Abstract
The equatorial spitting cobra, Naja sumatrana, is a distinct species of medically important venomous snakes, listed as WHO Category 1 in Southeast Asia. The diversity of its venom genes has not been comprehensively examined, although a few toxin sequences annotated to Naja sputatrix were reported previously through cloning studies. To investigate this species venom genes’ diversity, de novo venom-gland transcriptomics of N. sumatrana from West Malaysia was conducted using next-generation sequencing technology. Genes encoding toxins represented only 60 of the 55,396 transcripts, but were highly expressed, contributing to 79.22% of total gene expression (by total FPKM) in the venom-glands. The toxin transcripts belong to 21 families, and 29 transcripts were further identified as full-length. Three-finger toxins (3FTx) composed of long, short, and non-conventional groups, constituted the majority of toxin transcripts (91.11% of total toxin FPKM), followed by phospholipase A2 (PLA2, 7.42%)—which are putatively pro-inflammatory and cytotoxic. The remaining transcripts in the 19 families were expressed at extremely low levels. Presumably, these toxins were associated with ancillary functions. Our findings unveil the diverse toxin genes unique to N. sumatrana, and provide insights into the pathophysiology of N. sumatrana envenoming.
Keywords: venom-gland transcriptomics, Naja sumatrana, three-finger toxins, toxin genes
1. Introduction
Venom is a sophisticated and versatile weapon harnessed by venomous snakes for prey capture, and defense [1,2]. As the fundamental purpose of snake venom is predation, venom gene adaption is strongly driven by diet [3,4]. Over time, the choice and selection of prey may differ due to multiple factors, e.g., geographical and sexual variations, as well as ontogenic shifts [5,6,7,8,9]. Variations in snake venom composition has ramifications on snakebite management, as it may lead to unexpected clinical manifestations of envenomation. Moreover, variation in venom protein antigenicity can result in suboptimal antivenom efficacy or even treatment failure [10,11,12]. This is particularly relevant to snakes that are widely distributed such as cobras in Asia. Cobra venom compositions have been shown to vary widely between and within species, which can complicate the use of paraspecific antivenom, a common practice in many countries due to the unavailability or limited supply of species-specific antivenom [13,14].
In Southeast Asia, there are at least six to seven distinct cobra (Naja) species (http://reptile-database.reptarium.cz/) which can cause fatal and disabling envenomation. These include the equatorial spitting cobra, Naja sumatrana, a WHO Category 1 medically important venomous snakes [15]. This species is widely distributed throughout Peninsular Malaya (including West Malaysia and Singapore), Sumatra, southern part of Thailand, and part of Borneo Island [16]. The species was previously classified under a larger taxon, Naja naja sputatrix, and locally known as “Malayan spitting cobra”, without a clear distinction from other potentially different spitting cobras in the region such as those from the Indochina and the Java Island (Indonesia) [16].
The current systematics of Asiatic cobras have substantially resolved the taxonomic confusion pertaining to the taxon “Naja naja sputatrix”. In this context, three distinct spitting cobra species are recognized: Naja sumatrana (equatorial spitting cobra), Naja siamensis (Indochinese spitting cobra), and Naja sputatrix (Javan spitting cobra) [16]. Envenoming by these species often result in systemic neuromuscular paralysis and extensive local tissue necrosis. Local tissue necrosis can potentially lead to amputation and crippling disabilities in surviving victims [17]. The spitting cobras’ ability to spray venom, a well evolved defense strategy in these species, can cause venom ophthalmia and blindness [17,18,19]. Previous studies have shown that cobra venoms consist mainly of three-finger toxins and phospholipases A2, while their relative abundances, subtypes, and antigenicity can vary substantially between and within species [20,21]. Comprehensive understanding of the venom profile of individual species is very much dependent on the availability of species-specific databases with respect to venom gene sequences. This can be efficiently accomplished via high-throughput gene sequencing of tissues derived from authenticated specimens [6,22,23]. The present study aims to investigate the de novo venom-gland transcriptome of Naja sumatrana from Malaysia, to obtain a comprehensive profile of its venom genes using next-generation sequencing (NGS) technology. The findings will shed light on the diversity of venom genes specific to this unique species of spitting cobra in Malaysia, and provide deeper insights into the correlation of toxin composition and pathophysiology of cobra envenomation. In addition, the information obtained can be used to validate several toxin sequences annotated to “Naja naja sputatrix” available in the open database. These toxin sequences were reported from cloning studies in the late 1990’s and early 2000’s [24,25,26,27,28]. Unfortunately, the authenticity of the snake species used in the early days was difficult to be ascertained as the work was carried out at a time when confusion perhaps still existed in the systematics of spitting cobras.
2. Results and Discussion
2.1. Sequencing Output Statistics and De Novo Transcriptome Assembly
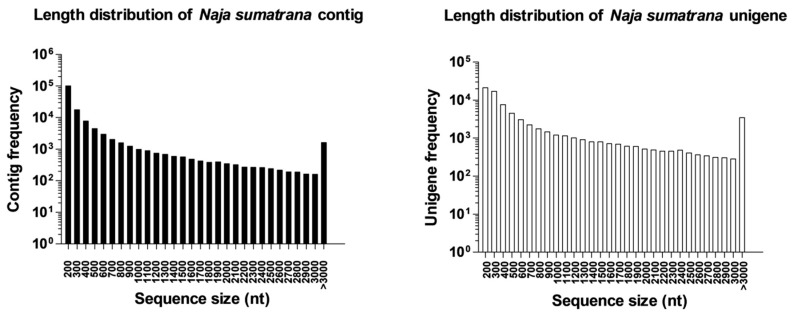
Sequencing of the cDNA libraries yielded a total of 46,878,172 clean reads for the Malaysian Naja sumatrana (herein NS-M) venom-gland transcriptome (Table 1). De novo assembly using the Trinity program created 148,475 contigs (N50 = 652) that were connected to form 75,387 Unigenes (N50 = 1702), with the length distribution shown in Figure 1. The high Q20 percentage of 97.94% indicated that the de novo assembly of NS-M venom-gland transcriptome was successful and of high quality. The 75,387 Unigenes assembled underwent filtering based on FPKM (fragments per kilobase per million) values, where transcripts with less than 1 FPKM mapped reads were removed. This reduced the number of Unigenes to 55,386. Following BLASTx alignment, the Unigenes—herein referred as transcripts—were assigned to three categories: (a) “unidentified” (transcripts whose gene/protein identities could not be identified during BLASTx alignment); (b) “non-toxin” (transcripts that encoded proteins which have no putative toxin role); and (c) “toxin” (transcripts that encoded known and putative toxins). The details of the results are summarized in Table 1.
Table 1.
Output statistics of de novo assembly of Naja sumatrana venom-gland transcriptome using Illumina HiSeq 2000 sequencing.
| Parameter | Sequencing Output |
|---|---|
| Total raw reads | 47,494,560 |
| Total clean reads | 46,878,172 |
| Contigs created | 148,475 |
| Q20 percentage | 97.94% |
| N percentage | 0.00% |
| GC percentage | 44.16% |
| Unigenes/transcripts assembled | 75,387 |
| Number of transcripts (FPKM > 1) | 55,386 |
| Unidentified | Abundance |
| Number of transcripts | 35,449 |
| Clean reads | 123,432.1986 |
| Total FPKM percentage (%) | 7.95% |
| Non-toxin | Abundance |
| Number of transcripts | 19,877 |
| Clean reads | 199,393.4726 |
| Total FPKM percentage (%) | 12.84% |
| Toxin | Abundance |
| Number of transcripts | 60 |
| Clean reads | 1,230,548.6634 |
| Total FPKM percentage (%) | 79.22% |
Figure 1.
Length distribution of contigs (left) and Unigenes (right) obtained following de novo transcriptome assembly.
2.2. Categorization of Transcripts and Gene Expression
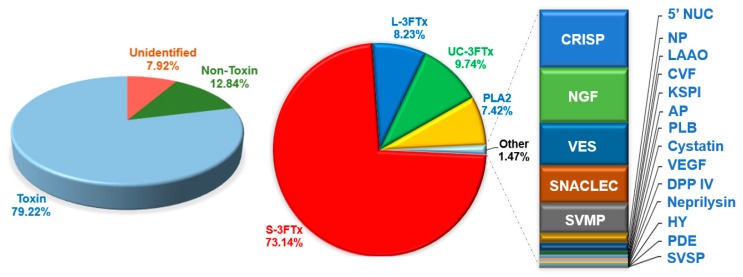
The “toxins” category consisted of transcripts that code for a great variety of toxin proteins. Although the toxin transcripts only accounted for 60 of the 55,396 transcripts obtained, they were highly expressed and contributed to 79.22% of total gene expression (by total gene FPKM) in the venom gland. Both “non-toxin” and “unidentified” groups were composed of very high numbers of genes but the gene expression levels were low, accounting for only 12.84% and 7.95%, respectively, of the total genes expressed (Figure 2). The “non-toxin” group mainly consisted of innocuous housekeeping genes, such as transcription factors, ribosomal proteins and miscellaneous proteins which are involved in cell metabolism. The expressions of toxin genes in NS-M venom glands were comparable to those reported for the Thai Naja kaouthia (82%) [6], Chinese Naja atra (70.24%) and Bungarus multicinctus (69.60%) [29]. However, the levels were much higher than those found in the Malaysian king cobra (Ophiophagus hannah; 35.30%), and the monocled cobra (Naja kaouthia) of Malaysia and China (41.20% and 54.42%, respectively) [6,30,31]. The variable venom gene expression could be due to inter- or intraspecific variation, or sampling times when there are differences in the snake’s physiological conditions during venom-gland harvesting. Nevertheless, collectively these findings show that toxin genes are highly expressed in venom-gland tissue, despite the limited number of toxin transcripts (Table 1).
Figure 2.
