Abstract
Five new (fusarisolins A–E, 1 to 5) and three known (6 to 8) polyketides were isolated from the marine-derived fungus Fusarium solani H918, along with six known phenolics (9 to 14). Their structures were established by comprehensive spectroscopic data analyses, methoxyphenylacetic acid (MPA) method, chemical conversion, and by comparison with data reported in the literature. Compounds 1 and 2 are the first two naturally occurring 21 carbons polyketides featuring a rare β- and γ-lactone unit, respectively. All isolates (1 to 14) were evaluated for their inhibitory effects against tea pathogenic fungus Pestalotiopsis theae and 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) synthase gene expression. Compound 8 showed potent antifungal activity with an ED50 value of 55 μM, while 1, 8, 13, and 14 significantly inhibited HMG-CoA synthase gene expression.
Keywords: ocean, microorganisms, β-lactones, HMG-CoA synthase, bioactivity
1. Introduction
Polyketides synthesized by polyketide synthases (PKS) are a large class of natural products with diverse structures and biological activities. The structural variations of polyketides are attributed to the post modification by tailoring enzymes and or a hybrid pathway, such as polyketide synthases−non-ribosomal peptide synthase (PKS-NRPS) [1]. β-Lactone featuring a strained four-membered oxygen heterocycle is a rare structural moiety in polyketides. To date, only 12 polyketides bearing β-lactone ring have been discovered in nature, including 1233A (also known as L-659,699 or F-244) [2,3,4], ebelactones A and B [5], obafluorin [6], belactosins A and C [7], lipstatin [8], salinosporamide A [9], vibralactone [10], vittatalactone [11], and cystargolides A and B [12]. Some of the polyketides have attracted wide attention owing to their potent pharmacological bioactivities [13,14,15]. For example, 1233A is a specific inhibitor of 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) synthase with an IC50 value of 0.12 µM [3]. Salinosporamide A has been shown to significantly inhibit proteasomal chymotrypsin-like proteolytic activity (IC50 = 1.3 nM) [9]. Lipstatin showed potent inhibitory effects against pancreatic lipase (IC50 = 0.14 µM) [8], and its derivative, tetrahydrolipstatin (orlistat), has been approved by the FDA to treat obesity [16].
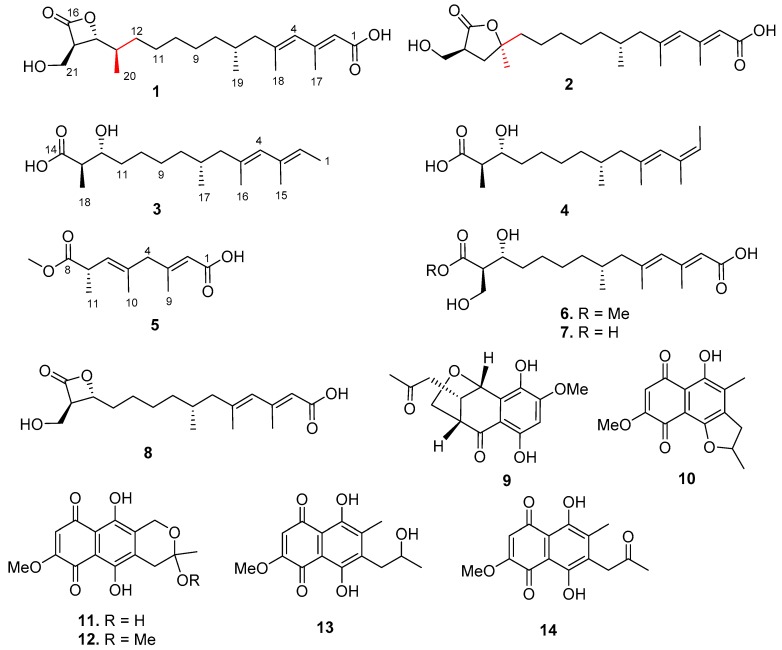
As part of our continuing discovery for structurally novel and biologically interesting secondary metabolites from marine microorganisms [17,18,19,20], the fungal strain Fusarium solani H918, isolated from mangrove sediments, was selected for a systematical chemical investigation due to its significant inhibitory activity against tea pathogenic fungus Pestalotiopsis theae. Extensive chromatographic separation of the EtOAc extract of the fermented cultures resulted in the isolation of eight polyketides (1 to 8) and six phenolic compounds (9 to 14) (Figure 1), of which compounds 1 and 2 are novel polyketides featuring a rare β- and γ-lactone ring, respectively. Herein, we report the isolation, structural elucidation, and bioactivities of these compounds.
Figure 1.
Chemical structures of 1 to 14 isolated from the Fusarium solani H918.
2. Results and Discussion
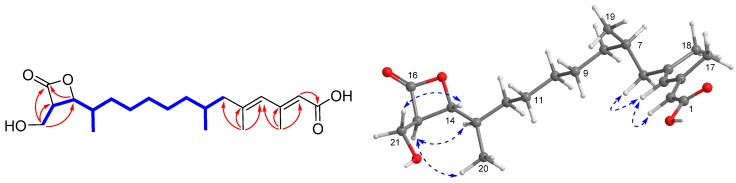
Compound 1 was isolated as a colorless oil. The protonated molecular ion peak at m/z 367.2446 [M + H]+ (calcd for C21H35O5, 367.2426) in the high resolution electron spray ionization mass spectrum (HRESIMS) indicated its molecular formula as C21H34O5, requiring five degrees of unsaturation. The infrared absorption (IR) absorption at 1818 cm−1 indicated the presence of a β-lactone moiety. Its 1H NMR spectrum showed two signals as doublets (δH 0.87 and 1.04) and two as singlets (δH 1.82 and 2.20) assigned to the four methyl groups, one hydroxymethyl (δH 3.77, 3.91), two olefinic protons (δH 5.65 and 5.76), and one oxymethine (δH 4.31) (Table 1), while the 13C NMR spectrum exhibited 21 carbon resonance signals, including four methyls, seven methylenes, six methines (two olefinics), and four nonprotonated sp2 carbons (two carbonyls) (Table 2). Among them, four olefinic carbons (δC 119.1, 130.6, 142.2, and 155.2) for two double bonds and two ester carbonyl carbons (δC 171.0 and 172.1) accounted for four indices of hydrogen deficiency. The remaining one degree of unsaturation was due to the presence of a monocyclic ring in the molecule, which was consistent with the presence of a β-lactone ring as indicated by the IR spectrum. Analyses of the heteronuclear single quantum correlations (HSQC), 1H-1H correlated spectroscopy (COSY), and heteronuclear multiple bond correlations (HMBC) spectra of 1 determined a 1,2-dialkylated β-lactone skeleton, structurally close the coexisted antibiotics 1233A (8) [2]. The COSY cross-peaks of H-14 (δH 4.31)/H-15 (δH 3.53)/H2-21 (δH 3.91, 3.77) and the HMBC interactions from H2-21 to C-14 (δC 80.2), C-15 (δC 58.2), and C-16 (δC 172.1) and from H3-18 (δH 1.04) to C-12 (δC 32.4)/C-13 (δC 38.1)/C-14 revealed a hydroxymethyl and a alkyl chain to be attached to C-15 and C-14 positions, respectively (Figure 2). The alkyl chain was expanded from C-12 to C-1 (δC 171.0) as evidenced by the contiguous COSY cross-peaks from H2-6 (δH 2.13, 1.88) to H2-12 (δH 1.44, 1.18), H-7 (δH 1.70)/H3-19 (δH 0.87), and H-13 (δH 1.82)/H3-20 (δH 1.04), together with the HMBC relationships originating from H-2 (δH 5.65), H3-17 (δH 2.20), and H3-18 (δH 1.82) to the corresponding carbons as shown in Figure 2. On the basis of the above data, the alkyl side chain with 13 carbons bearing four methyl groups was elucidated as a 3,5,7-trimethyl-tetradeca-2,4-dienoic acid. Therefore, the gross structure of 1 was determined as a novel 21 carbons polyketide featuring a rare β-lactone moiety.
