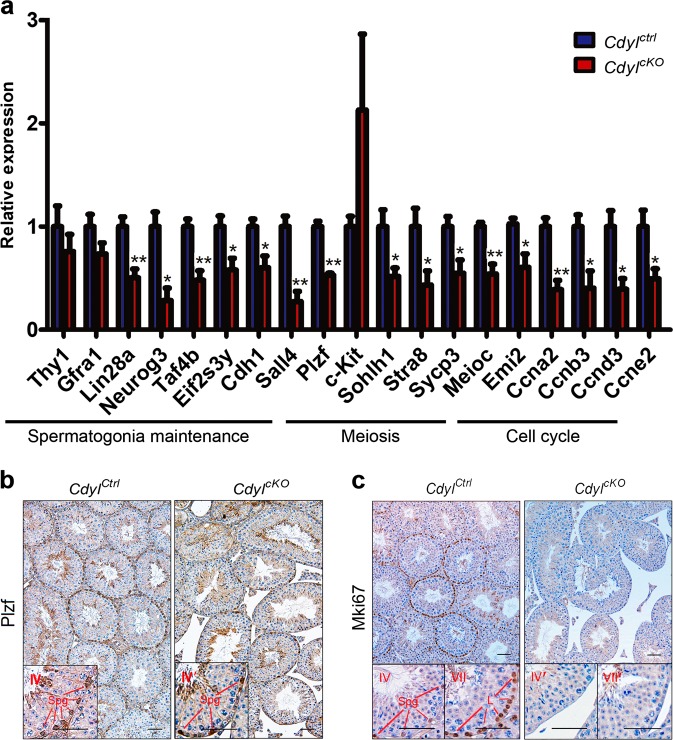
We then validated the RNA-seq results using qRT-PCR (n ≥ 3) (Fig. 6a). Among the target genes, Thy1 (Thy1 cell surface antigen), Gfra1 (GDNF family receptor alpha 1), Lin28a (Lin-28 homolog A), Neurog3 (neurogenin 3), Taf4b (TATA-box binding protein associated factor 4b), Eif2s3y (Eukaryotic translation initiation factor 2 subunit 3, Y-linked), Cdh1 (cadherin 1), Sall4 (spalt like transcription factor 4), and Plzf (promyelocytic leukemia zinc finger protein) have been reported to be expressed sequentially in testis, playing crucial roles for spermatogonial stem cell (SSCs) and spermatogonia maintenance29,31–34. Our results revealed that most of the tested genes were downregulated in CdylcKO testis. In another aspect, c-Kit (Kit proto-oncogene receptor tyrosine kinase), Sohlh1 (spermatogenesis and oogenesis specific basic helix-loop-helix 1), Stra8 (stimulated by retinoic acid 8), Sycp3 (synaptonemal complex protein 3), and Meioc (meiosis specific with coiled-coil domain) are involved in spermatogonial differentiation or meiosis progression32,35–37. Generally, these genes were downregulated in CdylcKO testis, except for c-Kit, which presented dramatic individual differences. For cell cycle regulator genes, Emi2 (Endogenous meiotic inhibitor 2), Ccna2 (cyclin A2), Ccnb3 (cyclin B3), Ccnd3 (cyclin D3), and Ccne2 (cyclin E2)38,39 were all obviously downregulated in CdylcKO testis. Thus, Cdyl conditional deletion significantly affected the testicular transcriptome, especially the expression of spermatogenesis-related genes.
Fig. 6. Expression of spermatogenesis-related genes was affected by Cdyl conditional deletion.
a Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) validation of dysregulated genes involved in spermatogenesis in CdylcKO testis (n ≥ 3). *p < 0.05; **p < 0.01. b Staining of spermatogonial marker Plzf in CdylcKO testis at 6 weeks. Bar = 50 μm. Spg, spermatogonia; L, leptotene spermatocytes; c Staining of proliferative marker Mki67 in CdylcKO testis at 6 weeks. Bar = 50 μm
This alteration of the transcriptome would profoundly interrupt the normal progress of spermatogenesis. Using immunohistochemistry, we investigated the protein levels of spermatogonial marker Plzf40 and proliferative marker Mki67 (marker of proliferation Ki-67)41. In detail, there were far fewer Plzf positive spermatogonia in 6-week-old CdylcKO testis compared with that in the control (Fig. 6b), implying deficiency of spermatogonia maintenance. In the 6-week-old control samples, a strong Mki67 signal was detected in all stages of spermatogonia, as well as in the daughter leptotene spermatocytes. Surprisingly, Mki67 staining completely disappeared in the CdylcKO testis, indicating the impediments of spermatogonial mitosis (Fig. 6c).
