Abstract
The isolation and structural elucidation of a structurally new desertomycin, designated as desertomycin G (1), with strong antibiotic activity against several clinically relevant antibiotic resistant pathogens are described herein. This new natural product was obtained from cultures of the marine actinomycete Streptomyces althioticus MSM3, isolated from samples of the intertidal seaweed Ulva sp. collected in the Cantabrian Sea (Northeast Atlantic Ocean). Particularly interesting is its strong antibiotic activity against Mycobacterium tuberculosis clinical isolates, resistant to antibiotics in clinical use. To the best of our knowledge, this is the first report on a member of the desertomycin family displaying such activity. Additionally, desertomycin G shows strong antibiotic activities against other relevant Gram-positive clinical pathogens such as Corynebacterium urealyticum, Staphylococcus aureus, Streptococcus pneumoniae, Streptococcus pyogenes, Enterococcus faecium, Enterococcus faecalis, and Clostridium perfringens. Desertomycin G also displays moderate antibiotic activity against relevant Gram-negative clinical pathogens such as Bacteroides fragilis, Haemophilus influenzae and Neisseria meningitidis. In addition, the compound affects viability of tumor cell lines, such as human breast adenocarcinoma (MCF-7) and colon carcinoma (DLD-1), but not normal mammary fibroblasts.
Keywords: desertomycin, macrolide, Streptomyces, seaweed associated actinobacteria, actinomycetes, intertidal seaweed
1. Introduction
Desertomycins are a family of macrolide natural products of pharmacological interest due to their diverse biological activities. Structurally, they belong to the marginolactones family; aminopolyol polyketides containing a macrolactone ring whose biosynthesis is the subject of active research [1]. After a report on the first member of the family, desertomycin A, with antibiotic activity against bacteria and fungi, [2], other structurally related compounds of the family such as the desertomycins B and D [3], E [4] and F [5] have also been identified. They are mainly produced by Streptomyces species isolated from soils, such as Streptomyces macronensis and Streptomyces flavofungini [6], but have also been found in cultures of Streptoverticillium baldaccii [7].
The marine environment has become a prime resource in the search for, and discovery of, novel natural products. Marine actinomycetes (Phylum Actinobacteria) have turned out to be important contributors [8]. Marine macroalgae (seaweeds) remain a relatively underexplored source in the search of Streptomyces producing bioactive compounds of pharmacological interest. Previous reports describe the isolation of actinomycetes of the Streptomyces genus from seaweeds on coastal ecosystems from temperate and cold waters of the North Atlantic Ocean, particularly from the Iberian Peninsula coasts [9] and from the Kiel Fjord in the Baltic Sea [10,11]. Previous research in the Cantabrian Sea (Biscay Bay, Northeast Atlantic) has revealed that bioactive Streptomyces species are sometimes associated with marine macro-algae [12,13,14]. Oceanic and atmospheric environments in this region [13,14,15] are emerging as novel sources for the discovery of new natural products with antibiotic properties and cytotoxic activities towards cancer cell lines [16,17,18,19,20].
We report herein the discovery of a new natural product, desertomycin G (1), obtained from Streptomyces althioticus MSM3, isolated from samples of the intertidal seaweed Ulva sp. (Phylum Chlorophyta) collected in the Cantabrian Sea. After growing this strain in R5A medium, extracts of the culture broth exhibited a strong antibiotic activity. A bioassay-guided identification of the active fractions after a semipreparative HPLC analysis of an extract pointed to a single peak as responsible for the activity. Following our LC-UV-MS and LC-HRMS chemical dereplication approaches for marine natural products [21], this peak turned out not to be included in our library and prompted us to perform isolation and identification. Its structure was elucidated by HRMS and 1D and 2D NMR analyses. Desertomycin G displays potent antibiotic activities against Gram-positive clinical pathogens and, to the best of our knowledge, is the first member of the family displaying strong antibiotic activity against Mycobacterium tuberculosis. Additionally, desertomycin G shows cytotoxic activity against two human tumor cell lines.
