Abstract
Background
Toll-like receptor 1 (TLR1) and TLR6 play important roles in the innate immune response against Mycobacterium tuberculosis (M.TB) via interactions with TIR domain-containing adaptor protein (TIRAP) and myeloid differentiation primary response 88 (MYD88). The aim of this study was to investigate the relationship of TLR1, TLR6, MYD88 and TIRAP polymorphisms with susceptibility to latent tuberculosis infection (LTBI) and tuberculosis (TB).
Methods
In total, 204 uninfected healthy controls (HC), 201 individuals with LTBI and 209 TB patients were enrolled. Two interferon-γ release assays were used to differentiate individuals with LTBI from uninfected controls. TagSNPs of the four genes were genotyped by the SNPscanTM Kit. The Haploview 4.2 and SHEsis software packages were combined to perform linkage disequilibrium (LD) and haplotype analyses. Multifactor dimensionality reduction (MDR) software was used to investigate gene-gene interaction. The Stata 12.0 software was used to perform meta-analysis of the relationship between rs5743557 and TB susceptibility.
Results
The AA genotype of rs5743557 was associated with reduced TB risk (P=0.006) and the AA/GA genotypes of TLR1 rs5743604 were associated with increased TB risk (P=0.017) when the LTBI group was compared with the TB group. The frequency of TLR1 haplotype rs4833095-rs5743604 CG was significantly higher in the LTBI group than in the TB group (P=0.019877). However, only the relationship between rs5743557 and TB susceptibility remained significant after 1000-fold permutation testing (P=0.023). The meta-analysis suggested that rs5743557_A was associated with decreased TB risk in the Chinese adult population (P<0.001, OR 0.80, 95% CI: 0.72–0.88). No significant gene-gene interactions were found.
Conclusions
The results of our study suggest that the tagSNP rs5743557 of TLR1 is associated with the risk of TB.
Keywords: Myeloid differentiation factor 88, Toll-like receptor 1 (TLR1), Toll-like receptor 6 (TLR6), TIR domain-containing adaptor protein (TIRAP), tuberculosis (TB)
Introduction
According to the 2016 Global Tuberculosis Report, there were an estimated 10.4 million new tuberculosis (TB) cases and 1.8 million TB deaths worldwide in 2015 (1). Although one third of the worldwide population has been infected with Mycobacterium tuberculosis (M.TB) (2), only approximately 5–10% of those infected develop clinical TB in their lifetime (3,4). In China, the TB infection rate was as high as 31.4% and there were 420 million individuals with latent tuberculosis infection (LTBI), as described in the 2000 National Epidemiological Sampling Survey of Tuberculosis (5).
Toll-like receptor 1 (TLR1) and Toll-like receptor 6 (TLR6) are members of the TLR2 subfamily (6) and play important roles in the innate immune response against M.TB by interacting with myeloid differentiation primary response 88 (MYD88) and TIR domain-containing adaptor protein (TIRAP). Variants in four genes (TLR1, TLR6, MYD88 and TIRAP) were reported to be associated with TB risk and to affect the function of their respective genes. The A allele of rs4833095 in TLR1 was shown to decrease susceptibility to TB in the Indian population, to increase the TNF response to M.TB lysates in mononuclear cells and to increase NF-κB expression in the HEK cell line (7). The T allele of rs5743810 in TLR6 was shown to decrease NF-κB signaling activity when HEK293 cells were stimulated by the di-acylated lipopeptide PAM2 and by M.TB lysates (8). The AG genotype of rs6853 in MYD88 was associated with reduced risk of pulmonary tuberculosis (PTB) and the allele A increased production of TNF-α, IFN-γ and NO (9). Heterozygosity for rs8177374 (S180L) in TIRAP was associated with reduced TB susceptibility (9) and the mutation reduced the affinity of TIRAP protein for the interferon-γ receptor and influenced responses to TB (10).
Results from numerous genetic association studies focused on TB risk have been inconsistent. Possible reasons for this lack of reproducibility include different phenotype definitions for TB cases and controls, differences in allele distribution and linkage disequilibrium (LD) among ethnic groups, varieties of M.TB strains, as well as complex gene-gene and gene-environment interactions (11,12).
Moreover, a meta-analysis of 16 published studies identified a 380 gene meta-signature which was uniquely expressed in active TB patients in nine or more datasets (13). In addition, a study revealed that Toll-like receptor-associated genes including MYD88 and TLR6 were differentially expressed in TB and LTBI patients (14). Therefore, we hypothesized that genetic and immunologic status are different between LTBI and TB groups and the Toll-like receptor related genes are involved in the pathogenesis of active TB. Furthermore, separating uninfected healthy controls from LTBI individuals might help to reduce discrepancies and further our understanding of the genetic determinants in TB and LTBI susceptibility.
A review for U.S. Preventive Services Task Force demonstrated that the T-SPOT.TB and QuantiFERON TB-GIT tests had higher sensitivity and specificity than the tuberculin skin test (TST) for identifying TB infection (15). Despite the difference in sensitivity between the two interferon-γ release assays (IGRAs), QuantiFERON TB-GIT assay and T-SPOT.TB, they are both widely used to recognize LTBI. Compared with the TST, IGRAs could differentiate TB infection from most nontuberculous mycobacterial (NTM) infection. In addition, Bacille Calmette-Guerin vaccination was common in China and therefore we utilized both IGRAs in this study to discriminate individuals with LTBI from uninfected controls in order to explore the relationship of tagSNPs of four genes (TLR1, TLR6, MYD88 and TIRAP) with susceptibility to LTBI and TB. In addition, a meta-analysis of the relationship between rs5743557 and TB risk was performed based on recently published reports.
Methods
Study population
Our study recruited 614 unrelated Han volunteers, including 209 TB patients, 201 LTBI subjects and 204 uninfected healthy controls (HC) from 2014 June to 2015 December in the West China Hospital, Chengdu, Sichuan. The confirmed active TB patients were identified based on syndromes, signs, computed tomography (CT) image, detection of acid-fast bacilli (AFB) and TB-DNA in samples such as sputum, biopsy tissues or bronchoalveolar lavage fluid and culture results as described in the diagnosis for PTB in China (16). In addition to culture, the finding of TB-DNA offers strong evidence to confirm TB infection instead of NTM infection. According to the Centers for Disease Control, patients with both positive Nucleic acid amplification test (NAAT) and AFB smear results, or with two or more specimens yielding positive NAAT results can be presumed to have TB (17). Moreover, patients with typical CT imaging and positive TB-DNA results can be diagnosed as confirmed TB in China (16).
