Abstract
Phospholipase A2 (PLA2) is a major component in snake venoms and it is found in many different isoforms. To identify transcripts encoding different PLA2 isoforms, we developed a simple, rapid procedure. Total RNA was extracted from the venoms of three cottonmouth snakes and two diamondback rattlesnakes, and further reverse-transcribed into complementary DNA (cDNA). Using one pair of cottonmouth PLA2-specific primers and Reverse Transcription Polymerase Chain Reaction (RT-PCR) technique, we identified 27 unique full-length PLA2 transcripts, including nine sequences identical to the previously documented ones in the database and one novel GIII-like PLA2. Two common transcripts respectively encoding Asp49 and Lys49 PLA2 isoforms were identified in all three cottonmouth venoms that contain more PLA2 transcripts than diamondback rattlesnake venoms. The placement of cloned PLA2 transcripts in snake venom PLA2s was further discussed by phylogenetic analysis. The procedure developed in this study paves the way for accelerated acquisition of transcriptome data on any other venom toxin families. The results obtained are crucial for insight into the structure and function of PLA2 isoforms for scientific and potential therapeutic purposes.
Keywords: snake venom, phospholipase A2 transcript, Bayesian inference
1. Introduction
Phospholipases (PLs) are ubiquitous enzymes present in all organisms and organs, and their primary function is to hydrolyze phospholipids. Depending on which ester linkage of phospholipid is hydrolyzed, PLs are classified into PLA1, PLA2, PLB, PLC and PLD [1]. Among these PLs, PLA2s constitute a superfamily. Based on the primary structures and the organs to be expressed, PLA2s are further divided into 16 groups [2,3]. Snake venom PLs are a secretory PLA2 and belong to at least group IA and II: IA (Elapidae), II (Viperidae). Group II venom PLA2s are further divided into at least IIA (Asp49) and IIB (Lys49) types [4,5,6,7].
In addition to the primary function of hydrolysis of the 2-acyl bond of phospholipids releasing arachidonic acid and lysophospholipids, snake PLA2s display a wide variety of physiological activities such as neurotoxicity, myotoxicity, cardiotoxicity, platelet aggregation induction or inhibition, edema, hemolysis, anti-coagulation and hypotension [8,9]. The variety of different activities is due to the related plural PLA2 isoforms encoded by the multi-gene family generated by gene duplication events occurring over evolutionary time [10,11]. For example, Protobothrops flavoviridis (Crotalinae) group II PLA2 genes form a multi-gene family of 16–32 copies per haploid and are located at two loci on a microchromosome [12,13]. In addition, Shibata et al. [14] observed extensive duplication and accelerated evolution of venom genes such as PLA2 families.
Transcriptome analysis by means of the generation and random sequencing of venom gland complementary DNA (cDNA) libraries has proven to be an effective way of identifying novel transcripts of toxin proteins [15,16,17]. However, the major drawbacks of this approach include (1) the necessity of sacrificing snakes, (2) a complicated procedure (costly and time-consuming), and (3) sequencing too many redundant clones. Intriguingly, snake venoms contain only a few major protein families (PLA2, metalloproteinase, serine, C-type lectins, etc.) and plenty of gene information of these protein families exists in databases. Therefore, to identify unknown transcripts in specific protein families, it is necessary to develop a simple, rapid procedure. Additionally, a common characteristic of venom toxin genes is that the 5’- and 3’- end untranslated regions (UTRs) are more conserved than the protein coding region for a specific toxin family. Thus, the aim of this study was to utilize this characteristic and develop a fast, efficient and inexpensive approach for the rapid identification of full-length transcripts coding different PLA2 isoforms from crude venoms. The venom-based PLA2 transcripts obtained in this study can be used to explore the potential pharmacological activities derived from their counterpart venom proteins and offer important insight into the molecular evolution within the PLA2 multi-gene superfamily.