Relative abundance of transcripts (in percentage of FPKM) expressed in the venom glands of Malaysian Naja sumatrana. Toxin transcripts dominate the overall expression (79.22%). Within toxin transcripts, three-finger toxin (3FTx) is the most abundantly expressed toxin family in the venom glands (91.11% of the toxin FPKM). Of these, the short three-finger toxins (S-3FTx) constitute 73.14%, whereas the long three-finger toxins (L-3FTx) constitute 8.23%. Non-conventional three-finger toxins (NC-3FTx) and phospholipases A2 (PLA2) constitute 9.74% and 7.42%, respectively. Abbreviations: S-3FTx, short three-finger toxin; L-3FTx, long three-finger toxin; UC-3FTx, non-conventional three-finger toxin; PLA2, phospholipase A2; CRISP, cysteine-rich secretory protein; NGF, nerve growth factor; VES, vespryn; SNACLEC, snake venom C-type lectin/lectin-like protein; SVMP, snake venom metalloproteinase; 5’NUC, 5’ nucleotidase; NP, natriuretic peptide; LAAO, L-amino acid oxidase; CVF, cobra venom factor; AP, aminopeptidase; PLB, Phospholipase B; KSPI, Kunitz-type serine protease inhibitor; VEGF, vascular endothelial growth factor; HY, hyaluronidase; DPP IV, dipeptidylpeptidase IV; PDE, phosphodiesterase; and SVSP, snake venom serine protease.
Exceptionally high redundancies were also observed in the toxin transcripts (20,509.14 FPKM/transcript). This is in sharp contrast to that observed in the non-toxin group of transcripts (10.03 FPKM/transcript). This indicates that toxins were highly expressed in multiple isoforms within a restricted set of gene families. The high redundancy of toxin gene expression in NS-M was consistent with findings from venom-gland transcriptomic studies of the Southeast Asian Naja kaouthia (monocled cobra), whereby the redundancy levels were reported to be 6,300–23,000 FPKM/transcript [6]. This is also in line with the theory behind the molecular diversity of snake venom proteins, where molecular adaptation is largely driven by repeated gene duplication followed by neofunctionalization of the proteins [1,2].
2.3. Complexity of N. sumatrana Venom-Gland Transcriptome
The 60 toxin transcripts derived from NS-M venom glands were classified into 21 gene families. A total of 29 transcripts were further identified as full-length (Table 2). The three-finger toxins (3FTx) inclusive of long, short, and non-conventional groups, constituted the majority of toxin transcripts (91.11% of total toxin FPKM), followed by phospholipase A2 (PLA2, 7.42%). The remaining transcripts in 19 families were expressed at extremely low levels, each constituting less than 0.4% of total FPKM (Figure 2). These were, in descending order of expression, cysteine-rich secretory protein (CRISP), nerve growth factor (NGF), vespryn (VES), snake venom C-type lectin/lectin-like protein (SNACLEC), snake venom metalloproteinase (SVMP), 5’ nucleotidase (5’ NUC), natriuretic peptide (NP), L-amino acid oxidase (LAAO), cobra venom factor (CVF), Kunitz-type serine protease inhibitor (KSPI), aminopeptidase (AP), phospholipase B (PLB), cystatin, vascular endothelial growth factor (VEGF), dipeptidyltidase IV (DPP IV), neprilysin, hyaluronidase (HY), phosphodiesterase (PDE), and snake venom serine protease (SVSP) (Table 3).
Table 2.
Full-length toxin transcripts derived from the venom-gland transcriptome of Malaysian Naja sumatrana (NS-M).
| Protein Family/Protein ID | Annotated Accession | Species | Amino Acid Chain | Mature Chain of Accession ID | Coverage (Mature Chain) | Coverage Percentage (%) | |
|---|---|---|---|---|---|---|---|
| Three-finger toxin | |||||||
| NSM_FTX01 | Cytotoxin 2a | Q9PST4 | N. sputatrix | 81 | 81 | 1–81 | 100 |
| NSM_FTX02 | Neurotoxin homolog NL1 | Q9DEQ3 | N. atra | 81 a | 86 | 6–86 | 93 |
| NSM_FTX06 | Long neurotoxin 7 | O42257 | N. sputatrix | 89 | 90 | 2–90 | 98 |
| NSM_FTX07 | Alpha-neurotoxin NTX-4 | O57327 | N. sputatrix | 83 | 83 | 1–83 | 100 |
| Phospholipase A2 | |||||||
| NSM_PLA01 | Acidic phospholipase A2 C | Q92086 | N. sputatrix | 146 | 146 | 1–146 | 100 b |
| Cysteine-rich secretory protein | |||||||
| NSM_CRP01 | Cysteine-rich venom protein natrin-1 | Q7T1K6 | N. atra | 239 a | 239 | 1–239 | 100 b |
| Nerve growth factor | |||||||
| NSM_NGF01 | Venom nerve growth factor 2 | Q5YF89 | N. sputatrix | 246 | 241 | 1–241 | 100 b |
| Vespryn | |||||||
| NSM_VES01 | Ohanin | P83234 | O. hannah | 190 a | 190 | 1–190 | 100 b |
| C-type/lectin-like protein | |||||||
| NSM_SCL01 | C-type lectin BfL-1 | Q90WI8 | B. fasciatus | 158 a | 158 | 1–158 | 100 b |
| Snake venom metalloproteinase | |||||||
| NSM_SMP07 | Zinc metalloproteinase-disintegrin-like kaouthiagin-like | D3TTC1 | N. atra | 593 a | 593 | 1–593 | 100 b |
| NSM_SMP08 | Zinc metalloproteinase-disintegrin-like atrase-B | D6PXE8 | N. atra | 613 a | 593 | 1–593 | 100 b |
| 5’ nucleotidase | |||||||
| NSM_NUC01 | Ecto-5’-nucleotidase 1 | U3FYP9 | M. fulvius | 569 a | 574 | 1–569 | 99 |
| L-amino-acid oxidase | |||||||
| NSM_LAO01 | L-amino-acid oxidase | A8QL58 | N. atra | 514 a | 449 | 1–449 | 100 b |
| Cobra venom factor | |||||||
| NSM_CVF01 | Cobra venom factor | Q91132 | N. kaouthia | 1646 a | 1642 | 1–1642 | 100 b |
| Kunitz-type serine protease inhibitor | |||||||
| NSM_KPI01 | Putative Kunitz-type serine protease inhibitor | B2BS84 | A. labialis | 249 | 252 | 1–252 | 98 |
| NSM_KPI02 | Kunitz-type protease inhibitor | U3FZD6 | M. fulvius | 513 | 511 | 1–511 | 100 b |
| Aminopeptidase | |||||||
| NSM_AP01 | Aminopeptidase | U3FZS8 | M. fulvius | 1000 | 993 | 1–993 | 100 b |
| Phospholipase B | |||||||
| NSM_PLB01 | Phospholipase-B 81 | F8J2D3 | D. coronoides | 553 a | 553 | 1–553 | 100 b |
| Vascular endothelial growth factor | |||||||
| NSM_VGF01 | Vascular endothelial growth factor 2 | U3FAK1 | M. fulvius | 421 | 421 | 1–421 | 100 b |
| Dipeptidylpeptidase IV | |||||||
| NSM_DPP01 | Dipeptidyl peptidase 4 | V8P9G9 | O. hannah | 753 | 754 | 1–753 | 99 |
| Neprilysin | |||||||
| NSM_NP01 | Neprilysin | A0A0B8RU83 | B. irregularis | 750 | 750 | 1–750 | 100 b |
| Hyaluronidase | |||||||
| NSM_HY01 | Hyaluronidase | A0A194APD1 | M. tener | 449 | 447 | 1–447 | 100 b |
| NSM_HY02 | Hyaluronidase | A0A194APD1 | M. tener | 449 | 447 | 1–447 | 100 b |
| NSM_HY03 | Hyaluronidase | A0A194APD1 | M. tener | 449 | 447 | 1–447 | 100 b |
| NSM_HY04 | Hyaluronidase | A0A194APD1 | M. tener | 449 | 447 | 1–447 | 100 b |
| Phosphodiesterase | |||||||
| NSM_PDE01 | Snake venom phosphodiesterase | A0A2D0TC04 | N. atra | 850 | 830 | 1–830 | 100 b |
| NSM_PDE02 | Snake venom phosphodiesterase | A0A2D0TC04 | N. atra | 848 | 830 | 1–830 | 100 b |
| Snake venom serine protease | |||||||
| NSM_SSP01 | Serine protease HTRA1 | A0A0B8RTL3 | B. irregularis | 471 | 489 | 18–488 | 96 |
| NSM_SSP02 | Serine protease 23 | V8N8N4 | O. hannah | 365 | 372 | 8–372 | 98 |
Notes: A., Austrelaps; B., Boiga/Bungarus; C., Crotalus; D., Drysdalia; M., Micrurus; N., Naja; O., Ophiophagus; and T., Trimeresurus. a Novel sequence reported in Naja sumatrana; b Protein sequence not identical to the annotated sequence.
Table 3.
