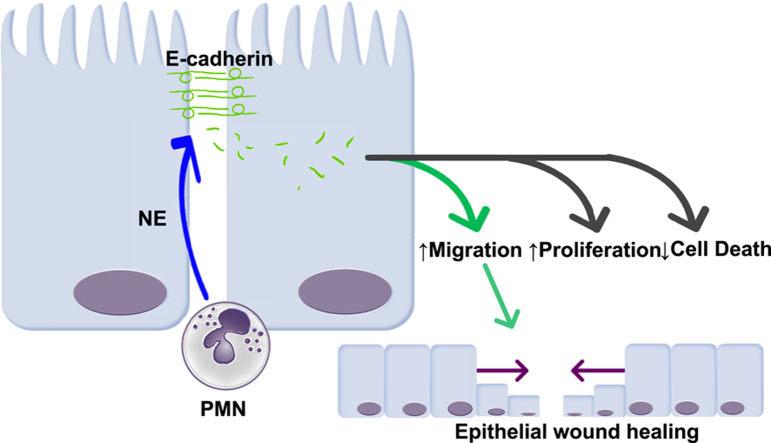
We report the novel observation that the inflammatory protease, neutrophil elastase (NE), present in high abundance in inflamed tissue in inflammatory bowel disease (IBD) patients, is capable of cleaving the cellular adherens junction protein, E-cadherin. Proteolysis of E-cadherin by NE generates a variety of short peptides, several of which were observed in patient tissue samples, showing biological activity to promote wound closure in an in vitro model system. This effect is independent of proliferation either in a wounded monolayer or under subconfluent conditions, suggesting a primarily migratory activity upon colonic epithelial monolayers. We report the novel observation that inflammatory proteases post-translationally modify cellular junction proteins to create signaling peptides that contribute to the wound healing response and identifies a new mechanism of mucosal healing to be examined further in the context of chronic inflammatory diseases.
IBDs, including ulcerative colitis and Crohn’s disease, comprise a spectrum of chronic inflammatory gastrointestinal diseases of complex etiology. Although there is no one defined cause or trigger for IBD, the unifying feature across the spectrum of IBD is the concept of chronic relapsing and remitting inflammatory disease, primarily in the colon (although in Crohn’s disease inflammation may occur anywhere along the gastrointestinal tract).1 Mucosal healing now is considered the current gold standard in assessing IBD therapeutic remission,2 however, our understanding of how and why many patients fail to achieve healing remains poorly elucidated. Upon an inflammatory stimulus, the intestinal epithelial monolayer is compromised by bacterial insult at the luminal surface as collateral damage from degranulation and an oxidative burst from lamina propria granulocytes, primarily neutrophils, and from cytokines released from leukocytes.3 Neutrophils are the first immune cells recruited to areas of inflammation in IBD and sustained high infiltration of activated neutrophils in inflamed tissue is a hallmark of disease.4 However, neutrophils now also are considered to be important players in the resolution phase of the inflammatory response.5, 6 Neutrophils can interact directly with epithelial cells by transmigrating through epithelia and interacting with apical intercellular adhesion molecule 1 to enhance wound healing through activation of the Akt and β-catenin pathways.7 Damage to the intestinal epithelium causes a shift from a tight barrier to a migratory/repair phenotype, a process that involves the proteolytic cleavage of junctional proteins such as E-cadherin. Proteases can cleave epithelial junctional proteins to generate peptides that have biological activity that can affect the intestinal mucosa,8 and recently NE was shown to cleave E-cadherin in bronchial epithelial cells,9 although the potential effects of E-cadherin degradation peptides was not assessed in that study. Of particular interest is the recent observation that NE can be internalized by cells and is thus capable of processing both intracellular and extracellular substrates.10, 11 In this context, we examined whether the inflammatory protease NE could proteolytically cleave the adherens junction protein E-cadherin and, specifically, whether the peptides resulting from this cleavage event could affect epithelial wound healing.