2. Results and Discussion
2.1. Cloning PLA2 cDNAs from Snake Venoms
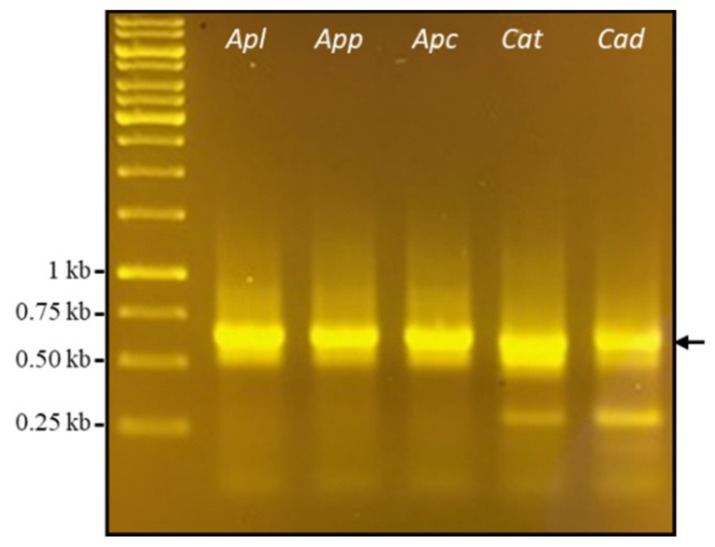
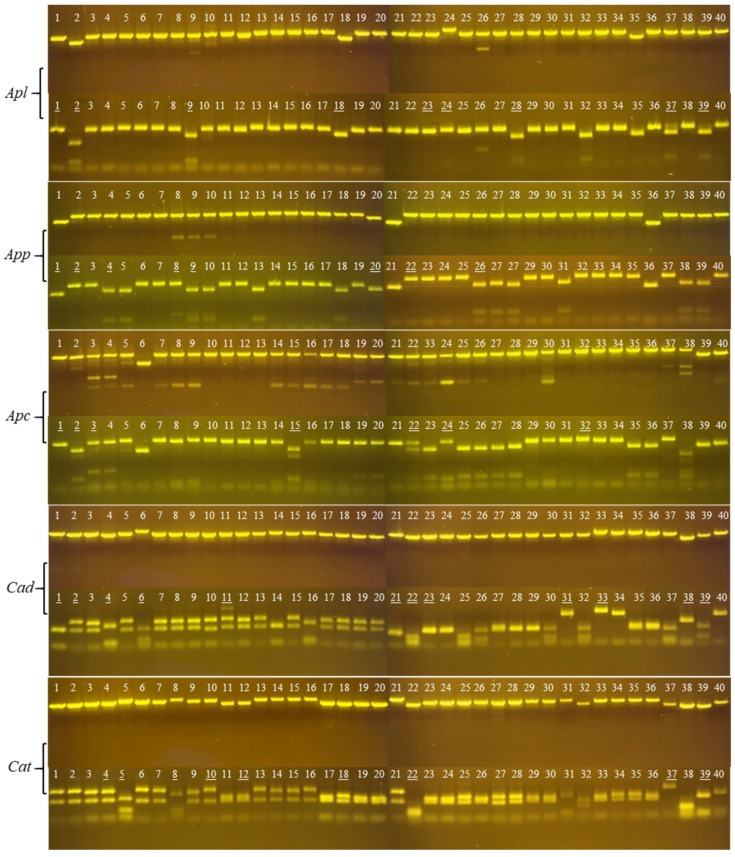
To investigate PLA2 transcripts, we selected a related group of cottonmouth snakes (three subspecies of Agkistrodon), and a related species from a different genus (two rattlesnakes of Crotalus). These species were selected based on an abundance of PLA2 in their crude venoms, and the previous description of PLA2 transcriptome in venom glands and PLA2 toxins in crude venom [17,18,19]. PLA2 was chosen as the focus of this study because it constitutes a very large venom protein superfamily that exhibits various pharmacological activities. One of the distinctive characteristics of this venom gene superfamily is that the variation in the coding regions is higher than that in the non-coding regions for the same gene family [12,20,21,22]. Thus, we utilized this characteristic and transcript sequences we previously generated from cottonmouth snake glands to design two conserved primers located in 5’- and 3’- end UTRs of cottonmouth PLA2 transcripts. Figure 1 shows the Reverse Transcription Polymerase Chain Reaction (RT-PCR) result using cottonmouth PLA2-specific primers and template cDNAs reverse-transcribed from five venoms—Agkistrodon piscivorus leucostoma (Apl), Agkistrodon piscivorus piscivorus (App), Agkistrodon piscivorus conanti (Apc), Crotalus atrox (Cat) and Crotalus adamentus (Cad). A strong band with the same size between 0.5 and 0.75 kb on five samples was successfully amplified after 35 cycles with a Polymerase Chain Reaction (PCR) annealing temperature of 55 °C. As we expected, cottonmouth PLA2-specific primers reacted strongly with the diamondback rattlesnake cDNAs (Cat and Cad), which further confirmed that a specific venom gene family in different species share conserved 5’- and 3’- end UTRs. After the gel purification of each of these bands, cDNAs were separately ligated into SmaI-digested pUC19 vectors by adding one unit of SmaI enzyme in a total ligation volume of 20 μL to prevent vector self-ligation, and we transformed each ligation into competent Escherichia coli cells by the heat shock method and obtained numerous white clones. We completed this process starting from crude venom to obtaining white colonies within 24 hours; using this procedure, cloning different toxin transcripts from crude venom is convenient, fast and economic. To our knowledge, since Chen et al. [23] cloned PLA2 cDNA from the venom of Crotalus durissus collilineatus, it has become a routine procedure to identify toxin transcripts from crude venom. Instead of a venom gland, using the crude venom (particularly the fresh venom) to obtain venom-derived mRNA possesses several advantages including (1) the same sample can be used for both transcriptomic and proteomic analyses, and (2) it avoids the necessity of sacrificing living snakes [24,25,26]. We randomly selected 80 white colonies for each ligation and screened for the expected molecular size (around 600 bp) of PLA2 full-length transcripts by PCR. Small molecules (less than 500 bp) were intentionally eliminated due to the possible 5’- or 3’- end fragments or immature transcripts. After eliminating short fragments, unamplified fragments and most of those with more than two bands, 40 clones with similar molecular weights for each venom (Figure 2, upper panel of each venom) were selected at the end for further restriction enzyme (AluI) digestion (Figure 2, lower panel of each venom). Theoretically, enzyme AluI with the recognition sequence AGCT will digest DNA frequently to produce an average size of 256 bp. From AluI digestion patterns, it can be concluded that:
-
a)
PLA2 transcript sequences are intraspecifically conserved;
-
b)
None or one AluI site is at the 5’- or 3’- end of cottonmouth PLA2 transcripts;
-
c)
One AluI site is near the middle of diamondback PLA2 transcripts.