Table 1.
1H NMR data for compounds 1 to 6 at 400 MHz (δ in ppm, J in parenthesis with Hz).
| No. | 1 a | 2 a | 3 a | 4 a | 5 b | 6 a |
|---|---|---|---|---|---|---|
| 1 | 1.67, d (7.0) | 1.50, dm (6.8) | ||||
| 2 | 5.65, br s | 5.65, br s | 5.29, q (7.0) | 5.30, qq (6.8, 1.2) | 5.74, br s | 5.65, br s |
| 4 | 5.76, br s | 5.76, br s | 5.59, br s | 5.53, br s | 2.85, s | 5.76, br s |
| 6 | 2.13, dd (13.2, 6.2) | 2.14, dd (13.2, 7.2) | 2.03, dd (12.8, 6.4) | 2.09, dd (13.2, 6.3) | 5.32, d (8.7) | 2.14, dd (13.1, 6.4) |
| 1.88, dd (13.2, 8.1) | 1.89, dd (13.2, 8.1) | 1.79, m | 1.86, ddd (13.2, 8.3, 0.8) | 1.89, dd (13.1, 8.2) | ||
| 7 | 1.70, m | 1.71, m | 1.65, m | 1.66, m | 3.39, m | 1.71, m |
| 8 | 1.35, m; 1.15, m | 1.34, m; 1.15, m | 1.36, m; 1.12, m | 1.38, m; 1.15, m | 1.36, m; 1.16, m | |
| 9 | 1.44, m | 1.34, m | 1.36, m | 1.38, m | 2.12, s | 1.37, m |
| 10 | 1.33, m | 1.34, m | 1.49, m; 1.37, m | 1.50, m; 1.38, m | 1.63, s | 1.48, m; 1.37, m |
| 11 | 1.33, m | 1.42, m | 1.52, m; 1.40, m | 1.53, m; 1.40, m | 1.26, d (6.8) | 1.53, m; 1.45, m |
| 12 | 1.44, m; 1.18, m | 1.72, m | 3.71, t (6.9) | 3.70, t (6.9) | 3.79, m | |
| 13 | 1.82, m | 2.48, m | 2.48, m | 2.67, ddd (8.5, 6.3, 5.4) | ||
| 14 | 4.31, dd (8.4, 4.2) | 2.16, br d (10.3) | ||||
| 15 | 3.53, q (4.2) | 3.02, m | 1.69, s | 1.70, m | 2.21, d (1.2) | |
| 16 | 1.71, s | 1.54, d (1.3) | 1.82, d (1.2) | |||
| 17 | 2.20, s | 2.21, s | 0.84, d (6.6) | 0.88, d (6.6) | 0.87, d (6.6) | |
| 18 | 1.82, s | 1.82, s | 1.13, d (6.9) | 1.13, d (7.0) | 3.82, dd (10.9, 8.7) | |
| 3.73, dd (10.9, 5.3) | ||||||
| 19 | 0.87, d (6.6) | 0.87, d (6.5) | ||||
| 20 | 1.04, d (6.5) | 1.38, s | ||||
| 21 | 3.91, dd (11.9, 4.5) | 3.87, dd (11.0, 4.6) | ||||
| 3.77, dd (11.9, 3.8) | 3.72, dd (11.0, 3.6) | |||||
| OMe | 3.70, s | 3.71, s |
a Measured in CD3OD; b Measured in CDCl3.
Table 2.
13C NMR spectroscopic data for 1 to 6 at 100 MHz (δ in ppm).
| No. | 1 a | 2 a | 3 a | 4 a | 5 b | 6 a |
|---|---|---|---|---|---|---|
| 1 | 171.0, C | 172.1, C | 13.7, CH3 | 15.3, CH3 | 171.7, C | 170.6, C |
| 2 | 119.1, CH | 119.1, CH | 123.7, CH | 121.8, CH | 116.6, CH | 118.8, CH |
| 3 | 155.2, C | 155.0, C | 134.9, C | 135.3, C | 160.1, C | 155.5, C |
| 4 | 130.6, CH | 130.6, CH | 131.6, CH | 126.8, CH | 51.1, CH2 | 130.6, CH |
| 5 | 142.2, C | 142.1, C | 135.1, C | 137.2, C | 133.5, C | 142.3, C |
| 6 | 49.9, CH2 | 49.9, CH2 | 49.8, CH2 | 48.8, CH2 | 128.0, CH | 50.0, CH2 |
| 7 | 32.1, CH | 32.2, CH | 32.1, CH | 32.1, CH | 38.9, CH | 32.1, CH |
| 8 | 37.8, CH2 | 37.9, CH2 | 38.0, CH2 | 38.0, CH2 | 175.7, C | 37.9, CH2 |
| 9 | 27.9, CH2 | 28.9, CH2 | 28.2, CH2 | 28.1, CH2 | 17.9, CH3 | 32.1, CH2 |
| 10 | 27.8, CH2 | 31.2, CH2 | 27.0, CH2 | 27.0, CH2 | 15.9, CH3 | 26.9, CH2 |
| 11 | 31.0, CH2 | 24.8, CH2 | 35.0, CH2 | 35.0, CH2 | 18.4, CH3 | 36.1, CH2 |
| 12 | 32.4, CH2 | 42.6, CH2 | 74.1, CH | 74.1, CH | 71.0, CH | |
| 13 | 38.1, CH | 86.5, C | 47.5, CH | 47.5, CH | 56.2, CH | |
| 14 | 80.2, CH | 36.5, CH2 | 179.7, C | 179.5, C | 175.1, C | |
| 15 | 58.2, CH | 44.6, CH | 17.0, CH3 | 24.2, CH3 | 19.9, CH3 | |
| 16 | 172.1, C | 179.5, C | 17.9, CH3 | 17.8, CH3 | 18.5, CH3 | |
| 17 | 19.8, CH3 | 19.8, CH3 | 20.0, CH3 | 19.9, CH3 | 19.8, CH3 | |
| 18 | 18.5, CH3 | 18.5, CH3 | 13.9, CH3 | 13.8, CH3 | 61.6, CH2 | |
| 19 | 19.9, CH3 | 19.9, CH3 | ||||
| 20 | 15.3, CH3 | 25.5, CH3 | ||||
| 21 | 58.3, CH2 | 61.0, CH2 | ||||
| OMe | 51.9, CH3 | 52.0, CH3 |
a Measured in CD3OD; b Measured in CDCl3.
