Summary
Elucidation of the role of different cell lineages in the liver could offer avenues to drive liver regeneration. Previous studies showed that SOX9+ hepatocytes can differentiate into ductal cells after liver injuries. It is unclear whether SOX9+ hepatocytes are uni- or bipotent progenitors at a single-cell level during liver injury. Here, we developed a genetic tracing system to delineate the lineage potential of SOX9+ hepatocytes during liver homeostasis and regeneration. Fate-mapping data showed that these SOX9+ hepatocytes respond specifically to different liver injuries, with some contributing to a substantial number of ductal cells. Clonal analysis demonstrated that a single SOX9+ hepatocyte gives rise to both hepatocytes and ductal cells after liver injury. This study provides direct evidence that SOX9+ hepatocytes can serve as bipotent progenitors after liver injury, producing both hepatocytes and ductal cells for liver repair and regeneration.
Keywords: lineage tracing, intersectional genetic strategy, clonal analysis, liver repair and liver regeneration, bipotent progenitors
Graphical Abstract
Highlights
-
•
SOX9+ hepatocytes respond distinctly to different liver injuries
-
•
Generation of a Confetti reporter responsive to dual recombinases
-
•
SOX9+ hepatocytes can serve as bipotent progenitors after liver injury
In this article, Zhou and colleagues developed an intersectional genetic strategy to specifically label SOX9+ hepatocytes. Using a dual recombinase-mediated Confetti reporter, they showed that a subset of SOX9+ hepatocytes, at single-cell level, are bipotent progenitors that differentiate into both hepatocytes and biliary epithelial cells during liver injury and repair.
Introduction
Under normal conditions the liver is an organ with slow parenchymal cell turnover, and the lifespan of hepatocytes can reach 200–300 days in a resting state. During homeostasis, it is generally believed that the liver lacks a multipotent stem cell population to maintain organ renewal, and thus ductal cells and hepatocytes are unipotent (Miyajima et al., 2014). Previous studies have indicated that facultative stem cells, also known as atypical ductal cells or oval cells, can differentiate into hepatocytes (Farber, 1956, Popper et al., 1957). Studies using Hnf1b or osteopontin (OPN) promoter-driven Cre fate mapping have suggested that liver progenitor cells or biliary epithelial cells could give rise to hepatocytes after liver injuries (Espanol-Suner et al., 2012, Rodrigo-Torres et al., 2014). However, recent lineage-tracing studies reported that virtually all hepatocytes are derived from pre-existing hepatocytes before injury rather than differentiating from stem cells (Grompe, 2014, Malato et al., 2011, Schaub et al., 2014, Wang et al., 2017, Yanger et al., 2014). Following severe chronic liver injury in which the proliferative potential of hepatocytes is significantly inhibited, a group of hepatic progenitor cells (HPCs) contribute significantly to the restoration of liver parenchyma, generating both hepatocytes and biliary epithelia (Lu et al., 2015). Indeed, following severe and chronic liver injuries that impair hepatocyte proliferation, committed ductal cells or cholangiocytes can act as facultative liver stem cells and generate hepatocytes for liver repair and regeneration (Deng et al., 2018, Raven et al., 2017). Conversely, hepatocytes can undergo hepatocyte-to-ductal cell transition after certain injuries (Yanger et al., 2013). In an animal model of human Alagille syndrome, hepatocytes could also convert into de novo mature biliary epithelial cells that form functional bile ducts (Schaub et al., 2018). Under chronic injury, mature hepatocytes could generate bipotential adult liver progenitors that give rise to both ductal cells and hepatocytes (Tarlow et al., 2014b). These studies indicated that mature hepatocytes or ductal cells could be reprogrammed into counterparts under certain conditions, as demonstrated in extremely severe liver injury models that promote cell-lineage conversion and cell plasticity. While these studies involved genetic lineage tracing at the population level, it remains unclear whether a single cell such as a hepatocyte is predetermined to give rise to hepatocytes, biliary epithelial cells, or both during injury. Unraveling the potency and plasticity of committed hepatocytes may provide evidence to help elucidate the liver progenitor cell hierarchy and their roles in liver repair and regeneration.
Sox9 (sry-related high mobility group-box gene 9) is a family gene homolog located on the male Y chromosome (Suzuki et al., 2015). In the liver, SOX9 regulates the development of intrahepatic bile ducts through a mode of tubulogenesis (Antoniou et al., 2009). Furuyama et al. (2011) reported that SOX9+ ductal epithelial cells are endogenous HPCs that contribute to hepatocytes during liver homeostasis and after injuries. Subsequent lineage-tracing studies using a multicolored fluorescent Confetti reporter showed that SOX9+ cells contribute only minimally (<1%) to hepatocytes (Tarlow et al., 2014a). Because SOX9 is also expressed in a subset of hepatocytes, albeit at a lower level compared with that in ductal cells (Font-Burgada et al., 2015, Yanger et al., 2013), the rare contribution of SOX9+ cells to hepatocytes could be due to prelabeled hepatocytes that express SOX9 (He et al., 2017). Indeed, these SOX9+ hepatocytes undergo extensive proliferation and replenish liver mass after chronic liver injuries without giving rise to hepatocellular carcinoma (Font-Burgada et al., 2015), indicating that SOX9+ hepatocytes could be an important source of hepatocytes with therapeutic potential. It remains unknown whether individual SOX9+ hepatocytes are unipotent (ductal cell or hepatocyte lineage) or bipotent (both ductal cell and hepatocyte lineages) during liver injury and repair.
The genetic lineage-tracing technique is an effective method for unraveling cell fate in development, disease, and regeneration (Tian et al., 2015). The conventional genetic tracing method depends on a singular gene marker that may show low efficacy in defining one particular cell population. For example, Sox9-CreER targets both periportal hepatocytes and biliary epithelial cells. To achieve more precise labeling of cell lineages and trace their cell fate in vivo, we recently incorporated a recombination system, Dre-rox, to supplement Cre-loxP for enhancing the precision of genetic lineage tracing (He et al., 2017). In this study, we generated an intersectional genetic strategy showing that a subset of SOX9+ hepatocytes, at the single-cell level, are bipotent progenitors that differentiate into both hepatocytes and biliary epithelial cells during liver injury and repair.
Results
Characterization of Sox9-CreER and Hnf4a-DreER Mouse Lines
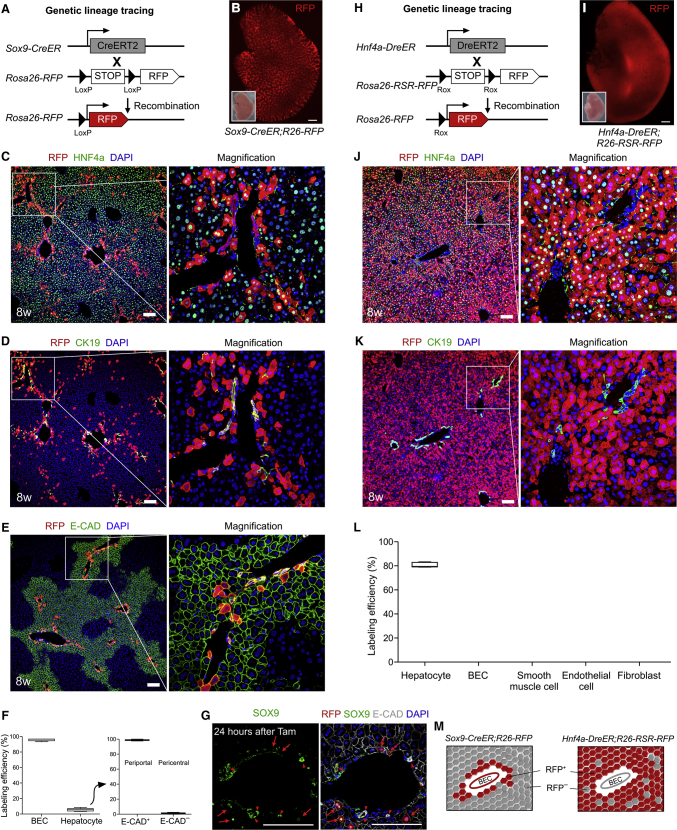
SOX9+ hepatocytes express both SOX9 and hepatocyte markers, such as HNF4a, but do not express the biliary epithelial cell marker CK19 (He et al., 2017). For lineage tracing of SOX9+HNF4a+ hepatocytes, we generated two distinct mouse lines that utilize two orthogonal recombinases: Sox9-CreER and Hnf4a-DreER. The Sox9-CreER mouse was crossed with the R26-RFP reporter mouse to generate the Sox9-CreER;R26-RFP mouse. Tamoxifen induction led to Cre-loxP recombination, which resulted in permanent labeling of SOX9+ cells and all their descendants (Figure 1A). Whole-mount fluorescence imaging of Sox9-CreER;R26-RFP livers showed that a substantial number of hepatic cells were labeled after tamoxifen induction (Figure 1B). Immunostaining for RFP, the hepatocyte marker HNF4a, or the ductal cell marker CK19 on Sox9-CreER;R26-RFP liver sections showed that RFP+ cells were HNF4a+ or CK19+ (Figures 1C and 1D), indicating hepatocytes and ductal cells/biliary epithelial cells (BECs), respectively. Notably, most RFP+ hepatocytes were close to the portal vein region where BECs were located, which is consistent with previous reports. Staining of the periportal hepatic zonation marker E-cadherin (E-CAD) verified that these SOX9+ hepatocytes were periportal hepatocytes (Figure 1E). Quantification of hepatocyte labeling efficiency showed that 96.51% ± 0.38% of BECs were RFP+ and 5.49% ± 1.93% of hepatocytes were RFP+ (Figure 1F). Of these positive hepatocytes, almost all were positive for E-CAD (>99%, Figure 1F), suggesting periportal hepatocytes. To confirm that SOX9 protein was indeed expressed in the hepatocytes in addition to BECs, we also collected the tissues 24 h after tamoxifen induction and stained them for RFP, SOX9, and E-CAD. We found that SOX9 was expressed in a subset of periportal hepatocytes (arrows) as well as BECs (arrowheads, Figure 1G).
