Abstract
Background & Aims:
The intestinal epithelium is maintained by long-lived intestinal stem cells (ISCs) that reside near the crypt base. Above the ISC zone, there are short-lived progenitors that normally give rise to lineage-specific differentiated cell types but can dedifferentiate into ISCs in certain circumstances. However, the role of epithelial dedifferentiation in cancer development has not been fully elucidated.
Methods:
We performed studies with Bhlha15-CreERT, Lgr5-DTR-GFP, Apcflox/flox, LSL-Notch (IC), and R26-reporter strains of mice. Some mice were given diphtheria toxin to ablate Lgr5 mRNA-positive cells, irradiated, or given 5-fluorouracil, hydroxyurea, doxorubicin, or dextran sodium sulfate to induce intestinal or colonic tissue injury. In intestinal tissues we analyzed the fate of progeny that expressed Bhlha15 mRNA. We used microarrays and reverse-transcription PCR to analyze gene expression patterns in healthy and injured intestinal tissues and in tumors. We analyzed gene expression patterns in human colorectal tumors using the TCGA dataset.
Results:
Bhlha15 identified Paneth cells and short-lived secretory precursors (including pre-Paneth label-retaining cells) located just above the ISC zone in the intestinal epithelium. Bhlha15+ cells had no plasticity after loss of Lgr5-positive cells or irradiation. However, Bhlha15+ secretory precursors started to supply the enterocyte lineage after doxorubicin-induced epithelial injury in a Notch-dependent manner. Sustained activation of Notch converts Bhlha15+ secretory precursors to long-lived enterocyte progenitors (EPs). Administration of doxorubicin and expression of an activated form of Notch resulted in a gene expression pattern associated with EPs, whereas only sustained activation of Notch altered gene expression patterns in Bhlha15+ precursors, towards that of ISCs. Bhlha15+ EPs with sustained activation of Notch formed intestinal tumors with serrated features in mice with disruption of Apc. In the colon, Bhlha15 marked secretory precursors that became stem-like, cancer-initiating cells following dextran sodium sulfate-induced injury, via activation of Src and YAP signaling. In analyses of human colorectal tumors, we associated activation of Notch with chromosome instability-type tumors with serrated features in the left colon.
Conclusion:
In mice, we found that short-lived precursors can undergo permanent reprogramming by activation of Notch and YAP signaling. These cells could mediate tumor formation, in addition to traditional ISCs.
Keywords: tumorigenesis, interconversion, colon cancer, Yes associated protein 1
Introduction
The intestinal epithelium is highly proliferative and rapidly turns over every 4–5 days1. Epithelial turnover is maintained by long-lived self-renewing intestinal stem cells (ISCs) that reside near the crypt base. ISCs divide continuously to give rise to their multi-lineage progeny throughout the crypt-villus unit. In the hierarchical model of intestinal epithelial cells, ISCs first produce the lineage-committed short-lived progenitors within the transit-amplifying (TA) cell zone, that after several cell divisions become committed to either the secretory or enterocyte lineage1. The secretory lineage includes goblet cells, enteroendocrine cells, and Paneth cells. Epithelial tuft cells are also presumed to be part of the secretory lineage, however, there remains some debate as to whether they are derived directly from either secretory or enterocyte progenitors, or generated from an independent precursor2.
Notch, Wnt, and Rspo signaling are critical for determining ISC cell fate and homeostasis3, 4. Wnt activation in ISCs enhances differentiation into the secretory cell lineage, while Notch activation in ISCs directs enterocyte differentiation with loss of the secretory lineage3, 5. Multiple markers of ISCs and progenitors have been described to date. Lgr5 is expressed in actively cycling crypt base columnar (CBC) cells distributed between Paneth cells6. In addition, there appears to be a distinct ISC pool near the “+4” position, just above Lgr5+ CBC cells, which comprises more quiescent ISCs. Interestingly, recent transcriptome analyses revealed that Bmi1-expressing cells also comprise a subset of enteroendocrine cells, which have been shown to interconvert to actively cycling ISCs in order to maintain epithelial homeostasis following acute injury7–12. Similarly, several short-lived progenitors and pre-Paneth label-retaining cells (LRCs) that express low levels of Lgr5, can also participate in the interconversion process through dedifferentiation, particularly when Lgr5+ ISCs are damaged13–15. It has been thought that such progenitors phenotypically and genetically dedifferentiate to Lgr5+ ISCs, and then contribute to regeneration11, 12.
ISCs are considered to be the primary cellular origin of intestinal tumors due to their longevity and self-renewing capacity. This is supported by mouse models, where oncogenic mutations in ISCs result in efficient dysplasia formation, whereas similar mutations in more differentiated cells fail to produce dysplasia16. To date, there is limited evidence that even short-lived progenitors can be a source of intestinal cancers, despite their potential to de-differentiate. Indeed, differentiated post-mitotic cells with genetic mutations can give rise to cancers in rare circumstances, such as in the presence of inflammation or aberrant NF-κB activation17, 18. We have recently reported that gastric stem cells expressing Bhlha15 (also known as Mist1), a basic helix-loop-helix transcriptional factor, can give rise to cancers19–22. Here, we report the presence of Bhlha15 expression in both the small intestinal and colonic epithelium, and investigate the identity and function of these cells.
Methods
Mice
Bhlha15-CreERT and Eef1a1-LSL-Notch1(IC) mice19 were described previously. LSL-KrasG12D and LSL-Trp53R172H mice were provided by Dr. Kenneth Olive (Columbia University). Apcflox and Lgr5-DTR-GFP mice were obtained from the National Cancer Institute and Genentech, respectively. R26 reporter mice were purchased from the Jackson Laboratory. Cre recombinase was activated by oral administration of TAM (2mg/0.2mL corn oil). Mouse and in vitro culture experiments were repeated at least 2 times, with at least 3 biological replicates, and representative results are shown. All animal studies and procedures were approved by the ethics committees at Columbia University and the University of Tokyo.
Treatment
To induce epithelial injury, hydroxyurea, 5-fluorouracil, and doxorubicin (Sigma) were administered intraperitoneally at a dose of 1g/kg, 150mg/kg, and 15mg/kg, respectively. To induce colonic injury, 2% dextran sodium sulfate (DSS) was given for 5 days. For Lgr5+ cell ablation, DT was administered via intraperitoneal injection at a dose of 50ug/kg. Whole body irradiation at a dose of 10 or 12 Gy was given to some mice. Src inhibitor (Saracatinib, Caymanchemicals) and γ-secretase inhibitor dibenzazepine (DBZ, Tocris BioScience) were administered at indicated time points orally at a dose of 50mg/kg and intraperitonially at a dose of 5 mg/kg, respectively. Bone marrow transplantation and EdU label retention assay was performed as described previously23, 24.
More detailed information is described in Supplementary Methods.
