Abstract
Objectives: This meta-analysis investigated the relationship between thyroid transcription factor-1 (TTF-1) expression and epidermal growth factor receptor (EGFR) mutations in non-small-cell lung cancer (NSCLC) to clarify whether TTF-1 can be a potential surrogate marker for EGFR mutation status in advanced NSLCL. Methods: A systematic searching of databases, including PubMed, EMBASE, Cochrane Library, and Google Scholar, was performed to identify studies assessing the correlation of TTF-1 expression with EGFR mutations. From 17 studies, 9764 patients were included in the combined analysis of odds ratio (OR) for the correlation between TTF-1 expression and EGFR mutations. Results: Compared with NSCLCs showing negative TTF-1 expression, tumors harboring TTF-1 overexpression showed a significantly higher rate of EGFR mutations (OR = 5.19, 95% confidence interval: 3.60–7.47, p < 0.00001). This correlation was observed in both subgroups of East Asian (OR = 4.33, 95% CI: 3.46–5.41, p < 0.00001) and European patients (OR = 4.64, 95% CI: 1.41–15.28, p < 0.01). In addition, TTF-1 expression was significantly associated with EGFR mutations in exon 19 (OR = 4.63, 95% CI: 2.89–7.41, p < 0.00001) as well as exon 21 (OR = 3.16, 95% CI: 1.04–9.60, p = 0.04). Conclusions: This meta-analysis demonstrates a significant correlation between TTF-1 expression and EGFR mutations in patients with NSCLC. The status of TTF-1 expression may be a biomarker to guide anticancer treatment in patients with NSCLC and unknown EGFR mutation status.
Keywords: thyroid transcription factor-1, EGFR mutation, non-small-cell lung cancer, biomarker, meta-analysis
1. Introduction
Lung cancer is the second most common malignancy in both genders worldwide [1]. It still remains the leading cause of cancer-associated deaths [1,2], although systemic chemotherapy or immune checkpoint inhibitors can significantly improve prognosis for patients with advanced non-small-cell lung cancer (NSCLC) [3,4,5]. For patients with epidermal growth factor receptor (EGFR)-mutant NSCLC, targeted therapy with tyrosine kinase inhibitors (TKIs) can significantly prolong survival [6,7].
Thyroid transcription factor-1 (TTF-1) is a regulatory transcription factor for tissue-specific genes [8]. It is expressed in the thyroid, forebrain, or lungs, playing a physiologic role during their development. In the normal lungs, TTF-1 helps to maintain functions of terminal respiratory unit cells [9]. High TTF-1 expression by immunohistochemistry (IHC) has been observed in 70–90% of primary lung adenocarcinomas (ADCs), while almost all squamous cell carcinomas are negative for TTF-1 IHC. Therefore, it has been considered a specific marker of ADCs of the lung. In addition, TTF-1 overexpression is a favorable prognostic factor not only in early-stage but also in advanced non-squamous NSCLC [10,11].
EGFR is a member of the hermaphrodite (HER) family of tyrosine kinase receptors, and somatic activating mutations in the adenosine triphosphate (ATP)-binding site of EGFR result in a more effective binding of EGFR TKIs [12,13]. EGFR gene mutations in four kinase domains (exons 18–21) comprise in-frame deletions, in-frame insertions/duplications, and point mutations [14,15]. NSCLC patients with EGFR mutations can achieve better progression-free survival and overall survival when treated with an EGFR TKI as first-line treatment rather than chemotherapy [6,16,17,18]. Therefore, it is essential to determine the EGFR mutation status of patients with advanced NSCLC when planning anticancer therapy.
However, for some patients, it is not easy to determine the EGFR mutation status because of inadequate tumor specimen or expense. Therefore, the identification of other pathologic markers that can predict EGFR mutation status may be very useful in clinical practice. In NSCLC, both TTF-1 expression and EGFR mutations are closely related to the female gender, non-smoking status, and ADC [13,19,20,21,22]. In addition, some studies suggested that TTF-1 expression had a significant positive correlation with EGFR mutations [21,22]. This meta-analysis assessed the relationship between TTF-1 expression and EGFR mutations in NSCLC to clarify whether TTF-1 can be a potential predictive biomarker for EGFR mutation status in patients with NSLCL.
2. Materials and Methods
2.1. Publication Search Strategy
This meta-analysis was done according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [23]. A systematic search of the databases including PubMed, EMBASE, Cochrane Library, and Google Scholar (up to December 2018) was performed to identify studies assessing the correlation of TTF-1 expression with EGFR mutations. The search used a combination of the following terms: “epidermal growth factor receptor” or “EGFR” AND “mutation” AND “thyroid transcription factor-1” or “TTF-1” AND “non-small-cell lung cancer” or “NSCLC” or “lung cancer.” All of the relevant articles identified by the related article function were also included in the analysis. The references reported in the identified articles were also reviewed to complete the search process.
2.2. Eligibility Criteria
Eligible studies should meet the following inclusion criteria: (i) patients with pathologically confirmed NSCLC; (ii) analysis of EGFR mutations in exons 19 and 21; (iii) IHC test for TTF-1 expression in lung cancer tissue; (iv) the use of adequate IHC methods and criteria for positive TTF-1 staining; and (v) prospective or retrospective cohort studies assessing the correlation of TTF-1 expression with EGFR mutations.