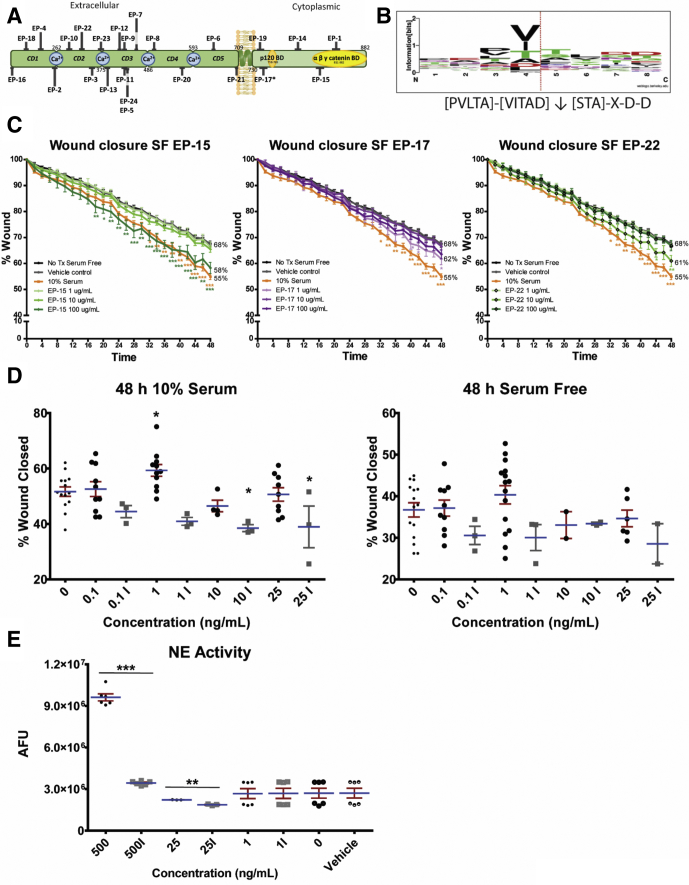
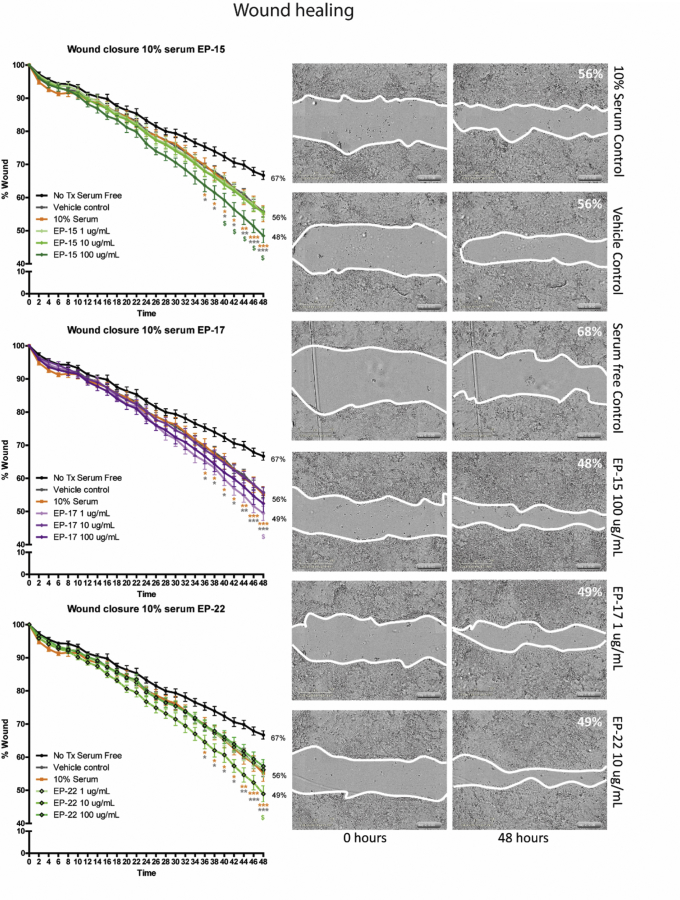
We first used a cell-free system to digest human recombinant E-cadherin with purified human NE and used liquid chromatography tandem mass spectrometry (LC-MS/MS) to identify the resulting peptides and cleavage events that occurred. We found that NE was capable of cleaving E-cadherin efficiently, with 48 peptides identified with high confidence using mass spectrometry. Of these 48 peptides, we focused on 24 of these based on their frequency of occurrence, P value scoring, and accessibility of peptide cleavage site to proteases (Supplementary Table 1 and Supplementary Figure 1A and B). To further focus on the most physiologically relevant peptides, we sought to confirm the presence of these peptides in patient tissues. By using a modified extraction protocol, we obtained protein/peptide fractions from banked formalin-fixed, paraffin-embedded IBD and control samples, and identified peptides originating from E-cadherin using mass spectrometry enriched in IBD samples. Six of these peptides showed substantial overlap with 6 peptides identified from our cell-free digest, and we chose these 6 peptides for biological activity screening (Table 1). First, we tested our peptides for effects on wound healing capacity using a scratch assay, under both 10% serum and serum-free conditions, over 48 hours at concentrations of 1, 10, and 100 μg/mL using a high-throughput protocol we designed for the Incucyte live cell imaging system (EssenBiosciences Inc, Ann Arbor, MI) (see the Supplementary Methods section for detailed methodologies). Three peptides, designated E-cadherin peptide (EP)-15, EP-17, and EP-22, at concentrations of 100, 1, and 10 μg/mL, respectively, showed increased wound closure compared with untreated and vehicle controls, in both 10% serum (Figure 1) and serum-free (Supplementary Figure 1C) conditions. The peptides appeared to have a synergistic effect with 10% serum.
