Abstract
Protein docking methods are powerful computational tools to study protein-protein interactions (PPI). While a significant number of docking algorithms have been developed, they are usually based on rigid protein models or with limited considerations of protein flexibility and the desolvation effect is rarely considered in docking energy functions, which may lower the accuracy of the predictions. To address these issues, we introduce a PPI energy function based on the Site-Identification by Ligand Competitive Saturation (SILCS) framework and utilize the fast Fourier transform (FFT) correlation approach. The free energy content of the SILCS FragMaps represent an alternative to traditional energy grids and they can be efficiently utilized to guide FFT-based protein docking. Application of the approach to 8 diverse test cases, including 7 from Protein Docking Benchmark 5.0, showed the PPI prediction using SILCS approach (SILCS-PPI) to be competitive with several commonly used protein docking methods indicating that the method has the ability to both qualitatively and quantitatively inform the prediction of PPI. Results show the utility of the SILCS-PPI docking approach for the determination of the probability distributions of PPI interactions over the surface of both partner proteins, allowing for identification of alternate binding poses. Such alternate binding poses are confirmed by experimental crystal contacts in our test cases. While more computationally demanding than available PPI docking technologies, we anticipate that the SILCS-PPI docking approach will offer an alternative methodology for improved evaluation of PPIs that could be used in a variety of fields from systems biology to excipient design for biologics-based drugs.
Keywords: Protein-protein interaction, docking, fast Fourier transform, biologics, protein-based drugs
Introduction
Protein-protein interactions (PPI) are macromolecular contacts through which the function and activity of proteins are often modulated in a range of biological contexts, including signal transduction, muscle contraction and cell metabolism.1,2 Accordingly, knowledge of PPIs is of utility for identifying targets for the development of both small-molecule and protein-based therapeutics.3,4,5 For example, protein-protein complex structures provide atomic level details of PPIs that are useful for guiding the engineering of proteins or the rational design of therapeutic molecules to enhance or weaken specific PPI in order to achieve a desired pharmacological outcome. In this regard, computational protein docking methods offer valuable alternatives to experimental structure determination methods, especially when experimental limitations or cost considerations are present.6–8
Similar to small ligand docking, protein docking is composed of two computational aspects; posing or sampling of the relative orientations of the proteins in the complex and scoring or ranking of the resulting complexes. However, protein docking involves additional technical issues due to the large contact region and high number of interactions formed between the proteins upon complex formation. In addition, large conformational changes in the protein conformations may occur upon binding, such that a significant amount of conformational space of the individual proteins need to be taken into account with respect to both posing and scoring in order to achieve a satisfactory protein docking result.
A variety of protein-docking algorithms have been developed and evaluated during last two decades.6–20 Some of these algorithms perform global searches to solve the posing problem and usually utilize fast Fourier transforms (FFT) to expedite the scoring of those generated protein.9 Examples include ZDOCK,10 FTDock,11 PIPER,12 and GRAMM13, among others6–8. Alternatively, a number of methods carry out local randomized searches in the context of Monte Carlo/Metropolis sampling including RosettaDock14 and ICM-DISCO.15 Scoring functions used by these docking methods vary from empirical, knowledge based potentials to physical force field based energy functions6–8. To date, most of the methods are based on rigid protein structures or protein models with limited flexibility. In addition, desolvation contributions are often not or only empirically included in the scoring function, which may limit the accuracy of PPI prediction.6–8 Accordingly, while significant progress has been made in solving the PPI problem, there is still need for more accurate PPI prediction methods, as reflected in the quality of results from the critical assessment of predicted interactions (CAPRI) competitions16 that aims to promote development of new PPI prediction algorithm and optimization of current methods.
The site identification by ligand competitive saturation (SILCS) approach21 is a functional group mapping method that utilizes explicit solvent all-atom Grand Canonical Monte Carlo (GCMC)/Molecular Dynamics (MD) simulations22 to map the functional group interaction pattern of a protein. In the SILCS approach, during the GCMC/MD simulation a selection of solutes or probe molecules are competing with water and with each other for binding to the protein. Information from the simulation trajectory can be extracted in the form of probability maps of the solute functional groups, termed FragMaps, by binning the residences of functional group atoms onto a three-dimensional grid that encompasses the entire protein and surrounding region. Accordingly, the FragMaps contain contributions from protein-functional group interactions, protein flexibility, and desolvation of both the functional groups and the protein. The FragMaps can then be Boltzmann weighted into a free energy representation, termed grid free energy (GFE) FragMaps, which can be used in various receptor-based computer-aided drug design methods such as identification of cryptic binding pockets,22 ligand docking and optimization23–26 or pharmacophore searching.27,28
As the SILCS GFE FragMaps include intrinsic contributions from protein flexibility and desolvation effects, their application in PPI prediction may help solve the main limitations discussed above. Many current protein docking methods employ the FFT based global search method since it only requires information on the protein structure itself, allowing for a near exhaustive global search by benefiting from the computation efficiency introduced by FFT. While highly efficient, it requires the scoring function to be expressed on 3D grids and in a correlation format, such that many FFT-based protein docking methods use very simple energy scoring functions combined with rigid protein structures. The SILCS FragMaps offer an ideal extension of these methods as they are grid-based as required for the FFT calculations and intrinsically include information on protein flexibility, protein and functional group desolvation effects as well as protein-functional groups interactions.
In this work, we explore the use of SILCS GFE FragMaps combined with FFT-based global docking for PPI prediction. The SILCS FragMaps were tested in combination with protein functional group-based probability maps to guide the docking. Results were compared with published values for a variety of docking methods as well as calculations performed as part of the present study using selected published methods. The method, designated SILCS-PPI, was shown to be competitive with or better than currently available methods though the approach requires a significant upfront computation to produce the required SILCS FragMaps and protein functional group-based probability maps.
Methods
FFT accelerated posing or sampling
FFT-based docking methods have been described in a number of previous publications.9–13 Briefly, the PPI posing problem may be solved by searching for the set of 3 rotational and 3 translational operations representing the relative orientation of the interacting proteins that yield the most favorable score based on the scoring function, as described in the following section. Searching of all possible rotational and translational operations is performed at a certain resolution with the translational part being accelerated through FFT in the present study. To take advantage of the Fourier transform, it is necessary that the energy function to be expressed on grids such that the overlapping distributions of terms defining the energy function (e.g., the electrostatic energy calculated from the partial atomic charges of the atoms mapped onto a grid) may be rapidly determined in Fourier space rather than explicitly calculating each grid point-grid point contribution to the energy. Recently, an extension of the FFT method to 5 degrees of freedoms has been presented29, though we only apply the FFT method to the 3 translation degrees of freedom in the present study.
Scoring scheme
SILCS GFE FragMaps describe how a protein interacts with functional groups of various types. Thus, if the complementary functional group from a binding partner occupies an energetically favorable region of the corresponding GFE FragMap, a favorable score will be obtained. In the context of a protein-protein complex, the energetically favorable regions of FragMaps from one protein should be occupied by the same type of functional groups of the other protein (e.g., a favorable apolar FragMap region would be occupied by an apolar side chain) for the favorable complex structures. Thus, we hypothesized that using FragMaps from one protein in combination with functional group probability maps from the other protein would be an effective PPI scoring scheme. Accordingly, a scoring function that uses FragMaps and the corresponding protein functional group probability distribution maps (or protein probability grid, PPG) is developed and its performance tested in this work. For each corresponding FragMap-PPG pair of functional type i, the score of the functional type can be written as:
| (1) |
where and represent the FragMap of protein A of and PPG of protein B of given functional group type i, respectively. x, y and z are grid coordinates in 3D and dx, dy, dz are possible translation operations in three directions. In the above equation, the FragMap of protein A is fixed on the origin and the protein B PPG is being translated, such that the FragMap for protein A and PPG for protein B are used for docking. In addition, equation 1 is defined in the context of the alternative scenario where the PPG is from protein A and the FragMaps for protein B are used.:
| (2) |
The score is again summed over all FragMap types and their corresponding PPG with the total scores based on the sum of the contributions from equations 1 and 2 in conjunction with the FFT algorithm.
