Abstract
Herbivory by fishes has been identified as a key ecological process shaping coral reefs through time. Although taxonomically limited, herbivorous reef fishes display a wide range of traits, which results in varied ecosystem functions on reefs around the world. Yet, we understand little about how these trait combinations and functions in ecosystems changed through time and across biogeographic realms. Here, we used fossils and phylogenies in a functional ecological framework to reveal temporal changes in nominally herbivorous fish assemblages among oceanic basins in both trait space and lineage richness among functions. We show that the trait space occupied by extant herbivorous fishes in the Indo-Pacific resulted from an expansion of traits from the ancestral Tethyan assemblages. By contrast, trait space in the Atlantic is the result of lineage turnover, with relatively recent colonization by lineages that arose in the east Tethys/Indo-Pacific. From an ecosystem function perspective, the Atlantic supports a depauperate fauna, with few extant herbivorous reef fish lineages performing each function. Indo-Pacific fishes support both more functions and more lineages within each function, with a marked Miocene to Pleistocene expansion. These disparities highlight the importance of history in explaining global variation in fish functional composition on coral reefs.
Keywords: parrotfishes, surgeonfishes, rabbitfishes, algal turf removal, macroalgae removal, bioerosion
1. Introduction
The diversity of extant ecological systems results from complex interactions between biotic and abiotic factors acting in space and time. While abiotic processes tend to influence biodiversity at larger spatial and temporal scales [1], biotic interactions are increasingly being recognized as important drivers of evolutionary change at smaller scales and in high-diversity systems [2]. As a consequence, our ability to describe how diversity has been built up through time depends on integrating distinct, but not independent, sources of information. This realization has stimulated recent calls to unite palaeontological and neontological data into a single comprehensive framework that facilitates a more integrative approach to biodiversity research [3,4]. At the core of this integration is the focus on a species' function rather than its taxonomic identity [4], reflecting its role in ecosystem dynamics [5] and evolution [6]. Species functional diversity is not only an important metric that can illuminate changes in ecosystem functioning through time [7], it can also reveal insights into larger scale biogeographic processes [8].
Although it is clear that integrating fossil and extant species information in a trait-based framework can promote a better understanding of biodiversity assembly through time (e.g. [9,10]), there have been few attempts, to date, to frame it in a historical biogeography context. Functional biogeography (sensu [9]) provides an important framework for analysing large-scale patterns in trait combinations [11,12]. However, the role of history remains largely unexplored, despite promising insights from high-diversity systems, such as coral reefs [13]. These high diversity environments have been shaped through time by major geological events that have left detectable traces in extant reef assemblages [14], fossil deposits [15], and molecular phylogenies [16,17].
While biodiversity in reef environments has a long history [18], the modern incarnation of coral-dominated reef systems is largely restricted to the last 60 million years (Myr) [19,20]. Throughout this time, fish and coral assemblages have displayed marked functional changes [20]. One of the most notable early changes was the expansion of herbivorous fishes [21]. Today, this group plays a critical role in reef ecosystems, mediating the competitive balance between corals and algae, and contributing to the resilience of coral reefs [22]. Without this top-down control, coral reef environments can shift to algal-dominated states [23,24] with detrimental effects on associated biodiversity [25]. Herbivorous fishes also play a pivotal role in carbonate dynamics by reworking and transporting calcareous sediments [26]. Thus, documenting how herbivorous fish traits and their associated ecosystem functions evolved through time and geographical space is essential if we wish to understand variation in ecosystem functions in present-day ecosystems.
The history of herbivorous reef fishes is tightly associated with the formation of modern coral reef systems [20]. Before the early Cenozoic, there is no clear evidence of herbivorous marine vertebrates [27]. Herbivory was performed primarily by invertebrates [28]. The Eocene fossil deposits of Monte Bolca in northern Italy, therefore, provide the first evidence of a fundamental change in the evolution of coral reef assemblages [29]; the first unequivocal shift in fish–benthos interactions [21]. Ancient surgeonfish and rabbitfish lineages from Monte Bolca laid the foundations for piscine herbivory on coral reefs and, along with later arising parrotfishes [30], form the main herbivorous components of present-day coral reefs [31]. Although Bolca fossils are extremely valuable for understanding the evolution of herbivorous fishes [10,21,32], it remains a unique assemblage. As a consequence, the only way to trace the functional history of herbivorous fishes on coral reefs in space and time is by combining fossils, molecular phylogenies, and extant species ecology in a holistic framework.
Recent efforts have been made to understand reef fish traits [33] and biogeography [17] through ancestral reconstructions, however, an integrative approach is still lacking. Although herbivorous fishes present a vast combination of traits and functions on coral reefs [34], their assemblages are unevenly distributed among biogeographic realms, with broad implications for ecosystem functioning [22]. Unfolding the historical factors that drive this disparity would shed light on the processes that shaped the assembly of essential functions. By combining information from fossils and extant species in a comparative phylogenetic framework, we provide an integrated functional and biogeographic overview of the global evolution of key herbivorous groups on tropical reefs. Furthermore, by specifically separating traits from ecosystem functions, we track: (i) how the herbivorous fish trait space has been occupied through time among major marine biogeographic realms since the Eocene; and (ii) the origin of lineages performing each of the key ecosystem functions of herbivorous fishes in space and time as indicated by extant taxa. Our framework provides a sequential view of the formation of one of the most important ecological groups on coral reefs, herbivores.
2. Methods
(a). Chronograms
We built the most comprehensive time-calibrated phylogenies to date for the three most important nominally herbivorous fish groups on modern coral reefs [31]: Acanthuridae (surgeonfishes), Siganidae (rabbitfishes), and Scarini (parrotfishes). The surgeonfishes and rabbitfishes are recognized as separate families, while the parrotfishes are a tribe within the Labridae [35], containing the subtribes Scarina and Sparisomatina. For each group, we downloaded sequences from GenBank and used Bayesian inferences in BEAST2 [36] to construct chronograms. The surgeonfish phylogeny incorporated two mitochondrial (Cox1 and Cytb) and seven nuclear genes (ENC1, myh6, plagl2, Rag1, Rh, zic1, and ETS2) and comprised 72 species (approx. 90% total diversity) from all extant genera. The rabbitfish phylogeny was based on two mitochondrial genes (Cytb and 16s) and one nuclear gene (ITS1). It contained 24 species (approx. 80% total diversity) in its single genus Siganus. Finally, the parrotfish phylogeny was based on five mitochondrial (Cox1, Cytb, 12s, 16s, and control region) and six nuclear markers (Bmp4, Dlx2, Otx1, Rag2, S7I1, and Tmo-4C4), for 87 species (approx. 87% total diversity) belonging to all extant genera.