Figure 1.
Fate Mapping of Hepatic Cell Lineages by Sox9-CreER or Hnf4a-DreER
(A and H) Schematic figure showing lineage-tracing strategy by Sox9-CreER;R26-RFP (A) or Hnf4a-DreER;R26-RSR-RFP (H).
(B and I) Whole-mount fluorescence views of livers from 8-week-old adult mice. Tamoxifen was induced 4 days later. Insets indicate bright-field images.
(C and J) Immunostaining for RFP and HNF4a on liver sections.
(D and K) Immunostaining for RFP and CK19 on liver sections.
(E) Immunostaining for RFP and periportal hepatocyte marker E-CAD on liver sections.
(F) Quantification of the percentage of labeled CK19+ biliary epithelial cells (BECs) or HNF4a+ hepatocytes (left panel). The percentage of E-CAD+ or E-CAD- cells in labeled hepatocytes is shown on the right panel. Data are mean ± SEM; n = 5.
(G) Immunostaining for SOX9, RFP, and periportal hepatocyte marker E-CAD on liver sections.
(L) Quantification of the percentage of labeled cells among different lineages. Data are mean ± SEM, n = 5.
(M) Cartoon image showing the fate mapping of hepatic cells by Sox9-CreER or Hnf4a-DreER. Scale bars, 1 mm (B and I) and 100 μm (C–E, G, J, and K).
We next generated an Hnf4a-DreER knockin mouse by placing the DreER cDNA in-frame with the initiating ATG of the Hnf4a gene (Figure S1A). The Hnf4a-DreER mouse was crossed with the R26-rox-stop-rox-RFP (R26-RSR-RFP) reporter to generate the Hnf4a-DreER;R26-RSR-RFP mouse. Tamoxifen treatment induced Dre-rox recombination, which removed the Stop cassette and led to RFP expression in HNF4a+ hepatocytes (Figure 1H). Whole-mount fluorescence imaging of Hnf4a-DreER;R26-RSR-RFP mouse liver showed RFP+ signals throughout the entire liver (Figure 1I). Immunostaining for RFP, HNF4a, CK19, or EpCAM on liver sections showed that HNF4a+ hepatocytes were RFP+, while CK19+ or EpCAM+ BECs were RFP– (Figures 1J, 1K, and S1B). We also stained other cell-lineage markers, such as desmin, α-smooth muscle actin (αSMA), VE-cadherin, platelet-derived growth factor receptor α (PDGFRα), and E-CAD, with RFP on liver sections and found that RFP+ cells were parenchymal epithelial cells but not endothelial cells, smooth muscle cells, hepatic satellite cells, or fibroblasts (Figures S1C–S1G). Quantification of labeled cells showed that 80.25% ± 2.03% of hepatocytes were RFP+ while other cell lineages did not express RFP in Hnf4a-DreER;R26-RSR-RFP (Figure 1L). Taken together, these data demonstrated that Hnf4a-DreER specifically and efficiently targeted hepatocytes in the liver. While Sox9-CreER labeled BECs and a subset of periportal hepatocytes, Hnf4a-DreER labeled only hepatocytes (Figure 1M); thus, SOX9+HNF4a+ hepatocytes could then be targeted by Sox9-CreER and Hnf4a-DreER dual recombinases.
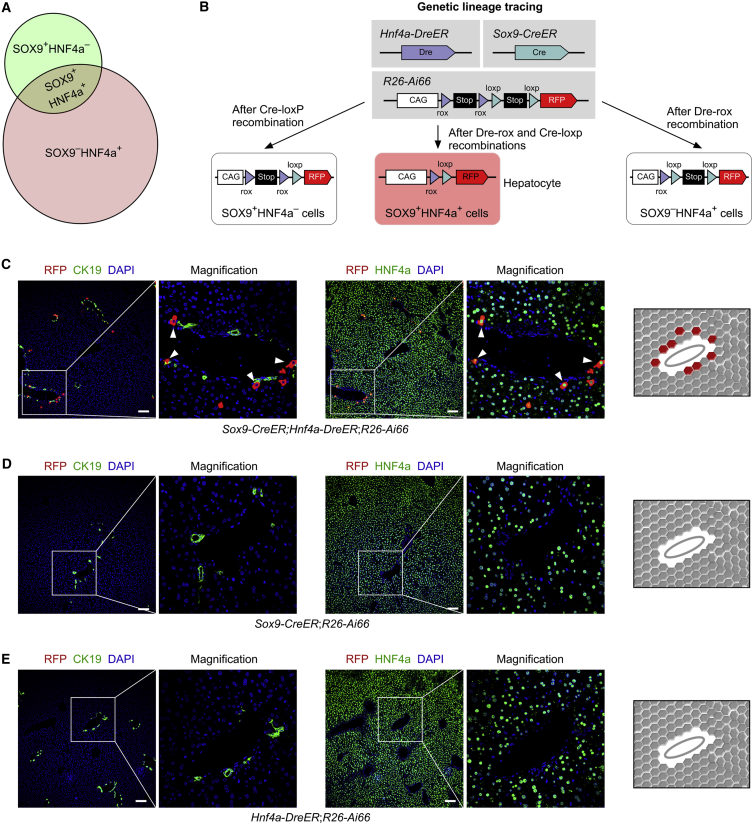
Specific Labeling of SOX9+ Hepatocytes by Dual Recombinases
To achieve specific labeling of SOX9+ hepatocytes, we needed to target SOX9+ HNF4a+ cells in the liver by intersectional genetics (Figure 2A) so that the double-positive cell population could be distinguished from single positive cells. We crossed the characterized Hnf4a-DreER and Sox9-CreER mice with a dual recombinase-mediated RFP reporter (R26-rox-Stop-rox-loxp-Stop-loxp-RFP, as R26-Ai66) to generate the Sox9-CreER;Hnf4a-DreER;R26-Ai66 triple-positive mouse line (Figure 2B). In this reporter line, activation of RFP required both Dre-rox and Cre-loxP recombination, which removed transcriptional Stop cassettes. Therefore, only SOX9+HNF4a+ hepatocytes that harbored both Cre and Dre recombinases could be genetically labeled (Figure 2B). With this genetic design, single positive cell populations, such as SOX9+HNF4a– cells (BECs) or SOX9–HNF4a+ hepatocytes, remained unlabeled (Figure 2B). One week after tamoxifen induction, livers were collected from Sox9-CreER;Hnf4a-DreER;R26-Ai66 mice for analysis. Immunostaining of liver sections for CK19 and HNF4a showed labeling of hepatocytes in the periportal zone of the liver lobule, and BECs were RFP– (Figure 2C), demonstrating selective labeling of SOX9+HNF4a+ hepatocytes. We examined the littermate controls Sox9-CreER;R26-Ai66 or Hnf4a-DreER;R26-Ai66 and performed the same tamoxifen induction strategy and immunostaining for subsequent analysis. In both groups, no RFP+ hepatocytes or BECs were detected (Figures 2D and 2E), demonstrating that RFP expression required both Cre-loxP and Dre-rox recombination. Taken together, these data demonstrated the successful generation of a mouse genetic tool for targeting SOX9+ hepatocytes in the liver.
