Abstract
The epidemiology and genetic diversity of transmissible gastroenteritis virus (TGEV) in the United States (US) was investigated by testing clinical cases for TGEV by real time RT-PCR between January 2008 and November 2016. Prevalence of TGEV ranged between 3.8–6.8% and peaked during cold months until March 2013, in which prevalence decreased to < 0.1%. Nineteen complete TGEV genomes and a single strain of porcine respiratory coronavirus (PRCV) from the US were generated and compared to historical strains to investigate the evolution of these endemic coronaviruses. Sixteen of our TGEV strains share 8 unique deletions and 119 distinct amino acid changes, which might greatly affect the biological characteristics of the variant TGEV, and resulted in a “variant” genotype of TGEV. The “variant” genotype shared similar unique deletions and amino acid changes with the recent PRCV strain identified in this study, suggesting a recombination event occurred between the ‘‘variant’’ TGEV and PRCV. Moreover, the results indicate the “variant” genotype is the dominant genotype circulating in the US. Therefore, this study provides insight into the occurrence, origin, genetic characteristics, and evolution of TGEV and PRCV circulating in the US.
Introduction
Transmissible gastroenteritis virus (TGEV) belongs to the genus Alphacoronavirus, family Coronaviridae and contains a single-stranded, positive-sense RNA genome of approximately 28.5 kb in length. The enteropathogenic coronavirus causes watery diarrhea, severe villous atrophy, high mortality in piglets, and severe morbidity in different stages of pig development. In the epidemic form, TGEV causes significant economic losses to the global pork industry1–4. In 1946, TGEV was first reported in the United States (US)5 and was later identified in Europe (Belgium6, England2, France7, Germany8, The Netherlands6, and Spain9), Asia (China10 and Japan11), Africa (South Africa4 and Uganda12), and South America (Brazil13). Recent TGEV epidemiological studies are lacking in the US while sporadic outbreaks have been reported from China14–16.
Harboring one of the largest RNA genomes, the TGEV genome includes four structural genes (spike [S], envelope [E], membrane [M] and nucleoprotein [N]) and five non-structural open reading frame (ORF): ORF1a and ORF1b, ORF3a, ORF3b, and ORF7. The single segment genome is organized as 5′UTR-ORF1a/1b-S-ORF3a-ORF3b-E-M-N-ORF7-3′UTR3. The ORF1a/b can be divided into 15 ORFs, encoding non-structural proteins (NSP)1-10 and NSP12-16. The NSPs contribute to regulation of host translation and localization of the RNA-dependent RNA polymerase (RdRp) during viral replication3. In coronavirus, NSP1 is a major factor associated with pathogenicity17. The NSP2, 3 and 8 of TGEV can incorporate into the viral articles18 while the NSP3 is also associated with protease and ADP-ribose 1”-monophosphatase activities19. The functions of other NSPs of TGEV are unknown. The S glycoprotein attaches to the host cellular receptor porcine aminopeptidase N (pAPN) or sialic acid, induces cellular fusion, stimulates neutralizing antibodies, and has hemagglutination activity20–23. The pAPN-binding domain of the S protein has two major antigenic sites, A and B24. In addition, deletions in the ORF3 gene led to attenuation and reduced pathogenesis of TGEV and in vivo25.
In 1984, porcine respiratory coronavirus (PRCV), which is believed to have evolved from TGEV since PRCV and TGEV shared a high nucleotide percent identity, was first identified in Belgium. Compared to the S gene of TGEV strains, the S gene of PRCV has 621–681 nt deletions at N-terminal, including in strains from Asia26 (UF-1 [n = 672 bp]), Europe7,27 (AR310 and LEPP [n = 621 bp] and RM4 [n = 672 bp]) and North America7,25,28 (OH7269 [n = 648 bp], IA1894 [n = 678 bp], and ISU-1 [n = 681 bp]). Thus, the S gene is used to differentiate PRCV from TGEV. Recently, a novel strain of PRCV (OH726928) was identified in the US, but the origin and evolutionary relationship to current TGEV strains in the US is unknown. While the decline of TGEV is believed to occur in response to partial immunity from PRCV infections29–32, the United States swine industry also made significant changes (increased biosecurity, 3 site production model, etc.) to raise healthier pigs, which may have contributed to the reduction of TGEV infections as well.
The porcine enteric coronaviruses (including TGEV, porcine epidemic diarrhea virus [PEDV], and porcine deltacoronavirus [PDCoV]) cause similar clinical presentation, and co-infection of these enteric coronaviruses can occur33. In 2010, a highly virulent PEDV emerged in China34 and later spread to the US in April 201335. Within less than a year, PDCoV was identified in the US36,37. PEDV quickly spread throughout the US38, and by March 2014, approximately 41% of the sow herds were infected with PEDV39. While the identification of the PEDV lead to an increased biosecurity measure within the US swine industry, prevalence of PEDV in the sow herds did not significantly decrease until July 201439. Within the past couple of years, a chimeric TGEV and PEDV virus (consisting of a TGEV backbone and the spike of PEDV) was identified in multiple countries in Europe40–42, illustrating the potential emergence of a chimeric TGEV and PEDV virus in the US.
The occurrence and genetic diversity of TGEV was investigated prior to and after the identification of PEDV and PDCoV in the US from the diagnostic cases submitted to the University of Minnesota Veterinary Diagnostic Laboratory since PEDV recently emerged in the US, and a chimeric TGEV and PEDV virus was identified in Europe. Nineteen TGEV strains (US, n = 18 and Mexico, n = 1) and a single PRCV strain from the US were sequenced, analyzed, and compared with other global TGEV and PRCV strains to characterize historical and currents strains. This research will further our understanding of the occurrence, genetic variability, and evolution of an endemic coronavirus in the US and will provide guidance for future efforts to prevent, monitor, and control endemic coronaviruses.