Supplementary Figure 1.
(A) Diagrammatic representation of peptide localizations of human E-cadherin generated by NE in vitro. (B) Sequence logo cleavage sites of E-cadherin by NE as generated by WebLogo Software.1 The height of the amino acid 1-letter code illustrates the relative observed frequency. The dotted red line indicates the NE cleavage site. The local cleavage site residue pattern is [PVLTA]-[VITAD]↓[STA]-X-D-D. (C) E-cadherin peptides (designated EP-15, 17, and 22) significantly and synergistically enhanced healing in scratch-wounded Caco-2 monolayers over 48 hours in the absence of serum. *P < .05, **P < .01, ***P < .001 compared with vehicle and untreated controls (using a 2-way analysis of variance with the Bonferroni post-test). (D) Low-dose NE enhances wound healing in Caco-2 cells. Purified human NE showed a concentration- and activity-dependent wound-healing effect in scratch-wounded Caco-2 monolayers over 48 hours in the presence of serum (left panel), but not in the absence of serum (right panel). (E) The proteolytic activity of NE was confirmed to be reduced significantly after heat denaturing. *P < .05, compared with vehicle and untreated controls (using a 1-way analysis of variance with the (D) Dunnett post-test or (E) unpaired t tests with Welch correction). (C–E) N = 3–10 independent experiments, each with 3 or more technical replicates. AFU, arbitrary fluorescence unit; SF, serum free.
Table 1.
Alignment of E-Cadherin Peptides Generated by Neutrophil Elastase In Vitro With Peptide Fragments Isolated From IBD Patient Tissue
| Peptide | Position | Overlap | |
|---|---|---|---|
| TAYFSLDTR | 66-74 | None | |
| VTEPLDR | 216-222 | None | |
| NTGVISVVTTGLDR | 332-335 | EP-22 | KNMFTINRNTGVISVVTTGLDRESFPTYTL |
| GQVPENEANVVITTLK | 382-397 | EP-13, partially EP-23 |
IFNPTTYKGQVPENEANVVITTLKVTDADAPN TVTDTNDNPPIFNPTTYKGQVPENEANVVITTLKVTDADAPN |
| DTANWLEINPDTGA | 528-541 | None | |
| ISTRAELDR | 542-550 | None | |
| TIFFCER | 559-565 | None | |
| MALEVGDYK | 656-664 | None | |
| EPLLPPEDDTR | 739-749 | EP-17 | LRRRAVVKEPLLPPEDDTRDNVYYYDE |
| GLDARPEVTR | 775-784 | Partially EP-14 | FDLSQLHRGLDARPEVTRNDVAPTLM |
| NDVAPTLMSVPR | 785-796 | EP-14 | DARPEVTRNDVAPTLMSVPRYLPRPANP |
| PANPDEIGNFIDENLK | 801-816 | Partially EP-15 | SVPRYLPRPANPDEIGNFIDENLKAADTDPTAPPYD |
NOTE. The left column indicates the peptides identified in patient tissue, the second column shows its position in the full E-cadherin protein, and the right columns indicate overlap with in vitro neutrophil elastase digestion of E-cadherin. Highlighted text indicates peptides identified in patient samples, and underlined text indicates peptides before trypsin treatment. Boxed text indicates E-cadherin peptides generated by NE cleavage.
Figure 1.
E-cadherin peptides enhance wound healing in Caco-2 cells. E-cadherin peptides (designated EP-15, EP-17, and EP-22) significantly and synergistically enhanced healing in scratch-wounded Caco-2 monolayers over 48 hours in the presence of 10% serum. *P < .05, **P < .01, and ***P < .001 compared with vehicle and untreated controls (using a 2-way analysis of variance with the Bonferroni post-test). N = 5 independent experiments, each with 3 or more technical replicates. Tx, treatment.