Both LTBI and HC participants were chosen from close contacts of bacteria-confirmed PTB patients. All LTBI and HC had neither TB history nor signs indicative of TB according to their chest X-ray results and symptoms, and participants diagnosed with TB during one-year follow up were excluded. LTBI was defined as a state of persistent immune response to M.TB antigens without evidence of clinically active TB (18). Provided that the negative and positive controls were appropriate, the IGRA result was considered positive if the IFN-γ value was >14 pg/mL after subtracting the value of the negative control in the QuantiFERON TB-GIT test (Wantai Biological Pharmaceutical Co., Beijing, China), or if the number of spots was ≥16 in the T-SPOT.TB test. Participants with positive IGRA results were identified as LTBI and participants with negative IGRA results were included as HC. Individuals with HIV infection, primary immune deficiency, autoimmune diseases, diabetes mellitus, malignant tumors, or being treated with immunosuppressive medications were excluded.
All study protocols were approved by the Ethics Committee of the West China Hospital of Sichuan University in China, as shown in File No. 131, approved in 2013. A 3 mL venous blood sample was obtained from each volunteer after acquiring informed consent. The study outcomes will never affect the future management of the patients.
SNP selection and genotyping
Thirteen tagSNPs were selected as previously described (19), including four SNPs in TLR1 (rs4833095, rs5743557, rs5743596 and rs5743604), four in TLR6 (rs1039559, rs3775073, rs5743808 and rs5743827), two in MYD88 (rs6853, rs7744) and three in TIRAP (rs595209, rs8177375 and rs3802813). Genomic DNA was extracted from whole blood using AxyPrep genomic DNA Mini kits (Axygen, USA). All SNPs were genotyped by custom-by-design 2x48-Plex SNPscanTM Kit (Cat#:G0104, Genesky Biotechnologies Inc., Shanghai, China). In addition, 5% duplicate samples were tested to evaluate genotyping repeatability and quality.
Meta-analysis of association between rs5743557 and TB susceptibility
The research strategy was the same as in our previously published report (20). Only case-control studies focusing on association between rs5743557 and TB susceptibility were included. After evaluating the Newcastle Ottawa Scale (NOS) of each included study, information including first author, publication year, population, sample size of the cases and controls, HIV status, Hardy-Weinberg equilibrium (HWE), allele frequency of cases and controls, OR and 95%CI was abstracted. Studies were excluded if the genotype distribution deviated from HWE or the NOS scores were <4.
Statistical analysis
The chi-squared test, non-paramateric test and binary logistic regression were performed using the Statistical Package for Social Sciences version 17.0 software (SPSS Inc., Chicago, IL, USA) for analysis of demographic characteristics and association under three genetic models (dominant, recessive and additive). It is well known that the Bonferroni correction is too conservative as it fails to take correlation among SNPs into account, which might lead to a high false negative rate. Therefore, we applied the permutation test instead, which is considered to be the gold standard of multiple testing correction in genome-wide association studies (21), to adjust the results in our study. Both the HWE and permutation test were calculated using the Plink 1.90 software (Shaun Purcell, Christopher Chang, www.cog-genomics.org/plink/1.9/) (22). Haploview 4.2 and the online software SHEsis were combined to perform LD and haplotype analysis, as well as permutation correction of the haplotype results (23,24). Nonparametric multifactor dimensionality reduction (MDR) was used to investigate gene-gene interactions. Statistical power was calculated with the PS: Power and Simple Size Calculation version 3.1.2 software (Dupont WD, Plummer WD, Nashville, Tennessee of USA, http://biostat.mc.vanderbilt.edu/). A significant P value was considered as a two-sided P<0.05. The Stata 12.0 software was used to accomplish the meta-analysis. We used a fixed effects model when the heterogeneity results showed P>0.1 and I2 <50%, otherwise a random effects model was applied. In addition, subgroup analysis was performed.
Results
Meta-analysis results of relationship between rs5743557 and TB risk
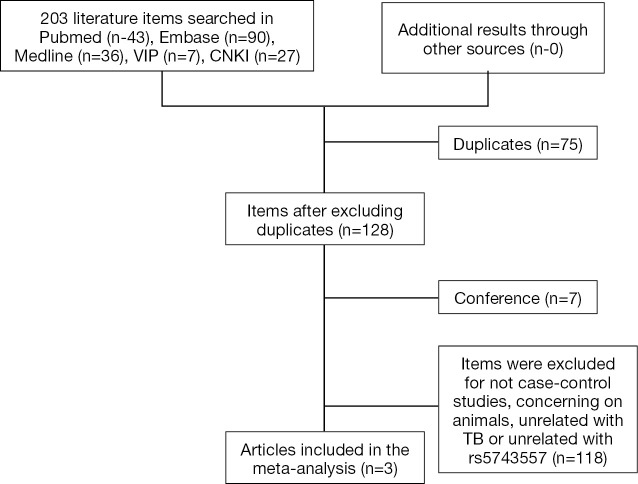
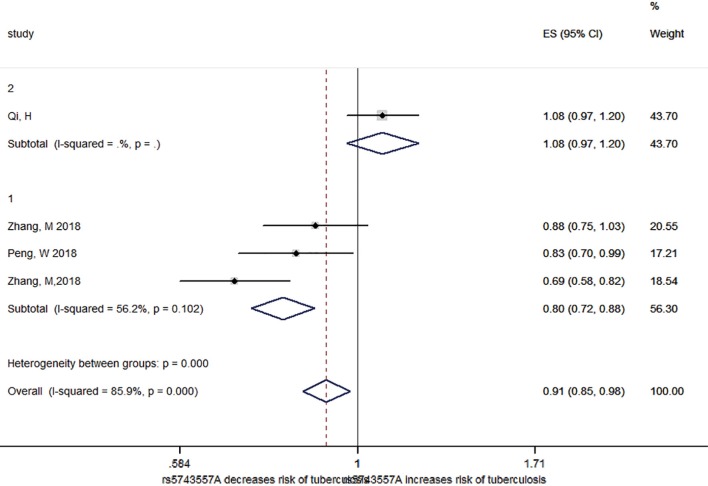
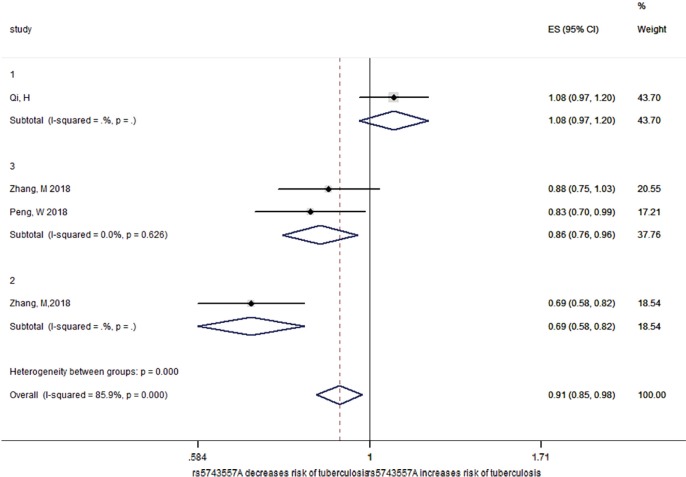
As shown in the flow diagram (Figure S1), only three reports including 2,179 TB patients and 2,049 healthy controls were enrolled (20,25,26). These reports contained of four studies, two of Chinese Han adults, one of Chinese Tibetan adults and one of Chinese Han children (Table S1). As demonstrated in Figure 1 (20,25,26), there was high degree of heterogeneity among the included studies (P<0.001 and I2=85.9%). After stratifying by age, no significant heterogeneity was observed (P=0.102), and rs5743557_A was associated with reduced TB risk and the odds ratio was 0.80 in the Chinese adult group (P<0.001, 95% CI: 0.72–0.88). As shown in Figure 2, when the Chinese adult population was grouped by Han and Tibetan nationality, no significant heterogeneity was observed (P=0.626, I2=0%)and the relationship between rs574355_ A and TB risk in Chinese Han adults was still present (P=0.01, OR 0.86, 95% CI: 0.76–0.96).