Progressive infertility of CdylcKO males owing to the deficiency in spermatogonia maintenance
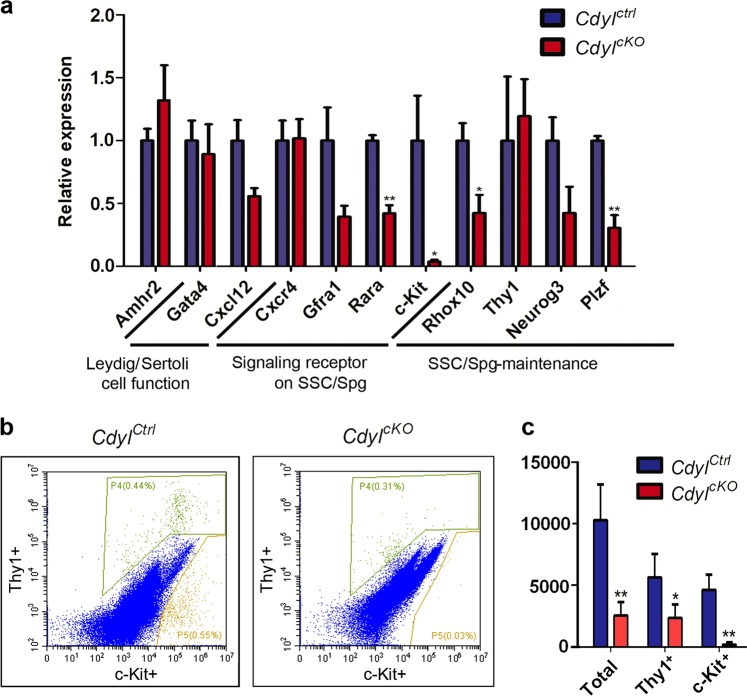
As shown in Fig. 6b, c, spermatogonial maintenance was already disrupted in CdylcKO testis by 6 weeks old. Therefore, we collected the testis from 1 week after birth, when meiosis has not started, and only SSCs/spermatogonia and Sertoli cells were present in the seminiferous tubules42. Amhr2 (anti-mullerian hormone receptor type 2) is the functional marker of Leydig cells43, whereas Gata4 (GATA-binding protein 4) and Cxcl12 (C-X-C motif chemokine ligand 12) are markers of Sertoli cells29,44,45. Using qRT-PCR (Fig. 7a), we detected no differences in the expression levels of Amhr2, Gata4, and Cxcl12 in CdylcKO samples. We then evaluated the critical signaling receptors expressed by SSCs/spermatogonia. Significantly, the RA receptor Rara46 and the SCF receptor c-Kit were downregulated, and the same inclination was observed for the GDNF family receptor Gfra1. Meanwhile, we observed decreased expression levels of Rhox10 (reproductive homeobox 10)47 and Plzf, which are essential for mouse SSC establishment.
Fig. 7. Spermatogonia maintenance was disrupted in CdylcKO male mice.
a Relative expression of important marker genes in 1-week-old CdylcKO testis (n = 3). SSC, spermatogonial stem cell; Spg, spermatogonia. b Representative histogram of quantitative flow cytometry for SSCs/ spermatogonia identification. The Thy1+ events were gated as P4, while the c-Kit+ events were gated as P5. c Changes in the absolute number of SSCs/ spermatogonia in CdylcKO males by 3 weeks (n = 4). *p < 0.05; **p < 0.01
Furthermore, we analyzed the absolute numbers of SSCs/spermatogonia in 3-week-old testis (n = 4) using quantitative flow cytometry (Fig. 7b, c, Figure S1). Thy1 has been recognized as a marker of undifferentiated spermatogonial stem cells48, whereas c-Kit is a hallmark for differentiating spermatogonia49. Strikingly, the absolute number of Thy1+ cells or c-Kit+ cells was much lower in the CdylcKO group (P < 0.05, P < 0.01, individually). Thus, the total amount of SSCs/spermatogonia was remarkably reduced in CdylcKO testis by 3 weeks old (P < 0.01).