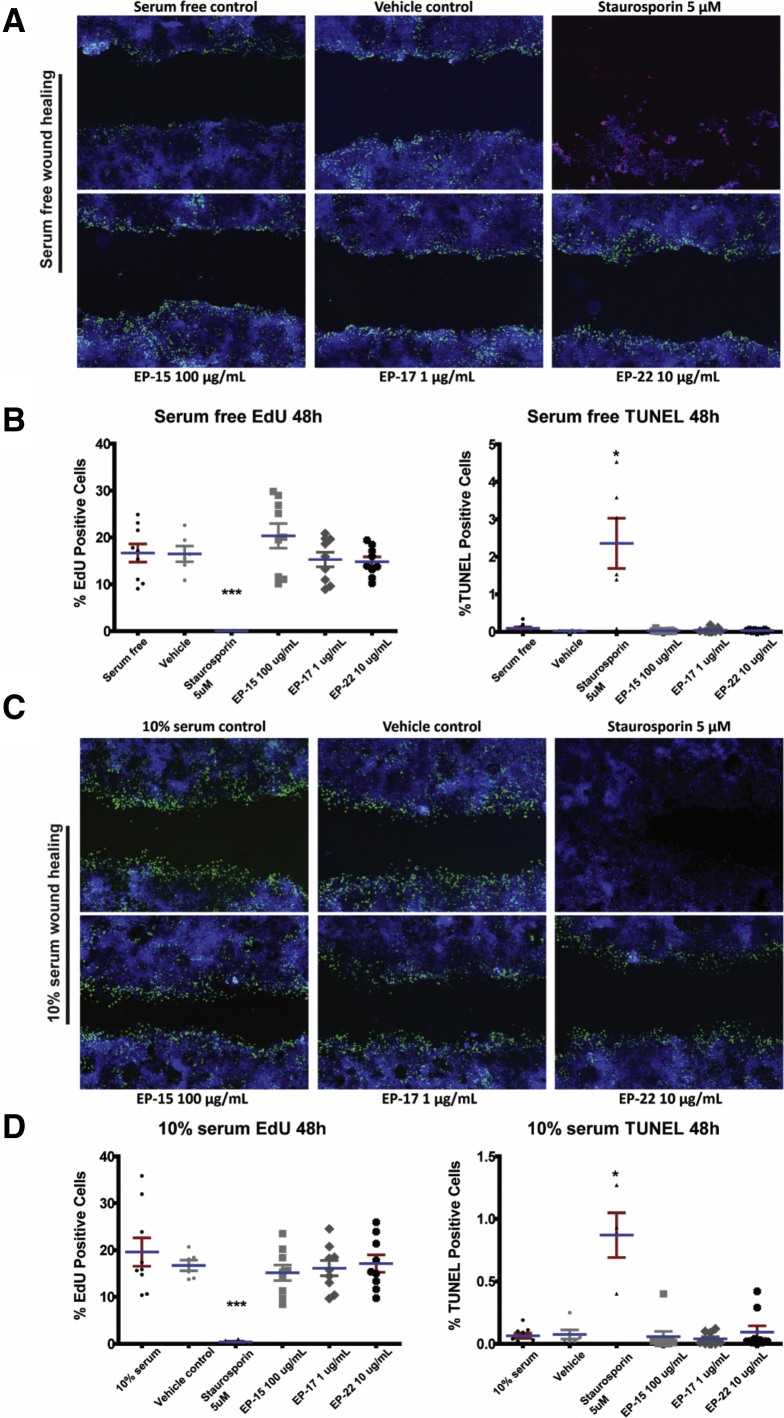
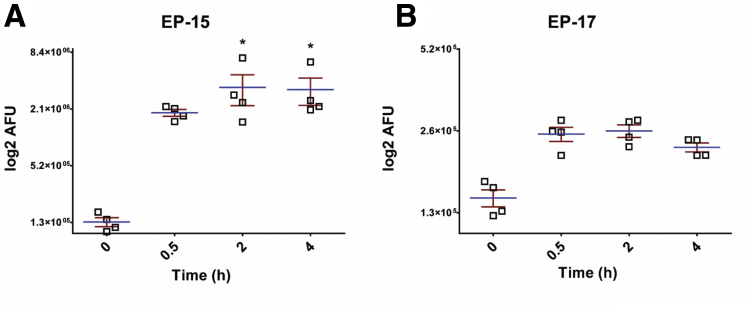
The pro-healing effect was replicated by NE in a concentration-dependent manner, with 1 ng/mL having a small but significant effect in the presence of serum (Supplementary Figure 1D). The effect of NE was dependent on its catalytic activity because the effect on wound healing was blocked with heat denaturation, which was confirmed to significantly reduce NE activity (Supplementary Figure 1E). To determine whether this effect was due to an increase in the proliferation of cells in response to the presence of E-cadherin peptides, we used the 5-Ethynyl-2′-deoxyuridine (EdU) system to identify actively dividing cells and found that there was no significant difference in proliferation under serum-free or serum conditions. Concurrently, we used the terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling enzyme to identify cell death to determine any cytotoxicity of our peptides at their effective doses and found no cytotoxicity of these peptides on Caco-2 monolayers (Supplementary Figure 2). To examine whether these peptides may have a mitogenic effect under nonwounding conditions, peptides also were tested for biological activity under subconfluent conditions. Caco-2 cells were transfected with a green fluorescent protein construct, seeded at medium density and exposed to the 6 peptides at the concentrations described earlier. Cell number and morphology were tracked over 48 hours. No significant changes in cell number or cell spreading were seen in response to any of the 6 peptides in either serum-free or 10% serum conditions (data not shown), suggesting that these peptides do not have mitogenic properties, and that the biological activity of the E-cadherin peptides is primarily to increase the migratory capacity under wound-healing conditions. To assess whether E-cadherin peptides could enter Caco-2 cells to potentially evoke intracellular signaling pathways, 7-amino-4-methylcoumarin (AMC) fluorescently tagged versions of EP-15 and EP-17 were synthesized and shown to transmigrate across the plasma membrane and into the cytosol (Supplementary Figure 3).
