Abstract
The protein abundances of phospholipases A2 in cobra venom proteomes appear to vary among cobra species. To determine the unique distribution of snake venom phospholipases A2 (svPLA2) in the cobras, the svPLA2 activities for 15 cobra species were examined with an acidimetric and a colorimetric assay, using egg yolk suspension and 4-nitro-3-octanoyloxy benzoic acid (NOBA) as the substrate. The colorimetric assay showed significant correlation between svPLA2 enzymatic activities with the svPLA2 protein abundances in venoms. High svPLA2 activities were observed in the venoms of Asiatic spitting cobras (Naja sputatrix, Naja sumatrana) and moderate activities in Asiatic non-spitters (Naja naja, Naja atra, Naja kaouthia), African spitters (subgenus Afronaja), and forest cobra (subgenus Boulengerina). African non-spitting cobras of subgenus Uraeus (Naja haje, Naja annulifera, Naja nivea, Naja senegalensis) showed exceptionally low svPLA2 enzymatic activities. The negligible PLA2 activity in Uraeus cobra venoms implies that PLA2 may not be ubiquitous in all snake venoms. The svPLA2 in cobra envenoming varies depending on the cobra species. This may potentially influence the efficacy of cobra antivenom in specific use for venom neutralization.
Keywords: svPLA2, acidimetric assay, colorimetric assay, venom enzymatics, immunoreactivity, cobra venom
1. Introduction
Phospholipases A2 (PLA2) (EC 3.1.1.4) are enzymes that hydrolyze glycerophospholipids to lysophospholipids and fatty acids. The first snake venom PLA2 enzymes were purified from the venoms of Naja naja and Naja tripudians as hemolysin due to their ability to lyse the phospholipid membranes of red blood cells [1]. Since then, various snake venom-derived PLA2 (svPLA2) have been characterized and shown to exist in virtually all venoms from the two major families of venomous snakes: Elapidae and Viperidae [2]. Homologous svPLA2 are especially abundant and diverse in the Asiatic elapids, including cobras, coral snakes, kraits, and some sea snake species [3,4,5,6], implying that the enzyme plays an essential role in the function of the venom.
Previous studies demonstrated that svPLA2 originated from ancestral physiological genes that have subsequently undergone several convergent and divergent evolutionary events crucial for the adaptation and survival of the snakes [7]. Typically, the snake venom PLA2 are single-chain polypeptides with 115–125 amino acid residues (13–15 kDa), and high degrees of sequence homology are observed across different cobra species [8]. Despite sequence similarity, svPLA2 can differ widely in their pharmacology, contributing to the diverse toxic activities in snakebite envenoming. In the pathophysiology of elapid snake envenoming, the svPLA2 are commonly associated with presynaptic neurotoxicity (kraits [9,10]), myotoxicity (sea snakes [3,11,12]) (suggest to remove citation 3 as the article do not contain information of sea snake myotoxicity) and possibly cardiotoxicity (king cobra [13]). In certain species of Asian cobras, such as the Javan spitting cobra (Naja sputatrix) and the equatorial spitting cobra (Naja sumatrana), the basic and neutral svPLA2 isoenzymes are known to contribute substantially to lethality in mice, with or without a synergistic action with the cobra venom cytotoxins/cardiotoxins [3,14,15]. Venom proteome data reported to date revealed that the PLA2 account for approximately 4−31% of venom protein content of various cobra species including Naja naja [16,17,18,19], Naja sputatrix [14], Naja atra [20,21], Naja kaouthia [20,22,23,24], Naja siamensis [20], Naja melanoleuca [25], Naja nigricollis, Naja katiensis, Naja pallida, Naja nubiae, Naja mossambica [26], Naja ashei [27], and Naja haje [28]. This indicates that svPLA2 is an important component in cobra venoms and likely plays an important role in envenoming of human, in addition to predatory and/or digestive functions. However, interspecific differences in the svPLA2 enzymatic activities of some cobra species had been reported. Particularly noteworthy is the extremely low level of PLA2 activity in two African cobra species, Naja nivea [29,30] and Naja haje, in comparison to several other Asian and African cobra species [30]. As such, the difference in the svPLA2 enzymatic activities could be a potential indicator for distinguishing the cobra venoms from different evolutionary clades of cobras.
Despite the findings reported decades ago, the taxonomic value of this information pertaining to the measurement of PLA2 enzymatic activities had since remained under-explored. This is a relevant research topic to be revisited, especially when the Naja cobra complex has undergone several taxonomic revisions since then. The Naja cobras are now known to include four subgenera: Naja, Afronaja, Boulengerina, and Uraeus [31,32]. The subgenus Naja represents the Asiatic lineage of both spitting and non-spitting cobras. The other three subgenera include the African species, where the spitting cobras are grouped under the subgenus Afronaja, whereas the non-spitting cobras are grouped under the subgenera Boulengerina and Uraeus. In total, there are no less than 20 distinct species under the genus Naja currently, compared to what were only recognized as six species half a century ago [32,33]. Thus, the profiling of PLA2 distribution in the vast cobra venoms is worthy of investigation to obtain valuable insights into the natural history of venom evolution. The knowledge gained is also important to increase the understanding of cobra venom toxicity in the context of the biogeography and phylogeny of cobras. In this study, we investigated the svPLA2 enzymatic activities for various cobra species from different geographical locales using two independent enzymatic assays. The findings were analyzed in correlation to the protein abundances of the PLA2 enzyme reported in various cobra venoms.
