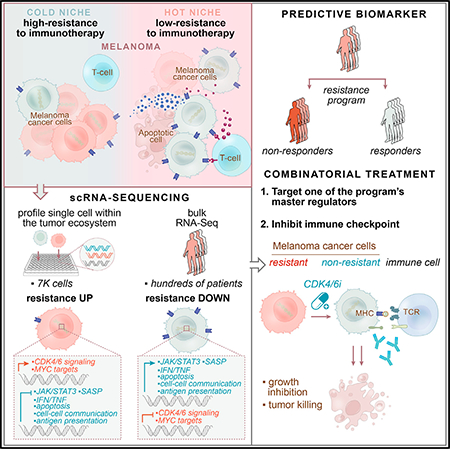
SUMMARY
Immune checkpoint inhibitors (ICIs) produce durable responses in some melanoma patients, but many patients derive no clinical benefit, and the molecular underpinnings of such resistance remain elusive. Here, we leveraged single-cell RNA sequencing (scRNA-seq) from 33 melanoma tumors and computational analyses to interrogate malignant cell states that promote immune evasion. We identified a resistance program expressed by malignant cells that is associated with T cell exclusion and immune evasion. The program is expressed prior to immunotherapy, characterizes cold niches in situ, and predicts clinical responses to anti-PD-1 therapy in an independent cohort of 112 melanoma patients. CDK4/6-inhibition represses this program in individual malignant cells, induces senescence, and reduces melanoma tumor outgrowth in mouse models in vivo when given in combination with immunotherapy. Our study provides a high-resolution landscape of ICI-resistant cell states, identifies clinically predictive signatures, and suggests new therapeutic strategies to overcome immunotherapy resistance.
In Brief
Single-cell sequencing of checkpoint-inhibitor-resistant melanomas identifies predictive signatures to guide therapeutic approaches to overcome immunotherapy resistance.
Graphical Abstract
INTRODUCTION
Immune checkpoint inhibitors (ICIs) have transformed the therapeutic landscape of several cancer types (Sharma and Allison, 2015), especially melanoma. Nonetheless, many patients manifest resistance, which is often intrinsic (Sharma et al., 2017). Because ICI targets cell-cell interactions, resistance can stem from different cells and their interactions in the tumor ecosystem. Recent studies illuminated ICI resistance with whole-exome sequencing (WES) and transcriptional profiles of bulk tumors (Hugo et al., 2016; Riaz et al., 2017; Van Allen et al., 2015) but had limited ability to dissect the cancer-immune interplay and generate reliable response biomarkers. Single-cell RNA sequencing (scRNA-seq) of patient tumors (Tirosh et al., 2016) can alleviate this limitation.
Infiltration of the tumor with T cells has been associated with patient survival and improved immunotherapy responses (Frid-man et al., 2012), but the determinants that dictate if a tumor will have high (“hot”) or low (“cold”) levels of T cell infiltration are only partially understood. Among multiple factors, malignant cells may play an important role in determining this phenotype. However, while current methods use bulk genomics to deconvolve the tumor’s composition (Newman et al., 2015), they cannot recover the salient intracellular programs of malignant cells. Thus, linking malignant cell states to T cell infiltration levels has been challenging.
Here, using an integrative data-driven approach (Figures 1A and 1B), we identified a malignant cell program that is associated with T cell exclusion and is predictive of ICI resistance. We demonstrated that CDK4/6 inhibitors repress this program and sensitize melanoma tumors to ICIs in mouse models. Our work provides a predictive biomarker for ICI response, suggests a new therapeutic modality that may sensitize melanoma tumors to ICIs, and provides a framework to study cell-cell interactions and drug effects in other tumor ecosystems.
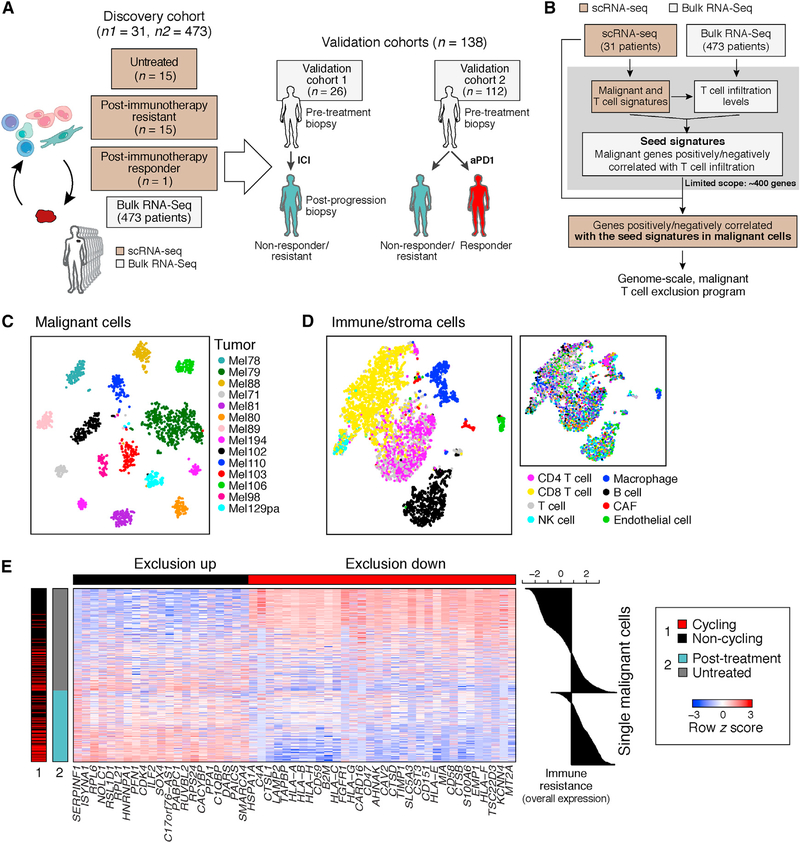
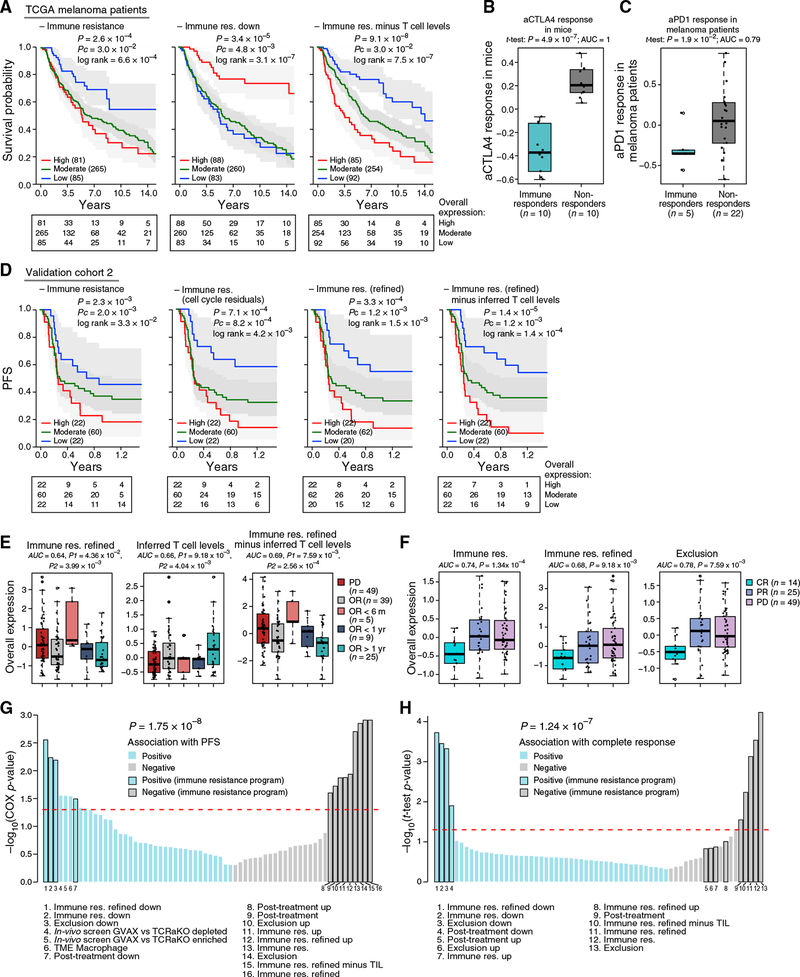
Figure 1. Identification of a T Cell Exclusion Program in Malignant Cells.
(A) Study overview.
(B) Method to discover malignant cell programs associated with immune cell infiltration or exclusion.
(C and D) Distinct profiles of malignant and non-malignant cells. t-stochastic neighbor embedding (t-SNE) of single-cell profiles (dots) from malignant (C) or non-malignant (D) cells, colored by post hoc annotation (D, left) or by tumor (C and D, right). In (C), only tumors with at least 50 malignant cells are shown.
(E) Exclusion program. Expression (centered and scaled, color bar) of the top genes (columns) in the exclusion program across malignant cells (rows) is sorted by untreated or post-treatment tumors (blue/gray color bar, left). Leftmost color bar: cycling (red) and non-cycling (black) cells. Right: overall expression (OE) (STAR Methods) of the exclusion program.
RESULTS
Systematic Approach to discover Cancer-Cell-Autonomous Programs Associated with T Cell Exclusion
To identify malignant cell programs that characterize cold tumors, we combined scRNA-seq and bulk RNA-seq data to relate the state of one cell type to the tumor composition (Figure 1B and STAR Methods). We describe the strategy in a specific context, but it can be applied to other pairs of cell types. First, we use scRNA-seq profiles to define cell-type-specific signatures of T cells and malignant cells in melanoma. Using the T cell signature and bulk RNA-seq profiles, we infer T cell infiltration levels in hundreds of tumors. We then define a “seed exclusion program”–genes from the malignant cell signature whose expression is strongly correlated (positively or negatively) with T cell abundance across those bulk tumors. To avoid mRNA contamination by the tumor microenvironment, we restrict the seed program only to a few hundred genes that are exclusively expressed by malignant cells. To recover potentially important genes that are also expressed by non-malignant cells (e.g., major histocompatibility [MHC] class I), we search for genes that are correlated with the seed program across the malignant cells in the scRNA-seq data irrespective of their expression in other cell types. Finally, we define a genome-scale, malignant-cell exclusion program consisting of genes induced or repressed by malignant cells in cold versus hot tumors. We then score each cell or tumor based on its overall expression (OE) of the program, defined as the overexpression of its induced part and underexpression of its repressed part (STAR Methods).
Analysis of Clinical scRNA-Seq Identifies a Malignant Cell Program Associated with T Cell Exclusion
We applied our approach to 7,186 high-quality scRNA-seq profiles from 33 human melanoma tumors (from 31 patients) comprised of 2,987 cells from 17 newly collected patient tumors and 4,199 cells from 16 patient tumors that we previously reported (Tirosh et al., 2016) (Figure 1A, Tables S1A and S2, and STAR Methods; one patient overlaps), along with 473 bulk RNA-seq melanoma profiles from the Cancer Genome Atlas (TCGA) (Akbani et al., 2015). 15 of the tumors in the single-cell cohort are post-ICI-resistant tumors (Table S1A).
We distinguished different cell subsets based on their expression profiles and inferred copy-number variation (CNV) profiles (STAR Methods), identifying malignant cells, CD8+ and CD4+ T cells, B cells, natural killer (NK) cells, macrophages, cancer-associated fibroblasts (CAFs), and endothelial cells (Figures 1C, 1D, and S1 and Table S3). Malignant cells primarily grouped by their tumor of origin (Figure 1C) and non-malignant cells by their cell type (Figure 1D), as we previously observed (Tirosh et al., 2016).
We applied our approach to delineate the relationship between malignant cell states and CD8+ T cell infiltration, identifying a T cell exclusion program (Figure 1E and Table S4A). The repressed part of the program was enriched for genes involved in antigen processing and presentation (e.g., B2M, CTSB, HLA-A/B/C, and TAPBP; p = 3.26 × 10−10, hypergeometric test), interferon (IFN-γ) signaling pathways (p = 2.94 × 10−9), response to the complement system (p = 1.13 × 10−8; e.g., CD59 and C4A), and immune modulation (p = 2.10 × 10−7; e.g., CD47 and CD58). The induced part included CDK4 and its downstream E2F targets (p = 3.97 × 10−5), transcriptional regulators (e.g., SOX4, SMARCA4), and numerous CDK7 and Myc targets (p < 1 × 10−17; Table S4A). Notably, the malignant cell programs associated with exclusion of different T cell subsets were highly overlapping (Figures S2A–S2C, Tables S3B and S4B, and STAR Methods).
The Exclusion Program Characterizes Malignant Cells from Patients who Progressed on Immunotherapy
To determine whether the exclusion program is associated with ICI resistance, we tested wheter it is more pronounced in malignant cells from ICI-resistant versus untreated patients in our scRNA-seq cohort. As clinical response rates to ICI vary, with up to ~57% responders to ICI combinations (Larkin et al., 2015), the untreated tumors we profiled likely include both ICI-sensitive and ICI-resistant ones, whereas the ICI-resistant tumors likely include mostly resistant malignant cells. Comparing malignant cells from post-ICI-resistant tumors to malignant cells from untreated tumors, we found a robust and generalizable post-treatment transcriptional program (cross-validation area under the curve [AUC] = 0.83; Figure 2A, Table S4A, and STAR Methods). This program might reflect both the overall impact of ICI therapy and intrinsic ICI resistance per se, but those cannot be distinguished based on our single-cell cohort, which includes neither matched samples from the same patient nor pre-treatment tumors from responders and non-responders. We address this later in two validation cohorts (Tables S1B and S1C).
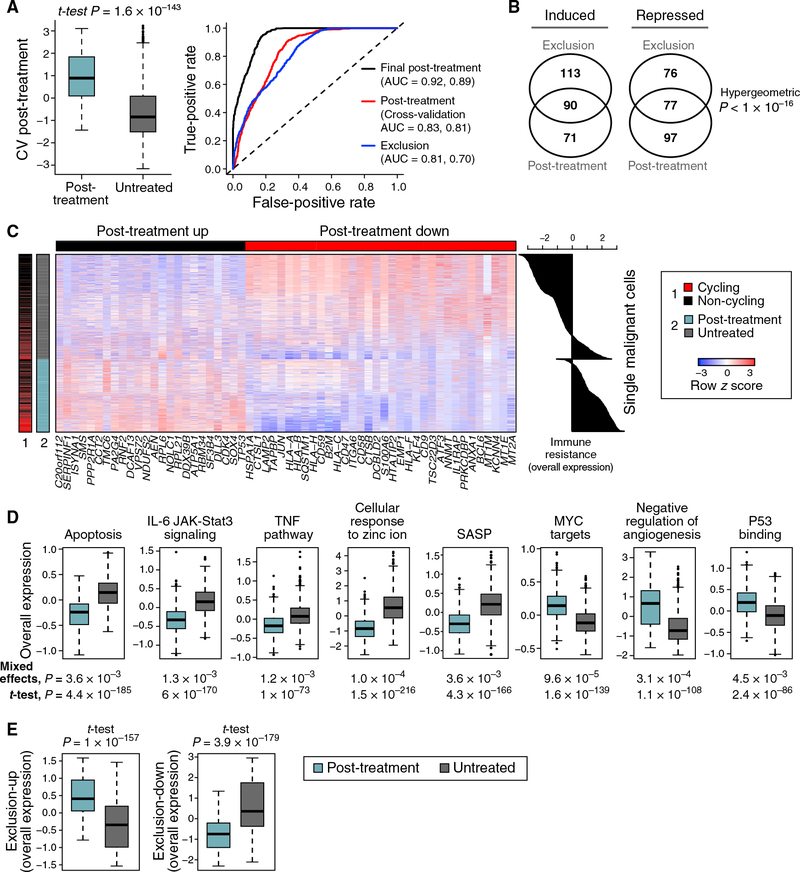
Figure 2. Exclusion and Resistance Programs Characterizing Individual Malignant Cells from Patients with Resistance to ICIs.