Figure S1.
Flow diagram of the meta-analysis.
Table S1. Data abstracted from studies included in the meta-analysis.
| First author | Year | Population | NOS | TB, n | HC, n | HIV | HWE | 0 (HC/TB) | 1 (HC/TB) | P | OR | 00 (HC/TB) | 01 (HC/TB) | 11 (HC/TB) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Zhang M | 2018 | Chinese Han adult | 7 | 580 | 605 | Negative | Yes | HC | 640/651 | 570/509 | 0.111 | 0.88 (0.75–1.03) | 175/187 | 290/277 | 140/116 |
| Zhang M | 2018 | Chinese Tibetan adult | 7 | 613 | 603 | Negative | Yes | HC | 767/881 | 437/345 | <0.001 | 0.69 (0.58–0.81) | 254/315 | 259/251 | 89/47 |
| Peng W | 2018 | Chinese Han adult | 7 | 646 | 475 | Negative | Yes | HC | 760/516 | 532/434 | 0.033 | 0.83 (0.70–0.99) | 230/134 | 300/248 | 116/93 |
| Qi, H | 2015 | Chinese Han children | 7 | 340 | 366 | Negative | Yes | HC | 365/314 | 367/366 | 94/81 | 177/152 | 95/107 |
TB, tuberculosis; HC, healthy controls; HIV, human immunodeficiency virus; HWE, Hardy-Weinberg equilibrium; 0 represents the major allele, 1 represents the minor allele; OR, odds ratio; CI, confidence interval; 00 represents the homozygote for the major allele, 01 represents the heterozygote, 11 represents the homozygote for the minor allele.
Figure 1.
Forest plot for the subgroup analysis of the association between rs5743557 and susceptibility to tuberculosis by age. 1 represents adults, 2 represents children. Each study cohort is represented by the name of the first author and publication year. The point estimate of the OR and 95% CI for each comparison are shown; the pooled OR and 95%CIs were derived from random-effects models. The I2 and P values are the heterogeneity test results. OR, odds ratio; CI, confidence interval.
Figure 2.
Forest plot for the subgroup analysis of the association between rs5743557 and susceptibility to tuberculosis by nationality. 1 represents Chinese Han children, 2 represents Chinese Tibetan adults, 3 represents Chinese Han adults. Each study cohort is represented by the name of the first author and publication year. The point estimate of the OR and 95% CI for each comparison are shown; the pooled OR and 95%CIs were derived from random-effects models. The I2 and P values are the heterogeneity test results. OR, odds ratio; CI, confidence interval.
Demographic characteristics
The demographic characteristics of all participants are summarized in Table 1. The LTBI group was older than the HC (P=0.027) and TB (P<0.001) groups. No significant difference was found in gender distribution. All uninfected controls were either negative for the QuantiFERON TB-GIT assay (n=180) or for the T-SPOT.TB test (n=24). All LTBI individuals were either positive for the QuantiFERON TB-GIT assay (n=180) or for the T-SPOT.TB test (n=21). As shown in Table 1, 51 (24.4%) active TB patients were culture positive, 64 (30.6%) had AFB positive biopsy results, 134 (64.1%) were positive for TB-DNA detection and 176 (84.2%) patients were found to be AFB positive in specimens. In addition, 142 (67.9%) active TB patients were either positive for TB culture or TB-DNA detection. According to a meta-analysis, the prevalence of NTM infections among suspected TB patients was 6.3% (5.4–7.4%) in mainland China and was 6.2% (4.6–8.2%) in the South-west region of China (27). Thus, NTM likely contributed little to our study results. Among all TB patients, 103 cases had satisfactory IGRAs with 90 positive for the QuantiFERON TB-GIT assay and two positive for the T-SPOT.TB test. The remaining 11 cases were negative for the QuantiFERON TB-GIT assay and the negative rate (10.7%) was comparable to that from a previous large epidemiological study (28).
Table 1. Demographic characteristics of the study populations.
| Characteristic | HC, n=204, n (%) | LTBI, n=201, n (%) | TB, n=209, n (%) | HC vs. LTBI, P value | LTBI vs. TB, P value |
|---|---|---|---|---|---|
| Age (mean ± IQR), year | HC vs. LTBI: 47.39±22.5; LTBI vs. TB: 43.84±29.5 | 0.027 | <0.001 | ||
| Gender | |||||
| Male | 93 (46.0) | 95 (47.0) | 107 (51.0) | ||
| Female | 111 (54.0) | 106 (53.0) | 102 (49.0) | 0.735 | 0.426 |
| TB culture positive, n (%) | 51 (24.4) | ||||
| TB-DNA positive, n (%) | 134 (64.1) | ||||
| Biopsy with positive acid-fast bacilli, n (%) | 64 (30.6) | ||||
| Acid-fast bacilli positive, n (%) | 176 (84.2) | ||||
| TB culture or TB-DNA positive, n (%) | 142 (67.9) | ||||