2. Results
2.1. PLA2 Enzymatic Activities (Acidimetric Assay)
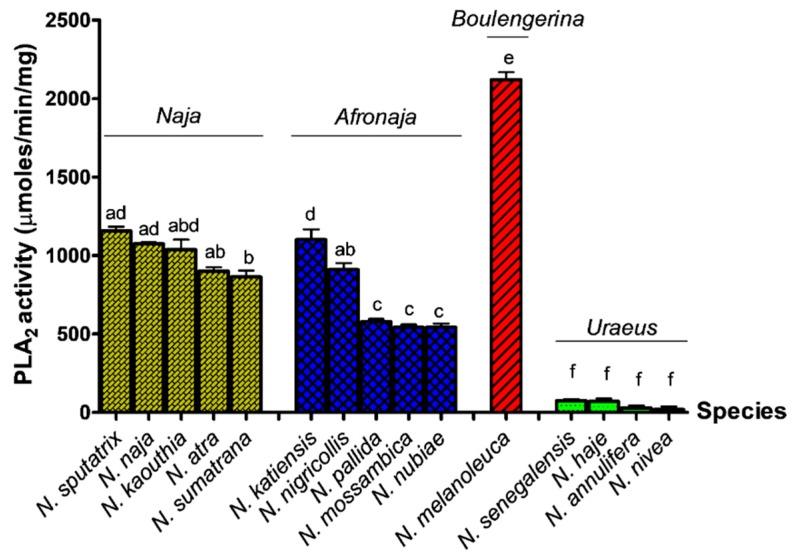
The pH of substrate (egg yolk suspension) generally reduced with time when reacting with the cobra venoms (Figure S1). The highest PLA2 activity in the acidimetric assay was noted in N. melanoleuca venom (subgenus Boulengerina) (rate = 2120.66 µmol/min/mg), followed by the venoms of Asian cobras (subgenus Naja) (rates = 864.04–1157.56 µmol/min/mg) and African spitting cobras (subgenus Afronaja) (rates = 543.01–1102.52 µmol/min/mg). The venoms of African non-spitting cobras (subgenus Uraeus) showed extremely low PLA2 enzymatic activity (rates = 20.18–75.21 µmol/min/mg). Figure 1 illustrates the PLA2 enzymatic activities of cobra venoms by subgenus tested in the acidimetric assay.
Figure 1.
Comparison of venom phospholipase A2 activities in acidimetric assay among the venoms of four subgenera of cobra. Values are expressed as mean ± S.E.M. of triplicates. Statistical analysis difference was performed by one-way analysis of variance (ANOVA) and Tukey’s post hoc test, where the statistical significance (p < 0.05) is indicated by different lower-case letters at the top of the bars. Bars without any common lowercase letter denote values that are significantly different (p < 0.05).
2.2. PLA2 Enzymatic Activities (Colorimetric Assay)
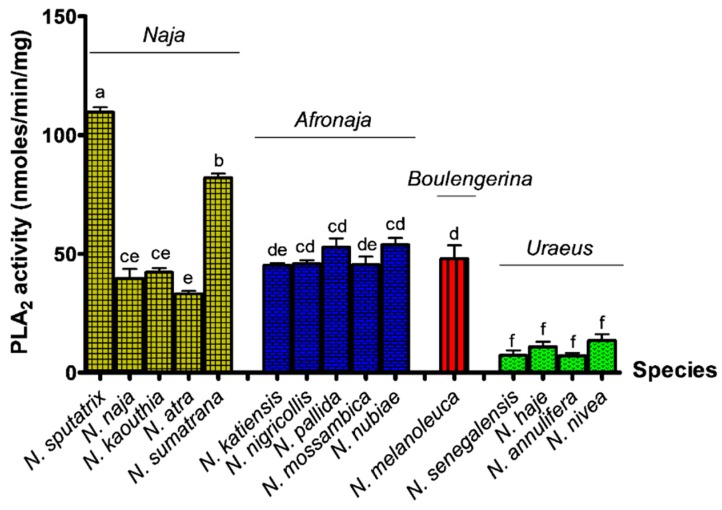
The PLA2 activities of the cobra venoms were measured using a colorimetric assay. The enzymatic activity of the venoms in hydrolyzing the non-micellar substrate (NOBA) over time is shown in Figure S2. High PLA2 activities were noted in the venoms of N. sputatrix (rate = 109.69 nmol/min/mg) and N. sumatrana (rate = 82.11 nmol/min/mg), followed by other species in the subgenera Naja (rate = 33.21–42.26 nmol/min/mg), Afronaja (rate = 45.15–53.82 nmol/min/g), and Boulengerina (rate = 48.03 nmol/min/mg). In comparison, cobra venoms of the Uraeus subgenus showed much lower PLA2 activities (rate = 7.12–13.52 nmol/min/mg) (Figure 2).
Figure 2.
Comparison of venom phospholipase A2 (PLA2) activities in colorimetric assay for the venoms of four subgenera of cobra. Values are expressed as mean ± S.E.M. of triplicates. Statistical analysis difference was performed by one-way ANOVA and Tukey’s post hoc test, where the statistical significance (p < 0.05) is indicated by different lower-case letters at the top of the bar. Bars without any common lowercase letter denote values that were significantly different (p < 0.05).
2.3. Correlation Between PLA2 Activities and PLA2 Abundances in Cobra Venoms
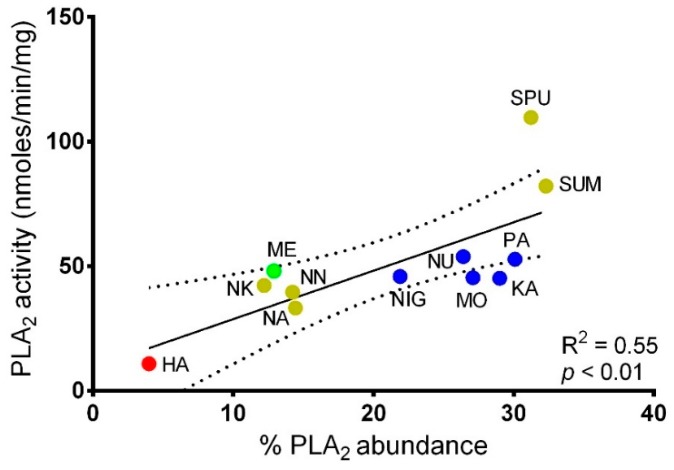
Figure 3 shows the correlation between PLA2 activities in the colorimetric assay and PLA2 protein abundances in cobra venoms. The PLA2 protein abundances of 12 cobra species were obtained from published studies that adopted a comparable quantitative approach, in which the protein abundances were estimated based on peak areas of reverse-phase high performance liquid chromatography (HPLC), followed by integration with the relative mass spectral intensity or relative gel band density of PLA2 eluted (Table S1) [14,19,20,22,25,26,28,34]. The PLA2 activities measured using the acidimetric assay showed a weak correlation with the PLA2 abundances (coefficient of determination, R2 = 0.01, p > 0.05). The colorimetric assay demonstrated a moderate to strong association between the PLA2 activities and PLA2 abundances of the cobra venoms studied (R2 = 0.55, p < 0.01) (Figure 3). The higher PLA2 enzymatic activities were observed in the venoms of spitting cobras under the subgenus Naja (N. sputatrix and N. sumatrana) and subgenus Afronaja (African spitters), whose PLA2 abundances were more than 20% of total venom proteins. The non-spitting Asiatic Naja cobras (N. kaouthia, N. naja and N. atra) had intermediate PLA2 abundances (12–14%) with moderate PLA2 enzymatic activities per unit venom. The venom of N. haje (African non-spitting cobra under the Uraeus subgenus), however, showed very low PLA2 enzymatic activity that was in line with its low PLA2 content (4%).
