Abstract
Despite recent advances, breast cancer (BrCa) still affects many women and the impact is disproportional in African Americans (AA) compared to European Americans (EA). Addressing socioeconomic and behavioral status has not been enough to reduce disparity, suggesting contribution of biological differences in BrCa disparity. Our laboratory was first to show involvement of CC chemokines in BrCa. In this study, using ONCOMINE, TCGA, bc-GenExMiner and KMplotter, we examined the association of CC chemokines in BrCa outcomes and disparity. We show over-expression of CCL5, -7, -11, -17, -20, -22 and -25 in BrCa tissues. High mRNA levels of CCL7, -8, -17, -20 and -25 predicted a decrease in overall survival (OS). CCL7 and CCL8 were associated with decreased relapse-free survival. Expression of CCL17 and CCL25 was associated with decreased OS in AA. In EA, CCL8 was associated with decreased OS. Expression of CCL5, -7, -8, -17, -20 and -25 was highest in TNBC. Expression of CCL11 and CCL22 was associated with HER2. CCL7, -8, -17, -20 and -25 were elevated in AAs. In conclusion, our analysis suggests significant association of CC-chemokines in BrCa progression, OS and disparate disease outcome in AA compared to EA patients.
Introduction
Breast cancer (BrCa) alone accounts for 30% of all new cancers diagnosed in women1 and is the second leading cause of cancer related deaths in women after lung cancer1. Complete etiology of BrCa is yet to be defined, however lifestyle, genetic and environmental factors are often associated with this multifactorial disease2. Undefined etiology and heterogeneity of BrCa are major challenges in developing definitive therapeutics. Among all BrCa types Triple Negative Breast Cancer (TNBC), which lacks estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) is the most lethal type. Moreover, this subtype is further elucidated into 6 subtypes identified by cluster analysis3. Despite higher incidence of BrCa in European Americans (EA), African Americans (AA) are more often diagnosed with TNBC and have a worst prognosis compared to EA with TNBC4,5. Studies have shown that BrCa progression and development is highly influenced by inflammation6 and the immune system7. It is crucial to identify biomarkers, which play a role in these processes, in order to develop novel personalized treatments for BrCa patients.
Chemokines are a large family of small cytokines, which are classified into 4 different subgroups (C, CC, CXC and CX3C), based on cysteine residues. These molecules are responsible for immune cell trafficking and shaping the immune system. They also play a role in inflammation. It is now well established that chemokines and chemokine receptors are expressed by cancer cells and play a significant role in cancer progression and therapeutic outcomes. Our laboratory and others have shown association of chemokines in various cancers8–15. Among all known chemokines and chemokine receptors, CXC chemokines are well studied in cancer8,9,11–16. However our laboratory was first to show association of CC chemokine receptor-9 (CCR9) and its natural ligand CCL25 in various cancers, including BrCa10,17–21.
Higher incidence of BrCa among AA at a younger age and higher mortality of AA with TNBC compared to EA indicate contribution of race specific biological differences to the disparity in disease and therapeutic outcome of the disease. Therefore, it is imperative to define these racial differences in BrCa molecular footprints to address the observed disparity. There are only a few studies available addressing this issue related to chemokines. Recent analysis, have shown association of CXC chemokines with BrCa22. In this study we have utilized large-scale bioinformatics to ascertain CC chemokine expression in BrCa patients based on clinical parameters, respective prognosis and have presented evidence suggesting association of CC chemokines with BrCa progression, TNBC and racial disparity in overall survival (OS).
Results
CC Chemokines are elevated in BrCa tissues
Using the ONCOMINE database (TCGA breast invasive carcinoma dataset), out of all known CC chemokines 7 CC chemokines’ [(CCL5 (FC = 1.6, p = 8.6e−05), CCL7 (FC = 4.5, p = 1.43e−14), CCL11 (FC = 5.5, p = 1.29e−31), CCL17 (FC = 2.0, p = 3.32e−09), CCL20 (FC = 2.2, p = 2.07e−06), CCL22 (FC = 1.44, p = 1.5e−04) and CCL25 (FC = 1.6, p = 1.33e−08)] mRNA were significantly (fold change ≥1.4 and p-value ≤ 0.04) elevated in BrCa compared to normal tissues (Table 1). Furthermore, we elucidated association of these chemokines with histological subtype of BrCa (Table 2). We used several datasets within ONCOMINE to establish the association of CC chemokines with histological subtypes. Curtis and Ginestier23,24 datasets showed that CCL5 was expressed in medullary BrCa with a fold change ranging from 3–8 between Curtis and Ginestier data sets, when compared to other BrCa histological types (p = 0.004 and p = 1.11e−07). The Bittner breast dataset showed CCL5 elevated in ductal (fold change of 1.4, p = 0.02) carcinoma. The TCGA dataset showed CCL7 to be 2 fold greater in ductal BrCa when compared to other BrCa types (p = 2.52e−06). The Radvanyi25 dataset reported CCL11 to have a 3 fold change in ductal carcinoma, compared to other types (p = 1.09e−04). CCL17 was 1.3 fold greater in lobular BrCa (p = 0.03) in the TCGA dataset. Lu dataset displayed 3.5 fold higher CCL20 (p = 9.88e−05) in ductal carcinoma26, whereas in Ginestier dataset 6 fold change in CCL20 was observed (p = 0.012) in medullary BrCa24. There was only 1 fold change in CCL22 expression in ductal carcinoma observed in Curtis dataset (p = 2.34e−09)23, while Radvanyi conveyed a 1.9 greater fold change (p = 0.035) in lobular BrCa for CCL2225. Perou dataset indicated 1.7-fold change in CCL25 (p = 0.034) in Lobular BrCa27 and as per TCGA this change was 1.4 fold (p = 0.015) in ductal carcinoma in situ.
Table 1.
Significant changes of CC- Chemokines among BrCa tissues in comparison to normal tissues using ONCOMINE.
| Fold Change | p-value | t-test | |
|---|---|---|---|
| CCL1 | 1.2 | 0.011 | 2.332 |
| CCL2 | −1.195 | 0.932 | −1.498 |
| CCL3 | 1.181 | 0.164 | 0.984 |
| CCL4 | 1.024 | 0.414 | 0.218 |
| CCL5 | 1.598 | 8.60E-05 | 3.865 |
| CCL7 | 4.522 | 1.43E-14 | 8.62 |
| CCL8 | −1.061 | 0.651 | −0.39 |
| CCL11 | 5.535 | 1.29E-31 | 15.426 |
| CCL13 | −1.393 | 0.986 | −2.226 |
| CCL14 | −1.12 | 0.989 | −2.325 |
| CCL15 | −1.12 | 0.989 | −2.325 |
| CCL16 | −1.161 | 0.996 | −2.655 |
| CCL17 | 1.957 | 3.32E-09 | 6.201 |
| CCL18 | 1.236 | 0.106 | 1.255 |
| CCL19 | −1.147 | 0.871 | −1.138 |
| CCL20 | 2.16 | 2.07E-06 | 4.804 |
| CCL21 | −1.957 | 1 | −6.17 |
| CCL22 | 1.438 | 1.47E-04 | 3.72 |
| CCL23 | −1.429 | 0.999 | −3.154 |
| CCL24 | |||
| CCL25 | 1.598 | 1.33E-08 | 5.909 |
| CCL26 | −1.209 | 0.989 | −2.329 |
| CCL27 | −1.204 | 1 | −3.648 |
| CCL28 | −6.33 | 1 | −9.015 |
Fold Change denotes the folds difference in CC chemokine mRNA expression in BrCa in comparison to normal tissues. The p value represents the statistical significance of the difference in the mean of the two groups as analyzed using t-test. Bold represents significant RNA expression with a cut off of: fold change ≥1.4 and p-value ≤ 0.04 using t-test.
Table 2.
