Abstract
Low back pain is a major cause of disability worldwide. Although numerous potential biomarkers for the early diagnosis or treatment of intervertebral disc degeneration (IDD) have been identified subsequent to the development of molecular biology technologies, the mechanisms of IDD remain unknown. Published studies found the unbalance of anabolism and catabolism of annulus fibrosus (AF) played an important role in it. The present study was aimed to identify the potential targets and signaling pathways of IDD, through the combined analysis of differential expression and based on the Gene Expression Omnibus (GEO) dataset from NCBI. PPI Networks Analysis indicated that MMP2 and AGE-RAGE signaling pathway and estrogen signaling pathway may play important roles in initiation and development of IDD. This study forecasted the pathogenesis molecular mechanism of IDD and the potential prognostic and diagnostic biomarkers, but we need to make further molecular biological experiments to confirm our assumptions.
1. Introduction
As a major cause of disability worldwide, low back pain (LBP) has increased the social and economic burden significantly [1]. According to the US national health interview survey, 28% of all people had experienced LBP that lasted one day or more during the past three months [2]. Almost half of them suffered from LBP over one year, with a quarter reporting “frequent” pain. In the USA, LBP has become the second frequent reason for visits to the physician and the fifth-ranking cause of admission to hospital and the third most common cause of surgical [3–5]. The total costs of LBP have exceed $100 billion per year in the United States alone according to WHO, which take the number one spot in all healthcare problems [6].
Several published studies have confirmed that LBP was commonly relevant to the intervertebral disc degeneration (IDD) [7, 8]. Although the etiology (ageing, living conditions, biomechanical loading, and genetic factors) [9] and biological process (apoptosis, inflammation) of IDD are difficult to define precisely, there will be similar phenomenon, including the unbalance of anabolism and catabolism of annulus fibrosus (AF) [10], which leads to decreased ability of resisting tension [11] and thus accelerating the process of IDD finally.
Removal of the degenerated or herniated tissue or even the partial or complete replacement of the disc with an artificial substitute is the current popular surgical therapy for IDD [7, 12]. However, this kind of surgery can bring about adjacent disc degeneration or the failure of intervertebral fusion [13, 14]. Therapeutic intervention (recovery the biomechanical and structural properties and restoration the biological behaviors of healthy AF cells) [10] could avoid highly invasive procedures at an early stage of IDD. Now, the multifactorial mechanisms of IDD have achieved substantial advancement, but its initiation and progression are still limited. So it is our key point to find signaling pathways to better understand the cascades of disc degeneration.
Our present study aimed to identify the potential targets and signaling pathways of IDD, through the combined analysis of differential expression and based on the Gene Expression Omnibus (GEO) dataset from NCBI. This may be helpful for the precise treatment of LBP, as well as early diagnosis.
2. Materials and Methods
2.1. Microarray Date
The gene expression (GSE34095) in the present study was deposited in GEO (http://www.ncbi.nlm.nih.gov/geo/), which was accumulated by Kazezian et al. [7]. GSE34095 datasets were based on Affymetrix GeneChip Human Genome U133A Array. A total of 6 samples were included, containing three nondegenerative disc annulus fibrosus (AF) cells samples and three degenerative disc AF cells samples. Pathway Analysis software system (IPA®, QIAGEN Redwood City) was used for the sequencing process.
2.2. Differential Expression Analysis
The GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/) is used to identify differentially expressed genes (DEGs) between degenerative disc AF cells and normal samples. GEO2R is based on R that comes with the GEO databases. Genes with fold change (logFC) >0.5 (upregulated) or <-0.5 (downregulated) and false discovery rate (FDR) adjusted for P<0.1 were considered to be differentially expressed. The Heml software was used to generate the heat maps.
2.3. Functional Enrichment Analysis
The differential expression genes were submitted to the Database for Annotation, Visualization, and Integrated Discovery (DAVID, Version6.8, https://david.ncifcrf.gov/) [15] for the analysis of enrichment of gene ontology (GO) terms [16], Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways [17], and Reactome pathways [18]. P<0.05 was considered to indicate a statistically significant difference for the screening of significant GO terms and KEGG pathways.
2.4. Protein-Protein Interaction (PPI) Networks Analysis
The differential expression genes were submitted to the Search Tool for the Retrieval of Interacting Genes/Proteins (String, Version 10.0, http://www.string-db.org/) for the analysis of differential protein. PPI score =0.4 was considered that the interacted protein nodes were all transcribed by differentially expressed genes. PPI networks were represented by Cytoscape Software (Version 3.2.0) [19–21].
