Abstract
Glioblastoma is the most common primary malignant brain tumor in adults and associated with poor survival. The Ivy Foundation Early Phase Clinical Trials Consortium conducted a randomized, multi-institution clinical trial to evaluate immune responses and survival following neoadjuvant and/or adjuvant therapy with pembrolizumab in 35 patients with recurrent, surgically resectable glioblastoma. Patients who were randomized to receive neoadjuvant pembrolizumab, with continued adjuvant therapy following surgery, had significantly extended overall survival compared to patients that were randomized to receive adjuvant, post-surgical PD-1 blockade alone. Neoadjuvant PD-1 blockade was associated with upregulation of T cell and interferon-γ-related gene expression, but downregulation of cell cycle-related gene expression within the tumor, which was not seen in patients that received adjuvant therapy alone. Focal induction of programmed death-ligand 1 (PD-L1) in the tumor microenvironment, enhanced clonal expansion of T cells, decreased PD-1 expression on peripheral blood T cells, and a decreasing monocytic population was observed more frequently in the neoadjuvant group than patients treated only in the adjuvant setting. These findings suggest that the neoadjuvant administration of PD-1 blockade enhances the local and systemic anti-tumor immune response and may represent a more efficacious approach to the treatment of this uniformly lethal brain tumor.
Introduction
Glioblastoma, with an incidence of 3.2 per 100,000 population, is the most common malignant central nervous system tumor, and carries an abysmal 3-year survival rate of just 10.1%.1 The median progression-free survival in primary glioblastoma is 6.9 months and median overall survival 14.6 months with standard of care surgery, radiation therapy and temozolomide.2 In recurrent glioblastoma, median overall survival is an estimated 24-44 weeks.3-5 New therapies are needed for patients diagnosed with this type of cancer.
Among the array of available cancer immunotherapeutics, PD-1 monoclonal antibody blockade has yielded promising results in patients with metastatic cancer.6-12 PD-1 inhibition is thought to disrupt the engagement of PD-1 with its inhibitory ligands, spurring cytotoxic T cell-mediated tumor elimination.10,13 Pembrolizumab, an anti-PD-1 monoclonal antibody, has demonstrated benefit as monotherapy in multiple cancer types,14,15 but primarily in the adjuvant setting.16 However, a preclinical metastatic breast cancer study suggested that neoadjuvant immune checkpoint inhibition could generate enhanced and sustained anti-tumor immune responses, resulting in a survival benefit over adjuvant therapy alone.17 Such concepts were recently validated by a small single-arm clinical study in resectable lung cancer,18 a small randomized trial in melanoma,19 as well as a phase II trial in melanoma comparing neoadjuvant nivolumab to neoadjuvant nivolumab with ipilimumab.20 All studies demonstrated enhanced T cell responses and a clinical benefit with neoadjuvant checkpoint inhibition.
To date, PD-1 blockade has demonstrated limited efficacy in patients with glioblastoma, except in isolated case reports associated with mismatch repair deficiency.21-24 Pre-clinical studies, however, have suggested that the PD-1/PD-L1 axis is immunologically relevant and a therapeutic window exists.25-30 To address the question of whether neoadjuvant PD-1 blockade would alter the functional immune landscape and extend survival in patients, the Ivy Consortium initiated a multi-institution, randomized, open-label pilot study of pembrolizumab in patients with recurrent, surgically resectable glioblastoma. We leveraged T cell receptor sequencing, gene expression profiling, mass cytometry and quantitative multiplex immunofluorescence to explore the intratumoral immune consequences of PD-1 monoclonal antibody administration and identify potential biomarkers of response.
Results
Trial patient characteristics
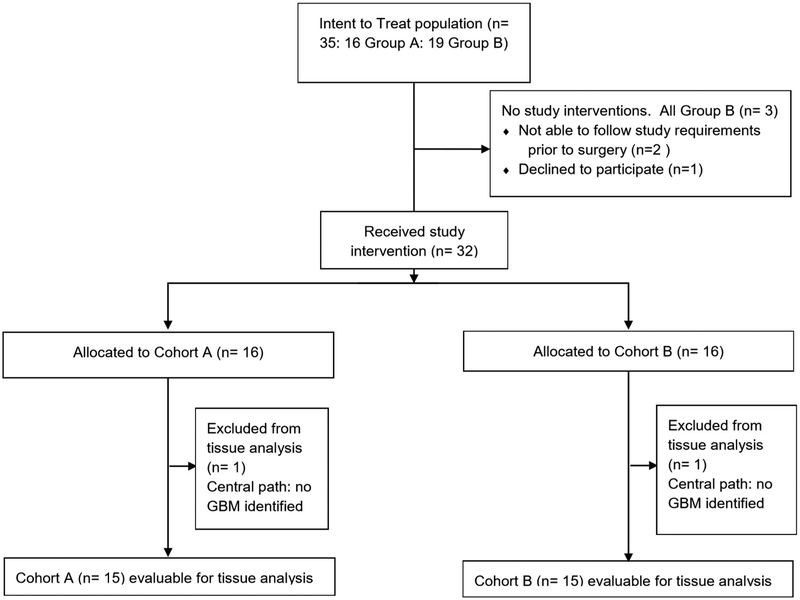
A total of 35 patients were enrolled and randomized between October 2016 and September 2017 at seven institutions and comprise the intention-to-treat population (Extended Data Figure 1). Sixteen patients were randomized into the neoadjuvant pembrolizumab group and nineteen into the adjuvant-only group. Three patients in the adjuvant-only group withdrew consent prior to surgery and two patients (one in the neoadjuvant group and one in the adjuvant-only group) were replaced following surgery according to the study protocol based on insufficient histological evidence of glioblastoma. These five patients were excluded from tissue studies but were included in the intention-to-treat efficacy analysis. The baseline patient characteristics are dichotomized by treatment group in Table 1. There were no statistically significant differences in age, sex, Karnofsky performance status, isocitrate dehydrogenase (IDH) mutation status, O6-methylguanine DNA methyltransferase (MGMT) methylation status, pre- or post-surgery tumor volume or steroid administration at registration. Furthermore, the extent of resection or the fraction of patients who received a gross total resection was not different between groups. As of the July 2, 2018 analysis cutoff date, 31 of 32 patients had discontinued pembrolizumab (two for unacceptable toxicity, 1 by investigator decision, 1 by withdrawal of consent, and 27 due to progressive disease). Of the 27 patients with progressive disease, 24 went on to receive a bevacizumab-containing regimen. Dexamethasone was also administered to individual patients at varying times at the site investigator’s discretion (Figure 1b). The Kaplan-Meier estimated median follow-up time was 476 days (15.6 months; interquartile range 414 to 522 days [13.6 to 17.2 months]).
Table 1.
Patient characteristics by group. IDH: isocitrate dehydrogenase. MGMT: O6-methylguanine DNA methyltransferase. SD: standard deviation. SEM: standard error of the mean.
| Patient Characteristics | |||
|---|---|---|---|
| Neoadjuvant (n=16) |
Adjuvant (n=16) |
Total (n=32) |
|
| Demographics | |||
| Age at enrollment - years, mean ± SD | 55.4 ± 13.5 | 59.3 ± 12.3 | 57.4 ± 12.8 |
| Gender, no. (%) | |||
| Male | 7 (44) | 5 (31) | 12 (38) |
| Female | 9 (56) | 11 (69) | 19 (63) |
| Karnofsky performance status, mean ± SD | 80 ± 8.9 | 86 ± 6.2 | 83 ± 8.2 |
| Steroid use at registration | |||
| No. of patients receiving steroids | 7 | 7 | |
| Daily dosage, median, mg/day (range) | 0 (0-4) | 0 (0-6) | |
| MGMT status, no. (%) | |||
| Methylated | 6 (38) | 11 (69) | 17 (53) |
| Unmethylated | 7 (44) | 4 (26) | 11 (34) |
| Unknown | 3 (19) | 1 (6) | 4 (13) |
| IDH status, no. (%) | |||
| Wild-type | 12 (75) | 13 (81) | 25 (78) |
| Mutant | 3 (19) | 2 (13) | 5 (16) |
| Unknown | 1 (6) | 1 (6) | 2 (6) |
| Pre-surgery tumor volume, cm3, mean ± SEM | 21.8 ± 6.2 | 18.9 ± 3.2 | |
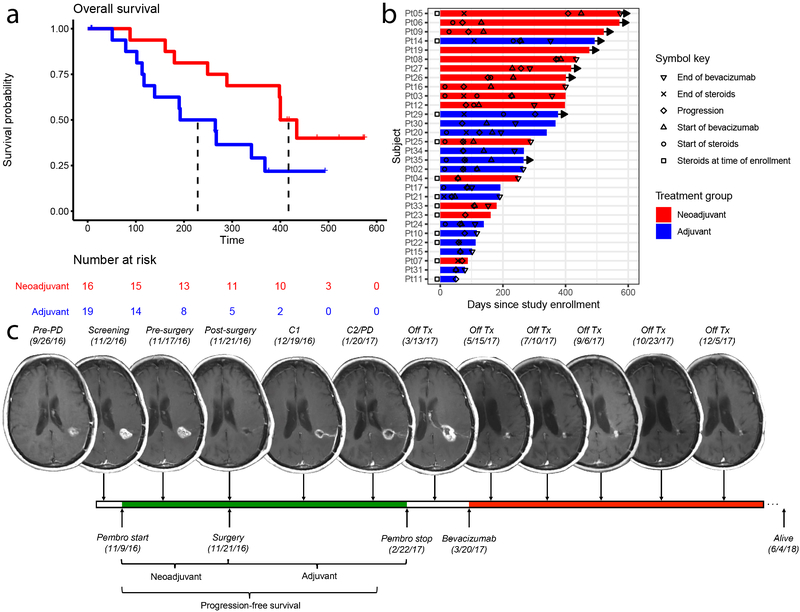
Figure 1. Neoadjuvant pembrolizumab confers significant improvement in overall and progression-free survival in patients with recurrent glioblastoma.