Supplementary Figure 2.
E-cadherin peptides do not significantly affect Caco-2 proliferation or apoptosis. Caco-2 cells were grown on plastic and scratch-wounded and stained with EdU as a marker of cell proliferation (green), terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling (TUNEL) stain for apoptotic cells (red), and 4′,6-diamidino-2-phenylindole to show nuclei (blue) under (A and B) serum-free and (C and D) 10% serum conditions. Representative images (A and C) of wounded monolayers and summary data (B and D) for EdU staining and TUNEL staining at 48 hours. E-cadherin peptides had no effect on either parameter. Similar results were seen at 30-minute and 24-hour treatment times (data not shown). *P < .05, ***P < .001 (using a 1-way analysis of variance with the Dunn post-test). N = 3 independent experiments, each with 3 or more technical replicates.
Supplementary Figure 3.
E-cadherin peptides are capable of crossing the lipid bilayer to enter the cytosol of Caco-2 cells. (A) EP-15 and (B) EP-17 entered the cytosol of scratch-wounded Caco-2 monolayers 5 days after confluency. *P < .01 compared with vehicle and untreated controls (using 1-way analysis of variance with the Dunnett post-test). N = 4 independent experiments, each with 3 or more technical replicates. AFU, arbitrary fluorescence unit.
In this research letter, we show the ability of an inflammatory protease, neutrophil elastase, to process the adherens junction protein E-cadherin to generate short peptide fragments with effects on epithelial function, and a novel role for low levels of NE being pro-resolution. These peptide fragments are present in IBD patient tissues and appear to enhance the wound-healing response of intestinal epithelial cell monolayers independently of cellular proliferation. Our study raises important questions about the cellular mechanism whereby NE-derived peptide fragments of E-cadherin stimulate an epithelial wound-healing response. Is the site of action of E-cadherin peptides on epithelial cells extracellular or intracellular, and what is the mechanism of transport across the cell membrane? What intracellular pathway(s) are altered by these peptides to modify epithelial cell behavior? Are other bioactive peptides proteolytically produced by NE or other inflammatory proteases as a resolution response, and what are the cellular targets of these peptides? Thus, our work provides the impetus for further research that will determine the signaling mechanisms underlying this phenomenon, identify potential new peptide biomarkers of inflammatory diseases, and develop new therapeutic targets. Overall, our discovery adds a new layer of complexity to our understanding of the signaling mechanisms underlying mucosal repair after inflammatory insult and suggests a new potential arm of repair signaling that may be dysregulated during IBD and other chronic inflammatory diseases.
Footnotes
Author contributions Marilyn H. Gordon was responsible for the study concept and design, acquisition and analysis of data, and writing the manuscript; Anaïs Chauvin acquired and analyzed the data; François-Michel Boisvert acquired and analyzed the data and edited the manuscript; and Wallace K. MacNaughton was responsible for the study concept and design and editing the manuscript.
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by grants from Crohn’s and Colitis Canada and the University Research Grants Committee at the University of Calgary (W.K.M.), and by grant 418404 from the Natural Sciences and Engineering Research Council of Canada (F.-M.B.). This research also was supported by the Live Cell Imaging Facility within the Snyder Institute for Chronic Diseases and the Southern Alberta Mass Spectrometry Facility at the University of Calgary.
Supplementary Methods
Cell-Free NE Digestion of E-Cadherin
A total of 5 μg of recombinant E-cadherin (H00000999-P01; Abnova, Neihu District, Tapei City, Taiwan) was incubated in modified Hank's balanced salt solution (14175; Gibco, Waltham, MA) with 0.02 U purified human NE (E8140; Sigma-Aldrich, St. Louis, MO) for 1 minute at 37°C. The enzymatic reaction then was quenched with ice-cold 0.1% trifluoroacetic acid and snap frozen at -80°C. The sample was processed directly using high-performance liquid chromatography (HPLC)–MS/MS without additional clean-up on an Orbitrap Fusion (Thermo Scientific, Waltham, MA), and peptides were identified using MASCOT software (Matrix Science, Boston, MA). The final modified Hank's balanced salt solution composition consisted of the addition of HEPES, MgCl2, and CaCl2 to Hank's balanced salt solution as follows: 400 mg/L KCL, 60 mg/L KH2PO4, 350 mg/L NaHCO3, 8000 mg/L NaCl, 48 mg/L Na2HPO4, 1000 mg/L dextrose, 10 mmol/L HEPES pH 7.0, 1.5 mmol/L MgCl2, and 1.5 mmol/L CaCl2.