2.5. Subnetworks Analysis
The biological process may interact with multiple genes to play a regulatory role. Generally, those genes perform the same or similar biological function in the subnetworks. The method of MCODE [22] was used to analyze the interacted gene of the significant clustering modules.
3. Results
3.1. Differentially Expressed Gene in Degenerative Intervertebral Discs Patients
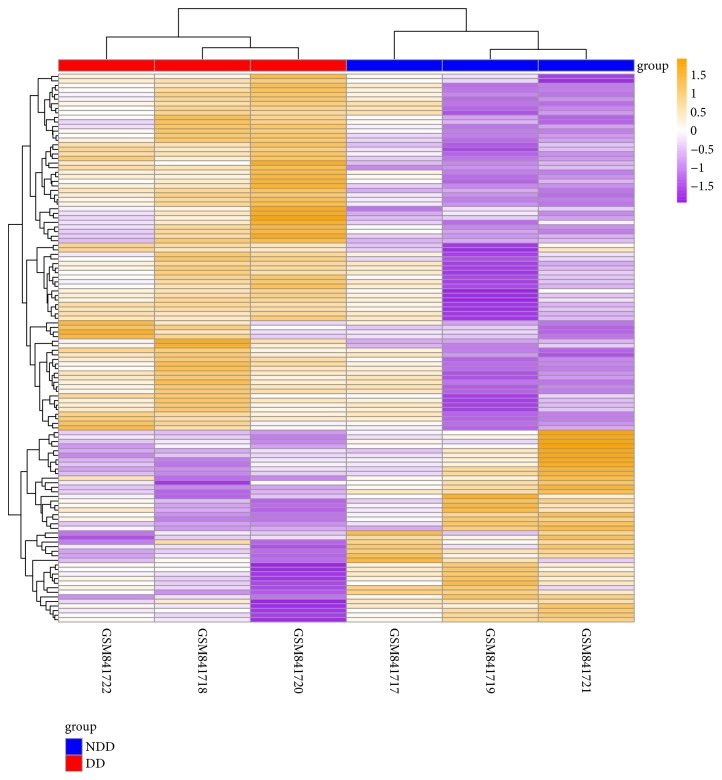
Raw read counts for a total of 22,215 genes were obtained for gene expression analysis. The date normalization of gene expression had no significant difference in 6 samples (Figure 1). Based on the criteria of |logFC| >0.5 and adj. P<0.1, numerous genes were revealed to be differential expression in degenerative samples compared with normal samples (Figure 2). Among them, 42 genes were downregulated and 78 genes were upregulated in degenerative disc AF cells compared to nondegenerative disc AF cells (Figure 2, Table 1).
Figure 1.
Box plot of GSE34095. Blue: nondegenerative samples; red: degenerative samples.
Figure 2.
Heat map of differentially expressed in GSE34095 datasets. Yellow: upregulated; purple: downregulated; red: degenerative samples; Blue: nondegenerative samples.
Table 1.
Top 10 differentially expressed genes in datasets GSE34095 according to degree.
| Node | HSP90AA1 | MMP2 | XPO1 | HSPD1 | COL3A1 | FN1 | POSTN | EIF2S2 | HSPA2 | TARS |
|---|---|---|---|---|---|---|---|---|---|---|
| Description | Up | Up | Up | Up | Up | Up | Up | Up | Down | Up |
| Degree | 15 | 10 | 9 | 7 | 7 | 6 | 6 | 5 | 5 | 5 |
3.2. Functional and Pathway Enrichment Analysis
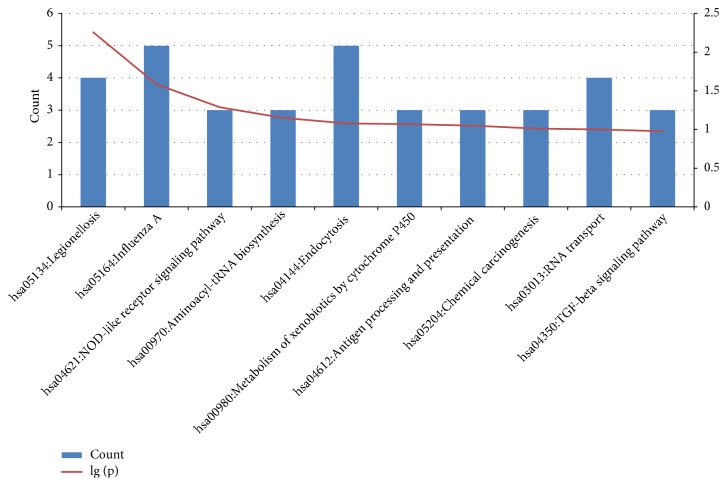
To learn more about the function of identified intersection DEGs, functional and pathway enrichment analysis was carried out using DAVID [15]. These DEGs were mainly enriched in pathway associated with endocytosis, influenza A, legionellosis, RNA transport, NOD-like receptor signaling pathway, aminoacyl-tRNA biosynthesis, metabolism of xenobiotics by cytochrome P450, antigen processing and presentation, chemical carcinogenesis, TGF-beta signaling pathway, and AGE-RAGE signaling pathway (Table 2, Figure 3).