2. Results
2.1. Taxonomy and Phylogenetic Analysis of the Strain MSM3
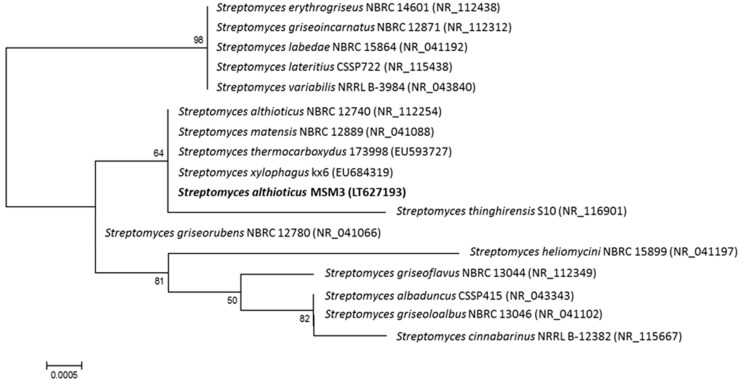
The 16 S rDNA of the producing strain MSM3 was polymerase chain reaction (PCR) amplified and sequenced as previously reported [14]. Sequence analysis showed a 99.9% identity to Streptomyces althioticus (AY999808). Thus, this strain was designated as Streptomyces althioticus MSM3 (EMBL Sequence number LT627193). Another strain of this species producing an unidentified antituberculous compound, Streptomyces althioticus (KCTC 9752), was isolated from the Thar Desert soil, in Rajasthan [22]. The phylogenetic tree generated by a neighbor-joining method based on 16S rRNA gene sequence clearly revealed the evolutionary relationship of the strain MSM3 with a group of known Streptomyces species (Figure 1).
Figure 1.
Neighbor-joining phylogenetic tree obtained by distance matrix analysis of 16S rDNA sequences, showing Streptomyces althioticus MSM3 position and its most closely related phylogenetic neighbors. Numbers on branch nodes are bootstrap values (1000 re-samplings; only values >50% are given). Bar indicates 0.05% sequence divergence.
2.2. Structure Determination
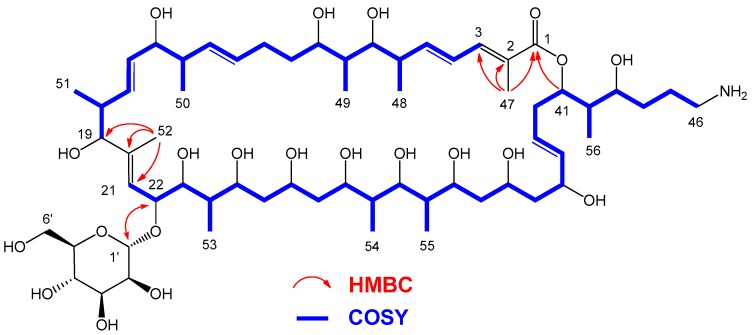
A molecular formula of C62H109NO21 was assigned to desertomycin G, according to a protonated ion observed at m/z 1204.7609 in its ESI-TOF spectrum (calcd. for C62H110NO21+ 1204.7565, ∆ 3.6 ppm). Analysis of its 1H, 13C (Table 1) and HSQC spectra revealed the presence in the molecule of 10 olefinic protons, 19 oxygenated methines, one doubly oxygenated methine, probably belonging to the anomeric position of a sugar residue, one oxygenated methylene, 8 aliphatic methines, 9 aliphatic and one nitrogenated methylenes and 10 aliphatic methyl groups, suggesting a polyketidic nature for the compound. Correlations observed in the COSY spectrum allowed us to establish the sequences from C-3 to C-19 and from C-21 to C-46 and also confirmed the attachment of methyl groups C-48, C-49, C-50, C-51, C-53, C-54, C-55 and C-56 to C-6, C-8, C-14, C-18, C-24, C-30, C-32 and C-42, respectively (Figure 2). On the other hand, HMBC correlations between H3-47 to C-1, C-2 and C-3, and H3-52 to C-19, C-20 and C-21 allowed completion of the linear sequence from C-1 to C-46. An additional HMBC correlation between H-41 and C-1 and the deshielded oxygenated proton H-41 (5.11 ppm) was also indicative of the existence of a lactone ring between C-41 and C-1 (Figure 2). Finally, the only nitrogen atom present in the molecule was associated with the presence of a primary amine group at C-46, as evidenced by the corresponding 1H and 13C NMR chemical shifts at this terminal position (δH 2.93 ppm and δc 40.8 ppm). A bibliographic search established that the planar structure of this macrocyclic moiety of the molecule was very similar to that found in desertomycin A, the two major differences between both molecules being the presence of an additional double bond at ∆4 and an additional methyl group (C-53), located at C-24 in the structure of desertomycin G. The E configuration of the ∆4 double bond was proposed based on the large JH4-H5 coupling constant measured (15.0 Hz). The remaining signals not yet assigned in the NMR spectra corresponded to one doubly oxygenated methine group, four oxygenated methines and one oxygenated methylene (C-1′ to -C-6′). These signals were in agreement with the presence in the molecule of an hexopyranose, identified as α-d-mannopyranose based on similar chemical shifts to those measured in the structure of desertomycin A [23]. Additionally, HMBC correlations from H-1′ to C-22 and from H-22 to C-1′ (Figure 2) confirmed the linkage of this sugar moiety to carbon C-22.