Results
Decline of TGEV positive cases from 2008–2016
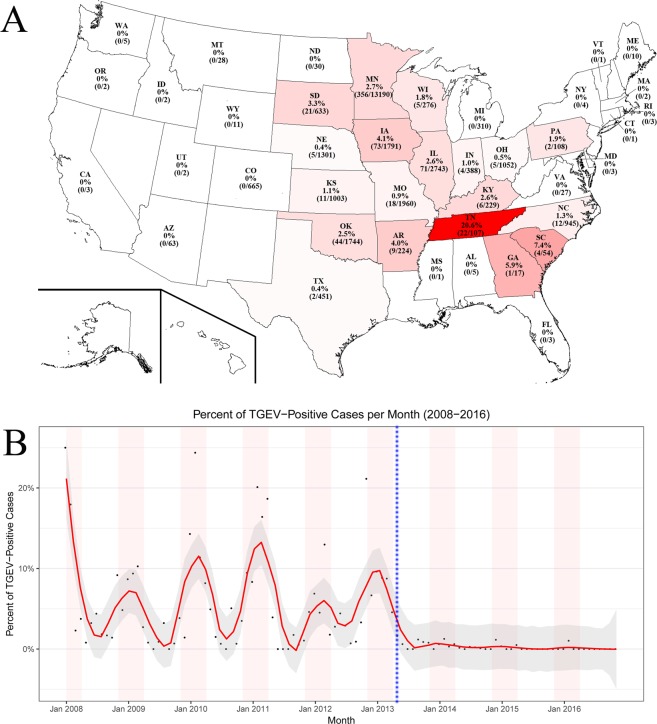
Between January 2008 and November 2016, 29,397 porcine enteric cases, distributed across 41 states in the US and Mexico, were tested for TGEV by real time RT-PCR, and 2.3% of the cases (n = 667) were positive for TGEV (Table 1). The percentage of TGEV positive cases was 4.0% in 2008, increased to 6.8% in 2010, and decreased to 0.4% in 2014 (Table 1). After the introduction of PEDV in the US, the prevalence of TGEV decreased further to less than 0.1%. Positive TGEV cases were detected in the main pig raising regions (Midwest, South-central, and Southeast) of the US (Fig. 1A) between January 2008 and November 2016. Most of the cases (n = 13,190) were from Minnesota where 2.7% (n = 356) were positive for TGEV. The percent of positive TGEV cases per state ranged between 0.4–20.6%, with the highest percentage found in Tennessee. A seasonality trend occurred with the positive cases between winter and spring (November to April) compared to summer and fall (May to October) (Fig. 1B).
Table 1.
Percentage of TGEV positive and negative cases between 2008 to 2016.
| Year | Positive Cases | Negative Cases | Total Count | ||
|---|---|---|---|---|---|
| Count | Percentage | Count | Percentage | ||
| 2008 | 56 | 4.0% | 1,346 | 96.0% | 1,402 |
| 2009 | 64 | 3.8% | 1,605 | 96.2% | 1,669 |
| 2010 | 134 | 6.8% | 1,827 | 93.2% | 1,961 |
| 2011 | 151 | 6.7% | 2,118 | 93.3% | 2,269 |
| 2012 | 119 | 6.1% | 1,839 | 93.9% | 1,958 |
| 2013 | 91 | 2.1% | 4,287 | 97.9% | 4,378 |
| 2014 | 48 | 0.4% | 11,185 | 99.6% | 11,233 |
| 2015 | 2 | 0.1% | 2,297 | 99.9% | 2,299 |
| 2016 | 2 | 0.1% | 2,226 | 99.9% | 2,228 |
| Total | 667 | 2.3% | 28,730 | 97.7% | 29,397 |
Figure 1.
Distribution of TGEV cases. (A) The geographical distribution of positive TGEV cases in the US. (B) The temporal distribution of positive TGEV cases per month between 2008 to 2016. The black dots represent the percentage of TGEV positive cases per month. A LOESS regression model was fitted to the data (red line), and the shaded area represents the 95% confidence interval. The light red shaded areas represent the cold months of November to April. The blue vertical dotted line represents the introduction of PEDV in April 2013.
Genomic characteristic, entropy and recombination analyses
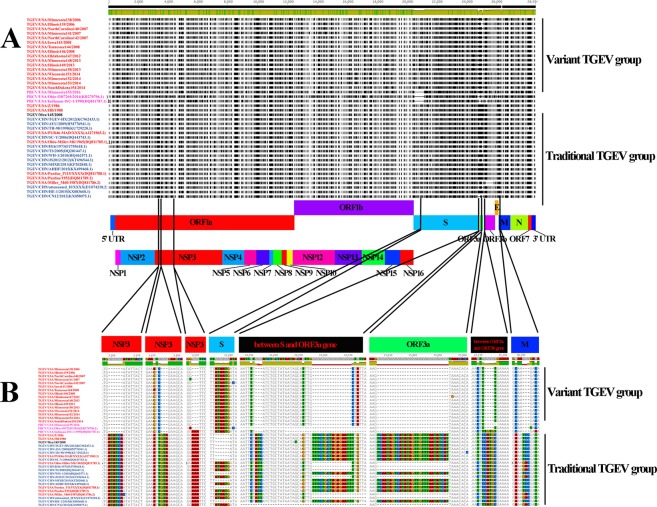
The genomic nucleotide sequence alignments of the 37 TGEV and 3 PCRV strains revealed two main genotypes (traditional and variant genotypes) (Fig. 2A). The traditional genotype shared a genome nucleotide identity of 98.26–100% and contained 21 TGEV strains: 5 US traditional TGEV strains, 2 historical US strains from this study (Z/1986 and HB/1988), the Mexican strain from our study (Mex/145/2008), and 13 Chinese traditional TGEV strains from 1973–2012. Among our 19 TGEV strains, 16 TGEV strains formed a new variant genotype and had a genome nucleotide identity of 98.68–99.97%. The traditional and variant genotypes shared only 95.76–97.13% nucleotide identity. The newly identified PRCV/USA/Minnesota155/2016 had a complete genome nucleotide percent identity of 96.6%, 95.5–96.2%, and 93.1–93.8% with the PRCV, variant TGEV strains, and traditional TGEV strains, respectively.
Figure 2.
Genomic insertion-deletions (INDELs) within TGEV and PRCV strains. (A) Variation within the 30 TGEV and 3 PRCV strains. Thin horizontal dash lines represent deletion in the alignment nucleotides. The vertical black bars represent the differences from consensus sequence. The organization of the genome is listed below (B). Visualization of insertion and deletion region of the 30 TGEV and 3 PRCV strains. The dashed lines represent absent nucleotides. The US TGEV strains are colored red. The US PRCV strains are colored purple. The Chinese TGEV strains are colored blue. The Mexican TGEV strain is colored black.