Figure 2.
Selected COSY ( ), HMBC (
), HMBC ( ), and NOESY (
), and NOESY ( ) correlations of 1.
) correlations of 1.
The geometries of double bonds at Δ2 and Δ4 were established as 2E and 4E configurations by the nuclear overhauser effect (NOESY) correlations from H-4 (δH 5.76) to H-2 and H2-6 (Figure 2). Additional NOESY relationships from H2-21 to H-14, in association with the similar coupling constants of 1 (JH-14/H-15 = 4.2 Hz), 1233A (J = 4.2 Hz), and vittatalactone (J = 4.0 Hz) indicated the trans-relationship of H-14 and H-15 in the β-lactone ring [11,21]. The same orientation of H-13 and H-15 was presumed by the NOESY cross-peaks from H-15 to H-13 and H3-20 (Figure 2). Absolute configuration of C-15 was determined by application of the MPA method used for establishing absolute configuration and optical purity of primary alcohols with chiral center at C-2 position [22,23]. The chemical shift difference between two methylene protons of H2-21 in (R)-MPA ester (1a) (ΔδH 0.08) is larger than that of (S)-MPA ester (1b) (ΔδH 0.05), revealing 15R configuration. Additionally, the absolute configuration of C-7 at linear side chain was presumed to be the same as that of 8 according to the biogenetic considerations, which was based on the fact that the isolate 8 was established as 1233A by comparison of their NMR data and specific rotation values between 8 ( 25.6, CHCl3) and 1233A ( 27.5, CHCl3) [2,24]. Therefore, the structure of 1 was then established as a novel (2E,4E,7R,13R)-13-((2R,3R)-3-(hydroxymethyl)-4-oxooxetan-2-yl)-3,5,7-trimethyltetradeca-2,4-dienoic acid, and given the name, fusarisolin A.
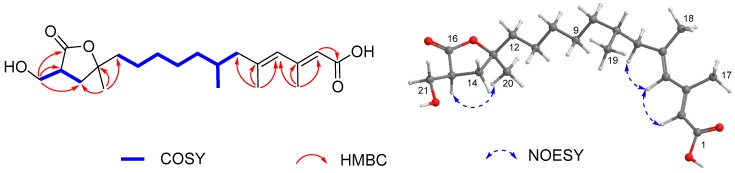
Compound 2 exhibited the same molecular formula as that of 1 according to its HRESIMS spectrum. Interestingly, it also showed nearly identical 13C NMR data to those of 1 revealing a structurally similar analogue. The difference was attributed to the methine of C-13 and oxymethine of C-14 of 1 being replaced by an oxygenated nonprotonated sp3 carbon (δC 86.5) and methylene (δC 36.5), respectively, suggesting the presence of a γ-lactone instead of a β-lactone unit in 2. This assumption was evidenced by the HMBC correlations from H3-20 (δH 1.38) to C-12 (δC 42.6)/C-13(δC 86.5)/C-14 (δC 36.5) and from H2-21 (δH 3.87, 3.72) to C-14/C-15 (δC 44.6)/C-16 (δC 179.5), and the COSY correlations of H2-14 (δH 2.16)/H-15 (δH 3.02)/H2-21 (Figure 3), in addition to the same molecular formula as well as the comparisons of the corresponding NMR data of C-13/C-14/C-15/C-16/C-21 (δC 61.0) of 2 with those of the structurally related compound 3-hydroxymethyl-5,5-dimethyl-dihydro-2(3H)furanone (corresponding to δC 83.2, 37.1, 43.3, 177.8, and 60.9, respectively) [25]. In the NOESY spectrum, H-15 was correlated to H3-20 indicating same orientation of these protons (Figure 3). Additionally, the configurations of the conjugated double bonds at Δ2 and Δ4 were determined to be same as those of 1 on the basis of the NOESY cross-peaks from H-4 (δH 5.76) to H-2 (δH 5.65) and H2-6 (δH 2.14, 1.89) (Figure 3). The absolute configuration of 2 was presumably assigned as 7R, 13S, and 15S on the basis of the biogenetic considerations between 2 and 1. Considering chemical rearrangement of a β-lactone (similar to compound 1) to a γ-lactone (similar to compound 2) catalyzed by EtMgBr [26], structure of 2 might be derived from 1 after chemical rearrangement. Therefore, the novel structure of 2 was elucidated to be (2E,4E,7R)-12-((2S,4S)-4-(hydroxymethyl)-2-methyl-5-oxotetrahydrofuran-2-yl)-3,5,7-trimethyldodeca-2,4-dienoic acid, and named, fusarisolin B.
Figure 3.
Key COSY, HMBC, and NOESY correlations of 2.
It is noteworthy that fusarisolins A (1) and B (2) are the first examples of 21-carbons polyketides bearing a rare β- and γ-lactone moiety, respectively.
Compound 3 exhibited the molecular formula of C18H32O3 as determined by negative HRESIMS spectrum (m/z 295.2256, [M − H]−). The 13C NMR spectrum showed 18 carbon resonance signals including five methyls, five methylenes, five methines (two olefinics), and three nonprotonated sp2 carbons (two olefinics and one carbonyl), which were similar to those of 12-epi-1233B (7) [2]. However, a close comparison of these two compounds indicated that the carbonyl of C-1 and the hydroxymethyl of C-18 in 7 were replaced by two methyls in 3. This assumption was evidenced by the COSY relationships from H3-1 (δH 1.67) to H-2 (δH 5.29) and between H-13 (δH 2.48) and H3-18 (δH 1.13), in addition to the HMBC interactions from H3-1 to C-2 (δC 123.7) and C-3 (δC 134.9) and from H3-18 to C-12 (δC 74.1), C-13 (δC 47.5), and C-14 (δC 179.7). In the NOESY spectrum, the cross-peaks from H-4 (δH 5.59) to H2-6 (δH 2.03, 1.79) confirmed the 4E geometry. It is noteworthy that the overlapped signals of H3-1 (δH 1.67) and H3-15 (δH 1.69) were unable to be used to determine the Δ2 configuration based on the NOESY correlations. Finally, the 2E geometry was assigned by the comparison of the 13C NMR data of C-1 (δC 13.7) and C-15 (δC 17.0) of 3 with those of a structurally partial identity of pteroenone (δC 13.6, C-1 and 16.6, C-15) [27]. Thus, 3 was elucidated to be 1-deoxo-1,18-dedioxy-12-epi-1233B, and named fusarisolin C.