Patients in the neoadjuvant arm (red) received 200 mg pembrolizumab 14±5 days prior to surgical resection; patients in the adjuvant-only arm (blue) did not; both groups received 200 mg adjuvant pembrolizumab every 3 weeks. (a) Kaplan-Meier plot of overall survival. Median overall survival for the patients receiving adjuvant treatment only was 228.5 days, whereas median survival in the neoadjuvant group was 417 days (hazard ratio (HR) 0.39 neoadjuvant/adjuvant; P = 0.04 by log-rank test, n = 35 patients). (b) Swimmer plot of individual patients depicting start and stop times of dexamethasone or equivalent (as recorded at study MRI visit) and/or bevacizumab, if applicable, as well as time to progression, in days after enrollment. Bars represent overall survival in days. Rightward arrow indicates that the patient was still alive at the time of final data collection. (c) Serial magnetic resonance imaging (MRI) scans of a representative patient from the neoadjuvant group. The patient, a 66-year old female in first recurrence, is IDH negative and MGMT unmethylated. IDH: isocitrate dehydrogenase. MGMT: O6-methylguanine DNA methyltransferase. PD: progression of disease. Pembro: pembrolizumab. Tx: treatment.
Safety
Pembrolizumab was generally well tolerated (Supplementary Table 1); the neoadjuvant administration of the antibody was not associated with any new, unreported toxicities in this patient population. As of the analysis cutoff date, 10 patients (67%) in the neoadjuvant group experienced grade 3-4 adverse events that were deemed unlikely, possible, probable or likely attributable to the study drug; this occurred in 7 patients (47%; two-sided P = 0.46, Fisher exact test, 95% confidence interval [CI] 0.42 to 12.95) in the adjuvant cohort. No patients in the neoadjuvant group had surgery delayed due to adverse events prior to tumor resection. Two patients, both in the neoadjuvant group, experienced toxicities leading to discontinuation of pembrolizumab; one patient developed grade 3 pneumonitis and the other a grade 4 elevation in alanine aminotransferase (ALT). The most common treatment-related toxicities were muscle weakness (50%), headache (47%), and hyperglycemia (37%). Such treatment-related adverse events are commonly seen in patients with central nervous system tumors and/or those who have received corticosteroids; indeed, several of these adverse events were deemed unlikely to be related to the study drug by the site investigator but were included for completeness of data reporting.
Neoadjuvant pembrolizumab confers significant improvement in overall and progression-free survival
As of the analysis cutoff date, there have been 9 deaths to date in the neoadjuvant arm and 12 in the adjuvant-only arm. In the intention-to-treat analysis, patients in the neoadjuvant arm demonstrated a statistically significant increase in overall survival, with a hazard ratio of 0.39 compared to the adjuvant-only group (95% CI 0.17 to 0.94; P = 0.04, log-rank test). Patients in the adjuvant-only group had a median overall survival of 228 days (7.5 months), whereas patients in the neoadjuvant arm had a median overall survival of 417 days (13.7 months; Figure 1a). Median progression-free survival was 72.5 days (2.4 months) in the adjuvant-only group and 99.5 days (3.3 months) in the neoadjuvant group (hazard ratio 0.43; 95% CI 0.20 to 0.90; P = 0.03, log-rank test; Figure 1b, Extended Data Figure 2); however, the establishment of progression-free survival may be complicated by pseudoprogression (Figure 1c), a finding observed before in malignant glioma patients treated with immunotherapy.31-33 In patients that received surgery and had histologic evidence of tumor (n = 15 patients per group), the median overall survival of the neoadjuvant treatment cohort was 400 days (13.2 months) from registration date, while that of the adjuvant treatment cohort was 192 days (6.3 months; hazard ratio = 0.35, 95% CI 0.14 to 0.88; P = 0.03, log-rank test).
Neoadjuvant PD-1 blockade induces distinct tumoral gene expression changes
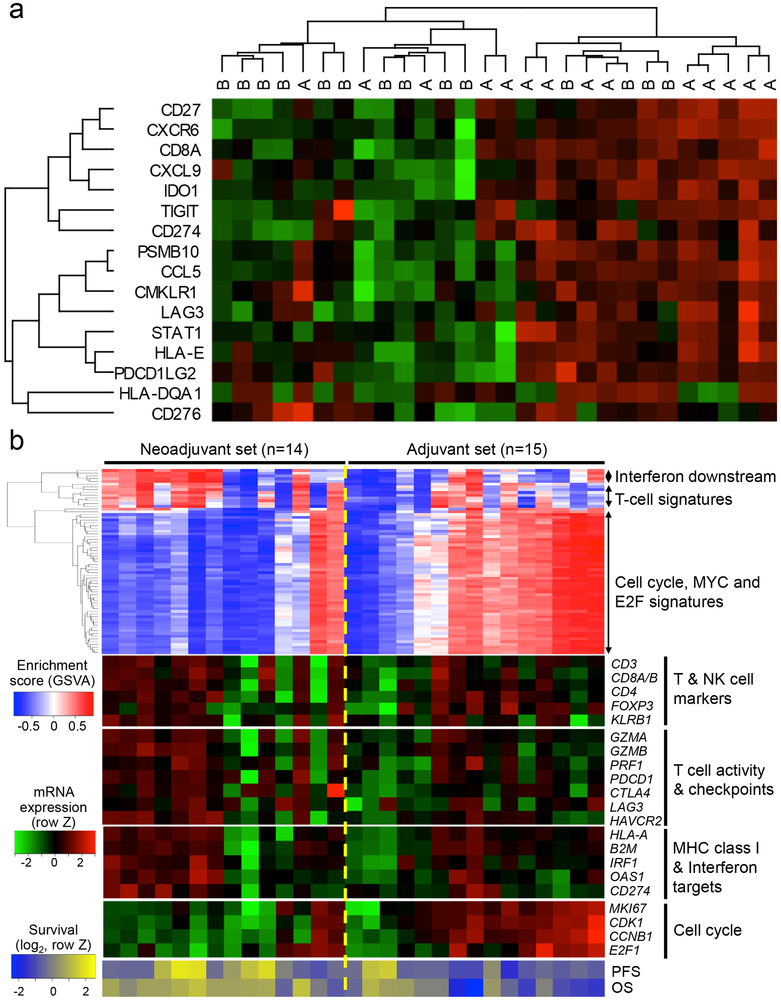
To evaluate whether neoadjuvant PD-1 blockade altered the gene expression profile of recurrent glioblastoma, we first analyzed transcriptional changes within patient tumors utilizing direct multiplexed mRNA quantification. We analyzed the differential gene expression between treatment groups, which revealed several statistically significant differences in tumor samples (Supplementary Table 2). Kyoto Encyclopedia of Genes and Genomes (KEGG) gene set enrichment analysis identified gene products associated with cytokine-cytokine interaction, the chemokine signaling pathway and toll-like receptor signaling pathway. To quantify the T cell-inflamed microenvironment, we compared an immune-related gene expression signature score (Supplementary Table 3)34 between the treatment groups. There was a significant transcriptional increase in genes related to interferon-γ responsiveness (P = 0.03, U = 49; Mann-Whitney U test; Figure 2a, Extended Data Figure 3). We also sought to determine whether neoadjuvant PD-1 blockade induced other transcriptional changes in these tumors. As such, we performed RNA sequencing on patient tumor samples and applied an unbiased screen for differentially expressed gene signatures with an interquartile range greater than 1. Our data demonstrated interferon- and T cell-pathway induction in 9 of 14 tumors in the neoadjuvant group and 5 of 15 in the adjuvant-only group, corroborating our mRNA quantification data. In addition, 3 out of 14 tumors in the neoadjuvant group demonstrated positive enrichment of cell cycle / cancer proliferation signatures, whereas this occurred in 11 of 15 tumors in the adjuvant group (Figure 2b; two-sided P = 0.01, Fisher exact test, 95% CI 0.01 to 0.70). We found no statistically significant correlation between the steroid dose at registration and representative interferon, T cell or cell cycle gene expression signature scores. The proportion of cell cycle signatures in our study as a whole was similar to that of publicly available RNA sequencing datasets from patients with recurrent glioblastoma35 (11 of 20 pre-treatment tumors; Extended Data Figure 4; two-sided P = 0.8, Fisher exact test, 95% CI 0.21 to 2.77) and The Cancer Genome Atlas36 (73 of 166 samples; two-sided P = 0.7, Fisher exact test, 95% CI 0.50 to 2.83; Extended Data Figure 5). Notably, only one of the samples with interferon pathway and T cell gene set enrichment in the neoadjuvant group demonstrated cell cycle pathway activation (Figure 2b), suggesting that neoadjuvant PD-1 blockade induced relevant immune cell activation within the tumor microenvironment that subsequently repressed the cell cycle-related transcriptional activity of tumor cells.
Figure 2. Tumor gene expression profile altered by neoadjuvant PD-1 blockade.
(a) Heat map of tumor mRNA expression of interferon-γ related gene panel for individual patients. Within this panel, “A” denotes patient in neoadjuvant group; “B” denotes patient in adjuvant-only group. Dendrograms represent unsupervised hierarchical clustering by Ward’s minimum distance. Green coloration represents decreased expression; red coloration represents increased expression. (b) (Top) Heatmap showing the gene set variation analysis (GSVA) enrichment scores of gene sets with interquartile range (IQR) ≥ 1. The gene sets can be grouped into the three categories: 1) interferon pathway induction, 2) T-cell activity and 3) cell cycle/proliferation. (Middle) The heatmap of mRNA expression of the representative genes corresponding to the gene set enrichments above. (Bottom) The heatmap of progression free- and overall survival of each patient (in log2 scale). MHC: major histocompatibility complex. OS: overall survival. PFS: progression-free survival.