Figure 1.
Reverse Transcription Polymerase Chain Reaction (RT-PCR) result. Snake venom phospholipase A2 (PLA2) complementary DNAs (cDNAs) were amplified from the reverse-transcribed cDNAs of each crude venom (Agkistrodon piscivorus leucostoma, Apl; Agkistrodon piscivorus piscivorus, App; Agkistrodon piscivorus conanti, Apc; Crotalus atrox, Cat; Crotalus adamanteus, Cad) using one pair of cottonmouth PLA2-specific primers. Arrow denotes the amplified PLA2 cDNAs.
Figure 2.
Selection of unique PLA2 transcripts. Polymerase Chain Reaction (PCR) amplicons (upper panels) were further digested by AluI enzyme (lower panels). The clones with unique patterns (underlined) were subjected to sequencing.
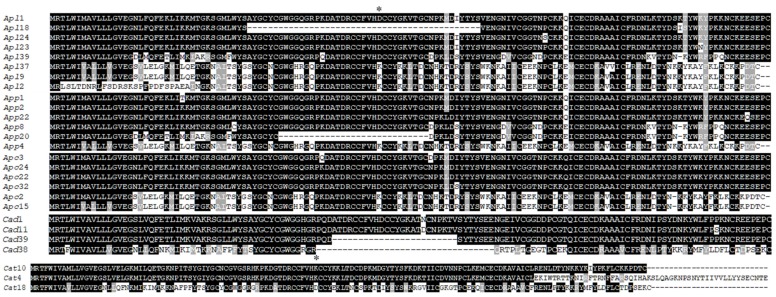
Based on the molecular size and enzyme digestion patterns, a total of 45 clones with unique patterns (Figure 2, underlined clones) from all five snake venoms were selected for sequencing. All obtained cDNA sequences were annotated by a BLASTX search against the database (NCBI) to ensure that they were all full-length transcripts encoding PLA2. Each verified PLA2 cDNA sequence was further translated into a PLA2 amino acid sequence via the ExPASy translate tool (https://web.expasy.org/translate/). Based on the protein sequence, the missense mutations (at least one amino acid difference among PLA2 protein sequences) or frameshift mutations of PLA2 proteins were perceived as unique PLA2 isoforms in each snake venom. In the venom of A. p. leucostoma (Apl), nine clones (underlined clones) were selected for sequencing, resulting in eight unique clones (Apl1, Apl2, Apl9, Apl18, Apl23, Apl24, Apl37 and Apl39) (clones 9 and 28 were identical). In A. p. piscivorus venom (App), eight clones were selected for sequencing, resulting in six unique clones (clones 4, 9 and 26 were identical). In A. p. conanti venom (Apc), seven clones were sequenced, resulting in six unique clones (clones 1 and 22 were identical). Interestingly, Apl1, App2 and Apc24 encoding Asp49 PLA2 were identical, while Apl9, App4 and Apc15 encoding Lys49 PLA2 were identical (Figure 3). The deduced amino acid sequences of these two Asp49 and Lys49 PLA2s transcripts were the same as the amino acid sequences of PLA2 proteins (C0HKC3 and C0HKC2 in the database, respectively) purified from crude venoms [18]. These results further confirmed that cDNA sequences derived from snake gland transcripts can be used to identify unknown transcripts in crude venom for a wide diversity of the PLA2 protein family. Contrastingly, only seven unique PLA2 transcripts were identified from the diamondback rattlesnake venoms after sequencing 21 clones. From C. adamanteus venom (Cad), 12 clones were selected for sequencing, resulting in only four unique clones including two Asp49 full-lengths (Cad1 and Cad11) and two truncated ones (Cad38 and Cad39) (Figure 3, Cad); Cad1 and Cad23 were identical, while Cad2, Cad4, Cad6, Cad21, Cad22, Cad31 and Cad33 did not encode PLA2 proteins (they were non-coding regions). Nine clones were sequenced from C. atrox venom (Cat), resulting in only three unique clones including two Lys49 (Cat4 and Cat10) and one Asp49 (Cat18) (Figure 3, Cat), while Cat12 and Cat18 were identical, and Cat5, Cat8, Cat22, Cat37 and Cat39 were not PLA2 transcripts. Interestingly, Cat4 was a distinct PLA2 transcript similar to GIII PLA2 in length and to GIIB PLA2 in N-terminal sequence, but the sequence close to the C-terminus had a frameshift mutation leading to a long C-terminal tail (27 extra amino acids). The amino acid sequences of Cad1, Cat10 and Cat18 were identical with those of PLA2 with accession numbers (AUS82451, Q8UVZ7 and APD70896) in the database, respectively. Evidently, it is worth mentioning that (1) there are likely many more unique PLA2 transcripts in cottonmouth venoms than in diamondback rattlesnake venoms, and this result is comparable with the previous report [19], and (2) many other sequences including non-coding regions were incidentally amplified from diamondback venoms, implying that 5’- and 3’- end UTRs of diamondback venom PLA2 transcripts are slightly different from those of cottonmouth PLA2 transcripts.