| QuantiFERON TB-GIT assay/T-SPOT.TB positive, n (%) | 180 (90.0)/21(10.0) | 90 (87.4)/2 (2.0) | |||
| QuantiFERON TB-GIT assay/T-SPOT.TB negative, n (%) | 180 (88.2)/24 (11.8) | 11 (10.7)/0 (0) | |||
HC, healthy controls; LTBI, latent tuberculosis infection; TB, tuberculosis; IQR, interquartile range
LD structure of all tagSNPs
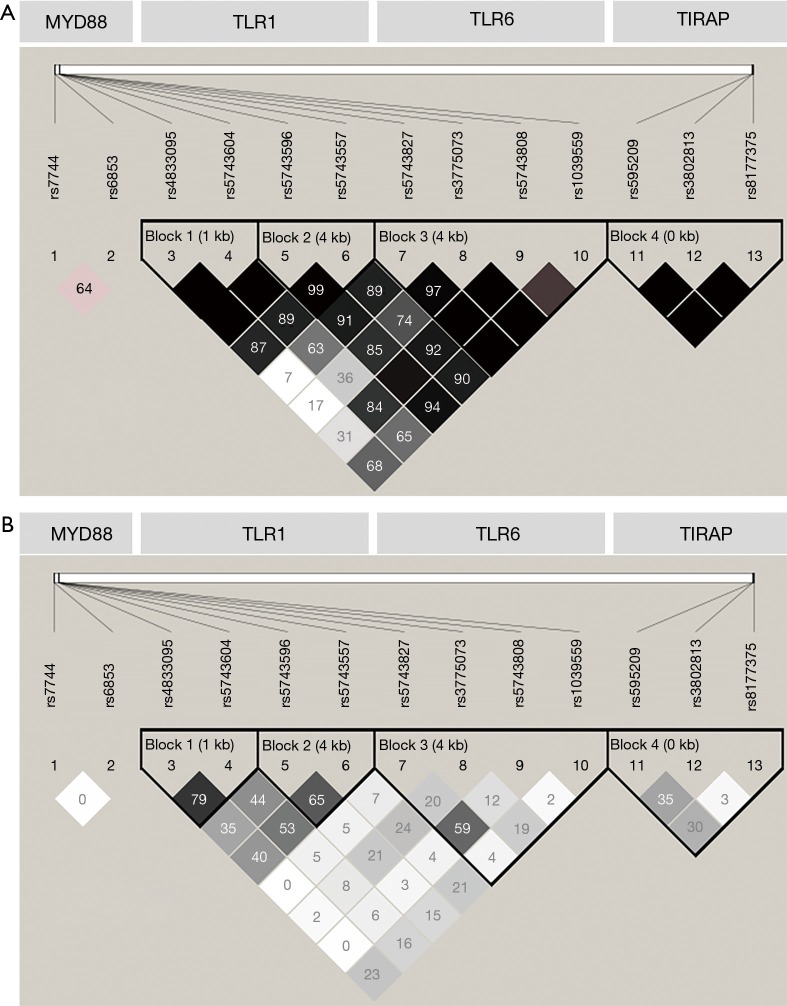
As shown in Table S2, the genotype distributions of rs4833095, rs5743557 and rs1039559 were not in HWE in the HC group. The genotype call rate of all SNPs was 99.79% and the consistency of the 5% repeat genotyping was 99.81%. Displayed in Figure 3A,B, two SNP pairs (rs4833095-rs5743604 and rs5743596-rs5743557) of TLR1 were in strong LD with r2≥0.65 and D’ approaching to 1. LD between SNP pairs of TLR6 and TIRAP were low (r2<0.36), and the two tagSNPs of MYD88 were not in LD at all (r2=0).
Table S2. Power of each tagSNP calculated by the PS: Power and Simple Size Calculation software and Hardy-Weinberg equilibrium (HWE) calculated with the Plink software.
| Gene | SNP | Power | HWE P value | ||||
|---|---|---|---|---|---|---|---|
| LTBI vs. HC | TB vs. LTBI | HC | LTBI | TB | |||
| TLR1 | rs4833095 | 0.778 | 0.518 | 0.034 | 0.230 | 1 | |
| rs5743557 | 0.66 | 0.789 | 0.030 | 0.477 | 0.021 | ||
| rs5743596 | 0.225 | 0.518 | 0.266 | 0.349 | 0.242 | ||
| rs5743604 | 0.54 | 0.762 | 0.092 | 0.667 | 0.166 | ||
| TLR6 | rs1039559 | 0.35 | 0.2 | 0.035 | 0.556 | 0.218 | |
| rs3775073 | 0.132 | 0.053 | 0.649 | 1 | 0.244 | ||
| rs5743808 | Very low | 0.156 | 0.066 | 0.242 | 0.371 | ||
| rs5743827 | 0.061 | 0.115 | 0.492 | 1 | 0.140 | ||
| TIRAP | rs3802813 | 0.124 | 0.414 | 0.612 | 1 | 1 | |
| rs595209 | 0.088 | 0.243 | 0.646 | 0.063 | 1 | ||
| rs8177375 | 0.05 | 0.135 | 0.382 | 0.773 | 0.322 | ||
| MYD88 | rs6853 | 0.07 | Very low | 1 | 1 | 1 | |
| rs7744 | 0.087 | 0.171 | 0.301 | 0.465 | 1 | ||
P<0.05 was significant for deviations from HWE. HC, healthy controls; LTBI, latent tuberculosis infection; TB, tuberculosis; HWE, Hardy-Weinberg equilibrium.
Figure 3.
Linkage disequilibrium (LD) of all tagSNPs of four genes. (A). Linkage disequilibrium (LD) of all tagSNPs of four genes displayed in form of D’. Each colored cell is related to the strength of LD between the corresponding two markers. The number in each cell represents the LD parameter D’ (×100), an empty cell indicates D’ (×100) =100. rs7744 and rs6853 are located in MYD88; rs4833095, rs5743604, rs5743596, and rs5743557 are located in TLR1; rs5743827, rs3775073, rs5743808 and rs1039559 are located in TLR6; rs595209, rs3802813 and rs8177375 are located in TIRAP. (B) LD of all tagSNPs of four genes displayed in form of r2 (×100). Each colored cell is related to the strength of LD between the corresponding two markers. The number in each cell represents the LD parameter r2 (×100). All SNPs are displayed as in (A).
Gene-gene interactions between four genes using MDR analysis
The MDR model was applied to screen for interactions among the thirteen SNPs in the four genes on LTBI and TB risk. Gene-gene interactions results of the MDR analysis are shown in Table S3. Between the HC and LTBI groups, rs4833095 of TLR1 formed the best model with 53.14% testing balanced accuracy and 10/10 cross-validation consistency and there was no gene-gene interaction. Between the LTBI and TB groups, rs5743604 of TLR1, rs1039559 of TLR6 and rs7744 of MYD88 formed the best model with 53.48% testing balanced accuracy and 7/10 cross-validation consistency. However, after 1000-fold permutation testing, no significant gene-gene interaction was found.
Table S3. Gene-gene interactions between TLR1, TLR6, MYD88 and TIRAP using MDR analysis.
| Group | Best model | Testing accuracy | CV consistency | P value* |
|---|---|---|---|---|
| HC vs. LTBI | rs4833095 | 0.5314 | 10/10 | 0.210–0.211 |
| LTBI vs. TB | rs5743604, rs1039559, rs7744 | 0.5348 | 7/10 | 0.428–0.429 |
*, P value of each best model was calculated after a 1,000-fold permutation test, applying testing accuracy and CV consistency in the MDR permutation test module. P<0.05 was significant. Testing accuracy: the ratio of correct classifications to the total classifications in the testing set; CV consistency (cross validation consistency): the number of times a combination model was selected as the best model in a 10-fold cross validation. HC, uninfected healthy controls; LTBI, latent tuberculosis infection; TB, tuberculosis.