Discussion
Although the human CDY gene has been identified as a candidate of YCM for decades, the biological functions of CDY or its autosomal homologs have not been thoroughly resolved6. Both human CDY/CDYL and mouse Cdyl proteins comprise an N-terminal chomodomain and a C-terminal crotonase-like fold14. Based on the properties of chromodomains, CDYL/Cdyl has been proven as an H3K9me3 and H3K27me3 reader, functioning as the mediator in various transcriptional repressive complexes26,27,50. This co-repressor effect of CDYL/Cdyl is important in neural development51–54, X chromosome inactivation55, and even the transformation of tumor cells26. Furthermore, human CDYL plays a vital role in the transmission/restoration of repressive histone markers and the maintenance of cell identity56.
However, the molecular functions of CDY/CDYL/Cdyl in spermatogenesis are not fully understood. Lahn et al.19 reported that, mouse Cdyl was co-localized with the acetyl-H4 in developing spermatids, possibly functioning as a histone acetyltransferase in spermatogenesis. This assumption was disproved by Franz et al.57, who failed to reproduce the histone acetyltransferase activity of CDY family proteins. Recently, Liu et al.28 demonstrated that human CDYL negatively regulates histone lysine crotonylation (Kcr) by acting as a crotonyl-CoA hydratase. In addition, using the overexpression technique, they verified the male subfertility in mice caused by the global upregulation of Cdyl, showing the importance of Cdyl in spermatogenesis. However, there had been no ideal animal model for CDY deletion in some of the human YCM cases.
To clarify the authentic function of CDY/CDYL/Cdyl in spermatogenesis and male reproduction, we established the Cdyl germline cKO mouse model. Compared with control littermates, the time span of fertility was significantly shortened in CdylcKO males: All the CdylcKO males we examined developed severe oligozoospermia by 5 months (n ≥ 15). This peculiar phenotype of progressive infertility was also observed in Map7 (microtubule associated protein 7)58, ERM (ermin)59, and Taf4b60-null mouse models, as well as in Rhox1047 cKO mice. In addition, the abnormal morphogenesis of spermatozoon was observed in CdylcKO males (Fig. 3), resembling human teratozoospermia and asthenozoospermia.
Importantly, we detected disordered patterns of given histone methylation and acetylation in CdylcKO testis (Fig. 4a–d), especially in haploid spermatids, where epigenetic reprogramming is active61. This suggested that mouse Cdyl is indispensable for the control of spermiogenesis, even the setups of paternal epigenetic imprinting. Among these histone modifications, H3K9me3 and H3K27me3 are usually recognized as markers of heterochromatin, whereas AcH3K27 and AcH4 indicate the open configuration of chromatin. The broad alteration of histone modifications would have an extensive impact on histone dynamics and chromatin architecture, and ultimately, transcriptional activity. As a result, the transcriptome was disrupted in CdylcKO testis, and the affected genes included those involved in SSC maintenance, spermatogonial differentiation, meiosis progression, spermiogenesis, and cell cycle control (Figs. 5, 6), which in turn led to the more complex spermatogenic defects. It put forward the question that, whether the inhibitors of histone methyltransferase and/or acetylase, such as G9a inhibitor UNC0642 or EZH2 inhibitor UNC1999 would rescue the phenotype of dyszoospermia in CdylcKO testis. In our study, global histone crotonylation seemed not affected by Cdyl germline cKO (Fig. 4e, f); therefore, phase and site-specific histone crotonylation should be analyzed.
In rodent testis, there are two distinct subsets of spermatogonia: one remains undifferentiated and establishes the SSC pool throughout adulthood; and the other differentiates directly to initiate the first wave of spermatogenesis62,63. By quantitative flow cytometry, we proved the decline in undifferentiated spermatogonia caused by Cdyl conditional deletion (Fig. 7c). It reflected the defects in the establishment and maintenance of the proliferative spermatogonia, which would lead to the exhaustion of spermatogonia and eventual infertility in CdylcKO mice. It could be the consequence of the intrinsic failure of SSC self-renewal, as well as the damaged crosstalk between the spermatogenic cells and their niche. To testify these hypotheses, spermatogonia could be enriched from CdylcKO testis and transplanted into the recipient seminiferous tubules, by which the role of Cdyl in spermatogenesis would be utterly evaluated. Since our data indicated the mouse Cdyl was expressed in E15.5 PGCs (Fig. 1a), and the disrupted transcription happened as early as 1-week old (Fig. 7a), the defects occurring in CdylcKO testis probably originated during the phase of embryonic gonads. Ongoing work in our laboratory is to trace the developmental trajectory of PGCs in CdylcKO mice.