There were 8 major regions of insertion or deletion (INDELs) between the traditional and variant TGEV strains. In the variant group, 3 deletion regions occurred within NSP3, 2 deletion regions occurred between the S and ORF3a genes, 1 deletion regions occurred in ORF3a genes, 1 deletion region occurred between the ORF3a and ORF3b genes and a single deletion occurred in the M genes (Table 2 and Fig. 2B). Interestingly, the same ORF3a and ORF3b deletions were present in some of the historical TGEV strains. However, these deletions were not present in the TGEV strain Purdue, which was used to create the attenuated TGEV vaccine in the US. The variant TGEV strain Illinois139/2006 had a 128-nucleotide deletion in ORF3a, which resulted in the truncated protein compared with our variant TGEV strains. The traditional TGEV strains Z/1986, HB/1988, and Mex/145/2008 shared the same 6 nucleotide deletions in the S gene that were present in the TGEV strain, Purdue. Surprisingly, the PCRV strains contained an assortment of these deletions in their genomes, and compared to the historical TGEV strains, the TGEV strains from our study (excluding Z/1986, HB/1988 and Mex/145/2008) contained 8 deletions and 119 amino acid changes similar to the recently reported PRCV strain OH7269 and the single PRCV strain Minnesota155/2016 from our study.
Table 2.
Description of the 8 INDELs between traditional and variant TGEV using TGEV/USA/Z/1986 as reference.
| No. | Nucleotide (nt) position | Number of nt deleted | Nt deleted | Correponding amino acid (aa) variation |
|---|---|---|---|---|
| 1 | 3259–3264 | 6 | ATATCA | Two aa deletion in the NSP3 protein |
| 2 | 3354–3356 | 3 | GTA | Single aa deletion in the NSP3 protein |
| 3 | 4249–4251 | 3 | AAA | Single aa deletion in the NSP3 protein |
| 4 | 24727–24729 | 3 | ATC | Deletion in the non-coding region between S and ORF3a genes |
| 5 | 24750–24765 | 16 | TTTCTGCTAGAGAATT | Deletion in the non-coding region between S and ORF3a gene |
| 6 | 25020–25048 | 29 | TCAATAGTCATATAGTTGTTTAATATCAT | Single deletion in the ORF3a protein |
| 7 | 25127–25130 | 4 | CATG | Deletion in the non-coding region between ORF3a and ORF3b genes |
| 8 | 26185–26187 | 3 | TCC | Single aa deletion in the M protein |
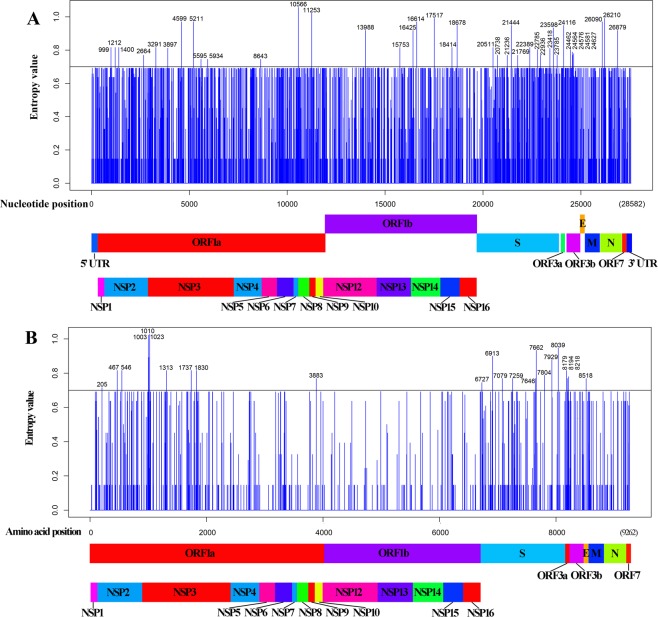
To determine the level of nucleotide or amino acid variation across the TGEV genomes, entropy analysis was conducted with an alignment of the complete genomes and concatenated amino acid sequences (Fig. 3). Based on the level of diversity in the dataset and previous work43, entropy values greater than 0.7 were considered highly variable for the nucleotide and amino acid sequence alignments. The ORF1 and S genes had the highest number of nucleotides with entropy levels above 0.7 (n = 20 and n = 12, respectively) while the ORF3b, E and, ORF7 genes lacked diversity in nucleotide positions (Fig. 3A). Within ORF1, the NSP2 and NSP3 had the highest number of nucleotide positions with entropy values greater than 0.7 (n = 4 and n = 6, respectively) while a single position was identified in NSP4, NSP6, NSP8, NSP12, NSP14, and NSP15. Within the amino acid alignment, the M, N and ORF7 genes lacked positions with entropy levels above 0.7 while ORF1 and S proteins (n = 10 and n = 9, respectively) had residues with entropy values higher than 0.7 (Fig. 3). There were 7 high-entropy positions at ORF1b gene in the nucleotide sequence entropy value analysis while entropy value higher than 0.7 were lacking in the amino acid sequence entropy value analysis.
Figure 3.
Nucleotide (A) and amino acid (B) entropy analysis of TGEV strains. The x-axis represents nucleotide or amino acid position while the y-axis represents entropy level. The TGEV non-structural and structural proteins are annotated below each plot. Black line represents the variance threshold of 0.7 for nucleotide and amino acid sequences.
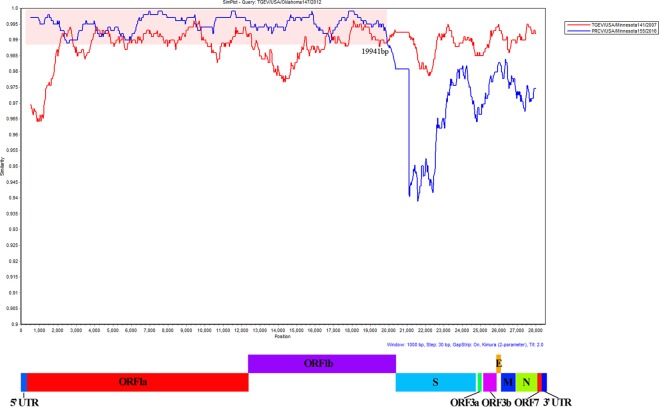
Recombination analysis was performed with the 37 TGEV and 3 PRCV strains. A single recombination event was detected by Chimera, BootScan, MaxChi, and SiScan and RDP within the dataset between the recently identified variant TGEV strains and the novel PRCV strain Minnesota155/2016 (Fig. 4). The TGEV variant Oklahoma147/2012 shares a high nucleotide identity of the first 19,941 nucleotides (breakpoint) with PRCV strain Minnesota155/2016 while the remainder of the genome shares a high nucleotide identity with TGEV strain Minnesota141/2007, indicating a complex nucleotide relationship between PRCV and TGEV.