2.3. Article Review and Data Extraction
Two authors (D.R.C. and B.H.) independently searched the databases and extracted data from the selected studies. The following data were extracted from each study: the first author, year of publication, study design, inclusion period, country, sample size, histology, disease stage, TTF-1 expression status, IHC criteria for positive expression, EGFR mutation status, and detecting method.
2.4. Quality Assessment
The methodological quality of included studies was scored based on the Newcastle–Ottawa System (NOS) with the score range of zero to nine [24]. Studies having a score ≥ six were considered to have a high quality.
2.5. Statistical Analysis
The strength of the association between TTF-1 expression and EGFR mutations was shown as odds ratios (ORs) with 95% confidence intervals (CIs). If the study did not report the OR or 95% CI directly, we calculated them from available data by using the Engauge Digitizer software. The heterogeneity of the individual ORs was estimated using the chi-squared test with significance being set at p < 0.1. The total variation among studies was estimated by anI2 inconsistency test, where I2 > 50% was considered to indicate significant heterogeneity. If there was heterogeneity among studies, we used the random effect model based on the DerSimonian–Laird method to pool the OR. Otherwise (p ≥ 0.1 and I2 ≤ 50%), the fixed effect model based on the Mantel–Haenszel method was selected. Subgroup analyses were planned according to the ethnicity and mutational types. The sensitivity analysis was performed to detect the influence of individual trials on the pooled results by removing one trial each time. Forest plots were produced to show a summary estimate of the combined results of all the studies. Each square represented the OR point estimate, and its size was proportional to the weight of the study. The location of the diamond represented the estimated effect size, and its width reflected the precision of the estimate.
The potential publication bias was assessed by visual inspection of the funnel plot [25]. For quantitative analyses, Egger’s test and Begg’s tests were performed using the statistical software packages R [26]. Statistical significances were considered when a p-value was less than 0.05.
3. Results
3.1. Search Results
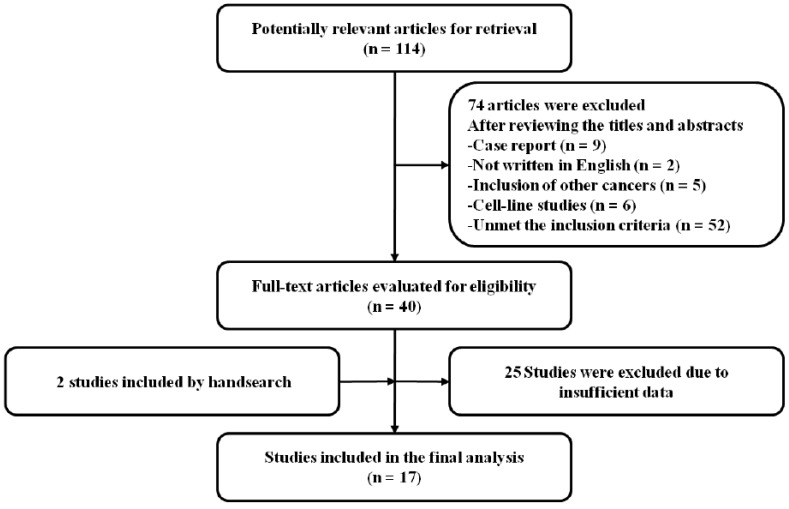
The search process identified 114 potentially relevant articles; however, 74 articles were excluded by screening of the titles and abstracts. Of the remaining 40 studies, 25 articles that failed to meet the inclusion criteria were further excluded. We manually searched the reference lists of the selected articles and found two more relevant articles. Eventually, a total of 17 studies were included in the meta-analysis [21,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42] (Figure 1).
Figure 1.
Flow diagram of search process.
3.2. Characteristics of the Included Studies
The main characteristics of the studies selected for this meta-analysis are summarized in Table 1. Except for two studies without any description about study design [35,40], seven recruited patients prospectively [21,27,30,32,34,39,41], and the remaining studies were performed retrospectively [28,29,31,33,36,37,38,42]. The NOS scores were more than seven in all of the included studies, suggesting a good methodological quality.
Table 1.
Summary of the 17 included studies.
