Abstract
In the recent few years, significant efforts have been undertaken for the development of different immunotherapeutic approaches against cancer. In this context, immune checkpoint inhibitors (ICIs), a novel class of immunotherapeutic drugs with the potential to unleash the immune system, have emerged as authentic game-changers for managing patients with various cancers, including gastrointestinal (GI) malignancies. Although the majority of GI cancers are generally considered poorly immunogenic, basic research findings and data from clinical trials have proven that subset(s) of patients with various digestive tract cancers are highly responsive to ICI-based therapy. In this context, a better understanding on the role of various DNA repair pathway alterations, especially the evidence supporting the significant importance of DNA mismatch repair (MMR) deficiencies and the efficacy of the antiprogrammed cell death 1 (PD-1) drugs, have led to FDA approval of two anti-PD-1 antibodies (pembrolizumab and nivolumab) for the treatment of patients with microsatellite instability. This review aims to provide a comprehensive and up-to-date summary for the role of DNA MMR deficiency in cancer, and its importance in the development of ICI therapy. In addition, we provide insights into the spectrum of various genetic alterations underlying ICI resistance, together with the important influence that the tumor microenvironment plays in mediating the therapeutic response to this new class of drugs. Finally, we provide a comprehensive yet succinct glimpse into the most exciting pre-clinical discoveries and ongoing clinical trials in the field, highlighting bench-to-beside translational impact of this exciting area of research.
Keywords: Gastrointestinal Cancers, DNA MMR, anti-PD-1, anti-CTLA-4, immune checkpoint inhibitors
Introduction
The enormous efforts undertaken to understand the interplay between cancer and immune cells in recent years has resulted in the development of one of the most exciting generation of drugs – the immune checkpoint inhibitors (ICIs). Ironically, even though most gastrointestinal (GI) cancers have historically been considered poorly immunogenic,1 the knowledge gained from different disciplines of basic science, together with the dramatic development of massively parallel sequencing technologies has paved the path for the successful and quite effective application of ICI treatment in several GI malignancies. Furthermore, neoplasms with a defective DNA damage repair system, a hallmark feature frequently observed in a subset of GI cancers, are positioned to be the best candidates for these kinds of therapies.2
In this review, we introduce basic concepts of DNA damage repair pathways and comprehensively summarize the historical evolution and up-to-date pre-clinical and clinical evidence in the development of ICI therapy in GI cancers.
DNA Mismatch Repair Pathway
Genomic instability and hyper-mutability are central hallmarks of cancer development.2 To maintain genomic integrity and stability, eukaryotic cells possess a plethora of genes encoding for proteins that coordinately function to repair different types of DNA damage during cancer progression. At least six DNA repair pathways are involved in repairing specific types of DNA damage:3,4 1) the DNA mismatch repair (MMR) pathway – repairs inappropriate nucleotide insertions/deletions (INDELs) and single nucleotide mismatches; 2) the base excision repair (BER) pathway – corrects single-strand breaks and homologous recombination (HR); 3) the non-homologous end joining (NHEJ) pathways ‒ repairs double-strand breaks; 4) the nucleotide excision repair (NER) pathway ‒ repairs DNA adducts; 5) the Fanconi anemia (FA) pathway ‒ fixes interstrand crosslinks; 6) the O6-methylguanine DNA methyltransferase (MGMT) pathway ‒ involved in repairing O6-methylguanine adducts. However, in the context of this article, we will specifically focus on the MMR pathway, and its clinical implications in the management of various GI malignancies.
The DNA MMR is an evolutionarily conserved pathway in which multiple protein complexes function in a coordinated manner to repair DNA damage. To date, several, functionally important, key MMR protein heterodimers have been described: MSH2 protein forms a heterodimer with either MSH6 (MutSα) or MSH3 (MutSβ) and MLH1 pairing with PMS2 (MutLα), MLH2 (MutLβ) or MLH3 (MutLγ) – all of which are responsible for repairing single nucleotide or INDEL mismatches.5,6 In addition, the proofreading activity of DNA polymerase-ε (POLE) or DNA polymerase-δ (POLD), has also a prominent role in the DNA MMR process.7
Microsatellite Instability in Gastrointestinal Malignancies
Short tandem repeat DNA sequences (1–6 or more base pairs), also known as microsatellites, are spread throughout the human genome. Due to their highly repetitive nature, these sequences have a higher propensity for acquiring mutations – a process tightly governed by an intact MMR system. Deficiency in DNA MMR activity results in a hypermutator phenotype, termed microsatellite instability (MSI), often characterized by the presence of single nucleotide substitutions or INDELs within these microsatellite repeats.7 The MMR deficiency resulting from germline mutations or epigenetic alterations (gene inactivation by promoter methylation) in any of the MMR genes (MLH1, MSH2, MSH6, and PMS2), as well as deletions in the EPCAM gene (which leads to constitutional repression of MSH2 gene expression through promoter methylation) is the principle cause of Lynch syndrome (LS) and its variants (Muir– Torre or Turcot’s syndromes).8 For the development of LS cancer, according to the Knudson’s ‘two-hit model’,9 somatic loss of function of the remaining wild-type allele of the germline altered MMR gene is mandatory.8 Homozygous germline mutations in any of the four aforementioned MMR genes can cause a constitutional MMR deficiency syndrome, which is one of the most aggressive, highly penetrant childhood cancer predisposition syndromes. In addition, LS also can result from mosaic germline MLH1 epimutations. In contrast, bi-allelic MLH1 promoter methylation is primarily the key somatic event responsible for the loss of MLH1 expression in ~75–80% of sporadic cancers with MSI.6,8
With the recent advent of immunotherapy during the last decade, tremendous efforts have been made to understand the biological mechanisms responsible for the observed clinical benefit in patients treated with ICIs.10 It was not until after the first clinical evidence suggested that patients with MSI-high (MSI-H) colorectal cancer (CRC) were more responsive to programmed cell death 1 (PD-1) blockade,11 the focus of the scientific community shifted towards DNA MMR-deficient (dMMR) tumors. Large-scale genomic studies have revealed that dMMR cancers, together with those bearing defects in the exonuclease domain of the catalytic subunits of the POLE or POLD1 genes, represent a hypermutator phenotype.12 A classical hallmark feature of the MSI-H CRCs is a prominent lymphocytic infiltrate, which correlates with a higher neoantigen load (resulting from the somatic mutations that produce more immunogenic peptides),13 as well as with a higher expression of various immune checkpoint molecules [PD-1, programmed death-ligand 1 (PD-L1), cytotoxic T-lymphocyte antigen 4 (CTLA4), LAG-3, and IDO].14,15
In view of these interesting discoveries, there is a growing interest in gaining a better understanding of the MSI landscape in different tumor types. Not surprisingly, nowadays, evaluation of the MSI status, either through PCR-based assays or immunohistochemically, has become a routine clinical practice for various cancers, particularly GI malignancies.8 With the emergence of the next-generation sequencing (NGS)-based technologies, others have developed alternate computational methods to infer MSI using targeted, whole exome or whole genome sequencing data (e.g., MSIsensor, mSINGS, and MANTIS).16–18 Table 1 presents the prevalence of MSI across different GI malignancies. The wide range in some of the less-classically characterized tumors is a reflection of methodologic issues as well as variability in tumor stages and other epidemiologic factors among the cohorts analyzed.6,19–34
Table 1.