Table 1.
1H and 13C NMR assignments for desertomycin G (1) (CD3OD, 500 MHz).
| 1H NMR | 13C NMR | 1H NMR | 13C NMR | ||
|---|---|---|---|---|---|
| Position | δ in ppm (mult, J in Hz) | δ in ppm | Position | δ in ppm (mult, J in Hz) | δ in ppm |
| 1 | 169.8 | 32 | 1.67 (m) | 41.7 | |
| 2 | 126.4 | 33 | 4.17 (br d, 9.7) | 70.1 | |
| 3 | 7.20 (br d, 11.1) | 140.8 | 34 | 1.63 (m); 1.40 (m) | 42.5 |
| 4 | 6.47 (dd, 14.4, 11.4) | 126.2 | 35 | 4.00 (m) | 66.4 |
| 5 | 6.23 (dd, 15.0, 7.5) | 148.9 | 36 | 1.51 (m) | 46.4 |
| 6 | 2.58 (m) | 41.2 | 37 | 4.26 (m) | 69.6 |
| 7 | 3.48 (dd, 8.7, 3.0) | 78.6 | 38 | 5.57 (dd, 15.5, 4.8) | 138.2 |
| 8 | 1.77 (m) | 43.0 | 39 | 5.63 (m) | 125.8 |
| 9 | 3.78 (m) | 74.6 | 40 | 2.46 (m); 2.31 (m) | 34.6 |
| 10 | 1.62 (m); 1.39 (m) | 34.0 | 41 | 5.11 (m) | 75.7 |
| 11 | 2.25 (m); 2.06 (m) | 30.4 | 42 | 2.00 (m) | 43.8 |
| 12 | 5. 48 (m) | 131.5 | 43 | 3.53 (m) | 72.5 |
| 13 | 5.48 (m) | 134.1 | 44 | 1.65 (m); 1.40 (m) | 30.3 |
| 14 | 2.19 (m) | 44.0 | 45 | 1.83 (m); 1.67 (m) | 25.6 |
| 15 | 3.87 (m) | 76.8 | 46 | 2.93 (m) | 40.8 |
| 16 | 5.49 (m) | 132.1 | 47 | 1.94 (br s) | 12.8 |
| 17 | 5.49 (m) | 134.6 | 48 | 1.06 (d, 6.6) | 12.8 |
| 18 | 2.35 (m) | 41.2 | 49 | 0.85 (d, 6.5) | 12.3 |
| 19 | 3.71 (m) | 83.6 | 50 | 0.96 (m) | 15.5 |
| 20 | 146.2 | 51 | 1.12 (d, 6.5) | 17.7 | |
| 21 | 5.40 (br d, 9.8) | 122.9 | 52 | 1.76 (br s) | 12.2 |
| 22 | 4.56 (dd, 9.9, 3.2) | 72.7 | 53 | 0.85 (d, 6.5) | 11.2 |
| 23 | 3.77 (m) | 77.3 | 54 | 0.94 (m) | 10.1 |
| 24 | 1.53 (m) | 41.0 | 55 | 0.78 (d, 6.8) | 11.5 |
| 25 | 4.29 (m) | 68.9 | 56 | 0.95 (m) | 10.6 |
| 26 | 1.71 (m); 1.40 (m) | 42.8 | 1′ | 4.86 (m) | 97.7 |
| 27 | 4.03 (m) | 69.4 | 2′ | 3.77 (m) | 72.3 |
| 28 | 1.72 (m) | 43.4 | 3′ | 3.75 (m) | 72.7 |
| 29 | 3.82 (m) | 75.1 | 4′ | 3.63 (dd, 9.2) | 68.8 |
| 30 | 1.63 (m) | 40.9 | 5′ | 3.56 (m) | 74.9 |
| 31 | 3.99 (m) | 73.5 | 6′ | 3.85 (m); 3.73 (m) | 62.9 |
Figure 2.
Key COSY and HMBC correlations determining the connectivity of 1.
2.3. Antimicrobial Activity of Desertomycin G
The antimicrobial activity of compound 1 was tested against a panel of human pathogens (Table 2). Some of them were isolated and identified in clinical microbiology laboratories from samples obtained from patients with clinical infections. The compound exhibited strong inhibitory activities against the pathogenic Gram-positive bacteria Corynebacterium urealyticum, Staphylococcus aureus, Streptococcus pneumoniae, Streptococcus pyogenes, Enterococcus faecium, Enterococcus faecalis, Clostridium perfringens and Mycobacterium tuberculosis; and moderate inhibition of the Gram-negative bacteria Bacteroides fragilis, Haemophilus influenzae, and Neisseria meningitidis.