| First Author (Year) [Ref.] | Country | Design | Sample Size | Inclusion Period | Pathology | Tumor Stage | IHC Criteria for TTF-1 (+): Nuclear Staining | Test for EGFR Mutations | TTF-1 Expression |
EGFR MT a/t TTF-1 (%) |
Significance | NOS Score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hiramatsu (2010) [27] | Japan | Pro | 93 | 1998–2001 | ADC | I–IV | Stronger than xenograft staining | PCR | (+): 74 (79.6%) (−): 19 (20.4%) |
48 (64.9%) 6 (31.6%) |
p = 0.017 | 6 |
| Vincenten (2012) [28] | Netherlands | Retro | 745 | 2004–2010 | NSCLC | I–IV | Percentage (0–100%) × intensity (0–3) ≥10 | PCR sequencing | (+): 508 (68.2%) (−): 237 (31.8%) |
105 (20.7%) 9 (3.8%) |
p < 0.00001 | 8 |
| Sun (2012) [29] | Korea | Retro | 190 | 2006–2010 | NSCLC | NA | Percentage (0–100%) × intensity (0–3) >100 | PCR sequencing | (+): 151 (79.4%) (−): 39 (20.6%) |
77 (60.0%) 6 (15.4%) |
p < 0.001 | 8 |
| Chung (2012) [30] | Taiwan | Pro | 496 | 2004–2009 | ADC | IIIB–IV | Any definite nuclear staining | PCR sequencing | (+): 443 (89.3%) (−): 53 (10.7%) |
274 (61.9%) 17 (32.1%) |
p < 0.001 | 8 |
| Vallee (2013) [31] | France | Retro | 1038 | 2010–2012 | NSCLC | I–IV | NA | PCR | (+): 820 (79.0%) (−): 218 (21.0% |
145 (17.7%) 3 (1.4%) |
p < 0.0001 | 8 |
| Gahr (2013) [32] | Germany | Pro | 854 | 2010 | NSCLC | Mainly IV | NA | Sanger sequencing | (+): 627 (73.4%) (−): 227 (26.6%) |
101 (16.1%) 7 (3.1%) |
p < 0.001 | 8 |
| Liu (2014) [21] | China | Pro | 139 | 2008–2011 | ADC | I–III | Intensity (0–3) x reactivity (0–100) > 100 | ARMS PCR | (+): 122 (87.8%) (−): 17 (12.2%) |
63 (51.6%) 2 (11.8%) |
p = 0.002 | 7 |
| Warth (2014) [33] | Germany | Retro | 418 | 2002–2008 | ADC | I–IV | NA | Sanger sequencing | (+): 366 (87.6%) (−): 52 (12.4%) |
56 (15.3%) 9 (17.3%) |
p = 0.685 | 8 |
| Shanzhi (2014) [34] | China | Pro | 664 | 2010–2013 | ADC | I–IV | >10% of tumor cells | PCR sequencing | (+): 654 (98.5%) (−): 10 (1.5%) |
261 (39.9%) 1 (10%) |
p < 0.001 | 8 |
| Somaiah (2014) [35] | USA | NA | 431 | NA | ADC | NA | NA | Allele-specific PCR | (+): 366 (84.9%) (−): 66 (15.3%) |
242 (66.1%) 3 (4.5%) |
p < 0.00001 | 8 |
| Shiau (2014) [36] | Canada | Retro | 1736 | 2010–2012 | Non-SQCC | Mainly III–IV | NA | PCR | (+): 1408 (81.1) (−): 328 (18.9) |
327 (23.2) 21 (6.4) |
p < 0.00001 | 8 |
| Elsamany (2015) [37] | Egypt | Retro | 80 | 2011–2012 | Non-SQCC | IIIB–IV | NA | NA | (+): 70 ((87.5%) (−): 10 (12.5%) |
20 (28.6%) 1 (10%) |
p = 0.28 | 6 |
| Zhao (2015) [38] | Taiwan | Retro | 200 | 2008–2013 | ADC | I–IV | ≥ 10% of tumor cells | EGFR liquid chip | (+): 163 (81.5%) (−): 37 (18.5%) |
83 (50.9%) 6 (16.2%) |
p = 0.000 | 7 |
| Zhang (2015) [39] | China | Pro | 1042 | 2008–2013 | ADC | I–III | Any positive nuclear staining | PCR | (+): 909 (87.26%) (−): 133 (12.8%) |
552 (60.7%) 50 (37.6%) |
p < 0.001 | 8 |
| Udupa (2015) [40] |
India | NA | 85 | 2009-2013 | ADC | I–IV | NA | ARMS real-time PCR | (+): 68 (80%) (−): 17 (20%) |
33 (48.5%) 1 (5.9%) |
p < 0.00001 | 6 |
| Wei (2016) [41] | China | Pro | 1083 | 2010–2016 | NSCLC | I–IV | Tan or brown nuclear staining | ARMS PCR | (+): 841 (77.7%) (−): 242 (22.3%) |
385 (45.8%) 28 (11.6%) |
p < 0.001 | 8 |
| Schilsky (2017) [42] | USA | Retro | 479 | 2009–2011 | ADC | IV | Any nuclear reactivity | NA | (+): 383 (80.0%) (−): 96 (20.0%) |
92 (24.0%) 6 (6.3%) |
p < 0.001 | 8 |
EGFR, epidermal growth factor receptor; NOS, Newcastle–Ottawa System; NSCLC, non-small-cell lung cancer; Pro, prospective; Retro, retrospective; pts, Patients; ADC, adenocarcinoma; SQCC, squamous cell carcinoma; TTF-1, thyroid transcription factor 1; ARMS, amplification refractory mutation system; PCR, polymerase chain reaction; IHC, immunohistochemistry; a/t, according to; NA, not available.
The criteria for the positivity of TTF-1 expression in IHC staining varied across studies; however, the rates of TTF-1 expression in most of the included studies were 80–90%. Six studies contained NSCLC [28,29,31,32,36,41], and 10 studies only included ADC [21,27,30,33,34,35,38,39,40,42]. Two studies included patients with stages I–III ADC [21,38], and five were conducted in patients with advanced disease (stages III–IV) [30,32,36,37,42].
The EGFR mutations were usually detected by polymerase chain reaction (PCR) and direct DNA sequencing methods, or by amplification refractory mutation system (ARMS) analysis. Most of the studies examined four kinase domains in exons 18–21; however, some screened for EGFR mutations in only exons 19 and 21 [21,31,36].