Figure 3.
Correlation plot of PLA2 enzymatic activities and PLA2 protein abundances in cobra venoms. Enzymatic measurement using colorimetric assay for 12 cobra venoms. The relative abundance of PLA2 (% by total venom proteins) in 12 cobra venoms were adopted from published literature for subgenus Naja: N. sputatrix [14], N. naja [19], Naja atra [20], N. kaouthia [22], Naja sumatrana [34]; subgenus Afronaja: N. katiensis, N. mossambica, N. pallida, N. nubiae, and N. nigricollis [26]; subgenus Boulengerina: N. melanoleuca [25]; and subgenus Uraeus: N. haje [28]. Abbreviations: R2: Coefficient of determination; NA, N. atra; NK, N. kaouthia; NN, N. naja; SUM, N. sumatrana; SPU, N. sputatrix; NIG, N. nigricollis; KA, N. katiensis; NU, N. nubiae; MO, N. mossambica; PA, N. pallida; ME, N. melanoleuca; HA, N. haje.
Figure 3 depicts the correlation between PLA2 activities exhibited by the whole venoms and relative abundances of PLA2 in the venoms. The PLA2 activities were measured using whole venoms and are expressed in nmol/min/mg venom proteins as previously established [29,35]. This allowed the determination of correlation between the PLA2 activities (measured per unit mass of venom) and the relative abundances of PLA2 in the cobra venoms. The higher the PLA2 relative abundance (% of total venom proteins) in a venom, the higher the snake venom PLA2 activity. The PLA2 activity of snake venom can also be expressed as “PLA2 specific activity” [36], where the PLA2 activity of a whole venom is normalized (divided) by the amount of PLA2 in the venom. This measurement is suitable for characterizing the enzymatic activity of purified or isolated PLA2, expressed in the unit of nmol/min/mg PLA2 isolated [36]. In this study, the PLA2 specific activity is included along with the relevant parameters used in calculating the values (Table 1). The findings showed that the cobra venoms tested could be generally classified into four groups: (1) Asiatic spitting cobras under the subgenus Naja (N. sputatrix and N. sumatrana), showing higher PLA2 activity with higher venom PLA2 abundance (>30%) and higher PLA2 specific activity (254–351 nmol/min/mg PLA2 protein); (2) African spitting cobras (subgenus Afronaja), showing intermediate PLA2 activities with high PLA2 abundances (>20%) and lower PLA2 specific activities (156–209 nmol/min/mg PLA2 protein); (3) non-spitting Asiatic Naja cobras (N. kaouthia, N. naja and N. atra) and African forest cobra N. melanoleuca (Boulengerina), showing intermediate PLA2 activities with moderate PLA2 abundances (12–14%) and higher specific activities (230–372 nmol/min/mg PLA2 protein); and (4) N. haje (African non-spitting cobra under the Uraeus subgenus), showing very low PLA2 enzymatic activity with low PLA2 content (4%) and higher specific activities (272 nmol/min/mg PLA2 protein) (Table 1).
Table 1.
Cobra venom PLA2 activities and PLA2-specific activities.
| Subgenus | Cobra Venom | Relative PLA2 Abundance (% Total Venom Proteins) |
PLA2 Activity of Venom (nmol/min/mg Venom Proteins) |
PLA2 Specific Activity (nmol/min/mg PLA2 Protein) |
|---|---|---|---|---|
| Naja | N. naja | 14.2 | 39.57 | 277.88 |
| N. kaouthia | 12.2 | 42.26 | 346.38 | |
| N. sputatrix | 31.2 | 109.70 | 351.13 | |
| N. atra | 14.4 | 33.21 | 230.12 | |
| N. sumatrana | 32.3 | 82.11 | 254.21 | |
| Afronaja | N. nigricollis | 21.9 | 45.85 | 209.36 |
| N. pallida | 30.1 | 52.78 | 175.36 | |
| N. nubiae | 26.4 | 53.82 | 203.84 | |
| N. mossambica | 27.1 | 45.35 | 167.36 | |
| N. katiensis | 29.0 | 45.15 | 155.69 | |
| Boulengerina | N. melanoleuca | 12.9 | 48.03 | 372.33 |
| Uraeus | N. haje | 4.0 | 10.87 | 271.71 |
2.4. Phylogenetics of Cobras in Relation to Venom PLA2 Activities
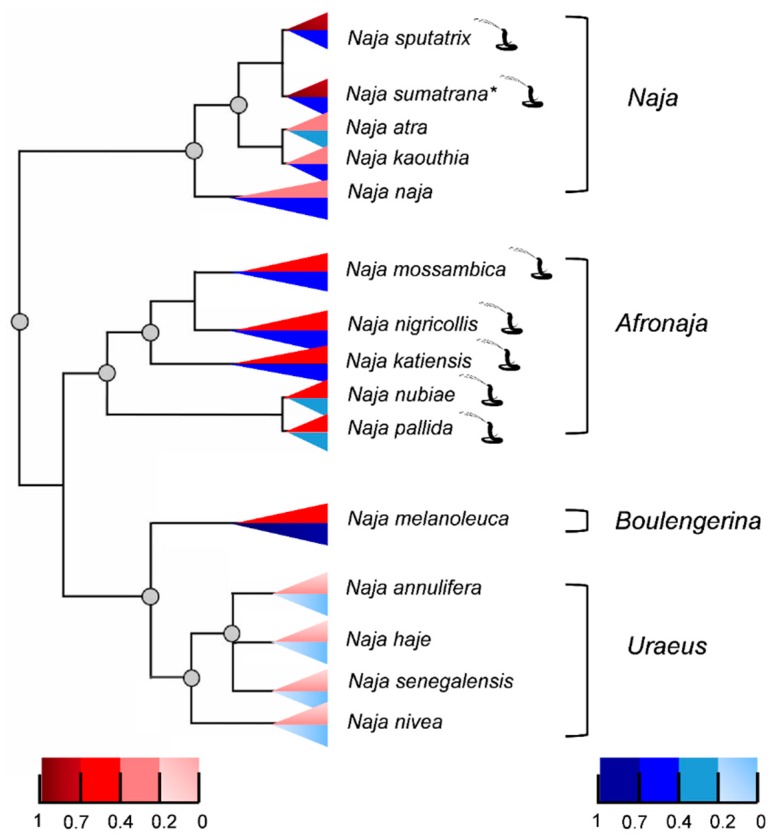
The relative PLA2 enzymatic activities of the 15 cobra venoms (by acidimetric and colorimetric methods) are related to the phylogeny of cobras in Figure 4. The venoms of Asian spitting cobras (N. sputatrix and N. sumatrana) exhibited the highest PLA2 enzymatic activity tested with the colorimetric method, whereas the highest PLA2 activity determined by the acidimetric method was observed in the African forest cobra N. melanoleuca venom. The venoms of other cobra species within the subgenera Naja (Asian cobras) and Afronaja (African spitting cobras) showed moderate to high levels of PLA2 activities in both enzymatic assays. The venoms of cobras within the Uraeus subgenus, representing a monophyletic group of non-spitting African cobras separated from the African forest cobras more recently, showed exceptionally low PLA2 activities in both acidimetric and colorimetric assays.
