Hepatitis B virus (HBV) infection affects 1.25 million persons in the United States and approximately 250 million persons worldwide.1 In persons exposed to HBV as adults, chronic infection is uncommon (<10%), but increases the risk for cirrhosis and hepatocellular carcinoma. The remainder of cases experience spontaneous clearance of HBV with acute self-limited hepatitis. However, in fewer than 1% of cases, acute infection with HBV can cause acute liver failure (ALF) with unfavorable outcomes.2 The factors that contribute to a chronic or fulminant course, rather than self-limited and resolving hepatitis, remain poorly understood. Efforts to identify unique molecular signatures in the hepatitis B genome responsible for HBV ALF have been unrewarding to date despite early evidence that unique precore mutations may be associated with ALF.3 Host genetic analysis of chronic HBV has been performed only in individuals of Asian descent and examination of exome arrays of patients with HBV ALF have not been published. By comparing 2 extreme phenotypes, HBV ALF with chronic HBV infection in adults of European descent, we evaluated exome arrays to explore the host factors underlying the wide-ranging response to HBV infection.
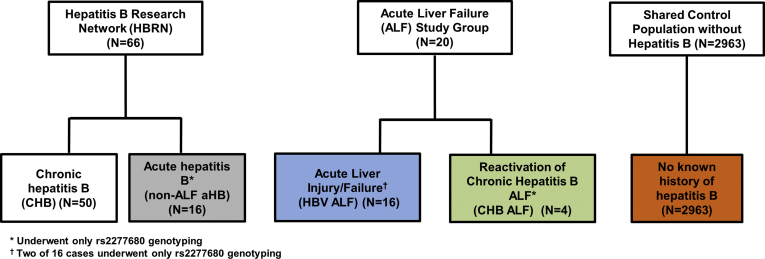
All of the study participants were white and of European descent. Fifty participants with chronic HBV were identified. Initially, 14 cases of HBV ALF were identified and underwent exome array testing. Two additional HBV ALF patients were identified later and only underwent polymerase chain reaction for rs2277680. The shared control population comprised 2963 patients with exome chip data. The additional populations explored with polymerase chain reaction genotyping for rs2277680 included 16 patients with acute hepatitis B without ALF (non-ALF aHB) and 4 acute-on-chronic HBV reactivations leading to HBV ALF (CHBV ALF) cases (Figure 1). The exome array results with P < 1 × 10-4 were reviewed to find exploratory genes for future study. No polymorphisms met the threshold for exome-wide significance (P < 5 × 10-7).
Figure 1.
Study population and parent cohorts.
When comparing the HBV ALF population with the general population controls, rs147992634 in LAMB4, rs9894429 in NPLOC4, and rs1195569 in RIMBP2 had increased minor allele frequencies in the HBV ALF patients, and rs2277680 and rs1050998 in chemokine ligand 16 (CXCL16) had decreased minor allele frequencies and met the prespecified threshold of P < 1 × 10-4, but not exome-wide significance (Supplementary Table 1).
When comparing the adult-acquired chronic hepatitis B (CHB) population with the control population, 14 loci were identified with a P < 1 × 10-4 for rs58022607, rs201682692, rs72669494, rs2199291, rs73134074, rs57168946, rs1000960, rs11539377, rs143596619, rs56195264, rs7192086, rs45602538, rs73365341, and rs9668527 (Supplementary Table 2), however, again, none met the threshold for exome-wide significance. When comparing the HBV ALF population with the CHB population only rs62417812 in MANEA and rs3849969 in SEC24C were associated with P < 1 × 10-4, but not exome-wide significance (Supplementary Table 3, Supplementary Table 4, Supplementary Table 5).
We evaluated the published literature regarding the aforementioned candidate genes. MANEA and SEC24C, which catalyze the release of oligosaccharides in the Golgi and are involved in vesicle trafficking, respectively, did not have a clear putative impact on HBV pathogenesis. CXCL16 is a chemokine that interacts with chemokine receptor CXCR6 and is induced by inflammatory cytokines. Natural killer T (NKT) cells are recruited to the hepatic sinusoids in response to the secretion of CXCL16, which is produced by multiple intrahepatic cells. We subsequently focused on the 2 polymorphisms in CXCL16, which were in complete linkage disequilibrium, and genotyped rs2277680 in 2 additional HBV ALF cases who were AG and GG. The aggregate P value was 8.8 × 10-5 for HBV ALF cases compared with controls in the population. We proceeded to evaluate rs2277680 in 4 patients with CHB ALF and identified that the allele frequency was similar to the general population: 50% A and 50% G (Table 1). Among 15 non-ALF aHB patients the genotype also was similar to the general population: 50% A and 50% G (Table 1).