Figure 2.
Lineage Tracing of SOX9+ Hepatocytes by Dural Recombination
(A) Schematic showing intersectional genetics for labeling of double-positive cells (SOX9+Hnf4a+).
(B) Schematic showing lineage-tracing strategy by Dre-rox and Cre-loxP recombinations. Dural recombination, but not singular one, labels SOX9+Hnf4a+ hepatocytes.
(C–E) Immunostaining for RFP, CK19, and HNF4a on liver sections shows periportal RFP+HNF4a+ hepatocytes (arrowheads) in Sox9-CreER;Hnf4a-DreER;R26-Ai66 (C) but not in Sox9-CreER;R26-Ai66 (D) or Hnf4a-DreER;R26-Ai66 (E) liver sections. Cartoon images on right panels show labeling result. Each figure is representative of five individual samples. Scale bars, 100 μm.
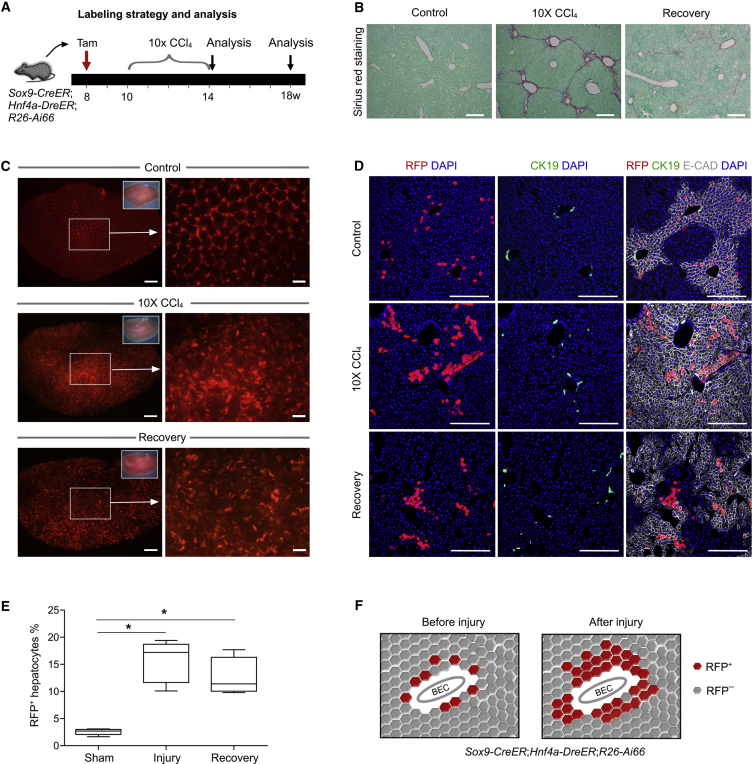
SOX9+ Hepatocytes Expand Significantly after CCl4-Induced Liver Injury
To examine the cell dynamics of SOX9+ hepatocytes after liver injuries, we injected Sox9-CreER;Hnf4a-DreER;R26-Ai66 mice with CCl4 or performed a partial hepatectomy (PHx) to induce liver injuries. CCl4 injury induces injury in the pericentral regions and stimulates the expansion of compensatory growth of periportal hepatocytes (Pu et al., 2016), while PHx induces more general hepatocyte growth throughout the liver, including hepatocyte hypertrophy and compensatory hepatocyte proliferation (Miyajima et al., 2014). Tamoxifen was induced at 8 weeks old, and after a 2-week washout period, mice were treated 10 times with CCl4 and liver samples collected at 14 weeks (after injury) and 18 weeks (recovery group) for analysis (Figure 3A). Sirius red staining of liver sections showed strong fibrosis in the injured liver compared with that of the oil-treated control group, and tissue fibrosis was reduced in the recovery group 4 weeks after the last CCl4 treatment (Figure 3B). Whole-mount fluorescent imaging of livers showed significant RFP signals in the injury and recovery group compared with the control group (Figure 3C). Immunostaining for CK19 and RFP showed that these RFP+ cells did not contribute to CK19+ BECs and remained as hepatocytes in the injured and recovered livers (Figure 3D). We also stained E-CAD in tissue sections of the control, CCl4 injury, and recovery groups. We found that RFP+ hepatocytes remained in zone 1 (E-CAD+) before injury. During CCl4 injury, the zone-1 marker E-CAD was expressed throughout the liver. After recovery the zonation was restored, with E-CAD expression restricted mainly to the periportal hepatocytes. Interestingly, a subset of the expanded RFP+ hepatocytes extended to the pericentral regions and no longer maintained E-CAD expression, indicating reprogramming of SOX9-derived hepatocytes after CCl4-induced liver injury (Figure 3D). Quantitatively, the percentage of RFP+ hepatocytes was significantly higher in the injured liver and recovered liver than in the control group (15.22% ± 1.57%, 12.99% ± 1.51%, and 2.55% ± 0.28%, respectively, Figure 3E), indicating expansion of SOX9+ hepatocytes in CCl4-induced injury (Figure 3F).
Figure 3.
SOX9+ Hepatocytes Contribute Parenchymal Restoration after CCl4-Induced Liver Damage
(A) Experimental strategy for tamoxifen treatment (Tam) and tissue analysis at different time points after injury.
(B) Sirius red staining of liver sections from oil (Control) or CCl4-treated mice.
(C) Whole-mount fluorescence view of Sox9-CreER;Hnf4a-DreER;Rosa26-Ai66 liver after oil (Control) or CCl4 treatment or recovery.
(D) Immunostaining for RFP, CK19, and E-CAD on liver sections shows the expansion of RFP+ hepatocytes following injuries.
(E) Quantification of RFP+ hepatocytes in control, injury, or recovery groups. Data are mean ± SEM, n = 5; ∗p < 0.05.
(F) Cartoon image showing expansion of SOX9+ hepatocytes after injury.
Scale bars, 200 μm (B), 1 mm (C, left), 500 μm (C, right), and 100 μm (D).
We also performed PHx in 10-week-old mice and analyzed liver tissues at 12–14 weeks of age (Figure S2A). Sirius red staining showed that there was no significant tissue fibrosis in the sham or recovery group 2 weeks or 4 weeks after the operation (R2W or R4W, Figure S2B). By whole-mount fluorescent imaging and sectional immunostaining for RFP, we found that SOX9+ hepatocytes did not expand preferentially after PHx compared with SOX9– hepatocytes (Figures S2C–S2E). By immunostaining with the BEC marker CK19, we found that these SOX9+ hepatocytes did not give rise to BECs after PHx and remained as hepatocytes after injury (Figure S2D). We observed an increase in Ki67+ hepatocytes after PHx injury compared with the sham control (Figure S2F). However, the percentage of RFP+ hepatocytes did not change significantly, indicating that PHx did not preferentially induce SOX9+ hepatocyte expansion. Thus, SOX9+ hepatocytes are a unique cell population that responds to a certain type of liver injury (such as CCl4) through significant self-expansion.