Associations between SNPs and LTBI or TB susceptibility
To investigate the relationship between the tagSNPs and susceptibility to LTBI and TB, we compared the HC group with the LTBI group and the LTBI group with TB group. As shown in Table 2, when comparing the HC group and LTBI group, TT /TC genotypes of rs4833095 and AA/GA genotypes of rs5743604 in TLR1 were associated with reduced susceptibility to TB infection (P=0.007 and P=0.034), while genotype AA of rs5743557 in TLR1was associated with increased risk of TB infection (P=0.012). In addition, when comparing the LTBI group and TB group, the AA genotype of both rs5743596 and rs5743557 was associated with decreased TB risk (P=0.036 and P=0.006), while the AA/GA genotypes of rs5743604 were associated with increased susceptibility to TB (P=0.017). None of the tagSNPs of the TLR6, MYD88 and TIRAP genes were associated with susceptibility to LTBI or TB (results shown in Table S4). However, only the relationship between rs5743557 and TB susceptibility remained significant after 1000-fold permutation (P=0.023).
Table 2. Associations between four tagSNPs of TLR1 and the risk of LTBI and TB.
| SNPs | Genetic models | Alleles/genotypes | HC, n (%) | LTBI, n (%) | TB, n (%) | HC vs. LTBI | LTBI vs. TB | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pa | OR (95% CI)a | P* | Pa | OR (95% CI)a | P* | |||||||
| rs4833095 | Dominant | TT + TC | 148 (72.6) | 119 (59.5) | 145 (69.4) | 0.007 | 0.56 (0.37–0.86) | 0.065 | 0.074 | 1.48 (0.96–2.28) | 0.343 | |
| CC | 56 (27.5) | 81 (40.5) | 64 (30.6) | |||||||||
| Recessive | TT | 32 (15.7) | 33 (16.5) | 42 (20.1) | 0.882 | 1.04 (0.61–1.78) | 1 | 0.386 | 1.27 (0.74–2.15) | 0.972 | ||
| TC + CC | 172 (84.4) | 167 (83.5) | 167 (79.9) | |||||||||
| Additive | CC/TC/TT | 0.074 | 0.77 (0.58–1.03) | 0.520 | 0.095 | 1.28 (0.96–1.71) | 0.445 | |||||
| rs5743557 | Dominant | AA + GA | 140 (68.6) | 138 (69.0) | 140 (67.0) | 0.837 | 1.05 (0.68–1.61) | 1 | 0.475 | 0.85 (0.55–1.32) | 1 | |
| GG | 64 (31.4) | 62 (31.0) | 69 (33.0) | |||||||||
| Recessive | AA | 26 (12.7) | 44 (22.0) | 24 (11.5) | 0.012 | 1.99 (1.17–3.39) | 0.111 | 0.006 | 0.45 (0.26–0.8) | 0.023 | ||
| GA + GG | 178 (87.3) | 156 (78.0) | 185 (88.5) | |||||||||
| Additive | GG/GA/AA | 0.120 | 1.26 (0.94–1.69) | 0.807 | 0.045 | 0.73 (0.54–0.99) | 0.485 | |||||
| rs5743596 | Dominant | AA + GA | 116 (56.8) | 110 (55.0) | 109 (52.2) | 0.813 | 0.95 (0.64–1.42) | 1 | 0.544 | 0.88 (0.58–1.33) | 1 | |
| GG | 88 (43.1) | 90 (45.0) | 100 (47.8) | |||||||||
| Recessive | AA | 18 (8.8) | 26 (13.0) | 14 (6.7) | 0.144 | 1.61 (0.85–3.06) | 0.842 | 0.036 | 0.47 (0.23–0.95) | 0.207 | ||
| GA + GG | 186 (91.1) | 174 (87.0) | 195 (93.3) | |||||||||
| Additive | GG/GA/AA | 0.605 | 1.08 (0.8–1.46) | 1 | 0.153 | 0.79 (0.58–1.09) | 0.783 | |||||
| rs5743604 | Dominant | AA + GA | 158 (77.5) | 136 (68.0) | 167 (79.9) | 0.034 | 0.62 (0.4–0.97) | 0.300 | 0.017 | 1.78 (1.11–2.85) | 0.056 | |
| GG | 46 (22.5) | 64 (32.0) | 42 (20.1) | |||||||||
| Recessive | AA | 43 (21.1) | 41 (20.5) | 52 (24.9) | 0.784 | 0.93 (0.58–1.52) | 1 | 0.273 | 1.32 (0.81–2.16) | 0.922 | ||
| GA + GG | 161 (78.9) | 159 (79.5) | 157 (75.1) | |||||||||
| Additive | GG/GA/AA | 0.126 | 0.8 (0.6–1.065) | 0.783 | 0.031 | 1.4 (1.03–1.87) | 0.166 | |||||
a, adjusting for age and gender when analyzing genotype models in a binary logistic regression analysis model; *, adjusting for multiple comparisons by 1,000-fold permutation testing. HC, uninfected healthy controls; LTBI, latent tuberculosis infection; TB, tuberculosis; OR, odds ratio; CI, confidence interval.
Table S4. Associations between tagSNPs of gene TLR6, TIRAP and MYD88 and the risk of LTBI and TB.