Identification of Peptides From Human Formalin-Fixed Paraffin-Embedded Tissues
Tissue samples were deparaffinized using a series of xylene–ethanol washes and dried completely before rehydration in high-performance liquid chromatography–grade water. Rehydrated samples then were homogenized/disrupted using a TissueRuptor (Qiagen, Venlo, Netherlands), and sodium dodecyl sulfate, Tris pH 7.5, and dithiothreitol added to the homogenized tissue to final concentrations of 2.5% sodium dodecyl sulfate, 50 mmol/L Tris pH 7.5, and 10 mmol/L dithiothreitol. Samples then were incubated at 98°C for 1 hour with vigorous vortexing of samples every 15 minutes. Samples then were centrifuged at 300 × g for 1 minute and the supernatants were collected into low-binding tubes. Samples then were concentrated using an Amicon Ultra 3K Spin column (UFC500324; Millipore, Burlington, MA), and the resulting concentrate was run on a 4%–12% sodium dodecyl sulfate–polyacrylamide electrophoresis gel. After staining with Coomassie Blue SimplyBlue SafeStain solution (LC6060; Thermo Scientific), lanes were excised from the gel and placed in new low-binding microtubes. Samples then were destained by washing with HPLC–grade water for 15 minutes with shaking, adding 1 volume acetonitrile and washing for 15 minutes with shaking and centrifuging for 1 minute at 300 × g. The wash supernatant then was removed from the samples and replaced with 20 mmol/L ammonium bicarbonate and washed with shaking for 15 minutes, followed by centrifugation for 1 minute at 300 × g. The wash supernatant then was removed from the samples and replaced with 20 mmol/L ammonium bicarbonate and acetonitrile (50:50 vol/vol), and washed for 15 minutes with shaking followed by centrifugation at 300 × g. The wash supernatant then was removed from the samples and replaced with acetonitrile to dehydrate samples until solid, and then dried using a Speedvac (Thermo Fisher Scientific). Samples then were digested using 12.5 ng/mL mass-spectrometry grade trypsin (90057; Pierce, Waltham, MA) reconstituted in ammonium bicarbonate 20 mmol/L at 30°C overnight. The peptide fraction then was recovered by adding 1 volume of acetonitrile and incubating at 30°C for 30 minutes with shaking followed by centrifugation at 300 × g for 1 minute. The supernatant was recovered to a new low-binding microtube and 1 volume of 1% formic acid was added to samples and incubated at 30°C for 20 minutes with shaking, followed by centrifugation at 300 × g for 1 minute. The supernatant was recovered and 1 volume was added and incubated at 30°C with shaking until solid, followed by centrifugation at 300 × g for 1 minute. Samples then were resuspended in 0.1% trifluoroacetic acid, and samples were cleaned using C18 Zip-Tips (ZTC18M960; Millipore) and run on HPLC-MS/MS. The Dionex Ultimate 3000 HPLC system was coupled to an OrbiTrap QExactive mass spectrometer via an EasySpray source (all Thermo Fisher Scientific, Inc). The spray voltage was used at 2.0 kV and the temperature of the column was 40°C. Full scan mass spectrometry survey spectra (m/z 350–1600) were acquired with a resolution of 70,000 after accumulation of 1,000,000 ions. The 10 most intense peptide ions then were fragmented by collision-induced dissociation at 35% energy with a resolution of 17,500, after accumulation of 50,000 ions. The maximal filling times were 250 ms for the full scans and 60 ms for the MS/MS scans. Singly, 7 and 8 charged precursor ions were rejected and a dynamic exclusion list was set to a maximum of 500 entries, a maximum retention period of 40 seconds, and a relative mass window of 10 ppm. The lock mass option was enabled for survey scans to improve mass accuracy.
Cell Culture
Caco2 human intestinal epithelial cells were grown in Dulbecco’s modified Eagle medium and Ham’s F12 (SH3002301; Hyclone, San Angelo, TX) supplemented with 10% fetal bovine serum (12483020; Gibco), 1% penicillin-streptomycin (SV30010; Hyclone), and 5 μg/mL Plasmocin (ANT-MPT; InvivoGen, San Diego, CA) maintained under standard cell culture conditions. Cells were maintained in a 37°C humidified incubator with 5% CO2. Media was changed in culture flasks every 2 days and cells were routinely passaged every 4 days at 80%–85% confluence using 1.5× trypsin-EDTA (T4174; Sigma-Aldrich), and cell numbers were quantified using a manual cytometer before plating. All experiments were performed using cells from passages 55–70.