Significant changes of CC chemokines among different histological types of BrCa.
| Data set | Tissue Type (No. of Cases) | CCL5 | CCL7 | CCL11 | CCL17 | CCL20 | CCL22 | CCL25 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FC | p | FC | p | FC | p | FC | p | FC | p | FC | p | FC | p | ||
| Bittner | Ductal (n = 260) | 1.4 | 0.02 | 1.5 | 0.002 | 1.4 | 0.008 | — | — | 1.5 | 0.008 | 1.1 | 0.3 | −1.1 | 0.7 |
| Lobular (n = 38) | −1.1 | 0.7 | −1.6 | 1 | 1.3 | 0.25 | — | — | −1.6 | 1 | −1.2 | 1 | −1.1 | 0.8 | |
| Boersma78 | Ductal (n = 83) | 1.1 | 0.4 | 1.2 | 0.014 | 1.3 | 0.003 | 1 | 0.3 | 1.1 | 0.3 | −1.2 | 0.7 | 1.3 | 0.008 |
| Lobular (n = 10) | 1.1 | 0.4 | −1.2 | 1 | −1.3 | 1 | −1 | 0.7 | −1.1 | 0.7 | 1.2 | 0.2 | −1.3 | 1 | |
| Curtis23 | Ductal (n = 1556) | 1 | 0.36 | 1 | 0.002 | 1 | 0.02 | — | — | 1 | 1.3e−4 | 1.1 | 2.3e−9 | 1 | 0.4 |
| Medullary (n = 32) | 3.3 | 1e −7 | 1.3 | 0.007 | 1 | 0.2 | — | — | 1.4 | 0.002 | 1.2 | 0.02 | 1.2 | 4.1e−4 | |
| Lobular (n = 148) | 1.1 | 0.14 | −1.1 | 1 | −1 | 1 | — | — | −1.1 | 1 | −1.1 | 1 | −1 | 1 | |
| Desmedt79 | Ductal (n = 158) | 1.5 | 0.06 | 1.3 | 0.09 | 1.1 | 0.3 | — | — | 1.2 | 0.2 | 1.1 | 0.3 | 1 | 0.4 |
| Lobular (n = 13) | −1.2 | 0.9 | −1.4 | 1 | −1.1 | 0.8 | — | — | −1.2 | 0.7 | −1.3 | 0.9 | −1 | 0.6 | |
| Esserman80 | Ductal (n = 105) | 1.1 | 0.4 | 1.9 | 0.04 | 1.3 | 0.05 | 1.1 | 0.4 | −1.1 | 0.7 | −1.1 | 0.7 | 1 | 0.4 |
| Lobular (n = 8) | 1.1 | 0.4 | −2.3 | 1 | −1.2 | 0.9 | −1.2 | 00.8 | 1.2 | 0.3 | 1.1 | 0.3 | 1 | 0.4 | |
| Ginester24 | Ductal (n = 45) | −3.8 | 1 | −1.3 | 0.8 | −1.3 | 0.74 | 1 | 0.23 | −3.2 | 1 | 1.2 | 0.84 | −1.1 | 0.7 |
| Medullary (n = 5) | 8.5 | 0.004 | 1.8 | 0.07 | 1.8 | 0.13 | 1 | 0.335 | 6 | 0.012 | 1.2 | 0.11 | 1.2 | 0.08 | |
| Lu26 | Ductal (n = 95) | 1.2 | 0.13 | 1.4 | 0.04 | 1.1 | 0.2 | — | — | 3.5 | 9.9e −5 | 1 | 0.5 | −1 | 0.5 |
| Lobular (n = 19) | −1.2 | 0.8 | −1.3 | 0.9 | −1.1 | 0.8 | — | — | −2.7 | 1 | 1.3 | 0.2 | −1 | 0.5 | |
| MA281 | Ductal (n = 32) | −1.6 | 0.9 | 1 | 0.3 | −1.1 | 0.8 | 1.1 | 0.14 | −1.1 | 0.6 | 1.1 | 0.4 | −1.1 | 0.7 |
| Lobular (n = 3) | 3.7 | 0.06 | −1 | 0.8 | −1.1 | 0.7 | −1 | 0.7 | −1.1 | 0.8 | −1.1 | 0.6 | −1 | 0.6 | |
| MA381 | Ductal (n = 51) | −1.2 | 0.7 | 1 | 0.4 | −1.2 | 0.8 | 1.1 | 0.3 | −1.1 | 0.6 | −1.2 | 0.9 | 1 | 0.4 |
| Lobular (n = 3) | 1.8 | 0.2 | 1 | 0.3 | 1.4 | 0.04 | 1.3 | 0.2 | −1.1 | 0.6 | 1.9 | 0.1 | 1.1 | 0.4 | |
| MA482 | Ductal (n = 18) | −1 | 0.61 | — | — | −1.1 | 0.8 | −1.1 | 1 | −1.2 | 0.6 | −1.2 | 1 | −1 | 0.5 |
| Ductal in situ (n = 20) | 1.9 | 0.04 | — | — | 1.1 | 0.2 | 1.1 | 0.04 | 1.2 | 0.4 | 1.2 | 0.07 | 1 | 0.5 | |
| Perou27 | Ductal (n = 54) | 1.1 | 0.4 | −1 | 0.5 | 1 | 0.4 | — | — | — | — | — | — | −1.7 | 1 |
| Lobular (n = 4) | −1 | 0.7 | 1 | 0.5 | −1 | 0.6 | — | — | — | — | — | — | 1.7 | 0.03 | |
| Pollack83 | Ductal (n = 105) | −1.1 | 0.63 | 1.6 | 0.1 | 1.3 | 0.3 | — | — | — | — | — | — | −2.5 | 0.8 |
| Lobular (n = 8) | 1.1 | 0.4 | −1.6 | 0.9 | −1.3 | 0.8 | — | — | — | — | — | — | 2.5 | 0.2 | |
| Radvanyi25 | Ductal (n = 26) | — | — | — | — | 2.8 | 1e−4 | 1.3 | 0.3 | 1.7 | 0.4 | 1.3 | 0.2 | −1.3 | 0.7 |
| Lobular (n = 4) | — | — | — | — | −1.7 | 0.9 | 1.5 | 0.2 | −1.1 | 0.8 | 1.9 | 0.04 | 1.1 | 0.3 | |
| Ductal in situ (n = 2) | — | — | — | — | −2.8 | 1 | −1.3 | 0.7 | −1.7 | 0.6 | 1.8 | 0.1 | 1.3 | 0.3 | |
| Schuetz84 | Ductal (n = 5) | 1.3 | 0.25 | — | — | 1.2 | 0.03 | −1.1 | 0.9 | −1.4 | 1 | −1.1 | 0.9 | 1 | 0.4 |
| Ductal in situ (n = 5) | −1.3 | 0.73 | — | — | −1.2 | 1 | 1.1 | 0.15 | 1.4 | 0.02 | 1.1 | 0.15 | −1 | 0.6 | |
| Sorlie85 | Ductal (n = 66) | 1.5 | 0.1 | 1.2 | 0.2 | 1.5 | 0.08 | — | — | — | — | — | — | −2 | 0.9 |
| Lobular (n = 5) | −1.2 | 0.6 | −1.2 | 0.8 | −1.5 | 0.83 | — | — | — | — | — | — | 2.8 | 0.2 | |
| Sorlie286 | Ductal (n = 132) | 1.2 | 0.3 | — | — | 1.4 | 0.05 | — | — | — | — | — | — | — | — |
| Lobular (n = 11) | −1 | 0.5 | — | — | −1.5 | 0.92 | — | — | — | — | — | — | — | — | |
| Tabchy87 | Ductal (n = 163) | −1.3 | 0.9 | 1.1 | 0.12 | −1 | 0.8 | −1 | 0.81 | 1.2 | 0.2 | −1.1 | 0.9 | −1.1 | 0.9 |
| Lobular (n = 7) | 1.5 | 0.11 | −1 | 0.8 | 1.1 | 0.09 | 1.1 | 0.3 | −1.2 | 0.7 | 1.1 | 0.1 | 1.2 | 0.1 | |
| TCGA | Ductal (n = 392) | 1.7 | 0.2 | 2.1 | 2.5e−6 | −2.4 | 0.8 | 1.5 | 0.2 | −1.4 | 0.7 | −1.1 | 0.6 | 1.2 | 0.2 |
| Lobular (n = 36) | 1.4 | 0.04 | −1.8 | 1 | −1 | 0.7 | 1.3 | 0.03 | −1.1 | 0.8 | 1 | 0.4 | −1 | 0.5 | |
| Ductal in situ (n = 3) | — | — | — | — | 2.1 | 0.1 | −1.3 | 0.7 | 2 | 0.2 | 2.6 | 0.1 | 1.4 | 0.02 | |
| Turashvili88 | Ductal (n = 5) | 1.5 | 0.3 | −1.6 | 0.8 | 2 | 0.2 | −1 | 0.5 | −1.1 | 0.6 | 1.1 | 0.4 | −1 | 0.5 |
| Lobular (n = 5) | −1.1 | 0.5 | 1.6 | 0.2 | −2 | 0.8 | 1 | 0.5 | 1.1 | 0.4 | −1.1 | 0.5 | 1 | 0.5 | |
| Zhao89 | Ductal (n = 39) | 1.3 | 0.01 | −1.2 | 0.81 | −1.1 | 0.6 | — | — | — | — | 1 | 0.3 | — | — |
| Lobular (n = 21) | −1.1 | 0.7 | 1.2 | 0.2 | 1.1 | 0.4 | — | — | — | — | −1 | 0.7 | — | — | |
Histological types of 7 CC chemokines over-expressed in BrCa compared to normal tissues were determined utilizing all datasets available in ONCOMINE.