3.3. Correlation of TTF-1 Expression and EGFR Mutations
3.3.1. Overall
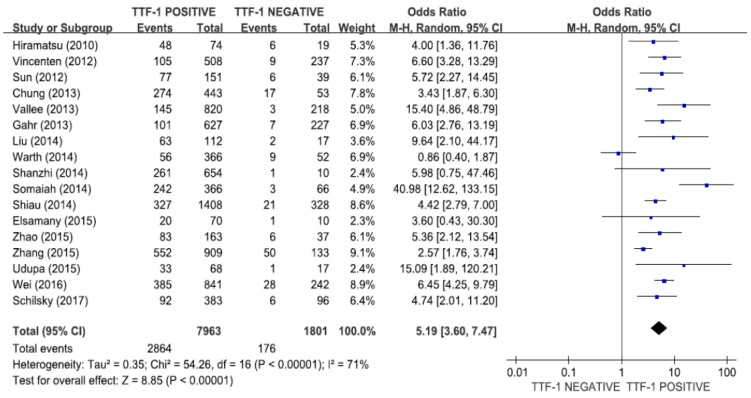
A total of 9764 patients from the 17 studies were included in the meta-analysis to determine the association of TTF-1 expression and EGFR mutations [21,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42]. The combined OR of 5.19 (95% CI: 3.60–7.47, p < 0.00001, random-effects model, Figure 2) indicated that NSCLCs with TTF-1 overexpression exhibited significantly higher rate of EGFR mutations. There was a significant heterogeneity among studies (X2 = 54.26, p < 0.00001, I2 = 71%).
Figure 2.
Forest plot for the correlation between TTF-1 expression and EGFR mutations in NSCLC.
3.3.2. Subgroup Analysis According to the Ethnic Group
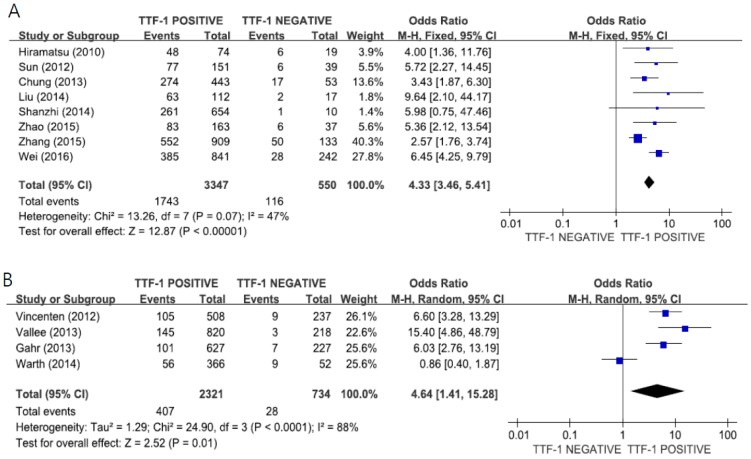
When we performed the subgroup analysis according to the region, the combined ORs were 4.33 (95% CI: 3.46–5.41, p < 0.00001, fixed-effects model) for patients in East Asia (Figure 3A) and 4.64 (95% CI: 1.41–15.28, p < 0.01, random-effects model) for patients in Europe (Figure 3B).
Figure 3.
Forest plots for the correlation of TTF-1 expression and EGFR mutations according to the ethnicity: East Asian (A) and European (B).
3.3.3. Subgroup Analysis According to the Gender
The subgroup analysis using data from three studies [30,34,38] indicated there was a significant correlation between TTF-1 expression and EGFR mutations in both female (OR = 4.87, 95%CI 2.27–10.45, fixed-effect model, Figure 4A, p < 0.0001) and male patients (OR = 3.34, 95%CI 1.43–6.82, fixed-effect model, Figure 4B, p = 0.0009).
Figure 4.
Forest plots for the correlation of TTF-1 expression and EGFR mutations according to the gender: female (A) and male (B).
3.3.4. Subgroup Analysis According to the Mutational Subtype
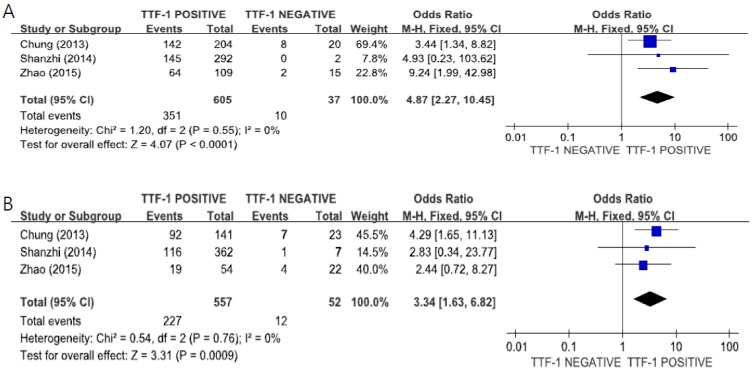
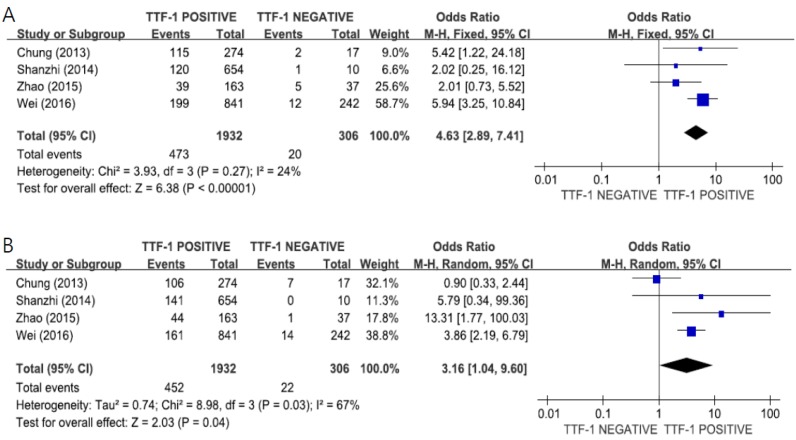
The pooled data from four studies [30,34,38,41] revealed a significant correlation between TTF-1 expression and EGFR mutations in both exon 19 (OR = 4.63, 95% CI: 2.89–7.41, p < 0.00001, fixed-effect model, Figure 5A) and exon 21 (OR = 3.16, 95% CI: 1.04–9.60, p = 0.04, random-effects model, Figure 5B).
