Figure 1. Intestinal RNA-seq analysis in WT and SHP-knockout mice.
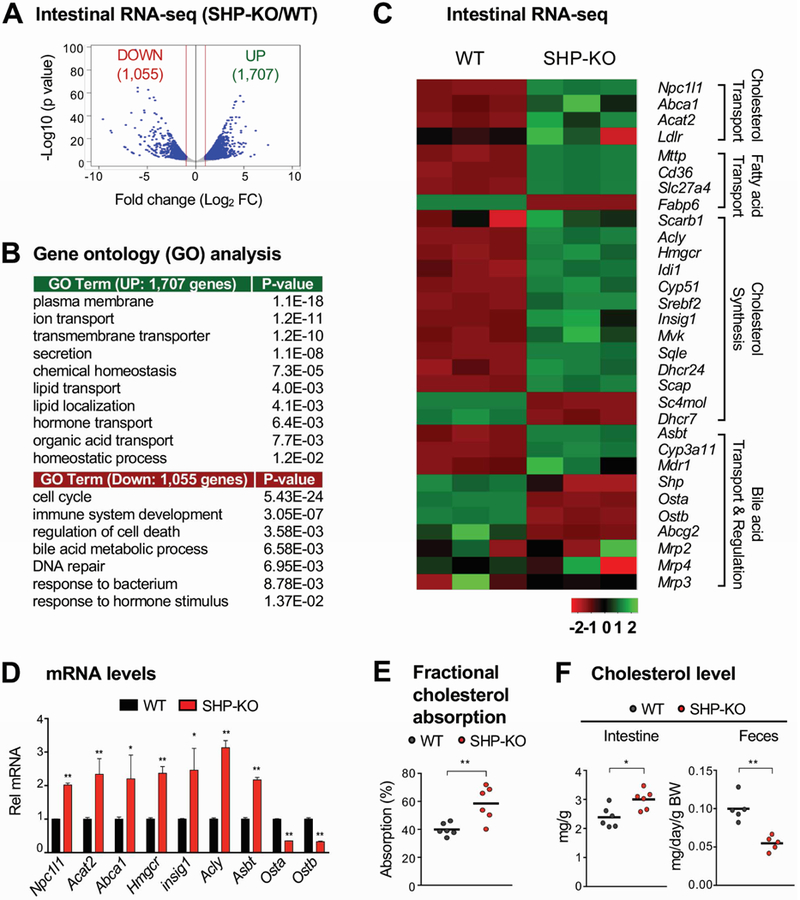
(A-C) WT (C57BL/6) and SHP-knockout (KO) mice were fed normal chow for 6 h after fasting overnight. Total RNA was isolated from the jejunum and ileum and analyzed by RNA-seq (n=3 mice). (A) Genome-wide changes in mRNA expression shown in a volcano plot. The numbers refer to the number of genes up- or down-regulated by 2-fold or more with a p-value < 0.01. (B) Gene ontology analysis of biological pathways using DAVID Tools (v 6.7). (C) Heatmap of the differential expression in three WT or SHP-knockout mice of selected genes in the functional categories indicated at the right. Normalized log2-counts-per-million (logCPM) gene counts are centered and z-scaled. (D) Levels of the mRNAs of the indicated genes were measured by RT-qPCR. (E-F) WT and SHP-knockout mice were individually caged, fasted overnight, and refed normal chow for 24 h. (E) [14C]cholesterol and [3H]sitosterol were administered by gavage at the time of refeeding. After 24 h, feces were collected and radioactivity in the feces was determined by scintillation counting and percent absorption was calculated as described in Methods. (F) Feces were collected 24 h after refeeding and cholesterol levels in the intestine and feces were measured and normalized to intestine weight or body weight, respectively. (D, E, F) Statistical significance was determined by the Student’s t-test, (SEM, n=3 to 6, *P <0.05, **P <0.01).