In practice, the FFT transform is initially applied to both the FragMap and the PPG for the larger protein, which is assigned as protein A (or the receptor). Subsequently for each rotation of protein B (or the ligand) relative to A the FFT transforms only needs be applied to the B FragMap grid and PPG. The correlations between the A FragMap grid-B PPG (Eq. 1) and A PPG-B FragMap grid are then performed and the inverse FFTs of the B grids performed. This yields the energy scores associated with equations 1 and 2 in a computationally efficient manner.
In addition to the FragMap-PPG pairs used in scoring, an exclusion-exclusion map correlation score is also considered. Exclusion maps from SILCS GCMC/MD simulations describe forbidden regions where no solute or water molecules access during the entire simulation (e.g., regions where water and solutes are excluded) and thus serve as an alternative to a rigid protein surface. As the exclusion maps omit regions of the protein that undergo conformational changes and allow either water or solutes under the protein surface they account for protein flexibility, while still allowing for steric overlap of the protein partners to be considered. The exclusion grid is present as a hard wall function with grids in the excluded regions assigned a value of one while exclusion grid points in the remaining region have a value of zero. The exclusion-exclusion correlation score is written as:
| (3) |
where EXCLA and EXCLB represent exclusion grid from protein A and B, respectively, and 𝝈 represents a scaling factor. The inclusion of the exclusion energy solution can prevent bad solutions with high favorable interaction scores (eq. 1 and 2) from being ranked favorably. Such bad solutions can occur in bound orientations where a large number of FragMaps of one protein overlap with the PPG of the partner protein, but the two protein structures have significant steric overlap. The exclusion map contribution may be considered analogous to the protein shape complementary score used by other docking methods.10–12 In addition, the SILCS scores based on the FragMap and PPGs themselves contain protein shape information associated with unfavorable regions of the FragMaps (e.g., where a grid point with a positive apolar GFE value is occupied by a Leucine side chain yielding an unfavorable contribution) but are not as “hard” as the exclusion maps. As defined in eq. (3), the exclusion-exclusion correlation score will either have zero contribution or contribute a very high positive value to the final total score, thereby helping to omit the solutions with highly unfavorable steric contacts. In this study, 𝝈 is set to either 0.1 or 1.0 for testing. When 𝝈 is set to 0.1, it is expected that the receptor and the ligand potentially have closer contact.
The final score used to rank the solutions is defined as:
| (4) |
and the total score is obtained by summing over all individual FragMap-PPG scores as well as the exclusion score. wi is a weighting factor for each type of interaction pair included to allow contributions from different type of interaction pair to be adjusted. The factor was treated as 1 in the current study for all interaction pairs.
Choice of map pairs for scoring
In principle, one can use as many pairs of the FragMap grids and PPGs as possible to include more energetic details obtained from SILCS. However, FFT calculations are required for each pair of energy grids and thus the computational cost, as well as the memory requirement, increases linearly with the number of grid pairs used for scoring. To balance computational cost and possible accuracy increases with additional energetic profiles, in the present study generic FragMaps were applied as previously adopted for SILCS small ligand docking studies.30,31 Generic FragMaps are defined based on combined solute probability distributions associated with all functional groups of a given class (e.g. hydrogen bond donor generic FragMap includes contributions from imidazole NH and formamide NH2 groups) in contrast to considering all possible specific FragMaps (eg. individual maps for imidazole NH or for formamide NH2). Six types of FragMaps, hydrophobic (APOLAR, benzene and propane carbons), hydrogen-bond donor (HBDON, formamide amino nitrogen and imidazole amide nitrogen) and acceptor (HBACC, formamide and acetaldehyde oxygens and imidazole nitrogen), positively charged donor (MAMN, methylammonium nitrogen) and negatively charged acceptor (ACEO, acetate oxygens) as well as hydroxyl group (MEOO, methanol oxygen) based maps, based on non-hydrogen atoms are considered for scoring. Correspondingly, the PPG maps are also classified into generic maps that include the same functional atom types as defined in Table 1.
Table 1.
Classification of protein functional group atoms in 22 amino acid typesa used to generate protein probability grid maps for correlation scoring with the FragMaps. Only non-hydrogen atoms were used to define the maps.
| Probability map type | Atoms | Amino acid |
|---|---|---|
| APOLAR | Side-chain carbons | ALA, VAL, LEU, ILE, MET, PRO, PHE, TYR, TRP |
| HBDON | Backbone and selected side-chain donor nitrogens | TRP, ASN, GLN, HSD, HSE |
| HBACC | Backbone and side-chain carbonyl oxygens or side-chain imido nitrogens | ASN, GLN, HSD, HSE |
| MAMN | Side-chain protonated nitrogens | LYS, ARG, HSP |
| ACEO | Side-chain deprotonated carboxylate oxygens | ASP, GLU |
| MEOO | Side-chain hydroxyl oxygens or thiol sulfurs | SER, THR, TYR, CYS |
The three different protonation states of HIS are considered individually.
SILCS-PPI workflow
Scheme 1 shows the overall SILCS-PPI workflow using SILCS FragMaps, functional-group PPGs, and exclusion maps with the FFT algorithm for protein docking. The only input files needed are the free energy grids (i.e., FragMaps), PPGs and exclusion maps from the SILCS Grand Canonical Monte Carlo/Molecular Dynamics (GCMC/MD) simulations (see below). Energy grids are first filtered to remove unnecessary information before being used to guide docking. For FragMaps, all voxels that are 5 Å away from the excluded area as defined by the exclusion map are removed since those grids are influenced by bulk phase behavior of the solutes rather than binding patterns on the protein surface. For the PPGs, all voxels that are overlapping with the exclusion map are removed since they are representing the repulsive core of the protein and only probability maps on the protein surface are useful for directing the matching with FragMaps from the other protein participating in docking.
Scheme 1.
Overall SILCS-PPI workflow using FragMaps and functional-group probability maps for protein-protein FFT docking. User inputs are shown in dashed boxes.
After all the energy grids are filtered, user input parameters are used to further tailor the energy grids for FFT. GFE cutoffs are used to control how much information from the FragMaps is used for scoring. This may be used to limit the extent of favorable or unfavorable contributions or one can use only favorable GFEs or both favorable and unfavorable GFEs to guide the docking. Protein docking can be performed either globally on the entire protein surface or locally when experimental binding site information is available by only using the corresponding portion of the maps. Weighting factors can be adjusted for the GFE based scoring as defined in eq. 4 to balance different contributions. In the present study a subset of possible parameters are tested; future studies will include tests of a wider range of parameter options, most notably the FragMap scaling factors.