Table 2.
Description of clinic bacterial pathogens and MIC values for compound 1.
| Clinical Pathogen | Isolate | Hospital | Year | Antibiotic Resistances | MIC (µg/mL) |
|---|---|---|---|---|---|
| Gram-positives | |||||
| M. tuberculosis H37Rv | ATCC 27294 | - | 16 | ||
| M. tuberculosis MDR-1 | 14595 | SNRL-Spain | 2013 | Multiresistance a | 16 |
| M. tuberculosis MDR-2 | 14615 | SNRL-Spain | 2013 | Multiresistance b | 16 |
| C. perfringens | 103281 * | HUCA | 2013 | - | 16 |
| C. urealyticum | 1492 * | Cabueñes | 2014 | Multiresistance c | <0.25 |
| E. faecalis | 10544 | Cabueñes | 2015 | Ery, clin, tet | 8 |
| E. faecalis | ATCC 51299 | - | - | 8 | |
| E. faecalis | ATCC 29212 | - | - | 8 | |
| E. faecium | 10701 | Cabueñes | 2015 | Amp, quin, ery | 4 |
| S. pneumoniae | 64412 * | HUCA | 2013 | Ery | |
| S. pyogenes | 81293 * | HUCA | 2013 | - | |
| S. aureus | 11497 | Cabueñes | 2015 | Methicillin sensitive | 4 |
| S. aureus | ATCC 43300 | - | - | 4 | |
| S. aureus | ATCC 25923 | - | - | 4 | |
| Gram-negatives | - | ||||
| B. fragilis | 61592 * | HUCA | 2013 | Amo, tet | 32 |
| B. fragilis | ATCC 25285 | - | - | 32 | |
| H. influenzae | 10996 | Cabueñes | 2015 | Amp, cot, quin | >64 |
| H.influenzae | ATCC 49247 | - | - | 64 | |
| N. meningitidis | 71327 | HUCA | 2013 | Clin | 64 |
* [14]; Amk: amikacin; amo: amoxicillin; amp: ampicillin; cap: capreomycin; cip: ciprofloxacin; clav: clavulanic acid; clin: clindamycin; cot: cotrimoxazole; emb: ethambutol; ery: erythromycin; fos: fosfomycin; inh: isoniazid; kan: kanamycin; nitro: nitrofurantoin; quin: quinolones; rif: rifampicin; str: streptomycin; tet: tetracycline. a Inh, rif, emb; b Inh, rif, emb, str, amk, kan, cap; c Amp, amo/clav, ery, cot, cip, fos, nitro.
2.4. Cytotoxic Activity of Desertomycin G
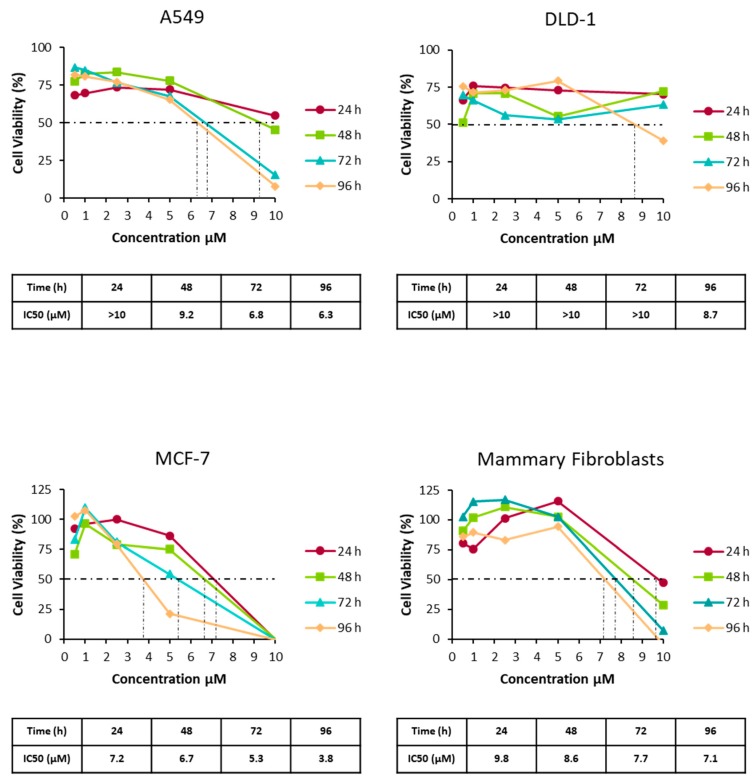
The cytotoxic activity of compound 1 was examined against the A549 human lung carcinoma, DLD-1 colon carcinoma and MCF-7 human breast adenocarcinoma cell lines, as well as healthy mammary fibroblasts. At day three, results indicated that DLD-1 and MCF-7 cell lines decreased their viability about 50% with respect to the control when employing 2,5 and 5 µM desertomycin G. A549 was more resistant and healthy mammary fibroblasts remained unaffected at these concentrations (Figure 3).