(A) Post-treatment program in malignant cells. Left: OE of the post-treatment program in malignant cells from post-treatment (blue) and untreated (gray) patients tested on withheld data (STAR Methods). Middle line: median; box edges: 25th and 75th percentiles; whiskers: most extreme points that do not exceed ± interquartile range (IQR) × 1.5; further outliers are marked individually. Right: the performances of different programs in classifying cells as post treatment or untreated; the first and second area under the curve (AUC) values are for classifying cells and samples, respectively.
(B) Significant overlap between the exclusion and post-treatment programs.
(C) Expression (centered and scaled, color bar) of the top genes (columns) in the post-treatment program across malignant cells (rows) sorted by untreated or post-treatment tumors (blue/gray color bar, left). Leftmost color bar: cycling (red) and non-cycling (black) cells. Right: OE of the post-treatment program.
(D) Distribution of OE scores (as in A) of differentially expressed gene sets in malignant cells from post-treatment (blue) and untreated (gray) tumors.
(E) Distribution of OE scores (as in A) of the exclusion program in malignant cells from post-treatment (blue) and untreated (gray) tumors.
The post-treatment and exclusion programs substantially overlapped (Figures 1E, 2B, and 2C and Table S4A; p < 10−16, hypergeometric test) and highlighted similar modules and pathways (Figure 2D and Table S5). Both programs were more pronounced in the post-treatment malignant cells and robustly classified malignant cells as untreated or post treatment (AUC = 0.83 and 0.81 for cross-validation post treatment and exclusion, respectively; Figures 2A and 2E). The upregulated components of both programs include 46 ribosomal protein (RP) genes, but their overlap remains significant even after removing RPs (p < 10−16,hyper-geometric test; Figure 2B). As Myc is a master regulator of ribosome biogenesis (Kress et al., 2015), the induced RP genes might reflect Myc activation. Indeed, the programs are enriched for Myc targets, even after removing RP genes (p < 7.18 × 10−10) and are predicted to be repressed by MYC knockdown according to the Connectivity Map (Subramanian et al., 2017).
In light of the congruence of the programs, we defined a unified immune resistance program as the union of the post-treatment and exclusion programs (Table S4A) and used it in all subsequent analyses unless indicated otherwise (STAR Methods).
The Immune Resistance Program Reflects a Coherent State of Immune Evasion
The immune resistance program manifests hallmarks of immune evasion, suppression, and exclusion. First, compared to cutaneous melanoma (where we initially identified it), the program is more pronounced in uveal melanoma (Figure 3A), which resides in an immune-privileged environment and has very low response rates to immunotherapy (Algazi et al., 2016). Second, inhibition of genes from the repressed component of the program in malignant melanoma cells conferred resistance to CD8+ T cells in a genome-wide CRISPR screen (p =1.67 × 10−3, hypergeometric test) (Patel et al., 2017). Third, the program marks the suppression of physical interactions between resistant malignant cells and other cell types in the tumor microenvironment, including MHC I:TCR (T cells), CD58:CD2 (T cells), and IL1RAP:IL1B (macrophages) (Figure 3B and STAR Methods), as well as the senescence-associated secretory phenotype (SASP) (p = 4.3 × 10−166 and 3.6 × 10−3, one-sided t test and mixed effects, respectively; Figure 2D), which has been shown to enhance T cell responses (Gordy et al., 2016).
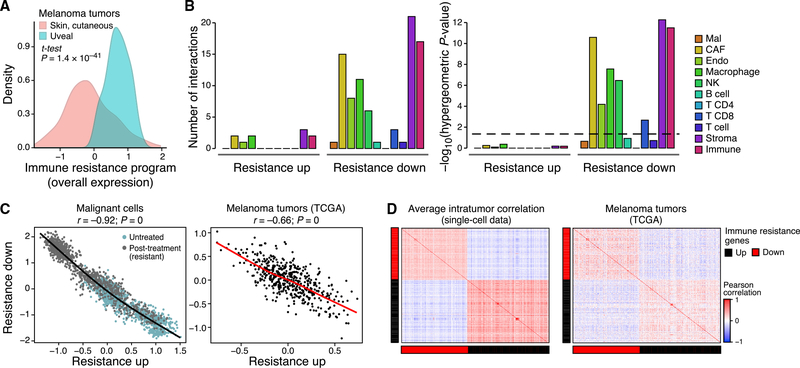
Figure 3. The Resistance Program Is a Coherently Regulated Module that Represses Cell-Cell Interactions.
(A) Distribution of program OE scores in cutaneous (pink) versus uveal (blue) melanoma from TCGA after filtering microenvironment contributions (STAR Methods).
(B) Right: Number of genes in each part of the program that mediate physical interactions with other cell types (color) and the significance of the corresponding enrichment. Dashed line: statistical significance.
(C and D) Co-regulation of the program.
(C) OE of the induced and repressed parts of the immune resistance programs in malignant cells (left, scRNA-seq data) and cutaneous melanoma tumors (right, TCGA RNA-seq data after filtering microenvironment signals). Pearson correlation coefficient (r) and p value are marked.
(D) Pearson correlation coefficients (color bar) between the program’s genes across malignant cells from the same tumor (left, average coefficient) or across cutaneous melanoma from TCGA (right, after filtering microenvironment effects).
See also Figure S3.
The program’s genes appear to be under shared control by a few master regulators. The expression of genes within each components (induced or repressed) is positively correlated, while the induced genes are anti-correlated with the repressed genes, both across single cells in one tumor and across TCGA tumors (Figures 3C, 3D, and S3). Thus, any given aspect of the program (e.g., underexpression of antigen presentation) is coupled to the state of the entire program. Moreover, there is a significant overlap between the perturbations that reverse the expression of the program’s repressed and induced components (p = 4.35 × 10−6, hypergeometric test), including the overexpression of IFN-γ and IFN-β and the knockdown of MYC and CDK7 (Subramanian et al., 2017). The latter mirrors the significantly large number of Myc and CDK7 (direct) targets (Oki et al., 2018; Subramanian et al., 2005) in the program (p < 1 × 10−17, hypergeometric test). Further supporting the role of CDKs as regulators, the program is more pronounced in cycling cells, though it is present and detectable in non-cycling cells (Figures 1E and 2C; p < 8.42 × 10−27, mixed effects). Notably, the association between cellular proliferation and the resistant program is only partial, and the two cell states can be decoupled (Figures S2D–S2H, Table S4B, and STAR Methods).
We compared the immune resistance program to 12 signatures (Table S6A) previously associated with the response to immunotherapy (Ayers et al., 2017; Hugo et al., 2016; Riaz et al., 2017) or targeted therapy (RAF and MEK inhibitors) (Hugo et al., 2015; Tirosh et al., 2016) in melanoma patients. Four of the six signatures that characterize immunotherapy-sensitive melanoma were enriched with one or more of our T cell signatures (p < 1 × 10−3, hypergeometric test), suggesting that they capture tumor composition and not malignant cell states. The induced component of our resistance program was not enriched in any of the previous 12 signatures. The repressed component was enriched in two signatures of immunotherapy sensitivity (p <2.65 × 10−3, hypergeometric test) and with signatures associated with sensitivity and resistance to targeted therapy (p <1.48 × 10−4) (Tirosh et al., 2016).
Malignant Cells in T Cell-Depleted Niches Express Features of the Resistance Program In Situ
To test if the resistance program in malignant cells is associated with T cell exclusion in situ, we used multiplexed immunofluorescence (t-CyCIF) (Lin et al., 2018). We stained histological sections of 19 tumors (472,771 cells per image on average) from our single-cell cohort for 14 proteins: six cell type markers (CD3, CD8, MHC-II, FOXP3, S100, and MITF) and seven resistance program members (induced: p53, Myc, and DLL3; repressed: HLA-A, c-Jun, SQSTM1, and LAMP2). Following cell segmentation and intensity quantification (STAR Methods), we assigned malignant cells (S100+, MITF+), T cells (CD3+), and cytotoxic T cells (CD8+); the rest were defined as uncharacterized.
The scRNA-seq and multiplex in situ protein profiles were congruent by cell-type assignment and by resistance program assessment. First, combining the two datasets using a variant of canonical correlation analysis (CCA) (Butler et al., 2018) (STAR Methods) successfully embedded and clustered cells primarily by type and not by method (Figures 4A, 4B, and S4). Second, the average immune resistance scores of the different tumors according to their in situ images were correlated with their scores according to scRNA-seq (R = 0.57, p = 0.041) (STAR Methods), even though the scRNA-seq and the in situ image of each tumor were obtained from separated specimens, measuring RNA versus protein, respectively. Additionally, in both cases, the inter-tumor variation of the program was significantly greater than its intra-tumor variation (p < 1 × 10−30, ANOVA).
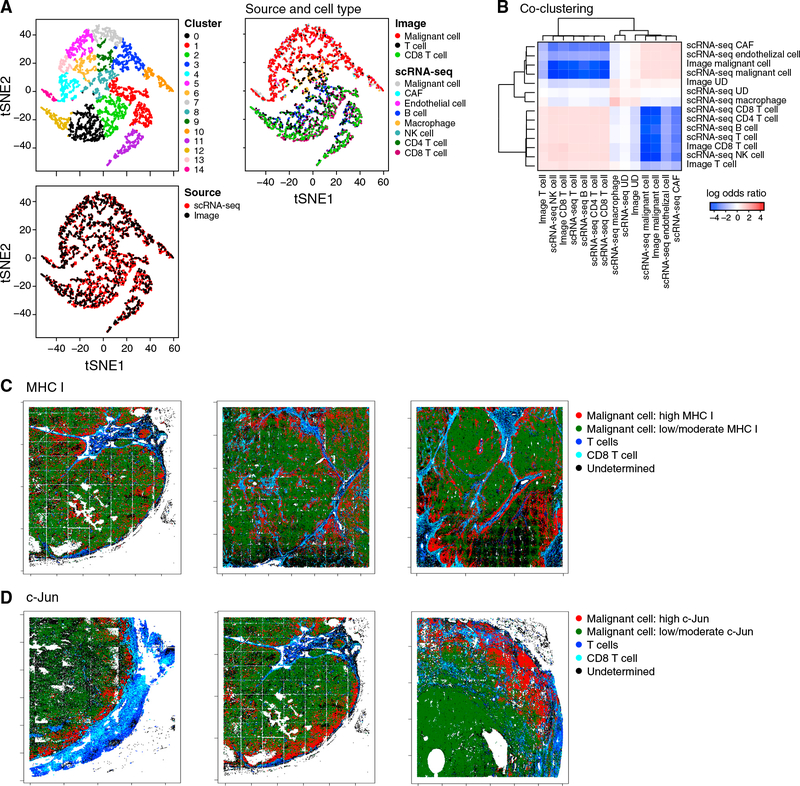
Figure 4. The Resistance Program Is Associated with the Cold Niche In Situ.
(A and B) Congruence of in situ multiplex protein and scRNA-seq profiles.
(A) Co-embedding of profiles from scRNA-seq and multiplex imaging of the Mel112 tumor (others in Figure S4), with cells colored by clusters (top left), data source (bottom left), or source and cell type (right).
(B) Log-odds ratio (color bar; STAR Methods) assessing for each pair of cell types (rows, columns) if they are assigned to the same cluster significantly more (>0, red) or less (<0, blue) than expected by chance.
(C and D) Multiplex imaging relates program genes to hot or cold niches. Malignant cells expressing high (red) or low/moderate (green) levels of the MHC class I (C) and c-Jun (D) proteins and their proximity to CD3+ T cells (blue) or CD3+CD8+ T cells (cyan) in three representative tumors.
See also Figure S4.
As predicted, the resistance score we computed from all seven program markers (STAR Methods) was significantly higher in malignant cells that reside in cold niches (p = 1.18 × 10−6, mixed effects). Aside from LAMP2, individual markers also showed the predicted trend: malignant cells in cold niches had significantly lower levels of markers repressed in the resistance program (Figures 4C and 4D; p = 1.41 × 10−13 and 8.35 × 10−18, mixed effects, for c-Jun and HLA-A, respectively), whereas p53 (induced in the resistance program) characterized cold niches (p = 5.25 × 10−3, mixed effects). Thus, our analysis validates our signature and its association with cold niches at the protein level in situ.
The Resistance Program Is Expressed Prior to Treatment and Is Enhanced following Immunotherapy in Resistant Lesions
We hypothesized that the immune resistance program, while more pronounced in the malignant cells of resistant patients after ICI, in fact reflects intrinsic resistance. Supporting this, the program is detected in untreated TCGA melanoma tumors and in a subset of malignant cells from untreated patients (Figures 1E and 2C, right plots).
To test this hypothesis, we analyzed an independent RNA-seq cohort of 90 specimens collected from 26 metastatic melanoma patients throughout the course of treatment. 14 patients received ICI therapy (anti-PD1 or anti-CTLA4) without prior targeted therapy, and 12 patients first received targeted therapy (BRAF/MEK inhibitors) followed by ICI after tumor progression (Figure 1A and Table S1B, validation cohort 1). The cohort has several partially overlapping sets (Table S1B): treatment naive (n = 18), on targeted therapy (n = 17), post targeted therapy (n = 25), on ICI (n = 35), and post ICI (n = 50). Given this composition, we used a mixed-effects model to determine the effects of the different treatments. As expected, we found a significant induction in the (inferred) T cell fraction of tumors on ICI treatment (p = 1.85 × 10−3, mixed-effects test), but not on targeted therapy.
The program was induced in on- and post-ICI samples compared to pre-ICI samples from the same patient (p = 7.41 × 10−3, mixed-effect test, controlling for tumor composition; STAR Methods). However, inter-patient variation in the program’s expression was significantly higher than these intrapatient changes (p < 4.98 × 10−14, ANOVA). This suggested that the major differences between the post-ICI and untreated tumors in the single-cell cohort reflect, at least in part, intrinsic differences between the two groups, which preceded the treatment.
We did not observe an induction of the program following RAF/MEK-inhibition. We confirmed this in another cohort of patient-matched melanoma tumors biopsied before MAPK-inhibition and during disease progression (Hugo et al., 2015) (p > 0.1, mixed effects).
The Resistance Program Predicts ICI Responses in Melanoma Patients
Next, we used the OE of the program (with and without an additional refinement; Table S4A and STAR Methods) to estimate the immune resistance level of a given tumor and tested its ability to predict clinical outcomes (Figures 5, S5, and S6).
Figure 5. The Resistance Program Is Prognostic and Predictive in Validation Cohorts.
(A) The program predicts melanoma patient survival in bulk RNA-seq from TCGA. Kaplan-Meier (KM) curves stratified by high (top 20%), low (bottom 20%), or intermediate (remainder) OE of the respective program. Number of subjects at risk indicated at the bottom of the KM curves for five time points. P, COX regression p value; Pc, COX regression p value that tests if the program enhances the predictive power of a model with inferred T cell infiltration levels as a covariate.