All energy grids are first translated to the origin and are mapped onto new grids that are large enough to encompass both sets of grids from the two proteins. As described above, forward FFT is then done once and stored for all maps of protein A since they are fixed throughout the full exploration of translational and rotational space. A full set of unique rotations are generated for a user-defined rotation step size (10˚ for the current study) based on the theory of uniform sampling of points on a sphere.32,33 For each rotation, forward FFT is conducted for all maps from protein B after the energy grids are rotated and mapped onto the new grids. The correlation calculations are then done for all pairs of energy grids in frequency space and inverse FFTs are subsequently performed to evaluate the total scores for all possible translations. Then the top scoring solutions based on a user-defined criterion are recorded per rotation. After all rotations are evaluated, all best solutions are output for final model construction and further analysis. In the current study, two-pass clustering was used to consolidate the solution list that will be discussed below. An in-house code written in C++ using the FFTW 3.3 library34, which is compatible with our available SILCS-based programs, was developed to realize the whole protocol.
Test sets for validation
During our initial test, a commonly used test system employed by other PPI methods was considered here, which is the Barnase and Barstar complex35. This was used to test our code and initially validate the whole protocol. Subsequently, we did thorough validations by including test cases from Protein Docking Benchmark 5.036, which is a widely used benchmark to evaluate protein-docking methods. Test cases were chosen to cover a range PPI types with different docking difficulties as defined by the benchmark. As listed in Table 2, the last seven test cases were selected from benchmark 5.0. They cover enzyme-inhibitor, antibody-antigen, antigen-bound antibody and G-protein PPI types. The seven cases have docking difficulties ranging from low to high based on their interface RMSD (iRMSD) values between apo-protein and complex structures as defined by the benchmark36.
Table 2.
List of test cases for SILCS-PPI validation.a
| Test Case | Complex PDB | Receptor Protein | Ligand Protein | Difficulty | Catagory |
|---|---|---|---|---|---|
| 1 | 1BRS | Barnase (PDB:1BRS) | Barstar (PDB:1BRS) | Low | Enzyme-inhibitor |
| 2 | 1AY7 | RNase Sa (PDB:1RGH) | Barstar (PDB:1A19) | Low | Enzyme-Inhibitor |
| 3 | 1KXQ | Alpha-amylase (PDB:1PPI) | Camel VHH (PDB:1KXQ) | Low | Antigen- Bound antibody |
| 4 | 1JTD | BLIP-II (PDB:3QI0) | Beta-lactamase (PDB:1BTL) | Low | Enzyme-Inhibitor |
| 5 | 1I9R | Fab (PDB:1I9R) | Cd40 ligand (PDB:1ALY) | Low | Antigen- Bound antibody |
| 6 | 3G6D | CNTO607 FAB (PDB:3G6A) | Interleukin-13 (PDB:1IK0) | Medium | Antibody-Antigen |
| 7 | 1ACB | Chymotrypsin (PDB:2CGA) | Eglin C (PDB:1EGL) | High | Enzyme-Inhibitor |
| 8 | 1BKD | Son of sevenless (PDB:2II0) | Ras GTPase (PDB:1CTQ) | High | Others, G-protein containing |
The first test case has been widely used to study PPI35 while the next seven test cases were taken from PPI benckmark 5.0.
SILCS simulations and FragMap preparation
SILCS simulations were performed using the previously described SILCS setup30 for the 16 target proteins from the 8 test cases. Crystal structures with access codes listed in Table 2 were obtained from the Protein Data Bank (PDB)37 to initialize the SILCS GCMC/MD simulations. Crystal waters or coordinated metal ions were retained when preparing the protein systems and the ligands were removed from holo structures. For protein structures with missing coordinates that affect stability during simulations if ignored, homology modeling was used to build the missing regions. For example, two small loops in the crystal structure of protein target Son of Sevenless (Table 2) have missing coordinates and may the affect PPI docking as the same protein in the complex crystal structure has full coordinates. Homology modeling was performed using the MODELLER program38 with the crystal structure from the complex used as the template. The best homology model based on the DOPE score38 was used for the SILCS simulations.
SilcsBio software package (SilcsBio LLC) was used with GROMACS tools39 to prepare the simulation systems involving protein, water and eight probe molecules including benzene, propane, methanol, formamide, acetaldehyde, imidazole, methylammonium, and acetate. Protonation states of ionizable residues were determined by GROMACS tools.39 Ten simulation systems with randomly positioned solutes at approximately 0.25 M each were simulated independently for better convergence. An iterative Grand-Canonical Monte Carlo (GCMC) and MD protocol22, where GCMC drives the sampling of the solutes and water with all solutes, water and protein atoms subsequently propagated in the MD simulations. Weak harmonic restraints with a force constant (k in 1/2 k δx2) of 0.12 kcal/mol/Å were kept on all Cα atoms of protein during MD simulations. The protein conformations and distributions of water and solutes were saved every 10 ps for analysis. The GCMC simulations are based on a previously described protocol where an oscillating excess chemical potential, μex, is applied to increase efficiency. The GCMC simulations were conducted using in-house developed code22 and MD simulations were performed using GROMACS,39 with the protein, solutes and water being described using the CHARMM36 protein force field,40 the CHARMM General force field (CGenFF)41,42 and the TIP3P water model modified for the CHARMM force field,43 respectively.
FragMaps were generated by binning selected solute atoms into voxels of a 1 Å spaced grid spanning the simulation system. 3D normalized probability distributions were obtained by normalizing the voxel occupancies computed in the presence of the protein by the respective values of the solutes alone in aqueous solution. The normalized distributions were Boltzmann-transformed to free energies for each FragMap type to yield GFE FragMaps. For SILCS-PPI, three generic FragMaps were generated including APOLAR, HBDON, and HBACC maps. In addition, three specific FragMaps were used including positive MAMN maps, negative ACEO maps, and alcohol (MEOO) maps. Specific alcohol (as well as thiol) maps are used as these functional groups can act as both hydrogen bond donors and acceptors complicating their inclusion in the generic HBDON or HBACC maps. Exclusion maps that represent the solute/water forbidden region during SILCS simulation were also generated. Protein functional group probability maps were generated based on the classifications listed in Table 1 and occupancy values were normalized by the maximum occupancy value for each type, resulting in PPG grid maps with 0 to 1 scale probability values.
Docking performance evaluation
To evaluate SILCS-PPI docking performance, coordinates of the complex were constructed directly using the crystal structures of the two proteins in each test case as listed in Table 2 according to the best scoring translation and rotation parameters. We note that additional refinement of the docked complexes, for example, using alternate protein structures from the SILCS MD simulation followed by local relaxation of complex models, will likely improve predictions;44 however no such refinements were conducted in the present study as the focus of the work is to show proof-of-concept. The top 10 solutions per rotation are saved and all coordinates for all solutions are constructed resulting in ~140,000 total solutions per docking run. Two-pass clustering is then used to cluster all models. First, center-of-mass (COM) based clustering is conducted to put all models whose COM distances are within 6 Å into the same cluster. Second, orientation-based clustering is performed using the members of a cluster found in the previous step. The distance in orientation clustering is measured by taking the Euler angles and applying the angular distance metric that preserve periodicity.45 The distance cutoff in orientation clustering is set to 0.5, which corresponds to about a 30˚ angle. After the clustering, the best scoring pose in each cluster is saved as the representative pose with the top 2000 representative poses as defined by SILCS-PPI score saved for further evaluation.