The chronograms were built using partitioned analysis, birth–death models with a relaxed lognormal clock prior, and fossil calibrations. Details on sequence alignment, model construction, and tree calibration can be found in the Supplementary methods in the electronic supplementary material.
(b). Herbivorous reef fish data
We categorized all species in the phylogenies according to seven traits related to feeding: two categorical morphological traits related to food processing (tooth and alimentary tract morphology), a continuous trait reflecting body size (maximum length), and four categorical behavioural traits (feeding mode, diet, feeding habitat, and schooling behaviour), summarizing how, what, and where species feed. Trait assignments were based on the literature, online datasets, and expert assessments. These seven traits, combined, provide a broad indication of each species' ecological role. However, the relationship between traits and ecosystem functions is complex. While a given function may depend on a single trait state, this is not always the case and, in many instances, functions may be correlated with numerous traits within and among states [37]. We therefore undertook a separate analysis, assigning ecosystem functions for all species, based on published literature and expert assessments. Details of trait and ecosystem function coding can be found in the Supplementary methods in the electronic supplementary material.
We also assembled a database for the putative herbivorous fossil fish species from the Eocene Lagerstätten of Monte Bolca. These fossils from a single locality (two deposits) in northern Italy, represent the richest and most well-preserved fossil record of present-day reef-associated fish families [29]. Because no parrotfish fossil has been recorded from the Eocene [38], we only included surgeonfishes and rabbitfishes in our fossil database. Moreover, we excluded fossil species described from incomplete or larval-stage specimens. We assigned the same traits to fossils as the extant species by correlating their morphology with modern analogues (cf. [10,39]). We also classified the fossils according to ecosystem functions based on their combination of traits. Most of our fossils show indications of being turf-algae croppers with some potential pair-forming species [10], however, others could potentially be macroalgae browsers or planktivores. To account for this uncertainty, we considered the ambiguous fossils as the most distinct states (browsers and planktivores); as a result, our fossil multidimensional space (see Methods section d) is most likely overestimated based on available evidence.
All extant species were also classified according to their geographical ranges based on data from the literature and International Union for Conservation of Nature's (IUCN) Red List [40]. We built a presence/absence matrix of species considering the six recognized biogeographic regions for reef-associated fishes [41]: Western-Indian (WI), Central-Indo-Pacific (CIP), Central-Pacific (CP), Tropical-Eastern-Pacific (TEP), Western-Atlantic (WA), and Eastern-Atlantic (EA). We also classified the fossil species biogeographically as being present in the Tethys Sea, since all were located in the ancient marine biodiversity hotspot in the Eocene [15].
(c). Ancestral states and biogeography
We retrieved the ancestral states in lineages present in four time-slices—20, 15, 10, and 5 Ma—chosen to encompass the changes that took place in the most important phases for coral reef fish diversification [20]. This was achieved by using the Bayesian framework of BayesTraits [42]. Within this software, we performed ancestral state reconstructions for each trait in our maximum clade credibility (MCC) trees using the VarRates model [43]. This model accounts for heterogeneity in the rates of trait evolution within the trees. For each discrete trait, we first set multistate models with uniform priors for the transition rates based on results from a maximum-likelihood analysis. We then ran three independent Markov Chain Monte Carlo (MCMC) chains of five million iterations each, sampling node state probabilities every 4000 iterations. For traits that had more than four states, we used the rjMCMC option that handles a higher number of parameters [42]. After discarding 20% burn-in, we obtained 1000 samples of node state posterior probabilities (PP) for each run. We assessed convergence within chains using the effective sample size (ESS) scores, and between chains using the marginal likelihoods, calculated using the stepping stone sampler [44] in BayesTraits. After ensuring convergence, we used the results of one chain per trait in downstream analysis. The same procedure was used for reconstructing the ecosystem functions. Finally, for the continuous trait (body size), we also used VarRates in BayesTraits with the same MCMC steps as the discrete analysis, but we applied the independent contrasts model [45] to assess ancestral state values.
The results from BayesTraits were used to retrieve the ancestral states in the four time-slices (20, 15, 10, and 5 Ma). We first assessed, for each internal node in the phylogenies, the states with the highest modal PP for the discrete traits and the reconstructed modal value for the continuous trait. With these states and values, we built a trait database for the branch points cut by the time-slices in our trees. Each of these intersecting points was classified according to the states of the closest node for the discrete traits and the proportional value of change between the two adjacent nodes for the continuous trait. These analyses were performed using R [46]. For the ecosystem functions, we applied the same approach as the discrete traits, however, we used more time-slices to assess the presumed origin of each function within ocean basins and to estimate the minimum number of lineages performing functions through time, based on extant taxa. We can only provide a minimum estimate of lineages in each function because, without the fossil record, molecular phylogenies are prone to a perception bias and the number of species can only increase or saturate [47,48]. Specifically, our reconstructions indicate the timing of origin of traits and functions that have led to the construction of modern faunas. They do not preclude earlier originations in extinct taxa.
To assess the ancestral ranges in our groups, we built biogeographic models using the ‘BioGeoBEARS’ R package [49]. We used this framework to build models according to time constraints from the past geological history of marine environments that are well known to influence coral reef fish biogeography [16,50]. Details of model constraints and selection can be found in the Supplementary methods in the electronic supplementary material. Based on results from range reconstructions, we split the lineages from each time-slice between two major oceanic realms (Atlantic and Indo-Pacific; electronic supplementary material, figures S4–S6). Lineages present in the TEP before 3.1 Myr were considered part of the Atlantic realm, given its connection with the WA prior to the closure of the Isthmus of Panama [17]. The extant species in our dataset were also biogeographically split between Atlantic and Indo-Pacific realms based on their current ranges.
(d). Multidimensional trait space
With the congregated trait dataset (including fossil and extant species, and lineages from time-slices) we were able to plot multidimensional convex hulls for each time period and biogeographic realm. Using the R package ‘FD’ [51], we calculated the Gower dissimilarity matrix between all lineages and extracted the axes of the principal coordinate (PCoA) analysis. We then assessed the number of axes that would adequately reflect our trait space using the ‘quality_funct_space’ R function [52]. As was expected from a mainly categorical space [52], the quality of representation increased with the number of axes (electronic supplementary material, figure S7), however, we kept only the first four axes for convenience of graphical representation and because they represent more than 70% of the explained variance in the data. Each principal coordinate axis is correlated differently with the classified traits (electronic supplementary material, figures S11–S13), therefore, changes in multidimensional space are better explained by the combination of traits rather than individual traits. Finally, we assessed the robustness of our results by performing ancestral reconstructions with a recently developed hidden Markov model [53] for discrete trait evolution. Moreover, we examined the effects of two issues that could potentially affect our results: phylogenetic uncertainties (topology and node dates) and uncertainty about reconstructed node states. Details of these sensitivity analyses can be found in the Supplementary methods in the electronic supplementary material.