Figure 4.
Recombination analysis of the variant TGEV strain Oklahoma147 with Minnesota141/2007 (red colored line) and Minnesota155/2016 (blue colored line). The x and y axes indicate the genomic nucleotide position and genetic identity, respectively. Possible recombination position is shaded with light red, and the breakpoint is marked with nucleotide position number (19,941).
Phylogenetic analyses of TGEV and PRCV strains
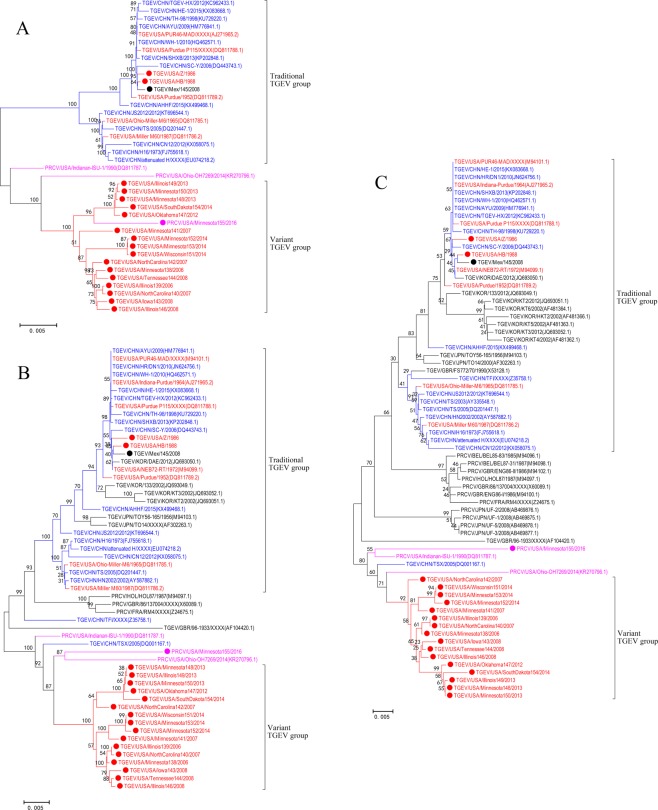
Phylogenetic trees of complete genome sequence (n = 40) and complete S gene (n = 56) revealed two distinct genotypes, representing the traditional and variant TGEV strains from the US (Fig. 5A,B, respectively). In the complete genome and S gene phylogenetic trees, the traditional genotype contained historical strains from the US, the recent strain from Mexico, and historical and recent strains from China. From our study, the 16 recent TGEV strains from the US formed the variant TGEV group, which share a common ancestor with the PRCV strain Ohio-OH7269/2014. Also, the variant TGEV strains were identified in the three main swine production regions of the US (Midwest, South-central, and Southeast) indicating substantial geographical distribution. Interestingly, whole genome phylogenetic analysis of PRCV strain Minnesota155/2016 clustered within the variant genotype and not with historical PRCV strains, indicating significant genetic diversity within PRCV strains circulating in the US.
Figure 5.
Phylogenetic trees of the TGEV and PRCV strains based on the whole genome (n = 40) (A), the whole S gene (n = 56) (B), and partial S gene (n = 70) (C). The scale bar represents nucleotide substitutions per site. The red color represents the US TGEV strains. The purple color represents the US PRCV strains. The blue color represents Chinese TGEV isolates. The black color represents strains from other countries. Our TGEV or PRCV strains were highlighted by solid circle.
Since additional partial S gene sequences (first 1383 nt) were available from geographically different locations, a partial S gene phylogenetic tree was constructed to further investigate the global evolutionary relationship between TGEV and PRCV strains (Fig. 5C). The traditional TGEV group consisted of TGEV strains isolated from the US, China, Japan, South Korea, England and Mexico. The variant TGEV group consisted our 16 US TGEV strains isolated in the US after 2006 and a Chinese strain (TSX/2005) from 2005. Interestingly, the Chinese strain TSX/2005 was closely related to the US PRCV strains.
Analysis of the receptor binding domain
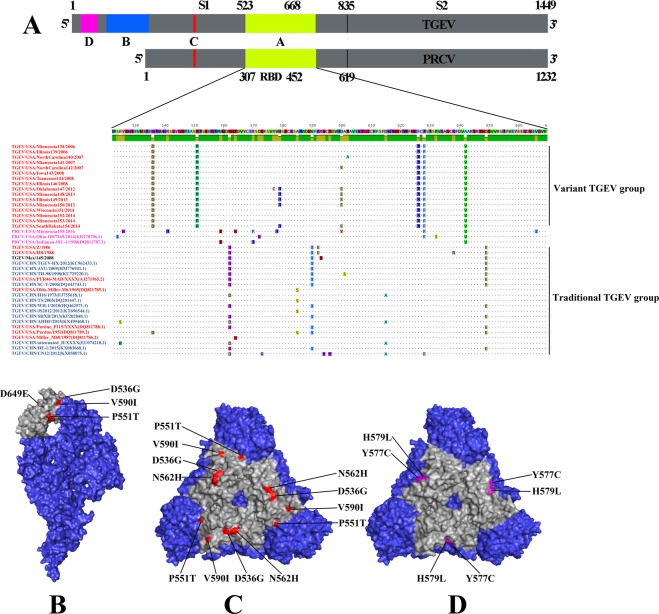
Given the important role of the S protein in pAPN-receptor binding of TGEV and PRCV, the receptor binding domain (RBD) was investigated for amino acid variation. When comparing the amino acid sequences of the RBD of the traditional to the variant TGEV genotype, eight amino acid substitutions (D536G, P551T, N562H, V590I, L626F, F628L, V642A and D649E) occurred in a majority of the strains while four additional amino acid substitutions (Y577C, H579L, D600E and T602A) were identified in the pAPN (Fig. 6A). The different amino acid between traditional and variant TGEV strains were highlighted in the predicted crystal structure of RBD (Fig. 6B–D). Four of eight amino acid substitutions within the RBD (L626F, F628L, V642A, and D649E) were exposed on the viral protein while two of the four amino acid changes in pAPN (D600E and T602A) were exposed on the viral protein (Fig. 6B–D).