Scratch Wound Assays
Caco2 cells were seeded into 96-well plates at a density of 5000 cells/well and grown under standard cell culture conditions, with media changes every 2 days. At 5 days after confluence, cells were wounded using the WoundMaker tool (Essen BioSciences). Wounded monolayers were washed twice with serum-free Dulbecco’s modified Eagle medium and Ham’s F12 to remove cellular debris and replaced with test media. Plates then were placed into the Incucyte live-cell imaging system (Essen BioSciences) maintained in a 37°C humidified incubator with 5% CO2, and whole-well phase images were taken every 2 hours for 48 hours. Confluency masks were generated using the Incucyte ZOOM software, and wound area over time was calculated using ImageJ (National Institutes of Health, Bethesda, MD). E-cadherin peptides were custom synthesized by NeoBiolabs (Woburn, MA) and reconstituted in sterile ddH2O. Purified human NE was obtained from Sigma Aldrich (#E8140).
NE Activity Assays
Purified human NE (E8140; Sigma Aldrich) was incubated at varying concentrations in modified Hank's balanced salt solution (see earlier) with the fluorogenic Elastase V substrate reporter (324470; Calbiochem, San Diego, CA) at a final concentration of 200 umol/L. To inactivate the catalytic activity of NE, the NE was heat-inactivated at 105°C for 1 hour, and loss of activity confirmed using the fluorogenic substrate. Substrate V fluorescence indicating proteolytic processing of the substrate was determined using a Victor X4 plate reader (Perkin Elmer, Waltham, MA), using a 360/480 excitation/emission program, reading for 0.5 seconds.
Peptide Internalization Assays
C-terminal–conjugated AMC-fluorogenic peptides were custom-synthesized by ABclonal Biotechnology (Woburn, MA) and reconstituted in sterile ddH2O. Caco-2 monolayers 5 days after confluence were wounded, washed twice with phenol-red–free Dulbecco’s modified Eagle medium and Ham’s F12 to remove cellular debris, and replaced with phenol-red–free test media/compounds and incubated for 0, 0.5, 2, and 4 hours. At the end of the experiment, supernatants were removed and retained, and cells were lysed and collected in T-PER lysis buffer (78510; Thermo-Fisher) and sonicated. Recovered supernatants and lysates were placed into a white-walled reading plate and AMC fluorescence signal was detected using a Victor X4 plate reader (Perkin Elmer), using a 360/480 excitation/emission program, reading for 0.5 seconds.
EdU and Terminal Deoxynucleotidyl Transferase–Mediated Deoxyuridine Triphosphate Nick-End Labeling Enzyme Immunofluorescence
Immunofluorescence of wounded Caco-2 monolayers was performed using a modified protocol with the Click-iT EdU Alexa Fluor 488 Kit (C10337; Invitrogen, Waltham, MA) and the terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling in situ cell death TMR Kit (12156792910; Roche, Basil, Switzerland). Briefly, cells were wounded and treated with test media as described earlier. Two hours before fixation, 10 μmol/L EdU was added to treatment wells to allow EdU to incorporate into dividing cells. At 30 minutes, 24 hours, and 48 hours, the wounded Caco2 monolayers were fixed in 4% paraformaldehyde for 15 minutes, washed with 3% bovine serum albumin in phosphate-buffered saline, and permeabilized with 0.5% Triton X-100 (Thermo Fisher Scientific) in phosphate-buffered saline at room temperature for 20 minutes. Monolayers were washed with 3% bovine serum albumin in phosphate-buffered saline and Click-iT reaction cocktail and terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling reaction cocktail was added to wells and incubated at 37°C for 1 hour while protected from light. Plates were washed 3 times with 3% bovine serum albumin in phosphate-buffered saline and monolayers were counterstained with 4′,6-diamidino-2-phenylindole. Images were taken using an Olympus IX71 inverted epi-fluorescence wide-field microscope (Olympus; Shinjuku, Tokyo, Japan). A total of 5 μmol/L Staurosporin (569397; Millipore) was used as a positive control for cell death.
Subconfluent Cell Growth and Morphologic Dynamics
Caco2 cells were transfected with a pEGFP-C1 plasmid to fluorescently label cells using the Lipofectamine LTX with Plus Reagent transfection protocol (Invitrogen). Stably transfected cell lines were obtained by selection using 400 μg/mL G418 for neomycin resistance. Labeled cells then were plated into 96-well plates at 5000 cells/well, allowed to adhere to the plates for 36–48 hours, treated with peptides, and imaged over 48 hours using the Incucyte live-cell imaging system to track the cell number by fluorescence. Cell spreading was determined by calculating the cell area changes in the same experiments.