Table 1.
CXCL16 rs2277680 Allele Frequencies by HBV Status
| N | Allele 1 (A) | Allele 2a (G) | |
|---|---|---|---|
| Chronic HBV | 50 | 43% | 57% |
| HBV ALFb | 16 | 9% | 91% |
| CHB ALF (reactivation) | 4 | 50% | 50% |
| Non-ALF acute hepatitis Bc | 16 | 50% | 50% |
| Controls shared from other studies | 2963 | 43% | 57% |
Genotype associated with increased NKT cell adhesion.
HBV ALF aggregate P = 8.8 × 10-5.
One participant had undetermined genotype by polymerase chain reaction.
By using multiple well-characterized cohorts encompassing the spectrum of acute and chronic hepatitis B infection, we evaluated the host exonic content using exome arrays and identified an association between rs2277680 in CXCL16 and HBV ALF that approached, but did not meet, the threshold for exome-wide significance. The trend toward increased frequency of the G allele was not present in the small group of patients with acute-on-chronic HBV ALF (n = 4), which likely represents a distinct phenotype.
Previous studies of factors associated with chronic HBV have been limited to Asian populations with primarily vertical transmission, and previously described loci associated with chronicity were not replicated in this white population of European descent with a spectrum of HBV infection. Although our study was limited in its ability to identify a locus meeting the threshold for exome-wide significance, the association between rs2277680 G in CXCL16 is supported by in vitro and in vivo studies of this polymorphism. CXCL16 is a transmembrane chemokine expressed on hepatic endothelial cells that interact with chemokine receptor CXCR6 and is integral to NKT cell patrolling of hepatic sinusoids.4 NKT cells make up less than 1% of lymphocytes in most organs but are highly expressed in the liver, representing approximately 30% of the lymphocyte population,5 and are critically involved in murine models of hepatitis.6 In addition, NKT cell activation in the livers of HBV transgenic mice resulted in rapid abolition of HBV replication.7 Activated macrophages also can express CXCL16, promoting recruitment of NKT cells that amplify the inflammatory response in murine models of hepatitis.8 Furthermore, pharmacologic CXCL16 blockade in animal models of hepatic injury decreased the inflammatory infiltrate and intrahepatic levels of inflammatory cytokines.9
The allele frequencies of the A and G allele in the general population resemble the shared controls with approximately 50% frequency for each allele. Importantly, the subset of patients with non-ALF aHB with rs2277680 genotyping had similar allele frequencies to the general population controls, supporting our use of the latter group as a control population. Patients with HBV ALF had over-representation of the G allele. The A allele, which is under-represented in HBV ALF patients, is a functional missense mutation that leads to decreased adhesion of CXCR6+ cells.10 Therefore, patients with HBV ALF over-represented a polymorphism that may lead to increased adhesion of NKT cells, which are critical mediators of inflammation in models of hepatitis. Our findings warrant further study of the role of CXCL16 in the pathogenesis of severe acute hepatitis.
Footnotes
Author contributions Veeral Ajmera was responsible for the study concept and design, analysis and interpretation of data, drafting the manuscript, and critical revision; Hailiang Huang was responsible for the study concept and design, statistical analysis, analysis and interpretation of data, and critical revision; Doan Dao was responsible for data collection, analysis, and interpretation; Jordan J. Feld was responsible for data collection, analysis, and interpretation, and critical revision; Daryl T. Lau was responsible for data collection, analysis, and interpretation, and critical revision; Keyur Patel was responsible for data collection, analysis, and interpretation, and critical revision; Jody Rule was responsible for data collection, analysis, and interpretation; Mark Daly was responsible for the study concept and design and interpretation of data; William M. Lee was responsible for the study concept and design, interpretation of data, and critical revision; and Raymond T. Chung was responsible for the study concept and design, interpretation of data, and critical revision.