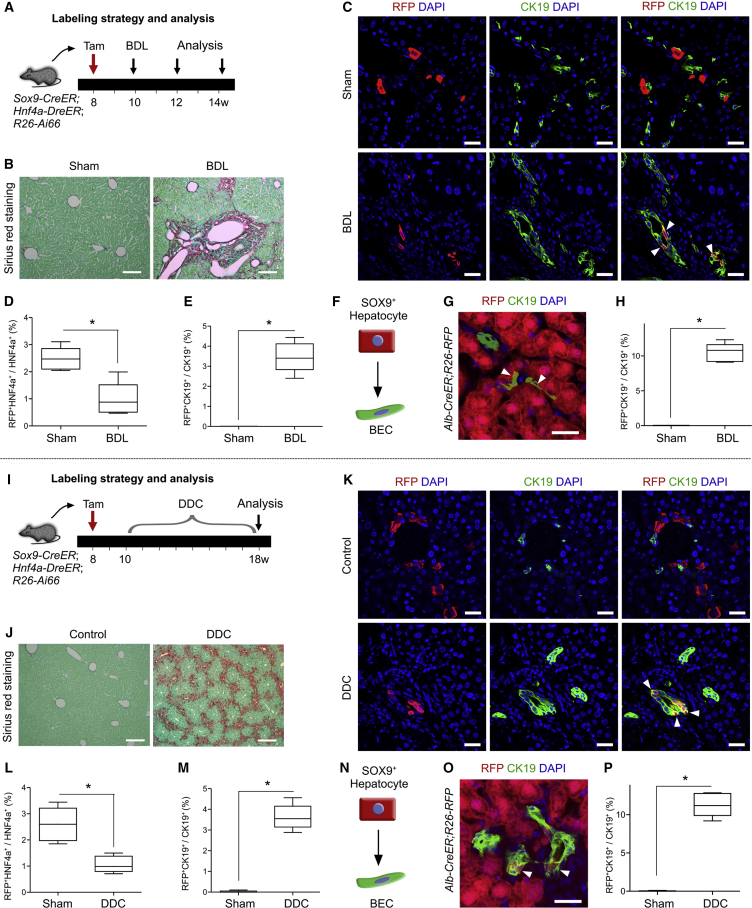
SOX9+ Hepatocytes Contribute to Cholangiocytes after Bile Duct Ligation or DDC-Induced Liver Injury
Previous work reported that bile duct ligation (BDL) or 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC) treatment induces hepatocytes to transition into BECs (Michalopoulos et al., 2005, Tarlow et al., 2014b). We next examined whether SOX9+ hepatocytes contribute to BECs after BDL- or DDC-induced liver injury. After tamoxifen treatment and a 2-week washout period, Sox9-CreER;Hnf4a-DreER;R26-Ai66 mice were subjected to BDL at 10 weeks old, and livers were collected at 12 or 14 weeks for analysis (Figure 4A). BDL resulted in significant liver fibrosis compared with the sham group (Figure 4B). In contrast to the CCl4 treatment, BDL injury did not stimulate significant expansion of SOX9+ hepatocytes (Figure 4C). Notably, a subset of RFP+ hepatocytes gave rise to BECs in the BDL-injured liver, while RFP+ BECs were not detected in the sham control group (Figure 4C). Quantitatively, 2.48% ± 0.19% and 0.99% ± 0.28% of hepatocytes were RFP+ in the sham and injury groups, respectively (∗p < 0.05; n = 5) (Figure 4D), indicating that BDL injury significantly reduced the SOX9+ hepatocyte population. The reduction in SOX9+ hepatocytes could be due to the lower competence of SOX9+ hepatocytes in expansion compared with SOX9– hepatocytes during liver injury. It is also likely that SOX9+ hepatocytes have an enhanced potential to develop into BECs. In the BDL liver, 3.46% ± 0.33% of BECs were RFP+, indicating that SOX9+ hepatocytes differentiated into BECs after BDL injury (Figures 4E and 4F). Tamoxifen induction in Alb-CreER;R26-RFP mice resulted in labeling of almost all hepatocytes (>99%, n = 5), and 10.49% ± 0.59% BECs were RFP+ in Alb-CreER;R26-RFP mouse livers after BDL injury (Figures 4G and 4H). In the liver, almost all hepatocytes contributed to approximately 10% of the BECs after BDL (99% hepatocytes to 10% BECs), while labeled SOX9+ hepatocytes that constituted less than 3% of all hepatocytes gave rise to over 3% of the BECs after BDL (∼3% hepatocytes to ∼3% BECs). Therefore, compared with the majority of hepatocytes that were negative for SOX9, this SOX9+ hepatocyte population had a greater propensity to differentiate into BECs after BDL.
Figure 4.
SOX9+ Hepatocytes Contribute to Cholangiocytes after BDL- or DDC-Induced Liver Injury
(A and I) Experimental strategies for tamoxifen treatment (Tam), liver injuries, and tissue analysis.
(B and J) Sirius red staining of liver sections from control and BDL-treated (B) or DDC-treated (J) mice.
(C and K) Immunostaining for RFP and CK19 on liver sections shows that SOX9+ hepatocytes contribute to BECs (arrowheads) following BDL-induced (C) or DDC-induced (K) liver injury.
(D and L) Quantification of the RFP+ hepatocytes percentage. ∗p < 0.05 (n = 5).
(E and M) Quantification of RFP+ BECs percentage. ∗p < 0.05 (n = 5).
(F and N) Cartoon images showing contribution of SOX9+ hepatocyte to BEC (arrowheads) after BDL-induced (F) or DDC-induced (N) liver injury.
(G and O) Immunostaining for RFP and CK19 on liver sections collected from Alb-CreER;R26-RFP after BDL-induced (G) or DDC-induced (O) injury.
(H and P) Quantification of RFP+ BECs percentage. ∗p < 0.05 (n = 5).
Scale bars, 200 μm (B and J) and 50 μm (C, G, K, and O).
Alternatively, we also used a DDC-induced cholestatic liver injury model. Two weeks after tamoxifen treatment, mice were treated with DDC and liver samples were collected 8 weeks later (Figure 4I). Compared with the sham group, significant fibrosis was detected in the DDC-treated mouse livers (Figure 4J). Immunostaining for RFP and CK19 identified RFP+CK19+ BECs in the DDC-treated liver but not in controls (Figure 4K). Quantitatively, 2.59% ± 0.29% and 1.07% ± 0.14% of hepatocytes were RFP+ in the sham and DDC-treated livers, respectively (∗p < 0.05; n = 5) (Figure 4L). In the DDC group, 3.63% ± 0.27% of BECs were RFP+, indicating the contribution of SOX9+ hepatocytes to BECs (Figures 4M and 4N). Likewise, we also induced Alb-CreER;R26-RFP mice with DDC and found that 11.31% ± 0.68% of BECs were RFP+ (Figures 4O and 4P). Quantitatively, >99% of hepatocytes labeled by Alb-CreER contributed to ∼11% of BECs. In comparison, less than 3% of hepatocytes marked by SOX9 contributed to over 3% of BECs, indicating that SOX9+ hepatocytes might have a greater propensity to give rise to BECs after DDC-induced injury. Taken together, these data demonstrated that while the SOX9+ hepatocyte number was reduced, these cells contributed to de novo BECs after BDL or DDC injuries.
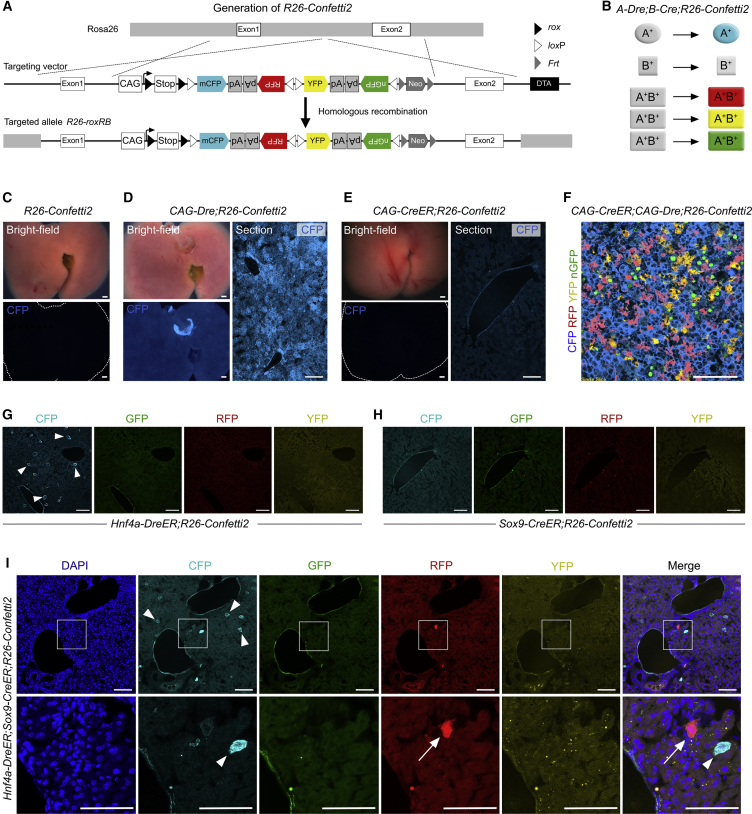
Generation of and Characterization of the R26-Confetti2 Mouse Line
The above data indicated that SOX9+ hepatocytes responded distinctly in different injury models. At the population level, SOX9+ cells could adopt both BEC and hepatocyte fates. We next asked whether some hepatocytes were predetermined to generate BECs or had the potential to generate both cell lineages during injury. To address this, we developed a strategy for single-cell clonal analysis of SOX9+HNF4a+ hepatocytes. The R26-Confetti reporter was previously generated to allow more precise clonal fate-mapping studies (Snippert et al., 2010). The conventional R26-Confetti is operated based on Cre-loxP recombination that leads to four different fluorescence readouts, permitting sparse labeling of cells in one color for more rigorous clonal analysis. To specifically label SOX9+HNF4a+ hepatocytes for clonal analysis, we generated a secondary version of R26-Confetti (R26-Confetti2) that was responsive to two orthogonal recombinases, Cre and Dre. A cDNA containing the CAG-rox-Stop-rox-Confetti cassette was knocked into the Rosa26 gene locus by homologous recombination (Figure 5A). When this R26-Confetti2 reporter was crossed with Cre and Dre mouse lines, there were distinct fluorescent readouts for cells: Cre+Dre– cells were negative for any fluorescence; Cre–Dre+ cells were positive for CFP but negative for other types of fluorescence; and Cre+Dre+ cells were positive for three fluorescent proteins, YFP, GFP, and RFP (Figure 5B). We next performed some control experiments to test whether R26-Confetti2 worked as designed. For R26-Confetti2 alone, there was no detectable fluorescence signal in the liver (Figure 5C). For CAG-Dre;R26-Confetti2 mice, cells in the liver expressed CFP in whole-mount or sectional imaging (Figure 5D). CFP, but not other fluorescence signals, was also detected in other organs of CAG-Dre;R26-Confetti2 mice (Figure S3A). For CAG-CreER;R26-Confetti2 mice treated with tamoxifen, there was no detectable fluorescence signal in the liver in whole-mount and sectional imaging (Figure 5E). We did not detect fluorescence signals in other organs or tissues (Figure S3B). For CAG-CreER;CAG-Dre;R26-Confetti2 mice treated with tamoxifen, we could detect CFP, GFP, YFP, and RFP signals in the liver (Figure 5F). Similarly, we detected these fluorescence signals in other organs, such as the heart (Figures S4A and S4B). Taken together, the above data indicated that R26-Confetti2 functioned as expected and could be used for clonal analysis of single SOX9+HNF4a+ hepatocytes during liver injury.