Figure 6.
Protein alignment and modeling of S1 domain receptor-binding domain RBD, S protein monomer and trimer. (A) The RBD alignment of the 37 whole genome strains and 3 PRCV strains. Identical residues to the consensus sequence are shown as dots while residues that differ from the consensus are highlighted in different colors. (B,C) The monomer and trimer theoretical protein model of the RBD region, with 4 of the 8 major substitutions highlighted in red. (D) Trimer theoretical structure model of RBD, with 2 of the 4 of the TGEV S protein trimer in pink. The body of S protein and the RBD region are highlighted in slate blue and grey.
Discussion
Porcine enteric coronaviruses (PEDV, PDCoV, and TGEV) are significant, emerging pathogens causing severe enteric diseases in the global swine industry. While global historical strains of TGEV caused severe enteritis and PRCV is endemic in Europe, Asia, and the US, the mortality rate due to TGEV infections has declined in these countries3,44,45. However, a virulent TGEV or PRCV strain could emerge since the epidemiology of TGEV and PRCV is different in China. In 1992, PRCV was first reported in China46 and was not identified or reported in China afterwards while TGEV is constantly reported as a significant viral pathogen for the Chinese pig industry10,15,16. The different clinical status of TGEV in China and the western countries could be due to the stringent biosecurity measures in western countries compared to China. TGEV was consistently detected in the US between 2008 and 2016, but after the introduction of PEDV into the US, the swine industry significantly increased their biosecurity to prevent and control PEDV infections and the prevalence of PEDV were reduced after March 2014 in US39,47,48, which indirectly may have reduced the prevalence of TGEV to less than 0.1%. Our study illustrates a seasonality pattern with TGEV in the US with infection peaking during cold months, which has been identified in our swine pathogens as well37,45,46.
While TGEV has been identified throughout Europe1,8,49,50, Asia11,15,16,45, and North America5,6, only the variant TGEV genotype was detected in US pig farms since 2006, suggesting that variant TGEV is the dominant genotype currently circulating in the main pig raising Midwest, South-central, and Southeast regions. In addition, the first TGEV genome from Mexico was characterized, which helps us to understand the evolutionary relationship of TGEV strains from different countries. Surprisingly, the Mex/148 strain is phylogenetically related to the traditional TGEV, and not the variant TGEV strain. Given the close geographical proximity of Mexico and US and that the Mexican and US PEDV strains are phylogenetically related51, the variant TGEV strains could be circulating in Mexico, but were not represented in our study since our study had only a single TGEV strain from Mexico. The circulation of TGEV and PEDV in the US indicates a chimeric virus could emerge in the US similar as the chimeric virus emerged in Europe. Characterizing of the current TGEV strains will aid in understanding the emergence of virulent or chimeric TGEV strains in the US, if such an event would occur in the future.
PRCV was first identified in Belgium in 198444 and has been identified multiple countries including Belgium52, China53, Japan26, Uganda12 and the US37. The evolutionary relationship between the recently identified PRCV strain (Ohio-OH7269/2014) and our PRCV strain (Minnesota155/2016) was unclear given the limited number of globally available PRCV strains. Ideally, additional PRCV strains would have been sequenced, but routine screen of PRCV does not occur since PRCV is not considered as a significant pathogen in swine. The lab accidently isolated the PRCV strain, which was added to the study since the genetic and evolutionary relationship of PRCV is unknown in the US. The variant TGEV strains shared the same nucleotide mutations and amino acid deletions with the novel PRCV strain Ohio-OH7269/2014, and a recombination event between PRCV and TGEV was identified, illustrating the complex evolutionary relationship between TGEV and PRCV strains in the US. Hopefully, future US studies will assess the genetic and evolutionary of PRCV strains to fully elicit the complex relationship with TGEV.
Historically, a partial gene fragment or a single gene of TGEV was sequenced6 to differentiate viral strains due to limitations in technology and economic constraints. In this study, NGS technology generated the whole TGEV genome, revealing a total of 8 unique INDELs located in NSP3, S, ORF3a, ORF3b and M protein, and these regions, excluding the M protein, had high entropy levels in our study (Fig. 3A). The NSP3, S, ORF3a and ORF3b proteins of TGEV are associated with enteric tropism, immunogenicity, neutralization, sialic acid and other receptor binding activity ability, virulence, protease and ADP-ribose 1 ″-monophosphatase activities18–25, and changes in these proteins might reduce INDELs efficacy to cause clinical disease. Mutations in the spike gene in other coronaviruses has impact on initiation of membrane fusion54, sialic acid binding activity22, confer resistance to virus neutralization55, and render trypsin independent for cell entry and fusion56, which could be contribution to a reduction of the overall fitness of the INDELs. The attenuation of murine coronavirus in the natural host occurred due to deletion of the HE, 2a/HE, and 4/5a genes57, epidemics of feline infectious peritonitis were contributed to deletions in the ORF7a and ORF7b of feline coronavirus58,59, and disruption of functional expression of accessory protein occurred due to deletion of ORF8 in human severe acute respiratory syndrome coronaviruses60. Given the results from the previous studies, we hypothesis the INDELs are significantly attenuated compared to the traditional strains of TGEV. Further studies are needed to confirm this hypothesis and investigate the effect of these deletions and mutations on the biological characteristics and fitness of the new TGEV genotype.
In conclusion, a variant genotype of TGEV is the predominant in the US and evolutionary relationship between TGEV and PRCV is complex. The decline of TGEV positive diarrhea cases after May 2013 in the US may be indirectly associated with the outbreak of PED in the US via increased biosecurity. Compared with the traditional TGEV strains, the unique amino acid INDELs might affect the biological characteristics of the variant TGEV, which could lead to changes in pathogenesis or chimeric virus in the future.