Overview of families and subtypes of toxin genes in the venom-gland transcriptome of Malaysian Naja sumatrana (NS-M).
| Protein Family/Protein Subtype | Accession/Species | Transcript Abundance % a |
|---|---|---|
| Three-finger toxin (3FTx) | 91.11 (10) | |
| S-3FTx | 73.14 (5) | |
| Cytotoxin 2a | Q9PST4 (N. sputatrix) | 72.83 (1) b |
| Neurotoxin homolog NL1 | Q9DEQ3 (N. atra) | 0.21 (1) b |
| Cardiotoxin 7 | Q91996 (N. atra) | 0.10 (2) |
| Muscarinic toxin-like protein 1 | P82462 (N. kaouthia) | 0.00 (1) |
| L-3FTx | 8.23 (4) | |
| Long neurotoxin 7 | O42257 (N. sputatrix) | 4.91 (1) b |
| Alpha-neurotoxin NTX-4 | O57327 (N. sputatrix) | 3.30 (1) b |
| Long neurotoxin-like OH-31 | Q53B55 (O. hannah) | 0.01 (2) |
| UC-3FTx | 9.74 (1) | |
| Weak neurotoxin 5 | O42255 (N. sputatrix) | 9.74 (1) |
| Phospholipase A2 (PLA2) | 7.42 (2) | |
| Acidic phospholipase A2 C | Q92086 (N. sputatrix) | 7.39 (1) b |
| Phospholipase A2 GL16-1 | Q8JFB2 (L. semifasciata) | 0.03 (1) |
| Cysteine-rich secretory protein (CRISP) | 0.33 (4) | |
| Cysteine-rich venom protein natrin-1 | Q7T1K6 (N. atra) | 0.32 (1) b |
| Cysteine-rich venom protein natrin-2 | Q7ZZN8 (N. atra) | 0.01 (2) |
| Cysteine-rich venom protein kaouthin-2 | P84808 (N. kaouthia) | 0.01 (1) |
| Nerve growth factor (NGF) | 0.31 (2) | |
| Venom nerve growth factor 2 | Q5YF89 (N. sputatrix) | 0.31 (1) b |
| Venom nerve growth factor 1 | Q5YF90 (N. sputatrix) | 0.00 (1) |
| Vespryn (VES) | 0.25 (1) | |
| Thaicobrin | P83234 (O. hannah) | 0.25 (1) b |
| Snake venom C-type/lectin-like protein (Snaclec) | 0.22 (2) | |
| C-type lectin BfL-1 | Q90WI8 (B. fasciatus) | 0.22 (1) b |
| C-type lectin BfL-2 | Q90WI7 (B. fasciatus) | 0.00 (1) |
| Snake venom metalloproteinase (SVMP) | 0.17 (10) | |
| Snake venom metalloproteinase-disintegrin-like morcarhagin | Q10749 (N. mossambica) | 0.03 (1) |
| Zinc metalloproteinase-disintegrin-like cobrin | Q9PVK7 (N. kaouthia) | 0.07 (3) |
| Carinatease-1 | B5KFV1 (Tr. carinatus) | 0.02 (1) |
| Zinc metalloproteinase-disintegrin-like atrase B | D6PXE8 (N. atra) | 0.03 (3) b |
| Zinc metalloproteinase-disintegrin-like kaouthiagin-like | D3TTC1 (N. atra) | 0.01 (1) b |
| Zinc metalloproteinase-disintegrin-like atrase A | D5LMJ3 (N. atra) | 0.01 (1) |
| 5’ nucleotidase (5’ NUC) | 0.05 (2) | |
| Ecto-5’-nucleotidase 1 | U3FYP9 (M. fulvius) | 0.05 (1) b |
| 5’ nucleotidase | A0A024AXW5 (M. ikaheca) | 0.00 (1) |
| Natriuretic peptide (NP) | 0.05 (2) | |
| Natriuretic peptide Na-NP | D9IX97 (N. atra) | 0.05 (2) |
| L-amino acid oxidase (LAAO) | 0.02 (1) | |
| L-amino-acid oxidase | A8QL58 (N. atra) | 0.02 (1) b |
| Cobra venom factor (CVF) | 0.02 (3) | |
| Cobra venom factor | Q91132 (N. kaouthia) | 0.02 (3) b |
| Kunitz-type serine protease inhibitor (KSPI) | 0.01 (2) | |
| Putative Kunitz-type serine protease inhibitor | B2BS84 (A. labialis) | 0.01 (1) b |
| Kunitz-type protease inhibitor | U3FZD6 (M. fulvius) | 0.00 (1) b |
| Aminopeptidase (AP) | 0.01 (1) | |
| Aminopeptidase | U3FZS8 (M. fulvius) | 0.01 (1) b |
| Phospholipase B (PLB) | 0.01 (1) | |
| Phospholipase-B 81 | F8J2D3 (D. coronoides) | 0.01 (1) b |
| Cystatin | 0.01 (5) | |
| Cystatin | V8NX38 (O. hannah) | 0.00 (1) |
| Cystatin | A0A098LYB6 (O. aestivus) | 0.00 (4) |
| Vascular endothelial growth factor (VEGF) | 0.01 (2) | |
| Vascular endothelial growth factor 2 | U3FAK1 (M. fulvius) | 0.01 (1) b |
| Vascular endothelial growth factor A | A0A098LYD7 (O. aestivus) | 0.00 (1) |
| Dipeptidylpeptidase IV (DPP IV) | 0.00 (1) | |
| Dipeptidyl peptidase 4 | V8P9G9 (O. hannah) | 0.00 (1) b |
| Neprilysin | 0.00 (1) | |
| Neprilysin | A0A0B8RU83 (B. irregularis) | 0.00 (1) b |
| Hyaluronidase (HY) | 0.00 (4) | |
| Hyaluronidase | A0A194APD1 (M. tener) | 0.00 (4) b |
| Phosphodiesterase (PDE) | 0.00 (2) | |
| Snake venom phosphodiesterase | A0A2D0TC04 (N. atra) | 0.00 (2) b |
| Snake venom serine protease (SVSP) | 0.00 (2) | |
| Serine protease HTRA1 | A0A0B8RTL3 (B. irregularis) | 0.00 (1) b |
| Serine protease 23 | V8N8N4 (O. hannah) | 0.00 (1) b |
A., Agkistrodon/Austrelaps; B., Boiga/Bothrops/Bungarus; C., Crotalus; D., Drydalia; E., Elaphe; L., Laticauda; M., Micrurus/Micropechis; N., Naja; O., Opheodrys/Ophiophagus; T., Trimeresurus; and Tr., Tropidechis. a Transcript expression in percentage (%) based on FPKM (fragments per kilobase of exon model per million mapped reads). Number in bracket refers to the number of non-redundant transcripts in the toxin family. b Full-length transcript with more than 90% length coverage of the annotated sequence.
Out of the 60 toxin transcripts, seven were annotated based on homology with sequences reported for Naja naja sputatrix (deposited previously in the open database). There is currently no toxin gene annotated by N. sumatrana in the database. The species of Naja sumatrana was erected and fully recognized only about 20 years ago [16]. Prior to that, toxinologists seldom distinguished between the three species of spitting cobras (N. sumatrana, N. sputatrix, and Naja siamensis) distributed in this region. Gene cloning studies on N. naja sputatrix by Jeyaseelan and colleagues [24,25,26,27,28] were based on venom glands extracted from spitting cobra(s) that appeared to originate from the Peninsular Malaya or Singapore. In view of the identical match of the sequences to N. sumatrana transcripts in the present study, it is very likely that the spitting cobra labeled as “N. naja sputatrix” previously was actually an N. sumatrana. The venom-gland transcriptome of NS-M hence can be used to validate the existing sequences annotated to N. naja sputatrix in the database.
Besides 3FTx and PLA2, the remaining 53 toxin transcripts were matched by at least 50% sequence similarity with toxins from other snake species of Naja (25 transcripts) and other genera (28 transcripts), probably because of the paucity of sequence database specific for the Southeast Asian spitting cobras.
2.4. Diversity of Toxin Transcripts and Major Venom Constituents
2.4.1. Three-finger Toxins (3FTxs)
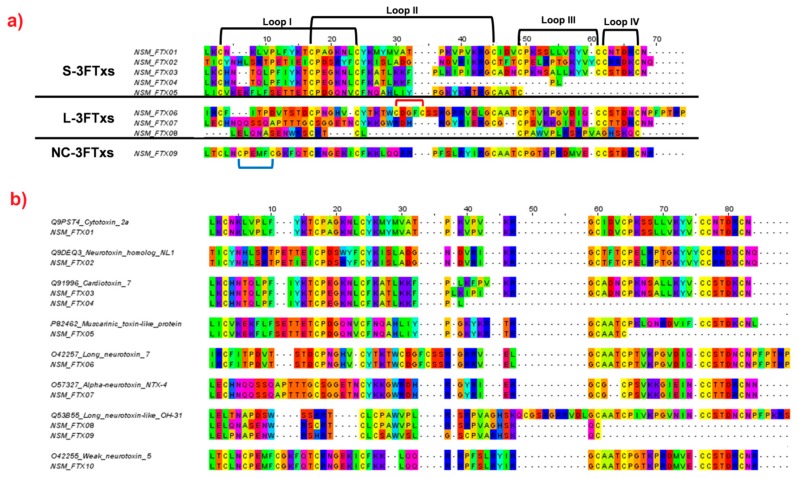
Three-finger toxins (3FTxs) are non-enzymatic polypeptides with 60–74 amino acids, stabilized by four or five disulfide cross-linkages [32]. The characteristic feature of 3FTx is their distinctive protein folding, containing a hydrophobic core with three extended beta-stranded loops. Its structural integrity is supported by the conserved peptide regions and four cross-linked disulfide linkages at the core [33,34,35]. The venom-gland transcriptomic results revealed a total of 10 distinct 3FTx transcripts. These transcripts altogether accounted for 91.11% of the total FPKM of all toxin transcripts. The transcripts were further categorized into short-chain (S-3FTxs; five transcripts; 73.14%), long-chain (L-3FTxs, four transcripts; 8.23%), and non-conventional (NC-3FTxs; one transcript; 9.74%) subgroups of 3FTx (Table 3). Organization of the 3FTx subtypes was based on the location and number of disulfide bonds that maintain the protein structure (Figure 3), where S-3FTxs possess four disulfide bonds, whereas L-3FTxs and UC-3FTxs carry a fifth disulfide bond at the lateral end of the second loop and first loop, respectively [32]. Among the ten 3FTx transcripts, four were annotated to Naja sputatrix (one cytotoxin “NSM_3FTX01”, two long neurotoxins “NSM_3FTX06; NSM_3FTX07”, one weak neurotoxin “NSM_3FTX09”). The amino acid sequences of the transcripts were found to be 100% identical to the sequences deposited as N. naja sputatrix toxins in the database. These four 3FTx sequences of N. sputatrix were derived from previous cloning and expression studies using tissues from spitting cobra(s) in Peninsular Malaya or Singapore (Q9PST4 [26]; O42257 [27]; and O57327 [25]). The 100% match with sequences obtained in the present transcriptomic study strongly suggests that the spitting cobra described as N. n. sputatrix or N. sputatrix used in earlier studies was most likely N. sumatrana of Malaysian origin. The other six transcripts of 3FTxs in this study were novel, showing distinct amino acid substitutions compared to the homologous sequences of other Naja cobra species.