Table 2.
Functional and pathway enrichment analysis of target genes in degenerative disc AF cells.
| Category | Term | Description | Count | P value |
|---|---|---|---|---|
| KEGG_PATHWAY | hsa04144 | Endocytosis | 5 | 0.0223 |
| KEGG_PATHWAY | hsa05164 | Influenza A | 5 | 0.0752 |
| KEGG_PATHWAY | hsa05134 | Legionellosis | 4 | 0.0045 |
| KEGG_PATHWAY | hsa03013 | RNA transport | 4 | 0.0908 |
| KEGG_PATHWAY | hsa04621 | NOD-like receptor signaling pathway | 3 | 0.0456 |
| KEGG_PATHWAY | hsa00970 | Aminoacyl-tRNA biosynthesis | 3 | 0.0632 |
| KEGG_PATHWAY | hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 3 | 0.0771 |
| KEGG_PATHWAY | hsa04612 | Antigen processing and presentation | 3 | 0.0807 |
| KEGG_PATHWAY | hsa05204 | Chemical carcinogenesis | 3 | 0.0882 |
| KEGG_PATHWAY | hsa04350 | TGF-beta signaling pathway | 3 | 0.0958 |
| KEGG_PATHWAY | hsa04933 | AGE-RAGE signaling pathway | 2 | 0.0801 |
| REACTOME_PATHWAY | R-HAS-2022870 | R-HAS-2022870 | 3 | 0.0051 |
| REACTOME_PATHWAY | R-HAS-379716 | R-HAS-379716 | 3 | 0.0073 |
| REACTOME_PATHWAY | R-HAS-2129379 | R-HAS-2129379 | 3 | 0.0177 |
| REACTOME_PATHWAY | R-HAS-1442490 | R-HAS-1442490 | 3 | 0.0466 |
| REACTOME_PATHWAY | R-HAS-300178 | R-HAS-300178 | 3 | 0.0618 |
| GOTERM_CC_DIRECT | GO:0005737 | Cytoplasm | 45 | 0.0001 |
| GOTERM_CC_DIRECT | GO:0070062 | Extracellular exosome | 37 | 5.6552E-08 |
| GOTERM_CC_DIRECT | GO:0005634 | Nucleus | 37 | 0.0457 |
| GOTERM_MF_DIRECT | GO:0005515 | Protein binding | 63 | 0.0005 |
| GOTERM_MF_DIRECT | GO:0005524 | ATP binding | 14 | 0.0493 |
| GOTERM_MF_DIRECT | GO:0044822 | Poly(A) RNA binding | 13 | 0.0152 |
| GOTERM_MF_DIRECT | GO:009864 | Cadherin binding involved in cell-cell adhesion | 8 | 0.0009 |
| GOTERM_MF_DIRECT | GO:0042803 | Protein homodimerization activity | 8 | 0.0904 |
| GOTERM_MF_DIRECT | GO:0005201 | Extracellular matrix structural constituent | 4 | 0.0053 |
| GOTERM_MF_DIRECT | GO:0051059 | NF-kappaB binding | 3 | 0.0109 |
| GOTERM_BP_DIRECT | GO:0007165 | Signal transduction | 12 | 0.0457 |
| GOTERM_BP_DIRECT | GO:0045944 | Positive regulation of transcription from RNA polymerase II promoter | 10 | 0.0795 |
| GOTERM_BP_DIRECT | GO:0098609 | Cell-cell adhesion | 9 | 0.0001 |
Note: BP, biological process; MF, molecular function; CC, cellular component; GO, gene ontology.