Neoadjuvant PD-1 antibody blockade is associated with focal upregulation of PD-L1 and CD8+ T cell infiltrate in recurrent glioblastoma
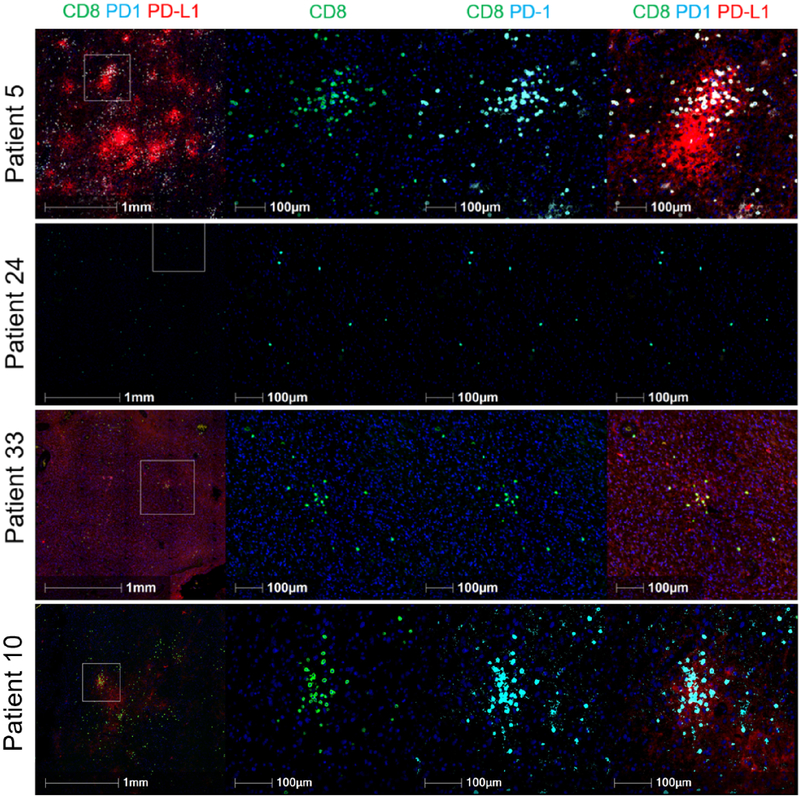
To address whether neoadjuvant PD-1 blockade increased the infiltration of T cells, we quantified the immune cell infiltrate in each patient’s tumor tissue utilizing multiplex immunofluorescence. The density of tumor infiltrating CD8+ T cells was not different between groups but demonstrated significant variability in the neoadjuvant cohort. Because PD-L1 is known to be upregulated in response to interferon-γ produced by infiltrating T cells,34,37-39 we evaluated whether PD-L1 was increased with neoadjuvant PD-1 blockade. Samples were classified as having either a constitutive, focal or negative PD-L1 expression pattern with varying degrees of CD8+ T cell infiltration. Seven neoadjuvant patients and 3 adjuvant patients exhibited a focal phenotype with a high CD8 infiltrate (Figure 3; Supplementary Table 4), suggesting that neoadjuvant treatment was associated with focal induction of PD-L1 expression. Qualitatively, the focal upregulation of PD-L1 in these adjuvant cases was not as striking, suggesting that the endogenous functional T cell response was not as dramatic.
Figure 3. Multiplex immunofluorescence imaging of tumor samples demonstrates varying degrees of PD-L1 expression and CD8+ T cell infiltration.
In the top panel, high CD8 infiltrate and focal PD-L1 expression are demonstrated in a patient tumor sample from the neoadjuvant group. In the second panel, low CD8+ T cell infiltrate and negative PD-L1 expression are seen in a patient tumor sample from the adjuvant-only group. In the third panel, a patient tumor sample from the neoadjuvant group demonstrates low CD8 infiltration with constitutive PD-L1 expression. In the bottom panel, a tumor sample from a patient in the adjuvant-only group demonstrates high CD8 infiltration with focal PD-L1 expression; however, the degree of PD-L1 expression is lower. Images on the left are depicted at lower magnification to demonstrate the difference between focal and constitutive PD-L1 expression and are representative of the entire tumor section/slide. Images second from the left show staining for CD8; second from the right, PD-1 stains are superimposed; on the right, the images depict co-staining for CD8, PD-1 and PD-L1. Multiplex staining was performed in one standardized run per patient. For each staining run, two sequential slides were used as duplicates for each patient. Computational analysis was performed two or more times per sample.
Neoadjuvant PD-1 blockade alters correlative relationships between blood and tumor repertoire features
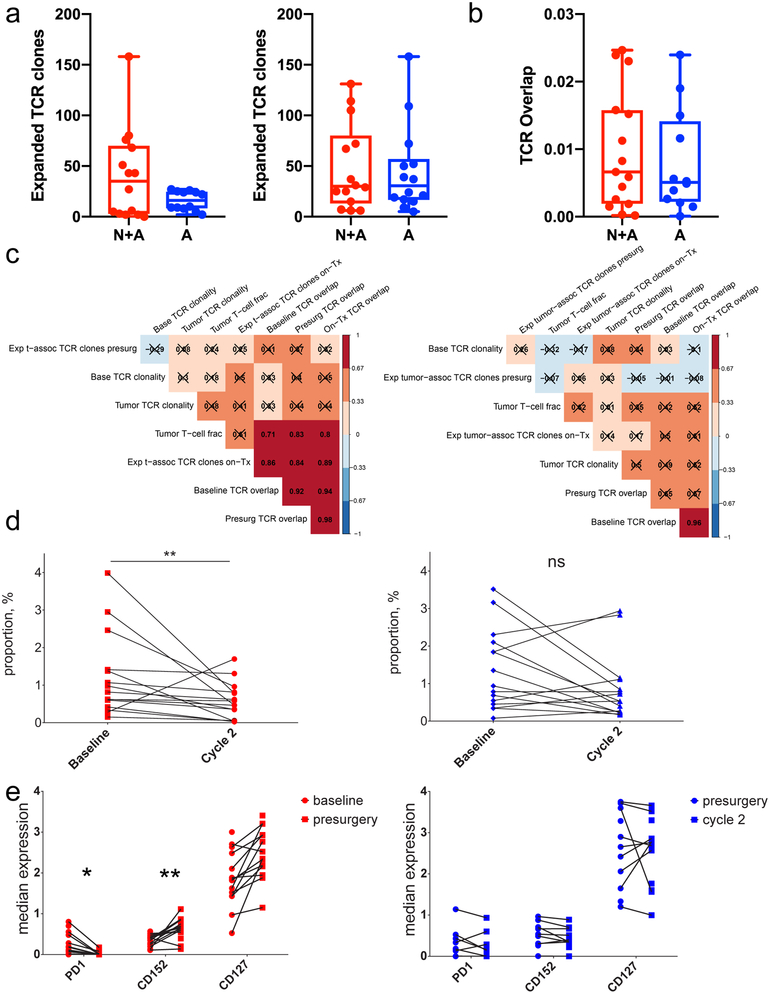
We hypothesized that the clinical effectiveness of neoadjuvant PD-1 blockade may occur because this specific timing resulted in a detectable local and systemic T cell response. To test this, we performed αβ T cell receptor sequencing on all patients’ peripheral blood mononuclear cells at baseline, the time of surgery, and approximately 6 weeks after surgery, as well as on all evaluable tumor samples. We statistically identified T cell receptor rearrangements that were differentially abundant in each patient’s peripheral blood mononuclear cells at on-treatment time points relative to baseline and quantified the number of clones that significantly increased in frequency (Figure 4a) as well as the proportion of T cell receptors shared between tumor and blood (Figure 4b). There was a trend toward an increased number of expanded clones between baseline and surgery in the neoadjuvant compared to the adjuvant group (two-sided P = 0.07, unpaired t test). We did not observe significant differences in the tumor infiltrating lymphocyte content, T cell receptor diversity or proportion of T cell receptors shared between tumor and blood by treatment group or clinical time point. However, in neoadjuvant patients, we noted commensurate increases in the fraction of expanded T cell clones following the post-surgery PD-1 blockade cycle and T cell receptor overlap between tumor and blood across all three collected time points (rs = 0.86, 0.84, 0.89; P = 0.0002, 0.0003, 0.00004, respectively, asymptotic t approximation, n = 25 patients). Similarly, the amount of T cell receptor overlap at the time of surgery was also significantly associated with the tumor infiltrating lymphocyte fraction, but only in the neoadjuvant treatment group (rs = 0.83; P = 0.0005, asymptotic t approximation, n = 25 patients; Figure 4c). These correlations were not found in the adjuvant only group, suggesting that neoadjuvant PD-1 blockade uniquely initiated a coordinated local and systemic T cell response.
Figure 4. Neoadjuvant PD-1 blockade alters correlative relationships between blood and tumor repertoire features and alters circulating immune cell phenotypes.