Results
Bhlha15 is expressed in short-lived secretory precursors in the small intestine
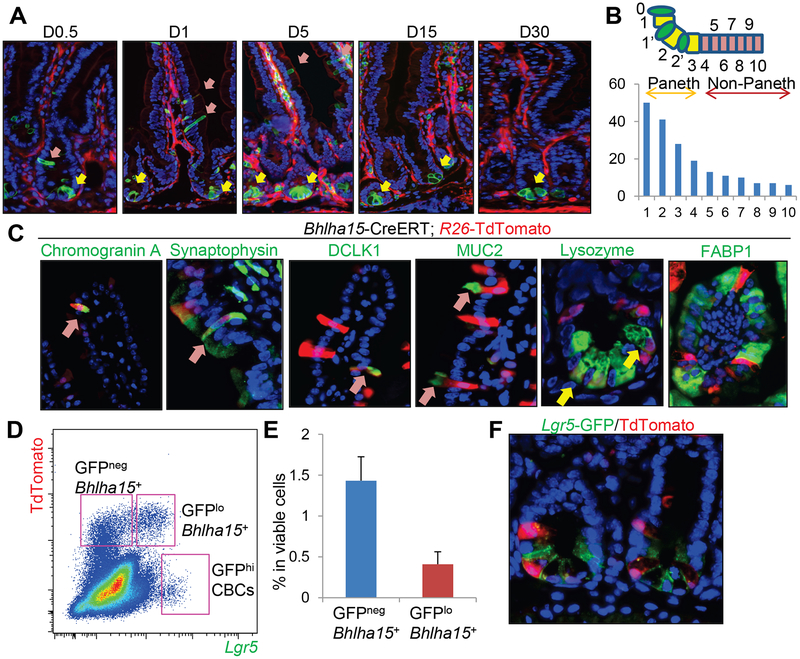
We performed lineage tracing experiments using Bhlha15-CreERT; R26-mTmG mice. We observed recombined GFP+ Paneth cells (at the crypt base below the +4 position) immediately following tamoxifen induction (Fig. 1A). We also detected another GFP+ cell population situated between positions +5 and +10 that was morphologically distinct from Paneth cells (Fig. 1B). We confirmed using an antibody that recognizes estrogen receptor that CreERT protein is expressed at the same locations, but was not expressed in the villus compartment (Fig. S1A). Analogous to this expression pattern, in situ hybridization and immunohistochemistry revealed that Bhlha15 mRNA and protein are strongly expressed in Paneth cells, and is moderately expressed in the TA cell zone (Fig. S1A). From day 1 to 5 after tamoxifen induction, there was an increase in the number of recombined non-Paneth cells that rapidly migrate upwards in the villus-crypt units, but these cells disappeared in around 10 days (Fig. 1A&S1B). Recombined Paneth cells persisted longer, but eventually turned over and disappeared after 90 days. This short-term supply for scattered cells appears to be similar to that reported in Dll1-CreERT mice13, which labels short-lived secretory progenitors. Indeed, cells in the Bhlha15 lineage include chromogranin A or synaptophysin-positive enteroendocrine cells, DCLK1+ tuft cells, MUC2+ goblet cells, and Lysozyme+ Paneth cells, but not FABP1+ enterocytes (Fig. 1C). More specifically, TdTomato expression is found predominantly in Paneth cells at the earliest time point, while a subset of TA and a few goblet cells are also labeled in the TA cell zone and lower villus (Fig. S1B). At 4 days after tamoxifen, the TdTomato+ population contains a greater proportion of goblet cells and newly includes tuft and enteroendocrine cells. However, most of these labeled cells, except for Paneth cells, rapidly disappear by day 7, suggesting that Bhlha15 marks secretory precursors that can supply goblet, enteroendocrine, and tuft cell lineages only for the short-term, as well as relatively long-lived Paneth cells.
Figure 1. Bhlha15 is expressed in the short-lived secretory precursors.
(A)Lineage tracing in Bhlha15-CreERT;R26-mTmG small intestine at indicated time points after tamoxifen. Yellow allows; Paneth cells, pink arrows; non-Paneth cells. (B)Cell position of Bhlha15+ cells in jejunum. Total 60 crypts from 3 mice were quantified. (C)Immunostaining in Bhlha15-CreERT;R26-TdTomato mice 5 days after tamoxifen. Arrows indicate double-positive cells. (D-F)FACS plot (D), percentage of indicated populations in viable cells (E, n=3), and image (F) of Bhlha15-CreERT;Lgr5-DTR-EGFP;R26-TdTomato mouse intestine 12 hours after tamoxifen. Mean±S.E.M.
Next, we compared the expression of the ISC marker Lgr5 to Bhlha15+ cells in Bhlha15-CreERT; Lgr5-DTR-EGFP; R26-TdTomato mice9. Based on FACS and histological analysis, we found no strong Lgr5-GFP expression in intestinal Bhlha15+ cells; only a subset of Bhlha15+ cells expressed low levels of Lgr5-GFP, which suggests that Bhlha15+ cells may contain the previously described Lgr5-expressing, pre-Paneth LRCs15, 24 (Fig. 1D–F&S1C). Indeed, when using the EdU label retention protocol, we found that 73% of LRCs express Bhlha15 (Fig. S1D–E). However, since we did not observe sustained, confluent lineage tracing, these Lgr5-expressing Bhlha15+ cells are likely distinct from Lgr5+ ISCs.
Transcriptome analysis of Bhlha15-expressing populations in the small intestine.
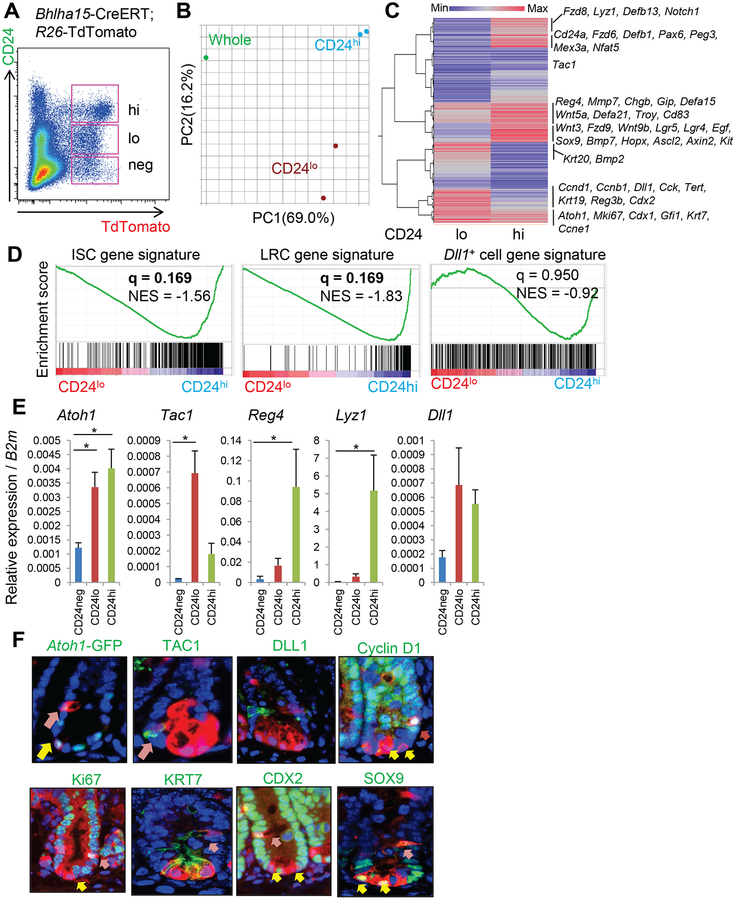
Bhlha15-expressing TdTomato+ cells isolated from induced Bhlha15-CreERT; R26-TdTomato mice can be divided into three distinct populations: CD24neg, CD24lo, and CD24hi (Fig. 2A). Bhlha15+Lgr5lo pre-Paneth precursors belong to the CD24hi population, together with Bhlha15+Lgr5neg mature Paneth cells (Fig. S1F–G). We isolated CD24lo and CD24hi populations by FACS, and performed gene expression analysis. Principal components analysis (PCA) of the microarray data showed that Bhlha15+CD24lo and Bhlha15+CD24hi cells are molecularly distinct populations (Fig. 2B). Hierarchical clustering indicated that Bhlha15+CD24hi cells express high levels of Paneth cell-specific genes, including defensin family genes, Lyz1, and Mmp7, suggesting Paneth cell enrichment (Fig. 2C). Bhlha15+CD24hi cells also highly express Wnt-target genes such as Sox9 or Axin2, and even stem cell marker genes including Lgr5, Troy, Hopx, and Ascl2. GSEA analysis also suggested that ISC-related genes are enriched in CD24hi population compared to CD24lo population (Fig. 2D). In addition, genes associated with LRCs, such as Mex3a, Cd83, and Nfat515, 24, are enriched in Bhlha15+CD24hi cells, confirming that Bhlha15+CD24hi cells contain pre-Paneth LRCs (Fig. 2C–D). In contrast, proliferation marker genes including the cyclin family and Mki67 as well as markers of TA progenitors such as Dll1, Tert, Krt19, and Alpi, are more prominently expressed in Bhlha15+CD24lo cells, suggesting that Bhlha15+CD24lo cells may represent a secretory precursor population.
Figure 2. Transcriptome analysis of Bhlha15-expressing populations.
(A)FACS analysis of Bhlha15-CreERT;R26-TdTomato mouse intestine 2 days after tamoxifen with CD24 immunostaining. Pink boxes indicate CD24neg, CD24lo, and CD24hi population among TdTomato+ cells. (B)PCA analysis of gene expression in TdTomato+CD24lo, TdTomato+CD24hi cells, and whole intestine. (C)Hierarchical clustering of average gene expression in TdTomato+CD24lo and TdTomato+CD24hi cells compared to whole intestine. (D)GSEA analysis for the comparison between TdTomato+CD24lo and TdTomato+CD24hi cells. (E)RT-PCR of sorted TdTomato+CD24neg, TdTomato+CD24lo, and TdTomato+CD24hi population (n=4). (F)Staining (green) in Bhlha15-CreERT;R26-TdTomato mice 1 day after tamoxifen. Yellow arrows; Paneth cells, pink arrows; non-Paneth secretory cells. Mean±S.E.M. *p<0.05.