Figure 5.
Generation and Characterization of R26-Confetti2 Mouse
(A) Schematic showing strategy for generation of R26-Confetti2 reporter allele by homologous recombination.
(B) Distinct cell labeling after Cre and Dre recombinations.
(C–E) Whole-mount fluorescent images of R26-Confetti2 (C), CAG-Dre;R26-Confetti2 (D), and CAG-CreER;R26-Confetti2 (E) mouse livers. CFP is detected in CAG-Dre;R26-Confetti2 liver and its section, but not in R26-Confetti2 or CAG-CreER;R26-Confetti2 livers. Tamoxifen was induced 2 days before tissue collection.
(F) Fluorescent image of liver section from CAG-CreER;CAG-Dre;R26-Confetti2 mouse. Tamoxifen was induced at 12.5 days before tissue collection.
(G and H) Fluorescent images of liver sections from Hnf4a-DreER;R26-Confetti2 (G) or Sox9-CreER;R26-Confetti2 (H) mouse. There are no GFP, RF,P or YFP signals detected in liver sections.
(I) Fluorescent image of Hnf4a-DreER;Sox9-CreER;R26-Confetti2 mouse liver sections. Arrows indicate sparse RFP+ hepatocytes, and arrowheads indicate hepatocytes that have recombined with DreER but not CreER.
Each image is representative of five individual samples. Scale bars, 100 μm.
We used R26-Confetti2 for the analysis of SOX9+HNF4a+ hepatocytes during liver homeostasis. Mouse crossing generated three different genotypes among mouse littermates: Hnf4a-DreER;R26-Confetti2, Sox9-CreER;R26-Confetti2, and Hnf4a-DreER;Sox9-CreER;R26-Confetti2. For these three groups, we injected tamoxifen at the adult stage (8–10 weeks old) and then collected liver samples for analysis after 10–12 weeks. Sectional imaging of Hnf4a-DreER;R26-Confetti2 liver tissues showed that hepatocytes were positive for CFP only (Figure 5G). There was no detectable fluorescence signal in Sox9-CreER;R26-Confetti2 mouse livers (Figure 5H). In Hnf4a-DreER;Sox9-CreER;R26-Confetti2 mice, Dre-rox recombination led to a subset of HNF4a+ hepatocytes labeled with CFP (arrowheads, Figure 5I). Both Dre-rox and Cre-loxP recombination led to one of the other three fluorescence signals, such as sparse RFP labeling in HNF4a+SOX9+ hepatocytes (arrow, Figure 5I). The sparsely labeled SOX9+ hepatocytes remained as single cells and did not contribute to any ductal cell during liver homeostasis.
Bipotency of Single SOX9+ Hepatocytes after Liver Injury
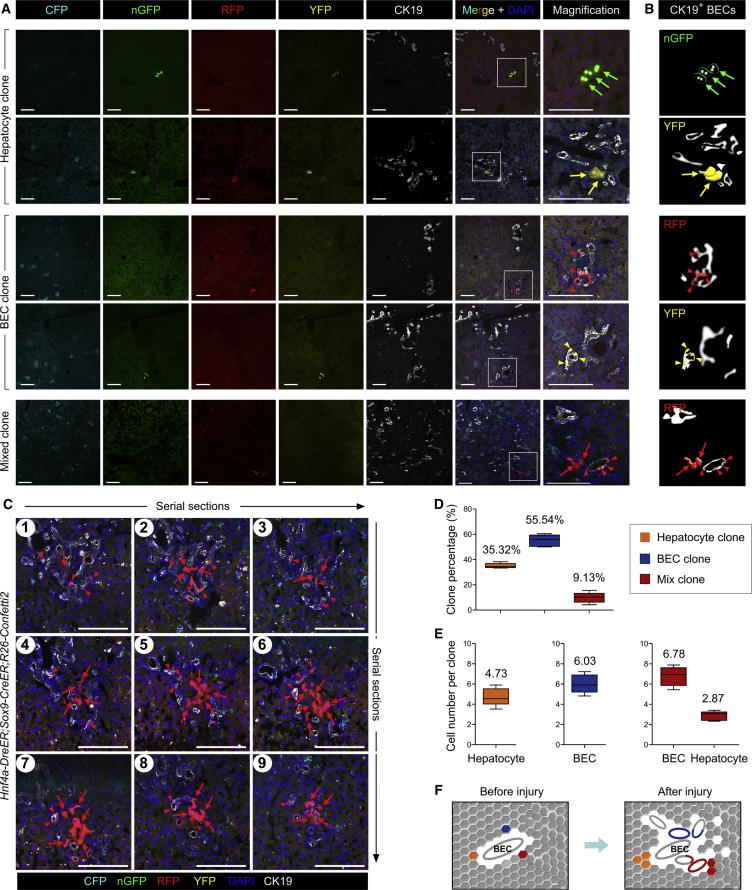
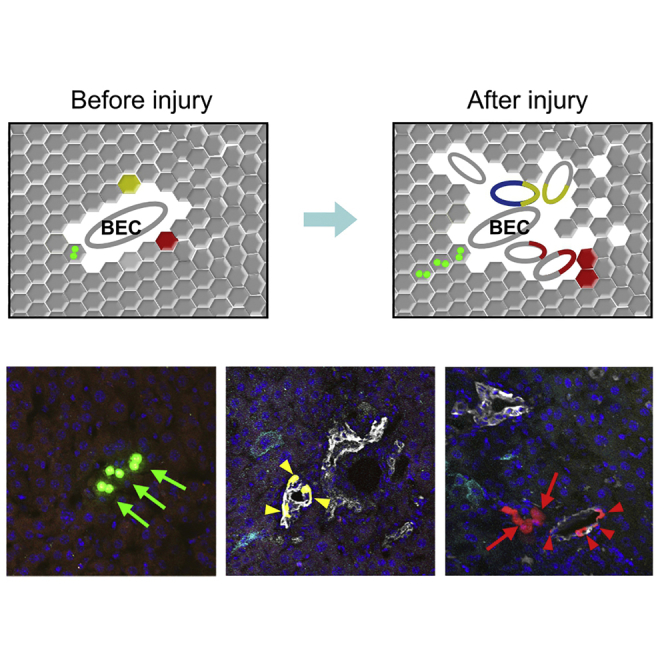
After labeling sparse SOX9+ hepatocytes after tamoxifen treatment, Hnf4a-DreER;Sox9-CreER;R26-Confetti2 mice were subjected to DDC-induced liver injury, and liver tissues were collected for analysis after 8–10 weeks. We observed very sparse labeling in the injured liver, and cell labeling could be categorized into three distinct clones: hepatocytes, BECs, and mixed clones that contained both hepatocytes and BECs (Figures 6A and 6B). Two or three labeled hepatocytes, such as nGFP+ or YFP+ hepatocytes, were detected (Figures 6A and 6B), indicating cell proliferation after liver injury. To determine whether some of the labeled SOX9+ hepatocytes generated BECs after injury, we stained the liver sections with CK19 and developed the signal in the far-red channel to avoid mixing with other fluorescence signals. In each field there was only one clone with a unique fluorescent color, such as GFP, YFP, or RFP. We found that a substantial number of clones exhibited CK19 expression, indicating that they were the BEC clones (arrowheads, Figures 6A and 6B). Fewer clones were hepatocytes in the injured liver, and these labeled hepatocytes in one clone were usually detected in a cluster (arrows, Figures 6A and 6B), indicating their proliferation after injury. Interestingly, we also detected mixed clones that contained both hepatocytes and BECs in one single-color clone (Figures 6A and 6B), suggesting that these two different cell lineages could be derived from a single SOX9+ hepatocyte. We next collected serial liver sections and stained them with CK19 for analysis of all the labeled cells in each clone (Figure 6C). Serially stained sections showed the clone clearly in the liver and confirmed the bipotency of a single SOX9+ hepatocyte during liver injury (Figures 6C, S5, and S6). Quantification of the three types of clones in Hnf4a-DreER;Sox9-CreER;R26-Confetti2 livers after injury showed hepatocyte clones, BEC clones, and mixed clones that constituted 35.32% ± 0.82%, 55.54% ± 2.10%, and 9.13% ± 1.91% of all clones, respectively (Figure 6D). We also quantified the cell number of each clone and found that there were 4.73 ± 0.39 hepatocytes in hepatocyte clones; 6.03 ± 0.41 BECs in BEC clones; and 2.87 ± 0.20 hepatocytes and 6.78 ± 0.42 BECs in mixed clones (Figure 6E). We confirmed the bipotency of SOX9+ hepatocytes by clonal analysis in the BDL model (Figure S7). Thus, clonal fate-mapping analysis of a single SOX9+ hepatocyte showed their uni- and bipotency during liver injury and repair (Figure 6F).
