Abstract
Dioecism has always been an issue in many plant species with its numerous disadvantages, especially in woody trees such as date palms. As one of the most important crops in the Middle Eastern countries, researchers are having problems identifying of sex of the plant in its early stages of development. Hence, proper population stands in the male: female ratio for maintenance is almost impossible in the field for better production. In this study, sex determination of date palm (Phoenix dactilyfera L.) were identified in regions of the Y chromosome (Date-SRY) gene, the pivotal gene that initiates sex determination, using a new technique and thus an economically desirable objective, which will significantly impact profits in seed based cultivations. Partial sequences of the Date-SRY were taken and amplified by nested polymerase chain reaction (PCR). According to the results, the exact sex of date palm was identified in all the tested plants, while amplified regions of the Date-SRY gene closely matched with the human and papaya sequences. In addition, a primer pair was designed to amplify the sequences of the SRY-date gene with confidence that it will identify male date palms. These primer sequences include SRY-date Forward 5′- cggccctctaagtatctgtgcgcaacg-3′ (SRY-date F) and the SRY-date Reverse 5′- gtttgcacttcgaagcagag-3′ (SRY-date R). The complete sequence of the DNA has been registered and deposited in GenBank (BankIt1598036 DPSRY1 KC577225 thenKJ873056).
Keywords: Date palm, Dioecious plants, Female, Male, Sex determination, SRY gene sequence
1. Introduction
Sex determination was usually identified with the physical separation of male and female plants from each other, and its mechanism in different crop species were documented in many reviews (Juarez and Banks, 1998, Geber et al., 1999, Ainsworth, 2000, Matsunaga and Kawano, 2001, Negrutiu et al., 2001, Barrett, 2002, Charlesworth, 2002, Heikrujam et al., 2015). It also plays a crucial role in the life cycle of plants that reproduce through sexual means. The system underlying sex determination in plants are largely unexplored, although recent studies of sex determination in many plant species - from ferns to maize - have been fruitful in identifying the diversity of genetic and epigenetic factors that are involved in determining the sex of the flower or individual. Furthermore, sex determination in non-crop plants is controlled by environmental factors; it comes with a pivotal role in population studies, the observation of the male and female ratios in population and the exploration of factors that manipulate sex distribution.
Due to its dioecious nature, breeding in date palm (Phoenix dactylifera) has not been practiced because the sex of the plant cannot be known until it reaches its reproductive stage (Bendiab et al., 1993). Until 1996 when the sex chromosomes were not distinguishable in plants, identification of differences between the heterochromatin region of chromosomes by chromomycin A3 staining of the root chromosome from male and female cells was the tool for male and female plant identification (Siljak-Yakovlev et al., 1996).
The sex of dioecious plants is usually tedious to identify, especially before flowering and during the early stages of its development. On the other hand, the identification of male and female flowers in fruit and seed bearing plants is becoming a priority with breeders, especially prior to propagation, since this would result in crop improvement and increased profits (Sarkar, et al., 2017). There are also a number of plant species where female flower representation is more important than males. These include Borassus flabellifer (George et al., 2007), Carica papaya (Parasnis et al., 2000), Actinidia deliciosa (Shirkot et al., 2002), Eucommia ulmoides (Xu et al., 2004), Myristica fragrans (Shibu et al., 2000), Piper longum (Manoj et al., 2005–2008), Pistacia vera (Hormaza et al., 1994), P. dactylifera (Younis et al., 2008), Hippophae rhamnoides (Persson and Nybom, 1998, Sharma et al., 2010), and Simmondsia chinensis (Agrawal et al., 2007, Sharma et al., 2008)
As one of the most important plantation crop in the palm family (Arecaceae), the date palm (Phoenix dactylifera L.) is an ornamental, dioecious, perennial, and monocotyledonous plant known for its distinct fruit. In dry tropical regions around the world such as western Asia and Africa, it plays a major role in socioeconomic conditions in terms of shelter, food, and fiber (Morton, 1987, Adawy et al., 2004, El Hadrami and El Hadrami, 2009, Jain et al., 2011, Srivashtav et al., 2013). However in Arab nations, it accounts for almost 60 percent of its production with more than 800 varieties (El-Juhany, 2010, Al-Abdoulhadi et al., 2011).
For decades of extensive date palm research, there have been numerous attempts to identify the sex of the plant, but most were unsuccessful, resulting to unpredictable number of female plants in the field that leads to uncertainty in the production potential of the crop. The number of female plants must be more than the male plants. Consequently, it is not possible to know the sex of the plant until it flowers and reaches the reproductive age spanning from five to ten years (Bendiab et al., 1993, Juarez and Banks, 1998, El Hadrami and El Hadrami, 2009). Breeding of the plant is also challenging due to the long phases of juvenility and dioecism. Because of the presence of low genetic diversity, methodologies that are simple and perfect for differentiation of the gender is absent even before the first flowering (Aberlenc-Bertossi et al., 2011). Thus, determining the sex of the date palm plant is necessary since the presence of a reliable method would significantly increase profitability in terms of seed based cultivation (Al-Khalifah et al., 2012).
There have been extensive efforts to understand the basis for sex identification and development using a wide range of methodologies that include biochemical and molecular markers that tend to be beneficial in the identification of male and female dioecious crop plants such as the date palm (Elmeer & Mattat, 2012; Al-Mahmoud et al., 2012; Heikrujam et al., 2015, Al-Ameri et al., 2016). In recent years, the Inter Simple Sequence Repeat (ISSR) marker is gaining usage that offers several advantages over other dominant markers which are widely utilized for plant genome analysis (Abdel-Mawgood, 2012, Gaafar et al., 2014, Maryam et al., 2016) but rarely used for sex identification in dioecious plants (Milewicz and Sawicki, 2011, Aleksandrov et al., 2011, Nanda et al., 2013, Heikrujam et al., 2014). Finally, research paper by Atia and Adawy (2015) show a novel set of sex-specific PCR-based markers reveals new and the most popular markers Hypothesis of sex determination in Date Palm.
Moreover, a wide range of popular molecular markers have also been used to show the differences between male and female plants that include the RAPD (Harvey et al., 1997, Shirkot et al., 2002, George et al., 2007, Sharma et al., 2010), AFLPs (Vos et al., 1995, Renganayaki et al., 2005, Danilova and Karlov, 2006, Wang et al., 2011), ISSR-SCAR (Korpelainen et al., 2008), RAPD_SCAR (Manoj et al., 2005–2008, Xu et al., 2004, Dhawan et al., 2013), ISSR and RAPD (Younis et. al., 2008), and SSR markers (Cherif et al.,2013).
Among the techniques, the most commonly used method for sex determination in plants is the RAPD-PCR. In date palm, Younis et al. (2008) used the ISSR and RAPD markers to identify sex-specific DNA markers for the selection and identification of good male pollinators for utilization in breeding programs, thus increasing the yield and improving some quality traits of fruits. Thirty RAPD were screened and 20 ISSR primers on seven date palm cultivars (four females and three males) collected from the Com Ambo farm at the Aswan Horticultural Services for Ministry of Agriculture in Egypt and reported two male-specific and three female-specific RAPD markers and five male-specific ISSR markers. However, the utility of these markers were not applicable when tested on other cultivars collected from other locations (Dhawan et al., 2013).