(B and C) Distribution of OE of the resistance program in bulk tumors from a lung cancer mouse model treated with anti-CTLA-4 therapy (Lesterhuis et al., 2015)
(B) or melanoma patients prior to pembrolizumab treatment (Hugo et al., 2016) (C). Middle line: median; box edges: 25th and 75th percentiles, whiskers: most extreme points that do not exceed ± IQR × 1.5; outliers are marked individually.
(D-F) The program predicts ICI responses in validation cohort 2.
(D) KM plots for PFS for the 104 patients in the cohort with available PFS data stratified by high (top 20%), low (bottom 20%), or intermediate (remainder) OE of the respective program (STAR Methods).
(E) OE of the resistance program (y axis) in the pre-treatment profiles of patients with intrinsic resistance (PD, n = 49) or objective response (OR, n = 39), with the latter further stratified by response duration. Patients with unknown response or stable disease are not shown. P1 and P2: one-tailed t test p value when comparing the PD patients to all the OR patients or to OR >1 year patients, respectively. AUC for predicting OR >1 year in all patients with a recorded response (n = 101) is denoted. Formatted as in (B).
(F) OE scores of the resistance program (y axis) in the pre-treatment bulk RNA-seq profiles of patients with complete response (CR, n = 14), partial response (PR, n = 25), or progressive disease (PD, n = 49). P: one-tailed t test p value comparing CR patients to PR and PD patients. AUC for predicting CR in all patients with a recorded response (n = 101).
(G and H) Predictive value (y axis) compared to alternative signature-based predictors. Blue/gray bars: signatures positively/negatively associated with response. Black outline of bars: subsets of the resistance program denoted with numbered legends at the bottom. Dashed line: p = 0.05.
(G) Predictive value for PFS (Pc as in D; STAR Methods).
(H) Predictive value for complete response.
The program’s expression was associated with poor survival in 473 TCGA melanoma patients (Figures 5A and S5) and also when controlling for tumor purity and inferred T cell infiltration. Combining the program with inferred T cell infiltration levels yielded significantly more accurate predictions than either alone (p = 9.1 × 10−8, COX regression; Figure 5A, right). Other proposed mechanisms, such as de-differentiation of melanoma cells reflected by an MITF-low state, and other malignant cell signatures (Tirosh et al., 2016) were not associated with survival, indicating that mere biological variation across malignant cells is insufficient for prognosis.
To test if the program can predict clinical responses to ICI, we analyzed five RNA-seq cohorts collected from melanoma patients prior to ICI treatment: validation cohort 2 collected by us across 112 patients (validation cohort 2; Figure 1A), validation cohort 1 (pre-ICI samples; Figure 1A), and three published cohorts of less than 50 patients each (Hugo et al., 2015; Riaz et al., 2017; Van Allen et al., 2015). We compared our predictors to 47 other gene signatures (Table S6A and STAR Methods), which we tested as alternative predictors, including commercial immune panels, the top hits of two CRISPR screens of resistance to T cells and anti-PD-1 (Manguso et al., 2017; Patel et al., 2017), and signatures generated by analyzing previous melanoma ICI cohorts (Hugo et al., 2016; Riaz et al., 2017).
The program’s expression distinguished ICI responders from non-responders (Figures 5B and 5C). In a lung cancer mouse model, its expression early on-treatment separated anti- CTLA-4 non-responders from responders (p = 4.89 × 10−7, one-sided t test; Figure 5B) (Lesterhuis et al., 2015). In 27 melanoma patients treated with pembrolizumab (anti-PD-1) (Hugo et al., 2016), underexpression of the program pre-treatment distinguished the five complete responders (p = 5.80 × 10−3 and 1.89 × 10−2, one-sided t test, refined and non-refined version, respectively; Figure 5C). In 42 melanoma patients treated with CTLA-4 blockade using ipilimumab (Van Allen et al., 2015), it was lower in the two complete-responders pre-treatment. In 43 melanoma patients profiled before and early on-treatment with nivolumab (anti-PD-1) (Riaz et al., 2017), it was downregulated early on-treatment specifically in responders (p < 9.60 × 10−6, hypergeometric test). However, in this specific cohort, the pre-treatment expression of the program (and of all other alternative predictors) was not associated with response, possibly due to the small number (three) of complete responders. In validation cohort 1, the program was underexpressed in the 7 pre-ICI samples from patients with an objective response (OR, including partial or complete response) compared to 12 pre-ICI samples from non-responders (progressive disease [PD]; p = 7.88 × 10−3, one-sided t test).
Finally, we tested the predictive value of the program in 112 patients with metastatic melanoma who underwent a pretreatment biopsy and RNA-seq followed by anti-PD-1 therapy (Figure 1A, validation cohort 2, and Tables S1C and S6B). We evaluated the program’s performance in predicting (1) progression-free survival (PFS, available for 104 patients; Figure 5D), (2) OR (including partial or complete response; Figure 5E), and (3) complete response (Figure 5F and STAR Methods).
The program was predictive of PFS (Figures 5D and S6), including when accounting for other potential biomarkers (T cell infiltration levels and PD-L1 expression) (Figure S6E). It outperformed all the alternative predictors we tested (Table S6A; p = 1.75 × 10−8, Wilcoxon rank-sum test), which were either not predictive or did not provide additional predictive value once accounting for T cell infiltration levels of the tested tumors (Figure 5G). Although the program was more pronounced in cycling cells, cell cycle alone was not associated with PFS (p > 0.25, COX regression), nor was a signature of malignant-specific genes that characterizes cycling malignant cells (p > 0.05, COX regression). Filtering the cell-cycle component from the resistance program score (STAR Methods) further improved PFS predictions (Figure 5D), suggesting that a tumor’s immune resistance profile should be evaluated considering its proliferation level.
The program was strongly underexpressed in patients with OR compared to those without response (PD) (Figure 5E). Moreover, patients with OR that nonetheless had high pre-treatment expression of the program were significantly more likely to rapidly (<6 months) develop PD (Figure 5E). Consistently, the program was most accurate in predicting complete responses (p =1.34 × 10−4, one-sided t test; Figure 5F), outperforming all other tested predictors (p = 1.24 × 10−7, Wilcoxon rank-sum test) (Figure 5H).
The Resistance Program Is Coherently Controlled by CDK4/6
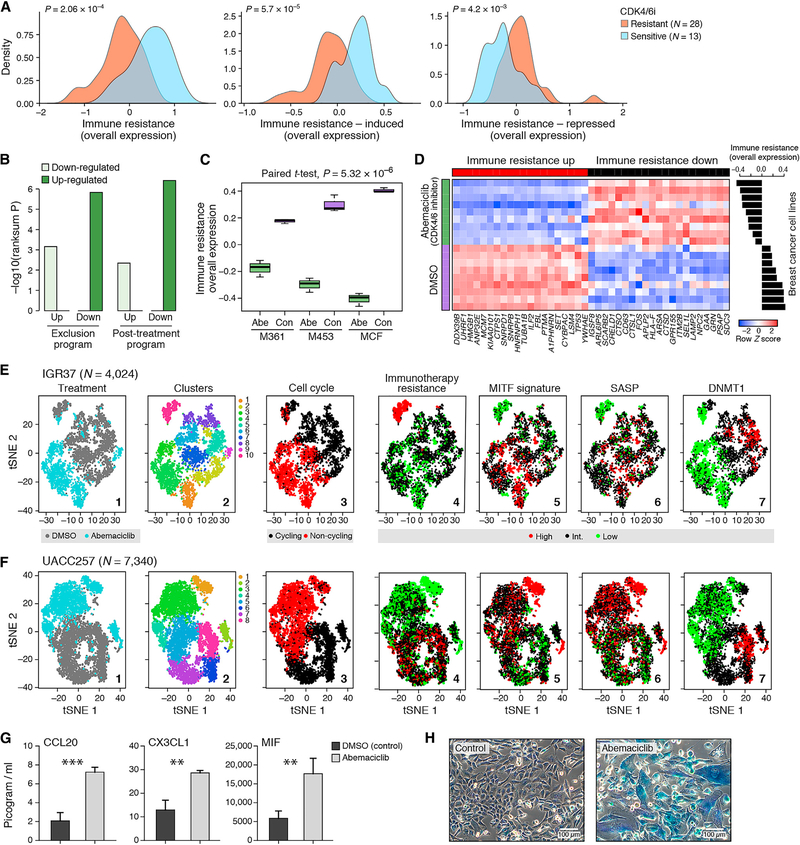
Next, we sought to pharmacologically target the program. We first identified drugs that were significantly more toxic to cell lines intrinsically overexpressing the program in a screen of 131 drugs across 639 human cell lines (STAR Methods) (Garnett et al., 2012). The third-highest-scoring drug was the CDK4/6 inhibitor (CDK4/6i) palbociclib (p =1.01 × 10−4, mixed effects). We confirmed this in another screen of two CDK4/6i (palbociclib and abemaciclib) across hundreds of cell lines (Gong et al., 2017) (p = 3.96 × 10−5, mixed effects; Figure 6A).
Figure 6.
The Resistance Program Can Be Reversed by CDK4/6 Inhibition
(A) OE of the resistance program across cancer cell lines that are resistant (orange) or sensitive (blue) to both abemaciclib and palbociclib.
(B-D) Impact of CDK4/6i on breast cancer tumors and cell line profiles.
(B) Significance (y axis, −log10(p value), Wilcoxon rank-sum test) of induction (dark) or repression (light) of the program subsets in tumors from abemaciclib-treated mice compared to vehicle (Goel et al., 2017).
(C) OE of the program in cell lines (M361, M453, and MCF) treated with abemaciclib (“abe”) or with DMSO vehicle (“con”). Middle line: median; box edges: 25th and 75th percentiles; whiskers: most extreme points that do not exceed ± IQR × 1.5; outliers are marked individually. P value: paired t test.
(D) Expression of 40 program genes (columns) that were most differentially expressed in abemaciclib-treated (green) versus control (purple) cell lines (rows) (STAR Methods). Expression is normalized in each cell line. Right: OE scores for each cell line.
(E–H) CDK4/6i reverses the program in RBI-sufficient melanoma cell lines and induces the SASP.
(E and F) tSNE of 4,024 IGR137 (E) and 7,340 UACC257 (F) melanoma cells colored by (1) treatment, (2) clusters, or (3) expression of cell-cycle signature, (4) resistance program, (5) MITF signature, (6) SASP signature, and (7) DNMT1.
(G) Concentration (pg/mL, y axis) of secreted chemokines in the supernatant of melanoma cells treated for 7 days with abemaciclib (500 nM) or with DMSO control. **p < 0.01, ***p < 0.001; t test (Table S7B).
(H) Senescence-associated β-galactosidase activity (blue) and morphological alterations in melanoma cells treated for 10 days with abemaciclib (500 nM, right) versus DMSO control (left).
We hypothesized that CDK4/6 may act as master regulators of the program. Multiple CDK target genes are members of the induced program (Table S4A), and CDK4 is a member of the induced component of both the exclusion and the post-treatment programs (Figures 1E and 2C). Three genes (CDKN2C/p18, CDKN1B/p27, and CDKN1A/p21) that inhibit CDK4 repress the program when overexpressed (Subramanian et al., 2017) (STAR Methods), and the program is more pronounced in cycling cells (Figures 1 E, 2C, and S2E), where CDK4/6 are active. Finally, analysis of published gene expression profiles of breast cancer cell lines and mouse models (Goel et al., 2017) showed that CDK4/6i represses the resistance program (Figures 6B–6D).
CDK4/6 Inhibitors Repress the Resistance Program in Melanoma Cells
We therefore tested if CDK4/6i could shift the malignant cell population to a less immune resistant state. We selected three melanoma cell lines that strongly expressed the program (Table S7A), two of which are RB1 sufficient (IGR37 and UACC257) and one that is RB1 deficient (A2058). We profiled each cell line with scRNA-seq before and after treatment with abemaciclib, analyzing >23,000 cells (Figures 6E and 6F and Table S2). The program’s expression varied between cells within each line, despite the absence of non-malignant cells, suggesting cell-intrinsic regulation (Figures 6E and 6F, panel 4). In the RB-sufficient cell lines, IGR37 and UACC257, 10% of cells had exceptionally strong expression of the program (“immune-resistant” cells) prior to treatment. Post treatment, the relative abundance of these cells decreased to 2% and 0.6% of the total population, respectively (Figures 6E and 6F, panel 4). In the RB1-deficient cell line A2058, the treatment did not repress the resistant state, consistent with the fact that the impact of CDK4/6i isRB1 dependent (Goel et al., 2017) (Figure S7A). In the two RB-sufficient cell lines, post-treatment cells showed substantial transcriptional changes linked to the resistance program. Abemaciclib induced the MITF program (Tirosh et al., 2016) (p < 1 × 10−17, hypergeometric test; Figures 6E and 6F, panel 5), which is repressed in immune-resistant cells and is associated with melanocytic differentiation. Abemaciclib also repressed the expression of DNMT1 (p < 2.23 ×10−106, likelihood-ratio test; Figures 6E and 6F, panel 7), which has been shown to confer an immunogenic phenotype (Goel et al., 2017).
Expression of the SASP—a repressed component in the resistance program—was induced in abemaciclib-treated cells (p < 3.33 × 10−16, hypergeometric test; Figures 6E and 6F, panel 6). Abemaciclib also increased cytokine secretion (Figure 6G and Table S7B), β-galactosidase activity, and morphological alterations that reflect cellular senescence (Figure 6H). It reduced proliferation rates compared to DMSO (0.3 versus 0.64 doublings per 24 hr, p < 1 × 10−4, t test; Figures 6E and 6F, panel 3) but was not cytotoxic.
Next, we tested abemaciclib effects on malignant cells in the presence of tumor-infiltrating T lymphocytes (TILs) in a patient-derived co-culture of melanoma cells and autologous ex vivo expanded TILs. While TIL exposure alone represses the immune resistance program in both conditions (p < 7.94 × 10−7, one-sided t test), this effect was smaller compared to the intrinsic variation between cells of the same cell line (IGR37; Figure 6E, panel 4; p < 8.17 × 10−12, F test). Treatment with abemaciclib alone or with additional TIL exposure significantly repressed the resistance program (p < 7.72 × 10−3, one-sided t test; Figure S7B). Overall, these results indicate that the resistance program is mostly intrinsically regulated and can be repressed by CDK4/6i.