Bold represents significant RNA expression with a cut off of: fold change (FC) ≥ 1.4 and p-value ≤ 0.04 using t-test.
CC Chemokine expression affects BrCa patient prognosis
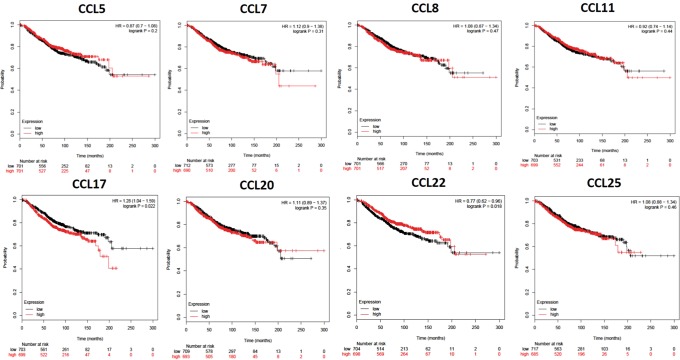
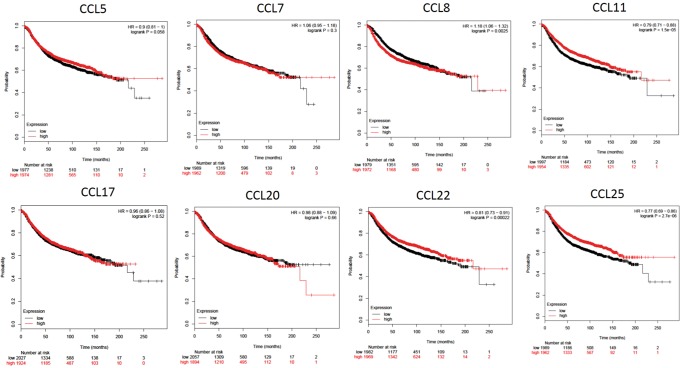
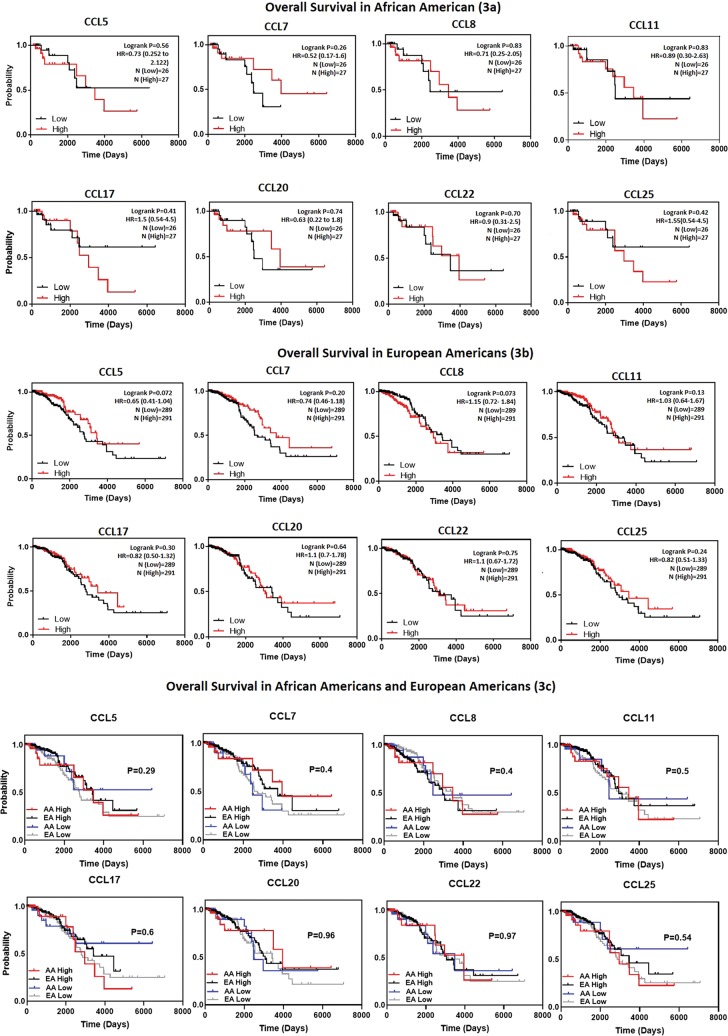
To elucidate the prognostic significance of all CC chemokines in BrCa in regards to metastatic relapse the Bc-GenExMiner-v4.1 online database was used (Table 3). Significance (p < 0.04) was seen in CCL4 (p = 0.004, HR = 0.91), CCL5 (p = 0.034, HR = 0.94), CCL8 (p = 0.0017, HR = 1.1), CCL19 (p = 0.001, HR = 0.9), CCL21 (p = < 0.0001, HR = 0.9), CCL22 (p = < 0.0001, HR = 0.9) and CCL23 (p = 0.0014, HR = 0.9). CCL8 was the only one significantly associated with an increase in metastatic relapse (HR > 1.0). Next, we determined the association of CC chemokines, which were higher in BrCa (CCL5, -7, -11, -17, -20, -22 and -25) compared to normal tissues, with OS and relapse free survival (RFS) (Figs. 1 and 2) using KMplotter. We also included CCL8 due to its association with increased metastatic relapse. A trend towards a decrease in OS (higher expression, red line) was seen in CCL7 (p = 0.31, HR = 1.12), CCL8 (p = 0.47, HR = 1.06), CCL20 (p = 0.3, HR = 1.11), CCL25 (p = 0.45, HR = 1.08) and significantly in CCL17 (p = 0.022, HR = 1.28). On the other hand, CCL5 (p = 0.2, HR = 0.87), CCL11 (p = 0.44, HR = 0.82) and CCL22 (p = 0.016, HR = 0.77) were correlated with an increase in OS overall survival but the correlation was found to be significant only with CCL22. Decreased RFS was associated with CCL7 (p = 0.3, HR = 1.05). Interestingly CCL8 expression, which was not significantly elevated in BrCa tissues compared to normal tissues, showed an association with decreased RFS (p = 0.0025, HR = 1.18). Increased CCL11 (p = 1.5e−05, HR = 0.77), CCL22 (p = 0.0002, HR = 0.81) and CCL25 (p = 2.7e−06, HR = 0.77) expression were associated with an increase in RFS. We also determined the effect of CC chemokine expression on OS based on race (Fig. 3a (AA), 3b (EA) and 3c (AA vs. EA)). When CC chemokines were analyzed among these races, higher expression of CCL17 (p = 0.41, HR = 1.5) and CCL25 (p = 0.42, HR = 1.55) were associated with decreased OS in AA, whereas CCL8 overexpression was associated with decreased OS in EA (p = 0.073, HR = 1.15). To the contrary, high CCL7 (p = 0.26, HR = 0.52), CCL11 (p = 0.83, HR = 0.89) and CCL20 (p = 0.74, HR = 0.63) expression in AA were associated with increased OS. However, higher CCL5 (p = 0.072, HR = 0.65), CCL7 (p = 0.20, HR = 0.74) and CCL17 (p = 0.30, HR = 0.82) were associated with better prognosis (increase in OS) is EA women. It is important to note that CCL25 that is associated with poor prognosis (decreased OS) in AA (p = 0.42, HR = 1.55), was an indicator of better prognosis in EA (p = 0.24, HR = 0.82). When comparing association of CC chemokine expression among races (Fig. 3c), high expression of CCL17 (p = 0.6) and CCL25 (p = 0.54) in AA was associated with decreased OS however no such association of CCL17 and CCL25 expression was observed in EA. Lastly, high expression of CCL8 was associated with decreased OS in EA but not in AA (p = 0.4).