Prevalence of MSI in various gastrointestinal cancers
| Tumor type | dMMR/MSI-H (%) |
|---|---|
| Esophageal carcinoma19–21 | 0–7 |
| GEJ adenocarcinoma21 | ~4 |
| Gastric cancer21–23 | 7–22 |
| SBA21,24,25 | 5–35 |
| CRC6,23 | ~15a |
| Anal cancer21 | 0 |
| HCC21,23,26 | 1.5–16 |
| Pancreatic cancer23,27–29 | 0–17 |
| Biliary tract cancers21 | ~2 |
| Cholangiocarcinoma30 | ~12.5 |
| Gallbladder carcinoma31,32 | 0–40b |
| Ampullary carcinoma23,33 | 6–10 |
~2.5% by germline mutations and ~12.5% by somatic DNA alterations (~75% by MLH1 promoter methylation).6,23
In anomalous pancreaticobiliary ductal junction patients: up to 80%.34
Abbreviations: GEJ, gastroesophageal junction; SBA, small-bowel adenocarcinoma; CRC, colorectal cancer; HCC, hepatocellular carcinoma.
ICI Development in Gastrointestinal Malignancies – The Current Landscape
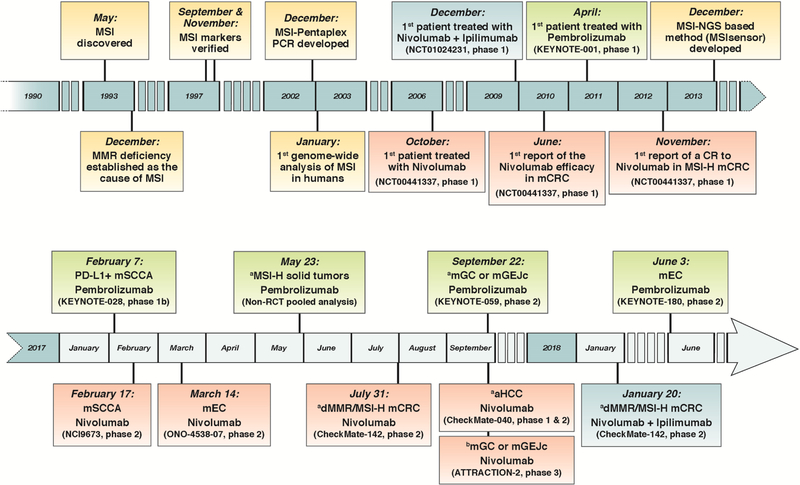
Since the discovery of the T-cell activity inhibition by the CTLA-4 and PD-1 proteins, several antibodies have been synthesized against these two receptors. With the premise of unleashing the ‘brakes’ of immune system, the development of ipilimumab (anti-CTLA-4), followed by nivolumab and pembrolizumab (anti-PD-1) marked a seminal milestone in the treatment of cancer. Following the promising efficacy of these ICIs in well-recognized, highly immunogenic tumors, such as melanoma and non-small cell lung cancer (NSCLC), over the past few years, several clinical trials have been launched to evaluate their efficacy in other tumor types.35 Some of the exciting data from these trials have paved the path for the accelerated approval by the US Food and Drug Administration (FDA) of ICI therapies in various GI malignancies (Fig. 1).
Figure 1:
Timeline of MSI-related discoveries and immune checkpoint inhibitors clinical development and regulatory approvals in different gastrointestinal cancers. aFDA accelerated approval. bJapanese Ministry of Health, Labor and Welfare approval. Abbreviations: MSI, microsatellite instability; MSI-H, MSI-high; CR, complete response; mSCCA, metastatic or locally advanced unresectable squamous cell carcinoma of the anal canal; mEC, metastatic or locally advanced unresectable esophageal carcinoma; dMMR, DNA mismatch repair deficient; mCRC, metastatic or locally advanced unresectable colorectal cancer; aHCC, advanced hepatocellular carcinoma; mGC, metastatic or locally advanced unresectable gastric cancer; mGEJc, metastatic or locally advanced unresectable gastro-esophageal junction cancer.
Color-coded boxes: Yellow: basic and translational milestones. Red: clinical milestones achieved with nivolumab. Blue: clinical milestones achieved with nivolumab plus ipilimumab. Green: clinical milestones achieved with pembrolizumab.
ICI Development in Microsatellite Unstable Malignancies
In an unprecedented move, in May 2017, the FDA granted pembrolizumab accelerated approval as the first drug for a tumor site-agnostic indication. This recommendation was based upon data reported from 149 patients with dMMR/MSI-H cancers enrolled across five uncontrolled, multiple cohort, multicenter, single-arm pembrolizumab-based clinical trials.36 Distribution of patients by tumor type and clinical efficacy are summarized in Supplementary Table S1. The relatively moderate to high frequency of dMMR/MSI-H prevalence in various GI malignancies (Table 1) emphasizes the potential magnitude of this approval in this clinical scenario.
Colorectal cancer: In spite of pembrolizumab being the first anti-PD-1 drug approved by the FDA for treatment of dMMR/MSI-H metastatic CRC (mCRC),36 the earliest insights into this paradigm were based on retrospective evaluation of the solitary CRC patient who responded to nivolumab in the first-in-human clinical trial, which turned out to be MSI-H on subsequent tumor analysis.11 In light of this finding, just within a matter for few months, two phase 2 clinical trials were initiated to test this promising hypothesis.37,38 Following evaluation of the results from the CheckMate-142 trial (Table 2), in July 2017, the FDA approved nivolumab for the treatment of dMMR/MSI-H mCRC patients, which otherwise demonstrated disease progression on cytotoxic chemotherapies (fluoropyrimidine, oxaliplatin, and irinotecan).39 More recently, in July 2018, almost a year later, the FDA has now allowed the use of nivolumab plus ipilimumab for treatment of the same subset of mCRC patients, based upon the promising anti-cancer activity with a manageable toxicity profile showed by this regimen in the combination therapy cohort of patients within the CheckMate-142 trial (Table 2).40
Table 2.