Similarly, the researchers screened 100 RAPD and 104 ISSR primers on 25 female and 20 male date palm plants and reported one RAPD primer that produced a 1000-bp band that was specific only to male plants. With less reproducibility characteristics of RAPD primers and after cloning and sequencing of this DNA band, a SCAR primer pair was designed and tested on 45 date palm plants. The report suggested an amplification of one 406-bp band that was specific to both female and male plants, and one 354-bp band that was specific to male plants only. Although the SCAR primer pairs show positive correlation with male-specificity, it does not confirm the location of the primer pair on date palm sex chromosome, specifically male-specific chromosome. To identify sex-linked markers, Cherifet al. (2013) studied 52 male and 55 female geographically diverged date palm genotypes using three microsatellite (SSR) markers and reported three genetically linked loci that are heterozygous only in males and thereby confirming the existence of an XY chromosomal system with a non-recombining XY-like region in the date palm genome. The existence of an XY chromosomal system in date palm suggests the possibility of discovering Y chromosome-specific DNA marker for identification of male plants.
The sex-determining region (SRY) gene, crucial in mammalian sex determination, was identified in recent years and has been extensively studied. An individual with a functional SRY is considered male, while an individual without it is female. It is a potential candidate for the mammalian testis-determining factor and a tool for gender identification in the early stages of fetus development, especially when ultrasound method is not suitable. SRY appears to be part of a family with several autosomal representatives. Generally, the X and Y chromosomes determine an individual’s sex. A female has two X chromosomes designated as XX, while a male comes with one X and one Y, designated as XY. However, there are cases where a female had XY chromosomes and a male with an XX blueprint. After the analysis of these individuals, it was revealed that some of the genes involved in sex determination included SRY as an important factor in the formation of testis, while Human SRY is located on the Y chromosome.
The SRY molecular factor is an attractive approach for sex determination and has already been successfully tested in mammals and in papaya. One of the existing molecular methods for sex determination is the polymerase chain reaction (PCR). Using two oligonucleotide primers that cross-hybrid into opposite strands and flank the region of interest in the target DNA, PCR is an in vitro and sterile method for the enzymatic synthesis of specific DNA sequences. The PCR-based sex determination identifies the presence (male) or absence (female) of the SRY gene. Although PCR-based sex determination of date palm is unattractive approach, suitable forward and reverse primers specific to Y chromosome were not available for identification in this species. The goal of this study is to test PCR amplification of SRY as a method for determining sex in different varieties of date palm trees. A new pair of primers was also designed with the capacity to target a section of the SRY gene; this resulted in the amplification of a male-specific PCR fragment using routine PCR procedure. Conventional PCR technology was used to select the male plant, as this will guarantee explicit and fast sex identification process. The legitimacy and its appropriateness, coupled with the specificity of these primers were confirmed with DNA from a number of known samples of date palms.
2. Materials and methods
2.1. Plant material
2.1.1. Isolation of genomic DNA
For DNA isolation, young leaf tissues from randomly selected female and male plants (3–5 plants) of the selected date palm cultivars Burhi, Khalas and Sukkari were collected from the Al-Ahsa Oasis Farm, Kingdom of Saudi Arabia. Their Genomic DNA was then isolated on a mini-prep scale using CTAB (cetyl trimethyl ammonium bromide) method as reported by Murray and Thompson (1980). The quantity and quality of the nucleic acid were determined by measuring the absorbance at 260 nm and 280 nm using a Spectrophotometer. DNA concentration was quantified using the formula A260 = 50 µg/ml, while the purity of the DNA was determined by calculating the ratio of A260/A280 for each sample.
2.1.2. Polymerase Chain Reaction (PCR) Materials
DreamTaq Polymerase, deoxynucleotide triphosphate (dNTP) and convergent primers used in this experiment were purchased from Fermentas, Thermo Fisher Scientific. Conventional method of PCR was used for amplification of the DNA fragment. Date palm male-specific PCR primers were designed based on the conserved SRY sequences of human and/or papaya Y chromosome using the Primer-BLAST tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). The primer pair(s) was tested on DNA of male and female date palm genotypes using PCR amplification. Each PCR reactions was carried out in a 50 μl of the reaction mix containing 1 × PCR buffer, 200 μM dNTP mix, 0.2 μM each of forward/reverse primer, 1.5 mM MgCl2, 1.5 U Taq DNA polymerase, and 100 ng of template DNA. The reaction conditions for PCR involved initial denaturation at 94 °C for 5 min followed by 30 cycles of denaturation at 940 °C for 30 s, annealing at 52 °C for 30 s and extension at 72 °C for 60 s, and a final extension at 72 °C for 5 min. To amplify male-specific SRY DNA fragment, SRY-150F (5-AGC AGT CAG GGA GGC AGATCA 3 V) was used as forward primer and SRY-245R was used as reverse primer as described by Lo et al. (1998).
The annealing temperature was standardized according to the specific Tm of the primer pair. After amplification, an aliquot of the reaction mixture was loaded onto a 0.8% agarose gel containing in 1X TBE buffer and checked by staining with ethidium bromide, and visualized under ultraviolet (UV) light. The gel picture was captured using Bio-Rad documentation system.
2.2. Isolation of SRY gene
2.2.1. Restriction digestion and End-filling with klenow
The genomic DNA of the date plant was digested overnight at 37 °C using 40 units of enzyme, making a final volume of 100 µl as described by Reddy et al. (2002). Enzymes used were BamHI and BglII Sou3A, while the digested DNA was partially end-filled by dGTP and dATP. The end-filling reaction mixture contained dNTPs, 10X reaction buffer, klenow (1 µl) in reaction volume of 25 µl and was incubated at 37 °C for 1 h. The purified DNA was extracted as described by Mohei El-Din, 2002, Mahmoud et al., 2009.
2.2.2. Adaptor annealing and ligation
The adaptors were generated according to the methods developed by Mohei El-Din, 2002, Mahmoud et al., 2009. For this purpose, oligonucleotides used were: 03301954 solliman-1 5′-AATACGACTCACTATAGGGCGGCCGCCCGGGC-3′ and solliman-2 (5′-CACTATACCCGCCGGCGGGCCCGCT-3′.
Oligonucleotides solliman-1 and solliman-2 (synthesized by Macrogen, Korea) were re-suspended in sterile double-distilled water at a concentration of 100 pM/µL. A volume of 20 µL for each adaptor was piped into a 0.5 mL Eppendorf microfuge tube and overlaid with mineral oil. The adaptors were then heated at 99 °C for 4 min in a beaker of water. The heat was removed, and the solution was allowed to cool for 1 h at room temperature. The annealed adaptors were decanted from under oil and stored at −20 °C.