Table 3.
Univariate Cox analysis of prognostic association of all 24 CC-Chemokine’s expression on metastatic relapse in BrCa based on the bc-GenExMiner v4.1 DNA gene chip database.
| Chemokine | p value | HR | 95% CI |
|---|---|---|---|
| CCL1 | 0.4894 | 0.98 | 0.92–1.04 |
| CCL2 | 0.6384 | 1.01 | 0.96–1.08 |
| CCL3 | 0.0785 | 0.91 | 0.83–1.01 |
| CCL4 | 0.0038 | 0.91 | 0.86–0.97 |
| CCL5 | 0.0335 | 0.94 | 0.88–0.99 |
| CCL7 | 0.173 | 1.05 | 0.98–1.11 |
| CCL8 | 0.0017 | 1.1 | 1.04–1.17 |
| CCL11 | 0.4707 | 0.98 | 0.92–1.04 |
| CCL13 | 0.9444 | 1 | 0.93–1.07 |
| CCL14 | 0.5541 | 0.94 | 0.76–1.15 |
| CCL15 | 0.1379 | 1.2 | 0.94–1.52 |
| CCL16 | 0.6384 | 1.01 | 0.96–1.08 |
| CCL17 | 0.8037 | 0.99 | 0.93–1.06 |
| CCL18 | 0.6961 | 1.01 | 0.95–1.08 |
| CCL19 | 0.001 | 0.9 | 0.85–0.96 |
| CCL20 | 0.8989 | 1 | 0.94–1.07 |
| CCL21 | <0.0001 | 0.88 | 0.82–0.93 |
| CCL22 | <0.0001 | 0.87 | 0.82–0.93 |
| CCL23 | 0.0014 | 0.88 | 0.81–0.95 |
| CCL24 | 0.9997 | 1 | 0.94–1.07 |
| CCL25 | 0.1064 | 1.05 | 0.99–1.12 |
HR-Hazard ratio. 95% CI- confidence interval. Bold represents a p value of ≤0.04.
Figure 1.
Kaplan-Meier curve of overall survival based on KMplotter mRNA gene chip expression of CC chemokines in BrCa patients. Red line represents higher expression and black line represents lower expression.
Figure 2.
Kaplan-Meier Curve of relapse free survival based on KMPlotter mRNA gene chip expression of CC chemokines in BrCa patients. Red line represents higher expression and black line represents lower expression.
Figure 3.
Kaplan-Meier curve of overall survival based on race using TCGA. In Panel a (AA) and Panel b (EA) red line represents higher expression and black line represents lower expression. Panel c shows AA vs. EA comparison. Red represents high expression in AA, black shows high expression in EA, blue represents low expression in AA and gray shows low expression in EA. Log-rank (Mantel-Cox) test was used to determine p-values and hazard ratio (HR).
Association of CC chemokine expression with clinical parameters in BrCa patients
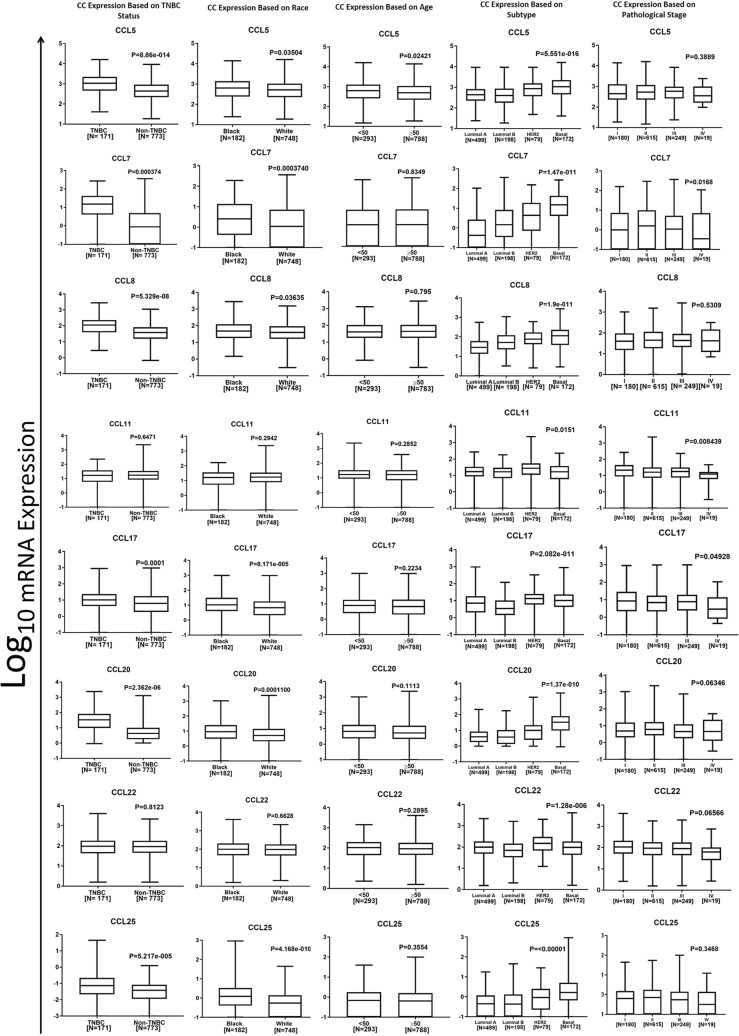
Using TCGA data, we determined the association of CC chemokine expression with clinical parameters of BrCa patients (Fig. 4). Expression of CCL5 (p = 8.86e−14), CCL7 (p = 0.004), CCL8 (p = 5.33e−08), CCL17 (p = 0.001), CCL20 (p = 2.36e−06) and CCL25 (p = 5.22e−05) were highest in TNBC tissues compared to non-TNBC tissues. Additionally, expression of chemokines i.e. CCL5 (p = 5.55e−016), CCL7 (p = 1.47e−011), CCL8 (p = 1.9e−11), CCL20 (p = 1.37e−010), and CCL25 (p = < 0.00001) were highest in the basal BrCa subtype, when compared to luminal A/B and HER2 status. In HER2 positive BrCa tissues expression of CCL11 (p = 0.015) and CCL22 (p = 1.28e−06) were significantly higher. Out of the CC chemokines we have analyzed, CCL5 expression was the only one associated with age, where there was higher expression at a younger age (p = 0.024). Furthermore, CCL7 (p = 0.0004), CCL17 (p = 8.17e−005), CCL20 (p = 0.0001) and CCL25 (p = 4.2e-01), were significantly elevated in AA BrCa tissue compared to EA tissues. CCL8 (p = 0.04) and CCL5 (p = 0.04) expression in AA tissues were marginally higher in comparison to EA. Additionally, higher expression of CCL7 (p = 0.017), CCL8 (p = 0.53) and CCL25 (p = 0.35) showed a trend towards an increase in stage (from I to III). Such association was not seen with stage-IV likely due to very small sample size in this category.
Figure 4.