Most relevant ICI clinical trials developed in GI cancers.
| Clinical trial | Phase | Drug(s) | Line of therapy | Tumor type(s) | Patients enrolled (n) | Clinical activity | Adverse events, % (n) |
|---|---|---|---|---|---|---|---|
| Colorectal cancer | |||||||
| CheckMate-142 | 2 (mc) | Nivolumab | ≥2 | mCRC | 74 | 12-mo OS rate: 73% | Grade 3–4: 21% |
| CheckMate-142 | 2 (mc) | Ipilimumab | ≥2 | mCRC | 119 | 9- & 12-mo OS rate: 87% / 85% | Grade 3–4: 32% |
| KEYNOTE-028 | 1b (mc) | Pembrolizumab | ≥2 | PD-L1+ mCRC | 23 | DCR: 22% | Grade 4–5:0% |
| KEYNOTE-016 | 2 (mc) | Pembrolizumab | ≥3 | CRC | mNon-CRC: 9 | mOS: NR | Grade 3–4: 41% |
| KEYNOTE-164 | 2 | Pembrolizumab | ≥3 | mCRC | 61 | 6-mo OS rate: 87% | Serious: 7% (4) |
| Hepatocellular carcinoma | |||||||
| CheckMate-040 | 1/2 | Nivolumab | ≥2 | aHCCC-PA/B7 | phase: 214 | 6- & 9-mo OS rate: 83% / 74% | Grade 3–4: 25% |
| Gastric cancer & Gastroesophageal junction cancer | |||||||
| ATTRACTION-2 | 3 | Placebo | ≥3 | mGC / mGEJc | 163 | 95% Cl 0.51–0.78) | Grade 5: 5c |
| KEYNOTE-059 | 2 (mc) | Pembrolizumab | ≥3 | mGC/ mGEJc | mGEJc) | mOS: 5.6 mo | Grade 3–5: 17.8% (2 grade 5)c |
| KEYNOTE-061 | 3 | vs Paclitaxel | 2 | mGC /mGEJc | Ll CPS > 1) | 95% Cl 0.66 −1.03) | 35% (1 grade 5)c |
| Esophageal carcinoma | |||||||
| ONO-4538–07 | 2 | Nivolumab | ≥2 | mEC | 64 (ESCC) | mOS: 10.8 mo | Grade 3–4: 17% |
| KEYNOTE-180 | 2 | Pembrolizumab | ≥3 | mEC | ESCC/58 EAC) | mOS: 5.8 mo | Grade 3–5: 12% (1 grade 5)c |
| Anal cancer | |||||||
| NCI9673 | 2 | Nivolumab | ≥2 | mSCCA | 37 | mOS: 11.5 mo | Grade 3:14% |
| KEYNOTE-028 | 1b (mc) | Pembrolizumab | ≥2a | PD-L1+ mSCCAb | 25 | mOS: 9.3 mo | Grade 3:16% |
Three patients included after disease progression within 6 months of completion of chemotherapy for limited-stage disease.
Because of a protocol violation, one perineal epidermoid carcinoma was included.
Grade 5 adverse events:
ATTRACTION-2: 5 with nivolumab (acute hepatitis, cardiac arrest, unknown, exertional dyspnea, and pneumonia) vs. 2 with placebo (GI perforation and sudden death).
KEYNOTE-059: 2 (acute kidney injury and pleural effusion).
KEYNOTE-061: 3 with pembrolizumab (colitis, interstitial lung disease, and unknown) vs. 1 with paclitaxel (pulmonary embolism).
KEYNOTE-180: 1 (pneumonitis).
Abbreviations: mc, multicohort; CRC, colorectal cancer; mCRC, metastatic or locally advanced unresectable CRC; mNon-CRC, metastatic or locally advanced unresectable non-CRC; HCC, hepatocellular carcinoma; aHCC, advanced HCC; C-P, Child–Pugh; mGC, metastatic or locally advanced unresectable gastric cancer; mGEJc, metastatic or locally advanced unresectable gastro-esophageal junction cancer; mEC, metastatic or locally advanced unresectable esophageal carcinoma; ESCC, esophageal squamous cell carcinoma; EAC, esophageal adenocarcinoma; mSCCA, metastatic or locally advanced unresectable squamous cell carcinoma of the anal canal; dMMR, DNA mismatch repair deficient; MSI-H, microsatellite instability-high; ORR, objective response rate; DCR, disease control rate, mDOR, median duration of response; mPFS, median progression free survival; mOS, median overall survival; NR, not reached; NA, not available; CPS, combined positive score.
ICI Development in GI Malignancies Independent of Microsatellite Instability Status
Independent of the MSI status, the development of ICI-based therapies is also garnering a lot of interest in other GI malignancies. Based on early-phase clinical trials, ICIs development is moving forward with anti-PD-1 drugs as realistic treatment options for patients previously treated for recurrent, locally advanced or metastatic disease.
Hepatocellular carcinoma: In September 2017, nivolumab was also approved for the treatment of HCC patients previously treated with sorafenib.39 This decision was based on the results of a subgroup of 154 patients enrolled in the CheckMate-040 - a multicenter, open-label phase 1/2 trial conducted in patients with HCC and Child–Pugh A cirrhosis who had either disease progression or were intolerant to sorafenib (Table 2).41 Very recently, in July 2018, FDA granted a Breakthrough Therapy Designation for the combination regimen of atezolizumab (anti-PD-L1) and bevacizumab for the treatment of patients with advanced or metastatic, treatment-naive HCC. This designation was based on the encouraging preliminary results reported in a phase 1b trial assessing the safety and activity of this regimen42.
Gastric cancer: Preliminary evidence for the activity of nivolumab and pembrolizumab in recurrent, locally advanced or metastatic gastric or gastroesophageal junction (GEJ) cancer was reported in phase 1/2 CheckMate-032 and phase 1 KEYNOTE-012 trials, respectively. These early encouraging findings were subsequently confirmed in phase 2 (KEYNOTE-059) and phase 3 (ATTRACTION-2) clinical trials (Table 2). In view of the significant positive clinical outcomes of the ATTRACTION-2 trial, which enrolled Asian patients previously treated with at least two lines of chemotherapy, the use of nivolumab in this setting was approved by the Japanese Ministry of Health, Labor and Welfare43. Currently, similar trials were underway in the Caucasian population, because insufficient data is yet available to its approval by the US FDA and the European Medicines Agency.
Based upon the results from 143 PD-L1 positive patients enrolled in the KEYNOTE-059 trial (a multicenter, non-randomized, open-label, 3-cohort, phase 2 trial), where pembrolizumab showed reasonable antitumor activity with a well-tolerated toxicity profile (Table 2),44 this ICI was approved by the FDA in September 2017 for the treatment of patients with recurrent, locally advanced or metastatic gastric or GEJ (Siewert type II/III), PD-L1– positive adenocarcinomas with disease progression after at least two prior lines of therapy.36 In spite of the negative results of the phase 3 KEYNOTE-061 trial, in which pembrolizumab did not further improve survival as second-line therapy in the same tumor subset of patients, this approval has not been revoked and multiple clinical trials continue to interrogate its clinical application as a first-line treatment option or its use in neoadjuvant/adjuvant settings.45
Esophageal cancer: Encouraging activity of nivolumab and pembrolizumab in previously treated, advanced or metastatic esophageal carcinoma or GEJ adenocarcinoma (Siewert type I) has been demonstrated in independent phase 2 clinical trials (ONO-4538–07 and KEYNOTE180; Table 2).46,47 To confirm these findings in a randomized scenario, phase 3 trials in patients who are either refractory or intolerant to standard chemotherapy are currently underway.
Anal cancer: Likewise, following disease progression in patients treated with a 5-FU plus platinum-based first-line therapeutic regimen, NCCN guidelines recommended the use of antiPD-1 drugs (pembrolizumab and nivolumab) as a treatment option for metastatic disease.48 This recommendation was based on the results of two clinical trials; a multi-cohort, multicenter, single-arm, open-label, phase 1b trial for pembrolizumab (KEYNOTE-028) and a single-arm, multicenter, phase 2 trial for nivolumab (NCI9673; Table 2).