(a) Box and whisker plots comparing the number of expanded T cell receptor clones between baseline and surgery (left; P = 0.07, two-tailed t test, t = 1.98, df = 14.33, 95% CI −1.94 to 50.7, n = 26 patients) and surgery and 1-2 cycles of pembrolizumab (right; P = 0.85, two-tailed t test, t = 0.19, df = 26.0, 95% CI −30.2 to 36.3, n = 28 patients). The Y axis denotes the number of expanded clones. (b) Box and whisker plot comparing the T cell receptor overlap between peripheral blood and tumor at the time of surgery (P = 0.59, two-tailed t test, t = 0.54, df = 24.8, 95% CI −0.01 to 0.008, n = 27 patients). On the Y axis, 0 indicates no clonal overlap and 1 indicates complete overlap. For panels (a) and (b), whiskers represent minima and maxima, boxes extend from 25th to 75th percentiles, middle line represents median. (c) Hierarchically ordered Spearman correlation plots of the T cell receptor sequencing data. Numbers indicate the Spearman correlation coefficient; boxes marked with an X had Benjamini-Hochberg adjusted, two-tailed P > 0.01 by asymptotic t approximation, n = 25 patients. Patients who received neoadjuvant PD-1 blockade (left) demonstrated significant relationships between multiple variables. (d) Scatter plot of the proportions of a peripheral cluster of intermediate monocytes (CD11b+CD11c+CD14+CD16+HLA-DRhi) at baseline and after 1-2 cycles of adjuvant therapy. The Y axis indicates percent of live mononuclear cells. n = 28 patients; Benjamini-Hochberg-corrected, two-sided P = 0.007 by general linear hypothesis test. (e) Scatter plot of selected cell surface markers on CD4+ T-cells before and after the first dose of pembrolizumab. Note the decreased PD-1 expression and increased CD152 (Benjamini-Hochberg corrected, two-sided P = 0.025 and 0.0015, respectively, general linear hypothesis test, n = 28 patients) in the neoadjuvant group (left). *: P < 0.05; **: P < 0.01; ns: non-significant; N+A: neoadjvant + adjuvant; A: adjuvant only.
Neoadjuvant PD-1 blockade is associated with monocytic and T cell phenotypic changes
To evaluate whether neoadjuvant PD-1 blockade altered the phenotype of systemic immune cell populations, we performed time-of-flight mass cytometry on peripheral blood samples collected from study subjects at baseline, prior to surgery and after receiving at least 1 adjuvant treatment cycle, enabling use of the adjuvant group as a concurrent control. After staining and sample acquisition, we concatenated a total of 84 samples. We utilized unsupervised clustering to conduct cell population identification (Extended Data Figure 6) and evaluated the characteristics of the identified clusters. There was a statistically significant decrease in an intermediate monocyte population in the neoadjuvant group between baseline and after at least 1 cycle of adjuvant PD-1 blockade, which was not observed in the adjuvant-only group (baseline mean proportion 1.35 ± 0.31% versus on-treatment mean proportion 0.56 ± 0.12%; Benjamini-Hochberg-corrected, two-sided P value = 0.007, general linear hypothesis test, n = 28]; Figure 4d). We then compared the expression of functional markers within clustered cell populations and noted statistically significant decreases in PD-1 and increases in CTLA4 (Benjamini-Hochberg-corrected, two-sided P values 0.025 and 0.0015, respectively, general linear hypothesis test, n = 28) on CD4+ T cell clusters (Figure 4e) solely in the neoadjuvant group between baseline and post-surgery. After correcting for multiple comparisons, no significant changes were observed in peripheral blood mononuclear cell populations from the adjuvant-only treatment group.
Cell cycle gene signature, baseline T cell receptor clonality and tumor infiltrating lymphocyte density may be associated with clinical response to neoadjuvant PD-1 blockade
Finally, we sought to determine which clinical and laboratory factors were directly associated with survival, utilizing an elastic net regularized Cox regression for variable selection. In the 30 patients with evaluable tumor, neoadjuvant PD-1 blockade therapy, IDH mutation status, lower baseline peripheral T cell receptor clonality and increased tumor infiltrating lymphocyte density were associated with increased overall survival. We then fit these variables, combined with age and sex, into a Cox proportional hazards regression, and confirmed that neoadjuvant treatment was associated with improved survival (hazard ratio 0.33, P = 0.045; Supplementary Table 5). Standardized baseline peripheral T cell receptor clonality trended toward a survival association, with a hazard ratio of 1.48 for each one standard deviation increase (P = 0.12). Given the significant treatment effect on the cell cycle signature in neoadjuvant patients, we sought to determine if a representative gene set40 could be utilized as a biomarker for overall survival, calculating the R2 statistic to evaluate the potential predictive power.41 We found that the cell cycle-related gene set variation analysis (GSVA) enrichment score explained 57% of the variance in overall survival (R2 = 0.57). When added to the previous Cox model, the R2 value increased from 0.36 to 0.62, suggesting that the cell cycle GSVA enrichment score may be one of the most significant predictors of overall survival.
Discussion
In this study, PD-1 monoclonal antibody blockade was associated with statistically significant improvements in overall survival and progression-free survival when administered in the neoadjuvant setting to patients with recurrent glioblastoma. We leveraged T cell receptor sequencing, mRNA expression profiling, quantitative multiplex immunofluorescence and mass cytometry on patient blood and tumor samples to understand the distinct biological effects induced by this neoadjuvant timing. Our data suggest that neoadjuvant PD-1 monoclonal antibody blockade induces functional activation of tumor infiltrating lymphocytes, producing an interferon response within the tumor microenvironment. Tumor infiltration with interferon-γ-producing, PD-1/PD-L1-suppressed T cells is likely essential for the systemic priming of tumor-specific T lymphocytes after neoadjuvant pembrolizumab. Indeed, tumor infiltrating lymphocyte density was one of the variables selected in our elastic net regression as being possibly related to survival; conversely, presurgical tumor volume, postsurgical tumor volume, percent resection, gross total resection (as a dichotomous variable) and dexamethasone dosage at time of registration were not. This suggests that the survival benefit derived from neoadjuvant PD-1 blockade is largely driven by the immune response. After neoadjuvant PD-1 blockade and resection, tumor-specific T cell clones maintain functionality with repeated anti-PD-1 monoclonal antibody administration in the adjuvant setting (Figure 5). We believe that the T cell-mediated interferon response induces downregulation of cell cycle-related gene expression within the tumor cells, enabling a therapeutic window and resulting in a survival benefit. It is already known that many interferon-regulated gene products block cell cycle activities and tumor cell proliferation,42-44 suggesting that the transcriptional suppression of such cell cycle-related genes is a key feature of the efficacy for PD-1 blockade. Of note, a single-arm phase II clinical trial by Melero and colleagues in this same issue, utilizing neoadjuvant nivolumab in newly diagnosed or relapsed glioblastoma (NCT02550249) demonstrated similar intratumoral and systemic immune changes as found by us.
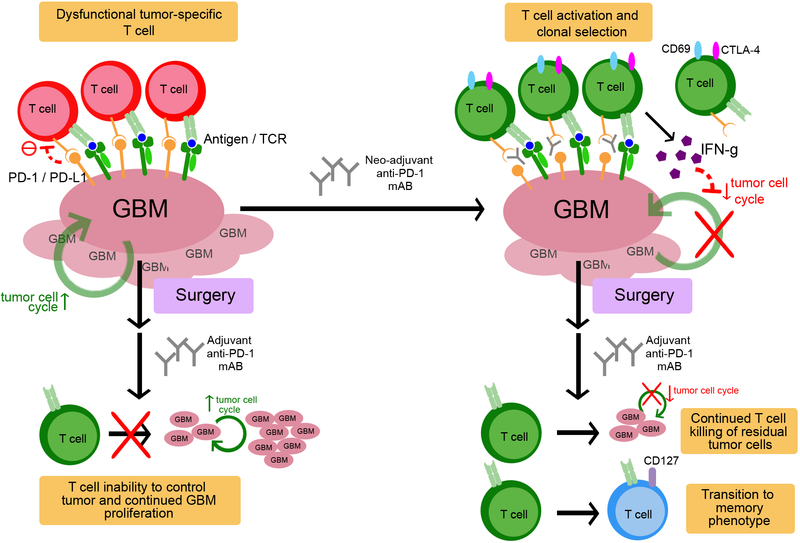
Figure 5. Proposed mechanism of neoadjuvant PD-1 blockade in recurrent GBM.
Tumor infiltrating lymphocytes, if present, are rendered ineffective through the PD-1/PD-L1 axis. Neoadjuvant PD-1 blockade releases this checkpoint, enabling modulation of the T cell receptor (TCR) clonotypes with systemic activation and clonal selection of tumor-specific T cells. Such T cell activation in turn upregulates interferon-γ-related signaling, while downregulating tumor cell cycle related genes. After surgery, and with continued adjuvant anti-PD-1 monoclonal antibody administration, tumor-specific T cells continue to eliminate residual tumor cells and begin transitioning toward a T memory phenotype. In the adjuvant-only group, surgery occurs before checkpoint blockade release. Because of the reduced residual antigenic burden, TCR modulation is less robust and fewer tumor-specific T cells are activated. With fewer tumor-specific T cells, the remaining tumor cells are able to proliferate at a more rapid pace.