After which, 10 µL of the genomic restriction digest was ligated to 1 µL of the annealed adaptors with 2 µL of T4 DNA ligase buffer and 2 µL of T4 DNA ligase (5units/µL) in a 20 µL reaction. The ligation was incubated overnight at 12 °C and heat inactivated at 65 °C for 10 min. Also, 180 µL volume of TE (pH 8) was added to the ligation mix called the adaptor library. Excess primer-adapter was removed by purification through QIAquik Columns (Qiagen).
2.3. Primary PCR amplification
The adaptor-ligated genomic DNA was diluted and used for the first PCR amplification in a 50 µl reaction volume. For amplification, the DNA was first denatured once for five minutes at 94 °C, followed by 35-cycles of PCR amplification. For each cycle, the DNA was denatured for 40 s at 94 °C, annealed for 40 s at 50 °C and elongated for two min at 72 °C. Adaptor primer T7 sequences are 5′- AATACGACTCACTATAGGGC-3′ and SRY-2 R (gene-specific primer, Date-SRY) was used for amplification. The sequence of the SRY-1 R primer is given: 5′- GGGCTGTAAGTTATCGTAAAAGGAGC-3′.SRY_2R; 5′-CCTAGCTGGTCACGTTGACCTTTTGTCC-3′. Genomic DNA amplified with T7 alone served as negative control as methods developed by Mohei El-Din, 2002, Mahmoud et al., 2009.
2.4. Secondary amplification
Amplified products (10 µl each) were electrophoresed on a 0.8% agarose gel and processed to check for amplification. The remaining PCR products of the primary amplification were used as template after purification by phenol extraction. After precipitation, the DNA was re-suspended in 20 µl T10E1. Various dilutions of the excised band were PCR amplified using T7 and SRY-2R primers. The same reaction composition and cycle parameters used were as described for the primary PCR amplification.
2.5. DNA sequencing
Amplified DNA fragments of different sizes resulted from primary and secondary amplifications were sequenced by an automated sequencing machine such as Applied Biosystems 3730xl and 9 ABI 3700 in Macrogen Company, Korea.
2.6. Analysis of nucleotide sequence by homology and structural comparison of SRY-date to other SRY genes
Most of the sequence (DNA) analyses were performed using CLC Vector program and Genbank database. Homology searches were done using FASTA, while multiple sequence alignment was done using CLUSTALW (Hobohm and Sander, 1995, Stultz et al., 1993).
3. Results
As a developmental process, the determination of sex especially in crop plants is of fundamental importance due to its economic implication. The present work reports on the molecular techniques used for determination of sex (male and female) in date palm cultivars. The emphases were on the strategy, which facilitate confirmation, and identification of the male and female plants.
3.1. - Development of a strategy to isolate date SRY gene
As a dioecious plant, date palm plant harbors XY sex chromosomes, thus has the potential to serve as an important model plant for sex determination. For the first time, the researchers investigated the existence of SRY-related sequences in Date palm cultivars. To isolate the date-SRY gene, PCR technique was used with the primers that were used to identify homologue of the conserved motif of the SRY gene from human. The analysts investigated the conserved sequences between forward and reverse primers of SRY for possible primers that produced smaller PCR products with the SRY primers in different regions of human and papaya genome. Henceforth, the investigators decided to use the same reverse primer SRY-1 R sequence as used in papaya (Yu et al., 2007, Yu et al., 2008). For forward primer, however, the researchers have chosen the adapter sequences as described in material and methods to the upstream from the reverse primers as mentioned by Mohei El-Din, 2002, Mahmoud et al., 2009.
The probers tested the amplification protocol using two annealing temperatures 48 °C and 55 °C. For male variety of date palm, no differences in amplification products between the two temperatures were observed (data not shown). In addition, the present protocol also utilized the use of a primer that enriched specific template prior to nested PCR. Also, one of the adopter strands was blocked at the 3′ end by attaching an amine group. Using this method, a number of unknown 3′ and 5′ regions of known SRY genes were isolated earlier from humans and papaya (Yu et al., 2007, Yu et al., 2008).
3.2. - Isolation of date palm genomic DNA and amplification of putative SRY-gene using PCR
The genomic DNAs isolated from both male and female date palm plants were measured for their quality and quantity. The ratio of A260/A280 for each sample was around 1.8 and quantity of DNA among 10 samples varied from 320 µg/ml to 805 µg/ml (data not shown).
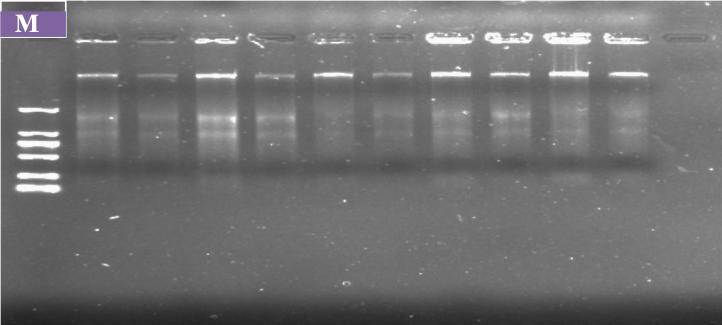
Quality undigested date palm genomic DNA, including Burhi, Khalas, and Sukkari were isolated from females and males of different cultivars grown in Al-Ahsa Oasis, King Saudi Arabia. The researchers ran it in 0.8% agarose gel in 1X TBE buffer stained with ethidium bromide as shown in Fig. 1. From visual observation of the DNA in the gel, quality and quantity of DNA looks great. DNA samples were digested with BamHI, BglII, or Sou3 A restriction enzymes (data not shown). The digested ends were partially filled with A and G nucleotides to prevent self-ligation of the DNA fragments. These partially filled genomic DNA fragments were ligated with adapter-primers (ADOP-32 and ADOP-27) as the adapter-primers were designed to anneal with partially filled genomic DNA. The ADOP-32 primer contained sequences of the T7 primer.
Fig. 1.
A Photograph of a gel showing DNA isolated from female and male plants of different cultivars of date palms growing in Al-Ahsa Oasis, KSA (Burhi, Khalas and Sukkari).Legend: f = female plant and m = male plant.
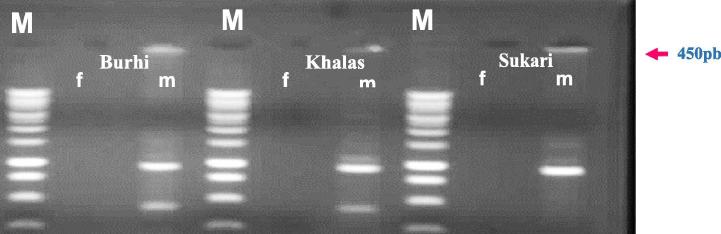
The 3′-end of the ADOP-27 primer was blocked to prevent extension during PCR amplification (Mohei El-Din, 2002, Mahmoud et al., 2009). The ligated DNA served as a template, while the SRY sequences were PCR-amplified with T7 and SRY-1R primers. Along this line, amplified DNA fragments of different sizes were obtained. The amplified DNA was used as template for the second PCR reaction with the T7 and SRY-2R primers (Fig. 2A). As an example, a band of amplified DNA of approximately 0.45 kb was observed in male plants of all three varieties of date palm tested in this investigation. The amplified DNA was purified and cloned into the pGEM-T vector to create pGEMT-SRY and verified through PCR method (Fig. 2B) (see Fig. 3A, Fig. 3B).