3. Results
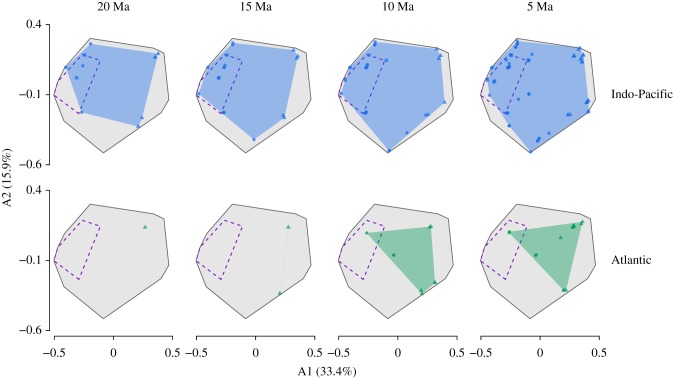
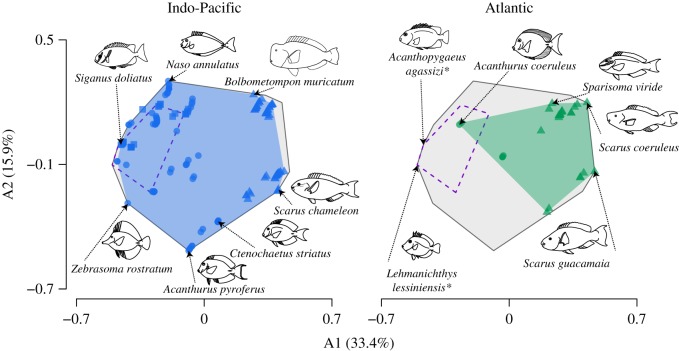
Our Bayesian phylogenetic inferences yielded well-supported trees for all taxa (electronic supplementary material, figures S1–S3). When we performed ancestral state and range reconstructions, we found remarkable differences in the multidimensional trait space occupied through time between biogeographic realms (figure 1). The trait space for fossil species overlaps with lineages retrieved from the Indo-Pacific in the 20 Ma time-slice (figure 1). From this time-slice onwards, the Indo-Pacific realm showed a marked expansion from the ancestral Tethyan trait space. This expansion is associated with the rise of parrotfishes between the Eocene and the Oligocene and the diversification of trait combinations of both surgeonfishes and parrotfishes in the Indo-Pacific in the last 15 Ma (electronic supplementary material, figures S5 and S6). By comparison, the 20–15 Ma Atlantic trait space exhibited a complete turnover when compared to the ancestral Tethyan assemblage (figure 1). The trait combinations retrieved from the Atlantic 20–15 Ma were not recorded from Tethys 50 Ma. After 15 Ma, the Atlantic trait space expanded with the initial diversification of sparisomatine parrotfishes and the dispersal of a surgeonfish lineage from the Indo-Pacific (electronic supplementary material, figures S5 and S6). This colonization event at 10 Ma represented the return of a trait combination to the Atlantic that is within the boundaries of the Tethyan trait space (figure 1). As a consequence of these historical differences, the total herbivorous reef fish trait space is occupied almost entirely by extant Indo-Pacific lineages, with space occupied by remnant Atlantic and Tethys lineages being largely nested within it (figure 2).
Figure 1.
Multidimensional trait space occupied by surgeonfish (circles), rabbitfish (squares), and parrotfish (triangles) lineages in two biogeographic regions through time. Plots show the first two axes (A1-A2) derived from a principal coordinate analysis (PCoA) performed on seven traits related to feeding. Each column represents a time-slice (20–5 Ma) in which we assessed the traits through ancestral reconstructions. Background grey area shows the total space occupied combining fossils, time-slices, and extant species. Convex hulls represent space occupied by Indo-Pacific (blue), Atlantic (green), and Tethys (50 Ma; purple dashed line) lineages in each time-slice. Symbols represent lineages present in each time-slice.
Figure 2.
Multidimensional trait space occupied by extant surgeonfish (circles), rabbitfish (squares), and parrotfish (triangles) species in two biogeographic regions. Plots show the first two axes (A1-A2) derived from a principal coordinate analysis (PCoA) performed on seven traits related to feeding. Background grey area shows the total space occupied combining fossils, time-slices, and extant species. Convex hulls represent space occupied by Indo-Pacific (blue), Atlantic (green), and Tethys (50 Ma; purple dashed line) species. Illustrations show representatives from each biogeographic realm and two fossil species (asterisk).
Our sample of posterior trees derived from the phylogenetic analyses showed that uncertainties in node dates and tree topology had very little effect on the patterns of trait space occupation through time (electronic supplementary material, figure S14). Moreover, the observed differences between biogeographic realms were consistently recovered even when we accounted for trait-dependent diversification and uncertainties in node states derived from the ancestral reconstructions (electronic supplementary material, figures S15–S17).
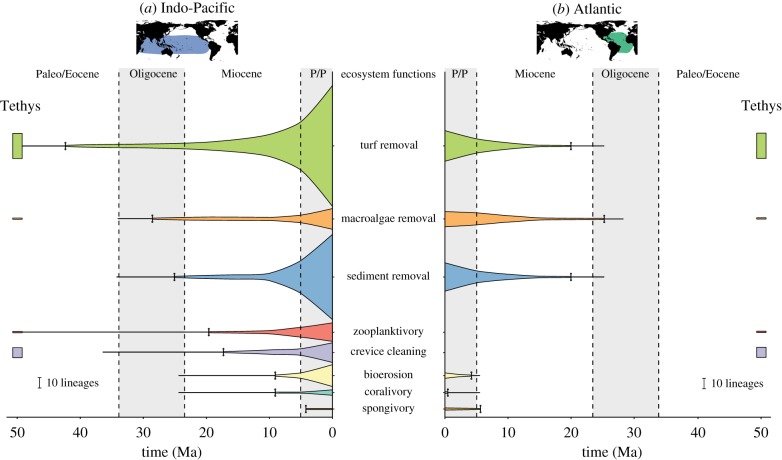
For the ecosystem functions, we identified marked differences between biogeographic realms in the origins of lineages performing each function through time (figure 3). The dominant functions present in the fossil assemblage were the removal of algal turfs and crevice feeding, which were performed by both surgeonfishes and rabbitfishes. Lineages retrieved from our reconstructions showed older origins for all extant ecosystem functions in the Indo-Pacific when compared to the Atlantic. In the Indo-Pacific, the removal of algal turfs and macroalgae by present-day lineages can be traced back to the Eocene/Oligocene, while the lineages performing other functions probably originated in the Oligocene/Miocene (figure 3a). By contrast, in the Atlantic, reconstructions suggested that removal of turf, macroalgae, and sediment by extant fish lineages started in the early Miocene, while bioerosion and corallivory are even more recent, with origins in the Pliocene/Pleistocene (figure 3b). Besides the absence of rabbitfishes in the Atlantic, this region also lacks herbivorous lineages that transitioned to zooplanktivory or are capable of feeding in crevices.