In our gene expression analysis of tumor samples, we noted an upregulation of interferon-γ-related gene products in patients who received neoadjuvant PD-1 blockade, whereas patients without this upregulation demonstrated enrichment of cell cycle pathway gene sets. In fact, in a murine neoadjuvant immunotherapy model for breast cancer, the neutralization of interferon-γ nullified the efficacy of neoadjuvant anti-PD-1 plus anti-CD137 antibody therapy, indicating a vital role in promoting anti-tumor activity.17 This, together with our data, strongly suggests that increased interferon-γ signaling is a consequence of anti-PD-1 therapy and mediator of the functional immunologic response. Our study indicates that, although increased interferon signaling occasionally occurs in the presence of recurrent glioblastoma (as demonstrated by patients in the adjuvant-only group with upregulated interferon-γ-related gene expression), checkpoint release in the presence of tumor (i.e., in the neoadjuvant setting) may be a crucial component of enhancing tumor-specific T cell effector function, a finding consistent with what Liu et al. observed in a murine model. In addition, mice that received no treatment and no surgery demonstrated increased tumor-specific T cells, but poor effector function compared to groups that received neoadjuvant therapy and tumor resection, suggesting that both timing and surgery may play a role in tumor-specific T cell expansion. A similar mechanism may account for the failure of anti-PD-1 therapy in recurrent, unresected glioblastoma.24
Quantitative multiplex immunohistochemistry revealed an increased proportion of patients in the neoadjuvant group that exhibited focal upregulation of PD-L1. However, 3 patients in the adjuvant-only group also demonstrated this finding, albeit with qualitatively limited PD-L1 intensity. Two of these three patients also demonstrated upregulation of cell cycle-related genes, suggesting that their endogenous T cell response was insufficient to disrupt tumor cell proliferation. The third patient with focal PD-L1 upregulation on multiplex immunofluorescence exhibited some CD8 T cell infiltration but did not have an elevated cell cycle signature. Additionally, the T cell- and interferon-related genes also were not increased, suggesting an aberrant mixture of low level activity. The overall survival of these three patients was below the median for all patients.
After analyzing the T cell receptor data, we found that expansion of tumor-associated T cell receptor clones in the neoadjuvant group after treatment was correlated with high overlap between tumor and blood. While the overall number of expanded clones did increase after the initial pembrolizumab dose in the adjuvant-only group, these additional correlations were not seen. This suggests that PD-1 blockade can lead to increases in T cell clones regardless of timing; however, pre-surgical checkpoint inhibition enables a selective, primary tumor-specific T-cell clonal modulation, driving systemic expansion of tumor-specific T lymphocytes.
Neoadjuvant pembrolizumab was not associated with a significant change in T cell receptor diversity, supporting previous findings that diversification of the T cell repertoire is not mediated via the PD-1 axis.45 However, as our data indicate that increased baseline T cell receptor clonality may be associated with reduced survival, it appears that greater initial T cell diversity may portend improved responsiveness to PD-1 blockade. Our data suggest a role for combination immunotherapy with CTLA-4 blockade, which was shown to increase the number and complexity of T cell receptor variants in patients with melanoma45 and has been demonstrated to be efficacious in other tumor types.12 Although this combination was evaluated in a small cohort of recurrent glioblastoma patients without an obvious survival signal,46 it has yet to be explored in a neoadjuvant investigation.
In our single cell peripheral blood analysis, no specific cell population appeared to consistently predict survival, contrasting with previous studies in other cancer types.47-49 In our evaluation of differentially expressed functional markers on various cell populations, CD4+ T cells demonstrated statistically significant increases in CD152 and CD127 and a decrease in PD-1, suggesting that the degree of phenotypic shift toward activation and memory may play a significant role in the post-surgical antitumor immune response.
Due to the established role of corticosteroid usage in the treatment of both CNS malignancies and immune-related adverse events, we evaluated the correlation between dexamethasone dosage at the time of registration as continuous and dichotomous variables and expression scores of interferon-, T cell- and cell cycle-related gene sets as well as overall survival. We found no statistically significant correlations, differing from published findings in patients with non-small cell lung cancer that received PD-1/PD-L1 blockade on high-dose baseline corticosteroids.50 This may be due to the fact that the protocol for this study excluded patients receiving high-dose dexamethasone at baseline greater than 4 milligrams per day. Many patients continued to receive or were started on low doses of dexamethasone during their treatment course. Our analyses corroborate previous findings that systemic corticosteroids can be utilized as management for adverse events without affecting objective response rates in patients receiving nivolumab for advanced melanoma.51
Given the noted improvement in survival, we intend to expand the current study and pursue further clinical trials with neoadjuvant combination immunotherapeutics. Although this was a randomized clinical study, it was powered for tissue analyses. As such, interpretation of efficacy outcomes is limited by small sample size. Similarly, the observed immune effects may be influenced by an imbalance in the number of pembrolizumab doses received by patients in each group; the nature of this clinical trial design as a multi-center study also limited tissue availability for in-depth biologic analyses. We also did not find statistically significant correlations between patients that exhibited large numbers of expanded T cell clones, focal upregulation of PD-L1 expression and the above gene expression signature score. This may be due to the small sample size and/or the heterogeneity in tumor sampling for quantitative multiplex immunofluorescence analysis. Notably, almost all patients in this study went on to receive bevacizumab. The precise role of bevacizumab is unclear, given the inter-patient heterogeneity in post-progression therapies and timing. Data suggest that bevacizumab in this setting is not associated with poorer survival outcomes.52 Overall, the consistency of the data from this study indicates an observable tissue-based and clinical treatment effect. As such, the neoadjuvant approach to immunotherapy and brain cancer may provide a unique clinical development path, while also providing a therapeutic window to study the immunobiology of malignant brain tumors.
Methods
Study design and patients
Patients were aged ≥18 years with recurrent glioblastoma that were candidates for surgical debulking. Key eligibility criteria included Karnofsky performance status ≥70, previous first line therapy with at least radiotherapy, first or second relapse with unequivocal evidence of tumor progression, adequate organ function, no high dose systemic corticosteroids (defined as greater than 4 milligrams per day of dexamethasone or bioequivalent for at least 3 consecutive days within 2 weeks of registration) and absence of previous anti-angiogenic or anti-vascular endothelial growth factor agents. All patients provided written informed consent; the study was approved by institutional review boards at all sites (Dana-Farber Cancer Institute; Huntsman Cancer Institute; M.D. Anderson Cancer Center; Massachusetts General Hospital; Memorial Sloan Kettering Cancer Center; University of California, Los Angeles; University of California, San Francisco) and conducted according to the Declaration of Helsinki.
After consent, patients randomized to the neoadjuvant group received pembrolizumab 200 mg intravenous infusions 14 ± 5 days prior to scheduled surgical resection. Tumor resection was performed according to institutional standards. After recovery from surgery, patients received pembrolizumab 200 mg intravenous infusions every 3 weeks until tumor progression or an adverse event requiring study drug discontinuation. Blood samples were obtained every two cycles (6 weeks). Patients were followed for magnetic resonance imaging (MRI) changes, clinical exams and steroid doses until death or second progression. After second progression, patients were followed every 3 months for vital status until death.
RNA isolation and quantification
RNA was isolated from tumor sections stored in AllProtect tissue reagent (Qiagen) at the time of surgery; peripheral blood mononuclear cells were isolated at baseline, time of surgery and at cycle 2 of therapy and lysed to obtain RNA and protein. The nCounter GX analysis system (NanoString) was utilized to quantify RNA and protein expression according to the manufacturer’s directions. The 770-gene nCounter PanCancer immune profiling panel was utilized (Nanostring; list of genes available from manufacturer).
RNA sequencing and gene set enrichment
Paired-end, 2×150bp transcriptome reads were mapped to the Genome Reference Consortium Human Build 38 (GRCh38) reference genome using HISAT2.53 These raw sequencing files have been uploaded to the Gene Expression Omnibus (GEO; GSE121810). The gene level counts are generated by the htseq-count54 program and we took log2 counts per million (CPM) as normalized gene expression values. To calculate single-sample gene set enrichment, we used the Gene Set Variation Analysis (GSVA) package55 to derive the absolute enrichment scores of the following perturbation gene sets from the Broad Institute’s Molecular Signatures Database:56
-
1)
c2.cgp.v6.0
-
2)
c6.all.v6.0
-
3)
c7.all.v6.0
-
4)
hallmark.v6.0
To ensure robustness, we chose only the gene sets with at least 15 genes. We also included curated immune/stromal cell signatures from the CIBERSORT program57 and from single cell RNA sequencing studies on patient tumors.58 To run GSVA, we first filtered out genes whose normalized expression’s interquartile range is less than two-fold (log2 CPM interquartile range ≥ 1). The shortlisted genes’ raw HTSeq-computed counts are then supplied to the GSVA program using the “kcdf=Poisson” mode. Once the GSVA scores of all gene sets were computed, we selected those with GSVA score interquartile range ≥ 1 for further analysis.
Tumors with GSVA score ≥ 0.2 in both the “T_generic” (reflecting general T cell activation markers), and “Ayers_et_al_IFN_genes” (interferon activation signatures reported by Ayers, et al34) and ≤ 0.2 in the “FARMER_BREAST_CANCER_CLUSTER_2” (reflecting cell cycle related genes) are defined to have high T cell infiltration and interferon pathway activation and decreased cell cycle. To evaluate for potential correlations between steroid administration and immune response, we calculated Spearman’s rho for steroid dose in milligrams at registration and the above representative gene sets for interferon-, T cell- and cell cycle-related signature scores.
Finally, in order to compare the enrichments of the shortlisted gene sets within our datasets and that of other glioblastomas, we combined the log2 CPM normalized expression of The Cancer Genome Atlas (TCGA) glioblastomas (based on HTSeq-counts data downloaded from the Genomic Data Commons59) and the pre-treatment RNAseq dataset GSE79671 (a study of patients with recurrent glioblastoma), with our own log2 CPM and removed potential batch effects across the two datasets using the RemoveBatchEffect function in the limma R package. The combined CPM is input into the GSVA program using the default “kcdf=Gaussian” option. We compared the statistical difference in the fraction of tumors with positive enrichment of each gene set, defined as having a GSVA score ≥ 0.2 for that gene set, in the neoadjuvant (n = 14), adjuvant (n = 15), GSE79671 (n = 20) and TCGA patient groups (n = 166) by one-sided Fisher exact test.