Fig. 2A.
PCR amplification while screening of the presence of SRY gene from different varieties of Date palm (Burhi, Khalas and Sukkari) using specific primers Sry1 F + Sry1 R lane M DNA 1 KB Marker.Legend: f = female plant and m = male plant.
Fig. 2B.
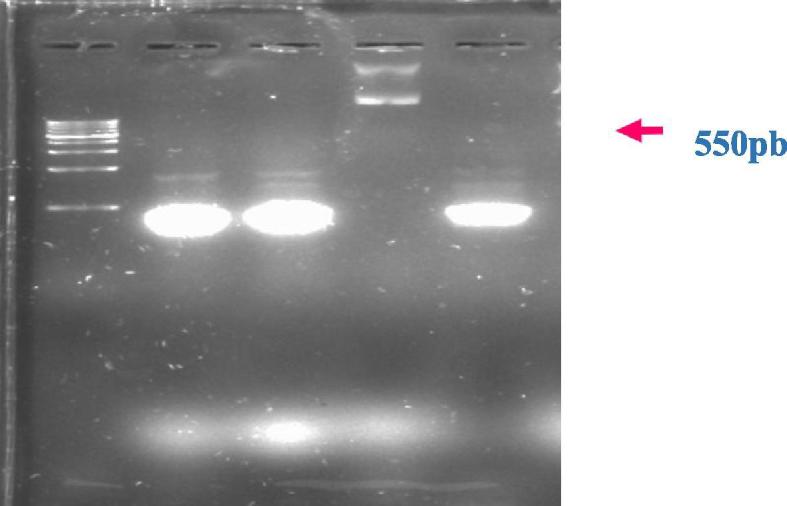
PCR Amplification products for clone of SRY full fragment after cloned in pGEMT vector (lane 2, 3 and 5 for Burhi, Khalas and Sukkari) and lane 4 for pGEMT vector only as control and lane M DNA 1 KB Marker.
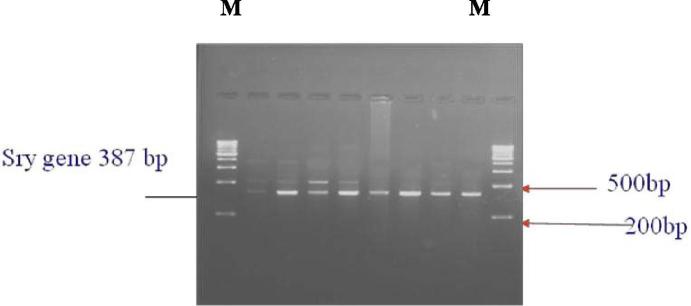
Fig. 3A.
PCR amplification for screening of the presence of SRY (387 bp) gene in lane (2–9) date palm male plants with specific primer for gene only SRY-date-F and SRY-date-R primers lane (1 and 10) M DNA 1 KB Marker.
Fig. 3B.
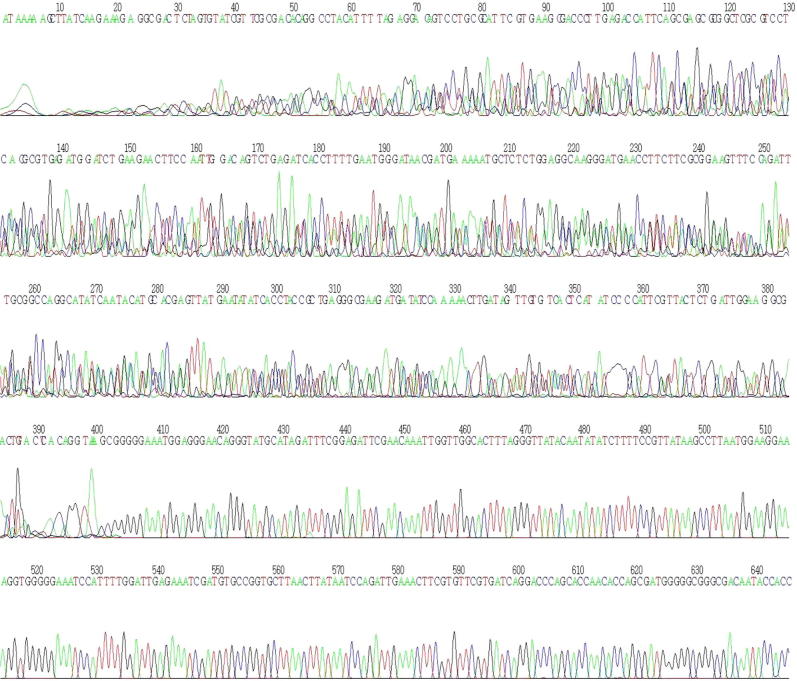
Reverse sequences of SRY gene, a sex determination gene from date palm (SRY-Date). The nucleotide sequence of the SRY full-length gene after isolated from Date palm and sequencing using reverse primer.
3.3. Sequencing of putative date palm SRY
Several primary amplified DNA fragments of different sizes were sequenced and the nucleotide sequence data of a 914-bp fragment is shown below.