Figure 3.
Multiple sequence alignments of the short, long, and non-conventional three-finger toxin (3FTx) transcripts of Malaysian N. sumatrana. (a) 3FTx transcripts were aligned based on the subgroups. (b) 3FTx transcripts were aligned and compared to annotated sequences obtained from public database. In (a): Black bracket represent disulfide linkages. Red and blue brackets highlight the fifth additional disulfide bridge located in the second and first loop of L-3FTx and NC-3FTx, respectively.
It is worthy to note that the four transcripts identical to 3FTx sequences of N. sputatrix (whose true identity should be N. sumatrana as explained above) were highly expressed, with NSM_3FTX01 (identical to cytotoxin 2a) accounting for 72.93% of total toxin FPKM. This was followed by NSM_3FTX09 (weak neurotoxin 5, 9.74%), NSM_3FTX06 (long neurotoxin 7, 4.91%), and NSM_3FTX07 (α-neurotoxin NTX-4, 3.30%). The remaining 3FTx subtypes that were matched to homologous sequences of other cobra species were expressed in extremely low abundance, each constituting far less than 1% of the total toxin FPKM.
Within the S-3FTx subfamily, the transcripts coded for two cytotoxins, a short neurotoxin and a muscarinic toxin-like protein. The expression of cytotoxin genes, in particular the major subtype (NSM_3FX01), far exceeded the abundance of transcripts for short neurotoxins and muscarinic toxin-like protein. The expression of cytotoxin in the venom glands of NS-M is in agreement with findings from the corresponding proteomic study whereby cytotoxins (also known as cardiotoxins) were reported as the main 3FTx subtype group of the venom proteins (44.41%) [36]. Cytotoxins are cytolytic proteins that are associated with necrotizing effect of cobra venom; the high abundance of CTX was consistent with extensive local tissue necrosis observed clinically at the wound site of cobra bite [37,38], and during in vitro cytotoxicity experiments [39]. On the other hand, short neurotoxins were expressed in a much lower abundance (0.21% by total toxin FPKM), although the proteomic study reported an abundance of 3.5% of short neurotoxins [36]. One muscarinic toxin-like protein transcript was detected at an extremely low FPKM; the presence of this protein has not been reported in the venom proteome of N. sumatrana. Meanwhile, the presence of alpha-neurotoxins in the venom-gland transcriptome corroborates the neurotoxicity of N. sumatrana envenomation. Alpha-neurotoxins bind to postsynaptic nicotinic acetylcholine receptors (nAChRs) at the skeletal neuromuscular junction, resulting in systemic paralysis and rapid death in cobra envenoming [40,41]. Three transcripts (NSM_FTX06, NSM_FTX07, and NSM_FTX08) detected were similar to long neurotoxins of the L-3FTx subgroup. NSM_FTX06 and NSM_FTX07 were expressed at similar levels (4.91% and 3.30%, respectively), whilst NSM_FTX08 was expressed at a very low level (0.01%).
There was only one NC-3FTx subtype detected in the NS-M venom-gland transcriptome, annotated as weak neurotoxin 5 (NSM_FTX09). Weak toxin has not been detected in the venom proteome of N. sumatrana [36]. The pathophysiological role of this toxin subtype in envenomation is poorly understood. Weak neurotoxin derived from monocled cobra venom has been shown previously as an antagonist of human and rat neuronal nicotinic receptors, but it has very low lethality (median lethal dose (LD50) of up to 5–80 μg/g) [42] compared to the highly lethal alpha-neurotoxins that have LD50 values of ~0.1–0.2 μg/g in mice [11,36,43,44].
The current transcriptomic findings on the diversity of principal toxins i.e. 3FTx correlated with the characteristic toxic activities of the venom. These include post-synaptic neuromuscular paralysis and severe tissue necrosis [17,18]. The presence of multiple distinct 3FTx transcripts also implies that this toxin group is highly evolved, and the role of 3FTx diversity in predation and envenomation deserves further investigation [45]. Furthermore, the current study revealed that the sequences of alpha-neurotoxins and cytotoxins specific to N. sumatrana exhibit marked consensus sequences with other cobra species such as Naja kaouthia and Naja atra.
The discrepancy between transcriptomic and proteomic expression has been shown in several other studies [6,30,46], although some studies reported conflicting findings [47,48]. The phenomenon could be attributed to a variety of factors including varying rates of mRNA expression for different toxins and post-translational modification of protein [49].
2.4.2. Phospholipase A2 (PLA2)
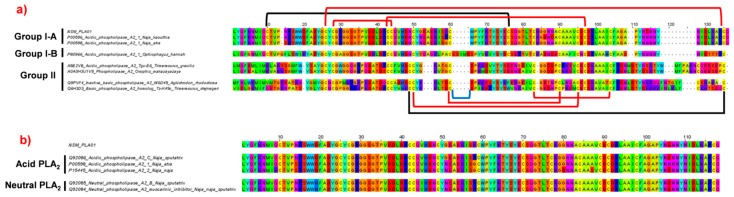
The secretory phospholipases A2 (PLA2) was the second most expressed toxin family of N. sumatrana venom gland. Two distinct PLA2 transcripts were detected, with the major transcript (NSM_PLA01) expressed at 7.39% of total toxin FPKM, followed by NSM_PLA02 (0.03%). The NSM_PLA01 is devoid of a pancreatic loop and is homologous with PLA2 of group I-A (Figure 4a). It is identical to the deposited acidic PLA2 sequence (UniprotKB: Q92086) (Figure 4b) reported previously for the Malayan spitting cobra labeled as N. sputatrix [24]. These findings again indicate that the spitting cobra specimen identified as N. sputatrix in the previous study was most likely N. sumatrana of Malaysian origin, and reaffirms the PLA2 sequence (UniprotKB: Q92086) specific to N. sumatrana. On the other hand, the other PLA2 transcript (NSM_PLA02) sequence was similar to a neutral PLA2 of the banded sea krait, Laticauda semifasciata.
Figure 4.
Multiple sequence alignment of phospholipase A2 (PLA2) transcript of the Malaysian N. sumatrana. (a) Multiple sequence alignment visualizing different groups of PLA2. (b) Sequence alignment of acid and neutral PLA2. In (a): Brackets in black represent disulfide bridges; red represents conservative disulfide bonds and blue represents pancreatic loop.
In contrast to the high PLA2 content (~31% by total protein) reported in the venom proteome [36], the expression of PLA2 transcripts by FPKM was much lower (~7.4%). The intense transcription of 3FTx genes (~91% of all toxins) could possibly reduce transcription levels of other toxin groups at the time of tissue sampling. The transcription of different genes probably proceeds at different rates, and transcripts with low abundance may have a long half-life that in a higher amount of translated protein.
Two PLA2 sequences from N. sumatrana, i.e., an acidic (Q92086) and a neutral PLA2 (Q92085) (previously labeled as for N. sputatrix), are readily accessible in the public database. These two sequences differ by only two amino acids, specifically on residues 20 and 46 (Figure 4). Substitution of the amino acids would have modified the isoelectric points (pI) of the PLA2 to 5.19 and 6.07, respectively, for the acidic and neutral PLA2s. In the present study, the acidic PLA2 transcript (NSM_PLA01) was 100% matched to Q92086 (acidic type), while the neutral PLA2 transcript (NSM_PLA02) was highly homologous to Q8JFB2, a neutral PLA2 (pI = 6.92) originating from L. semifasciata. Although a number of acidic PLA2 from Asian cobras and sea snakes were found to be non-lethal [11,44,50,51,52], the neutral PLA2 of N. sumatrana has been shown to be lethal in mice with an intravenous LD50 of 2.00 μg/g [43]. Clinically, the pathological effect of the neutral PLA2 is not known, but it has been shown to potentiate the toxic activity of cobra cardiotoxin/cytotoxin [53], and is hence a candidate toxin to be targeted for neutralization by antivenom. In addition, the high PLA2 enzymatic activity of N. sumatrana venom has been found to be comparable to other spitting cobras, implying that the PLA2 could have a local tissue effect related to venom ophthalmia [54].