CC chemokines’ mRNA expression based on clinical parameters using TCGA. Box and whisker plots represent minimum expression at the bottom whisker, maximum at the top whisker and median at the middle line with a Log10 axis scale showing mRNA epxression of 8 CC chemokines. Statistical analysis was conducted with non-parametric Mann-Whitney for t-test and global significance for ANOVA with Kruskal Wallis.
Discussion
Chemokines are predominantly known for their roles in immune cell trafficking and shaping the immune system. The role of chemokines and their corresponding receptors are well appreciated in cancer progression and metastasis28. Among all known chemokines, CXC chemokines are highly appreciated in cancer compared to CC chemokines. However, the CC chemokines are the largest family of chemokines and play an important role in inflammation29. Our laboratory was first to show association of CC chemokine receptor-9 in prostate, breast, ovarian and lung cancer10,18–20.
Studies have shown BrCa growth inhibition, after blocking CCR5 and CCR1, which are natural receptors for CCL5 and CCL730. Chemokine receptors CCR1 and CCR5 are expressed on monocytes and facilitate their recruitment under chemotactic gradient of CCL5 and CCL731 and play a crucial role in tumor progression. Our analysis showing higher CCL5 and CCL7 in BrCa tissues compared to normal tissues indicates that BrCa cells which produce high levels of CCL5 and CCL7 could be responsible for recruiting monocytes in the tumor microenvironment and supporting BrCa progression. In addition, production of CCL5 from mesenchymal stem cells (MSCs) results in BrCa production of colony-stimulating factor 1 (CSF1) under hypoxic conditions32. This causes the recruitment of myeloid-derived suppressor cells (MDSC) and tumor associated macrophages (TAM) to the tumor microenvironment32. Studies have shown that higher CCL5 promotes BrCa metastasis33. Our analysis also shows higher levels of CCL5 in medullary and ductal carcinoma and in TNBC compared to non-TNBC cases. Higher CCL5 may be involved in developing tumor tolerance resulting in the poor TNBC prognosis. As observed in our analysis, others have shown higher CCL5 in TNBC34. Non-remissive and later stage BrCa was reported to be correlated with CCL5 expression35,36, possibly due to its ability to promote pro-invasive factor MMP9 and monocyte migration to the BrCa tumor site, in which they undergo polarization allowing them to support tumor progression through angiogenesis37. This is not to our surprise as CCL5 was elevated in AA patients who often develop aggressive forms of BrCa. Furthermore, antagonizing CCR5 decreases CCL5 induced angiogenesis of BrCa cells38. CCL5 has been shown to promote BrCa progression in a p53 dependent manner through CCR539. Interestingly, endothelial cells have been shown to increase metastasis of TNBC cells through secretion of PAI-1 and CCL540. Hence our analysis showing higher CCL5 in BrCa (medullary and ductal carcinoma) compared to normal, higher CCL5 in TNBC compared to non-TNBC and higher CCL5 in AA compared to EA, suggests its potential contribution in shaping the tumor favoring microenvironment.
Our analysis also shows higher CCL7 in ductal breast carcinoma compared to normal tissues and other histological tissue types. It was also elevated in AA in comparison to EA patients. CCL7, that can bind and activate CCR1, CCR2 and CCR3, has been shown to promote metastasis by activating the MAPK cascade, promoting epithelial-mesenchymal transition (EMT) and CCR2+ TAM recruitment (enhancing vascular permeability41). Cancer associated fibroblast (CAF) derived CCL7 promotes BrCa proliferation42. Interestingly, CCL8 also known as monocyte chemo-attractant protein-2 (MCP-2), a natural ligand shared by CCR2, CCR3 and CCR5 was not elevated in BrCa tissues when compared to normal tissues, but was associated with poorer OS and RFS. It has been shown to drive BrCa metastasis. More specifically CCL8 stimulates fibroblasts generating a pro-tumor environment in the TNBC stroma, which was not seen in non-TNBC43. This was not to our surprise, as we found significantly higher CCL8 in TNBC tissues compared to non-TNBC tissues and it was associated with increased metastatic relapse. More importantly, when compared between AA and EA women, CCL8 was found higher in AA patients, who are frequently diagnosed with TNBC compared to EA. In contrast to its classical contribution on biology and outcome of BrCa, CCL8 was associated with poor OS in EA, suggesting race specific differences in CCL8 biology and immunity against BrCa.
Using tissue data we found that CCL11 (Eotaxin), which has a high affinity for CCR3, was higher in ductal breast carcinoma tissues when compared to healthy controls and other histological tissue types. Furthermore, a supporting study found that CCL11 was higher in the serum of BrCa patients when compared to serum of healthy individuals44. Bone colonization of BrCa cells is promoted by CCL1145, this is important to note as nearly 70% of BrCa cases show bone metastasis46. Co-culture studies showed an increase in CCL11 secretion by fibroblasts that enhanced chemoresistance and metastasis of BrCa cells47. In an allergic inflammation study, CCR3 antagonism prohibited chemotaxis of basophils and eosinophils48. Eosinophil degranulation has been reported in BrCa49, however this anti-tumor response is not quite understood. In pancreatic cancer basophil recruitment into tumor draining lymph nodes correlates with inflammation and poor survival50. Moreover, in hypoxic conditions tumor cells have been shown to secrete CCL11, recruiting CD206 expressing macrophages to the tumor, which in turn polarizes the classical macrophages to a M2 pro-angiogenic phenotype51. Clearly these facts and our data emphasize the need to study CCR3-CCL11 axis with respect to BrCa.
Furthermore CCR4 (receptor for CCL17 and CCL22) antagonists have been shown to decrease the tumor-promoting environment52. CCL17 recruits CCR4 positive regulatory T cells (Tregs) and promotes lung metastasis of BrCa by elimination of NK cells53,54. This coincided with our findings that higher CCL17 is expressed in BrCa and associated with decreased OS. Our data also shows CCL17 is overexpressed and is associated with poor prognosis in AA patients unlike EA. Its expression is higher in TNBC patients compared to non-TNBC patients. Moreover, CCR4 positively correlates with HER2 expression is BrCa cells54, supporting our result of higher expression of CCL22 in HER2 expressing tissues, when compared to basal and luminal types. We also found CCL22 to be significantly higher in BrCa (lobular) tissues compared to normal tissues, which agrees with a study reporting circulating CCL22 levels to be significantly higher in BrCa patients when compared to healthy controls55. As a prognostic factor in BrCa, tumor derived CCL22 has also been shown to activate and recruit CCR4 expressing Tregs56,57. Moreover, in prostate cancer, TAM promote tumor cell migration by activating CCL22-CCR4 signaling58. Additionally this axis has been shown to promote bone metastasis of lung cancer59. In contrast, our results showed that high CCL22 expression was associated with increased OS and RFS in BrCa.
The ligand for CCR6, CCL20 was higher in ductal and medullary breast carcinoma when compared to normal controls and other histological tissue types. Moreover, BrCa cells secrete CCL20, which recruits CCR6 expressing immune cells in the tumor vicinity60. Tumor cell produced CCL20/MIP3α allows them to attract CCR6 positive dendritic cell precursors61. Furthermore, CCL20 promotes migration and proliferation of surrounding breast cells62 and promotes BrCa initiation63. This could explain our finding showing higher expression of CCL20 in earlier stage I and stage II. Through paracrine signaling CCL20 promotes EMT in BrCa cells64. Studies have shown involvement of CCL20 in bone65 and lung8 metastasis of BrCa. We saw that higher expression of CCL20 associates with decreased OS. Moreover, expression of CCL20 was higher in AA with BrCa when compared to EA and in TNBC when compared to non-TNBC status. Chemotherapy has been shown to trigger higher production of CCL20 which in turn contributes to chemoresistance through ABCB1 drug efflux pump66, further suggesting involvement of CCL20 in disparities in therapeutic outcome.