Immunosequencing of the T-cell receptor β-chain
Genomic DNA (gDNA) was extracted from patient tumors and peripheral blood mononuclear cells (collected at baseline, prior to surgery, at the time of the first postsurgical pembrolizumab dose, every 6 weeks thereafter or at the end of treatment). gDNA was extracted using the Qiagen DNeasy blood extraction kit. Utilizing the immunoSEQ® Assay (Adaptive Biotechnologies, Seattle, WA) for PBMC samples and the immunoSEQ Assay for FFPE tumor samples, T cell receptor β (TCRβ) complementarity determining region 3 (CDR3) regions were amplified and sequenced from 2 μg of gDNA (or all available extracted DNA if less than 2 μg were available). Regions were amplified using a bias-controlled multiplexed polymerase chain reaction (PCR) method, followed by high-throughput sequencing. Sequences were collapsed and filtered to identify and quantitate the absolute abundance of each unique TCRβ CDR3 for further analysis as previously described.60-62
For measuring tumor infiltrating lymphocyte density, the immunoSEQ Assay for FFPE tumor samples amplifies both the TCRβ CDR3 as well as select reference genes, which quantitate the total number of T cells and nucleated cells, respectively. Tumor infiltrating lymphocyte density is calculated by dividing the total number of T cells by the total number of nucleated cells.
We assessed T cell receptor overlap between tumor infiltrating lymphocytes and peripheral blood mononuclear cells as previously described.63 To summarize, T cell receptor overlap between tumor and peripheral blood mononuclear cell samples from each patient was assessed by identifying the shared TCRβ CDR3 amino acid sequences found between two samples. The sum of shared sequences between two samples was then divided by the total number of unique TCRβ CDR3 sequences present across both samples. To evaluate T cell receptor diversity, we utilized the Daley-Smith Richness estimator, a Bayesian approach that enables characterization of the complexity of DNA sequencing data; this methodology provides an estimate of repertoire diversity that is robust to differences in input DNA between samples.64
No significant difference was observed in the number of expanded clones between treatment groups from baseline to surgery (two-sided P = 0.40, Wilcoxon rank sum test with continuity correction, W = 101, 95% CI −7.0 to 44.0, n = 26 independent biological samples) or from surgery to the second adjuvant cycle (two-sided P = 0.89, Wilcoxon rank sum test with continuity correction, W = 101.5, 95% CI −22.0 to 28.0, n = 28 independent biological samples). We also did not observe significant differences in the tumor infiltrating lymphocyte content (two-sided P = 0.41, Wilcoxon rank sum test, W = 133, 95% CI −0.006 to 0.019, n = 30 independent biological samples), T cell receptor diversity in tumor (two-sided P = 1, Wilcoxon rank sum test, W = 113, 95% CI −0.05 to 0.06, n = 30 independent biological samples) or peripheral blood (two-sided P = 0.35, Wilcoxon rank sum test, W = 83, 95% CI =0.15 to 0.05, n = 30 independent biological samples) or proportion of T cell receptors shared between tumor and blood by at baseline (two-sided P = 0.62, Wilcoxon rank sum test, W = 117, 95% CI −0.003 to 0.007, n = 29 independent biological samples), the time of surgery (two-sided P = 0.68, Wilcoxon rank sum test, W = 99, 95% CI −0.004 to 0.008, n = 27 independent biological samples) or at cycle 2 of adjuvant treatment (P = 0.27, Wilcoxon rank sum test, W = 131, 95% CI −0.002 to 0.009, n = 29 independent biological samples).
Mass cytometry
Peripheral blood mononuclear cells were collected at baseline, the time of surgery and after 1-2 cycles of adjuvant therapy and prepared for mass cytometry analysis according to the Maxpar cell surface staining protocol. Briefly, 0.5 to 3 × 106 cells were washed with PBS, then resuspended in 5 μM Cell-ID cisplatin (Fluidigm) as a live/dead marker for 5 minutes at room temperature. After quenching with cell staining buffer, the cells were incubated with a 24-marker panel (Supplementary Table 6) for 30 minutes at room temperature. After washing with cell staining buffer, cells were incubated overnight in 125 nM iridium intercalation solution (1000X dilution of 125 μM Cell-ID Intercalator-Ir in Maxpar Fix and Perm Buffer [Fluidigm]) to label intracellular DNA. Cells were then washed with cell staining buffer and distilled water.
Events were subsequently acquired on a Helios mass cytometer (Fluidigm) in the University of California, Los Angeles Jonsson Comprehensive Cancer Center flow cytometry core and analyzed the data as described previously.65 After acquisition, data were normalized utilizing EQ four element calibration beads (Fluidigm) and preprocessed to remove dead cells. Each dataset was then loaded into R with the flowCore package. The raw marker intensities were transformed utilizing hyperbolic inverse sine (arcsinh) with cofactor of 5. All samples were bead normalized and each marker intensity was scaled to values between 0 and 1. All markers were included in the analysis. We concatenated a total of 84 samples and utilized unsupervised clustering to conduct cell population identification (Extended Data Figure 6). Cell population identification was carried out using FlowSOM to assign cells into a 10×10 grid according to similarity utilizing the self-organizing map (SOM) algorithm. The resulting 100 vectors of marker expression (codes) were clustered using ConsensusClusterPlus hierarchical clustering with average linkage. Clustering the codes into 2 to 20 clusters, the delta area was then calculated and used to define the appropriate number of clusters based on the elbow criterion. No specific cell population appeared to consistently predict survival in our analyses, compared to studies in other cancer types.47-49
Multiplex fluorescent immunohistochemistry
Formalin-fixed paraffin-embedded tissue was stained using fluorescent multiplex immunohistochemistry to spatially visualize and quantify the following markers: CD8, PD-1, CD45, GFAP and PD-L1 using the Opal 4-Color Manual IHC Kit and protocol (PerkinElmer). Slides were first deparaffinized with xylene and rehydrated with an ethanol gradient. Heat induced antigen retrieval was them performed on slides prior to each antibody application using pH9 antigen retrieval buffer (AR9, PerkinElmer) for CD8, CD45, PD-1, and PD-L1 antibodies; pH6 antigen retrieval buffer (AR6, PerkinElmer) was used for GFAP. The following antibody clones and dilutions paired with Opal tyramide signal amplification reagent were used and applied in the following order for each panel: CD8 (C8/144B, 1:5000, Dako) with Opal 520, PD-L1 (SP142, 1:4000, Spring Biosciences) and Opal 570, PD-1 (EPR4877, 1:50000, Abcam) and Opal 650; CD45 (2B11+PD7/26, 1:1000, Dako) and Opal 520, PD-L1 (SP142, 1:4000). All slides were nuclear counterstained with Spectral DAPI (PerkinElmer), mounted (ProLong Diamond Antifade Mountant, Life Tech), and whole sections were imaged at 20× resolution (0.32μM/pixel) using a Leica Aperio Versa 200 Slide Scanning Microscope equipped with a 16-bit Andor Zyla 5.5-megapixel fluorescence camera (Translational Pathology Core Laboratory, University of California, Los Angeles). 8-bit images were captured using the equipped filters: DAPI 350/460 excitation/emission (ex/em), green 495/537 ex/em, red 580/625 ex/em, and Cy5 640/690 ex/em. Positive stain quantification and spatial cell analysis was performed using HALO image analysis software (Indica Labs). Positivity thresholds were determined using primary antibody-negative control slides for exclusion of auto-fluorescence and nonspecific background staining.
Magnetic Resonance Imaging Acquisition and Analysis
Anatomic MR images were acquired for all patients in the current study using a 1.5T or 3T clinical MR scanner using pulse sequences supplied by their respective manufacturers and according to their local standard of care protocols. Standard anatomic images were obtained with the axial T1-weighted fast spin-echo sequence or magnetization-prepared rapid acquisition gradient-echo (MPRAGE) sequence (repetition time (msec)/echo time (msec)/ inversion time (msec) = 400–3209/3.6– 21.9/0–1238; slice thickness = 1–6.5 mm; intersection gap = 0–2.5 mm; number of averages = 1-2; matrix size = 176–512 × 256–512; and field of view = 24–25.6 cm). Additionally, T2-weighted fast spin-echo and fluid-attenuated inversion-recovery (FLAIR) sequences were also obtained. In addition, parameter matched T1-weighted images enhanced with gadopentetate dimeglumine (Magnevist; Berlex), 0.1 mmol/kg, were acquired shortly after contrast material injection. All on trial scans were compliant with the consensus recommendations for the international standardized brain tumor imaging protocol.66
Linear registration was performed between all images (T2, FLAIR, pre-contrast T1, post-contrast T1) and post-contrast T1-weighted images at screening using a 12–degree-of-freedom linear transformation and a correlation coefficient cost function in FSL (FLIRT; FMRIB Software Library, Oxford, England; http://www.fmrib.ox.ac.uk/fsl/). Estimates of tumor volume were performed using contrast-enhanced T1-weighted digital subtraction maps to exclude areas of post-surgical blood products or other sources of T1 shortening as described previously.67-70 Initial segmentation was performed automatically using previously defined methodology67-70 and final segmented volumes were edited by an experienced investigator with more than 10 years of experience to exclude large vessels and any obvious non-tumor regions.
Statistics and reproducibility
For mRNA quantification data, expression values were normalized using positive and negative controls and housekeeping genes and analyzed using the nSolver analysis software 4.0 (NanoString). Differential expression based on treatment group was performed by utilizing a mixed negative binomial model for each gene with correction for multiple comparisons using the Benjamini-Hochberg adjustment. Gene set enrichment analysis was performed using the KEGG gene collection in the Molecular Signatures Database56,71 with P values determined by hypergeometric test and corrected using a false discovery rate of 0.05 with n of 28 independent biological samples. The interferon-γ associated gene expression profile score was calculated as previously described.34
In order to compute the differential abundance of T cell receptor clones, TCRβ rearrangements were detected by the immunoSEQ Assay for each patient’s peripheral blood mononuclear cell samples at baseline (n = 29 independent biological samples), the time of surgery (n = 27 independent biological samples) and cycle 2 (n = 29 independent biological samples) and compared to the previous time point. Rearrangements with a count of less than 10 between the two compared time points were excluded. For a TCRβ rearrangement to be considered significantly expanded or contracted, a two-sided binomial test with Benjamini-Hochberg adjustment was performed, using a false discovery rate of 0.01. To determine if there was a difference between the two groups, the number of expanded clones were compared using the Wilcoxon signed rank test.