RT-PCR analysis of these cells validated the microarray results (Fig. 2E). Although Reg4, a secretory progenitor marker25, appears to be predominantly expressed in Bhlha15+CD24hi cells, Bhlha15+CD24lo cells showed prominent expression of Tac1, consistent with the labeling of secretory precursors as reported5, 25. Analysis of Atoh1-GFP mice and TAC1 immunofluorescence revealed that Atoh1 is expressed in both populations, while Tac1 is specifically expressed in secretory progenitors (Fig. 2E–F). Dll1 expression was not significantly different between CD24lo and CD24hi populations, and we found using immunohistochemistry and in situ hybridization that the vast majority of Bhlha15+ cells did not express DLL1 (Fig. 2E–F&S2A–C). In fact, GSEA analysis showed that genes enriched in reported Dll1+ progenitors13 are not necessarily upregulated in Bhlha15+CD24lo cells (Fig. 2D), suggesting that while Dll1+ and Bhlha15+ progenitors exhibit some similarities, they are highly likely to be distinct populations. On the other hand, cyclin D1, CDX2, KRT7, and Ki67 are expressed mostly in Bhlha15+ secretory precursors rather than mature Paneth cells at the crypt base, while SOX9 is mainly expressed in Paneth cells but not in secretory precursors (Fig. 2F&S2D). Proliferation rates in TdTomato+ cells in the TA cell zone rapidly decreased by day 4, suggesting that the majority of initially labeled Bhlha15+ secretory precursors quickly generate progeny and turn over by this time point (Fig. S2E). It should be noted that approximately 30% of Ki67+TdTomato+ cells express MUC2, likely representing immature pre-goblet cells (Fig. S2F). Together, Bhlha15-expressing TA cells mark a short-lived secretory precursor population, which might include a subset of lineage-committed pre-goblet cells.
Bhlha15+ precursors are chemo-radioresistant, but do not convert to long-lived ISCs following injury
Dll1+ progenitors can revert to stem-like cells following irradiation-induced injury13. To test whether this is also true of Bhlha15+ precursors, we irradiated the Bhlha15-CreERT; R26-TdTomato mice and examined the subsequent cell fate mapping at various time intervals (Fig. 3A). When using a sensitive R26-TdTomato mouse line, we found quite rare stem cell-type lineage tracing, with labeling of the entire villus-crypt unit from the Bhlha15-lineage, but >99% of glands show secretory specific labeling as seen in crossed to the R26-mTmG line. Following 10 Gy irradiation, recombination occurs as the same pattern as in the unirradiated state without any apoptosis in Bhlha15+ cells, but there was no significant increase in stem cell-type lineage tracing events, suggesting that irradiation does not rigorously induce interconversion of Bhlha15+ lineage as was reported for Dll1+ cells (Fig. 3B&E, S3A). Given that ablation of Lgr5+ stem cells can alter the fate and induce lineage tracing from some facultative progenitors, we then treated Bhlha15-CreERT; Lgr5-DTR-EGFP; R26-TdTomato mice with diphtheria toxin (DT) in order to ablate Lgr5+ cells. Following Lgr5+ cell ablation, we observed no remarkable change in the Bhlha15+ cell recombination pattern above TA cell zone, while there was a dramatic decrease in labeled Paneth cell numbers. (Fig. 3B&E, S3B). Thus, while Bhlha15+ precursors can survive and supply secretory progeny following irradiation and Lgr5-DT induced injury, they do not interconvert into stem-like cells, indicating that Bhlha15+ cells are functionally distinct from other reserve stem cell populations, such as those expressing Dll1, Alpi, or Bmi1.
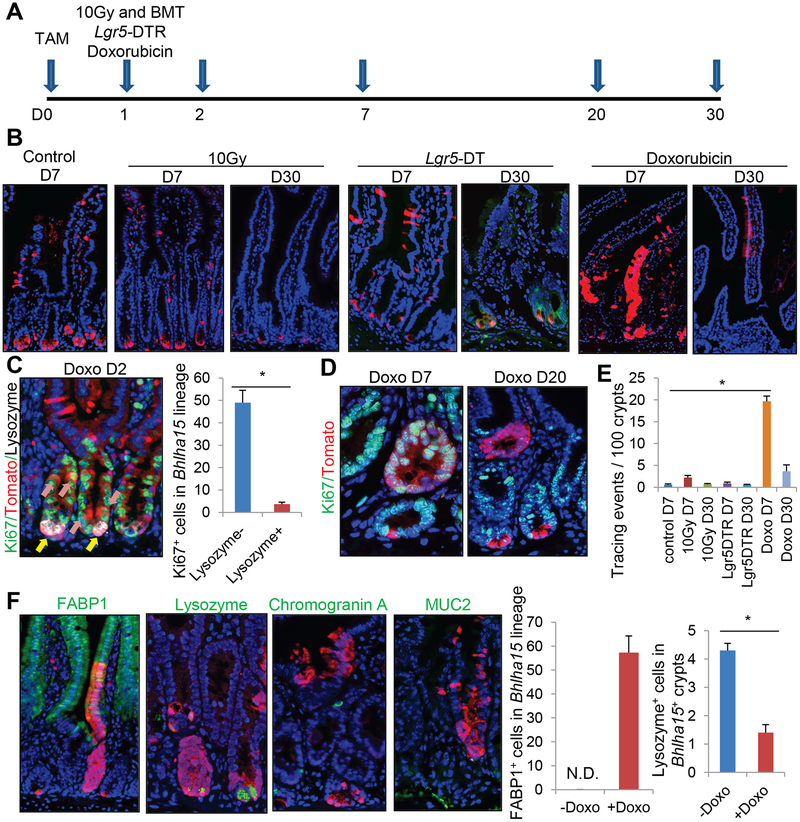
Figure 3. Bhlha15+ precursors show short-term lineage tracing following doxorubicin-induced injury, but do not convert to long-lived ISCs.
(A)Protocol. (B)Lineage tracing images of Bhlha15-CreERT;R26-TdTomato mouse intestine after 10-Gy whole body irradiation, Lgr5-DT ablation, or doxorubicin. WT bone marrow was transplanted after irradiation. (C)Ki67 (green), RFP (red), and Lysozyme (gray) staining on doxorubicin-treated Bhlha15-CreERT;R26-TdTomato mouse at day 2. Numbers of Ki67+ cells in 100 Tomato+Lysozyme+ or Lysozyme− cells are quantified. n=3/group. (D)Ki67 (green) and RFP (red) staining on doxorubicin-treated Bhlha15-CreERT;R26-TdTomato mice. (E)Lineage tracing events per 100 glands in Bhlha15-CreERT mouse intestine. n=3/group. (F)Immunostaining (green) of TdTomato-expressing clones in Bhlha15-CreERT;R26-TdTomato mice 7 days after doxorubicin. Numbers of FABP1+ and Lysozyme+ cells in 100 Tomato+ cells are quantified. n=3/group. Mean±S.E.M. *p<0.05.
It was also reported that cytotoxic agents can through cellular injury induce the dedifferentiation of pre-Paneth LRCs into stem-like cells15. While we observed no evidence of stem cell conversion following 5-fluorouracil or hydroxyurea treatment (not shown), lineage expansion and stem-like tracing from Bhlha15+ cells were observed following doxorubicin treatment (Fig. 3B). Given that proliferation was observed in Lysozyme− progenitors but not in Lysozyme+ mature Paneth cells after doxorubicin treatment (Fig. 3C–D), these clones are likely derived from the more undifferentiated precursor population rather than mature Paneth cells.
Nevertheless, after complete epithelial regeneration at 20–30 days post injury, lineage tracing events from Bhlha15+ cells gradually disappeared and only a few Paneth cells remained traced by the TdTomato reporter (Fig. 3B–E). Thus, Bhlha15+ progenitors can transiently contribute to crypt regeneration following doxorubicin-induced injury, but quickly lose this capacity and cease expanding, perhaps due to contributions from other stem or progenitor cells. Moreover, during transient lineage tracing from doxorubicin-treated Bhlha15+ cells, Bhlha15-derived clones contain FABP1+ enterocytes which are not normally derived from Bhlha15+ secretory precursors (Fig. 3F). On the other hand, the numbers of secretory cells such as enteroendocrine, goblet cells, and Paneth cells are decreased in the doxorubicin-treated Bhlha15+ lineage, suggesting that Bhlha15+ secretory precursors interconvert to short-lived enterocyte progenitors (EPs) and cease to supply secretory cells after doxorubicin-induced injury.