| Gene | SNPs | Genetic models | Alleles/genotypes | HC, n (%) | LTBI, n (%) | TB, n (%) | HC vs. LTBI, P valuea | LTBI vs. TB, P valuea |
|---|---|---|---|---|---|---|---|---|
| TLR6 | rs1039559 | Dominant | AA | 102 (50.0) | 116 (58.0) | 109 (52.2) | 0.101 | 0.434 |
| GA + GG | 102 (50.0) | 84 (42.0) | 100 (47.9) | |||||
| Recessive | GG | 9 (4.4) | 9 (4.5) | 11 (5.3) | 0.965 | 0.827 | ||
| AA + GA | 195 (95.6) | 191 (95.5) | 198 (94.8) | |||||
| Additive | AA/GA/GG | 0.157 | 0.461 | |||||
| rs3775073 | Dominant | TT | 83 (40.7) | 75 (37.5) | 74 (35.4) | 0.621 | 0.408 | |
| CT + CC | 121 (59.3) | 125 (62.5) | 135 (64.6) | |||||
| Recessive | CC | 24 (11.8) | 29 (14.5) | 27 (12.9) | 0.381 | 0.99 | ||
| TT + CT | 180 (88.2) | 171 (85.5) | 182 (87.1) | |||||
| Additive | TT/CT/CC | 0.426 | 0.557 | |||||
| rs5743808 | Dominant | AA | 178 (87.3) | 174 (87) | 174 (83.3) | 0.988 | 0.2 | |
| GA + GG | 26 (12.8) | 26 (13) | 35 (16.7) | |||||
| Recessive | GG | 3 (1.5) | 2 (1.0) | 0 (0) | 0.741 | 0.999 | ||
| AA + GA | 201 (98.6) | 198 (99.0) | 209 (100.0) | |||||
| Additive | AA/GA/GG | 0.935 | 0.338 | |||||
| rs5743827 | Dominant | CC | 160 (78.4) | 159 (79.5) | 164 (78.5) | 0.793 | 0.501 | |
| CT + TT | 44 (21.6) | 41 (20.5) | 45 (21.5) | |||||
| Recessive | TT | 4 (2.0) | 2 (1.0) | 0 (0) | 0.482 | 0.999 | ||
| CC + CT | 200 (98.0) | 198 (99.0) | 209 (100.0) | |||||
| Additive | CC/CT/TT | 0.669 | 0.675 | |||||
| TIRAP | rs3802813 | Dominant | AA + GA | 62 (30.4) | 58 (29.0) | 77 (36.8) | 0.614 | 0.114 |
| GG | 142 (69.6) | 142 (71.0) | 132 (63.2) | |||||
| Recessive | AA | 4 (2.0) | 5 (2.5) | 9 (4.3) | 0.657 | 0.951 | ||
| GA + GG | 200 (98.0) | 195 (97.5) | 200 (95.7) | |||||
| Additive | GG/GA/AA | 0.747 | 0.165 | |||||
| rs595209 | Dominant | AA + CA | 119 (58.4) | 121 (60.5) | 126 (60.3) | 0.375 | 0.293 | |
| CC | 85 (41.7) | 79 (39.5) | 83 (39.7) | |||||
| Recessive | AA | 23 (11.3) | 18 (9.0) | 28 (13.4) | 0.669 | 0.651 | ||
| CA + CC | 181 (88.8) | 182 (91.0) | 181 (86.6) | |||||
| Additive | CC/CA/AA | 0.929 | 0.87 | |||||
| rs8177375 | Dominant | GG + GA | 55 (27.0) | 53 (26.5) | 48 (23.0) | 0.903 | 0.376 | |
| AA | 149 (73.0) | 147 (73.5) | 161 (77.0) | |||||
| Recessive | GG | 2 (1.0) | 3 (1.5) | 1 (0.5) | 0.689 | 0.314 | ||
| GA + AA | 202 (99.0) | 197 (98.5) | 208 (99.5) | |||||
| Additive | AA/GA/GG | 0.983 | 0.293 | |||||
| MYD88 | rs6853 | Dominant | AA | 194 (95.1) | 193 (96.5) | 203 (97.1) | 0.401 | 0.786 |
| AG + GG | 10 (4.9) | 7 (3.5) | 6 (2.9) | |||||
| Recessive | GG | 0 (0) | 0 (0) | 0 (0) | ||||
| AA + AG | 204 (100.0) | 200 (100.0) | 209 (100.0) | |||||
| Additive | AA/AG/GG | 0.401 | 0.786 | |||||
| rs7744 | Dominant | AA | 74 (36.3) | 68 (34.0) | 82 (39.2) | 0.676 | 0.355 | |
| AG + GG | 130 (63.7) | 132 (66.0) | 127 (60.8) | |||||
| Recessive | GG | 26 (12.7) | 30 (15.0) | 29 (13.9) | 0.534 | 0.766 | ||
| AA + AG | 178 (87.3) | 170 (85.0) | 180 (86.1) | |||||
| Additive | AA/AG/GG | 0.534 | 0.418 |
a, adjusting for age and gender when analyzing genotype models in a binary logistic regression analysis model. HC, uninfected healthy controls; LTBI, latent tuberculosis infection; TB, tuberculosis; OR, odds ratio; CI, confidence interval.
After calculating the power for all tagSNPs, rs5743557 and rs5743604 had power approaching 80% when comparing the TB group with the LTBI group, and rs4833095 had similar power when comparing the HC group with the LTBI group (Table S2). Considering that rs4833095 deviated from HWE in the HC group, only results of rs5743557 and rs5743604 between TB group and the LTBI group should be taken into consideration.
Associations between haplotypes and LTBI or TB susceptibility
The rs4833095-rs5743604 CG haplotype of TLR1 was significantly higher in the LTBI group than in the TB group (P=0.019877). However, this result was no longer significant association after 1000-fold permutation testing (P=0.191, shown in Table 3). For the TLR1 gene, no significant haplotype associations were observed when the HC group was compared with the LTBI group.
Table 3. Haplotype analysis of four tagSNPs of TLR1 in the comparison of LTBI vs. TB.
| Haplotype | TB (freq) | LTBI (freq) | Chi2 | Pearson’s P | Odds ratio (95% CI) | P* |
|---|---|---|---|---|---|---|
| rs4833095-rs5743604 | ||||||
| CA | 32.00 (0.077) | 25.00 (0.062) | 0.623 | 0.429922 | 1.244 (0.723–2.139) | 1 |
| CG | 199.00 (0.476) | 223.00 (0.557) | 5.426 | 0.019877 | 0.721 (0.548–0.950) | 0.191 |
| TA | 187.00 (0.447) | 152.00 (0.380) | 3.823 | 0.050583 | 1.321 (0.999–1.746) | 0.396 |
| Global result | 400 | 418 | 5.445 | 0.066 | ||
| rs5743557-rs5743596 | ||||||
| AA | 123.00 (0.294) | 134.87 (0.337) | 1.819 | 0.177447 | 0.816 (0.607–1.097) | 0.874 |
| AG | 41.00 (0.098) | 47.13 (0.118) | 0.854 | 0.355397 | 0.812 (0.521–1.264) | 1 |
| GG | 254.00 (0.608) | 216.87 (0.542) | 3.418 | 0.064502 | 1.300 (0.984–1.717) | 0.475 |
| Global result | 400 | 418 | 3.454 | 0.178 |
*, P value after 1,000-fold permutation testing using the Haploview software. LTBI, latent tuberculosis infection; TB, tuberculosis; OR, odds ratio; CI, confidence interval.
Discussion
Although rs4833095 of TLR1 (7), rs5743810 of TLR6 (8), rs6853 of MYD88 (9) and rs8177374 of TIRAP (10) have been found to influence the host response to M.TB, we focused this study on the relationship between tagSNPs of these four genes and two stages of TB progression. Our study revealed that the AA genotype of rs5743557 was associated with reduced risk of TB compared with the LTBI group after permutation correction. The meta-analysis suggested rs5743557_A was associated with decreased TB susceptibility in the Chinese adult population.
It is well known that TLR1 is able to recognize triacyl lipoproteins of mycobacteria as a heterodimer with TLR2, as shown by the defective response to triacylated lipopeptides in TLR1-deficient mice (29). rs5743557, located 595 bp upstream of the TLR1 gene, might influence the promoter activity of this gene. rs5743557_A was reported to be significantly associated with decreased TB risk in Chinese Tibetans, while this relationship was only observed in Chinese Han women and no significant association was seen in Chinese Han children (20,25,26). After meta-analysis in our paper, rs5743557_A showed a relationship with decreased TB susceptibility in the Chinese adult population. Our study confirmed the meta-analysis results considering the association seen in the comparison of TB with LTBI. More information about rs5743557 and TB risk in other populations is needed to further confirm our results. As is well known, the immune system is quite different between children and adults, and children are more likely to develop active TB after infection (30,31). Thus, separating adults from children is necessary to better understand the mechanisms underlying TB pathogenesis in each group.
rs5743595 of TLR1, which was in strong LD with rs5743557 in the Chinese Han Beijing population, was reported to augment expression of TLR1 mRNA, TLR1 proteins and specific cytokines as determined by RT-PCR, flow cytometry and ELISA (32). However, whether rs5743557 participated in the regulation of TLR1 function is unclear. Instead, many reports have observed functional roles of rs4833095, which is a non-synonymous polymorphism and the amino acid change was reported to alter TLR1 folding or function (33). rs4833095 was also associated with lack of surface expression and impaired function of TLR1 (34). However, no association between rs4833095 and TB was observed in most studies and after meta-analysis (20). In consideration of the lack of relationship between rs4833095 and susceptibility to TB, the functional effects of rs4833095 might affect the risk of other diseases. Thus, special attention should be focused on rs5743557 or other linked polymorphisms, which may be the key variants in the relationship between TLR1 and TB susceptibility.