| 1 | ggggggatac cagttaccca aat cggccctctaagtatctgtgcgcaacg gccagacatc |
|---|---|
| 61 | tttagaggcc acttctgcga ttcttgaagc gacccttgag agcattcatc gagtggtctc |
| 121 | cgtcctcacg cgtggatggc tctcgagatc ccccaatgcg acactctgag atcacctatc |
| 181 | gactgggata ccggtgataa aatgctcact ggagccaagg gatggacctt cttcccggag |
| 241 | gttcagagat ttcaggccat gcacataaat aaatgcacga gttataaata tccacctcct |
| 301 | ctgaaggcga aaatactgcc gaagacttgc agtttgcttc ccgctcatcc cgcttcgtta |
| 361 | ctctgcttcg aagtgcaaac tggacaacag gtaagcgggg gaaatggagg gaacagggta |
| 421 | tgcatagatt tcggagattc gaacaaattg gttggcactt tagggttata caatatatct |
| 481 | tttccgttat aagccttaat ggaaggaaag gtgggggaaa tccattttgg attgagaaat |
| 541 | cgatgtgccg gtgcttaact tataatccag attgaaactt cgtgttcgtg atcaggaccc |
| 601 | agcaccaaca ccagcgatgg gggcgggcga caataccacc atagcggttc tagtagcagc |
| 661 | aaaaaatagt ggcataacaa acataagtag tagaacatga gtagcagaag tttacccggc |
| 721 | gcaataaccg gtggcactag tagcagaagc ataggctttt tgggtaccgg gcagaggcaa |
| 781 | ggcatcgaat ccctcatcaa gcccatccac atcaatagct cactacccac caacatcgaa |
| 841 | ataaggggat gagtcgactg gcgaatcagc tgcaataatt ggactcgacc agtgcaactg |
| 901 | gaacaacagg taaa |
3.4. - Putative SRY sequence analysis
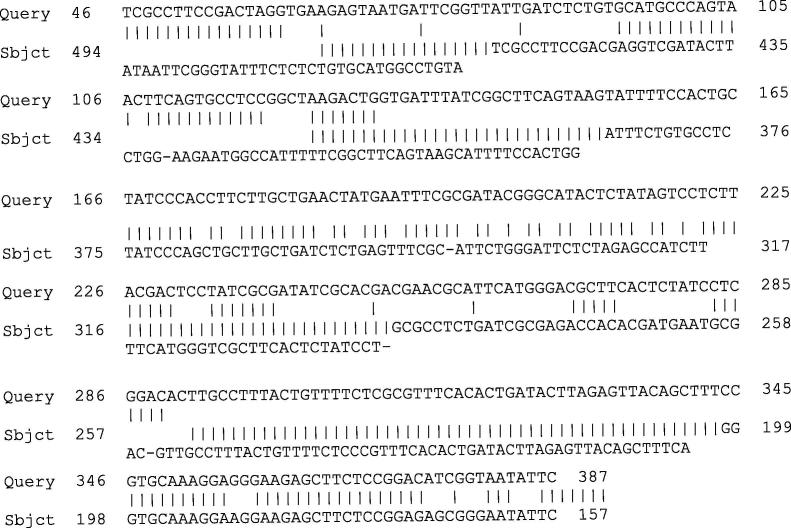
The SRY fragment was 355-bp in length as shown in Fig. 7. GenScan and CLC analysis showed that DNA fragment had a 3′end homology to 3′ends of human and papaya SRY. The full-length open reading frame (ORF) was 355 bp in length. The SRY fragment assemblies and identification of sequence were carried out with CLC Genomics Workbench (CLC bio). BLAST results showed that the predicted ORF had 84% similarity to the published sequence amplified with human primers, indicating a high degree of similarity with human SRY (Fig. 6). The full-length sequence of SRY was obtained with a modified genomic “walking” method, which combined vectorette and suppression PCR “walking” and amplification. Full-length SRY was 355-bp includes the putative transcription start site and open reading frame. The open reading frame encoded for 182 amino acids and had 86% similarity to the published sequence amplified with papaya primers (protein_id = “AAB58342.1″), indicating a high degree of similarity with papaya SRY (Fig. 5). The deduced DNA sequence also showed extensive homology with human SRY sequences (Fig. 6). The template is likely to be amplified in linear using this method, leading to a significant amount of noise. All the protocols of Reddy et al. (2002) was used in this study involving restriction digestion of genomic DNA followed by partial filling to prevent self-ligation between fragments. In addition, the present protocol also employed biotinylated primers to enrich for the specific template prior to performing nested PCR. One of the adopter strands was also blocked at the 3′ end by attaching an amine group (Reddy et al., 2002).
Fig. 7.
Forward sequences of SRY gene and comparison of the DNA sequences of SRY- primer with Phoenix dactylifera mitochondrion in complete genome Sequence ID: (Accession number: gb|JN375330.1|).
Fig. 6.
Comparison of the DNA sequences of SRY-date with SRY genes from human (Homo sapiens clone 15 SRY (SRY) gene, complete cds, Accession number: gb|JQ811918.1).
Fig. 5.
Comparison of the DNA sequences of SRY-date with SRY genes from papaya (Accession number: gb|AF000024.1|CPAF000024). The nucleotide sequence of the cloned SRY Sequence Alignment Editor.
Computer-aided sequence to identify date-SRY. A partial sequence of the SRY gene is shown in Fig. 5, Fig. 6. Computer-aided sequence analysis of Date-SRY revealed the ORF of SRY. Most of the sequence (DNA) analyses were performed with the CLC Vector program and Genbank database. Fig. 5 shows the homology searches with forward primers for SRY. Although there was a very low alignment, a very high degree of similarity was found when compared with P. dactylifera L., complete genome Sequence ID: (Accession number: gb|JN375330.1|). Homology searches were performed with BLAST. Multiple sequence alignments were performed with CLUSTALW.
3.5. - Deposition of isolated SRY DNA fragments into GenBank
Similar approach was adopted to isolate about 355-bp lengths DNA fragment of Date-SRY gene by walking the genomic DNA using primers SRY 1 R, SRY 2 R and T7 primer. The complete sequence of the DNA was presented in Fig. 3A, Fig. 3B and Fig. 4. The sequence information is also deposited in a public data base GenBank (BankIt1598036 DPSRY1 KC577225).
Fig. 4.
Forward sequences of SRY gene, a sex determination gene from date palm (SRY-Date).
A PCR primer pair according to this experiment and invention consists of: a forward PCR primer consisting of 20 consecutive nucleotides of the sequence of nucleotides flanking, in 5′, and a reverse PCR primer comprising, or consisting of, between 20 and 25 consecutive nucleotides of the sequence complementary to the sequence of at most nucleotides flanking, in 3′. The sequence of the SRY-F primer is SRY_F 5′- AAACTTGATAGTTGTGTCACTCAT-3′. The sequence of the SRY-1 R primer is 5′- GGGCTGTAAGTTATCGTAAAAGGAGC-3′ and SRY_2 R is 5′- CTAGCTGGTCACGTTGACCTTTTGTCC-3.
The amplified DNA fragment was sequenced using reverse primer (Figure 3) and forward primer (Fig. 4). Using the vector-specific T7 and SP6 primers, the cloning of the SRY gene was further confirmed with subsequent sequencing (Figs. 3 and 4).
The sex was identified as male after additional PCR reactions using primers specific for the Y chromosome (SRY and universal primer). The SRY marker has been previously shown to yield, a 350-bp to 400-bp PCR products that overlaps in size with human (Drobnić 2006) and papaya (Yu et al., 2007, Yu et al., 2008) sequences. Amplification of samples from the male plants had only one band of 355-bp, (Fig. 7). The sequence of the SRY forward primer is determined as SRY-date-F 5′- cggccctctaagtatctgtgcgcaacg - 3′. The sequence of the SRY-date-R reverse primer is determined as SRY-date R 5′- gtttgcacttcgaagcagag -3′.
A genomic fragment of approximately 355-bp was amplified from three date palm cultivars using specific primers identified from the human and papaya SRY sequence information.
The sequence was deposited in Genbank with the Accession number (BankIt1598036 DPSRY1 KC577225 and final released in GenBank: KJ873056.1).
/translation of the sry gene = “MALEIPQCDTLRSPIDWDTGDKMLTGAKGWTFFPEVQRFQAMHI
NKCTSYKYPPPLKAKILPKTCSLLPAHPASLLCFEVQTGQQVSGGNGGNRVCIDFGDS
NKLVGTLGLYNISFPL“
4. Discussion
In order to keep the desirable ratio of male and female plants in the field for better production, the identification of the sex of the plants will be economically beneficial to the date palm farmers. Early sex determination may prove useful to date palm breeding and will assist research programs for implementation of breeding programs of the palm trees.