Bhlha15+ precursors show long-term enterocyte-specific lineage tracing with sustained Notch activation
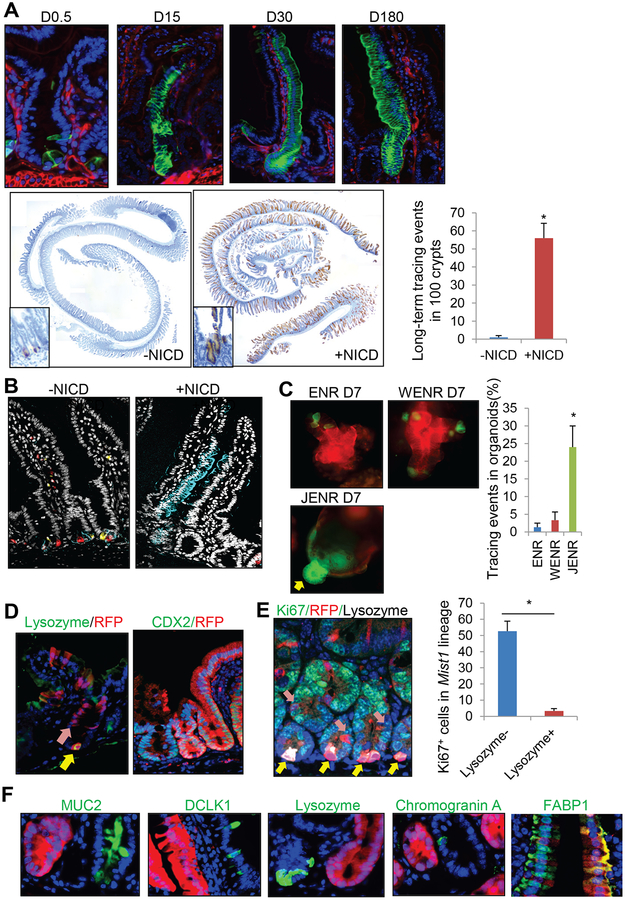
Notch signaling regulates intestinal epithelial cell differentiation by controlling stem cell fate. Indeed, a Notch downstream molecule HES1 is upregulated in Bhlha15+ precursor cells rather than in Paneth cells after doxorubicin treatment, whilst HES1 is not expressed in these cells in normal, irradiated, or Lgr5-ablated intestines (Fig. S3C). Cells in Bhlha15-derived clones are strongly positive for HES1 one week after doxorubicin treatment (Fig. S3D), suggesting that the Notch pathway in Bhlha15+ precursors is specifically activated by doxorubicin and contributes to regeneration. Therefore, we examined the effect of Notch activation in Bhlha15+ secretory precursors by generating Bhlha15-CreERT; LSL-Notch(IC) mice that enabled us to conditionally activate the Notch pathway in Bhlha15+ cells. After Notch activation, recombined Bhlha15+ cells initially showed strong HES1 expression in both precursor and Paneth populations, and later expanded and produced completely traced crypt-villus units that were long-lived (greater than 180 days), in contrast to normal Bhlha15 progeny that were restricted only to Paneth cells at later time points (Fig. 4A&S3C–E). Multicolor labeling of the Bhlha15 lineage using R26-Confetti mice revealed multicolor Paneth cells in the normal state, whereas single-color, monoclonal expansion of Bhlha15 lineage was seen with NICD expression (Fig. 4B). In organoid culture, the addition of Wnt3A to standard culture media containing EGF, Noggin and R-spondin1 (WENR) failed to induce expansion of the recombined Bhlha15 lineage, although we did observe GFP+ Paneth cells at sites of budding crypts (Fig. 4C&S3F–G). In contrast, addition of the Notch ligand Jagged-1 to culture (JENR) or induction of Notch(IC) expression induced lineage tracing from Bhlha15+ cells.
Figure 4. Bhlha15+ precursors show long-term enterocyte-specific lineage tracing with sustained Notch activation.
(A)Lineage tracing images of Bhlha15-CreERT;LSL-Notch1(IC);R26-mTmG (top) and RFP staining and quantification of Bhlha15-CreERT;R26-TdTomato and Bhlha15-CreERT;LSL-Notch1(IC);R26-TdTomato mouse intestine. Tracing events per 100 glands were quantified at 180 days after tamoxifen (n=3). (B)Bhlha15-CreERT;R26-Confetti and Bhlha15-CreERT;LSL-Notch1(IC);R26-Confetti mouse intestines 14 days after tamoxifen. (C)Organoid culture of Bhlha15-CreERT;R26-mTmG mouse intestine. Tamoxifen was given at day 0, and glands were taken after 12 hours. Crypts were cultured with indicated medium for 7 days, and tracing events ratio in total organoids were quantified (n=3). (D)Lysozyme (day5) and CDX2 staining (day30) on Bhlha15-CreERT;LSL-Notch1(IC);R26-TdTomato mice. (E)Ki67 (green), RFP (red), and Lysozyme (gray) staining 4 days after tamoxifen. Numbers of Ki67+ cells in 100 Tomato+Lysozyme+ or Lysozyme− cells are quantified. n=3/group. (F)Immunostaining (green) in Bhlha15-CreERT;LSL-Notch1(IC);R26-TdTomato mice. Mean±S.E.M. *p<0.05.
At 5 days after tamoxifen induction, we observed in Bhlha15-CreERT; R26-TdTomato; LSL-Notch(IC) murine crypts the beginning of red ribbon tracing from the +5 to +10 region, whereas Lysozyme+TdTomato+ Paneth cells did not show such clonal expansion at the crypt base (Fig. 4D). Proliferation was observed in Lysozyme− progenitors but not in Lysozyme+ mature Paneth cells (Fig. 4E). Moreover, traced TdTomato+ glands expressed CDX2, which is normally not expressed in the mature Paneth cell population (Fig. 4D). Thus, aberrant Notch activation in secretory precursors, rather than in mature Paneth cells, converts cells into stem-like cells. Nevertheless, we observed a complete loss of secretory cells including goblet, Paneth, enteroendocrine, and tuft cells in Notch-activated Bhlha15 lineage, and such villus-crypt units are completely replaced by the FABP1+ enterocyte lineage (Fig. 4F&3E). Thus, in the setting of Notch activation, Bhlha15+ secretory precursors interconvert into unipotent enterocyte progenitors, similar to what is observed following treatment with doxorubicin, although interconversion appears to be long-lived when sustained Notch activation is present. To confirm that Notch pathway is critical for doxorubicin-induced interconversion, we treated mice with DBZ, a Notch inhibitor. DBZ treatment significantly decreased Bhlha15-derived tracing events following doxorubicin, supporting the importance of Notch pathway in this process (Fig. S3H). In contrast to what is seen with Bhlha15-CreERT mice, NICD expression did not alter the frequency of lineage tracing in Lgr5-CreERT and Dclk1-CreERT mice (Fig. S4A–C), suggesting that Notch signaling does not convert all intestinal epithelial cells into more active progenitors, and that the tracing ability of Lgr5+ cells is not enhanced by additional forced Notch activation, which is not surprising given the constitutive Notch-active status in Lgr5+ cells.
Distinct gene profiles in activated Bhlha15+ precursors.
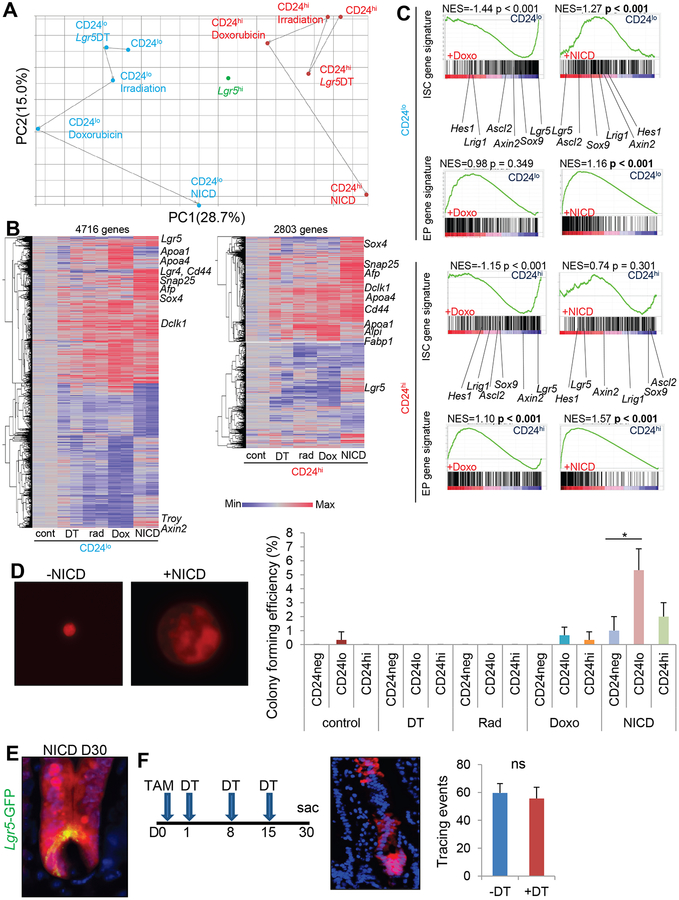
We performed transcriptome analysis of injury-treated and Notch-activated Bhlha15+ cell populations, as well as Lgr5hi ISCs. PCA analysis revealed that Lgr5-DT ablation induces minimal changes in both CD24lo and CD24hi populations, while irradiation and doxorubucin treatment induce more prominent changes (Fig. 5A). Following NICD expression, dramatic shifts of gene expression can be found in both CD24lo and CD24hi cells, while they nevertheless retain their clear and distinct gene signatures. NICD-expressing Bhlha15+ cells and Lgr5hi ISCs still display clearly distinct gene signatures, although both cell lineages appear to show longevity and tracing ability in mice. Hierarchical clustering supports these findings, demonstrating that irradiation and doxorubicin treatments induce similar gene alterations while changes in doxorubicin treatments are broader, and NICD expression produces a quite different pattern of transcriptional activation (Fig. S5A–B).