Figure 3.
Cell proliferation of A549, DLD-1, MCF-7 cancer cells lines and healthy mammary fibroblasts in the presence or absence of different concentrations of desertomycin G. Cell proliferation rates were determined on five consecutive days using an automated microtitre plate reader. *** Significant differences with a p value less than 0.005.
3. Discussion
Natural products of the desertomycin family were isolated in the last century from soil samples. A new member of the family, desertomycin G, was herein identified from cultures of the marine derived Streptomyces althioticus MSM3, associated with the intertidal seaweed Ulva sp. from the Cantabrian Sea. Its structure was established by analysis of its HRMS and 1D and 2D NMR spectra. Due to its potent antibiotic activities against clinically resistant pathogens and, given the medical needs for novel antibiotics against these microorganisms, desertomycin G deserves to be considered as a candidate for antibiotic chemotherapy, particularly against Mycobacterium tuberculosis resistant strains, but also against the rest of the indicated pathogens. Its cytotoxicity against tumor cell lines, but not against mammary fibroblasts, also highlights the use of desertomycin G as a potential antitumor agent. This is another example of the potential of marine Streptomyces species, particularly those isolated from the Cantabrian Sea, as new sources for the discovery of novel natural products with great potential as both antibiotic and antitumor agents.
4. Materials and Methods
4.1. General Experimental Procedures
Analytical and semipreparative HPLC analyses were conducted using a Waters Alliance (Waters Corporation, Milford, MA, USA) chromatographic system with a SunFire C18 column (10 µm, 10 × 250 mm, Waters). For UPLC analysis an Acquity UPLC equipment with a BEH C18 column (1.7 μm, 2.1 × 100 mm, Waters) was used. Optical rotations were measured using a Jasco P-2000 polarimeter (JASCO Corporation, Tokyo, Japan). UV spectra were obtained with an Agilent 1100 DAD (Agilent Technologies, Santa Clara, CA, USA). IR spectra were recorded on a JASCO FT/IR-4100 spectrometer (JASCO Corporation, Tokyo, Japan) equipped with a PIKE MIRacleTM single reflection ATR accessory (PIKE Technologies Inc., Madison, WI, USA). NMR spectra were recorded on a Bruker Avance III spectrometer (500 and 125 MHz for 1H and 13C NMR, respectively) equipped with a 1.7 mm TCI MicroCryoProbeTM (Bruker Biospin, Falländen, Switzerland), using the signal of the residual solvent as internal reference (δH 3.31 and δC 49.0 ppm for CD3OD). HRESIMS spectra were acquired using a Bruker maXis QTOF spectrometer (Bruker Daltonik GmbH, Bremen, Germany).
4.2. Microorganism and Fermentation Conditions
Streptomyces althioticus strain MSM3 was isolated from the macroalgae Ulva sp. collected in the Cantabrian Sea in Pedreña, Cantabria (coordinates 43°26′37″N, 3°46′5″W). Thirty Erlenmeyer flasks (250 mL), each containing 50 mL of R5A medium [12] were inoculated with spores of this strain and incubated in an orbital shaker at 28 °C and 250 rpm for 6 days.
4.3. Isolation and Purification of Desertomycin G
The cultures were then centrifuged and the pellets were discarded. The supernatants were filtered and applied to a solid-phase extraction cartridge (Sep-Pak Vac C18, 10 g, Waters). The retained material was eluted with a mixture of methanol and 0.05% TFA in water. A linear gradient from 0 to 100% methanol in 60 min, at 10 mL/min, was used. Fractions were collected every 5 min and analyzed by UPLC using chromatographic conditions previously described [12]. A peak corresponding to the desired compound was observed in fractions taken between 35 and 45 min. These fractions were pooled, partially dried in vacuo, and applied to a to a solid-phase extraction cartridge (Sep-Pak Vac C18, 2 g, Waters). The cartridge was washed with water; the retained compounds were eluted with methanol and dried in vacuo. The residue was subsequently re-dissolved in a small volume of methanol and DMSO (1:1). Purification was performed in two steps using a SunFire C18 column (10 µm, 10 × 250 mm, Waters). In the first step the extract was chromatographed with a mixture of acetonitrile and 0.1% TFA in water (32:68) in isocratic conditions and a flow rate of 7 mL/min. In the second step the mobile phase was a mixture of methanol and 0.1% TFA in water (67:33), at 6 mL/min. After every step, the solution containing the collected peak was partially evaporated under vacuum to reduce the concentration of the organic solvent and then applied to a solid-phase extraction cartridge (Sep-Pak C18, 500 mg, Waters). The cartridge was washed with water and the retained compound was eluted with methanol. The purified compound was finally re-dissolved in a mixture of tert-butanol and water (1:1) and lyophilized, resulting in 20.4 mg of pure product.