CCR9 is selectively expressed on T cells and it’s only natural ligand (CCL25) is expressed in the thymus and small intestines67. CCR9 is often expressed on immature double (CD4+ and CD8+) positive thymocytes68,69, but has been reported to be expressed on thymocytes of all stages70. Circulating CCR9/CD4+ T cells exhibit a Th1 cytokine profile71. We found that CCL25 expression was associated with increased RFS when looking at all BrCa patients. We previously reported that CCL25 promotes migration/invasion20 and chemo-resistance21 in BrCa. Another study confirmed its role in BrCa and liver cancer migration and invasion72. Our current analysis further confirms our previous studies showing higher expression of CCL25 in BrCa compared to normal controls. Additionally, our data shows higher levels of CCL25 in TNBC compared to non-TNBC status, AA women compared to EA women and that it is associated with decreased OS in AA. Additionally, there was a trend of increased expression of CCL25 from stage I to stage III. Hence our current analysis explains a potential role of CCR9-CCL25 in disparities associated with poor overall outcome of BrCa in AA women.
It has been shown that chemokine receptors alone do not play a role in disease free or OS in BrCa8 (which we confirmed using Kmplotter, data not shown), suggesting the importance of their ligands. To our knowledge, this manuscript is the first to consolidate patient tissue data on CC chemokine ligands in BrCa and show that CC chemokine ligands are associated with disease free and OS. This suggests that chemokines expressed in the BrCa tissues not only support cancer cells expressing their corresponding receptors, but also shapes tumor microenvironment to overcome immune attack. However, these results should be confirmed and used with caution due to the fact that there are many factors downstream that could alter the effects of the variations seen at the mRNA level. In conclusion, identifying the role of chemokines in BrCa patients showing prominent expression could provide patient or race specific immunotherapeutic targets.
Materials and Methods
Oncomine
ONCOMINE73, online database (www.oncomine.org) of RNA/DNA sequencing data was used to screen all 24- CC chemokines’ RNA expression in BrCa tissues in comparison to normal tissues using the TCGA dataset (Table 1). Significant overexpression in BrCa tissues compared to normal tissues was determined using a t-test, utilizing a cut off of: fold change ≥1.4 and p-value ≤ 0.04. Additionally, these cut offs were used to classify significance for all downstream analysis. Further elucidation of histological type of chemokines significantly expressed in BrCa (compared to normal tissues) was conducted with ONCOMINE utilizing all available datasets (Table 2). Using the referenced cutoffs, t-test were conducted comparing BrCa histological tissue types within each dataset.
Bc-GenExMiner-v4.1
Microarray DNA expression results from BrCa Gene-Expression Miner (bc-GenExMiner)74 were used to classify prognostic association of metastatic relapse in BrCa of all 24 CC chemokines using univariate cox analysis (Table 3).
Kaplan Meier Plotter-Breast Cancer
Association of CC chemokines on overall (Fig. 1) and relapse free (Fig. 2) survival was determined using Kaplan Meier (KM) Plotter (http://kmplot.com/analysis/)75,76. The two groups for plotting were high (red line) or low (black line) based on median mRNA gene chip expression.
cBioPortal and Analysis
The cBio Cancer Genomics Portal (http://cbioportal.org)77 open-access database was used to download The Cancer Genome Atlas (TCGA) Pan Cancer Atlas dataset (RNASeq V2 RSEM) of 1084 BrCa patients (normalization and batch correction description here: https://www.synapse.org/#!Synapse:syn4976363). Graphpad Prism was used to produce Kaplan-Meir OS curves based on race (AA (Fig. 3a) or EA (Fig. 3b)), whereas red represents high expression and black low expression based on expression median. For AA vs. EA (Fig. 3c) survival curves red represents high expression in AA, black is high expression in EA, blue is low expression in AA and gray is low expression in EA. Log-rank (Mantel-Cox) test was used to determine p-values and hazard ratio. Expression based on clinical parameters was graphed as Log10 axis scale using box and whisker plots with whiskers at the minimum and maximum values and the middle line as the median (Fig. 4). Statistical analysis was conducted with non-parametric Mann-Whitney for t-test and global significance for ANOVA with Kruskal Wallis (Graphpad Prism7, La Jolla, CA). All statistics were thoroughly confirmed and verified by a statistician using SAS.
Acknowledgements
The content of this manuscript benefited from many fruitful conversations with members of the Morehouse School of Medicine, Atlanta, GA, USA. This study was supported in part by the funds (R21 CA169716, SC1 CA180212, U54 CA118638, and U01 CA 179701) from the National Cancer Institute (NCI) and also by a RISE grant (GM05868) funded by the National Institute of General Medical Sciences, National Institute of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author Contributions
J.K.T. designed the study, conducted analysis and drafted the manuscript. H.M. and N.K. helped with manuscript preparations. S.S. conceptualized the study, supervised the authors throughout the study, and provided expertise in manuscript preparation. S.B. conducted statistical verification. All authors read and approved the final manuscript.
Data Availability
All data generated or analyzed during this study are included in this published article.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: A Cancer Journal for Clinicians. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Dignam JJ, Mamounas EP. Obesity and breast cancer prognosis: an expanding body of evidence. Annals of oncology: official journal of the European Society for Medical Oncology. 2004;15:850–851. doi: 10.1093/annonc/mdh241. [DOI] [PubMed] [Google Scholar]
- 3.Lehmann BD, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. The Journal of Clinical Investigation. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitman GJ, Albarracin CT, Gonzalez-Angulo AM. Triple-negative breast cancer: what the radiologist needs to know. Seminars in roentgenology. 2011;46:26–39. doi: 10.1053/j.ro.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 5.Ferlay J, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 6.DeNardo DG, Coussens LM. Inflammation and breast cancer. Balancing immune response: crosstalk between adaptive and innate immune cells during breast cancer progression. Breast cancer research: BCR. 2007;9:212. doi: 10.1186/bcr1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagarajan, D. & McArdle, S. E. B. Immune Landscape of Breast Cancers. Biomedicines6, 10.3390/biomedicines6010020 (2018). [DOI] [PMC free article] [PubMed]
- 8.Andre F, et al. Expression of chemokine receptors predicts the site of metastatic relapse in patients with axillary node positive primary breast cancer. Annals of oncology: official journal of the European Society for Medical Oncology. 2006;17:945–951. doi: 10.1093/annonc/mdl053. [DOI] [PubMed] [Google Scholar]
- 9.Muller A, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 10.Singh, S., Singh, U. P., Stiles, J. K., Grizzle, W. E. & Lillard, J. W. Jr. Expression and functional role of CCR9 in prostate cancer cell migration and invasion. Clinical cancer research: an official journal of the American Association for Cancer Research10, 8743–8750, doi:10.1158/1078-0432.ccr-04-0266 (2004). [DOI] [PubMed]
- 11.Singh S, et al. Clinical and biological significance of CXCR5 expressed by prostate cancer specimens and cell lines. International journal of cancer. 2009;125:2288–2295. doi: 10.1002/ijc.24574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh S, et al. Serum CXCL13 positively correlates with prostatic disease, prostate-specific antigen and mediates prostate cancer cell invasion, integrin clustering and cell adhesion. Cancer letters. 2009;283:29–35. doi: 10.1016/j.canlet.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El Haibi CP, et al. PI3Kp110-, Src-, FAK-dependent and DOCK2-independent migration and invasion of CXCL13-stimulated prostate cancer cells. Mol Cancer. 2010;9:85. doi: 10.1186/1476-4598-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Haibi CP, et al. Differential G protein subunit expression by prostate cancer cells and their interaction with CXCR5. Molecular Cancer. 2013;12:64. doi: 10.1186/1476-4598-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Haibi, C. P., Singh, R., Sharma, P. K., Singh, S. & Lillard, J. W. Jr. CXCL13 mediates prostate cancer cell proliferation through JNK signalling and invasion through ERK activation. Cell proliferation44, 311–319, doi:10.1111/j.1365-2184.2011.00757.x (2011). [DOI] [PMC free article] [PubMed]