A very important question being asked by Plant breeders and molecular biologists about the likely genes, those play a role in the determination of sex of Date palm? Our present results are the probable answer where we found Male-specific DNA sequence which provide new technologies, and a very important evidence of an XY chromosome, and allow the tracing of paternal lineages in date palm. Our result is consistent with the findings of Cherif et al. (2013).
The main aim of the study is to investigate on how sex could be precisely determined in date palm by isolating specific sequence from male date palms located in Y linked plant genes (Diade, 2014). This sequence is a special interest because dioecy appears to have evolved independently on two separate occasions: one is from a monoecious ancestor and this sequence, which is linked to SRY gene from humans.
The present research has resulted in a patent application on the invention by providing SRY date palm specific PCR primer pairs for the identification of the sex, wherein the PCR primer pairs are specific to the male plant. A primer pair according to the researcher’s invention consists of: a specific forward and a specific reverse PCR primer. The sequence of the SRY-F forward primer is given: SRY-date Forward 5′- TCTCGAGATCCCCCAATGC-3′. The SRY- date Reverse 5′- ACCCTAAAGTGCCAACCAAT -3′. These primer pairs should be considered as specific nested primers for SRY male date palm gene.
Moreover, the researchers revealed a new pair of primers designed to target a section of the SRY gene. This resulted in a 355-bp male-specific PCR fragment that can be amplified by a regular PCR procedure and speeding up the sex identification of date palms. The applicability and specificity of the primers were confirmed with DNA from a separate male and female date palm tree. Also, a novel gene was identified that have been amplified uniquely on the Y chromosome, and is homologous with that of a papaya and human Y-chromosome. A number of other researchers (Foote et al., 1992, Devlin et al., 1998, Shibata et al., 1999, Okada et al., 2001, Kimitsune et al., 2002) have reported that there are Y-chromosomes that harbor unique repeat sequences in various organisms. This indicates that the human SRY is similar to the SRY in mice and in date palm plants. It supports the claim of a new model and technique in plants for sex determination. The differences between the two sexes were confirmed by the presence of the SRY gene in males. The researchers have now accumulated additional information regarding molecular events that contribute to sex determination.
Skaletsky et al., 2003, Hawley, 2003 sequenced a small portion of Y-chromosome of Marchantia and shown the evidence that it is sufficient to make comparisons with the euchromatic male-specific region of the human Y-chromosome. The results are also similar to the research presented by Rozen et al., 2003, Tanurdzic and Banks, 2004 wherein the sequence similarity of the human MSY region with limited comparative sequencing of the MSY regions in great apes were demonstrated.
The present work relates to molecular techniques that are specific to identify the sex of date palm plants and to the use of these techniques for distinguishing male and female plants. The techniques have the advantage of allowing the early detection of male plants and therefore, limiting the plantation costs associated with the cultivation of too many undesirable non-productive male plants. Moreover, the use of these techniques are simple and unique. Likewise, these are “universal” in the sense that it can be used to identify male plants regardless of the origin, variety or cultivar of the date palm.
Acknowledgments
This work was financed by a grant NO.13006, from Deanship of Research at King Faisal University.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Mohei EL-Din Solliman, Email: msolliman@kfu.edu.sa, solliman@yahoo.com.
Heba Allah A. Mohasseb, Email: hebamohasseb77@yahoo.com.
Abdullatif A. Al-Khateeb, Email: aalkhateeb@kfu.edu.sa.
Suliman A. Al-Khateeb, Email: skhateeb@kfu.edu.sa.
Kamal Chowdhury, Email: kamalc54@yahoo.com.
Hany A. El-Shemy, Email: helshemy@yahoo.com.
Mohammed I. Aldaej, Email: maldaej@kfu.edu.sa.
References
- Abdel-Mawgood, A.L., 2012. DNA based techniques for studying genetic diversity. In: Caliskan, M. (Ed.), Genetic Diversity in Microorganisms, In Tech.
- Aberlenc-Bertossi F., Daher A., Chabrillange N., Tregear J.W., Mohamed N. Plant and animal genomes XIX conference, W519: sex chromosomes and sex determination. Town and Country Convention Center; San Diego, CA: 2011. Sex determination in date palm: new perspectives on an old theme; pp. 15–19. [Google Scholar]
- Adawy S.S., Hussein E.H.A., El-Khishin D., Saker M.M., Mohamed A.A., El-Itriby H.A. Genotyping Egyptian date palm cultivars using RAPD, ISSR, AFLP markers and estimation of genetic stability among tissue culture derived plants. Arab J. Biotechnol. 2004;8(1):99–114. [Google Scholar]
- Agrawal V., Sharma K., Gupta S., Kumar R., Prasad M. Identification of sex in Simmondsia chinensis (jojoba) using RAPD markers. Plant Biotechnol. Rep. 2007;1:207–210. [Google Scholar]
- Ainsworth C.H. Boys and girls come out to play the molecular biology of dioecious plants. Ann. Bot- London. 2000;86:211–221. [Google Scholar]
- Al-Abdoulhadi, I.A., Al-Ali, S., Khurshid, K., Al-Shryda, F., Al-Jabr, A.M., Abdallah, A.B., 2011. Assessing fruit characteristics to standardize quality norms in date cultivars of Saudi Arabia, Indian J. Sci. Technol., vol. 4, no. 10, pp. 1262–1266, 2011.