In a similar way to TLR1, TLR6 is known to recognize lipopeptides possessing a diacylglycerol group in the form of TLR6/TLR2 heterodimers (35), leading to activation of MYD88 dependent pathways. TIRAP, together with MYD88, is involved in TLR1, TLR2, and TLR4 signaling pathways and activates downstream NF-κB, leading to induction of cytokine secretion and the proinflammatory response. One study demonstrated that TLR6-deficient mice showed an impaired response to diacylated lipopeptides (29). MYD88 null mice were reported to exhibit increased susceptibility to virulent M.TB infection and rapidly succumb to TB (36). TIRAP-deficient mice also showed impaired MYD88-dependent pathways involving TLR2 and TLR4 (37).
rs5743808 of TLR6 was reported to associate with increased TB risk in the African American population (33). rs6853 of MYD88 was shown to have a relationship with TB risk in an Indian population and revealed functional effects (9). No other tagSNP of TLR6, MYD88 and TIRAP has been reported to significantly associate with TB risk in previous studies. Although a study suggested MYD88 and TLR6 were uniquely expressed in TB patients compared with LTBI subjects (14), the lack of significant results of the two genes in our study does not provide any confirmatory evidence to support a pathogenic role of these genes in TB. Nevertheless, in view of the important roles played by TLR6, MYD88 and TIRAP in the immune response against M.TB and the limited genetic epidemiologic information about them, further studies with larger sample size are required to verify their definitive relationship with LTBI and TB susceptibility.
Considering the large population of LTBI, early recognition and targeted treatment of high-risk LTBI patients will make a great contribution to reaching the End TB Strategy goal by the year 2035. Studies focused on finding a measureable way to predict the risk of TB infection and active TB from LTBI progression has recently begun. Histone deacetylase related genes were differentially expressed in the genome-wide transcriptional profiles between uninfected healthy controls and LTBI individuals (38). One study identified a 16 gene expression signature for predicting the risk of LTBI progression into active TB with a sensitivity of 66.1% and a specificity of 80.6% (39). Another study has shown that the TNFα-only T-cell phenotype was observed more frequently in recently acquired LTBI compared with remotely acquired LTBI (40). While great efforts have been made in predicting TB infection and progression, a meta-analysis revealed a 380 gene meta-signature with only five genes involved in all included studies (13). Thus, searching for genes and transcript markers to predict TB progression is still an active area of research.
Our study was designed to expand the knowledge of the relationship between four innate immunity genes and the risk of LTBI and TB, however, only one significant genetic association was identified. The main weaknesses of our study were the limited sample size and a lack of a replication study to verify our findings. However, our study also has some advantages. This is the first report to comprehensively investigate association between polymorphisms of four genes (TLR1, TLR6, MYD88 and TIRAP) and susceptibility to two stages of TB progression, namely LTBI and active TB. In addition, the enrolled Han participants were mostly from Sichuan Province, Southwestern China, and thus had minimal geographical and ethnic heterogeneity.
In conclusion, our study found a significant association between rs5743557 of TLR1 and risk of TB in the Chinese adult population. Validation of this association and experiments to determine the underlying mechanism are required. In addition, a larger study sample is still needed to investigate association between the other three critical genes (TLR6, MYD88 and TIRAP) and susceptibility to LTBI and TB. This study provides important clues to elucidate the relationship of innate immunity to the development of TB.
Acknowledgements
Funding: This work was supported by the Research Fund for the Doctoral Program of Higher Education of China (grant number 20130181110068), the National Natural Science Foundation of China (grant number 81170042, grant number 81370121), and the National Scientific and Technological Major Project of China (grant number 2012ZX10004-901, 2018ZX10715-003).
Ethical Statement: The study was approved by the Ethics Committee of the West China Hospital of Sichuan University in China (File No. 131) and written informed consent was obtained from all patients.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.CDC. Most Recent Asthma Data. Available online: https://www.cdc.gov/asthma/most_recent_data.htm. Accessed April 26, 2018. 2016.
- 2.Dye C, Scheele S, Dolin P, et al. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA 1999;282:677-86. 10.1001/jama.282.7.677 [DOI] [PubMed] [Google Scholar]
- 3.Comstock GW, Livesay VT, Woolpert SF. The prognosis of a positive tuberculin reaction in childhood and adolescence. Am J Epidemiol 1974;99:131-8. 10.1093/oxfordjournals.aje.a121593 [DOI] [PubMed] [Google Scholar]
- 4.Vynnycky E, Fine PE. Lifetime risks, incubation period, and serial interval of tuberculosis. Am J Epidemiol 2000;152:247-63. 10.1093/aje/152.3.247 [DOI] [PubMed] [Google Scholar]
- 5.Report on nationwide random survey for the epidemiology of tuberculosis in 2000. China J Antituberc 2002;24:65-107. [Google Scholar]
- 6.Takeuchi O, Kawai T, Sanjo H, et al. TLR6: A novel member of an expanding toll-like receptor family. Gene 1999;231:59-65. 10.1016/S0378-1119(99)00098-0 [DOI] [PubMed] [Google Scholar]
- 7.Dittrich N, Berrocal-Almanza LC, Thada S, et al. Toll-like receptor 1 variations influence susceptibility and immune response to Mycobacterium tuberculosis. Tuberculosis (Edinb) 2015;95:328-35. 10.1016/j.tube.2015.02.045 [DOI] [PubMed] [Google Scholar]
- 8.Shey MS, Randhawa AK, Bowmaker M, et al. Single nucleotide polymorphisms in toll-like receptor 6 are associated with altered lipopeptide- and mycobacteria-induced interleukin-6 secretion. Genes Immun 2010;11:561-72. 10.1038/gene.2010.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capparelli R, De Chiara F, Di Matteo A, et al. The MyD88 rs6853 and TIRAP rs8177374 polymorphic sites are associated with resistance to human pulmonary tuberculosis. Genes Immun 2013;14:504-11. 10.1038/gene.2013.48 [DOI] [PubMed] [Google Scholar]
- 10.Ni Cheallaigh C, Sheedy FJ, Harris J, et al. A Common Variant in the Adaptor Mal Regulates Interferon Gamma Signaling. Immunity 2016;44:368-79. 10.1016/j.immuni.2016.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stein CM. Genetic epidemiology of tuberculosis susceptibility: impact of study design. PLoS Pathog 2011;7:e1001189. 10.1371/journal.ppat.1001189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schurz H, Daya M, Moller M, et al. TLR1, 2, 4, 6 and 9 Variants Associated with Tuberculosis Susceptibility: A Systematic Review and Meta-Analysis. PLoS One 2015;10:e0139711. 10.1371/journal.pone.0139711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blankley S, Graham CM, Levin J, et al. A 380-gene meta-signature of active tuberculosis compared with healthy controls. Eur Respir J 2016;47:1873-6. 10.1183/13993003.02121-2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maertzdorf J, Repsilber D, Parida SK, et al. Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun 2011;12:15-22. 10.1038/gene.2010.51 [DOI] [PubMed] [Google Scholar]
- 15.Kahwati LC, Feltner C, Halpern M, et al. U.S. Preventive Services Task Force Evidence Syntheses, formerly Systematic Evidence Reviews. Screening for Latent Tuberculosis Infection in Adults: An Evidence Review for the U.S. Preventive Services Task Force. Rockville (MD): Agency for Healthcare Research and Quality (US); 2016. [Google Scholar]
- 16.NHFPC. Diagnosis for pulmonary tuberculosis in China 2017. 2017.