Four metrics are considered to evaluate the docking performances. COM RMSD between docking pose and crystal structure of ligand protein is evaluated. This metric indicates the ability of SILCS-PPI to identify the correct binding site on receptor protein surface. The remaining three metrics are commonly used by previous PPI docking studies and include ligand RMSD (L-RMSD), interface RMSD (iRMSD) and fraction of native contacts (fnat). These three metrics describe the quality of the docking pose in reproducing structural details at the receptor-binding site. iRMSD is defined as RMSD of all non-hydrogen backbone atoms of interface residues after structural alignments of the docking pose with crystal structure of the complex where interface residues are defined as residues from both proteins at the interface if any non-hydrogen residue atom of one protein is within 6 Å distance of the other protein. Consistent with the CAPRI evaluation,16 we define a model as acceptable by considering L-RMSD, iRMSD and fnat and we define a model as a hit if its iRMSD is within 4 Å. We note that L-RMSD, iRMSD and fnat include contributions from intramolecular conformational changes in the proteins that occur upon docking; such changes were omitted in the present study and will be considered in future efforts.
Comparison with other docking methods
The SILCS-PPI results were compared with those from other protein docking protocols. The previously published methods typically presented their docking results in unique ways and published results in the literature use different criteria to evaluate docking. Accordingly, the present SILCS-PPI results were analyzed using the corresponding approaches to allow direct comparison with previously published data for ZDOCK10 and RosettaDock.14 For ZDOCK,10 a hit is defined with an iRMSD less than 2.5 Å between the docked and crystal complex structure and only considered for the top 2000 ranked predictions. For RosettaDock,14 interface residues are defined as all residues with an intermolecular distance of at most 4.0 Å based on non-hydrogen atoms and iRMSD is calculated based on all backbone atoms of those residues. For ClusPro,20 instead of directly presenting docking structures from FFT, the method further clusters top predictions using a greedy clustering algorithm. To enable comparison with results from ClusPro, we run docking using the ClusPro server,20 and the SILCS-PPI results from our two-pass clustering are compared with ClusPro results using their balanced protocol20 and analyzed using the same criteria locally. The SILCS-PPI results were thus tailored using consistent criteria when compared with each individual method.
Results and Discussion
We present a computational protein-protein docking and scoring approach using FragMaps and protein side chain probability maps or PPG. The basic idea is that a protein-protein binding interface consists of various interaction partners, which can be captured by the high correlation between FragMaps from the receptor protein and side-chain probability maps from ligand protein. Such correlation can be computed in a computationally efficient manner via the FFT algorithm. Corresponding protein probability maps were correlated and fast evaluated using FFT. All individual scores from each pair of correlated FragMap-protein probability maps were summed and the total score was used to rank the solutions. This protocol, designated SILCS-PPI, fully utilizes information from both protein and functional groups probability distributions from multiple explicit-solvent MD simulation. The new method was tested against 7 test cases from protein docking benchmark 5.0 as well as an additional well-known PPI system, with the selected systems covering most PPI types as defined in the database. The result shows that SILCS-PPI has the ability to reproduce crystallographic solved PPI to a satisfactory level of accuracy. We also compared our results with docking results from other common docking methods such as ZDOCK, RosettaDock and ClusPro using the same validation criteria, showing that SILCS-PPI is competitive with the other methods in the majority of cases.
SILCS FragMaps can recapitulate the crystal binding modes of PPI.
FragMaps from SILCS were previously shown to be able to reproduce crystal binding modes of small ligands. Presently, the FragMaps were first examined for their ability to recapitulate the crystal binding modes of proteins. This serves as the first step to verify if the FragMap-PPG overlap approach can be used to guide protein docking.
Figure 1 shows the FragMaps on both proteins from RNase SA/Barstar and SOS/Ras GTPase complex test cases aligned with the corresponding complex crystal structures. For RNase SA/Barstar, RNase SA interacts with Barstar mainly through charged interactions as observed previously46. Panel A1 in Figure 1 shows the FragMaps for RNase SA (white cartoon representation) in the complex with Barstar (grey cartoon representation), including residues involved in the PPI interface. RNase SA ACEO FragMaps are observed at the interface overlapping with crystal binding positions of E76 and D39 from Barstar as indicated by the orange arrows. E76 forms a salt bridge with R40 while D39 interacts with Q32, R65, R69 and H85 on RNase SA. Panel A2 shows the FragMap for Barstar (grey cartoon representation) along with RNase SA (white cartoon representation). As shown by cyan arrows, MAMN FragMaps are observed on the surface of Barstar that reproduce the binding positions of positively charged residues R40, R65 and R69 on RNase SA. In addition, HBDON FragMaps overlap with positions of the amide group in Q32 and with H85 of RNase SA as shown by blue arrows. Also, HBACC FragMap overlap with the acceptor nitrogen in H85 that interacts with the backbone amide group of G31 of Barstar. Thus, FragMaps on both proteins are consistent with locations of functional groups on the partner proteins.
Figure 1.
SILCS FragMaps overlaid on crystal complex structures for (A1) FragMaps of Rnase SA (white cartoon representation) with Barstar (grey cartoon representation), (A2) FragMaps of Barstar (grey cartoon representation) with Rnase SA (white cartoon representation), (B1) FragMaps of Son of sevenless (white cartoon representation) with Ras GTPase (grey cartoon representation) and (B2) FragMaps of Ras GTPase (grey cartoon representation) with Son of sevenless (white cartoon representation). FragMaps are shown for Apolar (green), negative ACEO (orange), positive MAMN (cyan), H-bond donor (blue) and H-bond acceptor (red) FragMaps.
For the SOS/Ras GTPase complex, apolar FragMaps play a much larger role. Panel B1 of Figure 1 shows the FragMaps of Son of Sevenless (SOS) aligned with crystal complex structure47. Apolar FragMaps of SOS overlap with Ras GTPase residues I21 and Y32, which have nonpolar contacts with SOS residue G943. Apolar FragMap of SOS also overlap with Ras GTPase residue A59 and Y64 that are in contact with SOS residues L938 and F929, respectively. Binding positions of key residues of Ras GTPase that are involved in salt bridge interactions with SOS are captured by the corresponding FragMaps of SOS. These include overlaps between MAMN FragMap of SOS with Ras GTPase residues R102 and R68 and also between ACEO FragMap with Ras GTPase residue E63, which forms interactions with SOS residue R826. In addition, the location of the hydroxyl group of Ras GTPase Y64, which hydrogen bonds with the backbone amide group of G931, is captured by MEOO FragMap of SOS. For the FragMaps of Ras GTPase shown in panel B2, overlaps between apolar, MAMN and ACEO FragMaps with SOS residues F929, R826 and D1007, respectively, are observed further showing that the FragMaps capture key residues of SOS on the SOS-Ras GTPase interaction interface. Similar results are observed for other test cases as shown in Figures S1 of the supporting information, further indicating the FragMaps can qualitatively recapitulate the location of residues on the partner proteins in the complex and indicating that they can guide quantitative protein docking.