Figure 3.
Amino acid sequence alignment of PLA2 isoforms. Multiple sequence alignment was carried out by CLUSTALW followed by BoxShade manual adjustments. * denotes the Asp49 (D49) or Lys49 (K49) type of PLA2.
2.2. Placement of Cloned PLA2 Transcripts in Snake Venom PLA2s
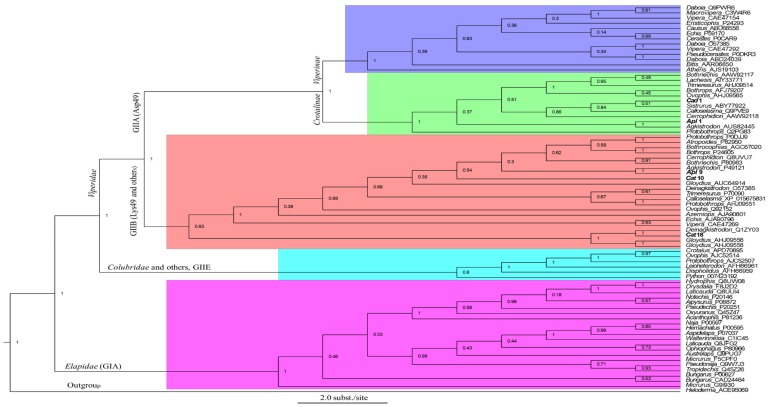
Using the common amino acid sequence of Asp49 PLA2 transcript (Apl1 = App2 = Apc24) (Figure 3) present in all three cottonmouth venoms, we performed the protein BLASTX search against both non-redundant protein sequences (nr) and Swiss-Prot databases in NCBI on October 15, 2018, resulting in 761 snake venom PLA2 amino acid sequences from six families (Viperidae, Elapidae, Colubridae, Hydrophiidae, Lamprophiidae and Pythonidae), 47 genera and 167 species. Most PLA2 were Asp49 types, then Lys49 and others including Ser49, Asn49, Arg49, etc. The top four genera where most PLA2s were obtained, either in the form of PLA2 transcripts or native PLA2 proteins purified from crude venoms, are Vipera (140), Trimeresurus (75), Crotalus (71) and Bothrops (57). We individually performed an amino acid sequence alignment in each genus and selected one to three of the most conserved representative sequences (either Asp49 or both Asp49 and Lys49, depending the availability). Consequently, a total of 74 representative PLA2 amino acid sequences from 47 snake genera and one PLA2 (ACE 95069) from a venomous lizard, the Gila monster (Heloderma suspectum cinctum), as a common outgroup were used for the analysis of evolutionary relationships (Figure 4). Bayesian phylogenetic analysis of these PLA2s showed that different PLA2 types (e.g., GIA, GIIE, GIIB and GIIA) were clustered into the respective monophyletic group with strong confidence (posterior probability of 1.0), which supports the single early recruitment event for each PLA2 type before the separation of snake families. Specifically, all type IA PLA2s (GIA) were grouped into the Elapidae family, while type II PLA2s were clustered into the Viperidae family, which were further divided into GIIA (Asp49) and GIIB (Lys49 and others). Type IIA (Asp49) PLA2s were further distributed into paraphyletic subfamilies: Crotalinae and Viperinae. Meanwhile, all type IIE PLA2s from different families (such as Crotalidae, Viperidae, Lamprophiidae and Pythonidae) were grouped in the same clade with the strong support of statistical probability (>0.8). It was hypothesized that type IIA PLA2 genes may be evolutionarily derived from the IIE PLA2 gene, and then diversified into multiple gene types with several toxic activities such as myotoxic, hemolytic, edema-inducing and neurotoxic activities [27]. This prediction was further confirmed in this study, as all GII PLA2s were nested into group IIE PLA2s (Figure 4). As expected, Asp49 PLA2s (Cad1 and Apl1) obtained in this study from eastern diamondback rattlesnake (C. adamanteus) and cottonmouth venoms, respectively, were grouped into the Crotalinae superfamily; particularly the Agkistrodon PLA2s (Apl1) and AUS82445 (isolated from Agkistrodon contortrix contortri) were in the well-supported same group (1.0 posterior probability). Both Lys49 PLA2s (Apl9 and Cat10), cloned