Desertomycin G (1): white solid; [α]20D + 15.6 (c 0.35, MeOH); IR (ATR) νmax 3367, 2970, 2927, 1683, 1637, 1455, 1433, 1387, 1255, 1232, 1203, 1136, 1105, 1070, 1053, 971 cm−1; for 1H and 13C NMR data see Table 1; HRESIMS m/z 1204.7609 [M + H]+ (calcd. for C62H110NO21+, 1204.7565), 593.8776 [M − H2O + 2H]2+ (calcd. for C62H109NO202+, 593.8774).
4.4. Phylogenetic Analysis (Taxonomy) of the Producer Microorganism
Strain Streptomyces althioticus MSM3 was subjected to phylogenetic analysis based on 16S rRNA sequences analysis. Phylogenetic analysis was performed using MEGA version 6.0 [24] after multiple alignment of data by CLUSTALO [25]. Distances (distance options according to the Kimura two-parameter model [26]) and clustering with the neighbor-joining [27] method were determined using bootstrap values based on 1000 replications [28].
4.5. Antimicrobial Activity of Compound against Clinic Pathogens
The antimicrobial activities of the compound were assessed and the minimum inhibitory concentrations (MIC) were determined against a panel of human pathogens, some of them multi-resistant to clinically-used antibiotics (Table 2). Some of them were isolated and identified in clinical microbiology laboratories from samples obtained in patients with clinical infections. Mueller-Hinton agar (Becton, Dickinson and Company, Le Pont de Cloix, France) was the culture medium in bioassays against E. coli, S. aureus, E. faecalis, E. faecium, M. luteus, H. influenzae, being supplemented according to the CLSI conditions for S. pneumoniae, S. pyogenes and N. meningitidis. Trypticasein soy agar (5% w/v) sheep blood DIFCOTM (Becton, Dickinson and Company) was used for C. urealyticum. Brucella Broth (SIGMA-ALDRICH, St. Louis, MO, USA) supplemented with hemin (5 µg/mL), vitamin K1 (1 µg/mL) and lysed horse blood (5% v/v) was used for B. fragilis and C. perfringens.
For most Gram-positive and Gram-negative bacteria, the antimicrobial assays were performed according to CLSI performance standards [29]. For M. tuberculosis susceptibility testing was done in Middlebrook 7H10 agar medium (Becton, Dickinson and Company) supplemented with 10% OADC and 0.5% glycerol according to the agar proportion method for slowly growing mycobacteria [30].
4.6. Cytotoxic Activity of Desertomycin G
Cell proliferation was measured using the CellTiter 96 Non-radioactive Cell Proliferation Assay kit purchased from Promega Promega Biotech Iberica, Alcobendas, Spain). To this end, 103 cells were seeded in 96-well plates and six replicates per condition and time point were assessed. Cell proliferation rates were determined for five consecutive days using an automated microtiter plate reader Power Wave WS (BioTek, Bad Friedrichshall, Germany)). The conditions studied were A549, DLD-1, MCF-7 cancer cells, and healthy mammary fibroblasts in the presence or absence of different concentrations of desertomycin G.
Acknowledgments
The authors want to thank the Universidad de Oviedo and Gobierno del Principado de Asturias for financial support.
Supplementary Materials
The following are available online at http://www.mdpi.com/1660-3397/17/2/114/s1, Figure S1: HPLC-UV trace and UV spectrum of compound 1; Figure S2: ESI-TOF spectrum of compound 1; Figure S3: 1H NMR spectrum (CD3OD, 500 MHz) of compound 1; Figure S4: 13C NMR spectrum (CD3OD, 125 MHz) of compound 1; Figure S5: COSY spectrum of compound 1; Figure S6: HSQC spectrum of compound 1; Figure S7: HMBC spectrum of compound 1; Figure S8: NOESY spectrum of compound 1; Video S1: Time-lapsed recording of MCF-7 cells (control); Video S2: Time-lapsed recording of MCF-7 cells treated with 50 mM desertomycin G.