Conflicts of interest The authors disclose no conflicts.
Funding Supported by the Advanced/Transplant Hepatology Fellowship, the Clinical and Translational Research Award from the American Association for the Study of Liver Diseases Foundation, and Brigham and Women’s Internal Medicine Residency Program Research grant (V.A.). Additional support was provided by U01 DK58369 to University of Texas Southwestern for the Acute Liver Failure Study Group. Samples were provided by the National Institute of Diabetes and Digestive and Kidney Diseases sample repository. Funding support was provided by the National Institute of Diabetes and Digestive and Kidney Diseases 1K01DK114379-01 and the Stanley Center for Psychiatric Research at the Broad Institute (H.H.), and National Institutes of Health K24DK078772 and U01DK082919 (R.T.C.). The Hepatitis B Research Network provided data and samples.
Supplementary Materials and Methods
Study Design and Participants
This was a cross-sectional study of adult participants of European ancestry recruited from 3 studies: the Hepatitis B Research Network, the Acute Liver Failure Study Group, and a shared control data set compiled from multiple studies and sources.
The Hepatitis B Research Network is a cooperative network of investigators from 28 clinical centers in North America funded by the National Institute of Diabetes and Digestive and Kidney Diseases. This study included participants from the adult cohort. Patients who were 18 years or older and hepatitis B surface antigen positive were eligible for enrollment. Patients with a history of hepatic decompensation, hepatocellular carcinoma, solid organ or bone marrow transplantation, hepatitis delta or human immunodeficiency virus co-infection, and those who were receiving antiviral therapy (with the exception of women who were pregnant at the time of enrollment) were excluded. Details of the Hepatitis B Research Network study population have been published previously.1 This study included 2 populations from the Hepatitis B Research Network: adult-acquired CHB not through vertical transmission and aHB. Participants were determined to have CHB infection if they had detectable HBV DNA on 2 or more occasions at least 6 months apart and had a known age of acquisition though medical history of age 18 or later. Fifty participants met inclusion and exclusion criteria and had available DNA. Participants were defined to have aHB by a total bilirubin level of 3 mg/dL or greater in the setting of acute onset of hepatitis B and were adjudicated by an expert committee to ensure the cases did not represent a flare of chronic HBV. Sixteen participants met inclusion and exclusion criteria and had available DNA and underwent only rs2277680 genotyping.
The Acute Liver Failure Study Group is a prospective National Institute of Diabetes and Digestive and Kidney Diseases–funded study including patients from 31 academic centers in the United States. ALF was defined as severe acute liver injury without known chronic liver disease, with a duration of illness of fewer than 26 weeks accompanied by hepatic encephalopathy and coagulopathy (prothrombin time ≥15 s or international normalized ratio ≥1.5). In addition, patients with acute liver injury secondary to HBV were included and defined by an international normalized ratio ≥2.0, alanine aminotransferase level ≥10× increased, and bilirubin level ≥3.0 mg/dL. The attribution of ALF or acute liver injury as caused by HBV required either IgM antibody to hepatitis B core antigen positive or hepatitis B surface antigen positive or both, in addition to the earlier-described definitions. Fourteen HBV ALF participants met criteria and had available DNA. Cases of acute-on-chronic or reactivation leading to HBV ALF (CHBV ALF, N = 4) were identified through review of each case (W.M.L.) as previously described2 and explored as a separate cohort to validate the association between CXCL16 and underwent only rs2277680 genotyping. In addition, 2 additional cases of HBV ALF were identified after the initial exome analysis and underwent only rs2277680 genotyping.
A shared general population of 2963 unselected controls of European ancestry was obtained from sources including the 1000 Genomes Project phase 1, Age-related Macular Degeneration Project, National Institute of Mental Health, Prospective Registry in Inflammatory Bowel Disease Study at Massachusetts General Hospital, International Serious Adverse Event Consortium, and Psychiatric Genomics Consortium. These general population controls were used to represent a surrogate for recovered HBV infection because this outcome would be expected in more than 90% of the population. A graphic description of the study population and parent cohorts is included as Figure 1.
Informed consent was obtained from all participants or the next of kin for patients with encephalopathy, and the studies were approved by the Institutional Review Boards at each clinical center. All authors had access to the data and reviewed and approved the final manuscript.