Protein docking performance of SILCS-PPI
As the first step towards applying SILCS FragMaps for protein docking, the current study is designed to provide proof of concept rather than undertake a comprehensive refinement of the entire protocol. Assumptions being applied in the current validation of SILCS-PPI, include the scoring function simply being the summation of all GFE contributions from each FragMap-PPG pair. Initial results from the SILCS-PPI runs will focus on the rotation and translation parameters applied to the FragMaps and their associated ranking based on the docked energy (Eq. 4). This allows the relative orientations of docked solutions to be analyzed and compared to experimental orientations. In addition, to compare the SILCS-PPI docking performance with previously published methods as well as with the protein conformations in complex crystallographic structures, the Cartesian coordinates of the predicted complex models are required. To obtain those we simply assemble the final Cartesian coordinates of the predicted complex structure by using the apo crystal structures of the two partner proteins and applying the translations and rotations of the docked solutions. Future studies will investigate using conformations of the proteins sampled in the SILCS simulations to assemble models followed by subsequent relaxation of the assembled models to improve predictions with respect to the intramolecular conformations of the proteins in the complexed state.
Test of the parameters used in the scoring function involved varying the range of GFE energy cutoff levels and the 𝝈, the exclusion map scaling parameter. The GFE cutoff for FragMaps controls the level of favorable vs. unfavorable energy information to be used for docking. Grid voxels having positive GFE values represent the regions that are unfavorable for the corresponding functional group. As implemented, setting the GFE cutoff to a negative value only includes voxels that have favorable GFE values. On the other hand, if a positive GFE cutoff value is used, both favorable and unfavorable voxels are considered. Typically, GFE values in the FragMaps range between −3 to 3 kcal/mol, and we chose two representative values, −1.2 kcal/mol and 3 kcal/mol, to test the effect of GFE cutoff level in docking performance. The favorable GFE cutoff of −1.2 kcal/mol means only FragMap grid points that have GFE value equal to or more favorable than −1.2 kcal/mol are used. The SILCS-PPI run conducted using a GFE cutoff of 3.0 kcal/mol includes all favorable grid points along with unfavorable grid points up to 3 kcal/mol.
In addition, the effect of exclusion map scaling parameter, 𝝈, was tested. Lowering the exclusion map scaling factor removes the penalty of having steric clashes, which could be useful for resolving hard test cases where the conformation of the binding interface adjusts significantly upon binding. We tested both 𝝈 = 0.1 and 𝝈 = 1.0 along with GFE cutoffs −1.2 kcal/mol and 3.0 kcal/mol, making a total of four trials. Among those, two trials, GFE cutoff of −1.2 kcal/mol with 𝝈 = 0.1 and GFE cutoff of 3.0 kcal/mol with 𝝈 = 1.0, performed better than the others and we will highlight only those two trials below. For clarity, the exclusion map scaling parameter is not mentioned below unless necessary.
Figure 2 shows the overall performance averaged over all the studied systems with respect to COM RMSD and L-RMSD for docking runs using −1.2 and 3.0 kcal/mol as GFE cutoffs. The successful hits are based on COM RMSD and L-RMSD cutoffs of 10 Å. The performance of the individual test cases is provided in Figures S2 and S3 of the Supporting Information. With respect to the COM RMSD and L-RMSD, overall the two GFE cutoff methods yield similar results. However, when the results for the individual proteins are analyzed in Figures S2 and S3 the use of GFE cutoff of 3.0 kcal/mol gives a significant number of successful solutions for all the test cases for COM RMSD, while with GFE cutoff −1.2 kcal/mol there were very few to no hits with the Beta-Lactamase and Fab-CD40 test cases. This indicates that when a positive GFE cutoff was used, the SILCS-PPI protocol could identify the correct binding pocket better than when a negative GFE cutoff was used, which is suggested to be due to the use of both favorable and unfavorable energetic information. However, the solutions with positive GFE cutoff using more information does not help to further improve predictions in terms of predicting the correct ligand orientation as indicated by the poor performance with respect to the L-RMSD metric. This may be due to the presence of too much detailed information from the combination of both favorable and unfavorable energy information which may introduce more background noise in the total energy profile during docking, thereby flattening the local energy score surface limiting its ability to identify near-native docking poses. Based on the improved COM RMSD analysis, using the combination of favorable and unfavorable FragMap grids in the context of the 3.0 kcal/mol GFE cutoff criteria gives consistent performance and, thus, is recommended for SILCS-PPI use in general. The utility of making a good COM prediction is that the ligand prediction can be refined and improved by using more computationally intensive methodologies once the general area of binding is identified. However, as may be seen by the results in the Supporting Information, the actual success of the two approaches is both system and analysis-type dependent.
Figure 2.
Comparing the SILCS-PPI performance with different GFE cutoff values, (A) GFE cutoff of −1.2 kcal/mol with 𝝈 = 0.1 and (B) GFE cutoff of 3.0 kcal/mol with 𝝈 = 1.0. The COM RMSD (left) and L-RMSD (right) were used to determine the number of successfully hits as a function of the number of predictions with the results averaged over the 8 test cases. The cutoffs for successful hits were 10 Å for both COM RMSD and L-RMSD.
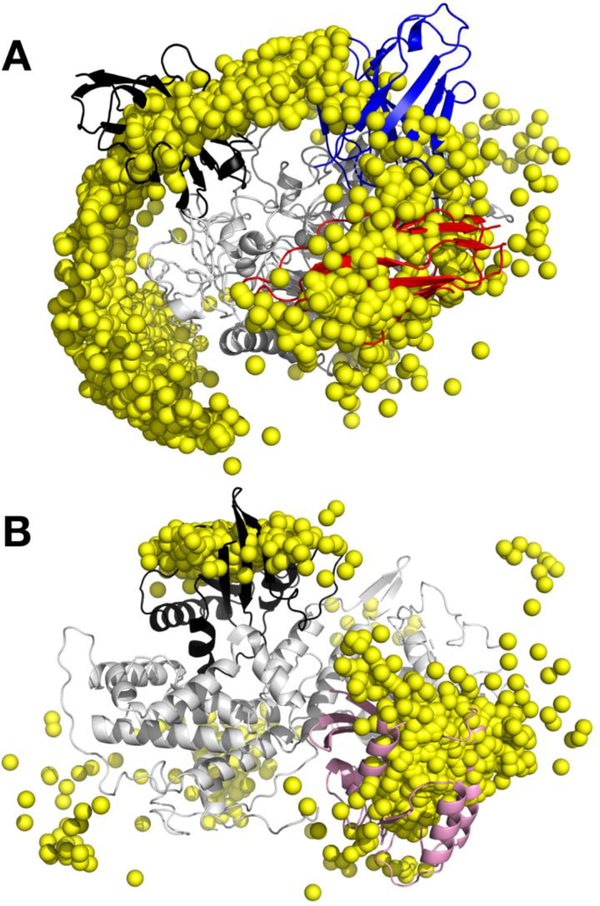
Figure 3 shows heat map style representations of the top 2000 predictions for each test case on the receptor protein surface along with COM positions of those predictions with GFE cutoff of 3.0 kcal/mol. The figure also contains the crystal binding orientation of ligand proteins. Images of the receptor heat maps alone along with the ligand proteins are presented in Figure S4 of the Supporting information. For all test cases, SILCS-PPI predictions encompass the correct binding site on the receptor surface. Quantitation of the level of agreement between the SILCS and experimental PPI orientations for the 8 targets is included in Table S1 of the supporting information. The results show that SILCS-PPI method to identify solutions based on COM and L-RMSD even for challenging test cases such as 3 and 5 (see additional discussion below). The overall docking performance is promising considering that only rigid crystal structures are used for model construction and no further refinement is applied prior to the predictions. In addition to the crystal binding sites, analysis of the SILCS-PPI predictions also covers other sites on the receptor protein surface and this may indicate alternate PPI sites.
Figure 3.