The results from various clinical trials described above clearly demonstrate that the clinical use of ICI-based therapy is moving forward in full swing in various GI malignancies. While several of these drugs have received an accelerated approval from the FDA for various tumor types are solely based upon objective response rate or other surrogate endpoints in early-phase trials, it is imperative that these early data are confirmed in randomized clinical trials to truly authenticate these favorable findings.49
ICI Predictive Biomarkers and Mechanisms of Resistance
ICI Predictive Biomarkers
Although quite encouraging, the results presented in the previous section highlight the need for an improved patient selection for using ICI-based therapies. The bulk of clinical evidence favoring the use of ICIs in MSI-H malignancies comes from mCRC patients where MSI-H tumors have shown a significant response to anti-PD-1 drugs.37,38,40 In other tumor types, the evidence is primarily anecdotal and stems from retrospective analysis of a small number of patients enrolled in nonrandomized clinical trials (Supplementary Table S1).36
However, as anticipated, the clinical strategy for the development of ICIs in GI malignancies is not solely hinging upon the MSI status. For instance, in gastric cancer, in spite of FDA’s approval for its use based on PD-L1 positivity, the KEYNOTE-059 trial patient selection did not rely on a specific biomarker. Intriguingly, the recent results from this clinical trial demonstrated no significant differences in progression-free survival (PFS) according to PD-L1 score, but showed a higher objective response rate (ORR) and duration of response (DOR) in patients with a positive PD-L1 score.36,44 This debate about PD-L1 status as a predictive biomarker for the use of ICI-based therapy has been ongoing for the past few years, and has yielded inconsistent results depending on tumor histology, anti-PD-1/PD-L1 drug, and even the line of therapy. Its temporospatial heterogeneity, together with lack of consensus on the technical issues and scoring systems, are potentially some of the factors underlying this inconsistency.50
With regards to gene expression-based predictive signatures for ICI, in the KEYNOTE059 trial, a high 18-gene T-cell–inflamed gene expression profiling score was significantly associated with a higher response rate and PFS.44 The response predictive value of another transcriptomic signature, the interferon-γ (IFN-γ)–related mRNA profile, has also been validated in gastric cancer.51 Likewise, the innate anti-PD-1 resistance (IPRES) signature in melanoma (with potential application in other cancers, such as CRC or pancreatic cancer)52 and the APOBEC3B expression in lung cancer,53 together with the other consensus molecular signatures developed in several GI malignancies, could be useful tools for patient selection.20,22,25,33,54–57
Several types of viral infections are associated with various GI malignancies: Epstein– Barr virus (EBV) with approximately 9% of gastric cancers, HPV with >90% of squamous cell carcinomas of the anal canal, and hepatitis C or B virus with HCC.22,58,59 Despite the fact that exploratory analysis obtained from GI cancers with viral-associations did not observe any clear correlation between infection status and therapeutic benefit from anti-PD-1 therapy,41,44,58,60–62 one can speculate that epitopes derived from viral open reading frames could potentially act as immunogenic neoantigens promoting T-cell tumor infiltration.13 In line with these results, Kim et al63 reported the response of all EBV positive metastatic gastric cancer patients (6 out of 61) involved in a pembrolizumab phase 2 trial. Interestingly and in the same cohort, they also evaluated the utility of ctDNA monitoring for therapy efficacy prediction and reported promising positive results.
The immunogenicity of mutation-associated antigens is precisely the concept behind tumor mutation load (TML) and neoantigen load as ICI-predictive biomarkers. By the years 2014 and 2015, the first evidence suggested the role of TML and neoantigen load as anti-CTLA4 and anti-PD-1 clinical benefit predictors in melanoma and NSCLC, respectively.64–66 The very same year, in addition to documenting for the very first time a higher sensitivity of MSI-H tumors (mainly mCRC), in an exploratory analysis, Le et al.37 revealed a significant correlation between TML and tumor neoantigen load with PFS, and intratumoral CD8+ T cell density with response. Several years later, a high TML also was evaluated as a predictive biomarker for the nivolumab plus ipilimumab regimen in NSCLC.67,68 In the GI malignancies field, it is known that almost every different tumor type includes subtypes that, because of their high TML and inflamed tumor microenvironment (TME), are good candidates to try this kind of drugs.20,22,25,33,54–57 The largest study published to date, involving 14 types of GI cancers, describes the highest average TML and the larger prevalence of high TML cancers between right-sided CRC and small-bowel adenocarcinomas (SBAs). As expected, high TML strongly correlated with MSI-H, which in turn, associated with a higher PD-L1 expression. Between MSS tumors, squamous cell anal and esophageal cancers presented the highest rate of high TML.21 Focusing specifically on CRC, neoantigen load also is positively correlated with TILs, Crohn’s-14 like reaction, and CD45RO+ T cells (but not with other T-cell subtypes). Besides, there is an approximately 1.3% rate of CRC harboring POLE exonuclease domain mutations and even a small percentage of POLE-mutated MSI-H tumors with high and extremely high TML, respectively.69 In 2015, Schumacher et al.13 postulated that neoantigen production is a probabilistic and individual process, and despite the fact that higher TML increases the likelihood of creating a pertinent neoantigen, it does not always happen. The opposite, by chance, can occur between any of the low TML tumors. Besides, and regarding tumor neoantigen load, it is important to note the higher influence of clonal neoantigens in producing an effective antitumor immunity compared with subclonal neoantigens.70
With the advent of these two closely related concepts (TML and tumor neoantigen load), which has been possible thanks to the enormous development of NGS platforms and computational analytical methods, reaching a consensus to validate the different platforms and algorithms (including cutoff to categorize both variables) in every single clinical context is becoming a necessity.