Exome Analysis
De-identified genetic material in the form of DNA was received and registered into the Broad Institute’s Biological Samples Platform from the CHB and HBV ALF cohorts. Upon receipt, the samples are registered into a secure database, quantified via Picogreen (Thermo Fisher Carlsbad, CA), and genotyped on a Fluidigm (Fluidigm San Francisco, CA) 96 snp fingerprint panel. Once quality control steps are complete, the samples were plated for the Genetic Analysis Platform for genotyping arrays. An aliquot of DNA of at least 1 uG at 50 ng/uL was used to allow for quantification reactions, fingerprinting genotyping, robotic plating of the sample, and re-dos that were required. Batches of up to 94 samples for Exome Chip were plated along with 1 HapMap control and 1 duplicate for internal quality assurance. Genotypes were called using standard clustering algorithms and more finely tailored rare variant clustering algorithms under development. All samples were genotyped on the Illumina Infinium Exome-24 Array (Illumina, San Diego, CA) with more than 240,000 variants covering the exonic content.
CXCL16 Genotyping in Validation Populations
The non-ALF acute hepatitis B and chronic hepatitis B ALF populations were used to validate the findings regarding CXCL16 and HBV ALF. Polymerase chain reaction on an iCycler (Bio-Rad, Hercules, CA) used 5 μL of genomic DNA (concentration 50 ng/μL) to evaluate rs2277680 with the following primers: forward-AACCTTGTGCAGACAACCTTGA and reverse-CCAGAAGCATTTACTTCCTACC designed using the National Center for Biotechnology Information Primer-BLAST tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast).
Statistical Analysis
We performed quality control using the RICOPILI pipeline with default parameters (https://sites.google.com/a/broadinstitute.org/ricopili). Cluster plots for HBV samples were inspected manually for variants with a P value < 1E-4 using Evoker (Sanger Institute, Hinxton, UK).3 The Bonferroni adjusted false discovery rate for the exome chip was P = 5 × 10-7. For each variant that passed the QC, we performed the Fisher exact test to compare its allele frequency in ALF patients vs shared controls, in CHB patients vs shared controls, and in ALF patients vs CHB patients.
Supplementary Table 1.
Exome Array Results for HBV ALF to Control
| Chromosome | Gene | SNP | Allelea | Allele frequencyb |
P value | |
|---|---|---|---|---|---|---|
| HBV ALF | Control | |||||
| 7 | LAMB4 | rs147992634 | C/A | 0.07 | 2 × 10-4 | 6.38 × 10-5 |
| 17 | NPLOC4 | rs9894429 | T/C | 0.82 | 0.44 | 7.59 × 10-5 |
| 12 | RIMBP2, STX2 | rs1195569 | G/A | 0.86 | 0.49 | 9.00 × 10-5 |
| 17 | CXCL16 | rs2277680 | A/G | 0.07 | 0.43 | 6.09 × 10-5 |
| 17 | CXCL16 | rs1050998 | G/A | 0.07 | 0.43 | 6.09 × 10-5 |
SNP, single-nucleotide polymorphism.
Minor allele/major allele.
Shown for minor allele.
Supplementary Table 2.