Figure 5.
Forest plots for the correlation of TTF-1 expression and EGFR mutation according to the mutational subtype: exon 19 (A) and exon 21 (B).
3.4. Publication Bias
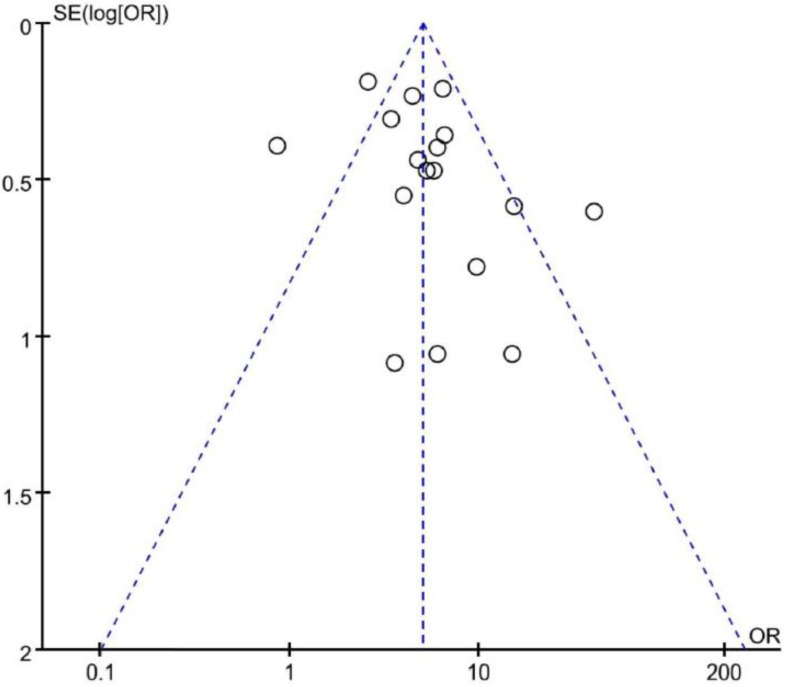
Visual inspection of the funnel plot for ORs showed symmetry, indicating that there was no substantial publication bias (Figure 6). Egger’s and Begg’s tests also indicated no evidence of substantial publication bias, with p-values of 0.172 and 0.152, respectively.
Figure 6.
Funnel plot for publication bias.
4. Discussion
Over the past two decades, several major progresses have been made toward the personalized treatment for patients with NSCLC. The first breakthrough was the discovery of EGFR mutations. TKIs are small molecular agents targeting EGFR mutations that have revolutionized the treatment of NSCLC, leading to improved survival in patients with advanced or metastatic EGFR-mutant tumor [6,7]. Therefore, it is essential to screen for EGFR mutations before introducing anticancer treatment for patients with advanced NSCLC.
Studies evaluating the prognostic value of TTF-1 expression in NSCLC reported TTF-1 expression to be an independent predictor of survival [10,11,43,44]. In the North East Japan 002 study, investigators reported that the rate of EGFR mutations was higher in patients with ADC positive for TTF-1 expression [6]. In particular, Asians, women, and non-smokers revealed a higher rate of EGFR mutations and TTF-1 positive expression. Based on these findings, some researchers hypothesized that TTF-1 expression is significantly correlated with EGFR mutations in patients with lung ADC [34].
In the current meta-analysis, we combined the data from 17 studies, including 9764 patients, to determine whether positive TTF-1 expression by IHC can be a surrogate marker for EGFR mutation status in NSCLC. The results revealed that EGFR mutations were significantly correlated with TTF-1 overexpression in patients with NSCLC (OR = 5.34, 95% CI: 3.54–8.04, p < 0.00001). Both high TTF-1 expression and EGFR mutations have been known to be significantly associated with Asian, women, and non-smokers. However, our subgroup analysis according to the ethnicity indicated that the correlation of TTF-1 expression and EGFR mutations was observed in European patients (OR = 4.64, 95% CI: 1.41–15.28, p < 0.01) as well as East Asian (OR = 4.33, 95% CI: 3.46–5.41, p < 0.00001). In addition, in the subgroup analysis according to the gender, the significant correlation between TTF-1 expression and EGFR mutations was observed not only in female (OR = 4.87, 95% CI 2.27–10.45, p < 0.0001), but also in male patients with NSCLC (OR = 3.34, 95% CI 1.43–6.82, p = 0.0009).