ICI Resistance Mechanisms
The question whether MSI status is a robust predictor for the use of ICI therapy in GI malignancies begs further clarity. For instance, in the monotherapy (nivolumab) and combination regimen (nivolumab plus ipilimumab) cohorts of the mCRC phase 2 CheckMate142, results showed a lack of disease control in 30% and 20% of patients respectively, even though they were selected based upon a positive predictive biomarker (dMMR/MSI-H).40 The mechanisms underlying this lack of efficacy remain unclear; however, several potential explanations might be viable. Recently, Grasso et al.69 reported the results of a large-scale genomic analysis (TCGA, NHS, and HPFS cohorts) comprising of 1,211 primary CRC tumor specimens. Mutations in genes involved in relevant immune modulatory pathways, but also in the neoantigen-presentation machinery (mainly B2M and HLA genes), showed a significant correlation with MSI-H. Along with JAK1/2 and IFNγ-receptor 1 mutations, similar alterations have been observed in melanoma, NSCLC, and CRC and deemed to be as genetic drivers of primary or acquired resistance to ICI therapy, reflecting their role as a mechanism of adaptive resistance against T-cell tumor infiltration.14,69,71–73 On the other hand, in both MSS and MSI-H tumors, an active WNT/β-catenin signaling was inversely associated with tumor T-cell infiltration, providing evidence of the existence of an anti-immune response mechanism beyond the MSI profile.69 Lastly, and also with potential applicability to different GI cancers, there is evidence from the metastatic melanoma setting regarding the association between PTEN loss and PI3K–AKT pathway activation and the resistance to anti-PD-1 therapy.74
Gut Microbiome, Host Factors, and Clinical Benefit
The latest evidence suggests that in addition to molecular alterations and TME, host factors exert their own significant influence on the clinical success of the immune checkpoint blockade. During the past few years, the gut microbiome, with a recognized role in driving inflammation and modulating the intestinal immune system during CRC pathogenesis,75 has emerged as an important mediator associated with responsiveness to ICI therapy. Following the initial evidence in preclinical animal models for the key role in mediating anti-CTLA-4 and anti-PD-L1 tumor responses,76,77 the importance of certain intestinal commensals has been subsequently substantiated in humans. In metastatic melanoma, a high diversity of the gut microbiome and abundance of Faecalibacterium were associated with an increased likelihood of response and prolonged PFS with anti-PD-1 therapy.78 A similar association was described with the abundance of Akkermansia muciniphila in NSCLC and renal cell carcinoma.79 This positive effect seems to manifest through a systemic and tumoral modulation of the immune system driven by a favorable gut microbiome. Besides, fecal microbiota transplantation has shown promising data in mice, opening up a new horizon to obviate primary resistance to ICIs through manipulation of the intestinal microbiome.78,79 Highlighting the relevance of host germline genetics, the HLA class I diversity has been associated with a better overall survival in melanoma and NSCLC patients treated with anti-PD-1/PD-L1 and/or anti-CTLA-4.80 Finally, a strong association was found between a higher pre-anti-PD-1 therapy percentage of classical monocytes and improved survival in melanoma patients, establishing a promising minimally invasive biomarker to treatment selection.81
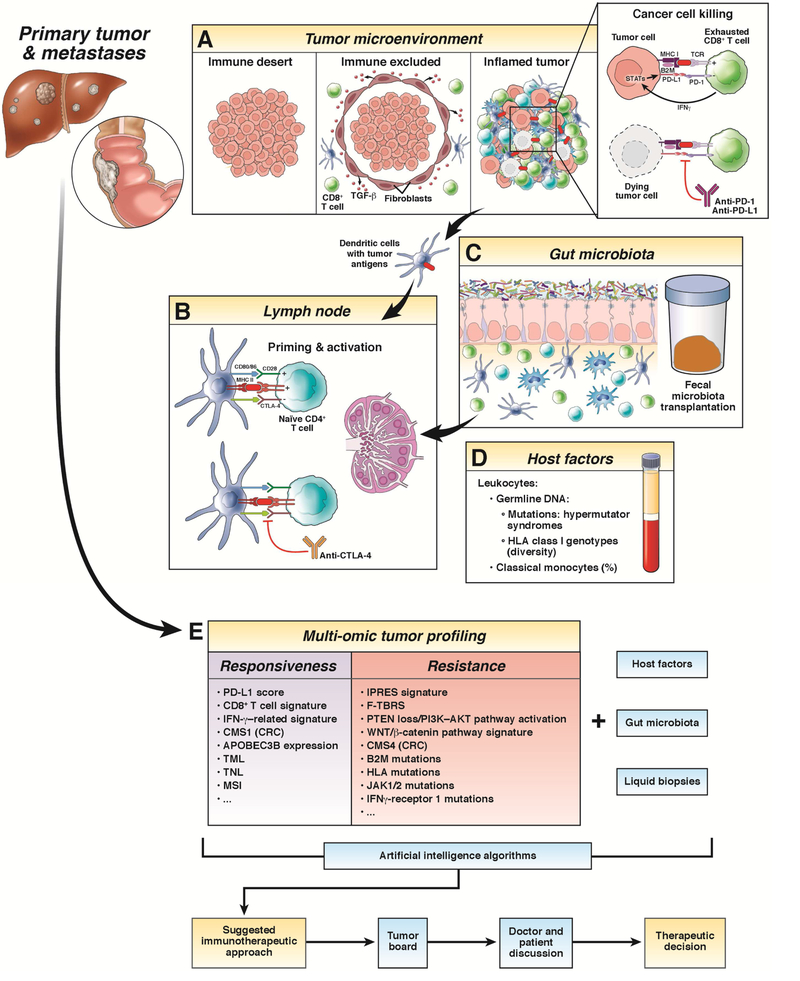
In view of the complexity of interplay between the immune system and cancer, considering only a single biomarker for selecting an immunotherapeutic strategy seems insufficient82. Following the ‘cancer immunogram’ approach, and considering the huge amount of data described previously, the integration of all factors involved in the success of immune checkpoint blockade using artificial intelligence algorithms will be a needed in near future (Fig. 2).
Figure 2:
This illustration provides a schematic and general overview of the different and most relevant mechanisms correlated with the efficacy of immune checkpoint inhibitors (ICIs). (a) There are three possible types of tumor microenvironment (TME) based on the presence of immune cells and their interplay with tumor cells: immune desert TME (characterized by the immunologic ignorance of tumor cells), immune excluded TME (characterized by the presence of immune cells in the tumor margin, due to the effect, among others, of TGF-β secreted by cancer-associated fibroblasts) and inflamed TME (characterized by the T-cell exhaustion in the context of immunosuppressive conditions). Expression of programmed death-ligand 1 (PD-L1) on the surface of tumor cells can be either constitutive by oncogenic activation (innate immune resistance) or induced by surrounding immune cells (adaptive immune resistance). Blockade of the programmed cell death 1 (PD-1)/PD-L1 interaction reverses T cell exhaustion and enables tumor cell killing. (b) Lymph nodes are where antigen-presenting cells, such as dendritic cells, participate in the priming and activation of naive or resting T cells. Cytotoxic Tlymphocyte antigen 4 (CTLA-4) expression is induced in the T cell surface at the time of the initial response to the tumor antigen (presented by the MHC II) to downregulate the response amplitude. CTLA-4 blockade interrupts its interaction with CD80 or CD86, avoiding attenuation of T cell stimulation. (c) The influence of different commensal bacteria of the intestinal microbiome (Faecalibacterium spp., A. muciniphilia, E. hirae, Bacteroides spp., and Bifidobacteria spp.) together with (d) several host factors, such as the percentage of classical monocytes or diversity in the HLA class I molecules, have been recently associated with ICI therapy outcomes. (e) Besides the multi-omic tumor profiling, which permits static evaluation of different molecular features associated with the efficacy of ICI, liquid biopsies can be considered a complement that offers spatial and temporal dynamic information. Integration of all these aspects using artificial intelligence algorithms, together with meticulous evaluation by a tumor board, and always considering patient’s desires, will help in the therapeutic decisionmaking process. Abbreviations: CMS, consensus molecular subtype; CRC, colorectal cancer; TML, tumor mutation load; TNL, tumor neoantigen load; MSI, microsatellite instability; IPRES, innate anti-PD-1 resistance; F-TBRS, pan-fibroblast TGF-β response signature.