Figure 4.
Phylogenic tree relating the venom phospholipase A2 activities of 15 cobra species by subgenera. Blue: acidimetric assay; red: colorimetic assay. The venom PLA2 activity is expressed in a ratio relative to the highest activity detected by acidimetric or colorimetric assay in this study (1.0 implies the highest activity). More intense color indicates higher PLA2 activity. The phylogenetic tree was redrawn with adaptation from phylogenetics of cobras [32]. Naja sumatrana is depicted here as a sister taxon of Naja sputatrix. Spitting cobras are marked with the snake symbol next to their species name.
3. Discussion
Phospholipase A2 (phosphatidylcholine 2-acylhydrolase) catalyzes the hydrolysis of phosphatidylcholine at the sn-2 ester bond to produce lysophospholipid and free fatty acids. We tested the activities of svPLA2 based on two different types of PLA2 assays [37]: an acidimetric assay that measured the release of proton from fatty acids during the hydrolysis of phosphate ester bond, and a colorimetric assay that measured the amount of chromogenic 4-nitro-3-hydroxybenzoic acid released by the cleavage of PLA2 at the ester bond between the octanol group of NOBA [38]. The activities tested on NOBA showed a better correlation with the PLA2 contents in the cobra venoms. The egg-yolk-based acidimetric assay probably contained less specific substrates (phospholipids, triglycerides) that could be targets of other lytic enzymes in the venoms, or atmospheric carbon dioxide could have interfered with the assay by dissolving into the suspension during the stirring process. On the whole, the enzymatic rates measured by both assays support that PLA2 enzymatic activities vary according to the svPLA2 composition in different cobra species. The enzymatic svPLA2 distribution is unique following a clustering trend among the four subgenera, and notably the remarkable lack of svPLA2 within the Uraeus subgenus.
The enzymatic activities measured in this study for the venoms of African spitting cobras (N. nubiae, N. nigricollis, N. mossambica, N. pallida, N. katiensis) and those from Asia (N. sumatrana, N. sputatrix) were high, consistent with the high abundances of svPLA2 reported previously in these venoms [14,26,34]. Some svPLA2 of Afronaja cobras were shown to be cytotoxic and to have dermonecrotic and myonecrotic activities [39,40]. Some African spitting cobra venoms possess coagulotoxic activity, which were inhibited by the use of phospholipase A2 inhibitor LY315920 in vitro [41]. In envenoming, bites from the African spitting cobras (Afronaja) are commonly associated with local tissue damages [42,43], which could be attributed to the svPLA2 and cytotoxins present abundantly in these venoms [26]. Previous studies also indicated that cobra svPLA2 enzymes worked synergistically with cytotoxins (cardiotoxins) to enhance venom toxicity [44,45] and the combination are probably responsible for causing venom ophthalmia (venom-induced conjunctivitis, chemosis, corneal erosions). Although the Asiatic non-spitters (N. naja, N. kaouthia, N. atra) also showed remarkably high PLA2 activities (correlated with the composition); their svPLA2 are mainly of acidic subtypes that lack lethal activity [46,47]. Two acidic PLA2 subtypes from Indian N. kaouthia venom were reported previously to exhibit anticoagulant activity [48]; this coagulopathic or hemotoxic effect, however, has not been commonly reported in clinical cobra envenoming. Similarly, the African forest cobra (N. melanoleuca, subgenus Boulengerina) is a non-spitting cobra species whose venom exhibited a strong PLA2 activity in this study; however, its svPLA2 had been shown to play no crucial role in the toxicity of the venom [25]. The pathophysiological role of these apparently non-toxic svPLA2 of cobras remain to be further elucidated, although these enzymes probably have more important ecological roles for the adaptation of the cobras to different niches. The lack of svPLA2 in the African non-spitting cobras of the subgenus Uraeus is a unique venom phenotype unveiled in this study. The finding implies that the svPLA2 is probably the least medically significant in the envenoming by this group of African cobras.
Snake venom PLA2s are isoenzyme products of multiple genes, and are further divided into distinct groups based on the differences in the number of disulfide bonds and the presence/absence of an N-terminal heptapeptide [49]. The svPLA2 of elapid snakes (including cobras) belongs to Group Ia PLA2 isoenzymes, which are homologous with the mammalian pancreatic PLA2 (Group Ib PLA2). Lynch [50] concluded that in Group I PLA2 enzymes, gene duplication and diversification occurred after speciation. This implies that the sequence homology and antigenicity of cobra svPLA2 are probably divergent between the different species of an individual subgenus. Hence, an antibody used against a PLA2 subtype of a specific cobra species may reveal variable immunoreactivity between different cobra venoms. From a practical point of view, the potentially diverse svPLA2 antigenicity and varying svPLA2 protein abundance among the different cobras pose challenges for antivenom production and usage in some regions. The phenomenon may variably affect the neutralizing efficacy of antivenoms that are produced and used for specific treatment against the envenoming by different cobra species in Asia and Africa [51,52].
From the phylogenetic perspective, the moderate-to-high enzymatic activity of svPLA2 is common in the Afronaja, Naja and Boulengerina lineages. Within the Asiatic Naja subgenus, the high PLA2 enzymatic activities along with the emergence of basic and neutral svPLA2 which are lethal, and the ability to ‘spit’ (to be exact, spray) venom, represents a more recently derived venom phenotype and defense trait unique to some of the Asiatic spitting cobras (N. sputatrix and N. sumatrana in this study). Although the Chinese/Formosan cobra (N. atra) and an unrecorded subpopulation of the Thai monocled cobra (N. kaouthia) were anecdotally reported to spit/spray venom on rare occasions, these two species are not considered accomplished spitters in this study due to their lack of formally documented specialized dental adaptations. The Asiatic spitting cobras, however, are known to be less accurate spitters compared with their African counterparts of the Afronaja subgenus that might have evolved a better defensive trait of venom spraying [53,54]. Some spitting cobras, e.g. N. sumatrana and N. sputatrix, produce lethal basic and/or neutral svPLA2 related to the pathophysiology of systemic envenoming and venom ophthalmia [2,3,14,55].