Materials and Methods
Veterinarians routinely send samples to the University of Minnesota Veterinary Diagnostic Laboratory (MNVDL) to determine potential pathogenic agents contributing to disease and to promote the health of swine herds. The samples may represent clinical outbreaks of diarrhea or are for routine monitoring of enteric pathogens in swine herds. Upon arrival, ownership of the samples belongs to the MNVDL and client(s) confidentiality is retained by removing identifiers associated with client(s) information. Between January 2008 and November 2016, a total of 29,397 samples, including fresh intestines and fecal samples from of clinical cases were submitted from 41 US states and Mexico and tested for TGEV by real time RT-PCR under Standard Operation Procedures (available upon request) and various bacterial and viral enteric pathogens dependent on the veterinarian’s request. Request may include beta-hemolytic Escherichia coli, non-beta-hemolytic Escherichia coli, rotavirus A, B, and C, PEDV, and PDCoV. Randomly selected historical positive TGEV samples from the MNVDL (n = 17) were saved from previous enteric studies, two TGEV samples from Ohio (Z and HB) were supplied from Dr. Linda Saif. Routine testing for PRCV does not occur since it is not considered a major swine pathogen. However, a PRCV isolate from MN was obtained from a nasal swab in 2016 during an attempted to isolate other viruses. All samples (n = 20) were selected for whole genome sequencing using next generation sequencing as previously described36. TGEV prevalence information was exported from the MNVDL database and analyzed at the case-level with R software61 using the ggplot262 and maps63.
To investigate the differences in TGEV strains and phylogenetic relationship with PRCV, our 20 sequences and available sequences from GenBank (Table S1) were aligned using ClustalW in Geneious v9.1.464. Nucleotide and amino acid entropy analyses of the concatenation ORFs (ORF1a/b, S, ORF3a/3b, envelope, membrane, nucleocapsid and ORF7) was performed using the MATLAB65 to determine regions of diversity within the alignment. Entropy values higher than 0.7 in the nucleotide and amino acid alignments were identified as high variation positions43. Recombination analysis was performed using Recombination Detection Program (RDP)66 v4 with RDP, BootScan, GEENECOV, SiScan and MAXCHI algorithms (Window size is 100 bp). Recombination event was represented using similarity plot, with Oklahoma147 as the query strain. The similarity plot was implemented in the SimPlot, v. 3.5.1 package67, using the two-parameter (Kimura) distance model with a sliding window of 1000 bp and step size of 30 bp.
Whole genome (n = 40), the whole S gene (n = 56), and the partial S gene (first 1383 nt) (n = 70) (Table S1) phylogenetic trees were constructed using the Maximum likelihood algorithm, with a GTR nucleotide substitution model (bootstrap analysis with 1,000 replicates) by MEGA v6.0668. The S protein receptor binding domains (RBDs) of TGEV and PRCV were modeled using the open-source modeling server SWISS-MODEL69 provided by the Swiss Institute of Bioinformatics. Predicted tertiary structure of the RBDs of TGEV and PRCV were modeled using PRCV RBD (PDB accession no. 5SZS) reported in the previous study. Spike monomer and trimer models were performed using human coronavirus NL63 model as a template to theatrically visualize the changes in the residues since TGEV and PRCV templates are lacking70. Illustrations were created using the Python-based molecular viewer PyMOL71.
Supplementary information
Acknowledgements
The study was funded by the MNVDL. The authors thank the faculty and personal at the University of Minnesota Veterinary Diagnostic Laboratory (MNVDL) for their technical services. Fangzhou Chen was supported by China Agriculture Research System (No. CARS-36) and China Scholarship Council No. 201606760012 and was a visiting scholar at the University of Minnesota College of Veterinary Medicine during the construction of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
Author Contributions
Conceived and designed the experiments: F. Chen, D. Marthaler. Performed the experiments: F. Chen, T. Knutson, D. Marthaler. Analyzed the data: F. Chen, T. Knutson, S. Rossow, L. Saif, D. Marthaler. Contributed reagents/materials/analysis tools: S. Rossow, L. Saif, D. Marthaler. Wrote the paper: F. Chen, D. Marthaler.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-40564-z.
References
- 1.Bohl, E. H. Transmissible gastroenteritis virus (classical enteric variant) and transmissible gastroenteritis virus (respiratory variant). In Virus Infections of Porcines (eds Pensaert, M. & Pensaert, M. B.) 139–165 (Elsevier Science, 1989).
- 2.Pritchard GC, Paton DJ, Wibberley G, Ibata G. Transmissible gastroenteritis and porcine epidemic diarrhoea in Britain. Vet. Rec. 1999;144:616–618. doi: 10.1136/vr.144.22.616. [DOI] [PubMed] [Google Scholar]
- 3.Saif, L., Pensaert, M. B., Sestak, K., Yeo, S. & Jung, K. Diseases of swine. in Coronaviruses (eds Zimmerman, J. J. & ebrary, I.) 10th ed., 501:524 (Chichester, West Sussex; Ames, Iowa: Wiley-Blackwell, 2012).
- 4.Williams R, Esterhuysen JJ, Robinson JT. Pseudorabies and transmissible gastroenteritis: a serological survey in South Africa. Onderstepoort J. Vet. Res. 1994;61:67–70. [PubMed] [Google Scholar]
- 5.Doyle LP, Hutchings LM. A transmissible gastroenteritis in pigs. J. Am. Vet. Med. Assoc. 1946;108:257–259. [PubMed] [Google Scholar]
- 6.Sanchez CM, et al. Genetic evolution and tropism of transmissible gastroenteritis coronaviruses. Virology. 1992;190:92–105. doi: 10.1016/0042-6822(92)91195-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasschaert D, Duarte M, Laude H. Porcine respiratory coronavirus differs from transmissible gastroenteritis virus by a few genomic deletions. J. Gen. Virol. 1990;71(Pt 11):2599–2607. doi: 10.1099/0022-1317-71-11-2599. [DOI] [PubMed] [Google Scholar]
- 8.Hess RG, Bollwahn W, Pospischil A, Heinritzi K, Bachmann PA. Current aspects in the etiology of viral diarrheas of swine: occurrence of infections with the epizootic viral diarrhea (EVD) virus. Berl. Munch. Tierarztl. Wochenschr. 1980;93:445–449. [PubMed] [Google Scholar]
- 9.Cubero MJ, et al. Transmissible gastroenteritis in pigs in south east Spain: prevalence and factors associated with infection. Vet. Rec. 1993;132:238–241. doi: 10.1136/vr.132.10.238. [DOI] [PubMed] [Google Scholar]
- 10.Hu X, et al. Molecular characterization and phylogenetic analysis of transmissible gastroenteritis virus HX strain isolated from China. BMC Vet. Res. 2015;11:72. doi: 10.1186/s12917-015-0387-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi K, Okada K, Ohshima K. An outbreak of swine diarrhea of a new-type associated with coronavirus-like particles in Japan. Nippon Juigaku Zasshi. 1983;45:829–832. doi: 10.1292/jvms1939.45.829. [DOI] [PubMed] [Google Scholar]
- 12.Muley, T. The possible role of transmissible gastroenteritis virus (TGE) and porcine respiratory corona virus (PRCV) in the mortality of Ugandan back-yard piglets. (Swedish University of Agricultural Sciences, 2012).