Distributions of COM of top ranked solutions (yellow dots) and their populations mapped onto the protein surface in heat map representation (red-blue-white, 1 to 0 scale). The crystal binding mode of the ligand protein is shown in magenta cartoon representation.
SILCS-PPI identify alternative binding sites
As protein docking methods sample the entire receptor surface, they may supply information about alternate protein-protein binding orientations. We here examined such potential using our SILCS-PPI results. Figure 4 shows the COM of representative ligand protein predictions for alpha amylase and SOS test cases along with ligand crystal bound orientations. For alpha-amylase there are also additional complex crystal structures beyond that with camel VHH AMD9, which is the ligand protein in the original test case. Complexes also occur with alternate camel VHH domains48 from different subfamilies (AMB7 and AMD10) as shown in Figure 4(A). Even though the three camel VHHs have different sequences at the binding site, the predictions using camel VHH AMD9 include solutions in the vicinity of the crystal binding modes of the other two camel VHHs. This partially shows that predictions from SILCS-PPI runs have information about alternative binding sites of similar proteins. A second example is the SOS test case that binds with Ras. In addition to the catalytic binding site on SOS, a study49 found that the Ras mutant Y64A binds to SOS at an alternative binding site that shown as the distal site in Figure 4(B). As may be seen, SILCS PPI identifies a large population of solutions in this region of the protein surface. While the alternate crystal binding orientations are associated with changes in protein sequence relative to that used for docking the present observation shows the potential utility of the full range of the predicted solutions in identifying alternative binding sites of similar ligand proteins.
Figure 4.
Ability of SILCS PPI to identify alternate PPI interactions. Shown are COM of top ranked solutions (yellow) for the receptor proteins (white color) alpha-amylase (A) and SOS (B). Crystal binding structures of the ligand proteins Camel VHH (A) and Ras GTPase (B) are shown in black. For (A) alpha-amylase alternate PPIs from crystal structures of AMB7 Camel VHH (blue) and for AMD10 (red) are shown. For (B) SOS, the Ras Y64A mutant (pink) binds to distal site.
SILCS-PPI performance compared with other docking methods
Given the large body of PPI docking methods it is essential that the SILCS-PPI docking method be compared with some commonly used docking methods. In the present study comparisons are performed with ZDOCK,10 RosettaDock14,50 and the ClusPro20 docking server. The docking result from above for each test case using the 3 kcal/mol GFE cutoff is used for the comparison. As discussed in the Methods section, consistent criteria are used for the comparisons with the other docking methods.
For ZDOCK and RosettaDock, docking results for test case 2, 3, 5, 7 and 8 are available from the literature.10,50 Table 3 shows SILCS-PPI docking results compared with ZDOCK 3.0.10 As a tighter iRMSD cutoff of 2.5 Å was used for hit definition with ZDOCK, no hits were found in three out of five test cases for both methods. ZDOCK 3.0 produced three hits for test case 3 while SILCS-PPI failed to give solutions with iRMSD less than 2.5 Å. For test case 2, ZDOCK produced three hits with the first hit ranked at 1041 with an iRMSD of 2.36 Å, whereas SILCS-PPI produced 15 hits with the first hit having a better iRMSD of 0.2 Å.
Table 3.
Docking performances of SILCS-PPI compared with ZDOCK 3.010 using the same evaluation criteria.
| Method | Evaluation criteria | Test case 2 | Test case 3 | Test case 5 | Test case 7 | Test case 8 |
|---|---|---|---|---|---|---|
| ZDOCK 3.0 | Nhitsa | 3 | 3 | 0 | 0 | 0 |
| Rank1stb | 52 | 2 | - | - | - | |
| iRMSD1stb | 2.36 | 1.30 | - | - | - | |
| SILCS-PPI | Nhitsa | 15 | 0 | 0 | 0 | 0 |
| Rank1stb | 64 | - | - | - | - | |
| iRMSD1stb | 0.2 | - | - | - | - | |
Nhits is the number of hits, which defined as solutions whose iRMSD is less than 2.5 A, among the top 2000 solutions.
Rank1st and iRMSD1st is the percentile rank (top %) and iRMSD of the first hit among top 2000 solutions.
Table 4 shows SILCS-PPI docking results compared with RosettaDock.50 Unlike SILCS-PPI where global docking is performed without information about the binding interface, the RosettaDock results14 were obtained from local docking searches that started from crystal complex structures. Of the five test cases, SILCS-PPI shows a similar level of agreement for three test cases when compared with RosettaDock. For test case 2, a slightly better result is seen in the SILCS-PPI with an iRMSD of 0.2 versus 1.55 Å for RosettaDock. For test cases 3 and 5, RosettaDock has superior performances with the top iRMSD of 1.25 and 0.52 Å, with this value being 5.0 and 5.5 Å for the best models from SILCS-PPI. In the case of test cases 7 and 8, the best solutions from SILCS-PPI have an iRMSD of 2.9 and 4.3 Å, respectively, which are much better than values of 4.56 and 5.62 Å from RosettaDock. Thus, the SILCS-PPI results are quite promising when considering that RosettaDock involves a significant amount of local refinement during the docking while the SILCS-PPI results are directly from the raw FFT-based global docking.
Table 4.
Docking performances of SILCS-PPI compared with RosettaDock using the same evaluation criteria.
| Method | Evaluation criteria | Test case 2 | Test case 3 | Test case 5 | Test case 7 | Test case 8 |
|---|---|---|---|---|---|---|
| RosettaDock | iRMSDbesta | 1.55 | 1.25 | 0.52 | 4.56 | 5.62 |
| SILCS-PPI | iRMSDbesta | 0.2 | 5.0 | 5.5 | 2.9 | 4.3 |
iRMSDbest is the iRMSD of the best near-native solution.
SILCS-PPI is also compared with docking results from the ClusPro server.20 The eight test cases were submitted to the ClusPro server using the default settings. The ClusPro docking results in the balanced mode were downloaded and compared with SILCS-PPI results using the same evaluation criteria. The ClusPro server performs docking using FFT based global docking algorithms followed by greedy clustering on solutions from the FFT to further filter the results. This is followed by energy minimization on representative solutions from each cluster to generate final predictions. In this work, the ClusPro results are compared to the 2000 solutions produced by SILCS-PPI without further refinement. It should be noted that ClusPro classifies input protein structures into different classes and different docking protocols can be chosen accordingly. Table 5 shows the SILCS-PPI to ClusPro comparison. Both SILCS-PPI and ClusPro found hits for 5 out of the 8 test cases. For test cases 1 and 4, similar performances are seen. For ClusPro the best iRMSD of 1.0 Å and 1.1 Å, respectively, are compared to slightly better iRMSD of 0.9 Å and 0.7 Å for SILCS-PPI. The best model from SILCS-PPI for test case 2 has a very small iRMSD of 0.2 Å compared to an iRMSD of 2.8 Å for the best model given by ClusPro. SILCS-PPI failed in test cases 3 while ClusPro has quite good predictions for this test case. Both methods failed test case 5, with the best model obtained from ClusPro having an iRMSD of 6.4 Å, compared to the slightly better iRMSD of 5.5 Å by SILCS-PPI. ClusPro failed to locate a hit for test case 6 while SILCS-PPI did a much better job and the best model has an iRMSD of 2.8 Å. ClusPro predicted a better model for test case 7 with an iRMSD of 1.8 Å, compared to the best model with an iRMSD of 2.9 Å given by SILCS-PPI. Though both methods failed test case 8, the best model predicted by SILCS-PPI has an iRMSD of 4.3 Å and is much better than ClusPro which predicted the wrong binding site as indicated by the large iRMSD of 14.9 Å. The present results are quite promising for SILCS-PPI since no further energy minimization is performed as is done with ClusPro.