- 16.Singh R, et al. CXCR6-CXCL16 axis promotes prostate cancer by mediating cytoskeleton rearrangement via Ezrin activation and alphavbeta3 integrin clustering. Oncotarget. 2016;7:7343–7353. doi: 10.18632/oncotarget.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma PK, et al. CCR9 mediates PI3K/AKT-dependent antiapoptotic signals in prostate cancer cells and inhibition of CCR9-CCL25 interaction enhances the cytotoxic effects of etoposide. International journal of cancer. 2010;127:2020–2030. doi: 10.1002/ijc.25219. [DOI] [PubMed] [Google Scholar]
- 18.Singh R, Stockard CR, Grizzle WE, Lillard JW, Jr., Singh S. Expression and histopathological correlation of CCR9 and CCL25 in ovarian cancer. International journal of oncology. 2011;39:373–381. doi: 10.3892/ijo.2011.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta P, et al. CCR9/CCL25 expression in non-small cell lung cancer correlates with aggressive disease and mediates key steps of metastasis. Oncotarget. 2014;5:10170–10179. doi: 10.18632/oncotarget.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson-Holiday C, et al. CCL25 mediates migration, invasion and matrix metalloproteinase expression by breast cancer cells in a CCR9-dependent fashion. International journal of oncology. 2011;38:1279–1285. doi: 10.3892/ijo.2011.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson-Holiday C, et al. CCR9-CCL25 interactions promote cisplatin resistance in breast cancer cell through Akt activation in a PI3K-dependent and FAK-independent fashion. World journal of surgical oncology. 2011;9:46. doi: 10.1186/1477-7819-9-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen E, et al. Identification of Potential Therapeutic Targets Among CXC Chemokines in Breast Tumor Microenvironment Using Integrative Bioinformatics Analysis. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2018;45:1731–1746. doi: 10.1159/000487782. [DOI] [PubMed] [Google Scholar]
- 23.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ginestier C, et al. Prognosis and gene expression profiling of 20q13-amplified breast cancers. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006;12:4533–4544. doi: 10.1158/1078-0432.ccr-05-2339. [DOI] [PubMed] [Google Scholar]
- 25.Radvanyi L, et al. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11005–11010. doi: 10.1073/pnas.0500904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu X, et al. Predicting features of breast cancer with gene expression patterns. Breast Cancer Res Treat. 2008;108:191–201. doi: 10.1007/s10549-007-9596-6. [DOI] [PubMed] [Google Scholar]
- 27.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 28.Singh R, Lillard JW, Jr., Singh S. Chemokines: key players in cancer progression and metastasis. Frontiers in bioscience (Scholar edition) 2011;3:1569–1582. doi: 10.2741/246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White GE, Iqbal AJ, Greaves DR. CC chemokine receptors and chronic inflammation–therapeutic opportunities and pharmacological challenges. Pharmacological reviews. 2013;65:47–89. doi: 10.1124/pr.111.005074. [DOI] [PubMed] [Google Scholar]
- 30.Robinson SC, et al. A chemokine receptor antagonist inhibits experimental breast tumor growth. Cancer research. 2003;63:8360–8365. [PubMed] [Google Scholar]
- 31.Weber C, et al. Specialized roles of the chemokine receptors CCR1 and CCR5 in the recruitment of monocytes and T(H)1-like/CD45RO(+) T cells. Blood. 2001;97:1144–1146. doi: 10.1182/blood.V97.4.1144. [DOI] [PubMed] [Google Scholar]
- 32.Chaturvedi P, Gilkes DM, Takano N, Semenza GL. Hypoxia-inducible factor-dependent signaling between triple-negative breast cancer cells and mesenchymal stem cells promotes macrophage recruitment. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E2120–2129. doi: 10.1073/pnas.1406655111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karnoub AE, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 34.Lv D, Zhang Y, Kim H-J, Zhang L, Ma X. CCL5 as a potential immunotherapeutic target in triple-negative breast cancer. Cellular And Molecular Immunology. 2013;10:303. doi: 10.1038/cmi.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niwa Y, et al. Correlation of tissue and plasma RANTES levels with disease course in patients with breast or cervical cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2001;7:285–289. [PubMed] [Google Scholar]
- 36.Luboshits G, et al. Elevated expression of the CC chemokine regulated on activation, normal T cell expressed and secreted (RANTES) in advanced breast carcinoma. Cancer research. 1999;59:4681–4687. [PubMed] [Google Scholar]
- 37.Azenshtein E, et al. The CC Chemokine RANTES in Breast Carcinoma Progression. Regulation of Expression and Potential Mechanisms of Promalignant Activity. 2002;62:1093–1102. [PubMed] [Google Scholar]
- 38.Sax MJ, et al. Cancer cell CCL5 mediates bone marrow independent angiogenesis in breast cancer. Oncotarget. 2016;7:85437–85449. doi: 10.18632/oncotarget.13387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manes S, et al. CCR5 expression influences the progression of human breast cancer in a p53-dependent manner. The Journal of experimental medicine. 2003;198:1381–1389. doi: 10.1084/jem.20030580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang W, et al. Endothelial cells promote triple-negative breast cancer cell metastasis via PAI-1 and CCL5 signaling. The FASEB Journal. 2018;32:276–288. doi: 10.1096/fj.201700237RR. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, Cai Y, Liu L, Wu Y, Xiong X. Crucial biological functions of CCL7 in cancer. PeerJ. 2018;6:e4928. doi: 10.7717/peerj.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rajaram M, Li J, Egeblad M, Powers RS. System-wide analysis reveals a complex network of tumor-fibroblast interactions involved in tumorigenicity. PLoS genetics. 2013;9:e1003789. doi: 10.1371/journal.pgen.1003789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farmaki E, Chatzistamou I, Kaza V, Kiaris H. A CCL8 gradient drives breast cancer cell dissemination. Oncogene. 2016;35:6309–6318. doi: 10.1038/onc.2016.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anber N, EL-Sebaie AH, Mousa SA, Elbaz O. Expression of Some Inflammatory Biomarkers in Patients with Breast Cancer. International Journal of Health Sciences and Research. 2016;6:172–181. [Google Scholar]
- 45.Sosnoski DM, Krishnan V, Kraemer WJ, Dunn-Lewis C, Mastro AM. Changes in Cytokines of the Bone Microenvironment during Breast Cancer Metastasis. International Journal of Breast Cancer. 2012;2012:9. doi: 10.1155/2012/160265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Irshad, I. & Varamini, P. Different Targeting Strategies For Treating Breast Cancer Bone Metastases. Current pharmaceutical design, 10.2174/1381612824666180619165728 (2018). [DOI] [PubMed]
- 47.Liu Y, Zhang J, Sun X, Su Q, You C. Down-regulation of miR-29b in carcinoma associated fibroblasts promotes cell growth and metastasis of breast cancer. Oncotarget. 2017;8:39559–39570. doi: 10.18632/oncotarget.17136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bryan SA, et al. Responses of Leukocytes to Chemokines in Whole Blood and Their Antagonism by Novel CC-Chemokine Receptor 3 Antagonists. American Journal of Respiratory and Critical Care Medicine. 2002;165:1602–1609. doi: 10.1164/rccm.200111-059OC. [DOI] [PubMed] [Google Scholar]
- 49.Samoszuk MK, Nguyen V, Gluzman I, Pham JH. Occult deposition of eosinophil peroxidase in a subset of human breast carcinomas. The American journal of pathology. 1996;148:701–706. [PMC free article] [PubMed] [Google Scholar]
- 50.De Monte L, et al. Basophil Recruitment into Tumor-Draining Lymph Nodes Correlates with Th2 Inflammation and Reduced Survival in Pancreatic Cancer Patients. Cancer research. 2016;76:1792–1803. doi: 10.1158/0008-5472.can-15-1801-t. [DOI] [PubMed] [Google Scholar]