Figure 3.
Surgeonfish, rabbitfish, and parrotfish lineages performing each ecosystem function through time in the Indo-Pacific (a) and Atlantic (b) realms. Funnel width through time was calculated based on the ancestral state reconstructions for ecosystem functions, therefore, numbers are limited to inferences from extant taxa. The split between regions was based on ancestral range reconstructions. The time of origin of each function (vertical dash) was based on the earliest reconstruction of an ancestral node performing that function, and the line traced from the funnels represent the length of the branch that lead to that node, indicating the possible origin of the function at any point along the branch. Rectangles at 50 Ma represent the number of fossil lineages from Monte Bolca performing each ecosystem function in the Tethys Sea. Dashed lines separate geological epochs (P/P: Pliocene/Pleistocene). (Online version in colour.)
Although the origin of lineages performing each ecosystem function varied among biogeographic realms, we found an expansion of lineages in the mid-Miocene continuing up to the Pliocene/Pleistocene in both the Atlantic and the Indo-Pacific (figure 3). Nevertheless, this expansion was much more pronounced in the Indo-Pacific, particularly for the removal of algal turfs and sediment (figure 3a), which reflects the diversification of the most speciose clades in these functional groups (e.g. Scarus and Acanthurus; electronic supplementary material, figures S2 and S3). In the Atlantic, the Pliocene/Pleistocene expansion in turf and sediment removing lineages (figure 3b) is related to the diversification of Sparisoma and Scarus parrotfishes, however, these lineages represent less than one-third of the number of parrotfish lineages present in the Indo-Pacific. Comparatively, the only ecosystem functions that have a similar number of extant lineages between biogeographic realms are the removal of macroalgae and sponges (figure 3). This reflects the fact that most Sparisoma species can feed on both turfs and macroalgae (with some also feeding on sponges), and that macroalgal browsers and spongivores are not very diverse in our focal taxa on Indo-Pacific coral reefs.
By mapping our classified traits with their respective ecosystem functions (electronic supplementary material, figures S18 and S19), we observed an intricate link between morphology, behaviour, and ecology. The removal of turfs, macroalgae, and sediment can be performed by multiple trait combinations (electronic supplementary material, figure S18) and species occupying similar areas of trait space tend to perform similar ecosystem functions irrespective of biogeography (electronic supplementary material, figure S19). However, there are specificities in each biogeographic realm. For example, most scraping parrotfishes in the Atlantic are also capable of removing macroalgae, which does not happen in the Indo-Pacific. There, the removal of macroalgae is exclusively performed by browsing species. Other ecosystem functions such as zooplanktivory and crevice cleaning are performed by lineages with more restricted combinations of traits. Since these functions are absent in lineages from the Atlantic (figure 3), this ocean basin also lacks associated traits (figure 1; electronic supplementary material, figures S18 and S19). Interestingly, spongivory is the only ecosystem function that is performed by lineages with very distinct trait combinations in different realms (i.e. Sparisoma spp. in the Atlantic and Siganus in the Indo-Pacific).
4. Discussion
Through a comprehensive framework combining fossils, phylogenies, and the ecology of extant species, we have revealed two very distinct scenarios for the evolution of herbivorous coral reef fishes between major marine biogeographic realms. The Indo-Pacific showed a clear history of continuity and expansion from the ancestral Tethyan herbivore assemblage. This result uncovered a trait-based component to the historical biogeography of coral reef fishes [16], with the fossil assemblage of the Tethys hotspot forming the core foundations of both taxa [15] and herbivorous trait combinations in the Indo-Pacific. By contrast, the Atlantic herbivore composition was marked by isolation from the marine diversity hotspots and the loss of ancient trait combinations. As a consequence, the extant Atlantic reef fish herbivore assemblage is a combination of a clade that arose there and a wider range of more recently derived lineages that have invaded from the east Tethys/Indo-Pacific (electronic supplementary material, figures S5 and S6). These different histories were also reflected in the lineages performing ecosystem functions within each realm. The Atlantic fauna contains only a small subset of the functions seen in the Indo-Pacific, with few lineages within each represented function. These two components of piscine herbivory in coral reefs, trait combinations and ecosystem functions, will be discussed separately below.
(a). Trait combinations in space and time
The major differences in herbivorous trait combinations between the two oceanic basins can be traced back to the earliest known herbivorous assemblage in marine environments. The absence of morphological features related to herbivory in Mesozoic marine fish fossils indicates that the early Cenozoic was likely a starting point for marine piscine herbivory [21]. However, it is hard to accurately place this origin in space and time. The first unequivocal assemblage of marine herbivorous fishes is in the Eocene fossils of Monte Bolca [21], with many groups already showing striking morphological similarities with their modern counterparts [10]. Considering the central location of this fossil assemblage and its connections with both the east and the west Tethys (presently Indo-Pacific and Atlantic), it is likely to represent a reference point from which global reef fish assemblages evolved [20]. From this reference point, the subsequent history of tropical extant herbivorous fishes mirrored that of other reef fish families [16] with two geographically independent components: the east and the west Tethyan provinces (sensu [19]) following different trajectories.
The eastern component was subject to biogeographic shifts in marine biodiversity. In the Eocene, the global marine biodiversity hotspot was located around the Monte Bolca deposits. Following tectonic events, it subsequently ‘hopped’ to the east during the Oligocene and Miocene, ultimately forming the current-day hotspot in the Indo-Australian-Archipelago (IAA) [15]. This biogeographic shift was reflected in our results, since surgeonfish and rabbitfish trait combinations that were present in the Eocene are maintained across time in the Indo-Pacific. However, as marine hotspots were shifting to the east, herbivorous fish lineages also diversified, resulting in the rise of new trait combinations. The Miocene (23–5.3 Ma) encompasses a period in which coral reefs were experiencing a marked restructuring with expansion of habitats occupied by reef organisms, diversification of lineages, trophic rearrangements, and new fish–coral interactions [20]. Herbivorous fishes were major players in this restructuring of reef ecosystems, particularly parrotfishes and surgeonfishes. These groups underwent evolutionary shifts that resulted in new traits related to the exploitation of detritus and epi/endolithic components of the marine benthos [32,54], which led to the trait space expansion that we detected in the Indo-Pacific. As an example of ecological opportunity driving faster diversification rates [55], these shifts may also have underpinned the high abundance of herbivorous fishes on modern Indo-Pacific coral reefs.