2.4.3. Phylogenetic Analysis of Cytotoxin and Phospholipase A2
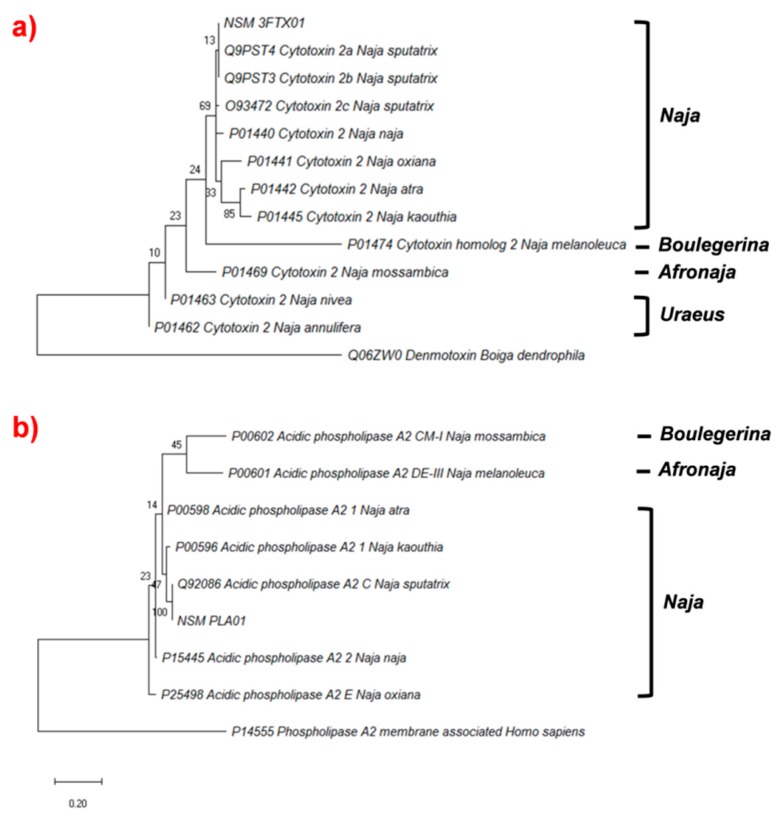
Phylogenetic analysis of cobra cytotoxins indicated that genes from the African non-spitting species (subgenus Uraeus) appeared to be basal (Figure 5a). The CTX 2 genes in Asiatic cobras (subgenus Naja) were further derived. The findings showed that sequences from the previously reported N. sputatrix (Q9PST3, Q9PST4) and those from N. sumatrana (this study) were virtually identical based on their undivided phylogenetic relationship. On the other hand, Figure 5b shows that acidic phospholipases A2 genes from the Asiatic cobras (subgenus Naja) were basal in the phylogenetic tree, but a similar unbranched relationship was again observed between the acidic PLA2 sequences from N. sputatrix (Q92086) and N. sumatrana (this study). These findings further support that N. sumatrana is phylogenetically diverged from other Afro-Asian cobras, and that toxin sequences previously reported as from N. sputatrix most likely originated from N. sumatrana in Malaysia. The phylogenetic relationship of the toxins between N. sumatrana and genuine N. sputatrix (Javan origin) remains to be further investigated, as little is known about the venom-gland transcriptome of the Javan N. sputatrix.
Figure 5.
Phylogenetic trees of cytotoxin and phospholipase A2 transcripts of Malaysian N. sumatrana. The tree was constructed using MEGA X (version 10.0.5), set to Maximum likelihood method, with bootstrap value of 100. (a) Phylogenetic tree of NSM_FTX01 with selected cytotoxin sequences retrieved from various Naja sp. (UniprotKB accession numbers of proteins selected: Q9PST4; Q9PST3; O93472; P01440; P01441; P01442; P01445; P01469; P01462; P01463, and P01463). (b) Phylogenetic tree of NSM_PLA01 with acidic phospholipases A2 from various Naja sp. (UniprotKB accession numbers of proteins selected: Q92086; P00596; P00598; P15445; P25498; Q5G290; P00601, and P00602).
2.5. Low Abundance Transcripts
2.5.1. Transcript Expression of Toxin Families Previously Reported in Venom Proteome
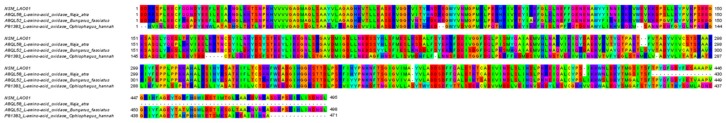
Other toxins transcripts were expressed at low abundance (below 0.4% of total FPKM) in NS-M venom glands. Toxin families whose proteins have been previously reported in the venom proteome include L-amino acid oxidase (LAAO), cobra venom factor (CVF), nerve growth factors (NGF), and vespryn. In the present study, one fully sequenced LAAO transcript was obtained (NSM_LAO01). The sequence contained the three well conserved domains of LAAO and was found to be highly homologous with LAAO sequences from different lineages including other cobra species (Figure 6). This enzyme was of low abundance (at transcript and protein levels) and it showed minimal sequence mutations, consistent with its rather conserved biological function in snake venoms [55,56]. LAAOs are flavoenzymes, found virtually in all front-fanged snake venoms including those of sea snakes [10,50,56,57], which were once thought to be devoid of LAAO. The enzymatic activity catalyzes oxidative deamination of L-amino acids to form alpha-keto acid, ammonia, and hydrogen peroxide; the pharmacological activities are diverse but in cobra venoms it is likely related to anti-microbial and digestive purposes.
Figure 6.
Multiple sequence alignment of L-amino acid oxidase (LAAO) transcript of Malaysian Naja sumatrana.
Three CVF (cobra venom factor) transcripts were uncovered in the NS-M venom-gland transcriptome. Amongst the three, a full-length sequence (NSM_CVF01) was obtained. NSM_CVF01 was almost indistinguishable from the CVF of N. kaouthia (UniprotKB: Q91132), with high homology at 99.82% (Figure 7). It has been suggested that the role of CVF is related to activation of complement system in the prey to increase vascular permeability and blood flow at the bite site [58]. Hence in envenoming, CVF action can promote rapid dissemination of toxin components into the blood circulation of the victim. This effect may also be facilitated by other protein components in the venom, such as nerve growth factor (NGF) that has been shown to cause plasma extravasation and histamine release, rendering the vascular tissue “leaky” to accelerate toxin diffusion [59,60]. Two transcripts of nerve growth factors (NSM_NGF01 and NSM_NGF02) were uncovered in this study with low expression (0.31% of total gene FPKM). This is consistent with the low abundance of NGF protein in the venom proteome [36]. On the other hand, the transcript NSM_NGF01 was highly homologous to the NGF reported for N. sputatrix (UniprotKB: Q5YF89) (Figure 8). The NGF reported has been shown to prevent metalloproteinase autodigestion of venom proteins, which is important for the stability of snake venom [59].
Figure 7.
Multiple sequence alignments of cobra venom factor (CVF) transcript of Malaysian Naja sumatrana.
Figure 8.
Multiple sequence alignment of nerve growth factor (NGF) transcript of Malaysian Naja sumatrana.
A full-length transcript (NSM_VES01) identical to thaicobrin (a vespryn protein) was also revealed in the current study. Similar to NGF, gene expression of vespryn was very low (0.25% of total toxin FPKM). This transcript was 93% homologous to ohanin of king cobra (Ophiophagus hannah), which has been shown to induce hyperalgesia and hypolocomotion in mice [61,62]. The vespryn in N. sumatrana venom may possess similar pharmacological actions and aid in prey immobilization.
2.5.2. Transcript Expression of Toxin Families Not Reported in Venom Proteome of N. sumatrana
Next generation sequencing is an efficient technology in snake venomic studies for exploring the diversity and novelty of toxin genes, even though the gene products (toxin proteins) may not be detected at proteomic level. The current transcriptomic study has successfully identified 15 distinct protein families not previously reported in N. sumatrana venom proteome [36]; these were CRISP, SNACLEC, SVMP, 5’NUC, NP, KSPI, AP, PLB, cystatin, VEGF, DPP IV, neprilysin, HY, PDE, and SVSP. These proteins were probably expressed at extremely low abundance levels, which were not detected by the mass spectrometry in the previous proteomic study. Notably, some proteins such as SNACLEC, SVSP, and SVMP were typically present abundantly in viperid snake venoms, contributing to hemotoxic effects such as thrombocytopenia, venom-induced consumptive coagulopathy, and hemorrhage [63]. However, none of these clinical effects have been reported in N. sumatrana envenomation. These proteins, if expressed, likely serve ancillary functions that aid in predation and digestion of prey.
It must be emphasized that the present transcriptomic study was based on the venom glands of a single adult specimen. The use of a single specimen is common in venom-gland transcriptomic studies due to the scarcity of specimen, and the need to reduce the number of animals sacrificed. This approach limits the interpretation of whether the transcriptional pattern observed is unique to an individual snake, or if it is representative of the entire species [6,22,29,30,45]. Factors such as ontogenic, sexual, and geographical differences may result in inter-individual variation of gene transcription [6,22]. Nonetheless, the transcriptomic data from the current study provide insights into the complexity of venom genes specific to the species N. sumatrana. Furthermore, the sequence database established is valuable for molecular and evolutionary characterization of snake toxins in the future.
3. Conclusions
The de novo venom-gland transcriptomic analysis revealed a unique profile of venom genes in N. sumatrana. The distribution and expression levels of the principal toxin components, i.e., the three-finger toxins and phospholipases A2, provide deep insights into the toxic syndrome and pathophysiology of N. sumatrana envenomation. Notably, highly expressed cytotoxins, whose action may be synergistically enhanced by phospholipases A2, are associated with local tissue necrosis and venom ophthalmia. The alpha-neurotoxins, composed of both short-chain and long-chain subtypes, correlate with post-synaptic neuromuscular paralysis observed in envenomation. Furthermore, full-length toxin sequences obtained from the study provide a reference for validating the origin of sequences previously deposited under the name of Naja sputatrix. These findings also consolidate the knowledgebase of venom genes of this medically important cobra species of Malaysian origin.
4. Materials and Methods
4.1. Preparation of Snake Venom-Gland Tissue
The adult Malaysian N. sumatrana (NS-M) snake was captured in the south-west region of Peninsular Malaysia. The snake was milked for venom and allowed to rest for four days to maximize transcription [64]. The venom glands were promptly removed after euthanasia and sectioned into dimensions of <5 × 5 mm. The slices were preserved in RNAlater® solution (Ambion, Texas, USA), at 4 °C overnight for the efficient penetration of solution into the sample before transferring to storage at −80 °C until further use. The study was conducted in accordance with the experimental protocol approved by the Institutional Animal Use and Care Committee (IACUC) of University of Malaya, Malaysia (code: #2013-11-12/PHAR/R/TCH, date of approval: 12 December 2013).