In humans, there are Y chromosome-specific CDY genes as well as autosomal CDYL genes. Current evidences showed that the deletion or low expression of CDY genes correlate with the human spermatogenic defects. In some cases, spermatogenesis was blocked at the stage of spermatid maturation, in spite of the existence of spermatocytes8,10,12. We supposed that the CDYL and CDY proteins could compensate mutually in human testis. Only if both of them were underexpressed would the SCOS phenotype develop, as observed in the CdylcKO mouse model. In summary, we revealed the comprehensive effect of mouse Cdyl on spermatogenesis in vivo. Our findings will aid the mechanism research and potential therapy for the human CDY-associated dyszoospermia.
Supplementary information
Acknowledgements
We thank Dr. Qiang Liu for his helpful discussions on the manuscript. We thank the faculty of Core Facility of Basic Medical Sciences, Shanghai Jiao Tong University for technical assistance. This work was supported by the National Natural Science Foundation of China (81471497, 81501308, 81771656, 81873857), the Shanghai Municipal Commission of Health and Family Planning (20134Y169), the Shanghai Natural Science Foundation Grant (12JC1405500), the Shanghai Municipal Hospital Appropriate Technology Project (SHDC12015122), the Shanghai Jiao Tong University Medicine-Engineering Fund (YG2017ZD11), and the Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant (20181803).
Authors’ contributions
C.X. and A.Z. conceived this project and supervised all the experiments and data analysis. X.X., Y.Q., and F.X. performed the mouse experiments. X.X. and X.Z. performed the cellular and molecular assays with assistance from Z.L. Y.H. performed histological and immunohistochemistry experiments. Y.Q. executed the statistical analysis. X.X. and B.X. analyzed the data and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by M Agostini
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xiaoyu Xia, Xiaowei Zhou, and Yanmei Quan
Contributor Information
Bufang Xu, Email: bufangxu@163.com.
Chen Xu, Email: chenx@shsmu.edu.cn.
Aijun Zhang, Email: zhaj1268@163.com.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-019-1455-y).
References
- 1.Krausz C, Casamonti E. Spermatogenic failure and the Y chromosome. Hum. Genet. 2017;136:637–655. doi: 10.1007/s00439-017-1793-8. [DOI] [PubMed] [Google Scholar]
- 2.Fu XF, et al. DAZ family proteins, key players for germ cell development. Int J. Biol. Sci. 2015;11:1226–1235. doi: 10.7150/ijbs.11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abid S, Sagare-Patil V, Gokral J, Modi D. Cellular ontogeny of RBMY during human spermatogenesis and its role in sperm motility. J. Biosci. 2012;38:85–92. doi: 10.1007/s12038-012-9281-8. [DOI] [PubMed] [Google Scholar]
- 4.Luddi A, et al. Spermatogenesis in a man with complete deletion of USP9Y. N. Engl. J. Med. 2009;360:881–885. doi: 10.1056/NEJMoa0806218. [DOI] [PubMed] [Google Scholar]
- 5.Nickkholgh B, et al. Y chromosome TSPY copy numbers and semen quality. Fertil. Steril. 2010;94:1744–1747. doi: 10.1016/j.fertnstert.2009.09.051. [DOI] [PubMed] [Google Scholar]
- 6.Lahn BT, Page DC. Retroposition of autosomal mRNA yielded testis-specific gene family on human Y chromosome. Nat. Genet. 1999;21:429–433. doi: 10.1038/7771. [DOI] [PubMed] [Google Scholar]
- 7.Kleiman SE, et al. Members of the CDY family have different expression patterns: CDY1 transcripts have the best correlation with complete spermatogenesis. Hum. Genet. 2003;113:486–492. doi: 10.1007/s00439-003-0990-9. [DOI] [PubMed] [Google Scholar]