from western cottonmouth and western diamondback rattlesnake venoms, respectively, were grouped into GIIB PLA2 (1.0 posterior probability). However, the analysis here placed the basic Asp49 PLA2s (including Cat18 obtained in this study from C. atrox, Q1ZY03 from Deinagkistrodon acutus [28], AHJ09556 and AHJ09558 from Gloydius [29]) as a sister group to the strongly supported clade (posterior probability of 93%) of other basic GIIB PLA2 homologs. In comparison, the phylogenetic tree topology in this study is strikingly similar to those generated by Modahl et al. [26] and Shibata et al. [14], and the higher-level relationships of snake venom PLA2s across these trees are (GIA (GIIE (GIIA + GIIB))). However, relationships among the snake families of Elapidae, Viperidae and Colubridae differ somewhat between the tree here and those generated by Zheng et al. [30] and Pyron et al. [31], probably due to the use of different genes: Elapidae is sister to Viperidae and Colubridae in this study, whereas Elapidae is nested inside of Viperidae in the trees constructed by Zheng et al. [30] and Pyron et al. [31]. Overall, the phylogenetic tree generated on the basis of amino acid sequences of PLA2 clearly grouped snake families or subfamilies as distinct clades (Figure 4), therefore, PLA2s can be used as a venom toxin gene in inferring the evolutionary history of snake species.
Figure 4.
Phylogenetic analysis of snake venom PLA2s. The tree was constructed by the Bayesian inference (BI) method using Bayesian Evolutionary Analysis Sampling Trees, v1.8.4 (BEAST) based on aligned amino acid sequences. Most PLA2 amino acid sequences used were retrieved from GenBank and are represented by their genus name and accession numbers. Representative PLA2 isoforms obtained from this study are in bold. Numbers on branches are posterior probability values. PLA2 from a lizard (Heloderma suspectum cinctum, ACE95069) was used as a common outgroup.
Collectively, a total of 27 unique PLA2 transcripts were identified after sequencing 45 clones using a simple, rapid technique; these 27 transcripts included eight, six, six, four and three transcripts from the crude venoms of A. p. leucostoma, A. p. piscivourus, A. p. conanti, C. adamanteus and C. atrox, respectively. Transcripts encoding Asp49 and Lys49 PLA2 isoforms were identified from each venom except C. adamanteus venom, in which only Asp49 PLA2 was obtained. The placement of some of the obtained PLA2 transcripts in snake venom PLA2s was phylogenetically analyzed. Importantly, snake venom toxins are encoded by relatively few (approximately 5–10) multi-locus gene families, with each gene family possessing conserved 5’- and 3’- end UTRs. Therefore, the use of conserved primers to identify unknown full-length transcripts for each toxin gene superfamily can be employed to screen transcript sequences within each species for toxins of interest, and to examine novel mutations within a venom protein family. The established technique and results are readily accessible to many researchers and provide a basis for future studies identifying the unique full-length transcripts for any other toxin superfamilies in unexplored venoms.