Statistical Analyses
Data analysis was performed using GraphPad Prism 5 software (La Jolla, CA). All analyses represent at least 3 independent experiments with at least 3 independent technical replicates and are expressed as means ± SEM. Wound closure over time was analyzed using a 2-way analysis of variance with Bonferroni post-tests. All other analyses were performed using a 1-way analysis of variance with the Tukey or Dunnett post-tests as appropriate. All N values presented in this report represent experimental replicates, with each individual experimental replicate including at least 3 technical replicates.
Supplemental Graphical Summary.
Supplementary Table 1.
Sequences and Characteristics of Peptide Fragments of Human E-Cadherin Generated by NE In Vitro
| Peptide name | Peptide sequence | Position | Domain |
|---|---|---|---|
| EcadP-1 | FDYEGSGSEAA | 833-843 | α β γ catenin binding domain |
| EcadP-2 | TVTDQNDNKPEFT | 251-263 | Between cadherin domains 1 and 2 (calcium binding) |
| EcadP-3 | QAADLQGEGLSTTAT | 346-360 | Cadherin domain 2 |
| EcadP-4 | SSNGNAVEDPMEI | 236-248 | Cadherin domain 1 |
| EcadP-5 | NNDGILKTA | 431-439 | Cadherin domain 3 |
| EcadP-6 | QYNDPTQESI | 641-650 | Cadherin domain 5 |
| EcadP-7 | SLTTSTATV | 465-473 | Cadherin domain 3 |
| EcadP-8 | SEDFGVGQEI | 496-505 | Cadherin domain 4 |
| EcadP-9 | TNPVNNDGILKT | 427-438 | Cadherin domain 3 |
| EcadP-10 | ATDADDDVNTYNAAI | 286-300 | Cadherin domain 2 (calcium binding) |
| EcadP-11 | TTNPVNNDGILKTA | 426-439 | Cadherin domain 3 |
| EcadP-12 | LNDDGGQFVV | 417-425 | Cadherin domain 3 |
| EcadP-13 | TYKGQVPENEANV | 378-391 | Cadherin domain 3 (calcium binding) |
| EcadP-14 | RNDVAPTLM | 784-792 | Cytoplasmic |
| EcadP-15 | KAADTDPTAPPYD | 816-828 | α β γ catenin binding domain |
| EcadP-16 | SGIQAELL | 133-140 | Propeptide |
| EcadP-17 | LPPEDDTRDNV | 741-752 | Cytoplasmic |
| EcadP-18 | TGQGADTPPVGV | 193-204 | Cadherin domain 1 |
| EcadP-19 | LLILILLLLL | 721-729 | Cytoplasmic |
| EcadP-20 | ALIIATDNGSPV | 563-574 | Cadherin domain 4 |
| EcadP-21 | EAGLQIPA | 702-709 | Ectodomain/transmembrane junction |
| EcadP-22 | NRNTGVISVV | 320-329 | Cadherin domain 2 |
| EcadP-23 | TVTDTNDNPPIFNPT | 364-377 | Between cadherin domains 2 and 3 (calcium binding) |
| EcadP-24 | NNDGILKT | 431-438 | Cadherin domain 3 |
NOTE. Samples were sequenced by mass spectroscopy to show 24 distinct peptides. Sequences are shown, as well as the regions of the full-length E-cadherin that they represent. Ecad, E-cadherin.
References
- 1.Zhang Y. World J Gastroenterol. 2014;20:91–99. doi: 10.3748/wjg.v20.i1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atreya R. Visceral Med. 2017;33:82–88. doi: 10.1159/000458006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parkos C.A. Am J Pathol. 2016;186:1404–1416. doi: 10.1016/j.ajpath.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brazil J. Inflamm Bowel Dis. 2013;19:1556–1565. doi: 10.1097/MIB.0b013e318281f54e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fournier B.M. Mucosal Immunol. 2012;5:354–366. doi: 10.1038/mi.2012.24. [DOI] [PubMed] [Google Scholar]
- 6.Colgan S.P. Semin Immunol. 2015;27:177–183. doi: 10.1016/j.smim.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sumagin R. Mucosal Immunol. 2016;9:1151–1162. doi: 10.1038/mi.2015.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nava P. Mol Biol Cell. 2007;18:4565–4578. doi: 10.1091/mbc.E07-05-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boxio R. Respir Res. 2016;17:129. doi: 10.1186/s12931-016-0449-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gregory AD, et al. J Biol Chem 2012;287:35341–35350. [DOI] [PMC free article] [PubMed]
- 11.Kerros C. J Biol Chem. 2017;292:10295–10305. doi: 10.1074/jbc.M116.773051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary Reference
- 1.Crooks G.E. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]