Table 5.
Docking performances of SILCS-PPI compared with ClusPro using the same evaluation criteria.
| Method | Docking criteria | Test Case 1 | Test Case 2 | Test Case 3 | Test Case 4 | Test Case 5 | Test Case 6 | Test Case 7 | Test Case 8 |
|---|---|---|---|---|---|---|---|---|---|
| ClusPro | Nhitsa | 2 | 4 | 2 | 2 | 0 | 0 | 2 | 0 |
| iRMSDbestb | 1.0 | 2.8 | 1.9 | 1.1 | - | - | 1.8 | - | |
| Rankbestc | 8 | 38 | 4 | 3 | - | - | 6 | - | |
| iRMSD1std | 1.0 | 3.1 | 1.9 | 1.1 | - | - | 1.8 | - | |
| Rank1ste | 8 | 23 | 4 | 3 | - | - | 6 | - | |
| iRMSDbest-2kf | - | - | - | - | 6.4 | 9.3 | - | 14.9 | |
| Rankbest-2kg | - | - | - | - | 93 | 70 | - | 61 | |
| SILCS-PPI | Nhitsa | 24 | 50 | 0 | 68 | 0 | 12 | 5 | 0 |
| iRMSDbestb | 0.9 | 0.2 | - | 0.7 | - | 2.8 | 2.9 | - | |
| Rankbestc | 75 | 64 | - | 76 | - | 68 | 21 | - | |
| iRMSD1std | 1.9 | 3.2 | - | 2.8 | - | 3.7 | 2.9 | - | |
| Rank1ste | 10 | 1 | - | 1 | - | 36 | 21 | - | |
| iRMSDbest-2kf | - | - | 5.0 | - | 5.5 | - | - | 4.3 | |
| Rankbest-2kg | - | - | 100 | - | 99 | - | - | 55 | |
Nhits is the number of hits, which defined as solutions whose iRMSD is less than 4 Å, among the top 2000 solutions.
iRMSDbest is the lowest iRMSD found in the top 2000 solutions.
Rankbest is the percentile rank (top %) of the hit with the lowest iRMSD found in the top 2000 solutions.
iRMSD1st is the iRMSD of the first hit found in the top 2000 solutions.
Rank1st is the percentile rank (top %) of the first hit with an iRMSD < 4 Å found in the top 2000 solutions.
iRMSDbest-2k is the lowest iRMSD found in the top 2000 solutions if no hit is found and
Rankbest-2k is the percentile rank (top %) of that solution. iRMSDbest-2k and Rankbest-2k are only reported when no hit is found among the top 2000 solutions.
Conclusion
In the present study, the SILCS-PPI framework for PPI prediction was established. The approach involves the computational demanding precomputation of the SILCS FragMaps, which is followed by efficient docking using the FFT approach. This allows the full range of translation and rotational search space to be sampled in the context of the distance and angle resolutions being applied. In the present study the generated solutions were directly from the FFT search with no additional inter- or intramolecular sampling performed. Using the approach allows for generation of full receptor protein PPI probability distributions. These distributions, in the form of heat maps or COM spheres, show the method to identify solutions in the vicinity of the crystallographic complexes. In addition, the distributions also identify alternate sites participating in PPIs of the receptor with proteins of structural similarity, but varying sequence with the ligand protein used for docking. Such information may be of utility for predicting regions of the protein surface that may contribute to aggregation, information that may be used in the formulation of biologics. We note that similar information is, in principle, available from other PPI docking methods though, to the best of our knowledge, analysis of this potential has not been presented.
Competitive performance by SILCS-PPI was obtained as compared with three other commonly used PPI docking methods. The results are very promising considering that only minimal refinement of the protocol was done to optimize SILCS-PPI performance in the current proof-of-concept study. Multiple improvements in the approach are possible. Examples include local intermolecular sampling of the top solutions using Monte Carlo or simulated annealing, and with the respect to intramolecular conformations, using conformations from the SILCS MD simulation followed by energy minimization in the context of an empirical energy function to construct the final predictions. With respect to scoring optimization of the different weighting factors to balance different energetic contributions to the scoring function, as done by other PPI docking methods,10–20 is anticipated to improve the identified solutions.
The current work presents the initial implementation and application of SILCS FragMaps as a scoring function for PPI prediction, representing the first extension of the approach beyond small ligand related drug design purposes. Notable are the promising results from the SILCS-PPI method as compared to selected commonly used protein docking methods despite the limited optimization of the approach. While this performance is promising it needs to be reiterated that the initial SILCS MD simulations required to generate the FragMaps are computational demanding and these need to be generated for both the receptor and ligand proteins. Accordingly, with respect to computational demands, the method is not competitive with the alternate methods tested in this study. However, once the FragMaps are available, the SILCS-PPI approach is computationally competitive such that studies of, for example, PPI of signaling proteins that interact with multiple partners such as ERK could take advantage of a library of FragMaps that could be readily accessed when new potential PPIs are to be studied.
Supplementary Material
Acknowledgment.
This work was supported by NIH grant R44GM109635 and the University of Maryland Center for Biomolecular Therapeutics. The authors acknowledge computer time and resources from the Computer-Aided Drug Design (CADD) Center at the University of Maryland, Baltimore.
Footnotes
Conflict of Interest. A.D.M. Jr. is co-founder and Chief Scientific Officer of SilcsBio, LLC. S.J. and S.K.L. were employees of SilcsBio LLC when the studies were performed.
Supporting Information. Additional Supporting Information can be found online in the supporting information tab for this article.