In marked contrast, in the Atlantic, the trait combinations found in herbivorous reef fishes reflect the isolation of the west Tethyan province. With the eastern migration of hotspots, the western side of the Tethys became increasingly isolated from the centre of marine biodiversity [16]. This isolation promoted a separate biogeographic dynamic in the Atlantic [56,57] that resulted in a high proportion of internally originated lineages [16]. Within the Atlantic, the Caribbean reefs are a hotspot for marine biodiversity [58], however, instead of showing a continuous history of connectivity with ancestral Tethys assemblages, like the IAA, the Caribbean was marked by profound faunal turnover as a result of extinction events [59,60]. Most of these extinction events are reported from the Pliocene [59]. However, for herbivorous reef fishes, the key extinction events were probably much earlier (Eocene and Oligocene) [20,61] and resulted in the loss of trait combinations that were present in the Tethys Sea. As a consequence, the present-day herbivore fish composition of the Atlantic represents a combination of traits that were derived from lineages that arose in the east Tethys/Indo-Pacific (Acanthurus and Scarus species) with only one parrotfish clade (Cryptotomus + Nicholsina + Sparisoma) that originated in the Atlantic. Although this clade presents some particular features when compared to other parrotfishes [62], the trait space occupied by these Atlantic parrotfishes is nested entirely within the Indo-Pacific parrotfish space. The recent invasion of the Mediterranean by rabbitfishes and their potential future expansion for Caribbean reefs [63] might represent yet another return of an ancient Tethys set of traits to the Atlantic.
Trait combinations provide an interesting picture of the evolution and global biogeography of herbivorous reef fishes, yet, they represent a partial view of the functional history of herbivory on tropical reefs. The morphological and behavioural traits used herein may be good proxies of their ecological role, which may, ultimately, drive ecosystem processes on coral reefs. However, to better understand how these organismal traits scale-up to larger scale ecological processes, we also have to consider the ecosystem consequences of such combinations [37]. The effect of a species on an ecosystem, as an ecosystem function, should be considered as a distinct measure when compared to the traditional views of functions based on organismal traits [64]. Thus, our analysis of herbivorous ecosystem functions provided a separate but complementary view of the global evolution of herbivory in the marine tropics.
(b). Ecosystem functions in space and time
The diversification of ecosystem functions among herbivorous fishes coincides with a period of major realignment of the marine hotspots that took place in the east Tethys/Indo-Pacific during the Miocene. This period not only marks the rise of most modern reef fish genera with increased subsequent lineage diversification [20,65], but coincides with the shift of the centre of tropical marine diversity to the IAA [15]. In parallel with the taxonomic diversification, this was also an important time for reef fish functional and trophic innovation [55,66,67], which was clearly reflected in our ecosystem functions results. Bellwood et al. [20] describe this as a distinct phase (phase 5) in the evolution of reef fishes, that was characterized by the formation of a high turnover, dynamic reef ecosystem. However, this important phase for functional expansion seems to be a phenomenon that is largely restricted to the Indo-Pacific. The Atlantic Ocean lags behind the Indo-Pacific both in the time of origin of most modern reef processes executed by extant herbivorous fishes and in the number of extant lineages performing each function.
Environmental changes have been a consistent feature in the Atlantic throughout the last 10 Myr, particularly in the Caribbean [59]. This has resulted in unstable conditions for corals and reefs through time [60,68], prompting the suggestion that lineages that could thrive in peripheral non-coral environments might have resisted periods of high extinction for coral reef-associated faunas [19]. Interestingly, most of the extant Atlantic herbivorous fish species still have macroalgae and seagrass as important components of their diet [69] and some of them are still associated with peripheral reef habitats in at least one of their life stages. These food habits and habitat associations might thus have been one of the main reasons for the persistence through time of the only Atlantic herbivorous fish clade restricted to the western Tethys (Sparisomatina). However, this may be a partial explanation given that some rabbitfish species are also not closely associated with coral reefs, yet, the family is no longer present in the Atlantic (its presence in Bolca presumably indicates it as an Atlantic component in the Eocene). Selective or not, extinction events shaped the Atlantic herbivore composition leaving the sparisomatine parrotfishes as major contributors to ecosystem functions through time in this ocean basin.
Our results from the trait approach were largely complementary with those from the ecosystem functions. The first has provided a clear picture of how ecological innovations have been a prevalent feature in the last 20 Ma for herbivorous fishes on coral reefs. The second has yielded insights about how these novel ecological features relate to ecosystem functions that enabled these lineages to play pivotal roles in modern coral reef ecosystems. For example, the initial diversification of parrotfishes and surgeonfishes in the Early Miocene resulted in the rise of new trait combinations in the Indo-Pacific (figure 1). However, it was not until the Late Miocene/Pliocene that these lineages started to expand in terms of functional roles, particularly in turf and sediment removal (figure 3). This showcases the complementarity of trait and function approaches. Parrotfishes and surgeonfishes have very distinct trait combinations reflecting different feeding strategies. However, when translated to ecosystem functions most lineages within these groups fit into only two processes: the removal of algal turfs and sediment (electronic supplementary material, figure S18). Interestingly, we see a similar pattern of lineage expansion within these functions in both biogeographic realms, although much more pronounced in the Indo-Pacific. Not coincidently, turf and sediment removal are critical functions that facilitate coral-dominated states on present-day reefs [23,24]. This suggests that trait innovations in herbivorous fishes from the Miocene might have been key to the evolution of coral reefs as we know today, and that recent human impacts might be shifting them back to pre-Miocene conditions before those traits evolved.
(c). Caveats
Although we found striking dissimilarities between biogeographic realms through time, our results are limited to what can be inferred from present-day faunas. It is important to note that the lack of trait combinations and ecosystem functions shown here at certain times or locations does not represent evidence of absence, merely absence of evidence. Our phylogenies indicate the presumed origins in terms of the time and location of traits and functions within extant lineages and are, therefore, blind to the attributes of extinct lineages unknown from the fossil record [47,48]. Indeed, the Eocene fossil evidence indicates that some traits and functions can predate these initial occurrences based on phylogenies. Moreover, extinctions may have erased the signal of trait and function trajectories that are not found in extant species [48]. Unfortunately, the incompleteness of the fossil record for herbivorous reef fishes after the Eocene leaves no alternatives other than using the only tool available to trace their history after Monte Bolca, molecular phylogenies. Finally, our inferences about the Tethyan traits and functions are limited to what is known from a single assemblage (Monte Bolca), which is likely to be an underestimate of the global Eocene reef fish fauna.