3. Materials and Methods
3.1. Cloning PLA2 cDNAs from Crude Venoms
Five different venoms, extracted from three cottonmouth snakes (Agkistrodon piscivorus leucostoma, Agkistrodon piscivorus piscivorus and Agkistrodon piscivorus conanti) and two diamondback rattlesnakes (Crotalus atrox and Crotalus adamanteus) in July 2015 were purchased from Miami Serpentarium Lab (Punta Gorda, FL, USA). Ten milligrams of each lyophilized venom mixed with 1 mL of TRIzol Reagents (ThermoFisher Scientific, Waltham, MA, USA) was used to isolate total venom RNA. The isolated total RNA was further reverse-transcribed into complementary DNA (cDNA) using a Maxima First Strand cDNA Synthesis kit (ThermoFisher Scientific) according to the instruction manual. Reverse Transcription Polymerase Chain Reaction (RT-PCR) was performed using a pair of cottonmouth PLA2-specific primers (Forward: 5’-CCGGCTTCTCCTTCTGATCCTT-3’, Reverse: 5’-GAGTGCAAAGCTGGCACCTGT-3’) and Phusion High-Fidelity PCR Master Mix (ThermoFisher Scientific). PCR products were separated in 1% agarose gel and further purified using a GenJET PCR Purification Kit (ThermoFisher Scientific). SmaI enzyme-digested pUC19 plasmid DNA (BioLabs, Ipswich, MA, USA) was ligated with purified PCR product using T4 DNA ligase (ThermoFisher Scientific) at room temperature for 1 hour. The ligation was transformed into competent E. coli cells (DH5α) and then spread on LB (Luria–Bertani) agar plates containing ampicillin (75 μg/μL), X-Gal (20 mg/mL) and IPTG (100 mM). The plates were then incubated at 37 °C overnight. At least 60 white colonies were selected and incubated in each colony in 300 μL of LB broth containing ampicillin overnight at 37 °C. Overnight culture (10 μL) mixed with 40 μL TE buffer was boiled in hot water for 3 minutes, then centrifuged for 3 minutes at top speed. The supernatant (1 μL) was used as the DNA template for PCR using M13 universal primers located on pUC19 vector and Classic Hot Start Taq DNA Polymerase (Tonbo Biosciences, San Diego, CA, USA) in a total volume of 20 μL. The PCR product (10 μL) was used to examine the expected molecular size for PLA2 full-length cDNA (approximately 600 bp) on 1% agarose gel, and the other 10 μL PCR product was used to detect the different sequences for clones with the same molecular size by AluI (BioLabs) restriction enzyme digestion. Based on molecular size and enzyme digestion patterns, the unique PLA2 cDNAs were selected for Sanger sequencing at the University of Texas at Austin (Austin, TX, USA). The identity of transcripts was verified by comparison of the translated cDNA sequences with previously characterized PLA2s using a BLASTX search against both non-redundant protein sequences (nr) and UniProtKB/Swiss-Prot (Swiss-Prot) databases in the National Center for Biotechnology Information search database (NCBI). Signal sequences of each deduced PLA2 proteins were ascertained using the SignalP v4.1 server (http://www.cbs.dtu.dk/services/SignalP/). Amino acid sequence alignment was performed by CLUSTALW (http://www.genome.jp/tools/clustalw/), followed by BoxShade (http://www.ch.embnet.org/software/BOX_form.html) modification. All full-length sequences were submitted to NCBI GenBank (accession numbers MK393884-MK393901).
3.2. Phylogenetic Analysis of Snake Venom PLA2s
To determine the placement of cloned PLA2 transcripts in snake PLA2 proteins, all amino acid sequences of snake PLA2s were retrieved from both non-redundant protein sequences (nr) and UniProtKB/Swiss-Prot (Swiss-Prot) databases by blasting the deduced mature amino acid sequence of common Asp49 PLA2 cDNAs cloned from all cottonmouth snake venoms in this work, and by setting up an expected threshold of 10−5 and max target sequences of 1000, based on the general rule that sequences with an E-value less than 10−5 are homologs of a query sequence [32,33]. A phylogenetic tree of Bayesian inference (BI) was constructed using Bayesian Evolutionary Analysis Sampling Trees, v1.8.4 (BEAST) [34] through the following procedure. A strong multiple alignment of amino acid sequences with an average p-distance of 0.473 and a maximus pairwise p-distance of 0.68 [33] was acquired by performing the MUSCLE program on the MEGA7 package [35] followed by visual inspection for errors and saved as a Nexus format through Seaview v4 [36]. The Tracer (v1.7.1) program [37] was used to eliminate the early low-probability trees from the final consensus tree by setting up the burn-in value of 100,000. We produced maximum clade credibility (MCC) trees using TreeAnnotator [38]. A deduced amino acid sequence of PLA2 obtained from a lizard (Heloderma suspectum cinctum) (Account number: ACE95069) was used as a common outgroup because both the snakes and lizard were revealed to be members of a clade (Toxicofera) whose venom systems are homologous [39]. The resulting phylogenetic tree was visualized in FigTree v1.4.3 [40] and the rotation was manually adjusted.
Acknowledgments
This work was supported by the UTRGV High Scholars Program and the Biology Department at the University of Texas Rio Grande Valley.