Author Contributions
M.S. and G.B. isolated the strain; A.F.B identified and performed the purification of the compound; A.S.-V. performed the taxonomic identification and phylogenetic analysis of the strain; I.P.-V., J.M., and F.R. performed the structural elucidation of the compound. L.O., J.J.P.-G. and J.F. performed the antibacterial assays. Y.M., T.F. and S.C. performed the cytotoxicity assays. A.F.B., F.R., S.C., L.A.G. and G.B. conceived and supervised the work. F.R. and G.B. wrote the paper which was revised and approved by all the authors.
Funding
This study was financially supported by the Universidad de Oviedo (UNOV-11-MA-02) and Gobierno del Principado de Asturias (SV-PA-13-ECOEMP-62). The polarimeter, IR, and NMR equipment used in this work were acquired via grants for scientific and technological infrastructures from the Ministerio de Ciencia e Innovaciόn [Grants No. PCT-010000-2010-4 (NMR), INP-2011-0016-PCT-010000 ACT6 (polarimeter and IR)].
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Hong H., Samborskyy M., Lindner F., Leadlay P.F. An Amidinohydrolase Provides the Missing Link in the Biosynthesis of Amino Marginolactone Antibiotics. Angew. Chem. Int. Ed. Engl. 2016;55:1118–1123. doi: 10.1002/anie.201509300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uri J., Bognar R., Bekesi I., Varga B. Desertomycin, a new crystalline antibiotic with antibacterial and cytostatic action. Nature. 1958;182:401. doi: 10.1038/182401a0. [DOI] [PubMed] [Google Scholar]
- 3.Bortolo R., Spera S., Cassani G. Desertomycin D, a new desertomycin related antibiotic. J. Antibiot. 1992;45:1016–1019. doi: 10.7164/antibiotics.45.1016. [DOI] [PubMed] [Google Scholar]
- 4.Ivanova V. New macrolactone of the desertomycin family from Streptomyces spectabilis. Prep. Biochem. Biotechnol. 1997;27:19–38. doi: 10.1080/10826069708001275. [DOI] [PubMed] [Google Scholar]
- 5.Dinya Z., Sztaricskai F., Horváth E., Schaág J.B., Varró K. Studies of the components of crude desertomycin complex by means of electrospray and matrix-assisted laser desorption/ionization mass spectrometric techniques. Rapid Commun. Mass Spectrom. 1996;10:1439–1448. doi: 10.1002/(SICI)1097-0231(199609)10:12<1439::AID-RCM625>3.0.CO;2-E. [DOI] [Google Scholar]
- 6.Dolak L.A., Reusser F., Baczynskyj L., Mizsak S.A., Hannon B.R., Castle T.M. Desertomycin: Purification and physical-chemical properties. J. Antibiot. 1983;36:13–19. doi: 10.7164/antibiotics.36.13. [DOI] [PubMed] [Google Scholar]
- 7.Mayer M., Thiericke R. Biosynthetic relationships in the desertomycin family. J. Chem. Soc. Perkin Trans. 1. 1993:2525–2531. doi: 10.1039/P19930002525. [DOI] [Google Scholar]
- 8.Ward A.C., Bora N. Diversity and biogeography of marine actinobacteria. Curr. Opin. Microbiol. 2006;9:279–286. doi: 10.1016/j.mib.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Genilloud O., Peláez F., González I., Díez M.T. Diversity of actinomycetes and fungi on seaweeds from the Iberian coasts. Microbiologia. 1994;10:413–422. [PubMed] [Google Scholar]
- 10.Staufenberger T., Thiel V., Wiese J., Imhoff J.F. Phylogenetic analysis of bacteria associated with Laminaria saccharina. FEMS Microbiol. Ecol. 2008;64:65–77. doi: 10.1111/j.1574-6941.2008.00445.x. [DOI] [PubMed] [Google Scholar]
- 11.Wiese J., Thiel V., Nagel K., Staufenberger T., Imhoff J.F. Diversity of antibiotic-active bacteria associated with the brown alga Laminaria saccharina from the Baltic Sea. Mar. Biotechnol. 2009;11:287–300. doi: 10.1007/s10126-008-9143-4. [DOI] [PubMed] [Google Scholar]
- 12.Braña A.F., Braña A.F., Fiedler H.-P., Nava H., González V., Sarmiento-Vizcaíno A., Molina A., Acuña J.L., García L.A., Blanco G. Two Streptomyces species producing antibiotic, antitumor, and anti-inflammatory compounds are widespread among intertidal macroalgae and deep-sea coral reef invertebrates from the central Cantabrian Sea. Microb. Ecol. 2015;69:512–524. doi: 10.1007/s00248-014-0508-0. [DOI] [PubMed] [Google Scholar]