4.2. Extraction of RNA and Purification of mRNA
The venom gland tissue was homogenized in a 1 ml glass homogenizer with TRIzol solution (Invitrogen, Calsbad, CA, USA) aseptically. Then, 20% chloroform was added, and the sample was centrifuged and treated with RNA-free DNAase I to separate RNA from the cellular debris and residual DNA. The separated RNA was then pelleted using isopropyl alcohol and washed with 75% ethanol. The polyadenylated mRNA (poly(A)+ mRNA) was purified with oligo (dT) magnetic beads from 20 μg of total RNA, as per manufacturer’s instructions (Illumina, San Diego, CA, USA). The quality of the purified RNA was assessed immediately using the Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany). The RNA integrity number (RIN) of the sample was determined to be 8.6, indicating that the RNA was in good condition for downstream transcriptomic analysis.
4.3. Construction of cDNA Library and Sequencing
Construction of the cDNA library was performed with the previously enriched poly(A)+ mRNA isolated from of the total venom-gland RNA. Following purification, the isolated mRNA was fragmented into short fragments by standard buffers containing divalent cations (Zn2+) to mediate the production of homogeneous fragments [65]. The fragments were then served as templates for cDNA synthesis. The first strand of cDNA was synthesized with random hexamer-primer (N6), followed by second strand cDNA synthesis using second strand buffers, RNase H, dNTPs, and DNA polymerase I. Purification of these short fragments was performed with QIAquick PCR extraction kit (Qiagen, Valencia, CA, USA). Purified short fragments are then dissolved with EB buffer for end repair, and the addition of a single adenine nucleotide to assist in the subsequent ligation of Illumina adaptors that contain a single thymine (T) base overhang at the 3’ ends. After the sequencing adaptors were ligated, these short fragments of cDNA were amplified via polymerase chain reaction (PCR) under the following conditions: an initial denaturation at 95 °C for 10 min, followed by 95 °C denaturation 30 s, 60 °C annealing 60 s, and 72 °C extension 60 s, for 40 cycles. The PCR products were then subjected to electrophoresis on 1.5–2% TAE (Tris base, acetic acid and EDTA) agarose gel. From the gel, fragments of 200–700 nucleotides were selected as templates for PCR amplification. PCR was carried out as follows: an initial denaturation at 95 °C for 12 min, 20 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and polymerization at 72 °C for 40 s, and a final extension cycle 72 °C for 5 min. The qualification and quantification of sample library were accessed with ABI StepOnePlus Real-time PCR system (Applied Biosystem, Foster, CA, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany). Sequencing of the amplified samples library was achieved in a single lane on the Illumina HiSeqTM 2000 platform (Illumina, San Diego, CA, USA) with a 100-base pair, pair-end reads.
4.4. Filtration of Raw Sequence Data
Sequence data generated from Illumina HiSeqTM 2000 were transformed into raw reads, stored in a FASTQ format. Prior to transcriptome assembly, filtration of the raw sequencing reads was performed to generate clean reads. This involved removal of low quality reads which possessed more than 5% ambiguous nucleotides, reads containing more than 20% bases with quality score of Q <10 and/or those containing adaptor sequences, using an in-house validated filtering program (Filter_fq, BGI, Yantian, ShenZhen, China).
4.5. Assembly of De Novo Transcriptome
The de novo ‘shot-gun’ transcriptome assembly was performed with Trinity, a short-reads assembly program [66]. The three independent software modules, i.e., Inchworm, Chrysalis, and Butterfly, constitute the Trinity program used to process the large volumes of RNA-seq reads based on De Bruijn graph construction that began by aligning k-mers (k = 25). Reads with a certain length of overlapping were joined to form linear contigs. By referring to pair-end reads, contigs of the same transcripts and the distance between them were determined. Contigs were then categorized into clusters, with each cluster possessing their own set of de Bruijn graphs as a representation of the transcriptional complexity of a given gene or locus. Full-length transcripts were obtained through individual processing of each generated graph, for alternatively spliced isoforms and to tease transcripts corresponding to paralogous genes. For quality control assessment, output statistics was performed with determining the Q20 percentage that serves as a benchmark. Unigenes and contigs were categorized and illustrated in Figure 1. The process was done based on the equation shown below:
4.6. Clustering of Transcripts
Transcript sequences that were generated from Trinity were labeled as Unigenes. Unigenes were further processed for sequence splicing and removal of redundant reads with TGI clustering tools (TGICL), version 2.1, to obtain long non-redundant (NR) transcripts at the longest possible length. The transcripts then underwent family clustering, to classify into: (1) singletons, ID with a prefix of Unigene (2) clusters, ID with the prefix CL and cluster ID are at the end as contigs. In each cluster, there were several transcripts possessing sequence similarities of more than 70% (containing various contigs); whereas singleton ‘Unigenes’ are lack of overlapping with other fragments at a given stringency.
Next, Unigenes were aligned with BLASTx protein database exclusive to NCBI non-redundant database (NR), with significance cut-off value E <10−5 High ranked proteins were then referred to determine the coding region of the Unigenes, followed by translation into amino acid sequences with the standard codon table, hence, both nucleotide sequences (both 5’ and 3’ end) and amino sequences of the Unigene-coding regions were obtained. Several procedures were performed for housekeeping and standardization of the data: redundancy was removed with the selection of the longest sequence present in each cluster as a transcript. Scaffold lengths were extended based on the overlapping sequences applying Phrap assembler (release 23.0) (http://www.phrap.org). The length distributions of the contigs, scaffolds and Unigenes were calculated. For assembly success, the N50 length statistics was set at N50 >500.
4.7. Quantifying the Expression Annotation of Transcripts
The FPKM method of Mortazavi et al. [67] was adapted to determine transcript abundances for the identified genes. The data was computed using RNA-seq with expectation maximization (RSEM) tool incorporated in Trinity, the assembly program according to the formula stated below:
The FPKM method was applied to eliminate the influence of different gene lengths and sequencing discrepancy on the calculation of gene expression. FPKM is defined as the expression of gene A; C is the number of fragments (i.e., reads) that are uniquely aligned to the gene A; N is the total number of fragments (i.e., reads) that are uniquely aligned to all genes; L is the base number in the coding sequence (CDS) of gene A.
4.8. Determination of Functional Annotation of Transcripts
Proteins derived in the process were aligned with the aid of BLASTx to obtain the most resembling sequences present in NR non-redundant protein database to provide proteins functional annotation. The annotation of transcripts provides information about mRNA expression (shown above) and the putative identity of the genes as illustrated in the Supplementary File S1.
4.9. Classification of Venom-Gland Transcripts Based on Toxinology
The transcripts (Unigenes) obtained in the preceding step were filtered to remove transcripts with FPKM <1. Post-filteration transcripts with expression >1 FPKM were broadly categorized into “toxins”, “non-toxins”, and “unidentified”. “Toxin” transcripts were derived by the search-and-find method for specific toxin-keywords against the annotations of the transcripts. Their identities were further validated by subjecting the amino acid sequences to BLASTp (Basic Local Alignment Search Tool-Protein) search in UniProt (Universal Protein Resource Knowledgebase) databank exclusively set to the taxonomy of Serpentes (as of 9th February 2018), applying the lowest E-score value and the highest similarity percentage for annotation purposes. Toxin transcripts were then categorized according to their respective protein families (Supplementary File S2). Toxin transcripts with high similarity to Viperidae (vipers and pit vipers), and FPKM <10 were excluded from the analysis for the possibility of trace contamination. The transcripts of cellular proteins and house-keeping genes were categorized as the “non-toxin” group, whereas those transcripts that could not be identified were classified as “unidentified”.
Relative gene expression (FPKM) of each group was summed and expressed in percentage over the total expression, followed by determining gene expression redundancy for genes by dividing the total transcript FPKM of each group by the total number of transcripts of their respective group. High redundancy indicates high expression level of gene group.
4.10. Multiple Sequence Alignment and Phylogenetic Tree Construction
Selected transcripts obtained in the current study were subjected for multiple sequence alignment. The amino acid sequences obtained from the current study were aligned with toxin sequences retrieved from the Elapidae database in UniprotKB depository (http://www.uniprot.org/). Multiple sequence alignment was achieved with Jalview software (version 2.10.5) [68] and MUSCLE (Multiple Sequence Comparison by Log-Expectation) [69]. Selected transcript sequences were also subjected to phylogenetic tree construction using Mega X (version 10.0.5), and the default statistical method of Maximum Likelihood with bootstrap value of 100 [70].
4.11. Supporting Data
Sequence data from the venom-gland transcriptome of Malaysian Naja sumatrana (NS-M) has been deposited in National Centre for Biotechnology Information (NCBI) Sequence Read Achieve (https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under submission ID: SUB4976990 (https://submit.ncbi.nlm.nih.gov/subs/sra/SUB4976990).
Acknowledgments
The authors are grateful to University of Malaya for funding and provision of laboratory facilities.
Supplementary Materials
The following are available online at http://www.mdpi.com/2072-6651/11/2/104/s1, Supplementary File S1: Categorization of venom-gland transcripts from Malaysian Naja sumatrana; Supplementary File S2: Venom-gland transcriptomic analysis of toxins from Malaysian Naja sumatrana.
Author Contributions
Conceptualization, K.Y.T. and C.H.T.; Formal analysis, H.P.C. and K.Y.T.; Funding acquisition, K.Y.T., N.H.T. and C.H.T.; Investigation, H.P.C., K.Y.T., C.H.T. and N.H.T.; Methodology, H.P.C., K.Y.T. and C.H.T.; Project administration, K.Y.T. and C.H.T.; Resources, C.H.T. and N.H.T.; Supervision, K.Y.T. and C.H.T.; Validation, H.P.C., K.Y.T., C.H.T.; Visualization, H.P.C. and K.Y.T.; Writing—original draft, H.P.C., K.Y.T., C.H.T.; Writing—review & editing, H.P.C., K.Y.T., N.H.T. and C.H.T.
Funding
This research was funded by research grants from University of Malaya with grant numbers FG020-17AFR and RF007C-2018.
Conflicts of Interest
The authors declare no conflicts of interest.
Key Contribution
The de novo venom-gland transcriptome of the Malaysian equatorial spitting cobra, Naja sumatrana, is reported for the first time. The diversity of toxin genes was investigated for deeper insights into the pathophysiology of N. sumatrana envenoming.