From Bench to Bedside and Back Again
Understanding the mechanism(s) of immune evasion and resistance to ICIs are of paramount significance as we move forward with the development of immunotherapeutic approaches. Currently, ongoing trials are exploring a variety of treatment regimens by combining ICIs with chemotherapeutic agents, targeted therapies, epigenetic modulators, and even other immunomodulatory drugs (Supplementary Table S2). Furthermore, coordinated efforts are underway for evaluating bench-based discoveries into various clinical settings. A fascinating example in this regard would be the recognition that increased levels of the transforming growth factor-β (TGF-β) within the TME of MSS CRCs represent a primary mechanism of immune evasion, which can be circumvented by blocking TGF-β signaling. This hypothesis is supported by the findings that combined therapy with galunisertib (a potent TGF-β receptor I inhibitor) and an anti-PD-L1 drug resulted in significant tumor regression in mice, through activation of tumor immune response.83 On similar lines, another group reported in a large cohort of metastatic urothelial cancer patients treated with atezolizumab, a lack of response in those patients with an immune excluded TME with a high pan-fibroblast TGF-β response signature (F-TBRS).84 Furthermore, it was recently demonstrated that inactivation of the MMR system in CRC cells, either by genomic editing or through pharmacological manipulations, resulted in improved immune surveillance and enabled tumor regression.85 Such effects were even more pronounced when restricted to a clonal population, providing an opportunity to evaluate this strategy individually, or in combination with ICIs in the adjuvant setting.
Similarly, in mouse pancreatic cancer models, the blockade of FAK, as well as dual inhibition with an anti-PD-1 antibody, led to tumor size reduction and prolonged survival in mice through modulation of the TME.86 In a similar tumor model, dual blockade of IL-6 and PDL1 elicited a significantly superior tumor regression compared to inhibition with each of the molecules separately.87 Lastly, mouse ovarian cancer models demonstrated that the ARID1A deficiency compromises MMR, promoting additional mutation burden, and hence potentiating the antitumor immunity, which subsequently can be unleashed by an anti-PD-L1 drug.88 These findings could have a potential application to several GI malignancies with known inactivating mutations in ARID1A (HCC, CRC, esophageal and pancreatic adenocarcinomas, SBA, ampullary carcinoma, and cholangiocarcinoma).14,20,25,33,57,89–91
Using development of ICI therapy in dMMR tumors as a role model, and following the bench-to-bedside-and-back again philosophy, the preclinical and translational results described above, represent important first steps for the exploration of promising and innovative approaches in the near future.
Conclusions
The significant advances made with the use of ICIs in a relatively short duration are unquestionably unprecedented. Nonetheless, the benefit derived from these regimens is only to a small proportion of patients with GI malignancies. Despite the fact that MMR-deficiency is clearly associated with clinical benefit in a bunch of cases, the molecular mechanisms underlying therapeutic response and resistance to ICIs remain unclear in the majority of cases. Although analysis of well-annotated tumor specimens is key to gaining insights into the clinical efficacy of ICIs, an effective translation of preclinical discoveries to early clinical phases, appropriate biomarker development and standardization processes are essential in appropriate selection of candidates for ICI therapies. However, the future seems bright- and the combination of the latest ICIs along with classical and emerging therapeutic strategies, in conjunction with the adoptive T-cell therapies, augurs frenetic and encouraging discoveries on the horizon in this chess game against cancer.
Supplementary Material
Acknowledgments
JR-B would like to thank Isabel Bañobre-Landeira for her inspiration and support.
Funding: This work was supported by CA72851, CA184792, CA202797, CA187956 and CA214254 grants from the National Cancer Institute, NIH; RP140784 from the Cancer Prevention Research Institute of Texas; grants from the Baylor Foundation and Baylor Scott & White Research Institute, Dallas, TX, USA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Wang J, Reiss KA, Khatri R, Jaffee E, Laheru D. Immune Therapy in GI Malignancies: A Review. J Clin Oncol 2015;33(16):1745–1753. doi: 10.1200/JCO.2015.60.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Dietlein F, Thelen L, Reinhardt HC. Cancer-specific defects in DNA repair pathways as targets for personalized therapeutic approaches. Trends Genet. 2014;30(8):326–339. doi: 10.1016/j.tig.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-Repair Gene MGMT and the Clinical Response of Gliomas to Alkylating Agents. N Engl J Med. 2000;343(19):1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 5.Kunkel TA, Erie DA. DNA MISMATCH REPAIR. Annu Rev Biochem. 2005;74(1):681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 6.Boland CR, Goel A. Microsatellite Instability in Colorectal Cancer. Gastroenterology. 2010;138(6):2073–2087.e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiricny J The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335 Available at: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 8.Giardiello FM, Allen JI, Axilbund JE, et al. Guidelines on Genetic Evaluation and Management of Lynch Syndrome: A Consensus Statement by the US Multi-Society Task Force on Colorectal Cancer. Am J Gastroenterol. 2014;109:1159 Available at: 10.1038/ajg.2014.186. [DOI] [PubMed] [Google Scholar]
- 9.Knudson AG. Hereditary Cancer, Oncogenes, and Antioncogenes. Cancer Res. 1985;45(4):1437 LP – 1443. Available at: http://cancerres.aacrjournals.org/content/45/4/1437.abstract. [PubMed] [Google Scholar]
- 10.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell. 2017;168(4):707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipson EJ, Sharfman WH, Drake CG, et al. Durable Cancer Regression Off-Treatment and Effective Reinduction Therapy with an Anti-PD-1 Antibody. Clin Cancer Res. 2013;19(2):462 LP – 468. Available at: http://clincancerres.aacrjournals.org/content/19/2/462.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nebot-Bral L, Brandao D, Verlingue L, et al. Hypermutated tumours in the era of immunotherapy: The paradigm of personalised medicine. Eur J Cancer. 2017;84:290–303. doi: 10.1016/j.ejca.2017.07.026. [DOI] [PubMed] [Google Scholar]
- 13.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015; 348(6230):69 LP – 74. Available at: http://science.sciencemag.org/content/348/6230/69.abstract. [DOI] [PubMed] [Google Scholar]
- 14.Giannakis M, Mu XJ, Shukla SA, et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep. 2016;15(4):857–865. doi: 10.1016/j.celrep.2016.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Llosa NJ, Cruise M, Tam A, et al. The Vigorous Immune Microenvironment of Microsatellite Instable Colon Cancer Is Balanced by Multiple Counter-Inhibitory Checkpoints. Cancer Discov. 2015;5(1):43 LP – 51. Available at: http://cancerdiscovery.aacrjournals.org/content/5/1/43.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niu B, Ye K, Zhang Q, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2014;30(7):1015–1016. doi: 10.1093/bioinformatics/btt755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salipante SJ, Scroggins SM, Hampel HL, Turner EH, Pritchard CC. Microsatellite Instability Detection by Next Generation Sequencing. Clin Chem. 2014;60(9):1192 LP – 1199. Available at: http://clinchem.aaccjnls.org/content/60/9/1192.abstract. [DOI] [PubMed] [Google Scholar]