The subgenus Uraeus broadly encompasses non-spitting cobras of the N. haje complex living in the open areas of Africa. The extremely low svPLA2 activities and the negligible PLA2 enzyme content are unique venom phenotypes in the Uraeus cobras, reflecting a less critical role of svPLA2 in envenomation. The negligible svPLA2 content could be correlated with weak cytotoxicity and low dermonecrotic activity of the venoms. These venoms are generally more neurotoxic among the African cobras. The loss of svPLA2 functions in the lineage probably followed a decelerated mode of evolution or pseudogenization of the svPLA2 as the cobras diverged from the common ancestor shared by their closest kin, the forest cobra of Boulengerina, which retained or evolved a venom with a high svPLA2 enzymatic activity. The underlying cause, mechanism, and implication of the evolutionary event await further investigation.
4. Conclusions
The present study demonstrated the correlation of svPLA2 enzymatic activities with the enzyme protein abundances in Afro-Asian cobras of different subgenus. The dominant presence of enzymatically active svPLA2 is in line with the emergence of venom-spitting (spraying) behavior, once in the African Naja (Afronaja) spp., and once in the more derived Asiatic spitters of Naja (Naja) spp. The African non-spitting cobras of the subgenera Boulengerina and Uraeus diverged with a distinctive svPLA2 distribution: the forest-dwelling species (Boulengerina) continued to use a venom rich in svPLA2, whereas the open-land species (Uraeus) adapted to a venom that has little or negligible svPLA2. The lack of svPLA2 in the venom phenotype of African non-spitters from the Uraeus subgenus is hence striking. This provides an alternative view on the commonly perceived ubiquitous presence of svPLA2 in cobra venoms, and implies that the significance of svPLA2 in cobra envenoming varies among different species.
5. Materials and Methods
5.1. Consumables and Reagents
All chemicals and reagents used in the studies were of analytical grade. Hydrogen peroxide and sulfuric acid were supplied by J. T. Baker (Phillipsburg, NJ, USA). Goat anti-horse IgG-horseradish peroxidase (HRP) conjugate was supplied by Bio-Rad Laboratories (Hercules, CA, USA). 4-nitro-3-octanoyloxy benzoic acid (NOBA) was supplied by Santa Cruz Biotechnology (Santa Cruz, CA, USA). The 15 cobra venoms (Naja species) used in this study were sourced from various localities. Venoms of Naja naja (Pakistan), Naja sputatrix (Indonesia), Naja pallida (Kenya), Naja katiensis (Burkina Faso), Naja mossambica (South Africa), Naja nigricollis (Cameroon), Naja nubiae (Egypt), Naja melanoleuca (Cameroon), Naja haje (Egypt), Naja annulifera (Mozambique), Naja senegalensis (Mali), and Naja nivea (South Africa) were supplied by Latoxan (Valence, France). Venoms of Naja kaouthia (Thailand), Naja sumatrana (Malaysia), and Naja atra (Taiwan) were pooled samples from multiple adult snakes of the respective regions.
5.2. PLA2 Assay (Acidimetric Method)
Phospholipase A2 activities of the cobra venoms were determined by the acidimetric method as described by Tan and Tan [56]. The egg yolk substrate was prepared in a suspension constituted of chicken egg yolk, 18 mM calcium chloride, and 8.1 mM sodium deoxycholate in a 1:1:1 ratio. The substrate suspension pH was adjusted to 8.0 using sodium hydroxide. To ensure good mixing, the substrate suspension was continuously stirred at room temperature. One hundred microliters of venom solution (containing 10 µg venom) was added to 5 mL of the substrate suspension. The rate of pH decrease was recorded using a pH meter. A decrease of 1 pH unit of the egg yolk suspension corresponded to 133 µmol of fatty acids released. The enzyme activity is expressed as µmoles of fatty acids released/min/mg. The values are expressed as means ± S.E.M. of triplicates.
5.3. PLA2 Assay (Colorimetric Method)
Phospholipase A2 activities of the cobra venoms were assayed according to the colorimetric method as described by Holzer and Mackessy [35] and modified for use in a 96-well plate [57] using the synthetic chromogenic substrate (NOBA). Briefly, the standard assay mixture contained 200 µL of buffer (10 mM Tris-HCl, 10 mM CaCl2, and 100 mM NaCl, pH 8.0), 20 µL of substrate (3 mM), 20 µL of water, 20 µL of PLA2, and a final volume of 260 µL. Sample (10 µg in 20 µL) was then added to the mixture and incubated at 37 °C for 40 min, with the reading of absorbance at kinetic interval of 5 min over a period of 60 min at 425 nm using Tecan i-control™ infinite M1000Pro microplate reader (Männedorf, Switzerland). The enzyme activity, expressed as the initial velocity of the reaction, was determined by calculating the increase in absorbance after 20 min at 425 nm. The enzyme activity is expressed as mean ± S.E.M of triplicates.
5.4. Statistical Analyses
The PLA2 activities of both acidimetric and colorimetric assays are expressed as means ± S.E.M. (standard error of mean) of triplicates. The statistical differences between the venom samples in individual assay were analyzed by one-way analysis of variance (ANOVA) (p < 0.05) followed by Tukey’s post hoc test using SPSS version 20 software (IBM, Armonk, NY, USA, 2016). Lower-case letters are labelled at the top of the bars to indicate if there is a significant difference between the values. Sharing of any common lower-case letters between bars indicates there was no significant difference (p > 0.05) between the values charted, whereas bars that do not share any common lowercase letters have values that are significantly different from one another (p < 0.05). The relationship between the PLA2 activity and the relative abundance of PLA2 in each venom sample was also studied by linear regression analysis, using Graphpad Prism 6 software (San Diego, CA, USA). The strength of the association was interpreted by the coefficient of determination (R2) and the cut-off value at p < 0.01 indicates a highly significant regression between the PLA2 activity and the PLA2 protein abundance.
Acknowledgments
The authors are grateful to the University of Malaya for research funding and laboratory facilities provided.