Compound 4 showed the same molecular formula as that of 3 by the HRESIMS spectrum and their NMR spectroscopic data were nearly identical, indicating a structurally related analogue. Comparison of the NMR data, as well as the analyses of 2D NMR spectra (HSQC, COSY, and HMBC), established the structure of 4 to be a 2Z isomer of 3. This assumption was recognized by the shielded chemical shifts of H3-1 (ΔδH −0.17), the deshielded shift of Me-15 (ΔδC +7.2), in association with the NOESY relationships from H-2 (δH 5.30) to H3-15 (δH 1.70) and from H-4 (δH 5.53) to H2-6 (δH 2.09, 1.86). The 2Z and 4E configuration of the double bond at Δ2 and Δ4 of 4 were unambiguously determined by the clear NOESY data, which further supported the 2E and 4E configurational assignments of 3. Accordingly, the structure of 4 was established as (2Z)-fusarisolin C, and named fusarisolin D.
Compound 5 exhibited the sodium adduct ion peak at m/z 249.1104 in the HRESIMS spectrum, consistent with the molecular formula of C12H18O4. The 1H NMR spectrum exhibited three methyls (δH 1.26, 1.63, and 2.12), one methoxy (δH 3.70), two olefinic protons (δH 5.32 and 5.74), one methylene (δH 2.85), and one methine (δH 3.39) (Table 1). The 13C NMR spectrum showed 12 carbons including four olefinic carbons for two double bonds, two carbonyl carbons, in addition to six sp3 carbons for three methyls, one methoxy, one methylene, and one methine (Table 2). The HMBC correlations originating from H-2 (δH 5.74), H3-9 (δH 2.12), H3-10 (δH 1.63), H3-11 (δH 1.26), and the OMe (δH 3.70) to the corresponding carbons of C-1−C-8 established the alkyl chained structure. The NOESY correlations from H-2 to H2-4 (δH 2.85) and from H3-10 to H-7 (δH 3.39) indicated 2E and 5E configurations. According to the similar optical rotation (OR) values of 5 ( +14.5), and the structurally related (S,E)-callosobruchusic acid ( +10.1) [28], the sole stereogenic center of C-7 was assigned as S configuration. Consequently, the structure of 5 was established as (2E,5E,7S)-8-methoxy-3,5,7-trimethyl-8-oxoocta-2,5-dienoic acid, and named fusarisolin E.
Compound 6 exhibited the molecular formula of C19H32O6 as established by the HRESIMS spectrum of the sodium adduct ion peak (m/z 379.2094). The NMR data were nearly identical to those of 7 except for the presence of additional methoxyl signals (δH 3.71, δC 52.0). The location of methoxyl group at C-14 (δC 175.1) was corroborated by the HMBC interaction from the methoxyl protons (δH 3.71) to the carbonyl carbon of C-14. The NOESY cross-peaks from H2-6 (δH 1.89, 2.14) to H-4 (δH 5.76) and from H-4 to H-2 (δH 5.65) revealed the 2E and 4E configurations of the conjugated double bonds. Therefore, the structure of 6 was elucidated as 14-O-methyl-12-epi-1233B. Although 6 is commercially available, no physicochemical data could be found. Accordingly, its 1H and 13C NMR data (Table 1 and Table 2) are reported here for the first time.
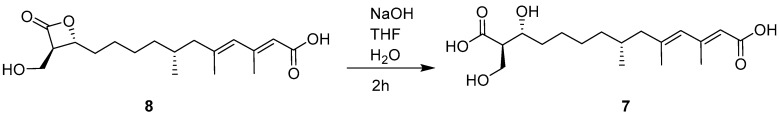
In order to establish the absolute configuration of C-12 in 7, compound 8 (1233A) was subjected to alkaline hydrolysis to provide 7 (Figure 4). According to the identical 1H- and 13C- NMR spectra and specific rotation data between hydrolysis product of 8 ( +7.2, MeOH) and 7 ( +6.7, MeOH) (Supplementary Materials Figure S8-1 and 8-2), 7 was concluded to have the opposite configuration at C-12 to that of 1233B, establishing 7 to be 12-epi-1233B [2]. Considering the absence of the 13C NMR spectroscopic data of 12-epi-1233B in the literature, the modern 1H- and 13C-NMR data of 7 are provided in Table S1 of the Supplementary Materials.
Figure 4.
Alkaline hydrolysis of 8 to yield 7.
By comparing NMR spectroscopic data and specific rotations with those published in the literature, six known phenolics were determined to be dihydronaphthalenone B (9) [29], 2,3-dihydro-5-hydroxy-8-methoxy-2,4-dimethylnaphthol[1,2-b]furan-6,9-dione (10) [30], fusarubin (11) [31], methyl ether fusarubin (12) [32,33], solaniol (13) [34,35], and javanicin (14) [31].
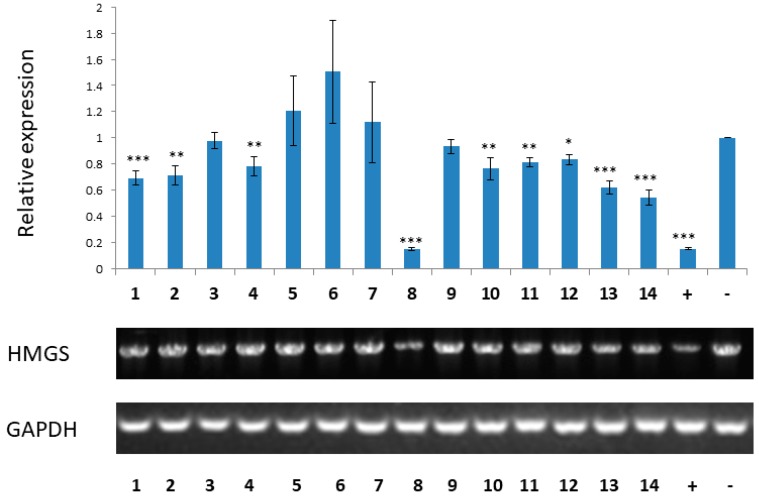
Compounds 1 to 14 were evaluated for antifungal activities against tea pathogenic fungus Pestalotiopsis theae. Only compound 8 exhibited a potent effect with an ED50 value of 55 ± 4.0 μM, which was stronger than that of the positive control hexaconazole (ED50 = 68 ± 5.7 μM). Previously, 1233A (8) was reported to be a specific inhibitor of HMG-CoA synthase [3]. However, currently there is no commercially available Pestalotiopsis theae HMG-CoA synthase enzyme. It is known that synthase gene expression is positively correlated to synthase expression, i.e., the down/up regulation of gene expression leads to the lower/higher expression of protein. Therefore, all isolates were tested for the down-regulation of Pestalotiopsis theae HMG-CoA synthase gene expression by real-time polymerase chain reaction (RT-PCR) at the concentration of 10 µM, and abscisic acid and dimethyl sulfoxide (DMSO) were used as positive and blank controls [36]. As a result, compounds 1, 8, 13, and 14 showed a significant effect, while 2, 4, 10, and 11 exhibited a moderate effect, and 12 showed a weak effect (Figure 5). As in the case of 8, compounds 1, 13, and 14 might also have potent inhibitory activity against HMG-CoA synthase, revealing their potential applications in regard to control of cholesterol biosynthesis.