- 13.Sarmiento-Vizcaíno A., Braña A.F., González V., Nava H., Molina A., Llera E., Fiedler H.-P., Rico J.M., García-Flórez L., Acuña J.L., et al. Atmospheric Dispersal of Bioactive Streptomyces albidoflavus Strains among Terrestrial and Marine Environments. Microb. Ecol. 2016;71:375–386. doi: 10.1007/s00248-015-0654-z. [DOI] [PubMed] [Google Scholar]
- 14.Sarmiento-Vizcaíno A., González V., Braña A.F., Palacios J.J., Otero L., Fernández J., Molina A., Kulik A., Vázquez F., Acuña J.L., et al. Pharmacological Potential of Phylogenetically Diverse Actinobacteria Isolated from Deep-Sea Coral Ecosystems of the Submarine Avilés Canyon in the Cantabrian Sea. Microb. Ecol. 2017;73:338–352. doi: 10.1007/s00248-016-0845-2. [DOI] [PubMed] [Google Scholar]
- 15.Sarmiento-Vizcaíno A., Espadas J., Martín J., Braña A.F., Reyes F., García L.A., Blanco G. Atmospheric Precipitations, Hailstone and Rainwater, as a Novel Source of Streptomyces Producing Bioactive Natural Products. Front Microbiol. 2018;9:773. doi: 10.3389/fmicb.2018.00773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braña A.F., Sarmiento-Vizcaíno A., Pérez-Victoria I., Otero L., Fernández J., Palacios J.J., Martín J., de la Cruz M., Díaz C., Vicente F., et al. Branimycins B and C, Antibiotics Produced by the Abyssal Actinobacterium Pseudonocardia carboxydivorans M-227. J. Nat. Prod. 2017;80:569–573. doi: 10.1021/acs.jnatprod.6b01107. [DOI] [PubMed] [Google Scholar]
- 17.Braña A.F., Sarmiento-Vizcaíno A., Osset M., Pérez-Victoria I., Martín J., de Pedro N., de la Cruz M., Díaz C., Vicente F., Reyes F., et al. Lobophorin K, a New Natural Product with Cytotoxic Activity Produced by Streptomyces sp. M-207 Associated with the Deep-Sea Coral Lophelia pertusa. Mar. Drugs. 2017;15:144. doi: 10.3390/md15050144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarmiento-Vizcaíno A., Braña A.F., Pérez-Victoria I., Martín J., de Pedro N., de la Cruz M., Díaz C., Vicente F., Acuña J.L., Reyes F., et al. Paulomycin G, a New Natural Product with Cytotoxic Activity against Tumor Cell Lines Produced by Deep-Sea Sediment Derived Micromonospora matsumotoense M-412 from the Avilés Canyon in the Cantabrian Sea. Mar. Drugs. 2017;15:271. doi: 10.3390/md15090271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ortiz-López F.J., Alcalde E., Sarmiento-Vizcaíno A., Díaz C., Cautain B., García L.A., Blanco G., Reyes F. New 3-Hydroxyquinaldic Acid Derivatives from Cultures of the Marine Derived Actinomycete Streptomyces cyaneofuscatus M-157. Mar. Drugs. 2018;16:371. doi: 10.3390/md16100371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodríguez V., Martín J., Sarmiento-Vizcaíno A., de la Cruz M., García L.A., Blanco G., Reyes F. Anthracimycin B, a Potent Antibiotic against Gram-Positive Bacteria Isolated from Cultures of the Deep-Sea Actinomycete Streptomyces cyaneofuscatus M-169. Mar. Drugs. 2018;16:406. doi: 10.3390/md16110406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pérez-Victoria I., Martín J., Reyes F. Combined LC/UV/MS and NMR Strategies for the Dereplication of Marine Natural Products. Planta Med. 2016;82:857–871. doi: 10.1055/s-0042-101763. [DOI] [PubMed] [Google Scholar]
- 22.Radhakrishnan M., Gopikrishnan V., Suresh A., Selvakumar N., Balagurunathan R., Kumar V. Characterization and phylogenetic analysis of antituberculous compound producing actinomycete strain D25 isolated from Thar Desert soil, Rajasthan. Bioinformation. 2013;9:18–22. doi: 10.6026/97320630009018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bax A., Aszalos A., Dinya Z., Sudo K. Structure Elucidation of the Antibiotic Desertomycin thorugh the Use of New Two-dimensional NMR Techniques. J. Am. Chem. Soc. 1986;108:8056–8063. doi: 10.1021/ja00285a029. [DOI] [Google Scholar]
- 24.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 27.Saitou N., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 28.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 29.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2014. Twenty-Fourth Informational Supplement. [Google Scholar]
- 30.Clinical and Laboratory Standards Institute . Susceptibility Testing of Mycobacteria, Nocardiae and Other Aerobic Actinomycetes: Approved Standard. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2011. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.