Among EGFR mutations, deletion in exon 19 and L858R mutation in exon 21 are common, accounting for more than 90% of all EGFR mutations. Some studies conducted subgroup analysis according to EGFR mutation subtype and reported that positive TTF-1 expression was only significantly correlated with EGFR mutation in exon 21, and not on exon 19 [34,38]. However, Zhao et al. also observed a significant correlation of TTF-1 positivity with EGFR mutation in exon 21 in Chinese patients with lung ADC [38]. In the current meta-analysis, the pooled data from four studies [30,34,38,41] indicated a significant relationship between TTF-1 expression and EGFR mutation in both exon 19 (OR = 4.63, 95% CI: 2.89–7.41, p < 0.00001) and exon 21 (OR = 3.16, 95% CI: 1.04–9.60, p = 0.04).
Several studies reported a high negative predictive value (88.2–97%) of TTF-1 for the presence of activating EGFR gene mutations [21,28,35,40]. In this study, only 9.8% (176 of 1801) of TTF-1–negative NSCLC cases had EGFR mutations. Overall, the negative predictive value of TTF-1 for EGFR mutations was 90.2%. However, when including only patients in Europe, where the rate of EGFR mutations is low, the negative predictive value of TTF-1 increased to 96.2%. Therefore, when the status of EGFR mutations cannot be tested in a timely manner because of inadequate tumor tissue or when treatment for NSCLC is urgent for clinical reasons, the IHC staining result of TTF-1 may be used to guide systemic anticancer therapy. In particular, negative TTF-1 expression can be a surrogate marker to recommend conventional chemotherapy.
This study has several limitations. First, the included studies showed considerable diversity in the methods used for TTF-1 staining and EGFR mutation detection. In addition, the cut-off criteria for positive TTF-1 staining also varied among studies. Second, the rate of TTF-1 overexpression and EGFR mutations are significantly higher with NSCLC patients with no smoking history. However, we could not perform a subgroup analysis according to the smoking status because of the limited data. Third, the substantial heterogeneity observed among studies could not be interpreted thoroughly, even though the random-effects model was selected. Finally, about half of the included studies were retrospective and therefore may carry the biases inherent to the study design.
5. Conclusions
This meta-analysis indicates a significant correlation between TTF-1 overexpression and EGFR mutations status in patients with NSCLC. In clinical practice, the status of TTF-1 expression may be a biomarker to guide anticancer therapy in advanced NSCLC patients with unknown EGFR mutation status. Especially, negative TTF-1 expression has such a high negative predictive value for EGFR mutations that it might be a surrogate marker to recommend conventional chemotherapy first when the status of EGFR mutations cannot be tested in a timely manner.
Author Contributions
J.H.K. designed the study. B.H. and D.R.C. searched the literatures and extracted the data. H.S.K. performed the statistical analyses. H.S.K. and J.H.K. wrote the manuscript.
Funding
This research was supported by Hallym University Research Fund (HURF-2018-37).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA. Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Jung K.W., Won Y.J., Kong H.J., Lee E.S. Community of population-based regional cancer registries. Cancer statistics in Korea: Incidence, mortality, survival, and prevalence in 2015. Cancer Res. Treat. 2018;50:303–316. doi: 10.4143/crt.2018.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.NSCLC Meta-Analyses Collaborative Group Chemotherapy in addition to supportive care improves survival in advanced non-small-cell lung cancer: A systematic review and meta-analysis of individual patient data from 16 randomized controlled trials. J. Clin. Oncol. 2008;26:4617–4625. doi: 10.1200/JCO.2008.17.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reck M., Rodriguez-Abreu D., Robinson A.G., Hui R., Csőszi T., Fülöp A., Gottfried M., Peled N., Tafreshi A., Cuffe S., et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N. Engl. J. Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 5.Horn L., Spigel D.R., Vokes E.E., Holgado E., Ready N., Steins M., Poddubskaya E., Borghaei H., Felip E., Paz-Ares L., et al. Nivolumab versus docetaxel in previously treated patients with advanced non-small-cell lung cancer: Two-year outcomes from two randomized, open-label, phase III trials (CheckMate 017 and CheckMate 057) J. Clin. Oncol. 2017;35:3924–3933. doi: 10.1200/JCO.2017.74.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maemondo M., Inoue A., Kobayashi K., Sugawara S., Oizumi S., Isobe H., Gemma A., Harada M., Yoshizawa H., Kinoshita I., et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N. Engl. J. Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 7.Bulbul A., Husain H. First-line treatment in EGFR mutant non-small cell lung cancer: Is there a best option? Front. Oncol. 2018:8. doi: 10.3389/fonc.2018.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guazzi S., Price M., De Felice M., Damante G., Mattei M.G., Di Lauro R. Thyroid nuclear factor 1 (TTF-1) contains a homeodomain and displays a novel DNA binding specificity. EMBO J. 1990;9:3631–3639. doi: 10.1002/j.1460-2075.1990.tb07574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L., Whitsett J.A., Stripp B.R. Regulation of Clara cell secretory protein gene transcription by thyroid transcription factor-1. Biochim. Biophys. Acta. 1997;1350:359–367. doi: 10.1016/S0167-4781(96)00180-7. [DOI] [PubMed] [Google Scholar]