Figure 5.
Inhibitory effects of 1 to 14 on the mRNA expression of HMG-CoA synthase. The gene expression level was determined by real-time polymerase chain reaction (RT-PCR). Dimethyl sulfoxide (DMSO) and abscisic acid were used as blank and positive controls, respectively. G lyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as reference gene. Values represent the mean ± SD of three independent experiments. * p < 0.05, ** p < 0.01, and *** p < 0.001.
3. Materials and Methods
3.1. General Experimental Procedures
Optical rotations were recorded on a Rudolph Autopol IV automatic polarimeter under 24 °C (Rudolph Research Analytical, Hackettstown, NJ, USA). The IR spectrum was recorded on a Bruker IFS-55 spectrometer (Bruker Optik BmbH, Ettlingen, Germany). The HRESIMS spectra were measured by a Waters Xevo G2 Q-TOF mass spectrometer (Waters, Milford, MA, USA). The NMR spectra were recorded on a Bruker Avance-400 FT MHz NMR spectrometer (Bruker, Fällanden, Switzerland). High performance liquid chromatography (HPLC) was carried out on an Alltech instrument equipped with UV detector (Series III, Alletch Inc., Nicholasville, Kentucky, USA). Thin-layer chromatography (TLC) analysis was performed on precoated silica gel plates (Jiangyou Silica Gel Development, Inc., Yantai, China). Column chromatography (CC) was performed on Sephadex LH-20, ODS, and silica gel, respectively.
3.2. Fungal Identification, Fermentation, and Extract
The fungus Fusarium solani H918 was isolated from mangrove sediments collected at the Zhangjiangkou Mangrove National Nature Reserve, Fujian, China. The internal transcribed spaces (ITS) region was amplified and sequenced by the general primers ITS1 and ITS4. The ITS region of the fungus was 576 bp DNA sequence (GenBank accession number: KY978584), which showed 99% identity to Fusarium solani. The strain was deposited at the China Center for Type Culture Collection (CCTCC) with the accession number of M 2017151.
Prior to the large-scale fermentation, the producing strain was incubated on a potato dextrose agar (PDA) plate medium under 25 °C for 3 days, and then the fresh mycelia were inoculated to 25 × 1 L Erlenmeyer flasks, each containing 80 g of rice and 120 mL of sea water. The fermentation was carried out under static conditions at 25 °C for 20 days. Following this, fermented cultures were extracted with EtOAc three times and concentrated under reduced pressure to get an organic extract. The extract was re-dissolved in MeOH and extracted with petroleum ether (PE) three times. The MeOH layer was evaporated under reduced pressure to get a defatted extract (6 g).
3.3. Isolation and Purification
The defatted extract was separated by an octadecylsilane (ODS) column using medium pressure liquid chromatography (MPLC) eluting with a gradient of MeOH-H2O (5:95→100:0) to get four fractions (Fr.1−Fr.4). Fraction Fr.2 (1.2 g) was chromatographed over silica gel CC using CHCl3-MeOH gradient elution (20:1→10:1) to get three subfractions (Fr.2-1−Fr.2-3). Subfraction Fr.2-1 was subjected to repeated silica gel CC eluting with PE-EtOAc (3:1) and PE-acetone (5:1) to yield 5 (2.2 mg) and 14 (2.7 mg). Subfraction Fr.2-2 was purified by CC over silica gel eluting with PE-EtOAc (3:1) to obtain 10 (11.2 mg) and 13 (9.8 mg). Subfraction Fr.2-3 was subjected to CC on silica gel eluting with CHCl3-MeOH-formic acid (20:1:0.1) to yield 7 (42.6 mg), 11 (1.7 mg), and 12 (5.5 mg). Fraction Fr.3 (625 mg) was fractionated by CC over Sephadex LH-20 (MeOH) to get four subfractions (Fr.3-1−Fr.3-4). Subfraction Fr.3-1 was purified by Prep. TLC using CHCl3-MeOH (10:1) elution to obtain 9 (8.0 mg). Subfraction Fr.3-2 was subjected to CC over silica gel (PE-acetone-formic acid, 1:1:0.01) to get 6 (25.0 mg). Compound 8 (18.2 mg) was separated from subfraction Fr.3-3 by CC over silica gel eluting with PE-acetone (3:1). Fraction Fr.4 (862 mg) was subjected to CC over Sephadex LH-20 (MeOH) to yield three subfractions (Fr.4-1−Fr.4-3). Compounds 1 (4.2 mg) and 2 (1.4 mg) were separated from subfraction Fr.4-2 by Prep. TLC (CHCl3-acetone, 3:1). Subfraction Fr.4-3 was purified by CC on silica gel (PE-acetone, 2:1), followed by semipreparative HPLC (MeOH-H2O, 3:1) to provide 3 (4.6 mg) and 4 (22.0 mg).
Fusarisolin A (1): Colorless oil; +0.8 (c 0.19, MeOH); UV (MeOH) λmax (log ε) 204 (3.80), 270 (4.11) nm; IR (KBr) νmax 2926, 2856, 1818, 1688, 1615, 1457, 1379, 1249, 1162, 1022 cm−1; 1H and 13C NMR data, see Table 1 and Table 2; HRESIMS m/z 367.2446 [M + H]+ (calcd for C21H35O5, 367.2426).
Fusarisolin B (2): Colorless oil; −5.3 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 206 (3.87), 268 (3.99) nm; 1H and 13C NMR data, see Table 1 and Table 2; HRESIMS m/z 365.2299 [M − H]− (calcd for C21H33O5, 365.2328).
Fusarisolin C (3): Colorless oil; −15.2 (c 0.04, MeOH); 1H and 13C NMR data, see Table 1 and Table 2; HRESIMS m/z 295.2256 [M − H]− (calcd for C18H31O3, 295.2273).
Fusarisolin D (4): Colorless oil; −19.8 (c 0.1, MeOH); 1H and 13C NMR data, see Table 1 and Table 2; HRESIMS m/z 295.2256 [M − H]− (calcd for C18H31O3, 295.2273).
Fusarisolin E (5): Colorless oil; +14.5 (c 0.03, MeOH); UV (MeOH) λmax (log ε) 216 (4.19), 314 (3.14) nm; 1H and 13C NMR data, see Table 1 and Table 2; HRESIMS m/z 249.1104 [M + Na]+ calcd for (C12H18O4Na, 249.1103).