Figure 3.
Significantly enriched KEGG pathways of DEGs in degenerative disc AF cells.
3.3. PPI Networks Analysis
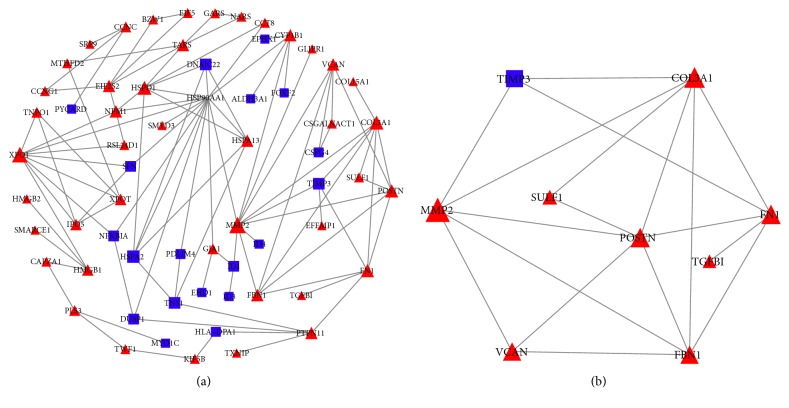
Cytoscape software was used to determine biological relationships among the 120 differentially expressed genes (Figure 4(a)). The top three module networks (Figure 4(b), Table 3), based on Fisher's exact test, were associated with immunological disease, cell signaling, and protein generation pathways. According to the value of degree (degree⩾5), we find that the key nodes of the PPI networks included HSP90AA1, COL3A1, MMP2, POSTN, and FN1.
Figure 4.
(a) PPI Networks Analysis with Cytoscape for the 120 genes that are differentially expressed between nondegenerative disc AF cells and degenerative disc AF cells samples; (b) module networks of Module A. The red color and triangle in network indicate a gene that is upregulated in degenerative disc AF cells compared to the nondegenerative disc AF cells; blue color indicates the genes that are downregulated in degenerative disc AF cells compared to the nondegenerative disc AF cells samples.
Table 3.
KEGG pathway enrichment analysis of target genes in the top three module networks.
| Term | Description | Count | P value | |
|---|---|---|---|---|
| Module A | hsa05134 | Legionellosis | 2 | 0.0156 |
| hsa04612 | Antigen processing and presentation | 2 | 0.0219 | |
| hsa04915 | Estrogen signaling pathway | 2 | 0.0285 | |
| hsa04141 | Protein processing in endoplasmic reticulum | 2 | 0.0483 | |
|
| ||||
| Module B | hsa05134 | Legionellosis | 2 | 0.0156 |
| hsa04612 | Antigen processing and presentation | 2 | 0.0219 | |
| hsa04915 | Estrogen signaling pathway | 2 | 0.0285 | |
| hsa04141 | Protein processing in endoplasmic reticulum | 2 | 0.0483 | |
|
| ||||
| Module C | hsa05205 | Proteoglycans in cancer | 3 | 0.0025 |
| hsa04512 | ECM-receptor interaction | 2 | 0.0373 | |
| hsa05146 | Amoebiasis | 2 | 0.0453 | |
| hsa04510 | Focal adhesion | 2 | 0.0868 | |
Functional enrichment analysis based on the DAVID identified a number of significantly enriched GO terms and KEGG pathways in module networks DEGs. The significant KEGG pathways of the top three module networks DEGs of degenerative disc AF cells samples are listed in Table 3. As shown in Table 3, KEGG pathways associated with protein processing in endoplasmic reticulum and estrogen signaling pathway were significantly enriched (P<0.05). The top five most significant GOTERM_BP of the module networks DEGs are listed in Table 4. Similar to the KEGG pathways, those GOTERM_BP were mainly involved in the protein processing (assembly, refolding, and stabilization) and degradation (extracellular matrix disassembly).
Table 4.