Figure 5. Distinct gene profiles in activated Bhlha15+ cells.
(A)PCA analysis of average gene expression (n=2/group) in TdTomato+CD24lo, TdTomato+CD24hi, and Lgr5hi cells after treatment. (B)Hierarchical clustering of gene expression in the individual TdTomato+CD24lo and TdTomato+CD24hi cells compared to the untreated cells. (C)GSEA analysis for the indicated comparison between untreated TdTomato+CD24lo or CD24hi cells and cells after doxorubicin or NICD expression. (D)Representative single cell culture images and colony forming efficiency of each population. Cells were sorted and collected from 3 mice/group, and 1000 singlet cells per well were cultured (3 wells/group). Colony formation rate at day 10 is shown. (E)Bhlha15-CreERT;LSL-Notch1(IC);Lgr5-DTR-EGFP;R26-TdTomato intestines 30 days after tamoxifen. (F)Bhlha15-CreERT;LSL-Notch1(IC);Lgr5-DTR-EGFP;R26-TdTomato intestines 30 days after tamoxifen and DT treatment. Tracing events per 100 glands were quantified. n=3/group. Mean±S.E.M. *p<0.05.
When compared with untreated CD24lo and CD24hi populations, the NICD-expressing CD24lo population exhibits an upregulation of stem cell-related markers such as Lgr5, Cd44, Troy, or Axin2, while the NICD+CD24hi population shows only mild upregulation of Lgr5 (Fig. 5B). Such upregulation of stem cell markers is not obvious in cells treated with Lgr5-DT, irradiation, or doxorubicin. GSEA curves suggest that irradiation and doxorubicin treatments are in fact negatively associated with ISC gene signatures in both CD24lo and CD24hi cells, while NICD expression shows a significant positive association with ISC markers only in CD24lo cells (Fig. 5C&S4C). Colony formation ability was predominantly limited to Bhlha15+CD24lo secretory precursors in which NICD expression was induced, compared to Bhlha15+CD24hi cells or cells sorted from injury models (Fig. 5D). Thus, short-term expansion induced by doxorubicin may not represent true stem cell interconversion either phenotypically or genetically, while Notch activation might induce permanent reprogramming in CD24lo secretory progenitors but not in CD24hi Paneth and pre-Paneth LRCs. Nevertheless, it should be noted that the “shift” towards ISC gene signature in Bhlha15+CD24hi cells likely represents the acquisition of only a few ISC properties, given the clearly distinct gene expression pattern from Lgr5hi ISCs, and this is likely due to the limited differentiation ability of Bhlha15+NICD+ clones, in contrast to the multipotency in ISCs.
At one month after tamoxifen treatment, we detected Lgr5-expressing CBCs that were derived from Notch-activated Bhlha15+Tomato+ clones (Fig. 5E&S5F), suggesting that Bhlha15+NICD+ cells are able to convert to Lgr5+ cells, consistent with our transcriptome analysis. Nevertheless, multiple DT treatments, resulting in complete ablation of Lgr5+ cells and even Paneth cell lineage10, did not diminish the lineage tracing ability of Bhlha15+NICD+ cells (Fig. 5F), suggesting that Lgr5+ cell populations may be dispensable for the initiation and maintenance of Bhlha15+ clones even following Notch activation.
Genes enriched in enterocyte progenitors (EPs), such as Sox426, Afp and Snap2511, are upregulated in both NICD-expressing CD24lo and CD24hi cells (Fig. 5B), consistent with the phenotypic conversion from secretory precursors to EPs by sustained Notch activation. Moreover, the enterocyte markers Alpi, Fabp1, Apoa1, or Apoa426 were upregulated in doxorubicin-treated CD24lo and CD24hi cells. Upon GSEA analysis, doxorubicin-treated CD24hi cells, NICD+CD24lo cells, and NICD+CD24hi cells all showed significant gene enrichment with EP gene signatures, although doxorubicin-treated CD24lo cells and cells from irradiated or Lgr5-ablated mice did not (Fig. 5C&S5C). Treatment with doxorubicin during EdU pulse administration increased label retention efficiency in Bhlha15+ cells (Fig. S1D–E), suggesting CD24hi pre-Paneth LRCs may contribute to doxorubicin-induced enterocyte clones in a greater extent than CD24lo precursors.
In fact, we observe gradual decrease and complete disappearance of Lgr5 expression by day 7 in the doxorubicin-treated Lgr5-CreERT-IRES-EGFP mice (Fig. S5D). Nevertheless, there are equivalent numbers of lineage tracing from Lgr5+ cells after doxorubicin treatment compared to untreated mice, likely produced by surviving Lgr5+ CBCs and interconverted Lgr5+ LRCs. Lgr5 ablation indeed increased the lineage tracing events from Bhlha15+ cells when combined with doxorubicin treatment (Fig. S5E). Thus, while Bhlha15+ secretory precursors normally do not show tracing events following Lgr5 ablation, once they give rise to EPs following doxorubicin, these cells readily respond to Lgr5 ablation to supply cryptic cells.
NICD+Bhlha15+ precursors contribute to regeneration and cancer.
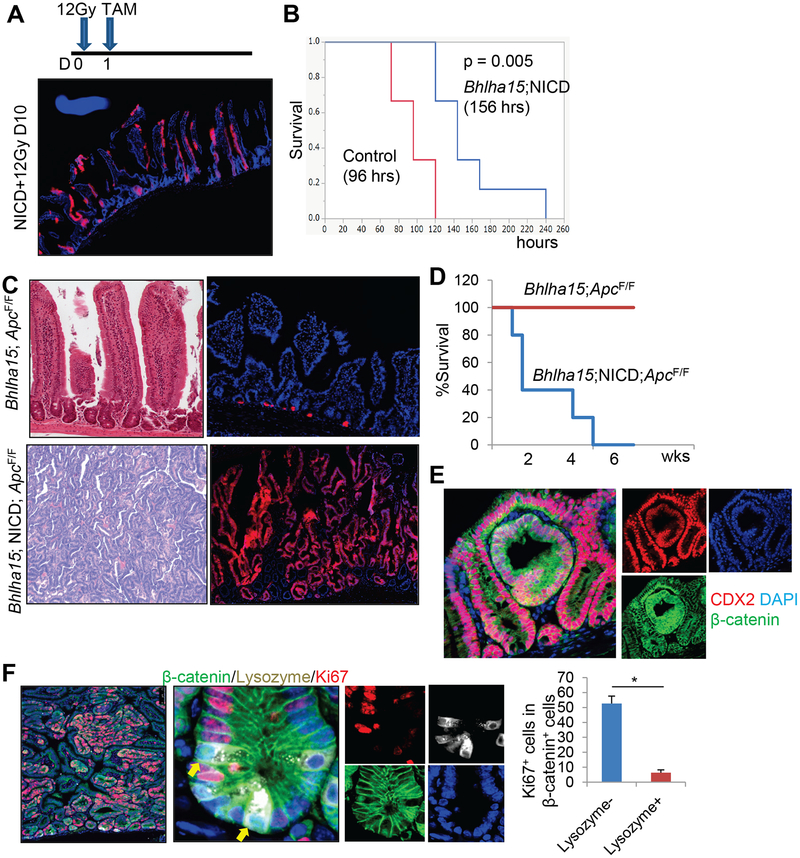
Given that Bhlha15+ precursors are radioresistant, we investigated whether Notch activation in Bhlha15+ progenitors may contribute to epithelial regeneration following lethal 12Gy whole body irradiation (Fig. 6A). Bhlha15-CreERT; LSL-Notch(IC); R26-TdTomato mice demonstrated abundant, fully lineage-traced intestinal glands, indicating that Notch-activated Bhlha15+ cells contribute to the maintenance of intestinal crypt-villus units (Fig. 6A). Accordingly, Bhlha15-CreERT; LSL-Notch(IC) mice display significantly longer survival than control mice (Fig. 6B).