- 13.Martins A, et al. Diagnosis to Detect Porcine Transmissible Gastroenteritis Virus (TGEV) by Optical and Transmission Electron Microscopy Techniques. Int. J. Morphol. 2013;31:706–715. doi: 10.4067/S0717-95022013000200059. [DOI] [Google Scholar]
- 14.Hou Y, et al. Complete genome of transmissible gastroenteritis virus AYU strain isolated in Shanghai, China. J. Virol. 2012;86:11935–12. doi: 10.1128/JVI.01839-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiwei H, et al. Complete genomic sequence of the coronavirus transmissible gastroenteritis virus SHXB isolated in China. Archives of Virology. 2014;159:2295–2302. doi: 10.1007/s00705-014-2080-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhenhui S, et al. Isolation and Identi cation of a New Strain of Porcine Transmissible Gastroenteritis Virus from Chongqing, Southwestern China. Israel Journal of Veterinary Medicine. 2015;707:22–30. [Google Scholar]
- 17.Züst R, et al. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 2007;3:e109. doi: 10.1371/journal.ppat.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nogales A, Márquez-Jurado S, Galán C, Enjuanes L, Almazán F. Transmissible gastroenteritis coronavirus RNA-dependent RNA polymerase and nonstructural proteins 2, 3, and 8 are incorporated into viral particles. J. Virol. 2012;86:1261–1266. doi: 10.1128/JVI.06428-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Putics A, Gorbalenya AE, Ziebuhr J. Identification of protease and ADP-ribose 1”-monophosphatase activities associated with transmissible gastroenteritis virus non-structural protein 3. J. Gen. Virol. 2006;87:651–656. doi: 10.1099/vir.0.81596-0. [DOI] [PubMed] [Google Scholar]
- 20.Ballesteros ML, Sánchez CM, Enjuanes L. Two amino acid changes at the N-terminus of transmissible gastroenteritis coronavirus spike protein result in the loss of enteric tropism. Virology. 1997;227:378–388. doi: 10.1006/viro.1996.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delmas B, Rasschaert D, Godet M, Gelfi J, Laude H. Four major antigenic sites of the coronavirus transmissible gastroenteritis virus are located on the amino-terminal half of spike glycoprotein S. J. Gen. Virol. 1990;71(Pt 6):1313–1323. doi: 10.1099/0022-1317-71-6-1313. [DOI] [PubMed] [Google Scholar]
- 22.Krempl C, Schultze B, Laude H, Herrler G. Point mutations in the S protein connect the sialic acid binding activity with the enteropathogenicity of transmissible gastroenteritis coronavirus. J. Virol. 1997;71:3285–3287. doi: 10.1128/jvi.71.4.3285-3287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sánchez CM, et al. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 1999;73:7607–7618. doi: 10.1128/jvi.73.9.7607-7618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godet M, Grosclaude J, Delmas B, Laude H. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol. 1994;68:8008–8016. doi: 10.1128/jvi.68.12.8008-8016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, et al. Complete genomic sequences, a key residue in the spike protein and deletions in nonstructural protein 3b of US strains of the virulent and attenuated coronaviruses, transmissible gastroenteritis virus and porcine respiratory coronavirus. Virology. 2007;358:424–435. doi: 10.1016/j.virol.2006.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Usami Y, et al. Virological and serological studies of porcine respiratory coronavirus infection on a Japanese farm. J. Vet. Med. Sci. 2008;70:929–936. doi: 10.1292/jvms.70.929. [DOI] [PubMed] [Google Scholar]
- 27.Vaughn EM, Halbur PG, Paul PS. Sequence comparison of porcine respiratory coronavirus isolates reveals heterogeneity in the S, 3, and 3-1 genes. J. Virol. 1995;69:3176–3184. doi: 10.1128/jvi.69.5.3176-3184.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Zhang Y. Genomic Characterization of a New PRCV Variant, United States, 2014. Transbound Emerg Dis. 2017;64:672–674. doi: 10.1111/tbed.12400. [DOI] [PubMed] [Google Scholar]
- 29.Brim TA, VanCott JL, Lunney JK, Saif LJ. Cellular immune responses of pigs after primary inoculation with porcine respiratory coronavirus or transmissible gastroenteritis virus and challenge with transmissible gastroenteritis virus. Vet. Immunol. Immunopathol. 1995;48:35–54. doi: 10.1016/0165-2427(94)05416-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cox E, Pensaert MB, Callebaut P. Intestinal protection against challenge with transmissible gastroenteritis virus of pigs immune after infection with the porcine respiratory coronavirus. Vaccine. 1993;11:267–272. doi: 10.1016/0264-410X(93)90028-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.VanCott JL, Brim TA, Lunney JK, Saif LJ. Contribution of antibody-secreting cells induced in mucosal lymphoid tissues of pigs inoculated with respiratory or enteric strains of coronavirus to immunity against enteric coronavirus challenge. J. Immunol. 1994;152:3980–3990. [PubMed] [Google Scholar]
- 32.Wesley RD, Woods RD. Induction of protective immunity against transmissible gastroenteritis virus after exposure of neonatal pigs to porcine respiratory coronavirus. Am. J. Vet. Res. 1996;57:157–162. [PubMed] [Google Scholar]
- 33.Marthaler D, et al. Rapid detection, complete genome sequencing, and phylogenetic analysis of porcine deltacoronavirus. Emerg. Infect. Dis. 2014;20:1347–1350. doi: 10.3201/eid2008.140526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun R-Q, et al. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerging Infect. Dis. 2012;18:161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevenson GW, et al. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Invest. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- 36.Marthaler, D., Jiang, Y., Collins, J. & Rossow, K. Complete Genome Sequence of Strain SDCV/USA/Illinois121/2014, a Porcine Deltacoronavirus from the United States. Genome Announc2, 10.1128/genomeA.00218-14 (2014). [DOI] [PMC free article] [PubMed]
- 37.Wang L, Byrum B, Zhang Y. Porcine coronavirus HKU15 detected in 9 US states, 2014. Emerg. Infect. Dis. 2014;20:1594–1595. doi: 10.3201/eid2009.140756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vlasova AN, et al. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013-February 2014. Emerg. Infect. Dis. 2014;20:1620–1628. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.pboyer. PED charts. College of Veterinary Medicine - University of Minnesota (2018). Available at: https://www.vetmed.umn.edu/centers-programs/swine-program/outreach-leman-mshmp/mshmp/ped-charts (Accessed: 15th August 2018).