- 10.Kim J.H., Kim H.S., Kim B.J., Han B., Choi D.R., Kwon J.H. Prognostic impact ofTTF-1 expression in non-squamous non-small-cell lung cancer: A meta-analysis. J. Cancer. 2018;9:4279–4286. doi: 10.7150/jca.26830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian H.H., Xu T.S., Cai X.Q., Ji T.L., Guo H.X. Prognostic value of TTF-1 expression in patients with non-small cell lung cancer: A meta-analysis. Clin. Chim. Acta. 2015;451:208–214. doi: 10.1016/j.cca.2015.01.023. [DOI] [PubMed] [Google Scholar]
- 12.Lynch T.J., Bell D.W., Sordella R., Gurubhagavatula S., Okimoto R.A., Brannigan B.W., Harris P.L., Haserlat S.M., Supko J.G., Haluska F.G., et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 13.Paez J.G., Janne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F.J., Lindeman N., Boggon T.J., et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 14.Shigematsu H., Gazdar A.F. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int. J. Cancer. 2006;118:257–262. doi: 10.1002/ijc.21496. [DOI] [PubMed] [Google Scholar]
- 15.Mitsudomi T., Yatabe Y. Mutations of the epidermal growth factor receptor gene and related genes as determinants of epidermal growth factor receptor tyrosine kinase inhibitors sensitivity in lung cancer. Cancer Sci. 2007;98:1817–1824. doi: 10.1111/j.1349-7006.2007.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mok T.S., Wu Y.L., Thongprasert S., Yang C.H., Chu D.T., Saijo N., Sunpaweravong P., Han B., Margono B., Ichinose Y., et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 17.Zhou C., Wu Y.L., Chen G., Feng J., Liu X.Q., Wang C., Zhang S., Wang J., Zhou S., Ren S., et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 18.Rosell R., Carcereny E., Gervais R., Vergnenegre A., Massuti B., Felip E., Palmero R., Garcia-Gomez R., Pallares C., Sanchez J.M., et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 19.Kosaka T., Yatabe Y., Endoh H., Kuwano H., Takahashi T., Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: Biological and clinical implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 20.Mitsudomi T., Kosaka T., Yatabe Y. Biological and clinical implications of EGFR mutations in lung cancer. Int. J. Clin. Oncol. 2006;11:190–198. doi: 10.1007/s10147-006-0583-4. [DOI] [PubMed] [Google Scholar]
- 21.Jie L., Li X.Y., Zhao Y.Q., Liu R.Q., Zhang J.B., Ma J., Chen L.J., Hu X.F. Genotype-phenotype correlation in Chinese patients with pulmonary mixed type adenocarcinoma: Relationship between histologic subtypes, TITF-1/SP-A expressions and EGFR mutations. Pathol. Res. Pract. 2014;210:176–181. doi: 10.1016/j.prp.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 22.Tang X., Kadara H., Behrens C., Liu D.D., Xiao Y., Rice D., Gazdar A.F., Fujimoto J., Moran C., Varella-Garcia M., et al. Abnormalities of the TITF-1 lineage-specific oncogene in NSCLC: Implications in lung cancer pathogenesis and prognosis. Clin. Cancer Res. 2011;17:2434–2443. doi: 10.1158/1078-0432.CCR-10-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moher D., Liberati A., Tetzlaff J., Altman D.G., PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 25.Sterne J.A., Sutton A.J., Ioannidis J.P., Terrin N., Jones D.R., Lau J., Carpenter J., Rücker G., Harbord R.M., Schmid C.H., et al. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ. 2011;343:d4002. doi: 10.1136/bmj.d4002. [DOI] [PubMed] [Google Scholar]
- 26.Eagger M., Davey Smith G., Schneider M., Minder C. Bias in meta-analysis detected by a simple graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiramatsu M., Ninomiya H., Inamura K., Nomura K., Takeuchi K., Satoh Y., Okumura S., Nakagawa K., Yamori T., Matsuura M., et al. Activation status of receptor tyrosine kinase downstream pathways in primary lung adenocarcinoma with reference of KRAS and EGFR mutations. Lung Cancer. 2010;70:94–102. doi: 10.1016/j.lungcan.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Vincenten J., Smit E.F., Vos W., Grünberg K., Postmus P.E., Heideman D.A., Snijders P.J., Meijer G., Kuik J., Witte B.I., et al. Negative NKX2-1 (TTF-1) as temporary surrogate marker for treatment selection during EGFR-mutation analysis in patients with non-small-cell lung cancer. J. Thorac. Oncol. 2012;7:1522–1527. doi: 10.1097/JTO.0b013e3182635a91. [DOI] [PubMed] [Google Scholar]
- 29.Sun P.L., Seol H., Lee H.J., Yoo S.B., Kim H., Xu X., Jheon S., Lee C.T., Lee J.S., Chung J.H. High incidence of EGFR mutations in Korean men smokers with no intratumoral heterogeneity of lung adenocarcinomas: Correlation with histologic subtypes, EGFR/TTF-1 expressions, and clinical features. J. Thorac. Oncol. 2012;7:323–330. doi: 10.1097/JTO.0b013e3182381515. [DOI] [PubMed] [Google Scholar]