Exome Array Results for CHB to Control
| Chromosome | Gene | SNP | Allelea | Allele frequncyb |
P value | |
|---|---|---|---|---|---|---|
| CHB | Control | |||||
| 2 | SCN9A | rs58022607 | T/C | 0.06 | 4.39 × 10-3 | 1.15 × 10-5 |
| 23 | NONE, NONE | rs201682692 | G/A | 0.03 | 2.04 × 10-4 | 2.86 × 10-5 |
| 1 | SGIP1 | rs72669494 | T/C | 0.03 | 3.37 × 10-4 | 4.07 × 10-5 |
| 4 | CCSER1,GRID2 | rs2199291 | T/G | 0.13 | 0.31 | 4.23 × 10-5 |
| 20 | GMEB2 | rs73134074 | T/C | 0.03 | 3.37 × 10-4 | 4.33 × 10-5 |
| 3 | TIMMDC1 | rs57168946 | A/G | 0.11 | 0.02 | 4.49 × 10-5 |
| 11 | TENM4 | rs1000960 | C/T | 0.48 | 0.28 | 4.62 × 10-5 |
| 3 | TIMMDC1 | rs11539377 | G/A | 0.11 | 0.02 | 5.70 × 10-5 |
| 23 | NONE | rs143596619 | T/C | 0.03 | 4.07 × 10-4 | 7.06 × 10-5 |
| 3 | POGLUT1 | rs56195264 | A/G | 0.11 | 0.03 | 7.13 × 10-5 |
| 16 | SHISA9 | rs7192086 | T/A | 0.42 | 0.24 | 7.25 × 10-5 |
| 11 | FOXR1 | rs45602538 | G/C | 0.06 | 6.41 × 10-3 | 7.60 × 10-5 |
| 10 | AFAP1L2 | rs73365341 | A/G | 0.09 | 0.02 | 8.26 × 10-5 |
| 12 | VSIG10 | rs9668527 | T/C | 0.03 | 5.06 × 10-4 | 8.55 × 10-5 |
SNP, single-nucleotide polymorphism.
Minor allele/major allele.
Shown for minor allele.
Supplementary Table 3.
Exome Array Results for HBV ALF to CHB
| Chromosome | Gene | SNP | Allelea | Allele frequncyb |
P value | |
|---|---|---|---|---|---|---|
| HBV ALF | CHB | |||||
| 6 | MANEA | rs62417812 | T/C | 0.36 | 0.05 | 8.15 × 10-5 |
| 10 | SEC24C | rs3849969 | A/G | 0.57 | 0.16 | 3.18 × 10-5 |
SNP, single-nucleotide polymorphism.
Minor allele/major allele.
Shown for minor allele.
Supplementary Table 4.
MANEA rs62417812 Allele Frequencies by HBV Status
| N | Allele 1 (T) | Allele 2 (C) | |
|---|---|---|---|
| Chronic HBVa | 50 | 5% | 95% |
| HBV ALF | 14 | 36% | 74% |
| Controls shared from other studies | 2963 | 11% | 89% |
P value for comparison with HBV ALF = 8.15 × 10-5.
Supplementary Table 5.
SEC24C rs3849969 Allele Frequencies by HBV Status
| N | Allele 1 (A) | Allele 2 (G) | |
|---|---|---|---|
| Chronic HBVa | 50 | 16% | 84% |
| HBV ALF | 14 | 57% | 43% |
| Controls shared from other studies | 2963 | 38% | 62% |
P value for comparison with HBV ALF = 3.18 × 10-5.
References
- 1.Schweitzer A. Lancet. 2015;386:1546–1555. doi: 10.1016/S0140-6736(15)61412-X. [DOI] [PubMed] [Google Scholar]
- 2.Reuben A. Ann Intern Med. 2016;164:724–732. doi: 10.7326/M15-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang T.J. N Engl J Med. 1991;324:1705–1709. doi: 10.1056/NEJM199106133242405. [DOI] [PubMed] [Google Scholar]
- 4.Geissmann F. PLoS Biol. 2005;3:e113. doi: 10.1371/journal.pbio.0030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsuda J.L. J Exp Med. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeda K. Proc Natl Acad Sci U S A. 2000;97:5498–5503. doi: 10.1073/pnas.040566697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kakimi K. J Exp Med. 2000;192:921–930. doi: 10.1084/jem.192.7.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wehr A. J Immunol. 2013;190:5226–5236. doi: 10.4049/jimmunol.1202909. [DOI] [PubMed] [Google Scholar]
- 9.Wehr A. PLoS One. 2014;9:e112327. doi: 10.1371/journal.pone.0112327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petit S.J. Arterioscler Thromb Vasc Biol. 2011;31:914–920. doi: 10.1161/ATVBAHA.110.220558. [DOI] [PubMed] [Google Scholar]
Supplementary References
- 1.Ghany M.G. Clin Gastroenterol Hepatol. 2015;13:183–192. doi: 10.1016/j.cgh.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dao D.Y. Hepatology. 2012;55:676–684. doi: 10.1002/hep.24732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morris J.A. Bioinformatics. 2010;26:1786–1787. doi: 10.1093/bioinformatics/btq280. [DOI] [PMC free article] [PubMed] [Google Scholar]