Supplementary Materials
The following are available online at http://www.mdpi.com/2072-6651/11/2/116/s1, Figure S1: Time-dependent pH changes in acidimetric assay for the venoms of four subgenera of cobra: (A) Naja, (B) Afronaja, (C) Boulengerina, and (D) Uraeus. Hydrolysis of phospholipids by phospholipase A2 released fatty acids that reduced the suspension pH in a time-dependent manner. Figure S2: Time-dependent absorbance changes in colorimetric assay for the venoms of four subgenera of cobra: (A) Naja, (B) Afronaja, (C) Boulengerina, and (D) Uraeus. Changes in absorbance were due to the hydrolysis of the synthetic chromogenic substrate (NOBA), corresponding to the enzymatic activity of phospholipases A2 in the venoms. Table S1: Relative abundances of snake venom phospholipase A2 of 12 cobra species (Genus: Naja).
Author Contributions
Conceptualization, C.H.T. and K.Y.T.; Data curation, C.H.T.; Formal analysis, K.Y.W. and K.Y.T.; Funding acquisition, K.Y.T., C.H.T, and N.H.T.; Investigation, C.H.T., K.Y.T., K.Y.W., and T.S.N.; Methodology, K.Y.W. and C.H.T.; Project administration, C.H.T.; Resources, C.H.T., K.Y.T., and N.H.T.; Validation, C.H.T., K.Y.T., and T.S.N.; Writing–original draft, C.H.T.; Writing–review & editing: C.H.T., K.Y.W., K.Y.T., T.S.N., and N.H.T.
Funding
This research was funded by research grants GPF009C-2018 and BKP041-2017 from the University of Malaya.
Conflicts of Interest
The authors declare no conflict of interest.
Key Contribution
This study reveals a distinctive distribution of secretory phospholipases A2 (PLA2) in the venoms of 15 cobra species according to their subgenera. The lack of svPLA2 activity in the venom phenotype of the African non-spitting cobras (subgenus Uraeus) is particularly remarkable. This provides an alternative view on the commonly perceived ubiquitous presence and pathophysiological role of svPLA2 in cobra venoms.
References
- 1.De S.S. Physicochemical studies on hemolysin. J. Indian Chem. Soc. 1944;21:290. [Google Scholar]
- 2.Tan C.H., Tan N.H. Toxinology of Snake Venoms: The Malaysian Context. In: Gopalakrishnakone P., Inagaki H., Mukherjee A.K., Rahmy T.R., Vogel C.-W., editors. Snake Venoms. Springer; Dordrecht, The Netherlands: 2015. pp. 1–37. [Google Scholar]
- 3.Leong P.K., Fung S.Y., Tan C.H., Sim S.M., Tan N.H. Immunological cross-reactivity and neutralization of the principal toxins of Naja sumatrana and related cobra venoms by a Thai polyvalent antivenom (Neuro Polyvalent Snake Antivenom) Acta Tropica. 2015;149:86–93. doi: 10.1016/j.actatropica.2015.05.020. [DOI] [PubMed] [Google Scholar]
- 4.Oh A.M.F., Tan C.H., Tan K.Y., Quraishi N.A., Tan N.H. Venom proteome of Bungarus sindanus (Sind krait) from Pakistan and in vivo cross-neutralization of toxicity using an Indian polyvalent antivenom. J. Proteomics. 2019;193:243–254. doi: 10.1016/j.jprot.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 5.Tan K.Y., Liew J.L., Tan N.H., Quah E.S.H., Ismail A.K., Tan C.H. Unlocking the secrets of banded coral snake (Calliophis intestinalis, Malaysia): A venom with proteome novelty, low toxicity and distinct antigenicity. J. Proteomics. 2019;192:246–257. doi: 10.1016/j.jprot.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Tan C.H., Fung S.Y., Yap M.K., Leong P.K., Liew J.L., Tan N.H. Unveiling the elusive and exotic: Venomics of the Malayan blue coral snake (Calliophis bivirgata flaviceps) J. Proteomics. 2016;132:1–12. doi: 10.1016/j.jprot.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 7.Tsai I.-H. Snake Venom Phospholipase A2: Evolution and Diversity. In: Gopalakrishnakone P., Calvete J.J., editors. Venom Genomics and Proteomics: Venom Genomics and Proteomics. Springer; Dordrecht, The Netherlands: 2014. pp. 1–13. [Google Scholar]
- 8.Kini R.M. Excitement ahead: Structure, function and mechanism of snake venom phospholipase A2 enzymes. Toxicon. 2003;42:827–840. doi: 10.1016/j.toxicon.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Dixon R.W., Harris J.B. Nerve Terminal Damage by β-Bungarotoxin: Its Clinical Significance. Am. J. Pathol. 1999;154:447–455. doi: 10.1016/S0002-9440(10)65291-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oh A.M.F., Tan C.H., Ariaranee G.C., Quraishi N., Tan N.H. Venomics of Bungarus caeruleus (Indian krait): Comparable venom profiles, variable immunoreactivities among specimens from Sri Lanka, India and Pakistan. J. Proteomics. 2017;164:1–18. doi: 10.1016/j.jprot.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 11.Ramalingam K., Karthigayan S., Muralimanoharan S., Vijayalakshmi S., Thangavel B. Histopathological changes induced in mice after inramuscular and intraperitoneal injections of venom from spine-bellied sea snake, Lapemis curtus (Shaw, 1802) J. Pharmacol. Toxicol. 2007;2:307–318. [Google Scholar]
- 12.Brook G.A., Torres L.F., Gopalakrishnakone P., Duchen L.W. Effects of phospholipase of Enhydrina schistosa venom on nerve, motor end-plate and muscle of the mouse. Q. J. Exp. Physiol. 1987;72:571–591. doi: 10.1113/expphysiol.1987.sp003098. [DOI] [PubMed] [Google Scholar]
- 13.Huang M.Z., Wang Q.C., Liu G.F. Effects of an acidic phospholipase A2 purified from Ophiophagus hannah (king cobra) venom on rat heart. Toxicon. 1993;31:627–635. doi: 10.1016/0041-0101(93)90117-2. [DOI] [PubMed] [Google Scholar]