5. Conclusion
Our study represents the first effort to collectively analyse the evolutionary processes behind the formation of major extant herbivorous coral reef fish groups among marine biogeographic realms. Character optimizations and fossil information showed that both trait space and the origin of lineages performing ecosystem functions differed between the Indo-Pacific and the Atlantic through time. While modern reef processes related to herbivory developed and expanded in the Indo-Pacific, the Atlantic was very likely shaped by extinction events. Present-day differences in the functional composition of herbivorous fish assemblages between the two major oceanic basins are, therefore, largely a result of their disparate evolutionary histories.
Supplementary Material
Supplementary Material
Acknowledgements
We thank J.H. Choat and L. Sorbini for inspiration; R. Morais for analytical guidance and helpful ideas; C. Hemingson, V. Huertas, M. Mihalitsis, R. Streit, and S. Tebbett for insightful discussions; L. Herrera-Alsina for helping with the SecSSE package; and C.E.L. Ferreira for helping with trait categorization.
Data accessibility
The codes and data to reproduce the results are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.h517t14 [70].
Authors' contributions
A.C.S., D.R.B., and P.F.C. conceived the study. A.C.S. collected and analysed the data. A.C.S. wrote the first draft of the manuscript and D.R.B. and P.F.C. contributed substantially to revisions.
Competing interests
We have no competing interests.
Funding
Funding was provided by the Australian Research Council (D.R.B. and P.F.C.) and James Cook University (Postgraduate Research Scholarship—A.C.S.).
References
- 1.Benton MJ. 2009. The Red Queen and the Court Jester: species diversity and the role of biotic and abiotic factors through time. Science 323, 728–732. ( 10.1126/science.1157719) [DOI] [PubMed] [Google Scholar]
- 2.Jablonski D. 2008. Biotic interactions and macroevolution: extensions and mismatches across scales and levels. Evolution 62, 715–739. ( 10.1111/j.1558-5646.2008.00317.x) [DOI] [PubMed] [Google Scholar]
- 3.Fritz SA, Schnitzler J, Eronen JT, Hof C, Böhning-Gaese K, Graham CH. 2013. Diversity in time and space: wanted dead and alive. Trends Ecol. Evol. 28, 509–516. ( 10.1016/j.tree.2013.05.004) [DOI] [PubMed] [Google Scholar]
- 4.Price SA, Schmitz L. 2016. A promising future for integrative biodiversity research: an increased role of scale-dependency and functional biology. Phil. Trans. R. Soc. B 371, 20150228 ( 10.1098/rstb.2015.0228) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mouillot D, Villéger S, Scherer-Lorenzen M, Mason NWH. 2011. Functional structure of biological communities predicts ecosystem multifunctionality. PLoS ONE 6, e17476 ( 10.1371/journal.pone.0017476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vermeij GJ. 1977. The Mesozoic marine revolution: evidence from snails, predators and grazers. Paleobiology 3, 245–258. ( 10.2307/2400374) [DOI] [Google Scholar]
- 7.Mouillot D, Graham NAJ, Villéger S, Mason NWH, Bellwood DR. 2013. A functional approach reveals community responses to disturbances. Trends Ecol. Evol. 28, 167–177. ( 10.1016/j.tree.2012.10.004) [DOI] [PubMed] [Google Scholar]
- 8.Violle C, Reich PB, Pacala SW, Enquist BJ, Kattge J. 2014. The emergence and promise of functional biogeography. Proc. Natl Acad. Sci. USA 111, 13 690–13 696. ( 10.1073/pnas.1415442111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villéger S, Novack-Gottshall PM, Mouillot D. 2011. The multidimensionality of the niche reveals functional diversity changes in benthic marine biotas across geological time. Ecol. Lett. 14, 561–568. ( 10.1111/j.1461-0248.2011.01618.x) [DOI] [PubMed] [Google Scholar]
- 10.Bellwood DR, Goatley CHR, Brandl SJ, Bellwood O. 2014. Fifty million years of herbivory on coral reefs: fossils, fish and functional innovations. Proc. R. Soc. B 281, 20133046 ( 10.1098/rspb.2013.3046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whittaker RJ, et al. 2014. Functional biogeography of oceanic islands and the scaling of functional diversity in the Azores. Proc. Natl Acad. Sci. USA 111, 13 709–13 714. ( 10.1073/pnas.1218036111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toussaint A, Charpin N, Brosse S, Villéger S. 2016. Global functional diversity of freshwater fish is concentrated in the Neotropics while functional vulnerability is widespread. Sci. Rep. 6, 1–9. ( 10.1038/srep22125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemingson CR, Bellwood DR. 2018. Biogeographic patterns in major marine realms: function not taxonomy unites fish assemblages in reef, seagrass and mangrove systems. Ecography 41, 174–182. ( 10.1111/ecog.03010) [DOI] [Google Scholar]
- 14.Pellissier L, et al. 2014. Quaternary coral reef refugia preserved fish diversity. Science 344, 1016–1019. ( 10.1126/science.1249853) [DOI] [PubMed] [Google Scholar]
- 15.Renema W, Bellwood DR, Braga JC. 2008. Hopping hotspots: global shifts in marine biodiversity. Science 321, 654–657. ( 10.1126/science.1155674) [DOI] [PubMed] [Google Scholar]
- 16.Cowman PF, Bellwood DR. 2013. The historical biogeography of coral reef fishes: global patterns of origination and dispersal. J. Biogeogr. 40, 209–224. ( 10.1111/jbi.12003) [DOI] [Google Scholar]
- 17.Cowman PF, Parravicini V, Kulbicki M, Floeter SR. 2017. The biogeography of tropical reef fishes: endemism and provinciality through time. Biol. Rev. 92, 2112–2130. ( 10.1111/brv.12323) [DOI] [PubMed] [Google Scholar]
- 18.Wood R. 1999. Reef evolution. Oxford, UK: Oxford University Press. [Google Scholar]
- 19.Bellwood DR, Wainwright PC. 2002. The history and biogeography of fishes on coral reefs. In Coral reef fishes: dynamics and diversity on a complex ecosystem (ed. Sale PF.), pp. 5–32. San Diego, CA: Academic Press; ( 10.1016/B978-012615185-5/50003-7) [DOI] [Google Scholar]