Top five most significantly enriched GOTERM_BP enrichment analyses of target genes in the top three module networks.
| Term | Description | Count | P value | |
|---|---|---|---|---|
| Module A | GO:0042026 | protein refolding | 3 | 2.23E-6 |
| GO:0009409 | response to cold | 3 | 1.34E-5 | |
| GO:0006986 | response to unfolded protein | 3 | 1.83E-5 | |
| GO:0009408 | response to heat | 3 | 2.40E-5 | |
| GO:0051131 | chaperone-mediated protein complex assembly | 2 | 0.0023 | |
|
| ||||
| Module B | GO:0042026 | protein refolding | 3 | 4.46E-6 |
| GO:0009409 | response to cold | 3 | 2.67E-5 | |
| GO:0006986 | response to unfolded protein | 3 | 3.65E-5 | |
| GO:0009408 | response to heat | 3 | 4.78E-5 | |
| GO:0050821 | protein stabilization | 3 | 3.87E-4 | |
|
| ||||
| Module C | GO:0030198 | extracellular matrix organization | 4 | 3.05E-5 |
| GO:0022617 | extracellular matrix disassembly | 3 | 3.00E-4 | |
| GO:0001501 | skeletal system development | 3 | 9.70E-4 | |
| GO:0048050 | post-embryonic eye morphogenesis | 2 | 0.0014 | |
| GO:0048048 | embryonic eye morphogenesis | 2 | 0.0036 | |
4. Discussion
The tear or partial injury of the intervertebral disc AF is one of the important factors leading to low back pain [23]. With the emerging of IDD studies, numerous potential biomarkers for the early diagnosis or treatment of IDD have been identified subsequent to the development of molecular biology technologies. However, the mechanisms of IDD remain unknown.
In this study, we analyzed the microarray data of degenerative disc AF cells from GEO database under the accession number GSE34095 by GEO2R to obtain DEGs and obtained their enriched GO terms and KEGG pathways. This study aimed to provide important clues for exploring the key genes and associated regulatory network in mechanisms of IDD resulted from AF. Based on the DEGs functional enrichment analysis, potential mechanisms and target gene for disc degeneration caused by AF were suggested as below.
The abnormal expression of MMP2 might cause disc degeneration by accelerating the matrix degradation. According to previous results, a total of 120 DEGs including 42 downregulated and 78 upregulated genes were identified. We selected 10 genes according to the value of degree. Among them, HSP90AA1, MMP2, XPO1, HSPD1, COL3A1, FN1, POSTN, EIF2S2, and TARS were significantly upregulated, and HSPA2 was significantly downregulated in degenerative disc AF cells samples in this study. Recent epidemiologic studies indicated that the key factor for disc degeneration was heredity [24]. The typical character of disc degeneration was the matrix degradation in the early. As an important member of matrix metalloproteinases (MMPs) family, MMP2 plays a critical role in the excessive breakdown of the extracellular matrix (ECM) during disc degeneration [25–27]. The increased expression and activity of MMP2 was responded for degenerative lesions in disc tissue. We reasonably surmise that MMP2 aberrantly expressed plays important roles in initiation and development of IDD.
The activation of AGE-RAGE signaling pathway might cause disc degeneration by accelerating the expression of MMP2. Multiple complexity factors, including age, injury, inflammation, and immunity, activate AGE-RAGE signaling pathway. Based on functional enrichment analysis, two genes (MMP2, FN1) were upregulated in AGE-RAGE signaling pathway. Notably, the two upregulated genes are downstream targets of AGE-RAGE signaling pathway [28]. What is more, AGE-RAGE signaling pathway was found over-expression in degenerative disc AF cells compared with nondegenerative disc AF cells. Taken together, it suggested that multiple complexity factors might lead to IDD by activating AGE-RAGE signaling pathway and then accelerating the expression of downstream targets-MMP2.
Estrogen signaling pathway plays an important role in the process of disc degeneration. Estrogen signaling pathway is ubiquitous in different tissues throughout the body, which participates in many pathological process, such as osteoporosis and osteoarthritis [29]. Recent evidence suggests that 17-beta-estradiol (E2) can promote the proliferation of AF cells by activating estrogen beta receptor [30]. Bai et al. [31] found that estrogen could delay the development of ovariectomized rabbit's IDD by reducing the expression of interleukin and MMPs, which could inhibit the degradation of matrix. Kato et al. [32] confirmed that E2 could stimulate the expression of COL2A1. However, estrogen secretion will decrease gradually with the increasing of age. It will lead to the activation of estrogen signaling pathway, the expression of inflammatory factors, and the degradation of matrix, which can accelerate the process of IDD.