Figure 6.
Identification of Bipotent SOX9+ Hepatocyte after Liver Injury
(A and B) Fluorescent sections stained with BEC marker CK19 shows three distinct clones: hepatocyte clone, BEC clone, and mixed clone that contains both hepatocyte and BEC. Arrows or arrowheads indicate nGFP+, RFP+, or YFP+ hepatocytes or BECs, respectively. Cartoon images in (B) denote the magnified images in (A).
(C) Serial sections of a bipotent clone that contains both BECs (arrowheads) and hepatocytes (arrows).
(D) Quantification of the percentage of three types of clones in injured liver. A total of 246 clones were analyzed. Data are mean ± SEM; n = 5.
(E) Quantification of cell number in each type of clones. A total of 246 clones were analyzed. Data are mean ± SEM; n = 5.
(F) Cartoon figure showing the cell fate of three types of clones after injury.
Scale bars, 100 μm.
Discussion
Two major epithelial cell populations, BECs and hepatocytes, have recently been reported to give rise to each other under specific injury or disease conditions (Deng et al., 2018, Raven et al., 2017, Schaub et al., 2018, Tarlow et al., 2014b, Yanger et al., 2013, Yanger et al., 2014). Whether a single hepatocyte is bipotent for both BECs and hepatocytes remains unknown. A recent study reported that SOX9+ hepatocytes exhibit a high proliferative potential depending on the condition of injury without tumorigenesis (Font-Burgada et al., 2015). In our study, we used a dual recombinase-based lineage-tracing strategy to genetically target SOX9+ hepatocytes. We found that a single SOX9+ hepatocyte differentiated into both BECs and hepatocytes after injury, providing genetic evidence for the existence of bipotent SOX9+ hepatocytes as progenitor cells for liver repair and regeneration.
This work mainly utilized genetic tools to specifically label SOX9+ hepatocytes. The combination of Cre-loxP and Dre-rox allowed another layer of regulation in the specificity in genetic targeting. Our recent study using dual recombinases and a nested reporter (NR1) permitted specific labeling of SOX9+ BECs without contamination of SOX9+ hepatocytes (He et al., 2017). Unlike a previous study reporting the differentiation of SOX9+ ductal cells to hepatocytes (Furuyama et al., 2011), our fate mapping of SOX9+ BECs showed their ductal cell fate but not hepatocyte fate via differentiation during liver homeostasis or after injury (He et al., 2017). The proposed ductal cell-specific Sox9-CreER used in a previous study (Furuyama et al., 2011) might not be specific for ductal cells, as SOX9 is also expressed in hepatocytes (Figures 1C and 1G), although the expression level of SOX9 in hepatocytes is lower than that that of BECs. As Sox9-CreER labels hepatocytes in addition to BECs, the contribution of SOX9+ progenitors to hepatocytes (Furuyama et al., 2011) could also be interpreted to indicate that SOX9+ hepatocytes labeled at the beginning contributed to more hepatocytes during liver regeneration after injury. Genetic lineage tracing with “duct-specific” Cre stains should be carefully examined to determine whether any hepatocyte was labeled.
With the advent of single-cell sequencing technology, more different subsets of hepatocytes (Halpern et al., 2017) or biliary epithelial cell populations (Tarlow et al., 2014b) could be elucidated, and their in vivo functions remain largely unknown. Our dual recombinase-mediated genetic systems reported here would be valuable to more precisely label a subset of liver cell populations and study their cell fate during liver homeostasis and injuries. Moreover, we recently developed a sequential intersectional genetics strategy whereby Dre-rox recombination mediates the release of Cre (Pu et al., 2018). This strategy would also allow gene functional analysis by crossing the Dre-mediated Cre mouse strain with the available flox gene allele for in vivo gain- or loss-of-function studies. Moreover, dual recombination systems could be used to manipulate two distinct cell populations in vivo to understand their function and cell plasticity during liver injuries.
SOX9 has been previously studied as a progenitor or stem cell marker for cell expansion and fate determination (Guo et al., 2012, Lincoln et al., 2007, Zhang et al., 2017). During injury, SOX9 could also be activated in hepatocytes (Cao et al., 2017, He et al., 2014). SOX9+ hepatocytes are located near ductal cells and could contribute to BECs during liver injuries (Yanger et al., 2013). These SOX9+ hepatocytes, compared with the remaining SOX9– hepatocytes, have a propensity to contribute to BECs after injuries. Quantitatively, the contribution of SOX9+ hepatocytes to BECs was 10-fold more efficient than SOX9– hepatocytes after BDL- or DDC-induced liver injuries. SOX9 may predispose hepatocytes to conditions that facilitate reprogramming into BECs (Yanger et al., 2013). Signaling pathways that regulate hepatocytes to BECs include Notch (Fan et al., 2012, Sekiya and Suzuki, 2012), Hippo-Yap (Yimlamai et al., 2014), and TGFβ (Schaub et al., 2018). Whether these signaling pathways were activated in SOX9+ hepatocytes to modulate their bipotency after liver injury merits investigation in the future. Alternatively, the adjacent ductal cells could directly or indirectly affect and reprogram the cell fate of SOX9+ hepatocytes. Whether ductal cells influence the cell fate of hepatocytes through paracrine signaling remains unknown, and niche signaling has now been reported to influence the function of cells rather than the hard-wired property of stem cells (Clevers and Watt, 2018). It is therefore of interest whether these SOX9+ hepatocytes, relocated away from the portal region, could maintain the robust ability to differentiate into BECs. Elucidation of the regulatory mechanism of SOX9+ hepatocyte progenitors may provide information for amplifying the bipotent progenitor population for potential applications in liver repair and regeneration. As SOX9+ hepatocytes are unique in their ability to differentiate into BECs and expand hepatocytes, they could be a potentially important drug target for treating liver diseases in the future.