- 20.Bellwood DR, Goatley CHR, Bellwood O. 2017. The evolution of fishes and corals on reefs: form, function and interdependence. Biol. Rev. 92, 878–901. ( 10.1111/brv.12259) [DOI] [PubMed] [Google Scholar]
- 21.Bellwood DR. 2003. Origins and escalation of herbivory in fishes: a functional perspective. Paleobiology 29, 71–83. () [DOI] [Google Scholar]
- 22.Bellwood DR, Hughes TP, Folke C, Nyström M. 2004. Confronting the coral reef crisis. Nature 429, 827–833. ( 10.1038/nature02691) [DOI] [PubMed] [Google Scholar]
- 23.Hughes TP, et al. 2007. Phase shifts, herbivory, and the resilience of coral reefs to climate change. Curr. Biol. 17, 360–365. ( 10.1016/j.cub.2006.12.049) [DOI] [PubMed] [Google Scholar]
- 24.Goatley CHR, Bonaldo RM, Fox RJ, Bellwood DR. 2016. Sediments and herbivory as sensitive indicators of coral reef degradation. Ecol. Soc. 21, 29 ( 10.5751/ES-08334-210129) [DOI] [Google Scholar]
- 25.Wilson SK, Graham NAJ, Pratchett MS, Jones GP, Polunin NVC. 2006. Multiple disturbances and the global degradation of coral reefs: are reef fishes at risk or resilient? Glob. Chang. Biol. 12, 2220–2234. ( 10.1111/j.1365-2486.2006.01252.x) [DOI] [Google Scholar]
- 26.Bellwood DR, Choat JH. 1990. A functional analysis of grazing in parrotfishes (family Scaridae): the ecological implications. Environ. Biol. Fishes 28, 189–214. ( 10.1007/BF00751035) [DOI] [Google Scholar]
- 27.Steneck RS, Bellwood DR, Hay ME. 2017. Herbivory in the marine realm. Curr. Biol. 27, R484–R489. ( 10.1016/j.cub.2017.04.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steneck RS. 1983. Escalating herbivory and resulting adaptive trends in calcareous algal crusts. Paleobiology 9, 44–61. ( 10.1017/S0094837300007375) [DOI] [Google Scholar]
- 29.Bellwood DR. 1996. The Eocene fishes of Monte Bolca: the earliest coral reef fish assemblage. Coral Reefs 15, 11–19. ( 10.1007/s003380050025) [DOI] [Google Scholar]
- 30.Choat JH, Klanten OS, Van Herwerden L, Robertson RD, Clements KD.. 2012. Patterns and processes in the evolutionary history of parrotfishes (Family Labridae). Biol. J. Linn. Soc. 107, 529–557. ( 10.1111/j.1095-8312.2012.01959.x) [DOI] [Google Scholar]
- 31.Choat JH. 1991. The biology of herbivorous fishes on coral reefs. In The ecology of fishes on coral reefs (ed. Sale PF.), pp. 120–155. San Diego, CA: Academic Press. [Google Scholar]
- 32.Bellwood DR, Hoey AS, Bellwood O, Goatley CHR. 2014. Evolution of long-toothed fishes and the changing nature of fish-benthos interactions on coral reefs. Nat. Commun. 5, 3144 ( 10.1038/ncomms4144) [DOI] [PubMed] [Google Scholar]
- 33.Floeter SR, Bender MG, Siqueira AC, Cowman PF. 2018. Phylogenetic perspectives on reef fish functional traits. Biol. Rev. 93, 131–151. ( 10.1111/brv.12336) [DOI] [PubMed] [Google Scholar]
- 34.Green AL, Bellwood DR. 2009. Monitoring functional groups of herbivorous reef fishes as indicators of coral reef resilience: a practical guide for coral reef managers in the Asia Pacific region. Gland, Switzerland: IUCN Resilience Science Group Working Paper Series No 7. [Google Scholar]
- 35.Westneat MW, Alfaro ME. 2005. Phylogenetic relationships and evolutionary history of the reef fish family Labridae. Mol. Phylogenet. Evol. 36, 370–390. ( 10.1016/j.ympev.2005.02.001) [DOI] [PubMed] [Google Scholar]
- 36.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537 ( 10.1371/journal.pcbi.1003537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bellwood DR, Streit RP, Brandl SJ, Tebbett SB. 2019. The meaning of the term ‘function’ in ecology: a coral reef perspective. Funct. Ecol. 1–14. ( 10.1111/1365-2435.13265) [DOI] [Google Scholar]
- 38.Bellwood DR, Schultz O. 1991. A review of the fossil record of the Parrotfishes (Labroidei: Scaridae) with a description of a new Calotomus species from the Middle Miocene (Badenian) of Austria. Ann. Des Naturhistorischen Museums Wien. Ser. A, Fur Mineral. und Petrogr. Geol. Und Palaontologie, Anthropol. und Prahistorie 92, 55–71. [Google Scholar]
- 39.Brandl SJ, Bellwood DR. 2013. Morphology, sociality, and ecology: can morphology predict pairing behavior in coral reef fishes? Coral Reefs 32, 835–846. ( 10.1007/s00338-013-1042-0) [DOI] [Google Scholar]
- 40.IUCN. 2017. IUCN Red List of Threatened Species. See www.iucnredlist.org
- 41.Kulbicki M, et al. 2013. Global biogeography of reef fishes: a hierarchical quantitative delineation of regions. PLoS ONE 8, e81847 ( 10.1371/journal.pone.0081847) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meade A, Pagel M. 2018. BayesTraits: a computer package for analyses of trait evolution. Version 3. See http://www.evolution.rdg.ac.uk/BayesTraits.html .
- 43.Venditti C, Meade A, Pagel M. 2011. Multiple routes to mammalian diversity. Nature 479, 393–396. ( 10.1038/nature10516) [DOI] [PubMed] [Google Scholar]
- 44.Xie W, Lewis PO, Fan Y, Kuo L, Chen M-H. 2011. Improving marginal likelihood estimation for Bayesian phylogenetic model selection. Syst. Biol. 60, 150–160. ( 10.1093/sysbio/syq085) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freckleton RP. 2012. Fast likelihood calculations for comparative analyses. Methods Ecol. Evol. 3, 940–947. ( 10.1111/j.2041-210X.2012.00220.x) [DOI] [Google Scholar]
- 46.R Development Core Team. 2018. R: a language and environment for statistical computing. See http://www.r-project.org
- 47.Quental TB, Marshall CR. 2010. Diversity dynamics: molecular phylogenies need the fossil record. Trends Ecol. Evol. 25, 435–441. ( 10.1016/j.tree.2010.05.002) [DOI] [PubMed] [Google Scholar]
- 48.Marshall CR. 2017. Five palaeobiological laws needed to understand the evolution of the living biota. Nat. Ecol. Evol. 1, 165 ( 10.1038/s41559-017-0165) [DOI] [PubMed] [Google Scholar]
- 49.Matzke N. 2013. BioGeoBEARS: BioGeography with Bayesian (and Likelihood) Evolutionary Analysis in R Scripts. R Package version 0.2.1.