- Al-Ameri, A.A., Al-Qurainy, F., Gaafar, A.Z., Khan, S., Nadeem, M., 2016. Molecular Identification of Sex in Phoenix dactylifera Using Inter Simple Sequence Repeat Markers. BioMed Res. Int. 2016, Retrieved from: 10.1155/2016/4530846. [DOI] [PMC free article] [PubMed]
- Aleksandrov O.S., Divashuk M.G., Karlov G.I. Development of a sex-specific molecular marker for Japanese hop Humulus japonicus Siebold & Zucc. Russian J. Genetics. 2011;47(8):1016–1020. [PubMed] [Google Scholar]
- Al-Khalifah N.S., Askari E., Shanavas Khan A.E. Molecular and morphological identification of some elite varieties of date palms grown in Saudi Arabia. Emirates J. Food and Agric. 2012;24(5):456–461. [Google Scholar]
- Atia A.M.M., Adawy S.S. Novel set of sex-specific PCR-based markers reveals new hypothesis of sex differentiation in date palm. J. Plant Sci. 2015;3(3):150–161. [Google Scholar]
- Barrett S. The evolution of plant sexual diversity. Nat. Rev. 2002;3:274–284. doi: 10.1038/nrg776. [DOI] [PubMed] [Google Scholar]
- Bendiab K., Baaziz M., Brakez Z., Sedra M. Correlation of isoenzyme polymorphism and Bayoud - disease resistance in date palm cultivars and progeny. Euphytica. 1993;65:23–32. [Google Scholar]
- Charlesworth D. Plant sex determination and sex chromosomes. Heredity. 2002;88:94–101. doi: 10.1038/sj.hdy.6800016. [DOI] [PubMed] [Google Scholar]
- Cherif E., Zehdi S., Castillo K., Chabrillange N., Abdoulkader S., Pintaud J.C., Santoni S., Salhi-Hannachi A. Male-specific DNA markers provide genetic evidence of an XY chromosome system, a recombination arrest and allow the tracing of paternal lineages in date palm. New Phytol. 2013;197:409–415. doi: 10.1111/nph.12069. [DOI] [PubMed] [Google Scholar]
- Danilova T.V., Karlov G.I. Application of inter simple sequence repeat (ISSR) polymorphism for detection of sex specific molecular markers in hop (Humulus lupulus L.) Euphytica. 2006;151:15–21. [Google Scholar]
- Devlin R.H., Stone G.W., Smailus D.E. Extensive direct tandem organization of a long repeat DNA sequence on the Y chromosome of chinook salmon (Oncorhynchus tshawytscha) J. Mol. Evol. 1998;46:277–287. doi: 10.1007/pl00006304. [DOI] [PubMed] [Google Scholar]
- Dhawan C., Kharb P., Sharma R., Uppal S., Aggarwal R.K. Development of male-specific SCAR marker in date palm (Phoenix dactylifera L.) Tree Genetics & Genomes. 2013;9:1143–1150. [Google Scholar]
- Diade, U.M.R., 2014. Meriem Kaid-Harche (Université des Sciences et de la Technologie d’Oran) and Stéphane Dussert. PhD thesis defense, December 2014.
- Drobnić K. A new primer set in a SRY gene for sex identification. Int. Congr. Ser. 2006;1288:268–270. [Google Scholar]
- El Hadrami, I., El Hadrami, A., 2009. Breeding date palm. In: Jain S.M., Priyadarshan, P.M. (Eds.), Breeding Plantation Tree Crops: Tropical Species, Springer Science Business Media. pp. 191–215.
- El-Juhany L.I. Degradation of date palm trees and date production in Arab countries: causes and potential rehabilitation. Aust. J. Basic Appl. Sci. 2010;4(8):3998–4010. [Google Scholar]
- Foote S., Vollrath D., Hilton A., Page D.C. The human Y chromosome: overlapping DNA clones spanning the euchromatic region. Science. 1992;258:60–66. doi: 10.1126/science.1359640. [DOI] [PubMed] [Google Scholar]
- Gaafar, A.R.Z., Al-Qurainy, F., Khan, S., 2014. Assessment of genetic diversity in the endangered populations of Breonadia salicina (Rubiaceae) growing in The Kingdom of Saudi Arabia using inter-simple sequence repeatmarkers. BMC Genetics, vol. 15, article 109. [DOI] [PMC free article] [PubMed]
- Geber M.A., Dawson T.E., Delph L.F., editors. Gender and Sexual Dimorphism in Flowering Plants. Springer; Berlin: 1999. [Google Scholar]
- George, J., Karun, A., Manimekalai, R., Rajesh M.K., Remya, P., 2007. Identification of RAP markers linked to sex determination in palmyrah (Borassus flabellifer L.). Curr Sci India, 93(8): 1075-1077.
- Harvey C.F., Gill G.P., Fraser L.G., McNeilage M.A. Sex determination in Actinidia. 1. Sex- linked markers and progeny sex ratio in diploid A. chinensis. Sex Plant Reprod. 1997;10:149–154. [Google Scholar]
- Hawley R.S. The human Y chromosome: Rumors of its death have been greatly exaggerated. Cell. 2003;113:825–828. doi: 10.1016/s0092-8674(03)00470-7. [DOI] [PubMed] [Google Scholar]
- Heikrujam M., Sharma K., Prasad M., Agrawal V. Review on different mechanisms of sex determination and sex-linked molecular markers in dioecious crops: a current update. Euphytica. 2015;201(2):161–194. [Google Scholar]
- Heikrujam M., Sharma K., Kumar J., Agrawal V. Generation and validation of unique male sex-specific sequence tagged sites (STS) marker from diverse genotypes of dioecious Jojoba-Simmondsia chinensis (Link) Schneider. Euphytica. 2014;199(3):363–372. [Google Scholar]
- Hobohm U., Sander C. A sequence property approach for sequencing protein databases. J Mol Biol. 1995;251:390. doi: 10.1006/jmbi.1995.0442. [DOI] [PubMed] [Google Scholar]
- Hormaza J.I., Dollo L., Polito V.S. Identification of RAPD marker linked to sex determination in Pistacia vera using bulked segregant analysis. Theor Appl Genet. 1994;89:9–13. doi: 10.1007/BF00226975. [DOI] [PubMed] [Google Scholar]
- Jain, S., Al-Khayri, M.J.M, Johnson, D.V., 2011. Date palm biotechnology: Springer Science & Business Media.
- Juarez C., Banks J.A. Sex determination in plants. Curr. Opin. Plant Biol. 1998;1(1):68–72. doi: 10.1016/s1369-5266(98)80130-1. [DOI] [PubMed] [Google Scholar]
- Kimitsune, I., Shimizu-Ueda, Y., Okada, S., Yamamoto, M., Fujisawa, M., Yamato, K., Fukuzawa, H., Ohyama, K., 2002. Multicopy genes uniquely amplified in the Y chromosome-specific repeats of the liverwort Marchantia polymorpha. Nucleic Acids Res., 2002, vol. 30 No. 21, 4675–4681. [DOI] [PMC free article] [PubMed]
- Korpelainen H., Bisang I., Hedenäs L., Kolehmainen J. The first sex-specific molecular marker discovered in the moss Pseudo calliergon trifarium. J. Hered. 2008;99(6):581–587. doi: 10.1093/jhered/esn036. [DOI] [PubMed] [Google Scholar]
- Lo Y.M.D., Tein M.S.C., Lau T.K., Haines C.J., Leung T.N., Poon P.M.K. Quantitative analysis of fetal DNA in maternal plasma and serum: implication for noninvasive prenatal diagnosis. Am. J. Hum. Genet. 1998;62:768–777. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud E.A., Mohei El-Din S.M., Aboul-Soud M.A.M., Aboul-Enein A.M., Sobhy G.A., El-Shemy H.A. Cloning of a Novel Antifungal Promoter from Phaseolus vulgaris and the Determination of its Activity in Stably Transformed Nicotiana tabacum plants. Curr. Issue Mol. Biol. 2009;11:55–63. [PubMed] [Google Scholar]
- Manoj, P., Banerjee N. S. & Ravichandran, P., 2005–2008. Development of sex specific molecular markers in dioecious Piper longum L. plants by differential display. JATIT, 459–465.