- 30.Chung K.P., Huang Y.T., Chang Y.L., Yu C.J., Yang C.H., Chang Y.C., Shih J.Y., Yang P.C. Clinical significance of thyroid transcription factor-1 in advanced lung adenocarcinoma under epidermal growth factor receptor tyrosine kinase inhibitor treatment. Chest. 2012;141:420–428. doi: 10.1378/chest.10-3149. [DOI] [PubMed] [Google Scholar]
- 31.Vallee A., Sagan C., Le Loupp A.G., Bach K., Dejoie T., Denis M.G. Detection of EGFR gene mutations in non-small cell lung cancer: Lessons from a single-institution routine analysis of 1,403 tumor samples. Int. J. Oncol. 2013;43:1045–1051. doi: 10.3892/ijo.2013.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gahr S., Stoehr R., Geissinger E., Ficker J.H., Brueckl W.M., Gschwendtner A., Gattenloehner S., Fuchs F.S., Schulz C., Rieker R.J., et al. EGFR mutational status in a large series of Caucasian European NSCLC patients: Data from daily practice. Br. J. Cancer. 2013;109:1821–1828. doi: 10.1038/bjc.2013.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warth A., Penzel R., Lindenmaier H., Brandt R., Stenzinger A., Herpel E., Goeppert B., Thomas M., Herth F.J., Dienemann H., et al. EGFR, KRAS, BRAF and ALK gene alterations in lung adenocarcinomas: Patient outcome, interplay with morphology and immunophenotype. Eur. Respir. J. 2014;43:872–883. doi: 10.1183/09031936.00018013. [DOI] [PubMed] [Google Scholar]
- 34.Shanzhi W., Yiping H., Ling H., Jianming Z., Qiang L. The relationship between TTF-1 expression and EGFR mutations in lung adenocarcinomas. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0095479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Somaiah N., Fidler M.J., Garrett-Mayer E., Wahlquist A., Shirai K., Buckingham L., Hensing T., Bonomi P., Simon G.R. Epidermal growth factor receptor (EGFR) mutations are exceptionally rare in thyroid transcription factor (TTF-1)-negative adenocarcinomas of the lung. Oncoscience. 2014;1:522–528. doi: 10.18632/oncoscience.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiau C.J., Babwah J.P., da Cunha Santos G., Sykes J.R., Boerner S.L., Geddie W.R., Leighl N.B., Wei C., Kamel-Reid S., Hwang D.M., et al. Sample features associated with success rates in population-based EGFR mutation testing. J. Thorac. Oncol. 2014;9:947–956. doi: 10.1097/JTO.0000000000000196. [DOI] [PubMed] [Google Scholar]
- 37.Elsamany S.A., Al-Fayea T.M., Alzahrani A.S., Abozeed W.N., Darwish W., Farooq M.U., Almadani A.S., Bukhari E.A. Thyroid transcription factor-1 expression in advanced non- small cell lung cancer: Impact on survival outcome. Asian Pac. J. Cancer Prev. 2015;16:2987–2991. doi: 10.7314/APJCP.2015.16.7.2987. [DOI] [PubMed] [Google Scholar]
- 38.Zhao Q., Xu S., Liu J., Li Y., Fan Y., Shi T., Wei S., Tang S.C., Liu H., Chen J. Thyroid transcription factor-1 expression is significantly associated with mutations in exon 21 of the epidermal growth factor receptor gene in Chinese patients with lung adenocarcinoma. OncoTargets Ther. 2015;8:2469–2478. doi: 10.2147/OTT.S90602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y., Wang R., Li Y., Pan Y., Hu H., Zhang Y., Li H., Shen L., Yu Y., Sun Y., et al. Negative thyroid transcription factor 1 expression defines an unfavorable subgroup of lung adenocarcinomas. J. Thorac. Oncol. 2015;10:1444–1450. doi: 10.1097/JTO.0000000000000626. [DOI] [PubMed] [Google Scholar]
- 40.Udupa K.S., Rajendranath R., Sagar T.G., Sundersingh S., Joseph T. Dual surrogate markers for rapid prediction of epidermal growth factor receptor mutation status in advanced adenocarcinoma of the lung: A novel approach in resource-limited setting. Indian J. Cancer. 2015;52:266–268. doi: 10.4103/0019-509X.176693. [DOI] [PubMed] [Google Scholar]
- 41.Wei W.E., Mao N.Q., Ning S.F., Li J.L., Liu H.Z., Xie T., Zhong J.H., Feng Y., Wei C.H., Zhang L.T. An Analysis of EGFR Mutations among 1506 Cases of Non-Small Cell Lung Cancer Patients in Guangxi, China. PLoS ONE. 2016;11 doi: 10.1371/journal.pone.0168795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schilsky J.B., Ni A., Ahn L., Datta S., Travis W.D., Kris M.G., Chaft J.E., Rekhtman N., Hellmann M.D. Prognostic impact of TTF-1 expression in patients with stage IV lung adenocarcinomas. Lung Cancer. 2017;108:205–211. doi: 10.1016/j.lungcan.2017.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X., Wan L., Shen H., Geng J., Nie J., Wang G., Jia N., Dai M., Bai X. Thyroid transcription factor-1 amplification and expressions in lung adenocarcinoma tissues and pleural effusions predict patient survival and prognosis. J. Thorac. Oncol. 2012;7:76–84. doi: 10.1097/JTO.0b013e318232b98a. [DOI] [PubMed] [Google Scholar]
- 44.Nakahara Y., Hosomi Y., Saito M., Ogawa M., Hishima T., Okamura T., Sasaki J., Masuda N. Predictive significance of thyroid transcription factor-1 expression in patients with non-squamous non-small cell lung cancer with wild-type epidermal growth factor receptor treated with erlotinib. Mol. Clin. Oncol. 2016;5:14–18. doi: 10.3892/mco.2016.870. [DOI] [PMC free article] [PubMed] [Google Scholar]