14-O-Methyl-12-epi-1233B (6): Colorless oil; +0.7 (c 0.67, MeOH); 1H and 13C NMR data, see Table 1 and Table 2; HRESIMS m/z 379.2094 [M + Na]+ (calcd for C19H32O6Na, 379.2097).
3.4. Preparation of MPA Esters of 1
Compound 1 (1.5 mg) and (R)-MPA (3.0 mg) were dissolved in CHCl3 (600 µL). Following this, N,N′-dicyclohexylcarbodiimide (DCC, 2.5 mg) and 4-dimethylaminopyridine (DMAP, 2.8 mg) were added and the mixture was stirred for 24 h. The final reaction products were purified by CC over silica gel (PE-acetone, 3:1) to give the (R)-MPA ester (1a, 1.8 mg). Similarly, (S)-MPA ester (1b, 2.0 mg) was obtained from the reaction mixture of 1 (1.5 mg) and (S)-MPA (3.0 mg).
(R)-MPA ester of 1 (1a): 1H NMR (CDCl3, 400 MHz) δH 7.36−7.44 (5H, m, phenyl protons), 5.73 (1H, br s, H-4), 5.70 (1H, br s, H-2), 4.81 (1H, s, CH of MPA), 4.47 (1H, dd, J = 12.0, 4.2 Hz, H-21a), 4.39 (1H, dd, J = 12.0, 6.0 Hz, H-21b), 4.06 (1H, dd, J = 7.8, 4.2 Hz, H-14), 3.42 (3H, s, OMe of MPA), 2.25 (3H, s, Me-17), 2.10 (1H, dd, J = 13.7, 4.9 Hz, H-6a), 1.94 (1H, m, H-12a), 1.87 (1H, dd, J = 13.7, 6.3 Hz, H-6b), 1.82 (3H, s, Me-18), 1.72 (1H, m, H-9a), 1.69 (1H, m, H-13), 1.66 (1H, m, H-7), 1.61 (1H, m, H-10a), 1.36 (1H, m, H-11a), 1.34 (1H, m, H-9b), 1.29 (1H, m, H-8a), 1.23 (1H, m, H-11b), 1.18 (2H, m, H-10b and 12b), 1.10 (1H, m, H-8b), 0.96 (3H, d, J = 6.6 Hz, Me-20), 0.84 (3H, d, J = 6.6 Hz, Me-19).
(S)-MPA ester of 1 (1b): 1H NMR (CDCl3, 400 MHz) δH 7.36−7.46 (5H, m, phenyl protons), 5.74 (1H, br s, H-4), 5.70 (1H, br s, H-2), 4.81 (1H, s, CH of MPA), 4.43 (1H, dd, J = 12.2, 3.7 Hz, H-21a), 4.38 (1H, dd, J = 12.2, 4.8 Hz, H-21b), 3.68 (1H, dd, J = 7.9, 4.2 Hz, H-14), 3.43 (3H, s, OMe of MPA), 2.26 (3H, s, Me-17), 2.09 (1H, dd, J = 13.2, 5.9 Hz, H-6a), 1.94 (1H, m, H-12a), 1.85 (1H, dd, J = 13.2, 4.6 Hz, H-6b), 1.82 (3H, s, Me-18), 1.72 (1H, m, H-9a), 1.65 (1H, m, H-7), 1.63 (1H, m, H-13), 1.61 (1H, m, H-10a), 1.35 (1H, m, H-9b), 1.30 (1H, m, H-11a), 1.27 (1H, m, H-8a), 1.21 (1H, m, H-11b), 1.18 (2H, m, H-10b and 12b), 1.08 (1H, m, H-8b), 0.89 (3H, d, J = 6.5 Hz, Me-20), 0.84 (3H, d, J = 6.6 Hz, Me-19).
3.5. Alkaline Hydrolysis of 8
A tetrahydrofuran (THF) solution (100 μL) of 8 (5.0 mg, 0.015 mmol) was mixed with NaOH (2.5 mg, 0.0625 mmol) and H2O (400 μL). This reaction mixture was left at room temperature for 2 h, and then the pH value was adjusted to 3–4 with hydrochloric acid (1 M). After being extracted with CHCl3 three times, the extract was subjected to CC over ODS eluting with MeOH/H2O (7:3) to yield 7 (3.0 mg).
3.6. Antifungal Assay
Antifungal assay against tea pathogenic fungus Pestalotiopsis theae HQ832793, isolated from foliar lesions of tea leaf, was performed in PDA Petri plates according to a previously described method [37]. In brief, a 0.6 cm diameter piece of tested fungal strains cylinder agar was placed on the center, and sterile blank paper discs (0.5 cm diameter) were placed at a distance of 2 cm away from the growing mycelial colony. The tested compounds (100 μg/mL, DMSO solution) were added to each paper disc. DMSO and hexaconazole were used as blank and positive controls, respectively. These plates were incubated at 28 °C until mycelial growth enveloped the discs including the control disc. The experiment was repeated three times.
3.7. ED50 Detection
As reported previously [38], different concentrations of the DMSO dissolved 1233A and hexaconazole were mixed with a PDA medium and poured into a set of PDA Petri plates. The Pestalotiopsis theae mycelial disk (5 mm) was placed in the center of each treated Petri dish and incubated at 28 °C. All treatments were quadruplicated against each fungus. DMSO and hexaconazole were used as blank and positive controls, respectively. The ED50 value was calculated statistically by Probit analysis.
3.8. Total RNA Isolation
Pestalotiopsis theae cells were cultured in a PDA medium for 3 days at 28 °C, then treated with tested compounds for 16 h. Cells were harvested by centrifugation at 6000 rpm for 5 min, and then homogenized in liquid nitrogen. Total RNA was extracted with Spin Column Fungal Total RNA Purification Kit (Sangon Biotech, Shanghai, China).
3.9. RT-PCR Analysis of HMG-CoA Synthase Gene Expression
The inhibitory effects of the tested compounds at 10 µM (DMSO dissolution) on the mRNA expression of HMG-CoA synthase in Pestalotiopsis theae cells were analyzed by RT-PCR. DMSO and abscisic acid (10 µM) were used as blank and positive controls, respectively. The expression of mRNA transcripts of HMG-CoA synthase (forward: TACTCG CTCACCTGCTACAC; reverse: GCGTACGACTTCTGGACGAC) and GAPDH (forward: CATGTCCATGCGTGTCCCTA; reverse: CAGTGGAGACAACCTCGTCC) was determined by RT-PCR. The cDNA was synthesized from total RNA using PrimeScript RT reagent kit with gDNA Eraser (Takara, Japan). TaKaRa SYBR® Premix Ex Taq™ II (Takara, Japan) and Stepone Real-Time PCR Detection System (Applied Biosystems, Foster City, CA, USA) were used for RT-PCR analysis. The values are expressed as the mean ± SD for three triplicate experiments.