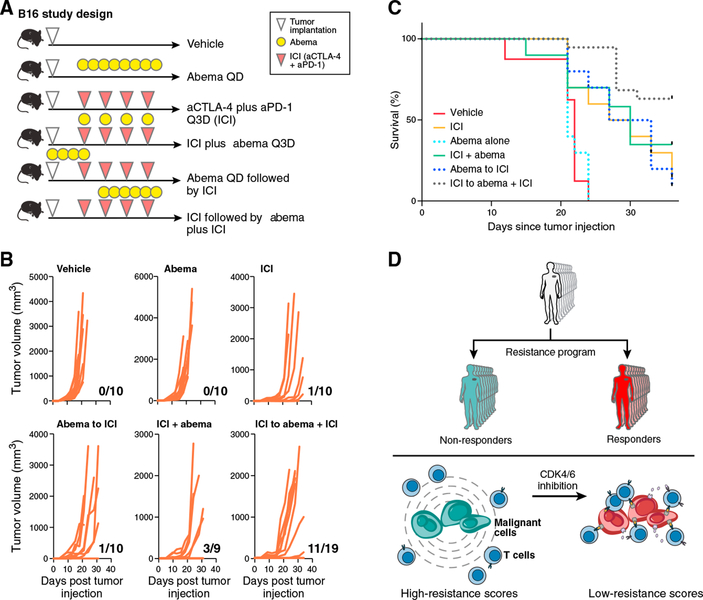
CDK4/6 Inhibition Enhances ICI Efficacy In Vivo
To determine abemaciclib’s efficacy in promoting anti-tumor activity in vivo, we tested its effect in the context of ICI therapy in the B16 melanoma model, which is relatively resistant to ICIs (Curran et al., 2010). scRNA-seq of B16, MC38, and CT26 cell lines in vitro showed that the resistance program is intrinsically expressed in most B16 cells but only in a portion of CT26 and MC38 cells (Figures S7C and S7D), corresponding to the responses of these models to ICI in vivo. We implanted B16 tumors in C57BL/6 mice and treated with different therapy arms (Figure 7A). Compared to vehicle, abemaciclib monotherapy had no impact on the rate of tumor outgrowth, but when used in a phased combination (ICI followed by ICI plus abemaciclib), there was a significant reduction in the rate of tumor outgrowth and improved survival (Figures 7B and 7C). Depletion of CD8+ T cells resulted in loss of the beneficial effect of phased combination therapy (Figures S7E and S7F) and the effect was at least in part RB dependent in the malignant cells (data not shown). Thus, abemaciclib may sensitize melanoma tumors to immunotherapies, even in the case of intrinsic resistance.
Figure 7. CDK4/6 Inhibition Combined with Immunotherapy Improves Response and Survival In Vivo.
(A) Study design. n = 9–19 per treatment group.
(B and ) Rate of tumor outgrowth (ratio for every graph) (B) is reduced in animals treated with phased combination (ICI followed by ICI plus abemaciclib) and results in higher survival rates compared to other treatments (C). p < 0.001, log-rank test.
(D) Immune resistance model.
See also Figure S7.
DISCUSSION
By leveraging clinical scRNA-seq data and multiple patient cohorts, we mapped malignant cell states associated with ICI resistance, revealing a coherent program that has a prognostic and predictive value and may be therapeutically targeted.
The program predicted ICI responses in several independent cohorts, outperforming other published signature-based biomarkers. Unlike our program, many of the existing biomarkers capture the tumor composition and do not have an additive predictive value once accounting for the inferred T cell levels. Our program predicts responses to anti-PD-1, and to some extent also to anti-CTLA-4 therapy, yet additional studies are required to dissect treatment-specific effects and predict ICI in all patients/cohorts.
The program is primarily associated with intrinsic ICI resistance. It is observed in bulk RNA-seq of untreated tumors and in a subset of ~24% of malignant cells of ~80% of untreated tumors. We predict that these malignant cells will have a selective advantage during ICI treatment. Indeed, the program is more pronounced after ICI failure, but not post targeted therapy, indicating that it does not merely reflect the impact of any therapeutic intervention.
By integrating scRNA-seq and in situ images of matched tissue slides, we showed that the program is robustly detected and consistent across data modalities and patient samples and is associated with cold niches within tumors. A key question is whether the program merely captures the response of malignant cells to immune infiltrates or marks an intrinsic mechanism that allows malignant cells to escape immunity and shape their microenvironment. As we show, the program expression varies across malignant human cell lines, which are not exposed to cues from non-malignant cells. Such intrinsic expression in vitro across mouse cell lines is aligned with their in vivo response to ICI.
Compounds that repress the program may sensitize malignant cells to immunotherapy and T cell-mediated killing (Figure 7D). We demonstrated that a CDK4/6i reverses the resistant cell state, induces components of SASP, and improves responses to ICI in vivo. These mechanisms are distinct from previously described immune-enhancing mechanisms of CDK4/6i (Deng et al., 2018; Goel et al., 2017) and indicate a potential role of CDK4/6, and specifically CDK4, as one of the master regulators of the program. Thus, CDK4/6i administered in a phased fashion could potentially alleviate ICI resistance in some melanoma patients, consistent with a recent observation (Schaer et al., 2018). More generally, the program’s repression in vitro could be a readout to screen for other compounds that sensitize melanoma tumors to ICI.
The program may be relevant in other tumor types. It is lower in some of the more ICI-responsive tumors (kidney, skin, lung) and higher in tumor types that are less responsive and/or arise from immune-privileged tissues (eye, testis) (Figures S7G and S7H). Synovial sarcoma, which is driven by a single genomic aberration in the BAF complex, has the highest resistance scores. The BAF complex plays a key role in ICI resistance (Pan et al., 2018), and one of its subunits (SMARCA4) is upregulated in the resistance program.
While we focused on malignant-cell-intrinsic mechanisms, we also tested for association of T cell abundance with the state of macrophages and B cells (Table S4C). We found a significant association only with macrophages: in cold niches/tumors, macrophages underexpress PD-L2, MHC class II genes (p < 1 × 10−17, hypergeometric test), and IFN-γ response genes (p = 9.76 × 10−10) and upregulate immunosuppressants, such as hypoxia genes (p = 4.55 × 10−6) and interleukin-8 (IL-8). Unlike the malignant cell program, the macrophage program was not associated with ICI resistance. Hence, it may represent the response of macrophages to T cell abundance rather than a cause of T cell exclusion.
Overall, our work sheds light on the interplay between cells and their microenvironment in tumors, uncovers improved biomarkers for patient selection, and reveals principles for new therapeutics.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Benjamin Izar (benjamin_izar@dfci.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human tumor specimen collection
For the discovery scRNA-seq cohort, tissue was procured under Institutional Review Board (IRB) approved protocols at Brigham and Women’s Hospital and Dana-Farber Cancer Institute, Boston, MA. Patients were consented to these protocols (11–104) in clinic visits prior to surgery/biopsy. Patients included in our earlier study (Tirosh et al., 2016), and those specimens newly collected here are annotated in Table S1A.
For validation cohorts (bulk-RNA-Seq), patient tissue was collected under IRB protocols of the University Hospital Essen, Germany and Massachusetts General Hospital, Boston, MA (protocol 11–181) and The Wistar Institute, Philadelphia, PA (Human subjects protocol 2802240).
Validation Cohort 1 included 90 samples from 26 patients, with multiple biopsies per patient, taken before, during, and/or after various treatment regimens, including both targeted therapies and immunotherapies. Clinical information, including the patient sex, age, and (ongoing/past) treatments when the sample was obtained are provided in Table S1B.
Validation Cohort 2 included 112 samples collected before treatment with pembrolizumab. Clinical information is provided in Tables S1C, including patient sex, and response to ICI (PFS, and RECIST). Information regarding the patient age was not available to us, and hence cannot be provided. Genes expression is provided in Table S6B.
Melanoma human cell lines
Established melanoma cell lines IGR39, A2058 and UACC62 were acquired from the Cancer Cell Line Encyplopedia (CCLE) from the Broad Institute. IGR39 was obtained from a 26 years old male patient. A2058 is an established cell line derived from a 43 years old male patient, UACC62 from a male patient (age unknown). Melanoma cell line 2686 and matched TILs ( = co-culture) were derived from a male patient (age unknown) and provided by MDACC (Peng et al., 2016).
Mouse cell line cultures
B16F10, MC38 and CT26 cell line were obtained from American Type Culture Collection (ATCC). B16F10 was derived from a male mouse, MC38 and CT26 were derived from female mice.
In vivo mouse model
B16F10 cells were subcutaneously injected into 6 weeks old female C57BL/6 mice (The Jackson Laboratory). All experiments were performed in accordance with the Dana-Farber Cancer Institute (DFCI) IACUC guidelines at the DFCI Longwood Center Animal Resource Facility per protocol 08–049.
METHOD DETAILS
scRNA-seq cohort data collection
Tissue handling and tumor disaggregation
Resected tumors were transported in DMEM (ThermoFisher Scientific, Waltham, MA) on ice immediately after surgical procurement. Tumors were rinsed with PBS (Life Technologies, Carlsbad, CA). A small fragment was stored in RNA-Protect (QIAGEN, Hilden, Germany) for bulk RNA and DNA isolation. Using scalpels, the remainder of the tumor was minced into tiny cubes < 1 mm3 and transferred into a 50 mL conical tube (BD Falcon, Franklin Lakes, NJ) containing 10 mL pre-warmed M199-media (ThermoFisher Scientific), 2 mg/mL collagenase P (Roche, Basel, Switzerland) and 10U/μl DNase I (Roche). Tumor pieces were digested in this media for 10 min at 37°C, then vortexed for 10 s and pipetted up and down for 1 min using pipettes of descending sizes (25 ml, 10 mL and 5 ml). As needed, this was repeated twice more until a single-cell suspension was obtained. This suspension was then filtered using a 70μm nylon mesh (ThermoFisher Scientific) and residual cell clumps were discarded. The suspension was supplemented with 30 mL PBS (Life Technologies) with 2% fetal calf serum (FCS) (Gemini Bioproducts, West Sacramento, CA) and immediately placed on ice. After centrifuging at 580 g at 4°C for 6 min, the supernatant was discarded and the cell pellet was re-suspended in PBS with 1% FCS and placed on ice prior to staining for FACS.
Fluorescence-activated cell sorting
Single-cell suspensions were stained with CD45-FITC (VWR, Radnor, PA) and live/dead stain using Zombie Aqua (BioLegend, San Diego, CA) per manufacturer recommendations. First, doublets were excluded based on forward and sideward scatter, then we gated on viable cells (Aqualow) and sorted single cells (CD45+ or CD45−) into 96-well plates chilled to 4°C, pre- prepared with 10μl TCL buffer (QIAGEN) supplemented with 1% beta- mercaptoethanol (lysis buffer). Single-cell lysates were sealed, vortexed, spun down at 3,700 rpm at 4°C for 2 min, placed on dry ice and transferred for storage at −80°C.
Library construction and sequencing
For plate-based scRNA-seq, we used a modified SMART-Seq2 protocol, as described previously (Trombetta et al., 2014). Briefly, cell lysates were thawed on ice, and RNA was purified using SPRI beads (Beckman Coulter, Brea, CA), followed by reverse transcription (RT). For RT, RNA species are incubated with the SMART CDS Primer IIA for 3 min at 72°C and placed on ice. Next., SMARTer IIA oligonucleotide and SMARTScribe RT were used for generating cDNA, and cleaned using SPRI beads (Beckman Coulter) per manufacturers’ instructions. For Whole- transcriptome amplification (WTA), we used Advantage 2 polymerase mix and IS PCR Primer (Clontech Ultra-low Input RNA-Kit). WTA products were cleaned with Agencourt XP DNA beads and 70% ethanol (Beckman Coulter) and Illumina sequencing libraries were prepared using Nextera XT kit (Illumina, San Diego, CA) The 96 samples of a multiwell plates were pooled, and cleaned with two 0.8X DNA SPRIs (Beckman Coulter). Library quality was assessed with a high sensitivity DNA chip (Agilent) and quantified with a high sensitivity dsDNA Quant Kit (Life Technologies).
For droplet-based scRNA-seq, experiments were performed on the 10x Genomics Chromium platform, with the Chromium Single Cell 3′ Library & Gel Bead Kit v2 and Chromium Single Cell 3′ Chip kitv2 according to the manufacturer’s instructions in the Chromium Single Cell 3′ Reagents Kits V2 User Guide. Briefly, ~6,000 cells were re-suspended in PBS supplemented with 0.04% BSA and loaded to each channel. The cells were then partitioned into Gel Beads in Emulsion in the GemCode instrument, where cell lysis and barcoded reverse transcription of RNA occurred, followed by amplification, shearing and 5′ adaptor and sample index attachment.
Barcoded single cell transcriptome libraries were sequenced with 38bp paired end reads on an Illumina NextSeq 500 Instrument.
RNA-Seq of validation cohorts 1 and 2
RNA extraction from formalin-fixed, paraffin-embedded (FFPE) tissue slides was performed by the Genomics Platform of the Broad Institute (Cambridge, MA). For cDNA library construction total RNA was assessed for quality using the Caliper LabChip GX2 (Perkin Elmer). The percentage of fragments with a size greater than 200nt (DV200) was calculated and an aliquot of 200ng of RNA was used as the input for first strand cDNA synthesis using Illumina’s TruSeq RNA Access Library Prep Kit. Synthesis of the second strand of cDNA was followed by indexed adaptor ligation. Subsequent PCR amplification enriched for adapted fragments. The amplified libraries were quantified using an automated PicoGreen assay (Thermo Fisher Scientific, Cambridge, MA). 200ng of each cDNA library, not including controls, were combined into 4- plex pools. Capture probes that target the exome were added, and hybridized for the recommended time. Following hybridization, streptavidin magnetic beads were used to capture the library-bound probes from the previous step. Two wash steps effectively remove any nonspecifically bound products. These same hybridization, capture and wash steps are repeated to assure high specificity. A second round of amplification enriches the captured libraries. After enrichment, the libraries were quantified with qPCR using the KAPA Library Quantification Kit for Illumina Sequencing Platforms (Illumina) and then pooled equimolarly. The entire process was performed in 96-well format and all pipetting was done by either Agilent Bravo or Hamilton Starlet. Pooled libraries were normalized to 2nM and denatured using 0.1 N NaOH prior to sequencing. Flowcell cluster amplification and sequencing were performed according to the manufacturer’s protocols using Illumina HiSeq 2000 or 2500 (Illumina). Each run was a 76bp paired-end with an eight-base index barcode read. Data was analyzed using the Broad Picard Pipeline (http://broadinstitute.github.io/picard/), which includes de-multiplexing and data aggregation.
In situ imaging
Formalin-fixed, paraffin-embedded (FFPE) tissue slides, 5 μm in thickness, were generated at the Brigham and Women’s Hospital Pathology Core Facility from tissue blocks collected from patients under IRB-approved protocols (DFCI 11–104). Multiplexed, tissue cyclic immunofluorescence (t-CyCIF) was performed as described recently (Lin et al., 2018). For direct immunofluorescence, we used the following antibodies: CEP170 (Abcam, ab84545), LAMP2 (R&D technologies, AF6228), MITF (Abcam, ab3201), DLL3 (Abcam, ab103102, Rab), MITF (Abcam, ab3201, Ms), S100α−488 (Abcam, ab207367), CD3–555 (Abcam, ab208514), CD8a-660 (eBioscience, 50–0008-80), cJUN-488 (Abcam, ab193780), cMyc-555 (Abcam, ab201780), HLAA-647 (Abcam, ab199837), TP53–488 (Cell Signaling, 5429), SQSTM1–555 (Abcam, ab203430). Stained slides from each round of CycIF were imaged with a CyteFinder slide scanning fluorescence microscope (RareCyte Seattle WA) using either a 10X (NA = 0.3) or 40X long- working distance objective (NA = 0.6). Imager5 software (RareCyte) was used to sequentially scan the region of interest in 4 fluorescence channels. Image processing and single-cell quantification was performed as previously described (Lin et al., 2018). Briefly, background subtraction was performed using the established rolling ball algorithm (with a 50-pixel radius) followed by registration in ImageJ. Nuclear stainings from each cycle were used to generate reference coordinates by Rigid-body transformation, and these coordinates were used to virtually hyper-stak all acquired 4-color-cycles. To obtain multiplex intensity measurements for single cells, images were first thresh-olded using the OTSU algorithm and binarized in the nuclear staining channels, and the Watershed algorithm (based on nuclear staining) was used to segment individual cells. For this purpose, the cytoplasm was captured by centripetal expansion (starting from the nucleus) of either of 3 pixels (10X objective) and 6 pixels (40X objective) toward the cells’ boundaries, defined as the cell membrane. The cytoplasm was defined as the region between defined as cell membrane and nucleus. Single-cell intensity measures were determined by projecting fluorescence-intensities to the coordinates of segmented individual cells. Additional details, protocols and code are deposited in https://www.cycif.org/.