- 18.Kautto EA, Bonneville R, Miya J, et al. Performance evaluation for rapid detection of pan-cancer microsatellite instability with MANTIS. Oncotarget. 2017;8(5):7452–7463. doi: 10.18632/oncotarget.13918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farris AB 3rd, Demicco EG, Le LP, et al. Clinicopathologic and molecular profiles of microsatellite unstable Barrett Esophagus-associated adenocarcinoma. Am J Surg Pathol. 2011;35(5):647–655. doi: 10.1097/PAS.0b013e31820f18a2. [DOI] [PubMed] [Google Scholar]
- 20.Network TCGA. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169 Available at: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salem ME, Puccini A, Grothey A, et al. Landscape of Tumor Mutation Load, Mismatch Repair Deficiency, and PD-L1 Expression in a Large Patient Cohort of Gastrointestinal Cancers. Mol Cancer Res. 2018;16(5):805 LP – 812. Available at: http://mcr.aacrjournals.org/content/16/5/805.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Network TCGA. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202 Available at: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dudley JC, Lin M-T, Le DT, Eshleman JR. Microsatellite Instability as a Biomarker for PD1 Blockade. Clin Cancer Res. 2016;22(4):813 LP – 820. Available at: http://clincancerres.aacrjournals.org/content/22/4/813.abstract. [DOI] [PubMed] [Google Scholar]
- 24.Aparicio T, Svrcek M, Zaanan A, et al. Small bowel adenocarcinoma phenotyping, a clinicobiological prognostic study. Br J Cancer. 2013;109:3057 Available at: 10.1038/bjc.2013.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.AB S, CE D, McWilliams R, et al. Genomic profiling of small-bowel adenocarcinoma. JAMA Oncol. 2017;3(11):1546–1553. Available at: 10.1001/jamaoncol.2017.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiappini F, Gross-Goupil M, Saffroy R, et al. Microsatellite instability mutator phenotype in hepatocellular carcinoma in non-alcoholic and non-virally infected normal livers. Carcinogenesis. 2004;25(4):541–547. Available at: 10.1093/carcin/bgh035. [DOI] [PubMed] [Google Scholar]
- 27.Nakata B, Wang YQ, Yashiro M, et al. Prognostic value of microsatellite instability in resectable pancreatic cancer. Clin Cancer Res. 2002;8(8):2536–2540. [PubMed] [Google Scholar]
- 28.Humphris JL, Patch A-M, Nones K, et al. Hypermutation In Pancreatic Cancer. Gastroenterology. 2017;152(1):68–74.e2. doi: 10.1053/j.gastro.2016.09.060. [DOI] [PubMed] [Google Scholar]
- 29.Hu ZI, Shia J, Stadler ZK, et al. Evaluating Mismatch Repair Deficiency in Pancreatic Adenocarcinoma: Challenges and Recommendations. Clin Cancer Res. 2018. Available at: http://clincancerres.aacrjournals.org/content/early/2018/01/24/1078-0432.CCR-17-3099.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu D, Momoi H, Li L, Ishikawa Y, Fukumoto M. Microsatellite instability in thorotrastinduced human intrahepatic cholangiocarcinoma. Int J cancer. 2002;102(4):366–371. doi: 10.1002/ijc.10726. [DOI] [PubMed] [Google Scholar]
- 31.Moy AP, Shahid M, Ferrone CR, et al. Microsatellite instability in gallbladder carcinoma. Virchows Arch. 2015;466(4):393–402. doi: 10.1007/s00428-015-1720-0. [DOI] [PubMed] [Google Scholar]
- 32.Silva VWK, Askan G, Daniel TD, et al. Biliary carcinomas: pathology and the role of DNA mismatch repair deficiency. Chinese Clin Oncol. 2016;5(5):62. doi: 10.21037/cco.2016.10.04. [DOI] [PubMed] [Google Scholar]
- 33.Gingras M-C, Covington KR, Chang DK, et al. Ampullary Cancers Harbor ELF3 Tumor Suppressor Gene Mutations and Exhibit Frequent WNT Dysregulation. Cell Rep. 2016;14(4):907–919. doi: 10.1016/j.celrep.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saetta AA, Gigelou F, Papanastasiou PI, et al. High-level microsatellite instability is not involved in gallbladder carcinogenesis. Exp Mol Pathol. 2006;80(1):67–71. doi: 10.1016/J.YEXMP.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 35.Baumeister SH, Freeman GJ, Dranoff G, Sharpe AH. Coinhibitory Pathways in Immunotherapy for Cancer. Annu Rev Immunol. 2016;34(1):539–573. doi: 10.1146/annurev-immunol-032414-112049. [DOI] [PubMed] [Google Scholar]
- 36.US Food and Drug Administration. Pembrolizumab FDA Label. 2017. https://www.accessdata.fda.gov/drugsatfda_docs/label/2017/125514s024lbl.pdf. Accessed May 22, 2018.
- 37.Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Overman MJ, McDermott R, Leach JL, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18(9):1182–1191. doi: 10.1016/S1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.US Food and Drug Administration. Nivolumab FDA Label. 2017. https://www.accessdata.fda.gov/drugsatfda_docs/label/2017/125554s041lbl.pdf. Accessed May 22, 2018.
- 40.Overman MJ, Lonardi S, Wong KYM, et al. Durable Clinical Benefit With Nivolumab Plus Ipilimumab in DNA Mismatch Repair–Deficient/Microsatellite Instability–High Metastatic Colorectal Cancer. J Clin Oncol. 2018;36(8):773–779. doi: 10.1200/JCO.2017.76.9901. [DOI] [PubMed] [Google Scholar]
- 41.El-Khoueiry AB, Sangro B, Yau T, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389(10088):2492–2502. doi: 10.1016/S0140-6736(17)31046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stein S, Pishvaian MJ, Lee MS, et al. Safety and clinical activity of 1L atezolizumab + bevacizumab in a phase Ib study in hepatocellular carcinoma (HCC). J Clin Oncol. 2018;36 (suppl; abstr 4074). [Google Scholar]
- 43.Kang Y-K, Boku N, Satoh T, et al. Nivolumab in patients with advanced gastric or gastrooesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538–12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390(10111):2461–2471. doi: 10.1016/S0140-6736(17)31827-5. [DOI] [PubMed] [Google Scholar]
- 44.CS F, Doi T, RW J, et al. Safety and efficacy of pembrolizumab monotherapy in patients with previously treated advanced gastric and gastroesophageal junction cancer: Phase 2 clinical keynote-059 trial. JAMA Oncol. 2018;4(5):e180013 Available at: 10.1001/jamaoncol.2018.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shitara K, Özgüroğlu M, Bang Y-J, et al. Pembrolizumab versus paclitaxel for previously treated, advanced gastric or gastro-oesophageal junction cancer (KEYNOTE-061): a randomised, open-label, controlled, phase 3 trial. Lancet. 2018. doi: 10.1016/S01406736(18)31257-1. [DOI] [PubMed] [Google Scholar]
- 46.Kudo T, Hamamoto Y, Kato K, et al. Nivolumab treatment for oesophageal squamouscell carcinoma: an open-label, multicentre, phase 2 trial. Lancet Oncol. 2017;18(5):631–639. doi: 10.1016/S1470-2045(17)30181-X. [DOI] [PubMed] [Google Scholar]
- 47.Shah MA, Kojima T, Enzinger PC, et al. Pembrolizumab for patients with previously treated metastatic adenocarcinoma or squamous cell carcinoma of the esophagus: Phase 2 KEYNOTE-180 study. J Clin Oncol. 2018;36(15_suppl):4049. doi: 10.1200/JCO.2018.36.15_suppl.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.National Comprehensive Cancer Network. Anal Carcinoma (Version 2.2018). https://www.nccn.org/professionals/physician_gls/pdf/anal.pdf. Accessed May 22, 2018. [DOI] [PMC free article] [PubMed]
- 49.Prasad V, Kaestner V, Mailankody S. Cancer drugs approved based on biomarkers and not tumor type—fda approval of pembrolizumab for mismatch repair-deficient solid cancers. JAMA Oncol. 2018;4(2):157–158. Available at: 10.1001/jamaoncol.2017.4182. [DOI] [PubMed] [Google Scholar]