5. Conclusion
Though we identified aberrantly expressed key gene (MMP2) from the GEO database and found the AGE-RAGE signaling pathway and estrogen signaling pathway in degenerative disc AF cells for IDD, which may benefit us in understanding the molecular mechanism of the pathogenesis of IDD and detecting potential prognostic and diagnostic biomarkers, it is still needed that we perform further molecular biological experiments to confirm our assumptions.
Acknowledgments
This research was funded by National Natural Science Foundation of China (Grant no. 30772816).
Contributor Information
Feng Minshan, Email: fengminshan@hotmail.com.
Zhu Liguo, Email: Zhlg95@aliyun.com.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Supplementary Materials
The highlights are used to describe the findings of this study.
References
- 1.Andersson G. B. J. Epidemiological features of chronic low-back pain. The Lancet. 1999;354(9178):581–585. doi: 10.1016/S0140-6736(99)01312-4. [DOI] [PubMed] [Google Scholar]
- 2.Summary Health Statistics for US Adults: National Health Interview Survey. Vol. 10. US Department of Health and Human Services; 2014. (Vital and Health Statistics Series 10. no. 260). http://www.cdc.gov/nchs/data/series/sr_10/sr10_260.pdf/ [PubMed] [Google Scholar]
- 3.Finch P. Technology insight: imaging of low back pain. Nature Clinical Practice Rheumatology. 2006;2(10):554–561. doi: 10.1038/ncprheum0293. [DOI] [PubMed] [Google Scholar]
- 4.Taylor V. M., Deyo R. A., Cherkin D. C., et al. Low back pain hospitalization: Recent united states trends and regional variations. The Spine Journal. 1994;19(11):1207–1212. doi: 10.1097/00007632-199405310-00002. [DOI] [PubMed] [Google Scholar]
- 5.Hart L. G., Deyo R. A., Cherkin D. C. Physician office visits for low back pain: frequency, clinical evaluation, and treatment patterns from a u.s. national survey. The Spine Journal. 1995;20(1):11–19. doi: 10.1097/00007632-199501000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Katz J. N. Lumbar disc disorders and low-back pain: socioeconomic factors and consequences. The Journal of Bone and Joint Surgery—American Volume. 2006;88(2):21–24. doi: 10.2106/jbjs.e.01273. [DOI] [PubMed] [Google Scholar]
- 7.Kazezian Z., Gawri R., Haglund L., et al. Gene expression profiling identifies interferon signalling molecules and IGFBP3 in human degenerative annulus fibrosus. Scientific Reports. 2015;5 doi: 10.1038/srep15662.15662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boos N., Semmer N., Elfering A., et al. Natural history of individuals with asymptomatic disc abnormalities in magnetic resonance imaging: predictors of low back pain-related medical consultation and work incapacity. The Spine Journal. 2000;25(12):1484–1492. doi: 10.1097/00007632-200006150-00006. [DOI] [PubMed] [Google Scholar]
- 9.Wang H., Zou F., Jiang J., et al. The correlation between ossification of the nuchal ligament and pathological changes of the cervical spine in patients with cervical spondylosis. The Spine Journal. 2014;39(26):B7–B11. doi: 10.1097/BRS.0000000000000430. [DOI] [PubMed] [Google Scholar]
- 10.Chu G., Shi C., Wang H., Zhang W., Yang H., Li B. Strategies for annulus fibrosus regeneration: from biological therapies to tissue engineering. Frontiers in Bioengineering and Biotechnology. 2018;6(90):1–13. doi: 10.3389/fbioe.2018.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teng Z., Feng Y. An animal experimental study on the repair of intervertebral disc annulus fi brosus with oxidized regenerated cellulose and TGF-β3. Medical Journal of Air Force. 2018;43(3):165–169. [Google Scholar]
- 12.Gibson J. N., Waddell G. Surgical interventions for lumbar disc prolapse: updated cochrane review. The Spine Journal. 2007;32(16):1735–1747. doi: 10.1097/brs.0b013e3180bc2431. [DOI] [PubMed] [Google Scholar]
- 13.Frei H., Oxland T. R., Rathonyi G. C., Nolte L.-P. The effect of nucleotomy on lumbar spine mechanics in compression and shear loading. The Spine Journal. 2001;26(19):2080–2089. doi: 10.1097/00007632-200110010-00007. [DOI] [PubMed] [Google Scholar]