- Maryam J.M.J., Awan F.S., Ahmad S., Khan I.A. Development of molecular method for sex identification in date palm (Phoenix dactylifera L.) plantlets using novel sex-linked microsatellite markers. Biotech. 2016;6(1):1–7. doi: 10.1007/s13205-015-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga S., Kawano S. Sex determination by sex chromosomes in dioecious plants. Plant Biol. 2001;3:481–488. [Google Scholar]
- Milewicz M., Sawicki, J., 2011. Molecular identification of sex in dioecious moss Nyholmiella obtusifolia (Orthotrichaceae) on the basis of ISSR markers. Casopis Slezsḱeho Zemsḱeho Muzea, vol. 60, no. 1, pp. 1–6.
- Mohei El-Din, S.M., 2002. Isolation and Characterization of plant promoters for high level and tissue specific expression of foreign genes in transgenic plants. PhD from Icgeb (International Cenetre For Genatic Engeneering and Biotechnology, 1998 - 2002), India.
- Morton, J. (1987). Dates. In: Julia F. (Ed.). Morton fruits of warm climates. Miami Fl, pp. 5–11.
- Murray M.G., Thompson W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acid Res. 1980;8:4321–4326. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda S., Kar B., Nayak S., Jha S., Joshi R.K. Development of an ISSR based STS marker for sex identification in pointed gourd (Trichosanthes dioica Roxb.) Sci. Hortic. 2013;150:11–15. [Google Scholar]
- Negrutiu I., Vyskot B., Barbacar N., Georgiev S., Moneger F. Dioecious plants: A key to the early events of sex chromosome evolution. Plant Physiol. 2001;127:1418–1424. [PMC free article] [PubMed] [Google Scholar]
- Okada, S., Sone, T., Fujisawa, M., Nakayama, S., Takenaka, M., Ishizaki, K., Ohyama, K., 2001. The Y chromosome in the liverwort Marchantia polymorpha has accumulated unique repeat sequences harboring a male-speci®c gene. Proc. Natl Acad. Sci. USA, 98, 9454±9459. [DOI] [PMC free article] [PubMed]
- Parasnis A.S., Gupta V.S., Tamhankar S.A., Ranjekar P.K. A highly reliable sex diagnostic PCR assay for mass screening of papaya seedlings. Mol Breeding. 2000;6:337–344. [Google Scholar]
- Persson H.A., Nybom H. Genetic sex determination and RAPD marker segregation in the dioecious species sea buckthorn (Hippophae rhamnoides L.) Hereditas. 1998;129:45–51. [Google Scholar]
- Reddy M.K., Nair S., Sopory S.K. A new approach for efficient directional genome walking using Polymerase Chain Reaction. Anal. Biochem. 2002;306:154–158. doi: 10.1006/abio.2002.5645. [DOI] [PubMed] [Google Scholar]
- Renganayaki K., Jessup R.W., Burson B.L., Hussey M.A., Read J.C. Identification of male-specific AFLP markers in dioecious texas bluegrass. Crop Sci. 2005;45:2529–2539. [Google Scholar]
- Rozen S., Skaletsky H., Marszalek J.D., Minx P.J., Cordum H.S., Waterston R.H., Wilson R.K., Page D.C. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- Sarkar S., Banerjee J., Gantait S. Sex-oriented research on dioecious crops of Indian subcontinent: an updated review. Biotech. 2017;7(2):93. doi: 10.1007/s13205-017-0723-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Zinta G., Rana S., Shirko P. Molecular identification of sex in Hippophae rhamnoides L using isozyme and RAPD markers. The China Study. 2010;12(2):62–66. [Google Scholar]
- Sharma K., Agrawal V., Gupta S., Kumar R., Prasad M. ISSR marker- assisted selection of male and female plants in a promising dioecious crop; jojoba (Simmondsia chinensis) Plant Biotechnol Rep. 2008;2:239–243. [Google Scholar]
- Shibata F., Hizume M., Kuroki Y. Chromosome painting of Y chromosomes and isolation of a Y chromosome-specific repetitive sequence in the dioecious plant Rumex acetosa. Chromosoma. 1999;108:266–270. doi: 10.1007/s004120050377. [DOI] [PubMed] [Google Scholar]
- Shibu M.P., Ravishankar K.V., Anand L., Ganeshaiah K.N., Shaanker U. Identification of sex-specific DNA markers in the dioecious tree, nutmeg (Myristica fragrans Houtt) PGR Newsletter. 2000;121:59–61. [Google Scholar]
- Shirkot P., Sharma D.R., Mohopatra T. Molecular identification of sex in Actinidia deliciosa var. deliciosa by RAPD markers. Sci Hortic- Amsterdam. 2002;94:33–39. [Google Scholar]
- Siljak-Yakovlev S.S., Benmalek M., Cerbah Coba, de la PeñaTBounaga N., Brown S., Sarr A. Chromosomal sex determination and heterochromatin structure in date palm. Sex Plant Reprod. 1996;9:127–132. [Google Scholar]
- Skaletsky H. The male-specific region of the human Y chromosome is a mosaic of discrete sequence clusters. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Srivashtav, V.S., Kapadia, C.V., Mahatma, M.K., Jha, S.K., Jha, S., Ahmad, T., 2013. Genetic diversity analysis of date palm (Phoenix dactylifera L.) in the Kutch region of India using RAPD and ISSR markers. Emirates J. Food and Agric., vol. 25, no. 11, pp. 907–915.
- Stultz C.M., White J.V., Smith T.F. Structure analysis based on state-space modelling. Protein Sci. 1993;2:305. doi: 10.1002/pro.5560020302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanurdzic, M., Banks, A.J., 2004. Sex-Determining Mechanisms in Land Plants. The Plant Cell June 2004 vol. 16 no. suppl. 1 S61-S71. [DOI] [PMC free article] [PubMed]
- Vos P., Hogers R., Bleeker M., Reijans M., Van de Lee T., Hornes M., Frijters A., Pot J., Peleman J., Kuiper M., Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.W., Li Y., Li Z.Q. Identification of a male specific amplified fragment length polymorphism (AFLP) and a sequence characterized amplified region (SCAR) marker in Eucommia ulmoides Oliv. Int J Mol Sci. 2011;12:857–864. doi: 10.3390/ijms12010857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W.J., Wang B.W., Cui K.M. RAPD and SCAR markers linked to sex determination in Eucommia ulmoides Oliv. Euphytica. 2004;136:233–238. [Google Scholar]
- Younis, R.A.A., Ismail, O.M., Soliman, S.S., 2008. Identification of sex- specific DNA markers for date palm (Phoenix dactylifera L.) using RAPD and ISSR techniques. Res. J. Agric. Biol. Sci., 4(4): 278–284.
- Yu Q., Hou S., Alex Feltus F. Chromosomal location and gene paucity of the male specific region on papaya Y chromosome. Mol. Genet. Genomics. 2007;278:177–185. doi: 10.1007/s00438-007-0243-z. [DOI] [PubMed] [Google Scholar]
- Yu Q., Hou S., Hobza R. Low X/Y divergence in four pairs of papaya sex-linked genes. Plant J. 2008;53(1):124–132. doi: 10.1111/j.1365-313X.2007.03329.x. [DOI] [PubMed] [Google Scholar]