- 40.Akimkin V, et al. New Chimeric Porcine Coronavirus in Swine Feces, Germany, 2012. Emerging Infect. Dis. 2016;22:1314–1315. doi: 10.3201/eid2207.160179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belsham GJ, et al. Characterization of a Novel Chimeric Swine Enteric Coronavirus from Diseased Pigs in Central Eastern Europe in 2016. Transbound Emerg Dis. 2016;63:595–601. doi: 10.1111/tbed.12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boniotti MB, et al. Porcine Epidemic Diarrhea Virus and Discovery of a Recombinant Swine Enteric Coronavirus, Italy. Emerging Infect. Dis. 2016;22:83–87. doi: 10.3201/eid2201.150544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Litwin, S. & Jores, R. Shannon Information as a Measure of Amino Acid Diversity. in Theoretical and Experimental Insights into Immunology (eds Perelson, A. S. & Weisbuch, G.) 2789–187, 10.1007/978-3-642-76977-1 (Springer Berlin Heidelberg, 1992).
- 44.Pensaert M, Callebaut P, Vergote J. Isolation of a porcine respiratory, non-enteric coronavirus related to transmissible gastroenteritis. Vet Q. 1986;8:257–261. doi: 10.1080/01652176.1986.9694050. [DOI] [PubMed] [Google Scholar]
- 45.Miyazaki A, Fukuda M, Kuga K, Takagi M, Tsunemitsu H. Prevalence of antibodies against transmissible gastroenteritis virus and porcine respiratory coronavirus among pigs in six regions in Japan. J. Vet. Med. Sci. 2010;72:943–946. doi: 10.1292/jvms.09-0377. [DOI] [PubMed] [Google Scholar]
- 46.Huang, S. A new virus associated with transmissible gastroenteritis virus-porcine respiratory coronavirus type. Chinese Journal of Veterinary Medicine 48–49 (1992).
- 47.Schulz LL, Tonsor GT. Assessment of the economic impacts of porcine epidemic diarrhea virus in the United States. J. Anim. Sci. 2015;93:5111–5118. doi: 10.2527/jas.2015-9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niederwerder MC, Hesse RA. Swine enteric coronavirus disease: A review of 4 years with porcine epidemic diarrhoea virus and porcine deltacoronavirus in the United States and Canada. Transbound Emerg Dis. 2018;65:660–675. doi: 10.1111/tbed.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang, S. A new virus associated with transmissible gastroenteritis virus-porcine respiratory coronavirus type. Chinese Journal of Veterinary Medicine. 1, 48–49 (1992).
- 50.Chasey D, Cartwright SF. Virus-like particles associated with porcine epidemic diarrhoea. Res. Vet. Sci. 1978;25:255–256. doi: 10.1016/S0034-5288(18)32994-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jarvis, M. C., Lam, H. C., Rovira, A. & Marthaler, D. G. Complete Genome Sequence of Porcine Epidemic Diarrhea Virus Strain COL/Cundinamarca/2014 from Colombia. Genome Announc4 (2016). [DOI] [PMC free article] [PubMed]
- 52.Pensaert M, Cox E, van Deun K, Callebaut P. A sero-epizootiological study of porcine respiratory coronavirus in Belgian swine. Vet Q. 1993;15:16–20. doi: 10.1080/01652176.1993.9694361. [DOI] [PubMed] [Google Scholar]
- 53.HaiE S, et al. Application of n-PCR for Detection and Differentiation of TGE Virus and Porcine Respiratory Coronavirus. Chinese Journal of Animal Quarantine. 2003;20:21–22. [Google Scholar]
- 54.Wang K, et al. PEDV ORF3 encodes an ion channel protein and regulates virus production. FEBS Lett. 2012;586:384–391. doi: 10.1016/j.febslet.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye S, et al. Porcine epidemic diarrhea virus ORF3 gene prolongs S-phase, facilitates formation of vesicles and promotes the proliferation of attenuated PEDV. Virus Genes. 2015;51:385–392. doi: 10.1007/s11262-015-1257-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu DX, Fung TS, Chong KK-L, Shukla A, Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Haan CAM, Masters PS, Shen X, Weiss S, Rottier PJM. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology. 2002;296:177–189. doi: 10.1006/viro.2002.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kennedy M, Boedeker N, Gibbs P, Kania S. Deletions in the 7a ORF of feline coronavirus associated with an epidemic of feline infectious peritonitis. Vet. Microbiol. 2001;81:227–234. doi: 10.1016/S0378-1135(01)00354-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin C-N, et al. Field strain feline coronaviruses with small deletions in ORF7b associated with both enteric infection and feline infectious peritonitis. J. Feline Med. Surg. 2009;11:413–419. doi: 10.1016/j.jfms.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oostra M, de Haan CAM, Rottier PJM. The 29-nucleotide deletion present in human but not in animal severe acute respiratory syndrome coronaviruses disrupts the functional expression of open reading frame 8. J. Virol. 2007;81:13876–13888. doi: 10.1128/JVI.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/ (2014).
- 62.Wickham, H. ggplot2., 10.1007/978-0-387-98141-3 (Springer New York, 2009).
- 63.Becker, R. A., Wilks, A. R., Brownrigg, R. & Minka, T. P. Maps: draw geographical maps. R package version 2.3-9 (2014).
- 64.Kearse M, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.MATLAB and Statistics Toolbox (The MathWorks, Inc).
- 66.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lole KS, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–8. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walls AC, et al. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2016;23:899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.