References
- 1.Protein-Protein Interactions and Networks, Panchenko A, Przytycka T, Ed. Springer, London, UK, 2008. [Google Scholar]
- 2.Protein-Protein Interactions in Drug Discovery, Dömling A, Ed. Wiley-VCH Verlag GmbH & Co. KGaA, 2013. [Google Scholar]
- 3.Keskin O; Tuncbag N; Gursoy A Chem. Rev, 2016, 116, 4884–4909. [DOI] [PubMed] [Google Scholar]
- 4.Yu W; Guvench O; MacKerell AD, Computational approaches for the design of protein–protein interaction inhibitors In Understanding and exploiting protein–protein interactions as drug targets, Zinzalla G, Ed. Future Science Ltd.: London, UK, 2013; pp 99–102. [Google Scholar]
- 5.Zhong S; Oashi T; Yu W; Shapiro P; MacKerell AD, Prospects of modulating protein–protein interactions In Protein-ligand interactions, Gohlke H, Ed. Wiley KGaA: Weinheim, Germany, 2012; pp 295–329. [Google Scholar]
- 6.Huang S Drug Discov. Today, 2014, 19, 1081. [DOI] [PubMed] [Google Scholar]
- 7.Gromiha MM; Yugandhar K; Jemimah S Curr. Opin. Struct. Biol, 2017, 44, 31. [DOI] [PubMed] [Google Scholar]
- 8.Kaczor AA; Bartuzi D; Stępniewski TM; Matosiuk D; Selent J, Protein–Protein docking in drug design and discovery In Computational drug discovery and design, Springer: New York, 2018; pp 285–305. [DOI] [PubMed] [Google Scholar]
- 9.Katchalski-Katzir E; Shariv I; Eisenstein M; Friesem AA; Aflalo C; Vakser IA Proc. Natl. Acad. Sci. USA, 1992, 89, 2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pierce BG; Hourai Y; Weng Z PLoS One, 2011, 6, e24657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabb HA; Jackson RM; Sternberg MJE J. Mol. Biol, 1997, 272, 106. [DOI] [PubMed] [Google Scholar]
- 12.Kozakov D; Brenke R; Comeau SR; Vajda S Proteins, 2006, 65, 392. [DOI] [PubMed] [Google Scholar]
- 13.Vakser IA Proteins, 1997, 29, 226. [PubMed] [Google Scholar]
- 14.Gray JJ; Moughon S; Wang C, Schueler-Furman O; Kuhlman B; Rohl CA; Baker D J. Mol. Biol 2003, 331, 281–299. [DOI] [PubMed] [Google Scholar]
- 15.Fermandez-Recio J; Totrov M; Abagyan R Proteins, 2003, 52, 113. [DOI] [PubMed] [Google Scholar]
- 16.Wodak SJ; Janin J Proteins 2017, 85, 357. [DOI] [PubMed] [Google Scholar]
- 17.Dominguez C; Boelens R; Bonvin AMJJ J. Am. Chem. Soc, 2003, 125, 1731. [DOI] [PubMed] [Google Scholar]
- 18.Zhang C; Lai L J. Comput. Chem, 2011, 32, 2598. [DOI] [PubMed] [Google Scholar]
- 19.Yan Y; Zhang D; Zhou P; Li B; Huang S Nucleic Acids Res, 2017, 45, W365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Comeau SR; Gatchell DW; Vajda S; Camacho CJ Nucleic Acids Res, 2004, 32, W96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guvench O; MacKerell AD Jr. PLoS Comput. Biol. 2009, 5, (7), e1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lakkaraju SK; Yu W; Raman EP; Hershfeld AV; Fang L; Deshpande DA; MacKerell AD Jr. J. Chem. Inf. Model. 2015, 55, 700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raman EP; Yu W; Guvench O; MacKerell AD J. Chem. Inf. Model. 2011, 51, (4), 877–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raman EP; Yu W; Lakkaraju SK; MacKerell AD J. Chem. Inf. Model. 2013, 53, 3384–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanning ME; Yu W; Yap JL; Chauhan J; Chen L; Whiting E; Pidugu LS; Atkinson T; Bailey H; Li W; Roth BM; Hynicka L; Chesko K; Toth EA; Shapiro P; MacKerell AD; Wilder PT; Fletcher S Eur. J. Med. Chem. 2016, 113, 273–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng H; Linhares BM; Yu W; Cardenas MG; Ai Y; Jiang W; Winkler A; Cohen S; Melnick A; MacKerell AD Jr.; Cierpicki T; Xue F J. Med. Chem. 2018, in production, DOI: 10.1021/acs.jmedchem.8b00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu W; Lakkaraju S; Raman EP; MacKerell AD Jr. J. Comput-Aided. Mol. Des. 2014, 28, (5), 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu W; Lakkaraju S; Raman EP; Fang L; MacKerell AD Jr. J. Chem. Inf. Model. 2015, 55, 407–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Padhorny D; Kazennov A; Zerbe BS; Porter KA; Xia B; Mottarella SE; Kholodov Y; Ritchie DW; Vajda S; Kozakov D Proc. Natl. Acad. Sci. U.S.A. 2016, 113, E4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cardenas MG; Yu W; Beguelin W; Teater MR; Geng H; Goldstein RL; Oswald E; Hatzi K; Yang S; Cohen J; Shaknovich R; Vanommeslaeghe K; Cheng H; Liang D; Cho HJ; Abbott J; Tam W; Du W; Leonard JP; Elemento O; Cerchietti L; Cierpicki T; Xue F; MacKerell AD; Melnick AM J. Clin. Invest. 2016, 126, 3351–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinzi GA; Huang W; Yu W; Giardina BJ; Zhou Y; MacKerell AD; Wilks A; Xue FJ Med. Chem. 2016, 59, 6929–6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lovisolo L; da Silva EA IEE Proc. Vis. Image Sign. 2001, 148, 187–193. [Google Scholar]
- 33.Solernou A; Fernandez-Recio J BMC Bioinformatics 2010, 11, 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frigo M; Johnson SG Proc. IEEE 2005, 93, 216–231. [Google Scholar]
- 35.Buckle AM; Schreiber G; Fersht AR Biochemistry 1994, 33, 8878–8889. [DOI] [PubMed] [Google Scholar]
- 36.Vreven T; Moal IH; Vangone A; Pierce BG; Kastritis PL; Torchala M; Chaleil R; Jimenez-Garcia B; Bates PA; Fernandez-Recio J; Bonvin AMJJ; Weng ZJ Mol. Biol. 2015, 427, 3031–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berman HM; Westbrook J; Feng Z; Gilliland G; Bhat TN; Weissig H; Shindyalov IN; Bourne PE Nucleic Acids Res. 2000, 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sali A; Blundell TL J. Mol. Biol. 1993, 234, 779–815. [DOI] [PubMed] [Google Scholar]
- 39.Hess B; Kutzner C; Van Der Spoel D; Lindahl EJ Chem. Theory Comput. 2008, 4, 435–447. [DOI] [PubMed] [Google Scholar]
- 40.Best RB; Zhu X; Shim J; Lopes PE; Mittal J; Feig M; MacKerell AD Jr J. Chem. Theory Comput. 2012, 8, 3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vanommeslaeghe K; Hatcher E; Acharya C; Kundu S; Zhong S; Shim J; Darian E; Guvench O; Lopes P; Vorobyov IJ Comput. Chem. 2010, 31, 671–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu W; He X; Vanommeslaeghe K; MacKerell AD J. Comput. Chem. 2012, 33, 2451–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Durell SR; Brooks BR; Ben-Naim AJ Phys. Chem. 1994, 98, 2198–2202. [Google Scholar]
- 44.Kozakov D; Beglov D; Bohnuud T; Mottarella SE; Xia B; Hall DR; Vajda S Proteins, 2013, 81, 2159–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaile GL; Burt JE, Directional Statistics. Concepts and Techniques in Modern Geography 25. Norwich: Geo Books, 1980. [Google Scholar]
- 46.Sevcik J; Urbanikova L; Dauter Z; Wilson KS Acta Crystallogr. Sect. D 1998, 54, 954–963. [DOI] [PubMed] [Google Scholar]
- 47.Boriack-Sjodin PA; Margarit SM; Bar-Sagi D; Kuriyan J Nature 1998, 394, 337–343. [DOI] [PubMed] [Google Scholar]
- 48.Desmyter A; Spinelli S; Payan F; Lauwereys M; Wyns L; Muyldermans S; Cambillau C J. Biol. Chem. 2002, 277, 23645–23650. [DOI] [PubMed] [Google Scholar]
- 49.Margarit SM; Sondermann H; Hall BE; Nagar B; Hoelz A; Pirruccello M; Bar-Sagi D; Kuriyan J Cell 2003, 112, 685–695. [DOI] [PubMed] [Google Scholar]
- 50.Chaudhury S; Berrondo M; Weitzner BD; Muthu P; Bergman H; Gray JJ PLoS ONE 2011, 6, e22477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.