References
- 1.Sunagar K., Moran Y. The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals. PLoS Genet. 2015;11:e1005596. doi: 10.1371/journal.pgen.1005596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casewell N.R., Wuster W., Vonk F.J., Harrison R.A., Fry B.G. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol. Evol. 2013;28:219–229. doi: 10.1016/j.tree.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 3.Daltry J.C., Wuster W., Thorpe R.S. Diet and snake venom evolution. Nature. 1996;379:537–540. doi: 10.1038/379537a0. [DOI] [PubMed] [Google Scholar]
- 4.Barlow A., Pook C.E., Harrison R.A., Wuster W. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc. R. Soc. Lond. B Biol. Sci. 2009;276:2443–2449. doi: 10.1098/rspb.2009.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Augusto-de-Oliveira C., Stuginski D.R., Kitano E.S., Andrade-Silva D., Liberato T., Fukushima I., Serrano S.M., Zelanis A. Dynamic Rearrangement in Snake Venom Gland Proteome: Insights into Bothrops jararaca Intraspecific Venom Variation. J. Proteome Res. 2016;15:3752–3762. doi: 10.1021/acs.jproteome.6b00561. [DOI] [PubMed] [Google Scholar]
- 6.Tan K.Y., Tan C.H., Chanhome L., Tan N.H. Comparative venom gland transcriptomics of Naja kaouthia (monocled cobra) from Malaysia and Thailand: elucidating geographical venom variation and insights into sequence novelty. PeerJ. 2017;5:e3142. doi: 10.7717/peerj.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zelanis A., Tashima A.K., Rocha M.M., Furtado M.F., Camargo A.C., Ho P.L., Serrano S.M. Analysis of the ontogenetic variation in the venom proteome/peptidome of Bothrops jararaca reveals different strategies to deal with prey. J. Proteome Res. 2010;9:2278–2291. doi: 10.1021/pr901027r. [DOI] [PubMed] [Google Scholar]
- 8.Tan C.H., Tan K.Y., Yap M.K., Tan N.H. Venomics of Tropidolaemus wagleri, the sexually dimorphic temple pit viper: Unveiling a deeply conserved atypical toxin arsenal. Sci. Rep. 2017;7:43237. doi: 10.1038/srep43237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan K.Y., Tan C.H., Fung S.Y., Tan N.H. Venomics, lethality and neutralization of Naja kaouthia (monocled cobra) venoms from three different geographical regions of Southeast Asia. J. Proteom. 2015;120:105–125. doi: 10.1016/j.jprot.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Oh A.M.F., Tan C.H., Ariaranee G.C., Quraishi N., Tan N.H. Venomics of Bungarus caeruleus (Indian krait): Comparable venom profiles, variable immunoreactivities among specimens from Sri Lanka, India and Pakistan. J. Proteom. 2017;164:1–18. doi: 10.1016/j.jprot.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 11.Wong K.Y., Tan C.H., Tan N.H. Venom and purified toxins of the spectacled cobra (Naja naja) from Pakistan: Insights into toxicity and antivenom neutralization. Am. J. Trop. Med. Hyg. 2016;94:1392–1399. doi: 10.4269/ajtmh.15-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faisal T., Tan K.Y., Sim S.M., Quraishi N., Tan N.H., Tan C.H. Proteomics, functional characterization and antivenom neutralization of the venom of Pakistani Russell’s viper (Daboia russelii) from the wild. J. Proteom. 2018;183:1–13. doi: 10.1016/j.jprot.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Alirol E., Sharma S.K., Bawaskar H.S., Kuch U., Chappuis F. Snake bite in South Asia: a review. PLoS Negl. Trop. Dis. 2010;4:e603. doi: 10.1371/journal.pntd.0000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan C.H., Liew J.L., Tan N.H., Khaldun I.A., Maharani T., Khomvilai S., Sitprija V. Cross reactivity and lethality neutralization of venoms of Indonesian Trimeresurus complex species by Thai Green Pit Viper Antivenom. Toxicon. 2017 doi: 10.1016/j.toxicon.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 15.WHO . Guidelines for the Production Control and Regulation of Snake Antivenomimmunoglobulins. World Health Organization; Geneva, Switzerland: 2010. [Google Scholar]
- 16.Wuster W. Taxonomic changes and toxinology: systematic revisions of the Asiatic cobras (Naja naja species complex) Toxicon. 1996;34:399–406. doi: 10.1016/0041-0101(95)00139-5. [DOI] [PubMed] [Google Scholar]
- 17.WHO . Guidelines for the Management of Snake Bites. Regional office for South-East Asia, World Health Organization; New Delhi, India: 2016. [Google Scholar]
- 18.Chew K.S., Khor H.W., Ahmad R., Rahman N.H. A five-year retrospective review of snakebite patients admitted to a tertiary university hospital in Malaysia. Int. J. Emerg. Med. 2011;4:41. doi: 10.1186/1865-1380-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reid H. Cobra-bites. Br. Med. J. 1964;2:540. doi: 10.1136/bmj.2.5408.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong K.Y., Tan C.H., Tan K.Y., Quraishi N.H., Tan N.H. Elucidating the biogeographical variation of the venom of Naja naja (spectacled cobra) from Pakistan through a venom-decomplexing proteomic study. J. Proteom. 2018;175:156–173. doi: 10.1016/j.jprot.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 21.Tan N.H., Wong K.Y., Tan C.H. Venomics of Naja sputatrix, the Javan spitting cobra: A short neurotoxin-driven venom needing improved antivenom neutralization. J. Proteom. 2017;157:18–32. doi: 10.1016/j.jprot.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 22.Margres M.J., Walls R., Suntravat M., Lucena S., Sanchez E.E., Rokyta D.R. Functional characterizations of venom phenotypes in the eastern diamondback rattlesnake (Crotalus adamanteus) and evidence for expression-driven divergence in toxic activities among populations. Toxicon. 2016;119:28–38. doi: 10.1016/j.toxicon.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sunagar K., Morgenstern D., Reitzel A.M., Moran Y. Ecological venomics: How genomics, transcriptomics and proteomics can shed new light on the ecology and evolution of venom. J. Proteom. 2016;135:62–72. doi: 10.1016/j.jprot.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 24.Armugam A., Earnest L., Chung M., Gopalakrishnakone P., Tan C., Tan N., Jeyaseelan K. Cloning and characterization of cDNAs encoding three isoforms of phospholipase A2 in Malayan spitting cobra (Naja naja sputatrix) venom. Toxicon. 1997;35:27–37. doi: 10.1016/S0041-0101(96)00071-2. [DOI] [PubMed] [Google Scholar]
- 25.Afifiyan F., Armugam A., Tan C.H., Gopalakrishnakone P., Jeyaseelan K. Postsynaptic α-neurotoxin gene of the spitting cobra, Naja naja sputatrix: structure, organization, and phylogenetic analysis. Genome Res. 1999;9:259–266. [PMC free article] [PubMed] [Google Scholar]
- 26.Jeyaseelan K., Armugam A., Lachumanan R., Tan C.H., Tan N.H. Six isoforms of cardiotoxin in Malayan spitting cobra (Naja naja sputatrix) venom: cloning and characterization of cDNAs1. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 1998;1380:209–222. doi: 10.1016/S0304-4165(97)00143-8. [DOI] [PubMed] [Google Scholar]
- 27.Jeyaseelan K., Poh S.L., Nair R., Armugam A. Structurally conserved α-neurotoxin genes encode functionally diverse proteins in the venom of Naja sputatrix. FEBS Lett. 2003;553:333–341. doi: 10.1016/S0014-5793(03)01039-1. [DOI] [PubMed] [Google Scholar]
- 28.Lay Poh S., Mourier G., Thai R., Armugam A., Molgó J., Servent D., Jeyaseelan K., Ménez A. A synthetic weak neurotoxin binds with low affinity to Torpedo and chicken α7 nicotinic acetylcholine receptors. Eur. J. Biochem. 2002;269:4247–4256. doi: 10.1046/j.1432-1033.2002.03113.x. [DOI] [PubMed] [Google Scholar]
- 29.Jiang Y., Li Y., Lee W., Xu X., Zhang Y., Zhao R., Zhang Y., Wang W. Venom gland transcriptomes of two elapid snakes (Bungarus multicinctus and Naja atra) and evolution of toxin genes. BMC Genom. 2011;12:1. doi: 10.1186/1471-2164-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan C.H., Tan K.Y., Fung S.Y., Tan N.H. Venom-gland transcriptome and venom proteome of the Malaysian king cobra (Ophiophagus hannah) BMC Genom. 2015;16:687. doi: 10.1186/s12864-015-1828-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu N., Zhao H.Y., Yin Y., Shen S.S., Shan L.L., Chen C.X., Zhang Y.X., Gao J.F., Ji X. Combined venomics, antivenomics and venom gland transcriptome analysis of the monocoled cobra (Naja kaouthia) from China. J. Proteom. 2017;159:19–31. doi: 10.1016/j.jprot.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 32.Kini R.M., Doley R. Structure, function and evolution of three-finger toxins: mini proteins with multiple targets. Toxicon. 2010;56:855–867. doi: 10.1016/j.toxicon.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 33.Antil S., Servent D., Menez A. Variability among the sites by which curaremimetic toxins bind to torpedo acetylcholine receptor, as revealed by identification of the functional residues of alpha-cobratoxin. J. Biol. Chem. 1999;274:34851–34858. doi: 10.1074/jbc.274.49.34851. [DOI] [PubMed] [Google Scholar]