To evaluate differences between the neoadjuvant and adjuvant-only groups in mass cytometry data, we employed the generalized linear hypothesis function with the R function glht to test for the difference in marker expression, fitting fixed and mixed models using the stats and lme4 packages in R. To compare cell population proportions between groups, we employed a generalized linear mixed model, using the sample identification and anonymized patient identifiers as random effects.72 For differential analysis of marker expression by cell population, we calculated the median expression of 12 signaling markers in each cell population and sample, using these as the response variable in the linear mixed model.
Progression-free survival (time from registration to disease progression per iRANO criteria or death) and overall survival (time from registration to death) were estimated using the PROC PHREG procedure in SAS. Multiple imputation with chained equations was utilized to account for missing biomarker data with 5 imputations of 50 iterations performed. Given the relatively low number of patients and large number of possible parameters with high correlation, we used an elastic net regularized regression for variable selection. Both clinical (including IDH mutation status, age, sex, MGMT methylation status, number of prior relapses, Karnofsky performance status, steroid dose in milligrams at time of registration) and laboratory data (including mass cytometry cluster percentages, T cell receptor overlap, tumor T cell density, expanded tumor-associated T cell clones, interferon-γ-related signature scores, presence or absence of inducible PD-L1 expression on multiplex immunofluorescence) were considered potential covariates. We used 5-fold cross-validation to obtain the value of λ that gave the minimum mean cross-validated error and determined the corresponding coefficients for each covariate. Variables with nonzero coefficients were then checked for collinearity and subsequently fitted into a Cox proportional hazards model, forcing in age and sex. Peripheral blood T cell receptor clonality and tumor infiltrating lymphocyte fractions were standardized before fitting into the Cox regression due to their wide ranges. As the tumor infiltrating T cell fraction was a measurement taken at the time of surgery, we ran both a two-sample t-test (P = 0.52, t = 0.65, df = 27.6) and Wilcoxon rank sum test (P = 0.41, W = 133, n = 30 patients) to ensure that there was no significant imbalance between the two groups at alpha = 0.05 that could potentially introduce confounding into the model. We performed a Cox-Snell residual plot, which did not suggest any lack-of-fit for the multivariate Cox model. We then analyzed a representative cell cycle signature-associated GSVA enrichment score (FARMER_BREAST_CANCER_CLUSTER_2) as a potential predictive biomarker. There was a clear imbalance between treatment groups (P = 0.02, t = −2.48, df = 27.8, two-sample t-test), indicating a correlation with the treatment variable; as such, it was not included on the initial multivariate Cox model to avoid confounding the treatment effect. We used a method previously described,41 which extends the R2 statistic to survival models with the R package PAmeasures, This R2 value can then be utilized as an estimate that measures the prediction function’s ability to capture the variability of the response, even if the model is possibly mis-specified.
The primary objective of the trial was to evaluate whether administration of pembrolizumab (MK-3475) would induce statistically significant increases in tumor infiltrating T lymphocyte density in recurrent/progressive GBM patients compared to an untreated concurrent control. The adjuvant-only group was considered an independent concurrent control to evaluate the tumor infiltrating lymphocyte density in the neoadjuvant group due to the infeasibility of multiple resections within a short time period for patients with recurrent glioblastoma. Based on our preliminary data, the mean tumor infiltrating lymphocyte density was estimated to be 0.4 T cells per nucleated cell (standard deviation = 0.5) in the control group. Fifteen patients per group was deemed sufficient to achieve 85% power to detect an increase of 0.5 in tumor infiltrating lymphocyte density comparing the neoadjuvant group against the adjuvant-only group at an alpha of 0.05 (one sided) using a two-sample t test. The three patients who withdrew consent prior to treatment and the two whose tumors were deemed non-evaluable based on pathology protocol were replaced with randomized subjects to achieve a total of 15 patients per group to power our primary objective. To test for imbalance in patient characteristics between the two groups in age, sex, Karnofsky performance status, IDH mutation status and MGMT methylation status, we performed the Fisher exact test for categorical variables and Student t-test for continuous variables. Statistical analysis was performed with SAS version 9.2, GraphPad Prism version 6.01 or R version 3.4.3 or higher. All P values are two-tailed unless otherwise specified. The false discovery rate was controlled at 5% with the Benjamini-Hochberg procedure unless otherwise specified.
Data availability statement
RNA sequencing data is available in the Gene Expression Omnibus under accession number GSE121810, which includes source data for Figure 2b and extended data figures 4 and 5. The remainder of data that support the findings of this study are available from the corresponding author upon reasonable request. Further information is available in the Life Sciences Reporting Summary.
Code availability statement
R code is available in packages as described in the manuscript.
Extended Data
Fig. 1. CONSORT diagram.
Flow diagram of disposition of patients enrolled in the study.
Fig. 2. Kaplan-Meier plot of progression-free survival.
Median progression-free survival (PFS) for patients who received pembrolizumab only in the adjuvant setting was 72.5 days; patients who received neoadjuvant and adjuvant pembrolizumab had a median PFS of 99.5 days (HR = 0.43, 95% CI 0.20 to 0.90; two-sided P = 0.03 by log-rank test).
Fig. 3. Eighteen gene interferon-γ-related signature scores in neoadjuvant versus adjuvant-only groups.
Line at middle of box represents the median; box extends from the 25th to 75th percentiles; whiskers represent minimum and maximum values; n = 28 independent biological samples; P = 0.025, U = 49 by two-sided Mann-Whitney U test. *: P < 0.05.
Fig. 4. RNAseq comparison to other recurrent glioblastoma samples.
We combined our RNAseq dataset with that of GSE79671 (an RNAseq dataset of recurrent glioblastoma pre- and post- bevacizumab treatment; only pre-treatment (Pre-Tx) samples were used) and The Cancer Genome Atlas (TCGA) glioblastoma samples. We applied appropriate batch correction on log transformed, normalized mRNA expression values using the removeBatchEffect function in the R package limma to estimate the fraction of glioblastoma patients with positive enrichment of the cell cycle/cancer proliferation signatures (GSVA score ≤ 0.2). The proportion of positive enrichment of the cell cycle/cancer proliferation signatures in our dataset as a whole is similar to GSE79671 (14 out of 29 (48%) vs. 11 out of 20 (55%)). The number of samples with positive enrichment in the TCGA GBM is lower at 41%. We observed that neoadjuvant PD-1 monoclonal antibody therapy group is associated with lower fraction of tumors with the cell cycle signatures. There were only 3 out of 14 tumors in the neoadjuvant group demonstrating positive enrichment while there were 11 of 15 tumors in the adjuvant group and 11 of 20 tumors in the GSE79671 set (one-sided Fisher exact test P = 0.01 and P = 0.05, respectively). GSVA: gene set variation analysis.
Fig. 5. RNAseq comparison to TCGA.
We combined our RNAseq dataset with that of TCGA glioblastoma dataset with appropriate batch correction to estimate the fraction of glioblastoma patients with positive enrichment of the cell cycle/cancer proliferation signatures (GSVA score ≥ 0.2). Three out of 14 tumors in the neoadjuvant group demonstrated positive enrichment while there were 11 of 15 tumors in the adjuvant group and 73 of 166 tumors in The Cancer Genome Atlas set. TCGA: The Cancer Genome Atlas. GSVA: gene set variation analysis.
Fig. 6. Mass cytometry dimension reduction.
(a) Diffusion map of peripheral blood mononuclear cells (PBMCs) sampled from n = 28 patients at baseline, the time of surgery and on-treatment. Phenotypically similar cells are depicted in an unsupervised manner along the same continuous axes in a pseudotemporal progression. (b) t-distributed stochastic neighbor embedding (tSNE) plot of PBMCs from n = 28 patients at all three time points. Phenotypically similar cells are clustered in an unsupervised manner. All represented cells in both panels are colored by algorithmically assigned cluster numbers using the FlowSOM package. CD3+CD4+, CD3+CD8+, CD3−CD19+ and CD3−CD14+CD16+CD11b+CD11c+ cells are labelled to demonstrate how clustered cells are plotted in close proximity to one another.
Supplementary Material
Acknowledgments
This study was funded in part by the National Institutes of Health SPORE in Brain Cancer (P50CA211015), the Parker Institute for Cancer Immunotherapy (grant number 20163828), the Cancer Research Institute, the Musella Foundation, the Ben and Catherine Ivy Foundation, the Uncle Kory Foundation, the Defeat GBM Program of the National Brain Tumor Society, the Ziering Family Foundation, and by Merck & Co., Inc. Research and/or financial support was also provided by Adaptive Biotechnologies. The authors would also like to thank A. Garcia, N. Akkad, M. Attiah, S. Khattab, J. Reynoso, M. Wong and M. Guemes for their contributions to the experiments.
Footnotes
Competing Interests Statement
J.A.R. and C.M.S. have a financial interest Adaptive Biotechnologies. T.F.C. and D.A.R. have received compensation from Merck as consultants on advisory boards. P.Y.W. and H.C. have received honoraria from Merck. J.F.d.G. has done consulting and/or received honoraria with Merck and Bristol-Myers Squibb. I.K.M. reports research funding from General Electric, Amgen and Lilly; advisory roles with Agios, Puma Biotechnology and Debiopharm Group; and honoraria from Roche for a presentation.