Experimental Procedures
Mice
All mouse studies were carried out strictly according to the guidelines of the Institutional Animal Care and Use Committee at the Institute of Biochemistry and Cell Biology and the Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. Hnf4a-DreER was generated by CRISPR/Cas9 through homologous recombination. A complementary cDNA encoding IRES-DreERT2 was inserted in-frame with the translation codon of the Hnf4a gene. The chimeric mice positive for targeted ESCs were germline transferred to F1 generation and bred on a C57BL/6 × ICR background. The Sox9-CreERT2 line was generated by the National Institute for Biological Sciences, Beijing, China. R26-Ai66 (Rosa26-rox-Stop-rox-loxP-Stop-loxP-tdTomato) was generated as reported previously (Madisen et al., 2015, Zhang et al., 2016). The Rosa26-Rox-Stop-Rox-tdTomato (R26-RSR-RFP) mouse line was generated by crossing ACTB-Cre with R26-Ai66 to excise the second loxP-flanked Stop cassette, and ACTB-Cre was not passaged to the subsequent mouse breeding. R26-RSR-RFP was responsive to Dre but not Cre recombinase. The Rosa26-rox-Stop-rox-Confetti (R26-Confetti2) was generated by targeting CAG-rox-Stop-rox-Confetti cassette into the Rosa26 gene locus by homologous recombination (Figure 5A). All experimental mice were maintained on a C57BL6/ICR mixed background. Tamoxifen (Sigma, T5648) was dissolved in corn oil (20 mg/mL) and administered by gavage at the indicated time points. We treated R26-RFP and R26-Confetti2 mice with 0.4 mg of tamoxifen per gram of mouse body weight (mg/g) and 0.15 mg/g tamoxifen, respectively.
Genomic PCR
Genomic DNA was prepared from mouse tails according to the standard protocols described previously (Wang et al., 2017). All mice were genotyped with specific primers that distinguish knockin allele from wild-type allele. Information on the primers can be found in Supplemental Information.
Injury Model
For the CCl4-induced chronic injury model, CCl4 was dissolved at 1:3 in corn oil and injected intraperitoneally at a dose of 4 μL/g body weight every 3 days, repeated 10 times. Partial hepatectomy (PHx) was generated by removing two-thirds of the liver to induce the injury (Wang et al., 2017). The BDL injury model was prepared according to established protocols described previously (Pu et al., 2016). For the DDC-induced chronic injury model, mice received mouse diet (Harlan Teklad, 5015) containing 0.1% DDC (Sigma-Aldrich).
Whole-Mount Fluorescence Microscopy
Collected mouse liver was washed in PBS and placed on agar for the whole-mount bright-field and fluorescence imaging using a Zeiss stereoscope (AxioZoom V16). To determine magnification of specific regions, we used the automated z-stack images acquired by the stereoscope (Zeiss AxioZoom V16).
Immunostaining
Immunostaining was performed according to the standard protocols described previously (Tian et al., 2013). The following antibodies were used: RFP (Rockland, 600-401-379, 1:200), HNF4a (Santa Cruz Biotechnology, sc-6556, 1:100), cytokeratin 19 (CK19, Developmental Studies Hybridoma Bank, TROMA-III, 1:100), VE-cadherin (R&D Systems, AF1002, 1:100), desmin (R&D, AF3844, 1:100), PDGFRα (R&D, AF1062, 1:100), E-cadherin (Cell Signaling Technology, 3195, 1:100), EpCAM (Abcam, ab92383, 1:100), αSMA (Sigma, F3777, 1:100), and Ki67 (Thermo Scientific, RM-9106-S0, 1:100). Signals were developed with Alexa fluorescence antibodies (Invitrogen), and nuclei were counterstained with 4′,6-diamidino-2-phenylindole (Vector Laboratories). For clonal analysis, CK19 was stained on the liver sections in the far-red channel. In total 246 clones were analyzed: 820 cholangiocytes in cholangiocyte clones; 412 hepatocytes in hepatocyte clones; 156 cholangiocytes; and 66 hepatocytes in mixed clones. There was bleed-through of fluorescence for YFP and GFP because the YFP signal could also be detected in the GFP channel. In addition, our GFP is nGFP (nuclear), so the pure GFP signal should be in the nucleus while the bleed-through signal in the GFP channel from YFP should not be a nuclear signal. Immunostaining images were acquired by an Olympus fluorescence microscope (BX53), a Zeiss stereomicroscope (AxioZoom V16), a Zeiss confocal laser scanning microscope (LSM510), and an Olympus confocal microscope (FV1200).
Sirius Red Staining
Sirius red staining was used to assess fibrotic tissue in chronic injury models, and was performed according to the standard protocols described previously (Wang et al., 2017).
Statistics
All data were collected from at least five independent experiments as indicated. Data for two groups were analyzed by a two-sided unpaired Student's t test, whereas comparison between more than two groups was performed using an analysis of variance followed by Tukey's multiple comparison tests. Significance was accepted when p < 0.05. All data are presented as mean values ± SEM.
Author Contributions
X. Han, Y.W., J.L., and B.Z. designed the study. X. Han and Y.W. performed experiments and analyzed the data. W.P., X. Huang, L.Q., Y.L., W.Y., H.Z., X.L., L.H., and L.Z. bred the mice, performed experiments, or provided material and valuable comments. Y.J., J.L., and K.O.L. provided reagents and intellectual input to this study and edited the manuscript. B.Z. supervised the study, analyzed the data, and wrote the manuscript.
Acknowledgments
We thank Shanghai Model Organisms Center for mouse generation; and Baojin Wu, Guoyuan Chen, Zhonghui Weng, and Aimin Huang for animal husbandry. We also thank the technical help from Wei Bian, Tengfei Zhang, and members of National Center for Protein Science Shanghai for assistance in microscopy. This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (CAS, XDA16020204, XDB19000000), National Key Research & Development Program of China (2018YFA0107900, 2018YFA0108100, 2016YFC1300600, 2017YFC1001303), National Science Foundation of China (31730112, 91639302, 31625019, 91849202, 81761138040, 81872241, 31701292), Youth Innovation Promotion Association of CAS (2060299), Key Project of Frontier Sciences of CAS (QYZDB-SSW-SMC003), Shanghai Science and Technology Commission (17ZR1449600), the Program for Guangdong Introduction Innovative and Enterpreneurial Teams (2017ZT07S347), Shanghai Yangfan Project, China Postdoctoral Science Foundation, China Postdoctoral Innovative Talent Support Program, China Young Talents Lift Engineering, Boehringer Ingelheim, Sanofi-SIBS Fellowship, AstraZeneca, Royal Society-Newton Advanced Fellowship, Research Council of Hong Kong (04110515, 14111916, C4024-16W), and Health and Medical Research Fund (03140346, 04152566).
Published: February 14, 2019
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and seven figures and can be found with this article online at https://doi.org/10.1016/j.stemcr.2019.01.010.
Contributor Information
Jie Lu, Email: lujie791111@tongji.edu.cn.
Bin Zhou, Email: zhoubin@sibs.ac.cn.
Accession Numbers
Data supporting the findings of this study are available within the article and its Supplemental Information, and from the corresponding author upon reasonable request.