Figure 6. NICD+Bhlha15+ progenitors contribute to regeneration and cancer.
(A)Bhlha15-CreERT;LSL-Notch1(IC);R26-TdTomato mice were irradiated at the dose of 12Gy and given tamoxifen at day 1. (B)Survival rate of 12Gy-irradiated Bhlha15-CreERT;R26-TdTomato (n=6) and Bhlha15-CreERT;LSL-Notch1(IC); R26-TdTomato mice (n=5). (C)H&E and TdTomato expression of Bhlha15-CreERT;Apcflox/flox;R26-TdTomato and Bhlha15-CreERT;LSL-Notch1(IC);Apcflox/flox;R26-TdTomato mouse intestine at day 20. (D)Survival curve of Bhlha15-CreERT;Apcflox/flox (red) and Bhlha15-CreERT;LSL-Notch1(IC);Apcflox/flox (blue) mice. (E-F)CDX2 (red) and β-catenin (green) staining (E) and Ki67 (red), Lysozyme (gray), and β-catenin (green) staining (F) of Bhlha15-CreERT;LSL-Notch1(IC);Apcflox/flox mice at day 10. Numbers of Ki67+ cells in 100 β-catenin+Lysozyme+ or β-catenin+Lysozyme− cells are quantified (n=3). Mean±S.E.M. *p<0.05.
Next, we analyzed the tumorigenic potential in Bhlha15+ cells in the intestine. We generated Bhlha15-CreERT; Apcflox/flox mice for conditional deletion of Apc in Bhlha15+ cells. Previous studies have shown that Apc loss in fast dividing Lgr5+ and Krt19+ stem cells leads to intestinal tumors and acute lethality7, 16. However, Apc loss in Bhlha15+ cells alone did not result in the spontaneous development of intestinal tumors nor acute lethality (Fig. 6C–D). Even when mice were treated with irradiation or doxorubicin, these mice did not develop tumors nor dysplasia (Fig. S6A). Thus, secretory precursors and differentiated secretory cells do not act as an origin of intestinal tumors in these settings. However, when Bhlha15-CreERT; Apcflox/flox mice were crossed to LSL-Notch(IC) mice, they rapidly developed intestinal tumors resulting in death within 6 weeks (Fig. 6C–D&S6B). These tumors show strong HES1 expression with serrated histological features, both of which are not evident in intestinal tumors in Lgr5-CreERT; Apcflox/flox mice (Fig. S6B–C). Simultaneous induction of mutant KrasG12D or Trp53R172H in the NICD+Bhlha15 lineage led to serrated-type hyperplasia (Fig. S6D). In contrast, simultaneous expression of mutant KrasG12D or Trp53R172H without Notch activation did not induce tumor formation even with loss of Apc. Thus, accumulation of multiple classical oncogenic mutations is not sufficient for the conversion from secretory progenitors into cancer-initiating cells, while activation of the Notch pathway is uniquely able to induce serrated-type malignant transformation through conversion to enterocyte lineage.
The tumors observed in Bhlha15-CreERT; LSL-Notch(IC); Apcflox/flox mice showed strong Ki67 positivity and nuclear translocation of β-catenin (Fig. 6E–F). Although nuclear β-catenin staining could be found both in Lysozyme+ mature Paneth cells and dysplastic cells in Bhlha15-CreERT; LSL-Notch(IC); Apcflox/flox mice, Ki67+ proliferation was restricted in dysplastic cells and was not evident in mature Paneth cells (Fig. 6F). In addition, the presence of abundant CDX2 expression and the absence of Lysozyme expression points to undifferentiated secretory precursors rather than mature Paneth cells, as the probable initial source of these tumors.
Bhlha15 marks colonic secretory precursors that can convert to stem-like, tumor-initiating cells following injury.
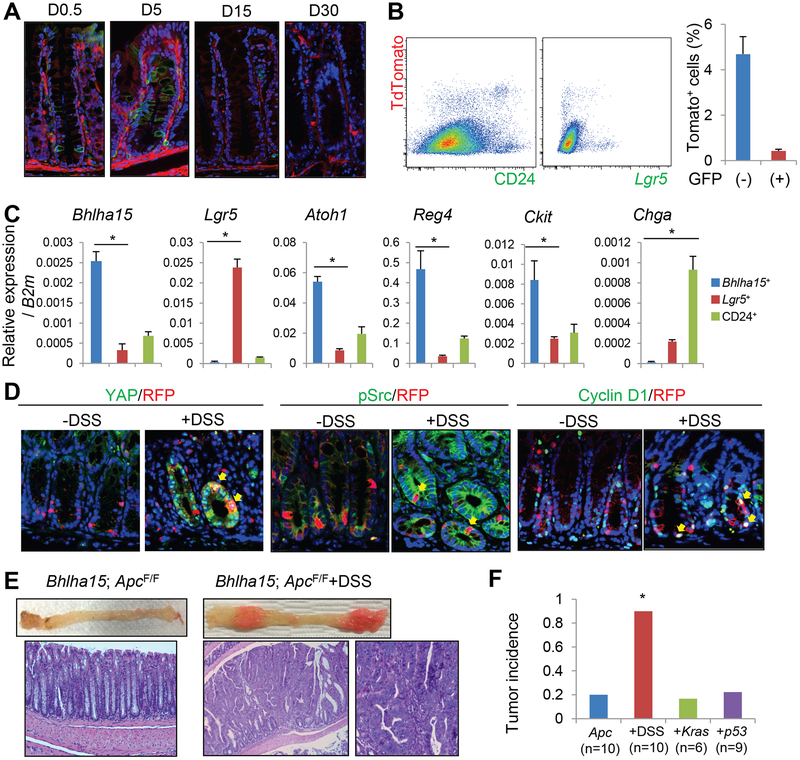
Next, we investigated the characters of Bhlha15+ cells in the colon. Tamoxifen induction of Bhlha15-CreERT; R26-mTmG mice revealed that Bhlha15+ cells were initially found at the +1 to +10 position with the peak at +4 or +5 near the colonic gland base (Fig. 7A&S7A). In situ hybridization and immunohistochemistry confirmed the endogenous expression pattern for this marker in the colon (Fig. S7B–C). Upon FACS analysis, the colonic Bhlha15+ cells overlapped with CD24+ cells (Fig. 7B). RT-PCR results from sorted Bhlha15+CD24+ cells revealed that these cells expressed high levels of Bhlha15 as well as Atoh1 (Fig. 7C). Furthermore, Bhlha15+CD24+ cells expressed high levels of Ckit and Reg4, which have been recently identified as colonic deep crypt secretory cell markers27, 28. However, Lgr5 was not highly expressed in Bhlha15+CD24+ cells, and ChgA expression was highest in Bhlha15−CD24+ cells that appeared to contain differentiated enteroendocrine cells.
Figure 7. Bhlha15 marks colonic secretary precursors that can lineage trace following injury.
(A)Lineage tracing of Bhlha15-CreERT;R26-mTmG mouse colon. (B)FACS analysis of Bhlha15-CreERT;Lgr5-DTR-EGFP;R26-TdTomato mouse colon 1 day after tamoxifen, gated by CD24 (left) or GFP (right). Percentages of GFP+Tomato+ and GFP−Tomato+ cells in viable cells are shown (n=3). (C)RT-PCR analysis of sorted TdTomato+CD24+, TdTomato−CD24+, and Lgr5-GFP+ cells (n=3). (D)YAP, pSrc, and cyclin D1 staining (green) of Bhlha15-CreERT;R26-TdTomato mouse colon with or without DSS. Tamoxifen was given 1 day after DSS treatment, then mice were analyzed on next day. (E)Macroscopic images and H&E staining of Bhlha15-CreERT;Apcflox/flox mouse colon with or without DSS treatment. (F)Colon tumor incidence in Bhlha15-CreERT;Apcflox/flox mice with DSS treatment, LSL-KrasG12D, or LSL-Trp53R172H. Mean±S.E.M. *p<0.05.