Other Disclaimers
immunoSEQ® Assays are for research use only and not for use in diagnostic procedures.
References
- 1.Ostrom QT, et al. CBTRUS Statistical Report: Primary brain and other central nervous system tumors diagnosed in the United States in 2010-2014. Neuro Oncol 19, v1–v88 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352, 987–996 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Lamborn KR, et al. Progression-free survival: an important end point in evaluating therapy for recurrent high-grade gliomas. Neuro Oncol 10, 162–170 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu W, et al. Joint NCCTG and NABTC prognostic factors analysis for high-grade recurrent glioma. Neuro Oncol 12, 164–172 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke JL, et al. Is surgery at progression a prognostic marker for improved 6-month progression-free survival or overall survival for patients with recurrent glioblastoma? Neuro Oncol 13, 1118–1124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brahmer JR, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 366, 2455–2465 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burki TK Pembrolizumab for patients with advanced melanoma. Lancet Oncol 16, e264 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Garon EB, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 372, 2018–2028 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Hamid O, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med 369, 134–144 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ribas A Tumor immunotherapy directed at PD-1. N Engl J Med 366, 2517–2519 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Topalian SL, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 366, 2443–2454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolchok JD, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med 369, 122–133 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ribas A Adaptive Immune Resistance: How Cancer Protects from Immune Attack. Cancer Discov 5, 915–919 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ribas A, et al. Association of Pembrolizumab With Tumor Response and Survival Among Patients With Advanced Melanoma. JAMA 315, 1600–1609 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Reck M, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 375, 1823–1833 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Redman JM, Gibney GT & Atkins MB Advances in immunotherapy for melanoma. BMC Med 14, 20 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, et al. Improved Efficacy of Neoadjuvant Compared to Adjuvant Immunotherapy to Eradicate Metastatic Disease. Cancer Discov 6, 1382–1399 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Forde PM, et al. Neoadjuvant PD-1 blockade in resectable lung cancer. N Engl J Med 378, 1976–1986 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blank CU, et al. Neoadjuvant versus adjuvant ipilimumab plus nivolumab in macroscopic stage III melanoma. Nat. Med. 24, 1655–1661 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Amaria RN, et al. Neoadjuvant immune checkpoint blockade in high-risk resectable melanoma. Nat. Med. 24, 1649–1654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blumenthal DT, et al. Pembrolizumab: first experience with recurrent primary central nervous system (CNS) tumors. J Neurooncol 129, 453–460 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Bouffet E, et al. Immune Checkpoint Inhibition for Hypermutant Glioblastoma Multiforme Resulting From Germline Biallelic Mismatch Repair Deficiency. J Clin Oncol 34, 2206–2211 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Johanns TM, et al. Immunogenomics of Hypermutated Glioblastoma: A Patient with Germline POLE Deficiency Treated with Checkpoint Blockade Immunotherapy. Cancer Discov 6, 1230–1236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reardon DA, et al. Randomized phase 3 study evaluating the efficacy and safety of nivolumab vs bevacizumab in patients with recurrent glioblastoma: Checkmate 143. Neuro-Oncology 19, 21–21 (2017). [Google Scholar]
- 25.Antonios JP, et al. Immunosuppressive tumor-infiltrating myeloid cells mediate adaptive immune resistance via a PD-1/PD-L1 mechanism in glioblastoma. Neuro Oncol 19, 796–807 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antonios JP, et al. PD-1 blockade enhances the vaccination-induced immune response in glioma. JCI Insight 1(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bloch O, et al. Gliomas promote immunosuppression through induction of B7-H1 expression in tumor-associated macrophages. Clin. Cancer. Res 19, 3165–3175 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reardon DA, et al. Glioblastoma Eradication Following Immune Checkpoint Blockade in an Orthotopic, Immunocompetent Model. Cancer Immunol Res 4, 124–135 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Wainwright DA, et al. Durable therapeutic efficacy utilizing combinatorial blockade against IDO, CTLA-4, and PD-L1 in mice with brain tumors. Clin. Cancer. Res 20, 5290–5301 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng J, et al. Anti-PD-1 blockade and stereotactic radiation produce long-term survival in mice with intracranial gliomas. Int. J. Radiat. Oncol. Biol. Phys 86, 343–349 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okada H, et al. Immunotherapy response assessment in neuro-oncology: a report of the RANO working group. Lancet Oncol 16, e534–e542 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollack IF, et al. Antigen-specific immune responses and clinical outcome after vaccination with glioma-associated antigen peptides and polyinosinic-polycytidylic acid stabilized by lysine and carboxymethylcellulose in children with newly diagnosed malignant brainstem and nonbrainstem gliomas. J Clin Oncol 32, 2050–2058 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prins RM, et al. Gene expression profile correlates with T-cell infiltration and relative survival in glioblastoma patients vaccinated with dendritic cell immunotherapy. Clin. Cancer. Res. 17, 1603–1615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ayers M, et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 127, 2930–2940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urup T, et al. Transcriptional changes induced by bevacizumab combination therapy in responding and non-responding recurrent glioblastoma patients. BMC Cancer 17, 278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J, et al. Interferon-gamma-induced PD-L1 surface expression on human oral squamous carcinoma via PKD2 signal pathway. Immunobiology 217, 385–393 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Spranger S, et al. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med 5, 200ra116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taube JM, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med 4, 127ra137 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farmer P, et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene 24, 4660–4671 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Li G & Wang X Prediction Accuracy Measures for a Nonlinear Model and for Right-Censored Time-to-Event Data. Journal of the American Statistical Association, 1–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Einat M, Resnitzky D & Kimchi A Close link between reduction of c-myc expression by interferon and, G0/G1 arrest. Nature 313, 597–600 (1985). [DOI] [PubMed] [Google Scholar]
- 43.Shearer M & Taylor-Papadimitriou J Regulation of cell growth by interferon. Cancer Metastasis Rev 6, 199–221 (1987). [DOI] [PubMed] [Google Scholar]
- 44.Yung WK, Steck PA, Kelleher PJ, Moser RP & Rosenblum MG Growth inhibitory effect of recombinant alpha and beta interferon on human glioma cells. J Neurooncol 5, 323–330 (1987). [DOI] [PubMed] [Google Scholar]
- 45.Robert L, et al. Distinct immunological mechanisms of CTLA-4 and PD-1 blockade revealed by analyzing TCR usage in blood lymphocytes. Oncoimmunology 3, e29244 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Omuro A, et al. Nivolumab with or without ipilimumab in patients with recurrent glioblastoma: results from exploratory phase I cohorts of CheckMate 143. Neuro Oncol 20, 674–686 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kamphorst AO, et al. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc Natl Acad Sci U S A 114, 4993–4998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang AC, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 545, 60–65 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krieg C, et al. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat. Med 24, 144–153 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Arbour KC, et al. Impact of baseline steroids on efficacy of programmed cell death-1 and programmed death-ligand 1 blockade in patients with non-small-cell lung cancer. J Clin Oncol 36, 2872–2878 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Weber JS, et al. Safety profile of nivolumab monotherapy: A pooled analysis of patients with advanced melanoma. J Clin Oncol 35, 785–792 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Hamza MA, et al. Survival outcome of early versus delayed bevacizumab treatment in patients with recurrent glioblastoma. J Neurooncol 119, 135–140 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim D, Langmead B & Salzberg SL HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anders S, Pyl PT & Huber W HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hanzelmann S, Castelo R & Guinney J GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grossman RL, et al. Toward a Shared Vision for Cancer Genomic Data. N Engl J Med 375, 1109–1112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carlson CS, et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun 4, 2680 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Robins H, et al. Ultra-sensitive detection of rare T cell clones. J. Immunol. Methods 375, 14–19 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robins HS, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood 114, 4099–4107 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emerson RO, et al. High-throughput sequencing of T-cell receptors reveals a homogeneous repertoire of tumour-infiltrating lymphocytes in ovarian cancer. J. Pathol 231, 433–440 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daley T & Smith AD Predicting the molecular complexity of sequencing libraries. Nat. Methods 10, 325–327 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nowicka M, et al. CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000Res 6, 748 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ellingson BM, et al. Consensus recommendations for a standardized Brain Tumor Imaging Protocol in clinical trials. Neuro Oncol 17, 1188–1198 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ellingson BM, et al. Volumetric response quantified using T1 subtraction predicts long-term survival benefit from cabozantinib monotherapy in recurrent glioblastoma. Neuro Oncol 20, 1411–1418 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ellingson BM, et al. Recurrent glioblastoma treated with bevacizumab: contrast-enhanced T1-weighted subtraction maps improve tumor delineation and aid prediction of survival in a multicenter clinical trial. Radiology 271, 200–210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ellingson BM, et al. Validation of post-operative residual contrast enhancing tumor volume as an independent prognostic factor for overall survival in newly diagnosed glioblastoma. Neuro Oncol (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ellingson BM, et al. Baseline pretreatment contrast enhancing tumor volume including central necrosis is a prognostic factor in recurrent glioblastoma: evidence from single and multicenter trials. Neuro Oncol 19, 89–98 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 72.Zhao K, Lu ZX, Park JW, Zhou Q & Xing Y GLiMMPS: robust statistical model for regulatory variation of alternative splicing using RNA-seq data. Genome Biol 14, R74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data is available in the Gene Expression Omnibus under accession number GSE121810, which includes source data for Figure 2b and extended data figures 4 and 5. The remainder of data that support the findings of this study are available from the corresponding author upon reasonable request. Further information is available in the Life Sciences Reporting Summary.