Author Contributions
Experimental design, results analysis and validation, and writing, Y.J.; preliminary experiment of two diamondback rattlesnakes, P.O.; preliminary experiments of three cottonmouth snakes, F.R., B.M., S.R.; All authors reviewed and approved the last version of the article.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Cullis P.R., Hope M.J. Physical properties and functional roles of lipids. In: Vance D., Vance J.E., editors. Biochemistry of Lipids and Membranes. Benjamin-Cummings; Menlo Park, CA: 1985. [Google Scholar]
- 2.Schaloske R.H., Dennis E.A. The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta. 2006;1761:1246–1259. doi: 10.1016/j.bbalip.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 3.Murakami M., Taketomi Y., Girad C., Yamamoto K., Lambeau G. Emerging role of secreted phospholipase A2 enzymes: Lessons from transgenic and knockout mice. Biochimie. 2010;92:561–582. doi: 10.1016/j.biochi.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Heinrikson R.L., Krueger E.T., Keim P.S. Amino acid sequence of phospholipase A2-α from the venom of Crotalus adamanteus. J. Biol. Chem. 1977;252:4913–4921. [PubMed] [Google Scholar]
- 5.Dennis E.A. Diversity of group types, regulation, and function of phospholipase A2. J. Biol. Chem. 1994;269:13057–13060. [PubMed] [Google Scholar]
- 6.Maraganore J.M., Merutka G., Cho W., Welches W., Kezdy F.J., Heinrikson R.L. A new class of phospholipases A2 with lysine in place of aspartate 49. Functional consequences for calcium and substrate binding. J. Biol. Chem. 1984;259:13839–13843. [PubMed] [Google Scholar]
- 7.Chakraborti S. Phospholipase A2 isoforms: A perspective. Cell. Signal. 2003;15:637–665. doi: 10.1016/S0898-6568(02)00144-4. [DOI] [PubMed] [Google Scholar]
- 8.Kini R.M. Excitement ahead: Structure, functional and mechanism of snake venom phospholipase A2 enzymes. Toxicon. 2003;42:827–480. doi: 10.1016/j.toxicon.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Angulo Y., Lomonte B. Biochemistry and toxicology of toxins purified from the venom of the snake Bothrops asper. Toxicon. 2009;54:949–957. doi: 10.1016/j.toxicon.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 10.Fry B.G., Wüster W. Assembling an arsenal: Origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences. Mol. Biol. Evol. 2004;21:870–883. doi: 10.1093/molbev/msh091. [DOI] [PubMed] [Google Scholar]
- 11.Casewell N.R., Wagstaff S.C., Wüster W., Cook D.A.N., Bolton F.M.S., King S.I., Pla D., Sanz L., Calvete J.J., Harrison R.A. Medically important differences in snake venom composition are dictated by distinct postgenomic mechanisms. PNAS. 2014;111:9205–9210. doi: 10.1073/pnas.1405484111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakashima K., Ogawa T., Oda N., Hattori M., Sakaki Y., Kihara H., Ohno M. Accelerated evolution of Trimeresurus flavoviridis venom gland phospholipase A2 isozyme. Proc. Natl. Acad. Sci. USA. 1993;90:5964–5968. doi: 10.1073/pnas.90.13.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda N., Chijiwa T., Matsubara K., Oda-Ueda N., Hattori S., Matsuda Y., Ohno M. Unique structural characteristics and evolution of a cluster of venom phospholipase A2 isozymes. Proc. Natl. Acad. Sci. USA. 2010;90:5964–5968. doi: 10.1016/j.gene.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Shibata H., Chijiwa T., Oda-Uedaa N., Nakamura H., Yamajuchi K., Hattori S., Matsubara K., Matsuda Y., Yamashita A., Isomoto A., et al. The habu genome reveals accelerated evolution of venom protein genes. Sci. Rep. 2018;8:11300. doi: 10.1038/s41598-018-28749-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Junqueira-de-Azevedo Ide L., Ho P.L. A survey of gene expression and diversity in the venom glands of the pitviper snake Bothrops insularis through the generation of expressed sequence tags (ESTs) Gene. 2002;299:279–291. doi: 10.1016/S0378-1119(02)01080-6. [DOI] [PubMed] [Google Scholar]
- 16.Wagstaff S.C., Harrison R.A. Venom gland EST analysis of the saw-scaled viper, Echis ocellatus, reveals novel alpha9beta1 integrin-binding motifs in venom metalloproteinases and a new group of putative toxins, renin-like aspartic proteases. Gene. 2006;377:21–32. doi: 10.1016/j.gene.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Jia Y., Cantu B.A., Sánchez E., Pérez J.C. Complementary DNA sequencing and identification of mRNAs from venomous gland of Agkistrodon piscivorus leucostoma. Toxicon. 2008;51:1457–1466. doi: 10.1016/j.toxicon.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia Y., Ermolinsky B., Garza A., Provenzano D. Phospholipase A2 in the venom of three cottonmouth snakes. Toxicon. 2017;135:84–92. doi: 10.1016/j.toxicon.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 19.Lomonte B., Tsai W.C., Ureña-Diaz J.M., Sanz L., Mora-Obando D., Sánchez E.E., Fry B.G., Gutiérrez J.M., Gibbs H.L., Sovic M.G., et al. Venomics of New World pit vipers: Genus-wide comparisons of venom proteomes across Agkistrodon. J. Proteomics. 