- 14.Eck J. C., Humphreys S. C., Hodges S. D. Adjacent-segment degeneration after lumbar fusion: a review of clinical, biomechanical, and radiologic studies. American Journal of Orthopedics (Belle Mead, N.J.) 1999;28(6):336–340. [PubMed] [Google Scholar]
- 15.Huang D. W., Sherman B. T., Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 16.Ashburner M., Ball C. A., Blake J. A., et al. Gene ontology: tool for the unification of biology. Nature Genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabregat A., Jupe S., Matthews L., et al. The reactome pathway knowledgebase. Nucleic Acids Research. 2018;46(1):D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software Environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li G., Zhang T., Zhang G., et al. Analysis of gene co-expression networks and function modules at different developmental stages of chicken breast muscle. Biochemical and Biophysical Research Communications. 2019;508(1):177–183. doi: 10.1016/j.bbrc.2018.11.044. [DOI] [PubMed] [Google Scholar]
- 21.Xiao Z. X., Chen R. Q., Hu D. X., Xie X. Q., Yu S. B., Chen X. Q. Identification of repaglinide as a therapeutic drug for glioblastoma multiforme. Biochemical and Biophysical Research Communications. 2017;488(1):33–39. doi: 10.1016/j.bbrc.2017.04.157. [DOI] [PubMed] [Google Scholar]
- 22.Bader G. D., Hogue C. W. V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4(1):1–27. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu W., Dianwen S., Lianshun J. Pathophysiological mechanism of low back pain. Orthopeic Journal of China. 2009;17(5):364–366. [Google Scholar]
- 24.Sambrook P. N., MacGregor A. J., Spector T. D. Genetic influences on cervical and lumbar disc degeneration: a magnetic resonance imaging study in twins. Arthritis & Rheumatology. 1999;42(2):366–372. doi: 10.1002/1529-0131(199902)42:2<366::AID-ANR20>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y., Gu Z., Qiu G. Association of the polymorphism of MMP2 with the risk and severity of lumbar disc degeneration in the Chinese Han population. European Review for Medical and Pharmacological Sciences. 2013;17(13):1830–1834. [PubMed] [Google Scholar]
- 26.Goupille P., Jayson M. I. V., Valat J.-P., Freemont A. J. Matrix metalloproteinases: the clue to intervertebral disc degeneration? The Spine Journal. 1998;23(14):1612–1626. doi: 10.1097/00007632-199807150-00021. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi M., Haro H., Wakabayashi Y., Kawa-uchi T., Komori H., Shinomiya K. The association of degeneration of the intervertebral disc with 5a/6a polymorphism in the promoter of the human matrix metalloproteinase-3 gene. The Journal of Bone & Joint Surgery. 2001;83(4):491–495. doi: 10.1302/0301-620X.83B4.11617. [DOI] [PubMed] [Google Scholar]
- 28.Ko S., Ko H., Shieh T., et al. Cell migration is regulated by AGE-RAGE interaction in human oral cancer cells in vitro. Plos One. 2014;9(10) doi: 10.1371/journal.pone.0110542.e110542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xin-meng J., Lei W., Jie L. Research progress of the relationship between estrogen receptor and disc degeneration. Orthopedic Journal of China. 2015;23(19):1783–1785. [Google Scholar]
- 30.Gruber H. E., Yamaguchi D., Ingram J., et al. Expression and localization of estrogen receptor-β in annulus cells of the human intervertebral disc and the mitogenic effect of 17-β-estradiol in vitro. BMC Musculoskeletal Disorders. 2002;3(4):1–5. doi: 10.1186/1471-2474-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.XiaoLiang B., Liang C., ChunMao C. Whether estrogen affects the expression of interleukin-20 and matrix metalloproteinase-3 in lumbar nucleus gelatinosus of ovariectomized rabbits. Journal of Clinical Rehabilitative Tissue Engineering Research. 2010;14(37):6955–6958. [Google Scholar]
- 32.Kato M., Takaishi H., Yoda M., et al. GRIP1 enhances estrogen receptor α-dependent extracellular matrix gene expression in chondrogenic cells. Osteoarthritis and Cartilage. 2010;18(7):934–941. doi: 10.1016/j.joca.2010.03.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The highlights are used to describe the findings of this study.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.