- 14.Tan N.H., Wong K.Y., Tan C.H. Venomics of Naja sputatrix, the Javan spitting cobra: A short neurotoxin-driven venom needing improved antivenom neutralization. J. Proteomics. 2017;157:18–32. doi: 10.1016/j.jprot.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 15.Tan N.-H. Isolation and preliminary characterisation of two toxic phospholipases A2 from the venom of the Malayan cobra (Naja naja sputatrix) BBA-Gen. Subj. 1982;719:599–605. doi: 10.1016/0304-4165(82)90250-1. [DOI] [PubMed] [Google Scholar]
- 16.Choudhury M., McCleary R.J.R., Kesherwani M., Kini R.M., Velmurugan D. Comparison of proteomic profiles of the venoms of two of the ‘Big Four’ snakes of India, the Indian cobra (Naja naja) and the common krait (Bungarus caeruleus), and analyses of their toxins. Toxicon. 2017;135:33–42. doi: 10.1016/j.toxicon.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Dutta S., Chanda A., Kalita B., Islam T., Patra A., Mukherjee A.K. Proteomic analysis to unravel the complex venom proteome of eastern India Naja naja: Correlation of venom composition with its biochemical and pharmacological properties. J. Proteomics. 2017;156:29–39. doi: 10.1016/j.jprot.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 18.Sintiprungrat K., Watcharatanyatip K., Senevirathne W.D., Chaisuriya P., Chokchaichamnankit D., Srisomsap C., Ratanabanangkoon K. A comparative study of venomics of Naja naja from India and Sri Lanka, clinical manifestations and antivenomics of an Indian polyspecific antivenom. J. Proteomics. 2016;132:131–143. doi: 10.1016/j.jprot.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 19.Wong K.Y., Tan C.H., Tan K.Y., Quraishi N.H., Tan N.H. Elucidating the biogeographical variation of the venom of Naja naja (spectacled cobra) from Pakistan through a venom-decomplexing proteomic study. J. Proteomics. 2018;175:156–173. doi: 10.1016/j.jprot.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 20.Liu C.-C., You C.-H., Wang P.-J., Yu J.-S., Huang G.-J., Liu C.-H., Hsieh W.-C., Lin C.-C. Analysis of the efficacy of Taiwanese freeze-dried neurotoxic antivenom against Naja kaouthia, Naja siamensis and Ophiophagus hannah through proteomics and animal model approaches. PLOS Negl. Trop. Dis. 2017;11:e0006138. doi: 10.1371/journal.pntd.0006138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang H.W., Liu B.S., Chien K.Y., Chiang L.C., Huang S.Y., Sung W.C., Wu W.G. Cobra venom proteome and glycome determined from individual snakes of Naja atra reveal medically important dynamic range and systematic geographic variation. J. Proteomics. 2015;128:92–104. doi: 10.1016/j.jprot.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 22.Tan K.Y., Tan C.H., Fung S.Y., Tan N.H. Venomics, lethality and neutralization of Naja kaouthia (monocled cobra) venoms from three different geographical regions of Southeast Asia. J. Proteomics. 2015;120:105–125. doi: 10.1016/j.jprot.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 23.Laustsen A.H., Gutierrez J.M., Lohse B., Rasmussen A.R., Fernandez J., Milbo C., Lomonte B. Snake venomics of monocled cobra (Naja kaouthia) and investigation of human IgG response against venom toxins. Toxicon. 2015;99:23–35. doi: 10.1016/j.toxicon.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Xu N., Zhao H.Y., Yin Y., Shen S.S., Shan L.L., Chen C.X., Zhang Y.X., Gao J.F., Ji X. Combined venomics, antivenomics and venom gland transcriptome analysis of the monocoled cobra (Naja kaouthia) from China. J. Proteomics. 2017;159:19–31. doi: 10.1016/j.jprot.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 25.Lauridsen L.P., Laustsen A.H., Lomonte B., Gutierrez J.M. Exploring the venom of the forest cobra snake: Toxicovenomics and antivenom profiling of Naja melanoleuca. J. Proteomics. 2017;150:98–108. doi: 10.1016/j.jprot.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 26.Petras D., Sanz L., Segura Á., Herrera M., Villalta M., Solano D., Vargas M., León G., Warrell D.A., Theakston R.D.G., et al. Snake venomics of African spitting cobras: Toxin composition and assessment of congeneric cross-reactivity of the Pan-African EchiTAb-Plus-ICP Antivenom by antivenomics and neutralization approaches. J. Proteome Res. 2011;10:1266–1280. doi: 10.1021/pr101040f. [DOI] [PubMed] [Google Scholar]
- 27.Hus K., Buczkowicz J., Petrilla V., Petrillová M., Łyskowski A., Legáth J., Bocian A. First Look at the venom of Naja ashei. Molecules. 2018;23:609. doi: 10.3390/molecules23030609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malih I., Ahmad rusmili M.R., Tee T.Y., Saile R., Ghalim N., Othman I. Proteomic analysis of Moroccan cobra Naja haje legionis venom using tandem mass spectrometry. J. Proteomics. 2014;96:240–252. doi: 10.1016/j.jprot.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 29.Mebs D. A comparative study of enzyme activities in snake venoms. Int. J. Biochem. 1970;1:335–342. doi: 10.1016/0020-711X(70)90077-7. [DOI] [Google Scholar]
- 30.Tan N.-H., Tan C.-S. A comparative study of cobra (Naja) venom enzymes. Comp. Biochem. Physiol. B Comp. Biochem. 1988;90:745–750. doi: 10.1016/0305-0491(88)90329-X. [DOI] [PubMed] [Google Scholar]
- 31.Uetz P., Freed P., Hošek J. The Reptile Database. [(accessed on 15 January 2019)]; Available online: http://www.reptile-database.org.