Abemaciclib treatment of melanoma cell lines
Established melanoma cell lines IGR39, UACC62 and A2058 were treated every 3 days with 500 nM abemacilib (LY2835219, Med-ChemExpress) or DMSO control. The doubling time of each cell line was established and lines were seeded such that cells collected for scRNA-seq were derived from culture dishes with ~50%–60% confluency on day 7 of treatment. Cells were lifted of culture dishes using Versene solution (Life Technologies), washed twice in 1x PBS, counted and resuspended in PBS supplemented with 0.04% BSA for loading for scRNA-seq with the 10X Genomics platform.
Abemaciclib cytostatic effects
5 × 104 human melanoma cells (cell line 2686) were seeded and treated with either DMSO or 500 nM abemaciclib (LY2835219, MedChemExpress) daily for a total of 6 days. Cell number and viability were determined using the Countess II FL Automated Cell Counter (ThermoFisher) on days 2, 4 and 6. All experiments were performed in triplicates. Cell doubling per 24 hours was determined based on cell numbers comparing cells seeded and on day 6 using the following formula: duration (in days) × log(2)/log(final cell number) – log(initial cell number).
Melanoma-TIL co-culture experiments
Melanoma cell line 2686 was pre-treated with 500 nM abemaciclib or DMSO control for 7 days followed by co-culture with autologous TILs (with an effector to target ratio of 5:1) for 48 hours. TILs were removed by pipetting of the supernatant, and the remaining melanoma cells were washed twice with PBS, lifted off the culture dish, and resuspended in PBS supplemented with 0.04% BSA for loading for scRNA-seq with the 10X Genomics platform.
Mouse cell line culture experiments
B16F10 cells were maintained in complete DMEM media (10% FBS and 50U/mL of Penicillin- Streptomycin). MC38 and CT26 were maintained in RPMI-1640 (10% FBS and 50U/mL of Penicillin-Streptomycin). For scRNA-seq, cells were washed twice with PBS, incubated with trypsin 0.05% for 2 min at 37°C, quenched with complete media, and resuspended in PBS supplemented with 0.04% BSA prior to loading onto the Chromium 10X platform and processed as described above. All cell lines were tested for Mycoplasma.
In vivo mouse experiments
5.0 * 105 B16F10 cells were subcutaneously injected into 6 weeks old female C57BL/6 mice (The Jackson Laboratory). Four or seven days after injection, animals were treated with either (1) isotype control antibody injected intraperitoneally (2A3 and polyclonal syrian hamster IgG, 200μg/mouse and 100μg/mouse, respectively) every 3 days for a total of 4 doses; (2) abemaciclib 90 mg/kg daily administered by oral gavage (LY2835219, MedChemExpress) for a total of 10 doses; (3) combination immune checkpoint blockade (ICI) with αPD-1 (clone 29F.1A12, 200μg/mouse) plus αCTLA-4 (clone 9H10, #BP0131,100μg/mouse) every 3 days for a total of 4 doses; (4) abemaciclib (90mg/kg) daily for 4 days followed by ICI every 3 days for a total of 4 doses; (5) ICI for one initial dose followed 3 days later by abemaciclib (90mg/kg) daily combined with ICI every 3 days for an additional 3 doses; or (6) upfront combination of ICI and abemaciclib (75mg/kg) every 3 days for a total of 4 doses. CD8+ T cell depletion was achieved using CD8b mAb (clone53–5.8, #BE0223, 100μg/mouse) that was administered on day-1, day 0 once a week until the end of the experiment. Tumor size was measured using digital calipers every 3 days. Tumor volume was determined by calculating (length × width2)/2. Growth curves and survival curves were generated using GraphPad Prism v8.
QUANTIFICATION AND STATISTICAL ANALYSIS
scRNA-seq cohort processing
Gene expression quantification
BAM files were converted to merged, demultiplexed FASTQ files. The paired-end reads obtained with the SMART-Seq2 protocol were mapped to the UCSC hg19 human transcriptome using Bowtie (Langmead et al., 2009), and transcript-per-million (TPM) values were calculated with RSEM v1.2.8 in paired-end mode (Li and Dewey, 2011). The paired-end reads obtained with the 10x Genomics platform were mapped to the UCSC hg19 human transcriptome using STAR (Dobin et al., 2013), and gene counts/TPM values were obtained using the 10x Genomics computational pipeline (cellranger-2.1.0).
For bulk RNA-Seq data, expression levels of genes were quantified as Ei,j = log2(TPMi,j+1), where TPMi,j denotes the TPM value of gene i in sample j. For scRNA-seq data, expression levels were quantified as Ei,j = log2(TPMi,j/10+1), where TPMi,j denotes the TPM value of gene i in cell j. TPM values were divided by 10 because the complexity of the single-cell libraries is estimated to be within the order of 100,000 transcripts. The 10−1 factoring prevents counting each transcript ~10 times, which would have resulted in overestimating the differences between positive and zero TPM values. The average expression of a gene i across a population of N cells, denoted here as P, was defined as
For each cell, we quantified the number of genes with at least one mapped read, and the average expression level of a curated list of housekeeping genes (Tirosh et al., 2016). We excluded all cells with either fewer than 1,700 detected genes or an average housekeeping expression (E, as defined above) below 3 (Table S2). For the remaining cells, we calculated the average expression of each gene (Ep), and excluded genes with an average expression below 4, which defined a different set of genes in different analyses depending on the subset of cells included. In cases where we analyzed different cell types together, we removed genes only if they had an average Ep below 4 in each of the different cell types that were included in the analysis. When analyzing CD45+ cells, we excluded genes as described above only after the assignment of cells to cell types in order to prevent the filtering of genes that were expressed by less abundant cell types.
Imputation and normalization
In all differential expression analyses of SMART-Seq2, we first modeled the read counts as a mixture of a negative binomial (NB) and Poisson components to estimate the expression levels, using SCDE (Fan et al., 2016) with the code provided in https://github.com/hms-dbmi/scde. The resulting normalized and imputed expression matrix, denoted as E’, was used in the differential expression analyses. Analysis of droplet-based scRNA-seq data (10X Genomics Chromium, above) was performed with the Seurat package (https://satijalab.org/seurat), using the likelihood-ratio test for differential gene expression analyses.
Classification of malignant and stromal cells
In the non-immune compartment (CD45− cells), we distinguished malignant and nonmalignant cells according to three criteria: (1) their inferred CNV profiles (Tirosh et al., 2016); (2) under- expression of different nonmalignant cell type signatures; and (3) higher similarity to melanoma tumors than to adjacent normal tissue, based on the comparison to bulk RNA-Seq profiles. Specifically: (1) to infer CNVs from the scRNA-seq data we used the approach described in (Tirosh et al., 2016) as implemented in the R code provided in https://github.com/broadinstitute/inferCNV with the default parameters. Cells with an average absolute CNV level that was below the 0.1 quantile of the entire CD45− cell population were considered as potentially nonmalignant according to this criterion. (2) We used signatures of endothelial cells, stromal cells, and CAFs, as provided in Table S3A. The signatures combine well-established markers from two sources (https://www.biolegend.com/cell_markers and (Tirosh et al., 2016)). We computed the OE of these three signatures in each of the CD45− cells, while controlling for the impact of technical cell quality (as described in section Gene sets overall expression). CD45− cells that expressed any one of these three signatures above the 0.95 quantile were considered as potentially nonmalignant according to this criterion. (3) We downloaded the pan-cancer TCGA RNA-SeqV2 expression data from https://xena.ucsc.edu/, and log2-transformed the RSEM-based gene quantifications. For each cell, we computed the correlation between its profile (in TPM) and each bulk profile (in TPM) of 473 skin cutaneous melanoma samples and 727 normal solid tissues. We then tested, for each cell, if it was more similar to the melanoma tumors compared to the normal tissues, by applying a one-sided Wilcoxon ranksum test on the correlation coefficients that were obtained for that cell. Cells that were more similar to the normal tissues (p < 0.05, Wilcoxon ranksum test) were considered as potentially nonmalignant according to this criterion.
The cell assignments that were obtained by these three different criteria were highly consistent (Figures S1A and S1B, hypergeometric p value < 10−17). Cells that were identified as potentially nonmalignant according to one or more of these three criteria were defined as nonmalignant, and were omitted from further analyses of the malignant cells. The nonmalignant CD45− cells were further classified into CAFs and endothelial cells, if they overexpressed only one of the corresponding gene signatures, and as unresolved cells otherwise.
Classification of immune cells
To classify immune cells, we first filtered CD45+ cells that were potentially malignant or doublets of immune and malignant cells based on their inferred CNV profiles. To this end, we defined the overall CNV level of a given cell as the sum of the absolute CNV estimates across all genomic windows. For each tumor, we generated its CNV profile by averaging the CNV profiles of its malignant cells, when considering only those with the highest overall CNV level (top 10%). We then evaluated each cell by two values: (1) its overall CNV level, and (2) its CNV-R-score, that is, the Spearman correlation coefficient obtained when comparing its CNV profile to the CNV profile of its tumor. These two values were used to classify cells as malignant, nonmalignant, and unresolved cells that were excluded from further analysis (Figures S1C-S1E).
Next, we applied two different clustering approaches to assign immune (CD45+) cells into cell types. In the first approach, we clustered the CD45+ cells according to 194 well-established markers of 22 immune cell subtypes (Table S3A; assembled from https://www.biolegend.com/cell_markers and (Tirosh et al., 2016)). The clustering was performed in three steps: (1) we computed the Principal Components (PCs) of the scRNA-seq profiles, while restricting the analysis to the 194 biomarker genes. We used the top PCs that captured more than 50% of the cell-cell variation. In the presented analyses, 10 PCs were used, but the results were robust and stable when using the first 5–15 PCs. (2) We applied t-SNE (t-Distributed Stochastic Neighbor Embedding) (van der Maaten and Hinton, 2008) to transform these first PCs to a two- dimensional embedding, using the R implementation of the t-SNE method with the default parameters, as provided in http://lvdmaaten.github.io/tsne/. (3) We applied a density clustering method, DBscan (Ester et al., 1996), on the two-dimensional t-SNE embedding that was obtained in (2). This process resulted in four clusters for which the top preferentially expressed genes included multiple known markers of particular cell types (Figures S1F and S1G).
To map between clusters and cell types we compared each cluster to the other clusters according to the OE of the different cell type signatures (one sided t test, Table S3A). The cell type signature that was most significantly (t test p value < 10−10) overexpressed in the cluster compared to all other clusters was used to define the cluster identity. In this manner, we annotated the clusters as CD8+ and CD4+ T cells, B cells, and macrophages (Figure 1D). Cells that clustered with the CD8+ T cells and did not express CD8A or CD8B were labeled as NK cells if they overexpressed NK markers, otherwise they were considered as unresolved T cells. T cells that were clustered together with the CD4 T cells and expressed CD8A or CD8B were also considered as unresolved T cells. Unresolved T cells were not used in further analyses.
To assess the robustness of our assignments, we applied another approach, and determined the concordance between the two assignments. In the second approach, we first made initial cell assignments based on the OE of well-established cell type markers: T cells (CD2, CD3D, CD3E, CD3G), B cells (CD19, CD79A, CD79B, BLK), and macrophages (CD163, CD14, CSF1R).
Across all the CD45+ cells, the OE levels of these signatures had bimodal distributions. We used the bimodal OE of each signature to assign cells to cell types (see Gene sets overall expression). Cells that were assigned to more than one cell type at this point were considered as unresolved. Cells that were defined as T cells according to this measure were further classified as CD8+ or CD4+ T cells if they expressed CD8 (CD8A or CD8B) or CD4, respectively. T cells that expressed both CD4 and CD8 were considered as unresolved. As a result, 67.3% of the cells had an initial cell type assignment.
Next, we clustered the cells with the Infomap algorithm (Rosvall and Bergstrom, 2008). Infomap decomposes an input graph into modules by deriving a compressive description of random walks on the graph. The input to the algorithm was an unweighted k-NN graph (k = 50) that we generated based on the expression of the 194 biomarker genes across the CD45+ cells. Infomap produced 22 clusters, separating the different CD45+ cells not only according to cell types but also according to various cell states. For each cluster, we examined if it was enriched with cells of a specific cell type, according to the initial assignments. Nineteen clusters were enriched with only one cell type. The cells within these clusters were assigned to the cell type of their cluster, unless their initial assignment was different, and in this case, they were considered as unresolved.
The cell type assignments that were obtained by the two approaches were highly concordant: 97% of the cells had the same assignment with both approaches.
Interactive visualization of the immune cells’ tSNE plots are provided in https://portals.broadinstitute.org/single_cell/study/melanoma-immunotherapy-resistance.
Gene sets overall expression
Gene modules are more robust to noise and provide more coherent signals than the expression of single genes. To compute the OE of a gene module or signature we used a scheme that filters technical variation and highlights biologically meaningful patterns. The procedure is based on the notion that the measured expression of a specific gene is correlated with its true expression (signal), but also contains a technical (noise) component. The latter may be due to various stochastic processes in the capture and amplification of the gene’s transcripts, sample quality, as well as variation in sequencing depth. The signal-to-noise ratio varies, depending, among other variables, on gene transcript abundance.
We therefore computed the OE of gene signatures in a way that accounts for the variation in the signal-to-noise ratio across genes and cells. Given a gene signature and a gene expression matrix E (as defined above), we first binned the genes into 50 expression bins according to their average expression across the cells or samples. The average expression of a gene across a set of cells within a sample is Ei,p (see: scRNA-seq cohort processing) and the average expression of a gene across a set of N tumor samples was defined as:. Given a gene signature S that consists of K genes, with kb genes in bin b, we sample random S-compatible signatures for normalization. A random signature is S-compatible with signature S if it consists of overall K genes, such that in each bin (b) it has exactly kb genes. The OE of signature S in cell or sample j is then defined as:
Where S is a random S-compatible signature, and Cij is the centered expression of gene i in cell or sample j, defined as . Because the computation is based on the centered gene expression matrix C, genes that generally have a higher expression compared to other genes will not skew or dominate the signal.
We found that 1,000 random S-compatible signatures are sufficient to yield a robust estimate of the expected value . The distribution of the OE values was normal or a mixture of normal distributions, and, unlike the expression of a single gene, fulfilled the assumptions of the mixed effects models or hierarchal linear models that we applied to study the differential expression of gene signatures (as described in the Differentially expressed gene sets section).
In cases where the OE of a given signature has a bimodal distribution across the cell population, it can be used to naturally separate the cells into two subsets. To this end, we applied the Expectation Maximization (EM) algorithm for mixtures of normal distributions to define the two underlying normal distributions. We then assigned cells to the two subsets, depending on the distribution (high or low) that they were assigned to.
We use the term a transcriptional program (e.g., the immune resistant program) to characterize cell states which are defined by a pair of signatures, such that one (S-up) is overexpressed and the other (S-down) is underexpressed. We define the OE of such cell states as the OE of S-up minus the OE of S-down.
To compute the OE of gene signatures in bulk tumors or single cell data one can apply the R code we provide via GitHub (https://github.com/livnatje/ImmuneResistance; ImmRes_OE.R).