- 17.Updated guidelines for the use of nucleic acid amplification tests in the diagnosis of tuberculosis. MMWR Morb Mortal Wkly Rep 2009;58:7-10. [PubMed] [Google Scholar]
- 18.WHO. WHO Guidelines Approved by the Guidelines Review Committee. Guidelines on the Management of Latent Tuberculosis Infection. Geneva: World Health Organization. Copyright (c) World Health Organization 2015; 2015. [Google Scholar]
- 19.Liu Q, Wang J, Sandford AJ, et al. Association of CYBB polymorphisms with tuberculosis susceptibility in the Chinese Han population. Infect Genet Evol 2015;33:169-75. 10.1016/j.meegid.2015.04.026 [DOI] [PubMed] [Google Scholar]
- 20.Zhang M, Tang X, Wang Y, et al. Variants of TLR1 associated with tuberculosis susceptibility in the Chinese Tibetan population but not in Han Chinese. Infect Genet Evol 2018;61:53-9. 10.1016/j.meegid.2018.02.021 [DOI] [PubMed] [Google Scholar]
- 21.Pahl R, Schafer H. PERMORY: an LD-exploiting permutation test algorithm for powerful genome-wide association testing. Bioinformatics 2010;26:2093-100. 10.1093/bioinformatics/btq399 [DOI] [PubMed] [Google Scholar]
- 22.Chang CC, Chow CC, Tellier LC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015;4:7. 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263-5. 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 24.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res 2005;15:97-8. 10.1038/sj.cr.7290272 [DOI] [PubMed] [Google Scholar]
- 25.Qi H, Sun L, Wu X, et al. Toll-like receptor 1(TLR1) Gene SNP rs5743618 is associated with increased risk for tuberculosis in Han Chinese children. Tuberculosis 2015;95:197-203. 10.1016/j.tube.2014.12.001 [DOI] [PubMed] [Google Scholar]
- 26.Peng W, Chen H, Zhao Z, et al. TLR1 polymorphisms are significantly associated with the occurrence, presentation and drug-adverse reactions of tuberculosis in Western Chinese adults. Oncotarget 2017;9:1691-704. 10.18632/oncotarget.23067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu X, Liu P, Liu G, et al. The prevalence of non-tuberculous mycobacterial infections in mainland China: Systematic review and meta-analysis. J Infect 2016;73:558-67. 10.1016/j.jinf.2016.08.020 [DOI] [PubMed] [Google Scholar]
- 28.Nguyen DT, Teeter LD, Graves J, et al. Characteristics Associated with Negative Interferon-gamma Release Assay Results in Culture-Confirmed Tuberculosis Patients, Texas, USA, 2013-2015. Emerg Infect Dis 2018;24:534-40. 10.3201/eid2403.171633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeda K, Takeuchi O, Akira S. Recognition of lipopeptides by Toll-like receptors. J Endotoxin Res 2002;8:459-63. 10.1177/09680519020080060101 [DOI] [PubMed] [Google Scholar]
- 30.Kollmann TR, Levy O, Montgomery RR, et al. Innate immune function by Toll-like receptors: distinct responses in newborns and the elderly. Immunity 2012;37:771-83. 10.1016/j.immuni.2012.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanden Driessche K, Persson A, Marais BJ, et al. Immune vulnerability of infants to tuberculosis. Clin Dev Immunol 2013;2013:781320. 10.1155/2013/781320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kormann MS, Depner M, Hartl D, et al. Toll-like receptor heterodimer variants protect from childhood asthma. J Allergy Clin Immunol 2008;122:86-92, 92.e1-8. [DOI] [PubMed]
- 33.Ma X, Liu Y, Gowen BB, et al. Full-exon resequencing reveals toll-like receptor variants contribute to human susceptibility to tuberculosis disease. PLoS One 2007;2:e1318. 10.1371/journal.pone.0001318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uciechowski P, Imhoff H, Lange C, et al. Susceptibility to tuberculosis is associated with TLR1 polymorphisms resulting in a lack of TLR1 cell surface expression. J Leukoc Biol 2011;90:377-88. 10.1189/jlb.0409233 [DOI] [PubMed] [Google Scholar]
- 35.Omueti KO, Beyer JM, Johnson CM, et al. Domain exchange between human toll-like receptors 1 and 6 reveals a region required for lipopeptide discrimination. J Biol Chem 2005;280:36616-25. 10.1074/jbc.M504320200 [DOI] [PubMed] [Google Scholar]
- 36.Fremond CM, Yeremeev V, Nicolle DM, et al. Fatal Mycobacterium tuberculosis infection despite adaptive immune response in the absence of MyD88. J Clin Invest 2004;114:1790-9. 10.1172/JCI200421027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto M, Sato S, Hemmi H, et al. Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature 2002;420:324-9. 10.1038/nature01182 [DOI] [PubMed] [Google Scholar]
- 38.Seshadri C, Sedaghat N, Campo M, et al. Transcriptional networks are associated with resistance to Mycobacterium tuberculosis infection. PLoS One 2017;12:e0175844. 10.1371/journal.pone.0175844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zak DE, Penn-Nicholson A, Scriba TJ, et al. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet 2016;387:2312-22. 10.1016/S0140-6736(15)01316-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halliday A, Whitworth H, Kottoor SH, et al. Stratification of Latent Mycobacterium tuberculosis Infection by Cellular Immune Profiling. J Infect Dis 2017;215:1480-7. 10.1093/infdis/jix107 [DOI] [PMC free article] [PubMed] [Google Scholar]