4. Conclusions
The present work reported five new (1 to 5) and nine known (6 to 14) compounds from the marine-derived fungus Fusarium solani H918. Fusarisolins A (1) and B (2), two novel 21-carbon polyketides featuring a rare β- and γ-lactone unit, respectively, were found for the first time in nature. Compounds 1, 8, 13, and 14 showed significant down-regulation HMG-CoA synthase gene expression. In addition, compound 8 exhibited potent inhibitory activity against tea pathogenic fungus Pestalotiopsis theae, revealing that it might be a potential lead compound for the development of an antifungal agrochemical after structural modification.
Supplementary Materials
The following are available online at http://www.mdpi.com/1660-3397/17/2/125/s1, Figures S1-1–S8-2 and Table S1: The 1D and 2D NMR spectra of 1–7, 1H and 13C NMR data for 7.
Author Contributions
X.-W.Y. designed and coordinated the project; S.N. and Z.F. performed the chemical experiments. X.-X.T. conducted the bioactive experiments. S.N., J.-M.X., and C.-L.X. analyzed the data. S.N. and X.-W.Y. wrote the paper, while critical revision of the publication was performed by all authors.
Funding
This research was funded by the Scientific Research Foundation of Third Institute of Oceanography, SOA (2017035 and 2018009), the National Natural Science Foundation of China (41606185 and 41676130), and the Xiamen Science and Technology Program (3502Z20172009).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Rateb M.E., Ebel R. Secondary metabolites of fungi from marine habitats. Nat. Prod. Rep. 2011;28:290–344. doi: 10.1039/c0np00061b. [DOI] [PubMed] [Google Scholar]
- 2.Aldridge D.C., Giles D., Turner W.B. Antibiotic 1233A: A fungal β-lactone. J. Chem. Soc. 1971;23:3888–3891. doi: 10.1039/J39710003888. [DOI] [PubMed] [Google Scholar]
- 3.Greenspan M.D., Yudkovitz J.B., Lo C.Y.L., Chen J.S., Alberts A.W., Hunt V.M., Chang M.N., Yang S.S., Thompson K.L., Chiang Y.C.P., et al. Inhibition of hydroxymethylglutaryl-coenzyme A synthase by L-659,699. Proc. Natl. Acad. Sci. USA. 1987;84:7488–7492. doi: 10.1073/pnas.84.21.7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomoda H., Kumagai H., Takahashi Y., Tanaka Y., Iwai Y., Omura S. F-244 (1233A), a specific inhibitor of 3-hydroxy-3-methylglutaryl coenzyme A synthase: Taxonomy of producing strain, fermentation, isolation and biological properties. J. Antibiot. 1988;41:247–249. doi: 10.7164/antibiotics.41.247. [DOI] [PubMed] [Google Scholar]
- 5.Umezawa H., Aoyagi T., Uotani K., Hamada M., Takeuchi T., Takahashi S. Ebelactone, an inhibitor of esterase, produced by actinomycetes. J. Antibiot. 1980;33:1594–1596. doi: 10.7164/antibiotics.33.1594. [DOI] [PubMed] [Google Scholar]
- 6.Wells J.S., Trejo W.H., Principe P.A., Sykes R.B. Obafluorin, a novel β-lactone produced by Pseudomonas fluorescens. Taxonomy, fermentation and biological properties. J. Antibiot. 1984;37:802–803. doi: 10.7164/antibiotics.37.802. [DOI] [PubMed] [Google Scholar]
- 7.Asai A., Hasegawa A., Ochiai K., Yamashita Y., Mizukami T. Belactosin A, a novel antitumor antibiotic acting on cyclin/CDK mediated cell cycle regulation, produced by Streptomyces sp. J. Antibiot. 2000;53:81–83. doi: 10.7164/antibiotics.53.81. [DOI] [PubMed] [Google Scholar]
- 8.Weibel E.K., Hadvary P., Hochuli E., Kupfer E., Lengsfeld H. Lipstatin, an inhibitor of pancreatic lipase, produced by Streptomyces toxytricini. I. Producing organism, fermentation, isolation and biological activity. J. Antibiot. 1987;40:1081–1085. doi: 10.7164/antibiotics.40.1081. [DOI] [PubMed] [Google Scholar]
- 9.Feling R.H., Buchanan G.O., Mincer T.J., Kauffman C.A., Jensen P.R., Fenical W. Salinosporamide A: A highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew. Chem. Int. Ed. 2003;42:355–357. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- 10.Liu D.Z., Wang F., Liao T.G., Tang J.G., Steglich W., Zhu H.J., Liu J.K. Vibralactone: A lipase inhibitor with an unusual fused β-lactone produced by cultures of the basidiomycete Boreostereum vibrans. Org. Lett. 2006;8:5749–5752. doi: 10.1021/ol062307u. [DOI] [PubMed] [Google Scholar]
- 11.Morris B.D., Smyth R.R., Foster S.P., Hoffmann M.P., Roelofs W.L., Franke S., Francke W. Vittatalactone, a β-lactone from the striped cucumber beetle, Acalymma vittatum. J. Nat. Prod. 2005;68:26–30. doi: 10.1021/np049751v. [DOI] [PubMed] [Google Scholar]
- 12.Gill K.A., Berrue F., Arens J.C., Carr G., Kerr R.G. Cystargolides, 20S Proteasome Inhibitors Isolated from Kitasatospora cystarginea. J. Nat. Prod. 2015;78:822–826. doi: 10.1021/np501060k. [DOI] [PubMed] [Google Scholar]
- 13.Böttcher T., Sieber S.A. β-Lactams and β-lactones as activity-based probes in chemical biology. Med. Chem. Commun. 2012;3:408–417. doi: 10.1039/c2md00275b. [DOI] [Google Scholar]
- 14.De Pascale G., Nazi I., Harrison P.H.M., Wright G.D. β-lactone natural products and derivatives inactivate homoserine transacetylase, a target for antimicrobial agents. J. Antibiot. 2011;64:483–487. doi: 10.1038/ja.2011.37. [DOI] [PubMed] [Google Scholar]
- 15.Pojer F., Ferrer J.L., Richard S.B., Nagegowda D.A., Chye M.L., Bach T.J., Noel J.P. Structural basis for the design of potent and species-specific inhibitors of 3-hydroxy-3-methylglutaryl CoA synthases. Proc. Natl. Acad. Sci. USA. 2006;103:11491–11496. doi: 10.1073/pnas.0604935103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerciolini R. Mode of action of orlistat. Int. J. Obes. Relat. Metab. Disord. 1997;21(Suppl. 3):S12–S23. [PubMed] [Google Scholar]