2014;96:103–116. doi: 10.1016/j.jprot.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogawa T., Oda N., Nakashima K., Sasaki H., Hattori M., Sakaki Y., Sakaki Y., Kihara H., Ohno M. Unusually high conservation of untranslated sequence in cDNA for Trimeresurus flavoviridis phospholipase A2 isozymes. PNAS. 1992;89:8557–8561. doi: 10.1073/pnas.89.18.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chijiwa T., Deshimaru M., Nobuhisa I., Nakai M., Ogawa T., Oda N., Nakashima K., Fukumaki Y., Shimohigashi Y., Hattori S., et al. Regional evolution of venom-gland phospholipase A2 isoenzymes of Trimeresurus flavoviridis snake in the southwestern islands of Japan. Biochem. J. 2000;347:491–499. doi: 10.1042/bj3470491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia Y., Pérez J. Molecular cloning and characterization of cDNAs encoding metalloproteinases from snake venom glands. Toxicon. 2010;55:2–3. doi: 10.1016/j.toxicon.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen T.B., Bjourson A.J., Orr D.F., Kwok H.F., Rao P.F., Ivanyi C., Shaw C. Unmasking venom gland transcriptomes in reptile venoms. Anal. Biochem. 2002;311:152–156. doi: 10.1016/S0003-2697(02)00404-9. [DOI] [PubMed] [Google Scholar]
- 24.Currier R.B., Calvete J.J., Sanz L., Harrison R.A., Rowley P.D., Wagstaff S.C. Unusual stability of messenger RNA in snake venom reveals gene expression dynamics of venom replenishment. Plos ONE. 2012;7:1–10. doi: 10.1371/journal.pone.0041888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H., Chen X., Wang L., Chen W., Zhou M., Chen T., Shaw C. Cloning and characterization of three novel disintegrin precursors from the venoms of three Atheris species: Atheris chlorechis, Atheris nitschei and Atheris squamigera. Toxicon. 2013;71:31–40. doi: 10.1016/j.toxicon.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 26.Modahl C.M., Mackessy S.P. Full-length venom protein cDNA sequences from venom-derived mRNA: exploring compositional variation and adaptive multigene evolution. PloS Negl. Trop. Dis. 2016;10:e0004587. doi: 10.1371/journal.pntd.0004587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamaguchi K., Chijiwa T., Ikeda N., Shibata H., Fukumaki Y., Oda-Ueda N., Hattori S., Ohno M. The finding of a group IIE phospholipase A2 gene in a specified segment of Protobothrops flavoviridis genome and its possible evolutionary relationship to group IIA phospholipase A2 genes. Toxins. 2014;6:3471–3487. doi: 10.3390/toxins6123471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y.M., Wang J.H., Tsai I.H. Molecular cloning and deduced primary structures of acidic and basic phospholipases A2 from the venom of Deinagkistrodon acutus. Toxicon. 1996;34:1191–1196. doi: 10.1016/0041-0101(96)00067-0. [DOI] [PubMed] [Google Scholar]
- 29.Malhotra A., Creer S., Thorpe R.S., Harris J.B., Stocklin R., Favreau P. Sequence submitted in Genbank. 2013. Unpublished. [DOI] [PubMed]
- 30.Zheng Y., Wiens J.J. Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptile (lizards and snakes) based on 52 genes and 4162 species. Mol. Phylogenet. Evol. 2016;94:537–547. doi: 10.1016/j.ympev.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 31.Pyron R.A., Burbrink F.T., Colli G.R., Montes de Oca A.N., Vitt L.J., Kuczynski C.A., Wiens J.J. The phylogeny of advanced snakes (Colubroidea), with discovery of a new subfamily and comparison of support methods for likelihood trees. Mol. Phylogenet. Evol. 2011;58:329–342. doi: 10.1016/j.ympev.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Pearson W.R. An introduction to sequence similarity (“homology”) searching. Curr. Protoc. Bioinf. 2013 doi: 10.1002/0471250953.bi0301s42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall B.G. Phylogenetic Trees Made Easy: A How-to Manual. 5th ed. Sinauer Associates, Inc.; Sunderland, MA, USA: 2018. pp. 57–62. [Google Scholar]
- 34.Drummond A.J., Suchard M.A., Xie D., Rambaut A. Bayesian phylogenetics with BEAUTi and BEAST 1.7. Mol. Biol. Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gouy M., Guindon S., Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 37.Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drummond A.J., Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fry G.G., Winter K., Norman J.A., Roelants K., Nabuurs R.J., van Osch M.J., Teeuwisse W.M., van der Weerd L., McNaughtan J.E., Kwok H.F., et al. Functional and structural diversification of the Anguimoupha lizard venom system. Mol. Cell. Proteomics. 2010;9:2369–2390. doi: 10.1074/mcp.M110.001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Figtree. Institute of Evolutionary Biology, University of Edinburgh; Edinburgh, UK: 2014. version 1.4.3. graphical viewer of phylogenetic trees. [Google Scholar]