Supplemental Information
References
- Antoniou A., Raynaud P., Cordi S., Zong Y., Tronche F., Stanger B.Z., Jacquemin P., Pierreux C.E., Clotman F., Lemaigre F.P. Intrahepatic bile ducts develop according to a new mode of tubulogenesis regulated by the transcription factor SOX9. Gastroenterology. 2009;136:2325–2333. doi: 10.1053/j.gastro.2009.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W., Chen K., Bolkestein M., Yin Y., Verstegen M.M.A., Bijvelds M.J.C., Wang W., Tuysuz N., Ten Berge D., Sprengers D. Dynamics of proliferative and quiescent stem cells in liver homeostasis and injury. Gastroenterology. 2017;153:1133–1147. doi: 10.1053/j.gastro.2017.07.006. [DOI] [PubMed] [Google Scholar]
- Clevers H., Watt F.M. Defining adult stem cells by function, not by phenotype. Annu. Rev. Biochem. 2018;87:1015–1027. doi: 10.1146/annurev-biochem-062917-012341. [DOI] [PubMed] [Google Scholar]
- Deng X., Zhang X., Li W., Feng R.X., Li L., Yi G.R., Zhang X.N., Yin C., Yu H.Y., Zhang J.P. Chronic liver injury induces conversion of biliary epithelial cells into hepatocytes. Cell Stem Cell. 2018;23:114–122.e3. doi: 10.1016/j.stem.2018.05.022. [DOI] [PubMed] [Google Scholar]
- Espanol-Suner R., Carpentier R., Van Hul N., Legry V., Achouri Y., Cordi S., Jacquemin P., Lemaigre F., Leclercq I.A. Liver progenitor cells yield functional hepatocytes in response to chronic liver injury in mice. Gastroenterology. 2012;143:1564–1575.e7. doi: 10.1053/j.gastro.2012.08.024. [DOI] [PubMed] [Google Scholar]
- Fan B., Malato Y., Calvisi D.F., Naqvi S., Razumilava N., Ribback S., Gores G.J., Dombrowski F., Evert M., Chen X., Willenbring H. Cholangiocarcinomas can originate from hepatocytes in mice. J. Clin. Invest. 2012;122:2911–2915. doi: 10.1172/JCI63212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber E. Similarities in the sequence of early histological changes induced in the liver of the rat by ethionine, 2-acetylamino-fluorene, and 3′-methyl-4-dimethylaminoazobenzene. Cancer Res. 1956;16:142–148. [PubMed] [Google Scholar]
- Font-Burgada J., Shalapour S., Ramaswamy S., Hsueh B., Rossell D., Umemura A., Taniguchi K., Nakagawa H., Valasek M.A., Ye L. Hybrid periportal hepatocytes regenerate the injured liver without giving rise to cancer. Cell. 2015;162:766–779. doi: 10.1016/j.cell.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama K., Kawaguchi Y., Akiyama H., Horiguchi M., Kodama S., Kuhara T., Hosokawa S., Elbahrawy A., Soeda T., Koizumi M. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat. Genet. 2011;43:34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- Grompe M. Liver stem cells, where art thou? Cell Stem Cell. 2014;15:257–258. doi: 10.1016/j.stem.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Guo W., Keckesova Z., Donaher J.L., Shibue T., Tischler V., Reinhardt F., Itzkovitz S., Noske A., Zürrer-Härdi U., Bell G. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern K.B., Shenhav R., Matcovitch-Natan O., Tóth B., Lemze D., Golan M., Massasa E.E., Baydatch S., Landen S., Moor A.E. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 2017;542:352–356. doi: 10.1038/nature21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Lu H., Zou Q., Luo L. Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology. 2014;146:789–800.e8. doi: 10.1053/j.gastro.2013.11.045. [DOI] [PubMed] [Google Scholar]
- He L., Li Y., Li Y., Pu W., Huang X., Tian X., Wang Y., Zhang H., Liu Q., Zhang L. Enhancing the precision of genetic lineage tracing using dual recombinases. Nat. Med. 2017;23:1488–1498. doi: 10.1038/nm.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln J., Kist R., Scherer G., Yutzey K.E. Sox9 is required for precursor cell expansion and extracellular matrix organization during mouse heart valve development. Dev. Biol. 2007;305:120–132. doi: 10.1016/j.ydbio.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W.Y., Bird T.G., Boulter L., Tsuchiya A., Cole A.M., Hay T., Guest R.V., Wojtacha D., Man T.Y., Mackinnon A. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat. Cell Biol. 2015;17:971–983. doi: 10.1038/ncb3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Garner A., Shimaoka D., Chuong A., Klapoetke N., Li L., van der Bourg A., Niino Y., Egolf L., Monetti C. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron. 2015;85:942–958. doi: 10.1016/j.neuron.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malato Y., Naqvi S., Schurmann N., Ng R., Wang B., Zape J., Kay M.A., Grimm D., Willenbring H. Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J. Clin. Invest. 2011;121:4850–4860. doi: 10.1172/JCI59261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima A., Tanaka M., Itoh T. Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell. 2014;14:561–574. doi: 10.1016/j.stem.2014.04.010. [DOI] [PubMed] [Google Scholar]
- Michalopoulos G.K., Barua L., Bowen W.C. Transdifferentiation of rat hepatocytes into biliary cells after bile duct ligation and toxic biliary injury. Hepatology. 2005;41:535–544. doi: 10.1002/hep.20600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper H., Kent G., Stern R. Ductular cell reaction in the liver in hepatic injury. J. Mt. Sinai Hosp. N. Y. 1957;24:551–556. [PubMed] [Google Scholar]
- Pu W., He L., Han X., Tian X., Li Y., Zhang H., Liu Q., Huang X., Zhang L., Wang Q.D. Genetic targeting of organ-specific blood vessels. Circ. Res. 2018;123:86–99. doi: 10.1161/CIRCRESAHA.118.312981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu W., Zhang H., Huang X., Tian X., He L., Wang Y., Zhang L., Liu Q., Li Y., Li Y. Mfsd2a+ hepatocytes repopulate the liver during injury and regeneration. Nat. Commun. 2016;7:13369. doi: 10.1038/ncomms13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven A., Lu W.Y., Man T.Y., Ferreira-Gonzalez S., O’Duibhir E., Dwyer B.J., Thomson J.P., Meehan R.R., Bogorad R., Koteliansky V. Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature. 2017;547:350–354. doi: 10.1038/nature23015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Torres D., Affo S., Coll M., Morales-Ibanez O., Millan C., Blaya D., Alvarez-Guaita A., Rentero C., Lozano J.J., Maestro M.A. The biliary epithelium gives rise to liver progenitor cells. Hepatology. 2014;60:1367–1377. doi: 10.1002/hep.27078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub J.R., Huppert K.A., Kurial S.N.T., Hsu B.Y., Cast A.E., Donnelly B., Karns R.A., Chen F., Rezvani M., Luu H.Y. De novo formation of the biliary system by TGFβ-mediated hepatocyte transdifferentiation. Nature. 2018;557:247–251. doi: 10.1038/s41586-018-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub J.R., Malato Y., Gormond C., Willenbring H. Evidence against a stem cell origin of new hepatocytes in a common mouse model of chronic liver injury. Cell Rep. 2014;8:933–939. doi: 10.1016/j.celrep.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya S., Suzuki A. Intrahepatic cholangiocarcinoma can arise from Notch-mediated conversion of hepatocytes. J. Clin. Invest. 2012;122:3914–3918. doi: 10.1172/JCI63065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert H.J., van der Flier L.G., Sato T., van Es J.H., van den Born M., Kroon-Veenboer C., Barker N., Klein A.M., van Rheenen J., Simons B.D. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Kanai-Azuma M., Kanai Y. From sex determination to initial folliculogenesis in mammalian ovaries: morphogenetic waves along the anteroposterior and dorsoventral axes. Sex. Dev. 2015;9:190–204. doi: 10.1159/000440689. [DOI] [PubMed] [Google Scholar]
- Tarlow B.D., Finegold M.J., Grompe M. Clonal tracing of SOX9+ liver progenitors in mouse oval cell injury. Hepatology. 2014;60:278–289. doi: 10.1002/hep.27084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarlow B.D., Pelz C., Naugler W.E., Wakefield L., Wilson E.M., Finegold M.J., Grompe M. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell. 2014;15:605–618. doi: 10.1016/j.stem.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Hu T., Zhang H., He L., Huang X., Liu Q., Yu W., He L., Yang Z., Zhang Z. Subepicardial endothelial cells invade the embryonic ventricle wall to form coronary arteries. Cell Res. 2013;23:1075–1090. doi: 10.1038/cr.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Pu W.T., Zhou B. Cellular origin and developmental program of coronary angiogenesis. Circ. Res. 2015;116:515–530. doi: 10.1161/CIRCRESAHA.116.305097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Huang X., He L., Pu W., Li Y., Liu Q., Li Y., Zhang L., Yu W., Zhao H. Genetic tracing of hepatocytes in liver homeostasis, injury, and regeneration. J. Biol. Chem. 2017;292:8594–8604. doi: 10.1074/jbc.M117.782029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanger K., Knigin D., Zong Y., Maggs L., Gu G., Akiyama H., Pikarsky E., Stanger B.Z. Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell. 2014;15:340–349. doi: 10.1016/j.stem.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanger K., Zong Y., Maggs L.R., Shapira S.N., Maddipati R., Aiello N.M., Thung S.N., Wells R.G., Greenbaum L.E., Stanger B.Z. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 2013;27:719–724. doi: 10.1101/gad.207803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yimlamai D., Christodoulou C., Galli G.G., Yanger K., Pepe-Mooney B., Gurung B., Shrestha K., Cahan P., Stanger B.Z., Camargo F.D. Hippo pathway activity influences liver cell fate. Cell. 2014;157:1324–1338. doi: 10.1016/j.cell.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Huang X., Liu K., Tang J., He L., Pu W., Liu Q., Li Y., Tian X., Wang Y. Fibroblasts in an endocardial fibroelastosis disease model mainly originate from mesenchymal derivatives of epicardium. Cell Res. 2017;27:1157–1177. doi: 10.1038/cr.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Pu W., Tian X., Huang X., He L., Liu Q., Li Y., Zhang L., He L., Liu K. Genetic lineage tracing identifies endocardial origin of liver vasculature. Nat. Genet. 2016;48:537–543. doi: 10.1038/ng.3536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.