Around 5 days after tamoxifen induction, there was an increase in the number of Bhlha15-derived GFP+ cells in the colonic glands, indicating that Bhlha15+ colonic cells rapidly generate progeny (Fig. 7A). These progenies contained MUC2+ goblet cells and DCLK1+ tuft cells, but not carbonic anhydrase 2+ absorptive cells, and almost all Bhlha15+-derived cells disappeared after 15 days, suggesting that Bhlha15+ colonic cells include short-lived secretory precursors (Fig. S7E).
We observed that the Bhlha15+ cells and Lgr5+ cells were located in close proximity but were nevertheless mostly distinct lineages in the colonic sections, although FACS analysis did show rare (9.93% in Lgr5+ cells) overlap (Fig. 7B&S7D). We found rare fully traced glands from Bhlha15+ cells even in normal homeostasis, suggesting that Bhlha15+ cells may occasionally overlap with the colonic stem cell population (Fig. S8A–B). However, we observed no apparent changes in lineage tracing pattern even after Lgr5 ablation (Fig. S7D&S8B). Analogous to the intestine, Notch activation in Bhlha15+ cells led to loss of the secretory lineage and a shift to absorptive lineage tracing (Fig. S7E). Nevertheless, the increase in Bhlha15-derived gland tracing events was not statistically significant, even with combined Lgr5 ablation (Fig. S8B), suggesting that interconversion is not efficiently induced by Notch activation in the colon. In contrast, dextran sodium sulfate (DSS)-induced injury significantly increased the lineage tracing ability in Bhlha15+ cells, while most of Lgr5+ cells disappeared (Fig. S8A–C).
Unlike doxorubicin-treated small intestinal Bhlha15+ cells, Bhlha15+ colonic precursors did not show HES1 expression even after DSS treatment (Fig. S8D). Recently, it has been recognized that the YAP/TAZ-mediated mechanosensory pathway plays a central role in colonic regeneration after DSS treatment29. Indeed, we found upregulation of YAP expression in regenerative glands after DSS, in which Bhlha15+ cells become positive for YAP, while no such upregulation following Lgr5-DTR ablation or forced Notch activation (Fig. 7D&S8E–F). Since YAP coordinates Wnt activity as a transcriptional coactivator, we examined Wnt signaling status in the sorted Bhlha15+ cells with or without DSS treatment. RT-PCR and immunohistochemical analysis revealed that DSS-treated Bhlha15+ cells showed upregulation of Wnt target molecules, including Lgr5, Olfm4, Cd44, Axin2, and cyclin D1, compared to normal Bhlha15+ cells (Fig. 7D&S8F–G). Following DSS treatment, we observed nuclear translocation of beta-catenin in cells in regenerative glands, including Bhlha15+ cells (Fig. S8H). We also found increased colony formation ability of Bhlha15+ cells following DSS treatment (Fig. S8I). Thus, DSS treatment leads to greater Wnt activation in Bhlha15+ cells, which likely contributes to greater stem cell potential. After DSS treatment, Bhlha15+ cells also start to express phosphorylated Src that activates YAP pathway29. When we treated mice with a Src inhibitor following DSS, tracing events from Bhlha15+ cells was significantly inhibited by the Src inhibitor (Fig. S8J–K). Thus, for colonic regeneration following DSS treatment, the activation of Src/YAP/Wnt pathway appears to be critical.
Next, we explored whether Bhlha15+ colonic precursors can give rise to tumor in the setting of Apc loss. The vast majority (80%) of Bhlha15-CreERT; Apcflox/flox mice did not develop macroscopic tumors in the colon, although on close inspection rare mice did show polypoid lesions that exhibited histologically intramucosal dysplasia (Fig. S9A). When we treated mice with DSS, we found macroscopic tumors with dysplastic histology and marked expansion of nuclear β-catenin+ cells in 90% of the mice (Fig. 7E–F&S9A). Addition of mutant Kras or Trp53 gene expression in Bhlha15-CreERT; Apcflox/flox mice did not increase tumor formation (Fig. 7F). As reported previously30, DSS treatment accelerates colonic tumor growth in Lgr5-CreERT; Apcflox/flox mice as well, however, histopathology of these tumors is quite similar to what was observed in DSS-treated Bhlha15-CreERT; Apcflox/flox mouse colon. Although DSS treatment initially activates the Wnt/YAP pathway but not the Notch pathway, all of established tumors and dysplastic lesions in both mouse lines display strong expression of YAP and HES1, without significant differences in the key Wnt/Notch target gene expression regardless of initial DSS treatment (Fig. S9B–C). These findings suggest that DSS-induced YAP/Wnt activation is responsible for initiating colonic tumors from additional cells-of-origins (e.g. progenitor cells beyond classical Lgr5+ stem cells), although established tumors appear to show vastly similar histological and molecular phenotypes when identical oncogenic mutations are induced.
Abnormal Notch signaling is associated with CIN-type, left-sided colon cancers with serrated histology in human.
Finally, we investigated BHLHA15 expression and Notch status in human patient samples. Similar to mice, BHLHA15 is normally expressed in Paneth and TA progenitor cells in the small intestine, as well as in rare cells near the base in the colon (Fig. S10A). The number of BHLHA15+ epithelial cells appears to be slightly increased in inflamed colonic glands, but almost completely lost in cancer, which may suggest that BHLHA15 or BHLHA15 is primarily a marker of differentiated secretory lineages, and that its expression disappears when cells undergo dedifferentiation. We next analyzed whether Notch signaling is activated in human colorectal cancers using a public TCGA dataset31. Among 55 predefined Notch-related genes, we found the most frequent mRNA upregulation or gene amplification (36%) in the ITCH gene, followed by RBPJL (16%) and LFNG (10%) genes. Approximately 44 % of cancer cases showed upregulation or amplification in one of these genes or the NOTCH1 gene (Fig. S10B). Notch-activated cancer cases are significantly associated with a chromosomal instability (CIN)-type signature31, particularly in the left-sided colon (Table S1). Notch-activated cancers are also positively correlated with the presence of serrated features on histology (Fig. S10C&Table S1–2). On the other hand, serrated features are not significantly correlated with other oncogenic mutations, while KRAS or BRAF mutants tend to show serrated histology more frequently (p = 0.058 and 0.051, respectively). Cases with RBPJL gene alterations show significantly worse outcome compared to cases without RBPJL abnormalities (Fig. S10D).
Among human colorectal adenomas, HES1 expression is found in 36 % of tubular adenoma cases, and 80 % of serrated polyps, suggesting the presence of Notch activation particularly in early serrated preneoplasia (Fig. S10E–F). Given that Itch, Rbpjl, and Lfng genes were upregulated in Notch-activated, Apc-deleted mouse tumors compared to tumors with Apc deletion alone (Fig. S10G), Notch activation may play an important role in human colorectal serrated-type carcinogenesis.
Discussion
Recent studies have suggested a greater degree of cellular plasticity in the gastrointestinal epithelium than previously expected, and multiple cell types appear to be capable of interconverting into stem-like cells. Our data and previous studies suggest that not all progenitors can equally behave as a reserve stem cell following unspecified injurious stimuli12–15. Studies in the past demonstrated that Bmi1+ cells and Alpi+ EPs can give rise to active stem-like cells following Lgr5-DT ablation11–15. Given the absence of tracing events from Bhlha15+ cells after Lgr5 ablation, Bhlha15+ precursors may play a secondary role compared to these immediate progenitors in the ISC hierarchy. Nevertheless, even the more reserved Bhlha15+ secretory precursors are able to become reprogrammed into long-lived cancer-initiating cells, primarily in response to sustained Notch activation.
The interconversion process appears to be accompanied by dynamic genetic and epigenetic alterations in these reserve progenitors, leading to a reversion towards an Lgr5+ cell identity11. In contrast, our microarray data revealed only minimal change in Lgr5-ablated Bhlha15+ precursors, and thus no phenotypic interconversion in this setting. However, following irradiation and even more so after doxorubicin treatment, there are numerous gene expression changes in Bhlha15+ precursors, although the changes did not correlate with Lgr5+ ISC gene signature contrasting with previous reports11. Nonetheless, we observed short-term enterocyte-specific tracing from doxorubicin-treated Bhlha15+ precursors. Lack of long-term lineage tracing in vivo, as well as clonogenic colony formation in vitro, suggest that such short-term conversion may not represent true dedifferentiation and stem cell interconversion. Careful and comprehensive analysis is required to evaluate the true cellular plasticity, which may not be reflected simply by short-term tracing or temporal proliferation.