- 50.Cowman PF, Bellwood DR. 2013. Vicariance across major marine biogeographic barriers: temporal concordance and the relative intensity of hard versus soft barriers. Proc. R. Soc. B 280, 20131541 ( 10.1098/rspb.2013.1541) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laliberté AE, Legendre P, Shipley B, Laliberté ME. 2014. FD: measuring functional diversity from multiple traits, and other tools for functional ecology. R Package 1.0.12.
- 52.Maire E, Grenouillet G, Brosse S, Villéger S. 2015. How many dimensions are needed to accurately assess functional diversity? A pragmatic approach for assessing the quality of functional spaces. Glob. Ecol. Biogeogr. 24, 728–740. ( 10.1111/geb.12299) [DOI] [Google Scholar]
- 53.Herrera-Alsina L, van Els P, Etienne RS. 2019. Detecting the dependence of diversification on multiple traits from phylogenetic trees and trait data. Syst. Biol. 68, 317–328. ( 10.1093/sysbio/syy057) [DOI] [PubMed] [Google Scholar]
- 54.Clements KD, German DP, Piché J, Tribollet A, Choat JH. 2017. Integrating ecological roles and trophic diversification on coral reefs: multiple lines of evidence identify parrotfishes as microphages. Biol. J. Linn. Soc. 120, 729–751. ( 10.1111/bij.12914) [DOI] [Google Scholar]
- 55.Lobato FL, Barneche DR, Siqueira AC, Liedke AMR, Lindner A, Pie MR, Bellwood DR, Floeter SR. 2014. Diet and diversification in the evolution of coral reef fishes. PLoS ONE 9, e102094 ( 10.1371/journal.pone.0102094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Floeter SR, et al. 2008. Atlantic reef fish biogeography and evolution. J. Biogeogr. 35, 22–47. ( 10.1111/j.1365-2699.2007.01790.x) [DOI] [Google Scholar]
- 57.Joyeux J, Floeter SR, Ferreira CEL, Gasparini JL. 2001. Biogeography of tropical reef fishes: the South Atlantic puzzle. J. Biogeogr. 28, 831–841. ( 10.1046/j.1365-2699.2001.00602.x) [DOI] [Google Scholar]
- 58.Briggs JC, Bowen BW. 2013. Marine shelf habitat: biogeography and evolution. J. Biogeogr. 40, 1023–1035. ( 10.1111/jbi.12082) [DOI] [Google Scholar]
- 59.O'Dea A, Jackson JBC, Fortunato H, Smith JT, D'Croz L, Johnson KG, Todd JA. 2007. Environmental change preceded Caribbean extinction by 2 million years. Proc. Natl Acad. Sci. USA 104, 5501–5506. ( 10.1073/pnas.0610947104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Budd AF. 2000. Diversity and extinction in the Cenozoic history of Caribbean reefs. Coral Reefs 19, 25–35. ( 10.1007/s003380050222) [DOI] [Google Scholar]
- 61.Tyler JC, Sorbini L. 1998. On the relationships of Eonaso, an Antillean fossil surgeon fish (Acanthuridae). Stud. e Ric. sui Giacimenti Terziari di Bolca, Mus. Civ. di Stor. Nat. di Verona 7, 35–42. [Google Scholar]
- 62.Streelman JT, Alfaro M, Westneat MW, Bellwood DR, Karl SA. 2002. Evolutionary history of the parrotfishes: biogeography, ecomorphology, and comparative diversity. Evolution 56, 961–971. ( 10.1554/0014-3820(2002)056) [DOI] [PubMed] [Google Scholar]
- 63.Bellwood DR, Goatley CHR. 2017. Can biological invasions save Caribbean coral reefs? Curr. Biol. 27, R13–R14. ( 10.1016/j.cub.2016.11.018) [DOI] [PubMed] [Google Scholar]
- 64.Violle C, Navas ML, Vile D, Kazakou E, Fortunel C, Hummel I, Garnier E. 2007. Let the concept of trait be functional! Oikos 116, 882–892. ( 10.1111/j.2007.0030-1299.15559.x) [DOI] [Google Scholar]
- 65.Cowman PF, Bellwood DR. 2011. Coral reefs as drivers of cladogenesis: expanding coral reefs, cryptic extinction events, and the development of biodiversity hotspots. J. Evol. Biol. 24, 2543–2562. ( 10.1111/j.1420-9101.2011.02391.x) [DOI] [PubMed] [Google Scholar]
- 66.Alfaro ME, Brock CD, Banbury BL, Wainwright PC. 2009. Does evolutionary innovation in pharyngeal jaws lead to rapid lineage diversification in labrid fishes? BMC Evol. Biol. 9, 255 ( 10.1186/1471-2148-9-255) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cowman PF, Bellwood DR, van Herwerden L.. 2009. Dating the evolutionary origins of wrasse lineages (Labridae) and the rise of trophic novelty on coral reefs. Mol. Phylogenet. Evol. 52, 621–631. ( 10.1016/j.ympev.2009.05.015) [DOI] [PubMed] [Google Scholar]
- 68.Johnson KG, Budd AF, Stemann TA. 1995. Extinction selectivity and ecology of Neogene Caribbean reef corals. Paleobiology 21, 52–73. ( 10.1017/S0094837300013075) [DOI] [Google Scholar]
- 69.Bonaldo RM, Hoey AS, Bellwood DR. 2014. The ecosystem roles of parrotfishes on tropical reefs. Oceanogr. Mar. Biol. An Annu. Rev. 52, 81–132. ( 10.1201/b17143-3) [DOI] [Google Scholar]
- 70.Siqueira AC, Bellwood DR, Cowman PF. 2019. Data from: the evolution of traits and functions in herbivorous coral reef fishes through space and time Dryad Digital Repository. ( 10.5061/dryad.h517t14) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Siqueira AC, Bellwood DR, Cowman PF. 2019. Data from: the evolution of traits and functions in herbivorous coral reef fishes through space and time Dryad Digital Repository. ( 10.5061/dryad.h517t14) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The codes and data to reproduce the results are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.h517t14 [70].