- 50.Büttner R, Gosney JR, Skov BG, et al. Programmed Death-Ligand 1 Immunohistochemistry Testing: A Review of Analytical Assays and Clinical Implementation in Non–Small-Cell Lung Cancer. J Clin Oncol. 2017;35(34):3867–3876. doi: 10.1200/JCO.2017.74.7642. [DOI] [PubMed] [Google Scholar]
- 51.Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ–related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hugo W, Zaretsky JM, Sun L, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2016;165(1):35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang S, Jia M, He Z, Liu X-S. APOBEC3B and APOBEC mutational signature as potential predictive markers for immunotherapy response in non-small cell lung cancer. Oncogene. 2018. doi: 10.1038/s41388-018-0245-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ally A, Balasundaram M, Carlsen R, et al. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017;169(7):1327–1341.e23. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47 Available at: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 57.Farshidfar F, Zheng S, Gingras M-C, et al. Integrative Genomic Analysis of Cholangiocarcinoma Identifies Distinct IDH-Mutant Molecular Profiles. Cell Rep. 2017;18(11):2780–2794. doi: 10.1016/j.celrep.2017.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morris VK, Salem ME, Nimeiri H, et al. Nivolumab for previously treated unresectable metastatic anal cancer (NCI9673): a multicentre, single-arm, phase 2 study. Lancet Oncol. 2017;18(4):446–453. doi: 10.1016/S1470-2045(17)30104-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Makarova-Rusher OV, Medina-Echeverz J, Duffy AG, Greten TF. The yin and yang of evasion and immune activation in HCC. J Hepatol. 2015;62(6):1420–1429. doi: 10.1016/j.jhep.2015.02.038. [DOI] [PubMed] [Google Scholar]
- 60.Seiwert TY, Burtness B, Mehra R, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol. 2016;17(7):956–965. doi: 10.1016/S1470-2045(16)30066-3. [DOI] [PubMed] [Google Scholar]
- 61.Ferris RL, Blumenschein G, Fayette J, et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N Engl J Med. 2016;375(19):1856–1867. doi: 10.1056/NEJMoa1602252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ott PA, Piha-Paul SA, Munster P, et al. Safety and antitumor activity of the anti-PD-1 antibody pembrolizumab in patients with recurrent carcinoma of the anal canal. Ann Oncol. 2017;28(5):1036–1041. Available at: 10.1093/annonc/mdx029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim ST, Cristescu R, Bass AJ, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med. 2018. doi: 10.1038/s41591-018-0101-z. [DOI] [PubMed] [Google Scholar]
- 64.Snyder A, Makarov V, Merghoub T, et al. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. N Engl J Med. 2014;371(23):2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207 LP – 211. Available at: http://science.sciencemag.org/content/350/6257/207.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hellmann MD, Nathanson T, Rizvi H, et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell. 2018;33(5):843–852.e4. doi: 10.1016/j.ccell.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hellmann MD, Ciuleanu T-E, Pluzanski A, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med. 2018. doi: 10.1056/NEJMoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grasso CS, Giannakis M, Wells DK, et al. Genetic mechanisms of immune evasion in colorectal cancer. Cancer Discov. 2018. Available at: http://cancerdiscovery.aacrjournals.org/content/early/2018/03/06/2159-8290.CD-171327.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McGranahan N, Furness AJS, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016. Available at: http://science.sciencemag.org/content/early/2016/03/02/science.aaf1490.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gao J, Shi LZ, Zhao H, et al. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell. 2016;167(2):397–404.e9. doi: 10.1016/j.cell.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zaretsky JM, Garcia-Diaz A, Shin DS, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med. 2016;375(9):819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peng W, Chen JQ, Liu C, et al. Loss of PTEN Promotes Resistance to T Cell–Mediated Immunotherapy. Cancer Discov. 2016;6(2):202 LP – 216. Available at: http://cancerdiscovery.aacrjournals.org/content/6/2/202.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tilg H, Adolph TE, Gerner RR, Moschen AR. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell. 2018. doi: 10.1016/j.ccell.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 76.Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti–PD-L1 efficacy. Science. 2015;350(6264):1084 LP – 1089. Available at: http://science.sciencemag.org/content/350/6264/1084.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vétizou M, Pitt JM, Daillère R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079 LP – 1084. Available at: http://science.sciencemag.org/content/350/6264/1079.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science. 2018;359(6371):97 LP – 103. Available at: http://science.sciencemag.org/content/359/6371/97.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1– based immunotherapy against epithelial tumors. Science. 2018;359(6371):91 LP – 97. Available at: http://science.sciencemag.org/content/359/6371/91.abstract. [DOI] [PubMed] [Google Scholar]
- 80.Chowell D, Morris LGT, Grigg CM, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science. 2018;359(6375):582 LP – 587. Available at: http://science.sciencemag.org/content/359/6375/582.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krieg C, Nowicka M, Guglietta S, et al. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat Med. 2018;24:144 Available at: 10.1038/nm.4466. [DOI] [PubMed] [Google Scholar]
- 82.Blank CU, Haanen JB, Ribas A, Schumacher TN. The “cancer immunogram.” Science. 2016;352(6286):658 LP – 660. Available at: http://science.sciencemag.org/content/352/6286/658.abstract. [DOI] [PubMed] [Google Scholar]
- 83.Tauriello DVF, Palomo-Ponce S, Stork D, et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 2018;554:538 Available at: 10.1038/nature25492. [DOI] [PubMed] [Google Scholar]
- 84.Mariathasan S, Turley SJ, Nickles D, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544 Available at: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Germano G, Lamba S, Rospo G, et al. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature. 2017;552:116 Available at: 10.1038/nature24673. [DOI] [PubMed] [Google Scholar]
- 86.Jiang H, Hegde S, Knolhoff BL, et al. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat Med. 2016;22:851 Available at: 10.1038/nm.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mace TA, Shakya R, Pitarresi JR, et al. IL-6 and PD-L1 antibody blockade combination therapy reduces tumour progression in murine models of pancreatic cancer. Gut. 2018;67(2):320 LP – 332. Available at: http://gut.bmj.com/content/67/2/320.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shen J, Ju Z, Zhao W, et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat Med. 2018;24(5):556–562. doi: 10.1038/s41591-018-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zucman-Rossi J, Villanueva A, Nault J-C, Llovet JM. Genetic Landscape and Biomarkers of Hepatocellular Carcinoma. Gastroenterology. 2015;149(5):1226–1239.e4. doi: 10.1053/j.gastro.2015.05.061. [DOI] [PubMed] [Google Scholar]
- 90.Network TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330 Available at: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Raphael BJ, Hruban RH, Aguirre AJ, et al. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017;32(2):185–203.e13. doi: 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.