- 8.Heydarian N, Favaedi R, Sadighi Gilani MA, Shahhoseini M. Expression level of chromodomain Y (CDY): potential marker for prediction of sperm recovery in non-obstructive azoospermia. Int J. Reprod. Biomed. (Yazd) 2016;14:383–388. doi: 10.29252/ijrm.14.6.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sen S, et al. Susceptibility of gr/gr rearrangements to azoospermia or oligozoospermia is dependent on DAZ and CDY1 gene copy deletions. J. Assist. Reprod. Genet. 2015;32:1333–1341. doi: 10.1007/s10815-015-0520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stahl PJ, Mielnik AN, Barbieri CE, Schlegel PN, Paduch DA. Deletion or underexpression of the Y-chromosome genes CDY2 and HSFY is associated with maturation arrest in American men with nonobstructive azoospermia. Asian J. Androl. 2012;14:676–682. doi: 10.1038/aja.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghorbel M, et al. Deletion of CDY1b copy of Y chromosome CDY1 gene is a risk factor of male infertility in Tunisian men. Gene. 2014;548:251–255. doi: 10.1016/j.gene.2014.07.042. [DOI] [PubMed] [Google Scholar]
- 12.Kleiman SE, et al. Expression of CDY1 may identify complete spermatogenesis. Fertil. Steril. 2001;75:166–173. doi: 10.1016/S0015-0282(00)01639-3. [DOI] [PubMed] [Google Scholar]
- 13.Li X, et al. Functional consequences of new exon acquisition in mammalian chromodomain Y-like (CDYL) genes. Trends Genet. 2007;23:427–431. doi: 10.1016/j.tig.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Wu H, Min J, Antoshenko T, Plotnikov AN. Crystal structures of human CDY proteins reveal a crotonase-like fold. Proteins. 2009;76:1054–1061. doi: 10.1002/prot.22472. [DOI] [PubMed] [Google Scholar]
- 15.Lei N, et al. Sex-specific differences in mouse DMRT1 expression are both cell type- and stage-dependent during gonad development. Biol. Reprod. 2007;77:466–475. doi: 10.1095/biolreprod.106.058784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmed EA, de Rooij DG. Staging of mouse seminiferous tubule cross-sections. Methods Mol. Biol. 2009;558:263–277. doi: 10.1007/978-1-60761-103-5_16. [DOI] [PubMed] [Google Scholar]
- 17.Xia X, Cai H, Qin S, Xu C. Histone acetylase inhibitor curcumin impairs mouse spermiogenesis-an in vitro study. PLoS ONE. 2012;7:e48673. doi: 10.1371/journal.pone.0048673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan X, Wu Q, Li H, Li B, Xiu R. Absolute circulating pericyte progenitor cell counts in mice by flow cytometry: comparison of 2 single-platform technologies. Int J. Biol. Markers. 2015;30:e434–e438. doi: 10.5301/jbm.5000156. [DOI] [PubMed] [Google Scholar]
- 19.Lahn BT, et al. Previously uncharacterized histone acetyltransferases implicated in mammalian spermatogenesis. Proc. Natl. Acad. Sci. USA. 2002;99:8707–8712. doi: 10.1073/pnas.082248899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pesce M, Wang X, Wolgemuth DJ, Scholer H. Differential expression of the Oct-4 transcription factor during mouse germ cell differentiation. Mech. Dev. 1998;71:89–98. doi: 10.1016/S0925-4773(98)00002-1. [DOI] [PubMed] [Google Scholar]
- 21.Gallardo T, Shirley L, John GB, Castrillon DH. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis. 2007;45:413–417. doi: 10.1002/dvg.20310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharpe RM, McKinnell C, Kivlin C, Fisher JS. Proliferation and functional maturation of Sertoli cells, and their relevance to disorders of testis function in adulthood. Reproduction. 2003;125:769–784. doi: 10.1530/rep.0.1250769. [DOI] [PubMed] [Google Scholar]
- 23.Wong EW, Mruk DD, Cheng CY. Biology and regulation of ectoplasmic specialization, an atypical adherens junction type, in the testis. Biochim Biophys. Acta. 2008;1778:692–708. doi: 10.1016/j.bbamem.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toure A, Rode B, Hunnicutt GR, Escalier D, Gacon G. Septins at the annulus of mammalian sperm. Biol. Chem. 2011;392:799–803. doi: 10.1515/BC.2011.074. [DOI] [PubMed] [Google Scholar]