- 32.Wallach V., Wüster W., Broadley D.G. In praise of subgenera: taxonomic status of cobras of the genus Naja Laurenti (Serpentes: Elapidae) Zootaxa. 2009;2236:26–36. [Google Scholar]
- 33.Wüster W. Taxonomic changes and toxinology: Systematic revisions of the Asiatic cobras (Naja naja species complex) Toxicon. 1996;34:399–406. doi: 10.1016/0041-0101(95)00139-5. [DOI] [PubMed] [Google Scholar]
- 34.Yap M.K., Fung S.Y., Tan K.Y., Tan N.H. Proteomic characterization of venom of the medically important Southeast Asian Naja sumatrana (Equatorial spitting cobra) Acta Trop. 2014;133:15–25. doi: 10.1016/j.actatropica.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 35.Holzer M., Mackessy S.P. An aqueous endpoint assay of snake venom phospholipase A2. Toxicon. 1996;34:1149–1155. doi: 10.1016/0041-0101(96)00057-8. [DOI] [PubMed] [Google Scholar]
- 36.Bacha A.B., Alonazi M.A., Elshikh M.S., Karray A. A novel bactericidal homodimeric PLA2 group-I from Walterinnesia aegyptia venom. Int. J. Biol. Macromol. 2018;117:1140–1146. doi: 10.1016/j.ijbiomac.2018.06.024. [DOI] [PubMed] [Google Scholar]
- 37.Reynolds L.J., Washburn W.N., Deems R.A., Dennis E.A. Methods in Enzymology. Volume 197. Academic Press; Cambridge, MA, USA: 1991. Assay strategies and methods for phospholipases; pp. 3–23. [DOI] [PubMed] [Google Scholar]
- 38.Cho W., Kezdy F.J. Chromogenic substrates and assay of phospholipases A2. Methods Enzymol. 1991;197:75–79. doi: 10.1016/0076-6879(91)97134-k. [DOI] [PubMed] [Google Scholar]
- 39.Gowda T.V., Middlebrook J.L. Effects of myonecrotic snake venom phospholipase A2 toxins on cultured muscle cells. Toxicon. 1993;31:1267–1278. doi: 10.1016/0041-0101(93)90400-D. [DOI] [PubMed] [Google Scholar]
- 40.Mebs D. Myotoxic activity of phospholipases A2 isolated from cobra venoms: Neutralization by polyvalent antivenoms. Toxicon. 1986;24:1001–1008. doi: 10.1016/0041-0101(86)90006-1. [DOI] [PubMed] [Google Scholar]
- 41.Bittenbinder M.A., Zdenek C.N., Op den Brouw B., Youngman N.J., Dobson J.S., Naude A., Vonk F.J., Fry B.G. Coagulotoxic cobras: Clinical implications of strong anticoagulant actions of African spitting Naja Venoms that are not neutralised by antivenom but are by LY315920 (Varespladib) Toxins. 2018;10:516. doi: 10.3390/toxins10120516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Habib A.G., Gebi U.I., Onyemelukwe G.C. Snake bite in Nigeria. Afr. J. Med. Med. Sci. 2001;30:171–178. [PubMed] [Google Scholar]
- 43.WHO . Guidelines for the Prevention and Clinical Management of Snakebite in Africa. WHO Regional Office for Africa; Brazzaville, Congo: 2010. [Google Scholar]
- 44.Tan N.H., Armugam A. In vivo interactions between neurotoxin, cardiotoxin and phospholipases A2 isolated from Malayan cobra (Naja naja sputatrix) venom. Toxicon. 1990;28:1193–1198. doi: 10.1016/0041-0101(90)90119-R. [DOI] [PubMed] [Google Scholar]
- 45.Bougis P.E., Marchot P., Rochat H. In vivo synergy of cardiotoxin and phospholipase A2 from the elapid snake Naja mossambica mossambica. Toxicon. 1987;25:427–431. doi: 10.1016/0041-0101(87)90076-6. [DOI] [PubMed] [Google Scholar]
- 46.Tan K.Y., Tan C.H., Fung S.Y., Tan N.H. Neutralization of the principal toxins from the venoms of Thai Naja kaouthia and Malaysian Hydrophis schistosus: Insights into toxin-specific neutralization by two different antivenoms. Toxins. 2016;8:86. doi: 10.3390/toxins8040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong K.Y., Tan C.H., Tan N.H. Venom and purified toxins of the spectacled cobra (Naja naja) from Pakistan: Insights into toxicity and antivenom neutralization. Am. J. Trop. Med. Hyg. 2016;94:1392–1399. doi: 10.4269/ajtmh.15-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mukherjee A.K., Kalita B., Thakur R. Two Acidic, Anticoagulant PLA2 isoenzymes purified from the venom of monocled cobra Naja kaouthia exhibit different potency to inhibit thrombin and factor xa via phospholipids independent, non-enzymatic mechanism. PLoS ONE. 2014;9:e101334. doi: 10.1371/journal.pone.0101334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doley R., Zhou X., Kini R.M. Snake venom phospholipase A2 enzymes. CRC Press; Boca Raton, FL, USA: 2009. [Google Scholar]
- 50.Lynch V.J. Inventing an arsenal: Adaptive evolution and neofunctionalization of snake venom phospholipase A2 genes. BMC Evol. Biol. 2007;7:2. doi: 10.1186/1471-2148-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ratanabanangkoon K., Tan K.Y., Eursakun S., Tan C.H., Simsiriwong P., Pamornsakda T., Wiriyarat W., Klinpayom C., Tan N.H. A simple and novel strategy for the production of a pan-specific antiserum against Elapid snakes of Asia. PLoS Negl. Trop. Dis. 2016;10:e0004565. doi: 10.1371/journal.pntd.0004565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan C.H., Liew J.L., Tan K.Y., Tan N.H. Assessing SABU (Serum Anti Bisa Ular), the sole Indonesian antivenom: A proteomic analysis and neutralization efficacy study. Sci. Rep. 2016;6:37299. doi: 10.1038/srep37299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wüster W., Thorpe R.S. Dentitional phenomena in cobras revisited: Spitting and fang structure in the Asiatic species of Naja (Serpentes: Elapidae) Herpetologica. 1992;48:424–434. [Google Scholar]
- 54.Panagides N., Jackson T.N., Ikonomopoulou M.P., Arbuckle K., Pretzler R., Yang D.C., Ali S.A., Koludarov I., Dobson J., Sanker B., et al. How the cobra got its flesh-eating venom: Cytotoxicity as a defensive innovation and its co-evolution with hooding, aposematic marking, and spitting. Toxins. 2017;9:103. doi: 10.3390/toxins9030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chong H.P., Tan K.Y., Tan N.H., Tan C.H. Exploring the diversity and novelty of toxin genes in Naja sumatrana, the Equatorial spitting cobra from Malaysia through de novo venom-gland transcriptomics. Toxins. 2019;11:104. doi: 10.3390/toxins11020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan N.-H., Tan C.-S. Acidimetric assay for phospholipase A using egg yolk suspension as substrate. Anal. Biochem. 1988;170:282–288. doi: 10.1016/0003-2697(88)90632-X. [DOI] [PubMed] [Google Scholar]
- 57.Calgarotto A.K., Damico D.C.S., Ponce-Soto L.A., Baldasso P.A., Da Silva S.L., Souza G.H.M.F., Eberlin M.N., Marangoni S. Biological and biochemical characterization of new basic phospholipase A2 BmTX-I isolated from Bothrops moojeni snake venom. Toxicon. 2008;51:1509–1519. doi: 10.1016/j.toxicon.2008.03.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.