Cell type signatures
To identify cell type signatures we performed pairwise comparisons between the eight cell types that we identified: malignant cells, CAFs, endothelial cells, NK cells, B cells, macrophages, and CD8+ and CD4+T cells. We performed these comparisons via one-sided Wilcoxon ranksum-tests on the normalized data E. Genes were considered as cell type specific if they were overexpressed in a particular cell type compared to all other cell types (Wilcoxon ranksum-test p value < 10−5). For T cell types (CD8+ and CD4+) we used more permissive cutoffs, as the transcriptional differences between these cell types are more subtle. To ensure that the genes are significantly higher in the specific T cell type across all pairwise comparisons, for each T cell type we: (1) computed the least significant (maximal) p value for each gene, when comparing its expression in the T cell type (CD8+ or CD4+) to its expression in each of the other cell types, (2) used the genes with the lowest maximal p values to construct the signature (at most 50 genes with maximal p < 2*10−3). To derive a Pan-T cell signature that characterizes both CD8+ and CD4+ T cells, we used genes that were overexpressed in CD8+ and CD4+ T cells (p < 10−5) compared to all other cell types, when disregarding T cells and NK cells.
To derive CD8+ T cell state signatures we: (1) classified CD8+ T cells as cytotoxic, exhausted, naive, or ‘undetermined’ based on well-established markers of these cell states (Table S3A); (2) performed pairwise comparisons between the different CD8+ T cell subsets to derive initial CD8+ T cell state signatures; (3) compared each CD8+ T cell subset to all other cell types (e.g., B cells, macrophages, etc.), and (4) filtered non-specific genes from the initial CD8+ T cell state signatures. We performed the same analysis with CD4+ T cells to derive signatures of cytotoxic, exhausted, naive, and regulatory CD4+ T cells. The cytotoxic CD4+ T cells signature included only four genes and thus was not used in subsequent analyses.
We also identified signatures of three broader cell type categories: immune cells, lymphocytes, and stromal cells. The immune cell signature includes genes that are overexpressed by all the immune cell types compared to all other non-immune cells, and likewise for the other two cell categories. The lymphocyte signature included only five genes and thus was not used in subsequent analyses.
The lists of cell subtype specific genes, which were identified as described above, are provided in Table S3B. The implementation of these analyses is provided via GitHub (https://github.com/livnatje/ImmuneResistance/wiki/Mapping-immune-resistance-in-melanoma; see ImmRes1_denovoCellTypeSig.R), and can be applied to reproduce the cell subtype signatures and generate cell subtype signatures based on other scRNA-seq cohorts.
Linking cell states to tumor composition
We combined scRNA-seq and bulk RNA-Seq data to characterize the state of a specific cell type in tumors with a specific cellular composition (Figure 1B). The method takes as input scRNA-seq data and a cohort of bulk RNA-Seq data, both collected from tumors of the same cancer type. The implementation of the approach is provided via GitHub (https://github.com/livnatje/ImmuneResistance/wiki/Mapping-immune-resistance-in-melanoma; ImmRes2_immuneResistanceProgram.R), and can be applied to regenerate the results reported here and facilitate new discoveries.
For clarity we describe the approach for malignant cells and T cells as applied here, although it can be applied to any pair of cell types, depending on data availability.
-
STEP 1.
Analyses of the input scRNA-seq data: (a) assign cells to cell types (see sections: Classification of malignant and stromal cells and Classification of immune cells); and (b) define a signature of malignant cells and a signature of T cells, consisting of genes which are primarily (specifically) expressed by malignant cells or T cells, respectively (see section: Cell type signatures).
-
STEP 2.
Analyses of the input bulk RNA-Seq data: (a) estimate the T cell infiltration level in each tumor by computing the overall expression (OE, see section: Gene sets overall expression) of the T cell signature in each bulk sample; (b) compute the Pearson correlation coefficient between the expression of the genes in the malignant signature and the OE of the T cell signature across the bulk tumors; and (c) define the seed exclusion-up (down) signature as the top 20 malignant genes that are significantly negatively (positively) correlated in (b) (adjusted p < 0.1, using Benjamini-Hochberg correction for multiple hypotheses testing (Benjamini and Hochberg, 1995)). If confounding factors in the bulk RNA-Seq data should be controlled for, partial Pearson correlation is used in (2.b).
-
STEP 3.
Analyses of the input scRNA-seq data of the malignant cells: (a) compute the OE of the seed exclusion signatures in each of the malignant cells; (b) compute the partial Spearman correlation coefficient between the expression of each gene and the OE of the seed exclusion signatures across the single malignant cells, while controlling for technical quality (the number of reads and genes that were detected in the cells). Other confounding factors, besides technical variation, can also be controlled for, for example, cell cycle (see below). Of note, to examine whether the seed signatures capture the two opposing transcriptional components of a malignant cell state, we test whether their OE across the malignant cells is negatively correlated.
-
STEP 4.
Derive the final genome-scale exclusion signatures, defined as: (i) exclusion-up: genes which were significantly positively correlated with the seed exclusion-up signature and significantly negatively correlated with the seed exclusion-down signature in the analysis described in (STEP 3); and (ii) exclusion-down: genes which were significantly positively correlated with the seed exclusion-down signature and significantly negatively correlated with the seed exclusion-up signature in the analysis described in (STEP 3). In this analysis, a gene is defined as significantly correlated with a signature if it was among the 200 topmost correlated genes, with Spearman correlation coefficient |r| > 0.1 and adjusted p value < 10−10 (Benjamini-Hochberg correction for multiple hypotheses testing (Benjamini and Hochberg, 1995)).
To generate the exclusion program reported in the manuscript, we applied the approach to our clinical scRNA-seq melanoma data and bulk RNA-Seq data of 473 Skin Cutaneous Melanoma (SKCM) tumors from TCGA (as provided in https://xenabrowser.net/datapages/). In (STEP 2) we used the CD8+ T cell signature we generated (Table S3B).
We also performed several additional analyses. First, for comparison, we also applied our approach to other T cell populations, by using other T cell signatures (Table S3B) in (STEP 2): pan-T cells (CD3+), CD4+ T cells; naive, cytotoxic, and exhausted CD8+ T cell subsets; and naive, exhausted and regulatory CD4+ T cell subsets. Second, to compute the relative abundance of CD8+ T cells among the non-malignant cells we used the CD8+ T cell signature and tumor purity that was previously assessed (Akbani et al., 2015) by genetic variations with ABSULOTE (Carter et al., 2012). Of note, in this setting the resulting “up” and “down” seed signatures were less anti- correlated with each other across the single malignant cells (Pearson r = −0.22, p = 1.40*10−24, compared to r = −0.58, p = 6.19*10−183, in the original setting). These findings indicate that the association between the state of the malignant cells and the relative proportion of T cells out of the non-malignant compartment may be more complex than the connection between malignant cells states and the proportion of T cells in the overall tumor. Tumor purity was provided in (STEP 2) as a confounding factor. Third, we applied our approach when controlling for tumor and cellular proliferation. To do so, we computed the OE of two cell cycle signatures (G1/S and G2/M, Table S3A (Tirosh et al., 2016)) across the bulk tumors and single malignant cells. We then provided these values as confounding factors in (STEP 2) and (STEP 3), to control for the tumor proliferation rate and cell cycling, respectively.
In cases where the approach is applied to two cell types whose abundance in the tumor is highly correlated, for example macrophages and T cells, the seed exclusion-up signature can be identified in a more permissive manner. Otherwise, it may include very few genes. In the case of macrophages and T cells, the (more permissive) seed exclusion-up signature includes macrophage specific genes that fulfill the following requirements: (1) in comparison to other macrophage specific genes, their expression across bulk tumors has a low correlation to T cell abundance (conditional probability < 0.1); and (2) their expression across macrophages in the scRNA-seq data is negatively correlated to the OE of the exclusion-down seed signature. The analysis then proceeds in the same manner as described above.
The post-treatment program
Differentially expressed genes
We identified genes differentially expressed between the malignant cells of untreated and post- treatment tumors by using a subsampling approach that mitigates the effects of outliers and prevents tumors with a particularly large number of sequenced malignant cells from dominating the results. In each subsample, we selected a subset of the tumors, subsampled at most 30 malignant cells from each tumor, and identified differentially expressed genes between the post- treatment and untreated cells. Differentially expressed genes were identified by applying SCDE (Kharchenko et al., 2014), a Bayesian method that was developed specifically to detect single-cell differential expression. As input to SCDE we used the normalized and imputed expression matrix E’ (see Imputation and normalization).
We repeated the subsampling procedure 500 times, and computed for each gene g the fraction of subsamples in which it was found to be significantly under (Fdown,g) or over (Fup,g) expressed in the post-treatment population compared to the untreated population (|z-score| > 1.96). Genes with Fdown,g values larger than the 0.9 quantile of the Fdown distribution were considered as potentially downregulated in the post-treatment malignant cells. Likewise, genes with Fup,g values larger than the 0.9 quantile were considered as potentially upregulated in the respective post-treatment malignant cells.
We further filtered the signatures with two additional statistical tests that we applied on the full scRNA-seq data (E’) of the malignant cells. The first test was SCDE followed by multiple hypotheses correction (Holm-Bonferroni (Holm, 1979)). The second was a nonparametric empirical test, where we performed a Wilcoxon ranksum test to examine if a given gene is differentially expressed in the post-treatment versus untreated cells, based on E’. We corrected for multiple hypotheses testing using the Benjamini-Hochberg False Discovery Rate (FDR) (Benjamini and Hochberg, 1995), and obtained empirical p values to ensure the differences in expression were not merely reflecting differences in cell quality (i.e., the number of aligned reads per cell). To this end, we generated 1,000 random permutations of the gene expression matrix E’, such that each permutation preserves the overall distribution of each gene, as well as the association between the expression of each gene and cell quality. We performed the Wilcoxon ranksum test on the permuted E’ matrixes to compute empirical p values.
To assemble the final post-treatment signatures, we selected genes that fulfilled the subsampling criteria described above and were most significantly differentially expressed according to both the SCDE and empirical tests (top 200 genes with corrected p < 0.05).
The implementation of these analyses is provided via GitHub (https://github.com/livnatje/ImmuneResistance/wiki/Mapping-immune-resistance-in-melanoma; ImmRes2_immuneResistanceProgram.R), and can be applied to regenerate the posttreatment signatures from our data.
Differentially expressed gene sets
To test the ability of a given gene signature to distinguish between the malignant cells collected from post-treatment versus untreated patients we modeled the data with a mixed-effects model that accounts for the dependencies and structure of the data. The model had overall five covariates and two levels: (1) a cell-level, and (2) a sample-level. Level-1 covariates controlled for cell quality by providing the number of reads (log-transformed) and the number of genes that were detected in each malignant cell, and denoted which cells were cycling, based on the bimodal OE of the cell cycle signatures defined in (Tirosh et al., 2016). Level-2 covariates were the patient’s gender, age, and treatment group, and a binary covariate that denotes if the sample was a metastatic or a primary lesion. The sample-level controlled for the dependency between the scRNA-seq profiles of malignant cells that were obtained from the same patient, having a sample-specific intercept. Using this model we quantified the significance of the association between the treatment covariate and the OE of a given signature across the malignant cells. We implemented the mixed-effects model in R, using the lme4 and lmerTest packages (https://cran.r-project.org/web/packages/lme4/index.html, https://cran.r-project.org/web/packages/lmerTest/index.html).
We applied this approach to test the post-treatment and exclusion programs defined here, as well as annotated pre-defined gene sets downloaded from MSigDB v6.0 (Subramanian et al., 2005). The results are provided in Table S5 and Figure 2D.
Cross-validation
To examine the generalizability of the post-treatment signatures we performed a cross-validation procedure. In each cross-validation round the test set consisted of all the cells of one patient, and the training set consisted of the data from all the other patients in our cohort. In each round, we used only the training data to generate post-treatment signatures (as described in Differentially expressed genes), and computed the OE of the resulting post-treatment program. To center the expression matrix for the computation of the OE values, we used all the malignant cells in the data, such that the OE scores of one patient were relative to those of the other patients. Finally, we computed Receiver Operating Characteristic (ROC) curves based on the resulting post-treatment OE scores, obtained for the test set.
The immune resistance program
We combined the post-treatment and exclusion programs with a simple union of the matching signatures, into the immune resistance program (Table S4A). To give more weight to genes that are included in both signatures we compute the OE of the resistance program by averaging the OE of the exclusion and post-treatment programs.
We further refined the immune resistance program by integrating the scRNA-seq data with the results of a genome-scale CRISPR screen that identified gene KOs which sensitize malignant melanoma cells to T cell killing (Patel et al., 2017). We defined our single malignant cells as putatively “resistant” if they underexpressed (lowest 1%) one of the top hits of the screen: B2M, CD58, HLA-A, MLANA, SOX10, SRP54, TAP2, TAPBP. This underexpression did not reflect low cell quality, because these “resistant” cells had a higher number of genes and reads. These cells had significantly higher immune resistance scores (p = 2.24*10−18 and 1.59*10−3, t test and mixed- effects, respectively), and were enriched with cycling cells (p = 1.74*10−13, hypergeometric test). We derived a functional resistance program that consists of differentially expressed genes when comparing the “resistant” cells to other malignant cells (Table S4A). We then refined the resistance score by adding to it also the OE of this functional program (note that this purposely increases the contribution to the score of genes that are both in the original resistance program and are identified by this additional comparison).
We report the performances of all the resistance program subsets: exclusion, post-treatment, and their (weighted) union, with and without the functional refinement (Figures S5 and S6). As comparators, we used the hits of the co-culture screen along with other potentially prognostic signatures, to generate competing predictors of patient survival and response (Figures 5G and 5H, Table S6A, see section Alternative ICI-response predictors).
The immune resistance program versus cell cycle
We applied two approaches to examine the association between the immune resistance program and cell cycle: (1) we detected immune resistance genes when using only non-cycling malignant cells, and (2) we used all the data after filtering cell cycle effects. We start by computing the OE of two cell cycle signatures (G1/S and G2/M, Table S3A) (Tirosh et al., 2016) across the malignant cells and bulk melanoma TCGA tumors.
In the first approach we classified malignant cell as cycling or non-cycling, and confirmed that the immune resistance program can be recovered also when excluding all cycling cells from consideration. More specifically, when analyzing only the non-cycling malignant cells, all but two of the genes in the post-treatment signatures were still significantly differentially expressed between the untreated and post-treatment tumors, and all genes from the exclusion program were still significantly associated with T cell exclusion.
In the second approach, we re-generated the program based on all the data while controlling for cell cycle as a potential confounder. In the case of the exclusion signatures, we provided these cell cycle scores as confounding factors in the partial correlation analyses (see steps 2 and 3 in Linking cell states to tumor composition). In the case of the post-treatment signatures, we filtered out the cell cycle features from the data using PAGODA (Fan et al., 2016) (using pagoda.subtract.aspect) and regenerated the posttreatment signature. This regression successfully masked the differences between cycling and non-cycling cells, for example, when considering cell cycle genes (Figure S2F). However, it did not mask the association between cycling cells and the expression of the immune resistance genes (Figures S2G and S2H).
The resistance program that we generated after filtering cell cycle effects (Table S4B) was very similar to the original one (p < 1*10−17, hypergeometric test, Jaccard index = 0.56 and 0.66, induced and repressed signatures, respectively; Figure S2D). Lastly, the OE of the two programs was highly correlated across the malignant cells (r = 0.99, p < 1*10−17), such that cycling cells overexpressed both programs (p < 3.48*10−7, mixed-effects; Figure S2E).