- 17.Yang X.W., Peng K., Liu Z., Zhang G.Y., Li J., Wang N., Steinmetz A., Liu Y. Strepsesquitriol, a rearranged zizaane-type sesquiterpenoid from the deep-sea-derived actinomycete Streptomyces sp. SCSIO 10355. J. Nat. Prod. 2013;76:2360–2363. doi: 10.1021/np400923c. [DOI] [PubMed] [Google Scholar]
- 18.Xie C.L., Liu Q., Xia J.M., Gao Y., Yang Q., Shao Z.Z., Liu G., Yang X.W. Anti-allergic compounds from the deep-sea-derived actinomycete Nesterenkonia flava MCCC 1K00610. Mar. Drugs. 2017;15:71. doi: 10.3390/md15030071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niu S., Zhou T.T., Xie C.L., Zhang G.Y., Yang X.W. Microindolinone A, a novel 4,5,6,7-tetrahydroindole, from the deep-sea-derived actinomycete Microbacterium sp. MCCC 1A11207. Mar. Drugs. 2017;15:230. doi: 10.3390/md15070230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niu S., Liu Q., Xia J., Xie C., Luo Z., Shao Z., Liu G., Yang X. Polyketides from the deep-sea-derived fungus Graphostroma sp. MCCC 3A00421 showed potent antifood allergic activities. J. Agric. Food Chem. 2018;66:1369–1376. doi: 10.1021/acs.jafc.7b04383. [DOI] [PubMed] [Google Scholar]
- 21.Chiang Y.C.P., Yang S.S., Heck J.V., Chabala J.C., Chang M.N. Total synthesis of L-659,699, a novel inhibitor of cholesterol biosynthesis. J. Org. Chem. 1989;54:5708–5712. doi: 10.1021/jo00285a017. [DOI] [Google Scholar]
- 22.Yasuhara F., Yamaguchi S. Use of shift reagent with MTPA derivatives in 1H NMR spectroscopy. III. Determination of absolute configuration and enantiomeric purity of primary carbinols with chiral center at the C-2 position. Tetrahedron Lett. 1977;18:4085–4088. doi: 10.1016/S0040-4039(01)83433-X. [DOI] [Google Scholar]
- 23.Sugimoto Y., Tsuyuki T., Moriyama Y., Takahashi T. Application of the MTPA method to determination of abusolute stereochemistry. The hydroxymethyl-substituted chiral carbon of carbocycles. Bull. Chem. Soc. Jpn. 1980;53:3723–3724. doi: 10.1246/bcsj.53.3723. [DOI] [Google Scholar]
- 24.Wovkulich P.M., Shankaran K., Kiegiel J., Uskokovic M.R. Total synthesis of 1233A. J. Org. Chem. 1993;58:832–839. doi: 10.1021/jo00056a013. [DOI] [Google Scholar]
- 25.Franot C., Benezra C., Lepoittevin J.P. Synthesis and interaction studies of 13C labeled lactone derivatives with a model protein using 13C NMR. Bioorg. Med. Chem. 1993;1:389–397. doi: 10.1016/S0968-0896(00)82146-X. [DOI] [PubMed] [Google Scholar]
- 26.White J.D., Johnson A.T. Synthesis of the aliphatic depside (+)-bourgeanic acid. J. Org. Chem. 1994;59:3347–3358. doi: 10.1021/jo00091a022. [DOI] [Google Scholar]
- 27.Yoshida W.Y., Bryan P.J., Baker B.J., McClintock J.B. Pteroenone: A defensive metabolite of the abducted Antarctic Pteropod Clione antarctica. J. Org. Chem. 1995;60:780–782. doi: 10.1021/jo00108a057. [DOI] [Google Scholar]
- 28.Nanda S., Scott A.I. Asymmetric synthesis of (E)- and (Z)-3,7-dimethyl-2-octene-1,8-diol and callosobruchusic acid. Tetrahedron Asymmetry. 2004;15:963–970. doi: 10.1016/j.tetasy.2004.01.032. [DOI] [Google Scholar]
- 29.Kornsakulkarn J., Dolsophon K., Boonyuen N., Boonruangprapa T., Rachtawee P., Prabpai S., Kongsaeree P., Thongpanchang C. Dihydronaphthalenones from endophytic fungus Fusarium sp. BCC14842. Tetrahedron. 2011;67:7540–7547. doi: 10.1016/j.tet.2011.07.078. [DOI] [Google Scholar]
- 30.Kimura Y., Shimada A., Nakajima H., Hamasaki T. Structures of naphthoquinones produced by the fungus, Fusarium sp., and their biological activity toward pollen germination. Agric. Biol. Chem. 1988;52:1253–1259. doi: 10.1271/bbb1961.52.1253. [DOI] [Google Scholar]
- 31.Chilton W.S. Isolation and structure of norjavanicin. J. Org. Chem. 1968;33:4299–4301. doi: 10.1021/jo01275a074. [DOI] [Google Scholar]
- 32.Tatum J.H., Baker R.A. Naphthoquinones produced by Fusarium solani isolated from citrus. Phytochemistry. 1983;22:543–547. doi: 10.1016/0031-9422(83)83042-8. [DOI] [Google Scholar]
- 33.Kurobane I., Zaita N., Fukuda A. New metabolites of Fusarium martii related to dihydrofusarubin. J. Antibiot. 1986;39:205–214. doi: 10.7164/antibiotics.39.205. [DOI] [PubMed] [Google Scholar]
- 34.Arsenault G.P. Fungal metabolites. III. Quinones from Fusarium solani D2 purple and structure of (+)-solaniol. Tetrahedron. 1968;24:4745–4749. doi: 10.1016/S0040-4020(01)98671-5. [DOI] [Google Scholar]
- 35.Hashimoto J., Motohashi K., Sakamoto K., Hashimoto S., Yamanouchi M., Tanaka H., Takahashi T., Takagi M., Shin-ya K. Screening and evaluation of new inhibitors of hepatic glucose production. J. Antibiot. 2009;62:625–629. doi: 10.1038/ja.2009.93. [DOI] [PubMed] [Google Scholar]
- 36.Alex D., Bach T.J., Chye M.L. Expression of Brassica juncea 3-hydroxy-3-methylglutaryl CoA synthase is developmentally regulated and stress-responsive. Plant J. 2001;22:415–426. doi: 10.1046/j.1365-313X.2000.00751.x. [DOI] [PubMed] [Google Scholar]
- 37.Woo J.H., Kitamura E., Myouga H., Kamei Y. An antifungal protein from the marine bacterium Streptomyces sp. Strain AP77 is specific for pythium porphyrae, a causative agent of red rot disease in Porphyra spp. Appl. Environ. Microb. 2002;68:2666–2675. doi: 10.1128/AEM.68.6.2666-2675.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kundu A., Saha S., Walia S., Shakil N.A., Kumar J., Annapurna K. Cadinene sesquiterpenes from Eupatorium adenophorum and their antifungal activity. J. Environ. Sci. Health B. 2013;48:516–522. doi: 10.1080/03601234.2013.761921. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.