- 34.Servent D., Antil-Delbeke S., Gaillard C., Corringer P.J., Changeux J.P., Menez A. Molecular characterization of the specificity of interactions of various neurotoxins on two distinct nicotinic acetylcholine receptors. Eur. J. Pharmacol. 2000;393:197–204. doi: 10.1016/S0014-2999(00)00095-9. [DOI] [PubMed] [Google Scholar]
- 35.Antil-Delbeke S., Gaillard C., Tamiya T., Corringer P.J., Changeux J.P., Servent D., Menez A. Molecular determinants by which a long chain toxin from snake venom interacts with the neuronal alpha 7-nicotinic acetylcholine receptor. J. Biol. Chem. 2000;275:29594–29601. doi: 10.1074/jbc.M909746199. [DOI] [PubMed] [Google Scholar]
- 36.Yap M.K., Fung S.Y., Tan K.Y., Tan N.H. Proteomic characterization of venom of the medically important Southeast Asian Naja sumatrana (Equatorial spitting cobra) Acta Trop. 2014;133:15–25. doi: 10.1016/j.actatropica.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 37.Gasanov S.E., Dagda R.K., Rael E.D. Snake venom cytotoxins, phospholipase A2s, and Zn2+-dependent metalloproteinases: mechanisms of action and pharmacological relevance. J. Clin. Toxicol. 2014;4:1000181. doi: 10.4172/2161-0495.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan C.H., Tan N.H. Cytotoxicity of Snake Venoms and Toxins: Mechanisms and Applications. In: Utkin Y.N., editor. Snake Venoms and Envenomation: Modern Trends and Future Prospects. Nova Science Publishers; Hauppauge, NY, USA: 2016. pp. 215–254. [Google Scholar]
- 39.Panagides N., Jackson T.N.W., Ikonomopoulou M.P., Arbuckle K., Pretzler R., Yang D.C., Ali S.A., Koludarov I., Dobson J., Sanker B., et al. How the Cobra Got Its Flesh-Eating Venom: Cytotoxicity as a Defensive Innovation and Its Co-Evolution with Hooding, Aposematic Marking, and Spitting. Toxins. 2017;9:103. doi: 10.3390/toxins9030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barber C.M., Isbister G.K., Hodgson W.C. Alpha neurotoxins. Toxicon. 2013;66:47–58. doi: 10.1016/j.toxicon.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 41.Tan C.H., Tan N.H. Toxinology of Snake Venoms: The Malaysian Context. In: Gopalakrishnakone P., Inagaki H., Mukherjee A.K., Rahmy T.R., Vogel C.-W., editors. Snake Venoms. Springer; Dordrecht, Switzerland: 2015. pp. 1–37. (Toxinology Series). [Google Scholar]
- 42.Utkin Y.N., Kukhtina V.V., Kryukova E.V., Chiodini F., Bertrand D., Methfessel C., Tsetlin V.I. “Weak toxin” from Naja kaouthia is a nontoxic antagonist of alpha 7 and muscle-type nicotinic acetylcholine receptors. J. Biol. Chem. 2001;276:15810–15815. doi: 10.1074/jbc.M100788200. [DOI] [PubMed] [Google Scholar]
- 43.Leong P.K., Fung S.Y., Tan C.H., Sim S.M., Tan N.H. Immunological cross-reactivity and neutralization of the principal toxins of Naja sumatrana and related cobra venoms by a Thai polyvalent antivenom (Neuro Polyvalent Snake Antivenom) Acta Trop. 2015;149:86–93. doi: 10.1016/j.actatropica.2015.05.020. [DOI] [PubMed] [Google Scholar]
- 44.Tan K.Y., Tan C.H., Fung S.Y., Tan N.H. Neutralization of the Principal Toxins from the Venoms of Thai Naja kaouthia and Malaysian Hydrophis schistosus: Insights into Toxin-Specific Neutralization by Two Different Antivenoms. Toxins. 2016:8. doi: 10.3390/toxins8040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vonk F.J., Casewell N.R., Henkel C.V., Heimberg A.M., Jansen H.J., McCleary R.J., Kerkkamp H.M., Vos R.A., Guerreiro I., Calvete J.J., et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. USA. 2013;110:20651–20656. doi: 10.1073/pnas.1314702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Durban J., Juarez P., Angulo Y., Lomonte B., Flores-Diaz M., Alape-Giron A., Sasa M., Sanz L., Gutierrez J.M., Dopazo J., et al. Profiling the venom gland transcriptomes of Costa Rican snakes by 454 pyrosequencing. BMC Genom. 2011;12:259. doi: 10.1186/1471-2164-12-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aird S.D., Watanabe Y., Villar-Briones A., Roy M.C., Terada K., Mikheyev A.S. Quantitative high-throughput profiling of snake venom gland transcriptomes and proteomes (Ovophis okinavensis and Protobothrops flavoviridis) BMC Genom. 2013;14:790. doi: 10.1186/1471-2164-14-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rokyta D.R., Margres M.J., Calvin K. Post-transcriptional Mechanisms Contribute Little to Phenotypic Variation in Snake Venoms. G3 Genes Genomes Genet. 2015;5:2375–2382. doi: 10.1534/g3.115.020578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J.J., Bickel P.J., Biggin M.D. System wide analyses have underestimated protein abundances and the importance of transcription in mammals. PeerJ. 2014;2:e270. doi: 10.7717/peerj.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan C.H., Tan K.Y., Lim S.E., Tan N.H. Venomics of the beaked sea snake, Hydrophis schistosus: A minimalist toxin arsenal and its cross-neutralization by heterologous antivenoms. J. Proteom. 2015;126:121–130. doi: 10.1016/j.jprot.2015.05.035. [DOI] [PubMed] [Google Scholar]
- 51.Tan C.H., Tan K.Y., Ng T.S., Sim S.M., Tan N.H. Venom Proteome of Spine-Bellied Sea Snake (Hydrophis curtus) from Penang, Malaysia: Toxicity Correlation, Immunoprofiling and Cross-Neutralization by Sea Snake Antivenom. Toxins. 2019;11:3. doi: 10.3390/toxins11010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan C.H., Wong K.Y., Tan K.Y., Tan N.H. Venom proteome of the yellow-lipped sea krait, Laticauda colubrina from Bali: Insights into subvenomic diversity, venom antigenicity and cross-neutralization by antivenom. J. Proteom. 2017;166:48–58. doi: 10.1016/j.jprot.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 53.Tan N.-H., Armugam A. In vivo interactions between neurotoxin, cardiotoxin and phospholipases A2 isolated from Malayan cobra (Naja naja sputatrix) venom. Toxicon. 1990;28:1193–1198. doi: 10.1016/0041-0101(90)90119-R. [DOI] [PubMed] [Google Scholar]
- 54.Tan C.H., Wong K.Y., Tan N.H., Ng T.S., Tan K.Y. Distinctive distribution of secretory phospholipases A2 in the venoms of Afro-Asian cobras (subgenus: Naja, Afronaja, Boulengerina and Uraeus. Toxins. 2019;11:116. doi: 10.3390/toxins11020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Du X.-Y., Clemetson K.J. Snake venom l-amino acid oxidases. Toxicon. 2002;40:659–665. doi: 10.1016/S0041-0101(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 56.Tan N.H., Fry B.G., Sunagar K., Jackson T.N.W., Reeks T., Fung S.Y. L-amino acid oxidase enzymes. In: Fry B.G., editor. Venomous Reptiles and Their Toxins: Evolution, Pathophysiology, and Biodiscovery. Oxford University Press; New York, NY, USA: 2015. pp. 291–298. [Google Scholar]
- 57.Tan K.Y., Tan N.H., Tan C.H. Venom proteomics and antivenom neutralization for the Chinese eastern Russell’s viper, Daboia siamensis from Guangxi and Taiwan. Sci. Rep. 2018;8:8545. doi: 10.1038/s41598-018-25955-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vogel C.W., Fritzinger D.C. Cobra venom factor: Structure, function, and humanization for therapeutic complement depletion. Toxicon. 2010;56:1198–1222. doi: 10.1016/j.toxicon.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 59.Lavin M.F., Earl S., Birrel G., St Pierre L., Guddat L., de Jersey J., Masci P. Snake venom nerve growth factors. In: Mackessy S.P., editor. Handbook of Venoms and Toxins of Reptiles. Taylor and Francis Group, CRC Press; Boca Raton, FL, USA: 2009. pp. 377–391. [Google Scholar]
- 60.Kostiza T., Meier J. Nerve growth factors from snake venoms: chemical properties, mode of action and biological significance. Toxicon. 1996;34:787–806. doi: 10.1016/0041-0101(96)00023-2. [DOI] [PubMed] [Google Scholar]
- 61.Pung Y.F., Kumar S.V., Rajagopalan N., Fry B.G., Kumar P.P., Kini R.M. Ohanin, a novel protein from king cobra venom: its cDNA and genomic organization. Gene. 2006;371:246–256. doi: 10.1016/j.gene.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 62.Pung Y.F., Wong P.T., Kumar P.P., Hodgson W.C., Kini R.M. Ohanin, a novel protein from king cobra venom, induces hypolocomotion and hyperalgesia in mice. J. Biol. Chem. 2005;280:13137–13147. doi: 10.1074/jbc.M414137200. [DOI] [PubMed] [Google Scholar]
- 63.Tan C.H., Tan K.Y., Ng T.S., Quah E.S.H., Ismail A.K., Khomvilai S., Sitprija V., Tan N.H. Venomics of Trimeresurus (Popeia) nebularis, the Cameron Highlands pit viper from Malaysia: Insights into venom proteome, toxicity and neutralization of antivenom. Toxins. 2019;11:95. doi: 10.3390/toxins11020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rotenberg D., Bamberger E.S., Kochva E. Studies on ribonucleic acid synthesis in the venom glands of Vipera palaestinae (Ophidia, Reptilia) Biochem. J. 1971;121:609–612. doi: 10.1042/bj1210609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wery M., Descrimes M., Thermes C., Gautheret D., Morillon A. Zinc-mediated RNA fragmentation allows robust transcript reassembly upon whole transcriptome RNA-Seq. Methods. 2013;63:25–31. doi: 10.1016/j.ymeth.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 66.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 68.Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.