It is important to note that the expression of most reported stem and progenitor markers is not restricted to one cell types, but can be observed in a number of lineages and thus a broader heterogeneous cell population4, 12, 15, 25. Along these lines, we show here that Bhlha15 expression in the intestine is not confined to mature Paneth cells, but is also detected in secretory precursors including pre-Paneth LRCs that functionally behave in a manner distinct from Paneth cells. Previous reports suggested that these Lgr5lo pre-Paneth precursors represent slowly cycling LRCs, which do not behave as other Lgr5+ CBCs but instead can dedifferentiate into active ISCs following injury15, 24. However, it is not entirely clear whether such LRCs are one entity, or also represent a heterogeneous population. Our comparison between Bhlha15+ cells and DLL1+ cells suggests that secretory precursors also may be heterogeneous and contain several cell types, and that not all secretory progenitors behave in an equivalent manner, either phenotypically or genotypically. Although our data suggest that CD24lo TA progenitors, rather than CD24hi Paneth cell lineage, preferentially contribute to long-lived clones, it is certainly possible that both populations participate to some extent in the regeneration process, as recently reported32, 33. Nonetheless, our careful observations in conjunction with previous studies15, 22 supports the notion that the ability of such interconversion or dedifferentiation is predominantly restricted to cells in undifferentiated state.
Serrated dysplasia or adenoma can be classified two main subtypes, traditional serrated adenoma (TSA) and sessile serrated adenoma (SSA). The former predominantly arises in the left-sided colon, while the latter is more frequently seen in the right-sided colon. Both adenomas are thought to be precursors of colorectal cancer, but their molecular signatures suggest that SSA-derived cancers likely represent MSI/CIMP-type accompanied with frequent BRAF mutation, while TSA can progress to cancers carrying classical gene mutations such as KRAS, TP53, or APC by following adenoma-carcinoma sequence34. However, it has not been understood how serrated histological signatures are created within these tumors. Based on our analysis, Notch signaling may contribute to serrated histology, particularly CIN-type left-sided cancers that likely include TSA-derived cancers, and our mouse model may phenocopy this particular subtype. Additionally, given that most of Lgr5+ cells are ablated and Bhlha15+ precursors can act as stem-like cells in the colon following injury, injury-resistant cells such as Bhlha15+ progenitors or DCLK1+ cells18 may be important sources of inflammation-associated colorectal cancers.
Supplementary Material
Acknowledgements
Authors received the NIH grants (R35CA210088 (T.C.W.) and 5U01DK103155-04 (T.C.W. & C.G.). Y.Hayakawa is supported by the KAKENHI Grant-in-Aid for Scientific Research, 17K09347 and 17H05081, P-CREATE from AMED, the Mitsubishi Foundation, Natural Sciences, the research grant of Astellas Foundation for Research on Metabolic Disorders, the Yasuda Medical Foundation, and the Takeda Science Foundation Visionary Research Grant, the Princess Takamatsu Cancer Research Fund, and the Advanced Research and Development Programs for Medical Innovation (PRIME). R.N. is supported by the KAKENHI Grant-in-Aid for Scientific Research 17K15928.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures:
The authors disclose no conflicts.
Transcript profiling:
References
- 1.Clevers H The intestinal crypt, a prototype stem cell compartment. Cell 2013;154:274–84. [DOI] [PubMed] [Google Scholar]
- 2.Middelhoff M, Westphalen CB, Hayakawa Y, et al. Dclk1-expressing tuft cells: critical modulators of the intestinal niche? Am J Physiol Gastrointest Liver Physiol 2017;313:G285–G299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tian H, Biehs B, Chiu C, et al. Opposing activities of Notch and Wnt signaling regulate intestinal stem cells and gut homeostasis. Cell Rep 2015;11:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan KS, Janda CY, Chang J, et al. Non-equivalence of Wnt and R-spondin ligands during Lgr5+ intestinal stem-cell self-renewal. Nature 2017;545:238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shroyer NF, Helmrath MA, Wang VY, et al. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology 2007;132:2478–88. [DOI] [PubMed] [Google Scholar]
- 6.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007;449:1003–7. [DOI] [PubMed] [Google Scholar]
- 7.Asfaha S, Hayakawa Y, Muley A, et al. Krt19(+)/Lgr5(−) Cells Are Radioresistant Cancer-Initiating Stem Cells in the Colon and Intestine. Cell Stem Cell 2015;16:627–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet 2008;40:915–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian H, Biehs B, Warming S, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 2011;478:255–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metcalfe C, Kljavin NM, Ybarra R, et al. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell 2014;14:149–59. [DOI] [PubMed] [Google Scholar]
- 11.Jadhav U, Saxena M, O’Neill NK, et al. Dynamic Reorganization of Chromatin Accessibility Signatures during Dedifferentiation of Secretory Precursors into Lgr5+ Intestinal Stem Cells. Cell Stem Cell 2017;21:65–77 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan KS, Gevaert O, Zheng GXY, et al. Intestinal Enteroendocrine Lineage Cells Possess Homeostatic and Injury-Inducible Stem Cell Activity. Cell Stem Cell 2017;21:78–90 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Es JH, Sato T, van de Wetering M, et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol 2012;14:1099–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tetteh PW, Basak O, Farin HF, et al. Replacement of Lost Lgr5-Positive Stem Cells through Plasticity of Their Enterocyte-Lineage Daughters. Cell Stem Cell 2016;18:203–13. [DOI] [PubMed] [Google Scholar]
- 15.Buczacki SJ, Zecchini HI, Nicholson AM, et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature 2013;495:65–9. [DOI] [PubMed] [Google Scholar]
- 16.Barker N, Ridgway RA, van Es JH, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 2009;457:608–11. [DOI] [PubMed] [Google Scholar]
- 17.Schwitalla S, Fingerle AA, Cammareri P, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 2013;152:25–38. [DOI] [PubMed] [Google Scholar]
- 18.Westphalen CB, Asfaha S, Hayakawa Y, et al. Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J Clin Invest 2014;124:1283–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayakawa Y, Ariyama H, Stancikova J, et al. Mist1 Expressing Gastric Stem Cells Maintain the Normal and Neoplastic Gastric Epithelium and Are Supported by a Perivascular Stem Cell Niche. Cancer Cell 2015;28:800–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayakawa Y, Sakitani K, Konishi M, et al. Nerve Growth Factor Promotes Gastric Tumorigenesis through Aberrant Cholinergic Signaling. Cancer Cell 2017;31:21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakitani K, Hayakawa Y, Deng H, et al. CXCR4-expressing Mist1(+) progenitors in the gastric antrum contribute to gastric cancer development. Oncotarget 2017;8:111012–111025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinoshita H, Hayakawa Y, Niu Z, et al. Mature gastric chief cells are not required for the development of metaplasia. Am J Physiol Gastrointest Liver Physiol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayakawa Y, Hirata Y, Nakagawa H, et al. Apoptosis signal-regulating kinase 1 regulates colitis and colitis-associated tumorigenesis by the innate immune responses. Gastroenterology 2010;138:1055–67.e1–4. [DOI] [PubMed] [Google Scholar]
- 24.Barriga FM, Montagni E, Mana M, et al. Mex3a Marks a Slowly Dividing Subpopulation of Lgr5+ Intestinal Stem Cells. Cell Stem Cell 2017;20:801–816 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grun D, Lyubimova A, Kester L, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 2015;525:251–5. [DOI] [PubMed] [Google Scholar]
- 26.Haber AL, Biton M, Rogel N, et al. A single-cell survey of the small intestinal epithelium. Nature 2017;551:333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothenberg ME, Nusse Y, Kalisky T, et al. Identification of a cKit(+) colonic crypt base secretory cell that supports Lgr5(+) stem cells in mice. Gastroenterology 2012;142:1195–1205 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sasaki N, Sachs N, Wiebrands K, et al. Reg4+ deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon. Proc Natl Acad Sci U S A 2016;113:E5399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yui S, Azzolin L, Maimets M, et al. YAP/TAZ-Dependent Reprogramming of Colonic Epithelium Links ECM Remodeling to Tissue Regeneration. Cell Stem Cell 2018;22:35–49 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tauriello DVF, Palomo-Ponce S, Stork D, et al. TGFbeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018;554:538–543. [DOI] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas N Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu S, Tong K, Zhao Y, et al. Paneth Cell Multipotency Induced by Notch Activation following Injury. Cell Stem Cell 2018;23:46–59 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitt M, Schewe M, Sacchetti A, et al. Paneth Cells Respond to Inflammation and Contribute to Tissue Regeneration by Acquiring Stem-like Features through SCF/c-Kit Signaling. Cell Rep 2018;24:2312–2328 e7. [DOI] [PubMed] [Google Scholar]
- 34.JE IJ, Vermeulen L, Meijer GA, et al. Serrated neoplasia-role in colorectal carcinogenesis and clinical implications. Nat Rev Gastroenterol Hepatol 2015;12:401–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.