Cell-cell interaction network
We generated genome-scale cell-cell interactions networks by integrating (1) protein-protein interactions that were previously assembled by (Ramilowski et al., 2015) as cognate ligand- receptor pairs, with (2) cell subtype specific signatures from the singlecell profiles, identified as described above in Cell type signatures. The resulting network maps the physical interactions between the different cell subtypes that we characterized. Each cell subtype and protein are represented by a node in the network. An edge between a cell subtype node and a ligand or receptor node denotes that this protein is included in the cell-subtype signatures. An edge between two proteins denotes that they can physically bind to each other and mediate cell-cell interactions. A path from one cell subtype to another represents a potential route by which the cells can interact. For each cell subtype, we defined a ‘communication signature’, which includes all the surface proteins that can bind to this cell subtype signature proteins. To examine if the immune resistant malignant cells suppress their interactions with other cell subtypes we examined if the different immune resistance signatures were enriched (hypergeometric test) with genes from the different immune and stroma ‘communication signatures’ (Figure 3B). An interactive map of the cell-cell interaction network is provided in https://portals.broadinstitute.org/single_cell/study/melanoma-immunotherapy-resistance#study-download, and can be explored with Cytoscape https://cytoscape.org/.
Clinical longitudinal analyses
Validation cohort 1
We used a mixed-effects model to represent the longitudinal treatment data (Figure 1A, validation cohort 1) and examine the association between the expression of various gene signatures and different treatment categories. The model included two levels: a sample-level and a patient-level. The sample-level had overall 8 covariates: three treatment covariates and five tumor microenvironment covariates. The sample-level treatment covariates denote in this case whether the sample was exposed to: (1) targeted therapy (on/post RAF/MEK-inhibitors), (2) ICI (on/post), or (3) non-ICI immunotherapy (NK antibodies, IL2, IFN, or GM CSF) without ICI. The five sample-level tumor microenvironment covariates control for potential changes in the tumor composition by providing the OE of the different nonmalignant cell type signatures that we identified (Table S3B). The patient-level controlled for the dependency between the scRNA-seq profiles of samples that were obtained from the same patient, having a patient-specific intercept. The dependent variable was the OE of a given signature, which enabled us to quantify the association between the immune resistance program (or any other signature) and the exposure to ICI or targeted therapy (the second and first sample-level covariates, respectively). The implementation of this model is provided via GitHub (https://github.com/livnatje/ImmuneResistance/wiki/Predicting-immunotherapy-resistance; ImmRes3_longitudinal.R).
Targeted therapy cohort
To test if the immune resistance program is related to resistance to MAPK inhibitors we analyzed a published clinical cohort of patient-matched melanoma tumors biopsied before MAPK inhibitor therapy and during disease progression (Hugo et al., 2015). We used the same mixed-effects model described above (Validation cohort 1), except that instead of three treatment covariates we had only one, denoting if the sample was pre or post MAPKi treatment. The implementation of this analysis and its application to this published cohort is provided via GitHub (https://github.com/livnatje/ImmuneResistance/wiki/Predicting-immunotherapy-resistance; ImmRes3_longitudinal.R).
In situ imaging analyses
Mapping cell-cell interactions
Given the processed imaging data, we assigned cells into cell types by discretizing the log- transformed expression levels of the cell type markers (S100, MITF, CD3, and CD8). We applied the EM algorithm for mixtures of normal distributions to characterize the two normal distributions for each of these cell type marker intensities. S100+/MITF+/CD3−/CD8− cells were defined as malignant cells; S100−/MITF−/CD3+/CD8− cells were defined as T cells, and S100−/MITF−/CD3+/CD8+ cells were defined as CD8+ T cells; other cells were defined as uncharacterized.
For each malignant cell, we computed an immune resistance score based on the in situ protein levels of the immune resistance markers. First, we centered and scaled the log-transformed expression of each protein across the malignant cells from all images (to have a zero average and a standard deviation of 1). Using this normalized data, we computed the resistance-up score as the sum expression of p53, DLL3, and Myc, the resistance-down score as the sum of HLA-A, Jun, LAMP2, and SQSTM1, and the final resistance score by subtracting the resistance-down score from the resistance-up score. The average resistance score of the malignant cells in each image was used as the in situ resistance score of the corresponding tumor. We then compared these tumor resistance scores to those computed based on the scRNA-seq data from the same patient.
Next, we examined the association between the expression of the individual markers and the overall resistance score in the malignant cells and the level of T cell infiltration. Each image in our data was composed of a few hundred frames (119–648 frames/image), where each frame consists of 1,502 cells on average. In each frame, we computed: (1) the fraction of T cells, (2) the average expression of the individual markers in the malignant cells, and (3) the average immune resistance (as above) across the malignant cells. We used a hierarchical logistic regression model to quantify the associations between the expression of the individual markers (or overall resistance score) in the malignant cells (2–3) and the fraction of T cells (1). The independent variables included the average expression of the marker (or the average immune resistance score) in the malignant cells of the frame (level-1), the average expression of normalization markers in the malignant cells of the frame (level-1), and the image the frame was sampled from (level-2). The dependent variable was the discretized T cell infiltration level of the frame, defining frames with high/low lymphocyte-fraction as “hot”/”cold,” respectively. We used different cutoffs to define hot/cold frames, such that a frame with a T cell fraction below the Q quantile was defined as cold, and a T cell fraction above the 1-Q quantile was defined as hot. We report only markers that showed a consistent association with the “hot” or “cold” niche, when starting with a cutoff of Q = median (0.5), and then using increasingly more stringent cutoffs (0.4, 0.3, 0.2 and 0.1). We provide the p- values obtained with Q = 0.2.
Integrating scRNA-seq and in situ data
We integrated the scRNA-seq and multiplexed immunofluorescence (t-CyCIF) data via a variant of Canonical Correlation Analysis (CCA), using the code provided in the R toolkit Seurat (Butler et al., 2018). CCA aims to identify shared correlation structures across datasets, such that each dataset provides multiple measurements of a gene-gene covariance structure, and patterns which are common to both datasets are identified. Cells from both sources are then represented in an aligned-CCA space (Butler et al., 2018).
In our application, each cell in the t-CyCIF data was represented by the log-transformed intensities of 14 markers. Each cell in the scRNA-seq data was represented by the imputed expression of the genes encoding the same 14 proteins. To impute the scRNA-seq data we identified a signature for each marker, consisting of the top 50 genes which were mostly correlated with the marker expression across the cell population in the scRNA-seq data. We then used the OE of the marker signature as a measure of its activity in the scRNA-seq data.
The cells from both sources were represented in the resulting aligned-CCA space. Next, we used the first five aligned-CCA dimensions to cluster the cells and represented them in a 2D t-SNE embedding (van der Maaten and Hinton, 2008). Clustering was preformed using a shared nearest neighbor (SNN) modularity optimization based clustering algorithm, which calculates k-nearest neighbors, constructs an SNN graph, and optimizes the modularity function to determine clusters (Waltman and van Eck, 2013).
To examine if cells clustered according to cell type or according to source we computed the expected number of cells from each two categories to be assigned to the same cluster by chance, assuming a random distribution of cells into clusters. We then used the observed versus expected co- clustering ratio to quantify the deviation from the random distribution, and used the binomial test to compute the statistical significance of this deviation from random.
Predicting clinical outcomes
Predicting survival and ICI-response
To test if a given signature predicts survival or progression free-survival (PFS) we first computed the OE of the signature in each tumor based on the bulk RNA-Seq data. Next, we used a Cox regression model with censored data to compute the significance of the association between the OE values and prognosis. To examine if the signature’s predictive value was significant beyond T cell infiltration levels we computed for each sample the OE of our CD8+ T cell signature (above), used this as another covariate in the Cox regression model, and computed another p value for each signature, based on its association with survival or PFS in this two-covariate model.
To visualize the predictions of a specific signature in a Kaplan Meier (KM) plot, we stratified the patients into three groups according to the OE of the signature: high or low expression correspond to the top or bottom 20% of the population, respectively, and intermediate otherwise. We used a one-sided log-rank test to examine if there was a significant difference between these three patient groups in terms of their survival or PFS rates.
OR was defined according to RESICT criteria, such that patients with a complete or partial response were defined as OR patients. Patients with progressive disease were defined as PD, and patients with more ill-defined response, such as stable disease or marginal responses, were excluded from this analysis. We further stratified the OR patients according to the duration of the response: (1) less than 6 months, (2) more than 6 months and less than a year, and (3) more than a year (long-term OR). We applied one-sided t tests to examine if the OE of the different signatures were differentially expressed in the OR versus PD patients, or in the long-term OR patients compared to the PD patients. Finally, we tested the ability of the different signatures to predict complete response by comparing (t test) between the complete responders and all other patients with a RECIST annotation (n = 101, Figure 5H), and computing the Area Under the Curve (AUC) of the resulting ROC curve.
To reproduce this analysis and results see ImmRes_valCohort2.R provided in https://github.com/livnatje/ImmuneResistance/wiki/Predicting-immunotherapy-resistance (relevant functions are included in ImmRes5_valCohort2.R).
Controlling for tumor proliferation
The single-cell data demonstrated that cycling cells have higher expression of the immune resistant program. Since the tumor proliferation rate may be a dynamic and context-dependent property, it might be advisable to compare between tumors based on their basal resistance level, namely, after controlling for the cell cycle effect. To this end, we compute for each tumor the OE of two cell cycle signatures (G1/S and G2/M signatures in Table S3A). We then fitted a linear model to estimate the expected OE of the resistance signature, when using the OE of the two cell cycle signatures as covariates. The residuals of this linear model, which quantify the deviation from the expected resistance OE values, were considered as the basal resistance level.
Examining association with sex
We confirmed that our results are not confounded by the patient sex: (1) the overall expression of the immune resistance program is not associated with the patient sex, both in the single-cell data and in the bulk RNA-Seq data of validation cohort 2 (p > 0.2, mixed-effects and t test respectively); (2) sex is not associated with better response to ICI, as we demonstrate by analyzing validation cohort 2 (p > 0.4, log-rank and hypergeometric tests for association with PFS and OR).
Alternative ICI-response predictors
To compare the predictive value of our resistance signatures to that of other signatures, we repeated the prediction process, as describe in Predicting survival and ICI-response, for each of the following gene signatures (Table S6A): (1) cell type specific signatures identified from our scRNA-seq data (as described in the Cell type signatures section); (2) signatures that characterize melanoma cell states (the AXL-high, MITF-high, and cell cycle states from (Tirosh et al., 2016)); (3) six sets of genes whose CRISPR knockout in melanoma cells conferred resistance or sensitivity (FDR < 0.05) to different types of immune killing according to a genome-scale in vivo screen (Manguso et al., 2017); (4) genes whose CRISPR knockout in melanoma cells conferred resistance to T cell killing (top 10 and top 50) in a genome-scale co-culture screen (Patel et al., 2017); (5) immune-related signatures that were identified based on the analysis of multiple pembrolizumab clinical datasets, and were shown to predict the response to pembrolizumab in an independent cohort (Ayers et al., 2017); (6) the Fluidigm Advanta Immuno-Oncology Gene Expression signatures (https://www.fluidigm.com/applications/advanta-immuno-oncology-gene-expression-assay); (7) immunotherapy resistance signatures identified in a clinical cohort of pre and post nivolumab treated melanomas (Riaz et al., 2017); (8) immune related signatures identified in a clinical melanoma cohort (Hugo et al., 2015) (9) immunotherapy resistance signatures identified in a clinical cohort of pre-anti-PD1 melanoma tumors (Hugo et al., 2016) and (10) PD-L1 expression.
Searching for immune sensitizing drugs
We performed the following analysis to identify drugs that could selectively eradicate malignant cells with a high expression of the resistance program, using efficacy measures of 131 drugs across 639 human cancer cell lines (Garnett et al., 2012). For each drug, we defined sensitive cell lines as those with the lowest (bottom 25%) IC50 values. We then used the gene expression provided in (Garnett et al., 2012), computed the OE of the resistance program in each of the 639 cells, and defined “resistant” cell lines as those with the highest OE values (top 25%). Next, for each drug we built a hierarchical logistic regression model, where the dependent variable is the cell line’s (drug-specific) binary sensitivity assignment, and the independent variables are the cell lines’ “resistance” assignments (level-1) and cancer types (level-2). Drugs then were ranked based on the one-tailed p values that quantify the significance of the positive association between the drug sensitivity (dependent) variable and the immune resistance (independent) variable.
To reproduce this analysis and results see https://github.com/livnatje/ImmuneResistance/wiki/Repressing-the-immune-resistance-program.
DATA AND SOFTWARE AVAILABILITY
Data availability
Processed scRNA-seq data generated for this study, and the Overall Expression of the different signatures that were used in the analysis of the two clinical validation cohorts, are provided through the Single Cell Portal along with interactive plots at the following URL: https://portals.broadinstitute.org/single_cell/study/melanoma-immunotherapy-resistance. The expression of the immune resistance program genes in Validation Cohort 2 is provided in Table S6B. The processed scRNA-seq data is provided via the Gene Expression Omnibus (GEO), accession number GEO: GSE115978. The raw scRNA-seq data is being deposited in dbGAP.
Code availability
All the relevant code, with instructions is provided via GitHub https://github.com/livnatje/ImmuneResistance. The code enables one to repeat the analyses that were performed, reproduce the results, figures, and tables, and apply the computational approaches to other datasets.
Supplementary Material
Figure S1. Cell-Type Assignments, Related to Figure 1
(A) Inferred large-scale CNVs distinguish malignant from nonmalignant cells. The inferred CNVs (red, amplification; blue, deletion) of two representative tumors (Mel79 and Mel102) are shown along the chromosomes (x axis) for cells (y axis) that were classified as malignant (left) or non- malignant (right) according CD45 sorting and transcriptional features.
(B-E) Congruence between different assignment methods (STAR Methods).
(B) Each plot shows the distribution of scores (y axis) by one scheme, when assigning the CD45− cells to malignant and nonmalignant cells according to anther scheme. Each scheme evaluates the likelihood that a given cell is malignant in a different way: CNV inference (as in A); the overall expression of melanoma and stroma cell signatures; differential similarity to bulk melanoma tumors compared to normal tissue. Middle line: median; box edges: 25th and 75th percentiles, whiskers: most extreme points that do not exceed ± IQR*1.5; further outliers are marked individually.
(C) Distribution of CNV-R-scores for cells classified as malignant (red) or non-malignant (blue). The CNV-R-score of a cell is the Spearman correlation coefficient (is) between the cell’s CNV profile and its tumor’s inferred CNV profile (STAR Methods).
(D) The distribution of CNV-R-scores across each identified cell subset. Boxplots as in (B).
(E) The CNV-R-score (y axis) versus the overall CNV signal (x axis, STAR Methods) for malignant (red) and non-malignant (blue) cells; Nonmalignant cells with values that exceed the dashed lines were considered unresolved and were omitted from further analyses.
(F and G) tSNE plots of all non-malignant cells (dots), colored by (F) overall expression (color bar) of de novo cell type signatures (Table S3B), and (G) detection (red) of T cell markers CD4 or CD8 (CD8A or CD8B), and B cell markers (CD19 and CD22).