- 51.Tripathi C, et al. Macrophages are recruited to hypoxic tumor areas and acquire a Pro-Angiogenic M2-Polarized phenotype via hypoxic cancer cell derived cytokines Oncostatin M and Eotaxin. Oncotarget. 2014;5:5350–5368. doi: 10.18632/oncotarget.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berlato C, et al. A CCR4 antagonist reverses the tumor-promoting microenvironment of renal cancer. J Clin Invest. 2017;127:801–813. doi: 10.1172/jci82976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olkhanud PB, et al. Breast cancer lung metastasis requires expression of chemokine receptor CCR4 and regulatory T cells. Cancer research. 2009;69:5996–6004. doi: 10.1158/0008-5472.can-08-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li JY, et al. The chemokine receptor CCR4 promotes tumor growth and lung metastasis in breast cancer. Breast Cancer Res Treat. 2012;131:837–848. doi: 10.1007/s10549-011-1502-6. [DOI] [PubMed] [Google Scholar]
- 55.Jafarzadeh A, et al. Higher circulating levels of chemokine CCL22 in patients with breast cancer: evaluation of the influences of tumor stage and chemokine gene polymorphism. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:1163–1171. doi: 10.1007/s13277-014-2739-6. [DOI] [PubMed] [Google Scholar]
- 56.Li YQ, et al. Tumor secretion of CCL22 activates intratumoral Treg infiltration and is independent prognostic predictor of breast cancer. PloS one. 2013;8:e76379. doi: 10.1371/journal.pone.0076379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gobert M, et al. Regulatory T Cells Recruited through CCL22/CCR4 Are Selectively Activated in Lymphoid Infiltrates Surrounding Primary Breast Tumors and Lead to an Adverse Clinical Outcome. Cancer research. 2009;69:2000–2009. doi: 10.1158/0008-5472.can-08-2360. [DOI] [PubMed] [Google Scholar]
- 58.Maolake A, et al. Tumor-associated macrophages promote prostate cancer migration through activation of the CCL22-CCR4 axis. Oncotarget. 2017;8:9739–9751. doi: 10.18632/oncotarget.14185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakamura ES, et al. RANKL-induced CCL22/macrophage-derived chemokine produced from osteoclasts potentially promotes the bone metastasis of lung cancer expressing its receptor CCR4. Clinical & Experimental Metastasis. 2006;23:9–18. doi: 10.1007/s10585-006-9006-1. [DOI] [PubMed] [Google Scholar]
- 60.Osuala KO, Sloane BF. Many Roles of CCL20: Emphasis on Breast Cancer. Postdoc journal: a journal of postdoctoral research and postdoctoral affairs. 2014;2:7–16. [PMC free article] [PubMed] [Google Scholar]
- 61.Thomachot MC, et al. Breast carcinoma cells promote the differentiation of CD34+ progenitors towards 2 different subpopulations of dendritic cells with CD1a(high)CD86(−)Langerin- and CD1a(+)CD86(+)Langerin+ phenotypes. International journal of cancer. 2004;110:710–720. doi: 10.1002/ijc.20146. [DOI] [PubMed] [Google Scholar]
- 62.Marsigliante S, Vetrugno C, Muscella A. CCL20 induces migration and proliferation on breast epithelial cells. J Cell Physiol. 2013;228:1873–1883. doi: 10.1002/jcp.24349. [DOI] [PubMed] [Google Scholar]
- 63.Boyle ST, Faulkner JW, Mcoll SR, Kochetkova M. The chemokine receptor CCR6 facilitates the onset of mammary neoplasia in the MMTV-PyMT mouse model via recruitment of tumor-promoting macrophages. Molecular Cancer. 2015;14:115. doi: 10.1186/s12943-015-0394-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marsigliante S, Vetrugno C, Muscella A. Paracrine CCL20 loop induces epithelial-mesenchymal transition in breast epithelial cells. Molecular carcinogenesis. 2016;55:1175–1186. doi: 10.1002/mc.22360. [DOI] [PubMed] [Google Scholar]
- 65.Lee SK, et al. Human antigen R-regulated CCL20 contributes to osteolytic breast cancer bone metastasis. Scientific Reports. 2017;7:9610. doi: 10.1038/s41598-017-09040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen W, et al. CCL20 triggered by chemotherapy hinders the therapeutic efficacy of breast cancer. PLoS biology. 2018;16:e2005869. doi: 10.1371/journal.pbio.2005869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Uehara S, Grinberg A, Farber JM, Love PE. A Role for CCR9 in T Lymphocyte Development and Migration. The Journal of Immunology. 2002;168:2811–2819. doi: 10.4049/jimmunol.168.6.2811. [DOI] [PubMed] [Google Scholar]
- 68.Uehara S, Song K, Farber JM, Love PE. Characterization of CCR9 expression and CCL25/thymus-expressed chemokine responsiveness during T cell development: CD3(high)CD69+ thymocytes and gammadeltaTCR+ thymocytes preferentially respond to CCL25. Journal of immunology (Baltimore, Md.: 1950) 2002;168:134–142. doi: 10.4049/jimmunol.168.1.134. [DOI] [PubMed] [Google Scholar]
- 69.Wurbel MA, et al. The chemokine TECK is expressed by thymic and intestinal epithelial cells and attracts double- and single-positive thymocytes expressing the TECK receptor CCR9. European journal of immunology. 2000;30:262–271. doi: 10.1002/1521-4141(200001)30:1<262::aid-immu262>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 70.Carramolino L, et al. Expression of CCR9 β-chemokine receptor is modulated in thymocyte differentiation and is selectively maintained in CD8+ T cells from secondary lymphoid organs. Blood. 2001;97:850–857. doi: 10.1182/blood.V97.4.850. [DOI] [PubMed] [Google Scholar]
- 71.Papadakis KA, et al. CC Chemokine Receptor 9 Expression Defines a Subset of Peripheral Blood Lymphocytes with Mucosal T Cell Phenotype and Th1 or T-Regulatory 1 Cytokine Profile. The Journal of Immunology. 2003;171:159–165. doi: 10.4049/jimmunol.171.1.159. [DOI] [PubMed] [Google Scholar]
- 72.Zhang Z, et al. CCL25/CCR9 Signal Promotes Migration and Invasion in Hepatocellular and Breast Cancer Cell Lines. DNA and cell biology. 2016;35:348–357. doi: 10.1089/dna.2015.3104. [DOI] [PubMed] [Google Scholar]
- 73.Rhodes DR, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia (New York, N.Y.) 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jézéquel P, et al. bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Research and Treatment. 2012;131:765–775. doi: 10.1007/s10549-011-1457-7. [DOI] [PubMed] [Google Scholar]
- 75.Gyorffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 76.Lanczky A, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160:439–446. doi: 10.1007/s10549-016-4013-7. [DOI] [PubMed] [Google Scholar]
- 77.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–404. doi: 10.1158/2159-8290.cd-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boersma BJ, et al. A stromal gene signature associated with inflammatory breast cancer. International journal of cancer. 2008;122:1324–1332. doi: 10.1002/ijc.23237. [DOI] [PubMed] [Google Scholar]
- 79.Desmedt C, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13:3207–3214. doi: 10.1158/1078-0432.ccr-06-2765. [DOI] [PubMed] [Google Scholar]
- 80.Esserman LJ, et al. Chemotherapy response and recurrence-free survival in neoadjuvant breast cancer depends on biomarker profiles: results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657) Breast Cancer Research and Treatment. 2012;132:1049–1062. doi: 10.1007/s10549-011-1895-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ma XJ, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 82.Ma XJ, Dahiya S, Richardson E, Erlander M, Sgroi DC. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast cancer research: BCR. 2009;11:R7. doi: 10.1186/bcr2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pollack JR, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proceedings of the National Academy of Sciences. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schuetz CS, et al. Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer research. 2006;66:5278–5286. doi: 10.1158/0008-5472.can-05-4610. [DOI] [PubMed] [Google Scholar]
- 85.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sorlie T, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tabchy A, et al. Evaluation of a 30-gene paclitaxel, fluorouracil, doxorubicin, and cyclophosphamide chemotherapy response predictor in a multicenter randomized trial in breast cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16:5351–5361. doi: 10.1158/1078-0432.ccr-10-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Turashvili G, et al. Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC cancer. 2007;7:55. doi: 10.1186/1471-2407-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao H, et al. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Molecular biology of the cell. 2004;15:2523–2536. doi: 10.1091/mbc.e03-11-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.