- 25.Fischle W, Franz H, Jacobs SA, Allis CD, Khorasanizadeh S. Specificity of the chromodomain Y chromosome family of chromodomains for lysine-methylated ARK(S/T) motifs. J. Biol. Chem. 2008;283:19626–19635. doi: 10.1074/jbc.M802655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulligan P, et al. CDYL bridges REST and histone methyltransferases for gene repression and suppression of cellular transformation. Mol. Cell. 2008;32:718–726. doi: 10.1016/j.molcel.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, et al. Corepressor protein CDYL functions as a molecular bridge between polycomb repressor complex 2 and repressive chromatin mark trimethylated histone lysine 27. J. Biol. Chem. 2011;286:42414–42425. doi: 10.1074/jbc.M111.271064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu S, et al. Chromodomain protein CDYL acts as a crotonyl-CoA hydratase to regulate histone crotonylation and spermatogenesis. Mol. Cell. 2017;67:853–66 e5. doi: 10.1016/j.molcel.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 29.Chen SR, Liu YX. Regulation of spermatogonial stem cell self-renewal and spermatocyte meiosis by Sertoli cell signaling. Reproduction. 2015;149:R159–R167. doi: 10.1530/REP-14-0481. [DOI] [PubMed] [Google Scholar]
- 30.Song HW, Wilkinson MF. Transcriptional control of spermatogonial maintenance and differentiation. Semin Cell Dev. Biol. 2014;30:14–26. doi: 10.1016/j.semcdb.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oatley JM, Brinster RL. Regulation of spermatogonial stem cell self-renewal in mammals. Annu Rev. Cell Dev. Biol. 2008;24:263–286. doi: 10.1146/annurev.cellbio.24.110707.175355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mei XX, Wang J, Wu J. Extrinsic and intrinsic factors controlling spermatogonial stem cell self-renewal and differentiation. Asian J. Androl. 2015;17:347–354. doi: 10.4103/1008-682X.148080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tokuda M, Kadokawa Y, Kurahashi H, Marunouchi T. CDH1 is a specific marker for undifferentiated spermatogonia in mouse testes. Biol. Reprod. 2007;76:130–141. doi: 10.1095/biolreprod.106.053181. [DOI] [PubMed] [Google Scholar]
- 34.Matsubara Y, et al. TALEN-mediated gene disruption on Y chromosome reveals critical role of EIF2S3Y in mouse spermatogenesis. Stem Cells Dev. 2015;24:1164–1170. doi: 10.1089/scd.2014.0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abby E, et al. Implementation of meiosis prophase I programme requires a conserved retinoid-independent stabilizer of meiotic transcripts. Nat. Commun. 2016;7:10324. doi: 10.1038/ncomms10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan L, et al. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol. Cell. 2000;5:73–83. doi: 10.1016/S1097-2765(00)80404-9. [DOI] [PubMed] [Google Scholar]
- 37.Filipponi D, et al. Repression of kit expression by Plzf in germ cells. Mol. Cell Biol. 2007;27:6770–6781. doi: 10.1128/MCB.00479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gopinathan L, et al. Emi2 is essential for mouse spermatogenesis. Cell Rep. 2017;20:697–708. doi: 10.1016/j.celrep.2017.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolgemuth DJ, Manterola M, Vasileva A. Role of cyclins in controlling progression of mammalian spermatogenesis. Int J. Dev. Biol. 2013;57:159–168. doi: 10.1387/ijdb.130047av. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buaas FW, et al. Plzf is required in adult male germ cells for stem cell self-renewal. Nat. Genet. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- 41.Takagi M, Nishiyama Y, Taguchi A, Imamoto N. Ki67 antigen contributes to the timely accumulation of protein phosphatase 1gamma on anaphase chromosomes. J. Biol. Chem. 2014;289:22877–22887. doi: 10.1074/jbc.M114.556647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zindy F, et al. Control of spermatogenesis in mice by the cyclin D-dependent kinase inhibitorsp18(Ink4c) and p19(Ink4d) Mol. Cell Biol. 2001;21:3244–3255. doi: 10.1128/MCB.21.9.3244-3255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orvis GD, et al. Functional redundancy of TGF-beta family type I receptors and receptor-Smads in mediating anti-Mullerian hormone-induced Mullerian duct regression in the mouse. Biol. Reprod. 2008;78:994–1001. doi: 10.1095/biolreprod.107.066605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanatsu-Shinohara M, et al. Reconstitution of mouse spermatogonial stem cell niches in culture. Cell Stem Cell. 2012;11:567–578. doi: 10.1016/j.stem.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 45.Chen SR, et al. Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget. 2015;6:37012–37027. doi: 10.18632/oncotarget.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peer NR, et al. Germ cell-specific retinoic acid receptor alpha functions in germ cell organization, meiotic integrity, and spermatogonia. Endocrinology. 2018;159:3403–3420. doi: 10.1210/en.2018-00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song HW, et al. The homeobox transcription factor RHOX10 drives mouse spermatogonial stem Cell Establishment. Cell Rep. 2016;17:149–164. doi: 10.1016/j.celrep.2016.08.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bai Y, et al. Differential gene expression in mouse spermatogonial stem cells and embryonic stem cells. Int J. Mol. Med. 2016;38:423–432. doi: 10.3892/ijmm.2016.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshinaga K, et al. Role of c-kit in mouse spermatogenesis: identification of spermatogonia as a specific site of c-kit expression and function. Development. 1991;113:689–699. doi: 10.1242/dev.113.2.689. [DOI] [PubMed] [Google Scholar]
- 50.Shi Y, et al. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 51.Wan L, et al. Generation and neuronal differentiation of induced pluripotent stem cells in Cdyl-/- mice. Neuroreport. 2013;24:114–119. doi: 10.1097/WNR.0b013e32835cf179. [DOI] [PubMed] [Google Scholar]
- 52.Qi C, et al. Coordinated regulation of dendrite arborization by epigenetic factors CDYL and EZH2. J. Neurosci. 2014;34:4494–4508. doi: 10.1523/JNEUROSCI.3647-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qin R, et al. CDYL deficiency disrupts neuronal migration and increases susceptibility to epilepsy. Cell Rep. 2017;18:380–390. doi: 10.1016/j.celrep.2016.12.043. [DOI] [PubMed] [Google Scholar]
- 54.Liu Y, et al. CDYL suppresses epileptogenesis in mice through repression of axonal Nav1.6 sodium channel expression. Nat. Commun. 2017;8:355. doi: 10.1038/s41467-017-00368-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Escamilla-Del-Arenal M, et al. Cdyl, a new partner of the inactive X chromosome and potential reader of H3K27me3 and H3K9me2. Mol. Cell Biol. 2013;33:5005–5020. doi: 10.1128/MCB.00866-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y, et al. Chromodomain protein CDYL is required for transmission/restoration of repressive histone marks. J. Mol. Cell Biol. 2017;9:178–194. doi: 10.1093/jmcb/mjx013. [DOI] [PubMed] [Google Scholar]
- 57.Franz H, Mosch K, Soeroes S, Urlaub H, Fischle W. Multimerization and H3K9me3 binding are required for CDYL1b heterochromatin association. J. Biol. Chem. 2009;284:35049–35059. doi: 10.1074/jbc.M109.052332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Komada M, McLean DJ, Griswold MD, Russell LD. Soriano P. E-MAP-115, encoding a microtubule-associated protein, is a retinoic acid-inducible gene required for spermatogenesis. Genes Dev. 2000;14:1332–1342. [PMC free article] [PubMed] [Google Scholar]
- 59.Schlesser HN, et al. Effects of ETV5 (ets variant gene 5) on testis and body growth, time course of spermatogonial stem cell loss, and fertility in mice. Biol. Reprod. 2008;78:483–489. doi: 10.1095/biolreprod.107.062935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Falender AE, et al. Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev. 2005;19:794–803. doi: 10.1101/gad.1290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng J, et al. Erasure of the paternal transcription program during spermiogenesis: the first step in the reprogramming of sperm chromatin for zygotic development. Dev. Dyn. 2008;237:1463–1476. doi: 10.1002/dvdy.21499. [DOI] [PubMed] [Google Scholar]
- 62.Yoshida S, et al. The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development. 2006;133:1495–1505. doi: 10.1242/dev.02316. [DOI] [PubMed] [Google Scholar]
- 63.Niedenberger BA, Busada JT, Geyer CB. Marker expression reveals heterogeneity of spermatogonia in the neonatal mouse testis. Reproduction. 2015;149:329–338. doi: 10.1530/REP-14-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.