Figure S5. The Immune Resistance Program Predicts Survival of TCGA Melanoma Patients, Related to Figure 5
(A-D) Kaplan-Meier (KM) plots stratified by high, intermediate or low Overall Expression of the respective signature in bulk RNA-Seq of TCGA tumors. P: COX regression p value; Pc: COX regression p value that tests if the program further enhances the predictive power of a model with inferred CD8+ T cell infiltration levels as a covariate compared to its prognostic value alone
(E) The number of subjects at risk are indicated for each group at the bottom of the KM curves for five different time points.
Figure S6. The Immune Resistance Program Predicts Response to Anti-PD-1 Therapy in an Independent Cohort, Related to Figure 5
(A-E) KM plots of progression-free survival (PFS) for the 104 of 112 patients in validation cohort 2 with PFS data, with patients stratified by high, intermediate and low Overall Expression values of the respective signature (labeled on top, see Tables S1C and S6B), after controlling for cell cycle as a confounding factor (STAR Methods). Pc p values test if the signature further enhances the predictive power of models with inferred CD8+ T cell infiltration levels as a covariate. The number of subjects at risk are indicated for each group at the bottom of the KM curves for six different time points.
Figure S7. The Immune Resistance Program in Cell Cultures, Mouse Cell Lines, and Various Tumor Types, Related to Figures 6 and 7
(A) The distribution of the immune resistance scores across three human melanoma cell lines, in control conditions (gray) and following abemaciclib treatment (cyan).
(B) The transcriptional state of patient-derived melanoma cells grown in monoculture or in co-culture with autologous ex vivo expanded TILs, with and without abemaciclib; right: tSNE plots; left: the fraction of immune resistant and sensitive cells in each condition.
(C and D) Differences in expression of the resistance program are aligned with the different level of resistance of mouse cell line models to ICI.
(C) Distribution of Overall Expression of the resistance program (x axis) across single cells of three mouse cell lines (B16, CT26 and MC38) grown in in vitro monocultures.
(D) Heatmap (bottom) shows genes (rows) from the immune resistance program that are differentially expressed between MC38 (blue bar), CT26 (green bar) and B16 (red bar) cells (columns) by the comparisons indicated on the left color bar and legend; cells are first sorted by model and then by Overall Expression of the resistance program (top); the corresponding cell cycle score are shown in the middle panel.
(E and F) To test the effect of CD8+ T cell depletion on response to phased combination of CDK4/6i and immunotherapy. B16cellswere implanted in C7BL/6 mice and treated with either vehicle control (n = 5), phased combination of ICI and abemaciclib (n = 10) or phased combination following CD8+ T cell depletion (n = 9, STAR Methods).
(E) The reduction in tumor outgrowth rate seen with phased therapy is lost in animals without CD8+ T cells. (F) KM survival curves of animals in (E).
(G and H) The resistance program across tumor types. Distribution of Overall Expression scores of the resistance program (x axis) in 9,559 tumors from 33 cancer types (y axis) from TCGA, either scored as-is (G) or with a regression-based approach that controls for tumor microenvironment-related signals (H, STAR Methods). Middle line: median; box edges: 25th and 75th percentiles, whiskers: most extreme points that do not exceed ± IQR*1.5; further outliers are marked individually.
Figure S2. The Immune Resistance Program Is Robust to Varying Definitions of T Cell Infiltration and Can Be Decoupled from Cell Cycle, Related to Figures 1 and 2
(A-C) Congruence between the transcriptional programs in malignant that are associated with the exclusion of different T cell subsets.
(A) The inferred abundance of different T cell subsets is highly correlated across TCGA melanoma tumors. Pearson correlation coefficient (color bar) between the inferred proportions of T cells of different subsets.
(B and C) Overlap (B, Jaccard similarity coefficient) and correlation in expression across melanoma cells (C, left) or bulk tumors (C, middle and right) between malignant cell programs associated with the exclusion of different T cell subsets (rows, columns).
(D) Congruence between exclusion programs derived with and without controlling for the cell cycle. Venn diagrams show the number of genes shared in up- regulated (left) and downregulated (right) portions of indicated programs when they are derived with (top) and without (bottom) regressing out cell cycle from the scRNA-seq and bulk RNA-Seq data.
(E) Resistance program is stronger in cycling cells even when filtering cell cycle effects. The distribution of Overall Expression scores of the resistance program (y axis) derived with (right) and without (left) filtering out cell cycle effects (STAR Methods). Solid line: mean of the respective distribution; dashed line: mean across all malignant cells.
(F and G) Regression of cell cycle effects masks differences between cycling and non-cycling cells, but preserves differences in the resistance program.
(F) Expression of cell cycle genes (columns) across the malignant cells (rows), with (left) and without (right) regressing out cell cycle. Cells are sorted as cycling or non-cycling (red/black, respectively; leftmost color bar).
(G) Expression of genes in the exclusion (right) and post- treatment (left) programs (columns), after regressing out cell cycle from the data. Cells (rows) are sorted by untreated and post-treatment tumors (right color bar) and clustered within each set; rightmost color bar: cycling status.
Figure S3. Co-variation of the Immune Resistance Program Genes across Single Cells within Each Tumor, Related to Figure 3
Gene-gene Pearson correlation coefficients (color bar) between the genes in the resistance program, across individual malignant cells from each specific tumor (as labeled). Genes are sorted in the same order in all heatmaps (and in Figure 3D). The consistent intra-tumor correlation suggests shared regulation.
Figure S4. Integrative Analysis of scRNA-Seq and In Situ Immunofluorescence Data, Related to Figure 4
(A-D) Integrative analysis of scRNA-seq and CyCIF multiplex protein data (STAR Methods) from each of four representative tumors: (A) Mel79, (B) Mel80, (C) Mel74, and (D) Mel89. Left: tSNE plots of co-embedding of cells from scRNA-seq and images of each tumors, with cells colored by (from left): clusters, data source, or combined source and cell type. Right: Log-odds ratio (color bar, STAR Methods) assessing for each pair of cell types (rows, columns) if they are assigned to the same cluster significantly more (> 0, red) or less (< 0, blue) than expected by chance.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD45 | VWR | cat#ABNOMAB12230 |
| Zombie live/dead | BioLegend | cat#423101 |
| MITF | Abcam | cat#ab3201 |
| S100α | Abcam | cat#ab207367 |
| CD3 | Abcam | cat#ab208514 |
| CD8 | eBioscience | cat#50-0008-80 |
| cJun | Abcam | cat#ab193780 |
| InVivoMAb anti-mouse CTLA-4 (Clone 9H10) | Bioxcell | cat#BE0131 |
| InVivoMAb anti mouse CD279 (PD-1) (Clone 29F.1A12) | Bioxcell | cat#BE0273 |
| InVivoMAb anti-mouse CD8β (Clone53–5.8) | Bioxcell | cat#BE0223 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Abemaciclib (LY2835219) | MedChemExpress | cat#HY-16297 |
| Deposited Data | ||
| scRNA-seq | This paper | https://portals.broadinstitute.org/single_cell/study/melanoma-immunotherapy-resistance |
| Multiplexed immunofluorescence images | This paper | https://portals.broadinstitute.org/single_cell/study/melanoma-immunotherapy-resistance |
| scRNA-seq | This paper | GEO: GSE115978 |
| RNA-Seq (expression of the immune resistance genes), Validation Cohort 2 | This paper | https://portals.broadinstitute.org/single_cell/study/melanoma-immunotherapy-resistance |
| Raw scRNA-seq | This paper | dbGAP; https://duos.broadinstitute.org/#/home, dataset ID: DUOS-000001 |
| TCGA RNA-SeqV2 | TCGA | https://xena.ucsc.edu/ |
| Breast cancer gene expression following abemaciclib | Goel et al., 2017 | GEO: GSE99063 |
| Human reference genome NCBI build 37, GRCh37 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| Pan-cancer drug efficacies | Garnett et al., 2012 | https://www.nature.com/articles/nature11005 |
| Efficacies of CDK4/6 inhibitors across cancer cell lines | Gong et al., 2017 | https://www.sciencedirect.com/science/article/pii/S1535610817305093?via%3Dihub |
| Experimental Models: Cell Lines | ||
| IGR37 | Broad Institute | n/a |
| UACC257 | Broad Institute | n/a |
| A2058 | Broad Institute | n/a |
| Melanoma cell line 2686 | MD Anderson Cancer Center | n/a |
| Software and Algorithms | ||
| Code generated for this study | This paper | https://github.com/livnatje/ImmuneResistance |
| Seurat | Butler et al., 2018 | https://satijalab.org/seurat/ |
| SCDE | Fan et al., 2016 | http://hms-dbmi.github.io/scde/ |
| Bowtie | Langmead et al., 2009 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| RSEM v1.2.8 | Li and Dewey, 2011 | http://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-12-323 |
| Other | ||
| CyteFinder microscope | RareCyte | n/a |
Highlights.
Single-cell RNA-seq identifies an immune resistance program in malignant cells
Multiple immune resistance mechanisms are co-regulated in the program
The program predicts clinical responses to immunotherapy in melanoma patients
CDK4/6 inhibitors repress the program and may sensitize melanoma to immunotherapy
ACKNOWLEDGMENTS
We thank Meromit Singer for discussions; Leslie Gaffney for help with artwork; and Laura Dellostritto, Karla Helvie, Nichole Straub, and the CCPM members at DFCI. L.J.-A. is a CRI Irvington Fellow supported by the CRI and a fellow of the Eric and Wendy Schmidt postdoctoral program. B.I. was supported by NCI grant K08-CA222663, DFCI Claudia Adams Barr Program for Innovative Cancer Research, the Burroughs Wellcome Fund Career Award for Medical Scientists, and the SITC Translational Fellowship. A. Regev is an HHMI Investigator. Work was supported by the Klarman Cell Observatory, STARR Cancer Consortium, NCI grants 1U24CA180922 and R33-CA202820, Koch Institute NCI Support (core) grant P30-CA14051, Ludwig Centers at Harvard and MIT, AMRF, and the Broad Institute. Processed scRNA-seq data are available at https://portals.broadinstitute.org/single_cell/study/melanoma-immunotherapy-resistance and under accession number GEO: GSE115978. Raw scRNA-seq data will be deposited in dbGAP.
Footnotes
DECLARATION OF INTERESTS
A. Regev is an SAB member for ThermoFisher Scientific, Syros Pharmaceuticals and Driver Group and a founder of Celsius Therapeutics. A. Regev, L.J.-A., B.I., O.R.-R., and A. Rotem are co-inventors on provisional patent application filed by the Broad Institute relating to this manuscript. L.A.G. is now an employee of Eli Lilly and company.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and seven tables and can be found with this article online at https://doi.org/10.1016/j.cell.2018.09.006.
REFERENCES
- Akbani R, Akdemir KC, Aksoy BA, Albert M, Ally A, Amin SB, Arach-chi H, Arora A, Auman JT, Ayala B, et al. ; Cancer Genome Atlas Network (2015). Genomic Classification of Cutaneous Melanoma. Cell 161,1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algazi AP, Tsai KK, Shoushtari AN, Munhoz RR, Eroglu Z, Piulats JM, Ott PA, Johnson DB, Hwang J, Daud AI, et al. (2016). Clinical outcomes in metastatic uveal melanoma treated with PD-1 and PD-L1 antibodies. Cancer 122, 3344–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, Albright A, Cheng JD, Kang SP, Shankaran V, et al. (2017). IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest 127, 2930–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, and Hochberg Y (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Stat. Methodol 57, 289–300. [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. (2012). Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol 30, 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran MA, Montalvo W, Yagita H, and Allison JP (2010). PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc. Natl. Acad. Sci. USA 107, 4275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Wang ES, Jenkins RW, Li S, Dries R, Yates K, Chhabra S, Huang W, Liu H, Aref AR, et al. (2018). CDK4/6 Inhibition Augments Antitumor Immunity by Enhancing T-cell Activation. Cancer Discov. 8, 216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ester M, Kriegel H-P, Sander J, and Xu X (1996). A density-based algorithm for discovering clusters a density-based algorithm for discovering clusters in large spatial databases with noise. In Proceedings of the Second International Conference on Knowledge Discovery and Data Mining, pp. 226–231. [Google Scholar]
- Fan J, Salathia N, Liu R, Kaeser GE, Yung YC, Herman JL, Kaper F, Fan J-B, Zhang K, Chun J, and Kharchenko PV (2016). Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nat. Methods 13, 241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman WH, Pages F, Sautès-Fridman C, and Galon J (2012). The immune contexture in human tumours: impact on clinical outcome. Nat. Rev. Cancer 12, 298–306. [DOI] [PubMed] [Google Scholar]
- Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, et al. (2012). System atic identification of genomic markers of drug sensitivity in cancer cells. Nature 483, 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel S, DeCristo MJ, Watt AC, BrinJones H, Sceneay J, Li BB, Khan N, Ubellacker JM, Xie S, Metzger-Filho O, et al. (2017). CDK4/6 inhibition triggers anti-tumour immunity. Nature 548, 471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Litchfield LM, Webster Y, Chio L-C, Wong SS, Stewart TR, Dowless M, Dempsey J, Zeng Y, Torres R, et al. (2017). Genomic Aberrations that Activate D-type Cyclins Are Associated with Enhanced Sensitivity to the CDK4 and CDK6 Inhibitor Abemaciclib. Cancer Cell 32, 761–776. [DOI] [PubMed] [Google Scholar]
- Gordy JT, Luo K, Zhang H, Biragyn A, and Markham RB (2016). Fusion of the dendritic cell-targeting chemokine MIP3α to melanoma antigen Gp100 in a therapeutic DNA vaccine significantly enhances immunogenicity and survival in a mouse melanoma model. J. Immunother. Cancer 4, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm S (1979). A Simple Sequentially Rejective Multiple Test Procedure. Scand. J. Stat 6, 65–70. [Google Scholar]
- Hugo W, Shi H, Sun L, Piva M, Song C, Kong X, Moriceau G, Hong A, Dahlman KB, Johnson DB, et al. (2015). Non-genomic and Immune Evolution of Melanoma Acquiring MAPKi Resistance. Cell 162, 1271–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G, et al. (2016). Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell 165, 35–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Silberstein L, and Scadden DT (2014). Bayesian approach to single-cell differential expression analysis. Nat. Methods 11, 740–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress TR, Sabò A, and Amati B (2015). MYC: connecting selective transcriptional control to global RNA production. Nat. Rev. Cancer 15, 593–607. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P, et al. (2015). Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N. Engl. J. Med 373, 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesterhuis WJ, Rinaldi C, Jones A, Rozali EN, Dick IM, Khong A, Boon L, Robinson BW, Nowak AK, Bosco A, et al. (2015). Network analysis of immunotherapy-induced regressing tumours identifies novel synergistic drug combinations. Sci. Rep 5, 12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J-R, Izar B, Wang S, Yapp C, Mei S, Shah PM, Santagata S, and Sorger PK (2018). Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, Collins NB, Bi K, LaFleur MW, Juneja VR, et al. (2017). In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547, 413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, and Alizadeh AA (2015). Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki S, Ohta T, Shioi G, Hatanaka H, Ogasawara O, Okuda Y, Kawaji H, Nakaki R, Sese J, and Meno C (2018). Integrative analysis of transcription factor occupancy at enhancers and disease risk loci in noncoding genomic regions.
- Pan D, Kobayashi A, Jiang P, Ferrari de Andrade L, Tay RE, Luoma AM, Tsoucas D, Qiu X, Lim K, Rao P, et al. (2018). A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 359, 770–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SJ, Sanjana NE, Kishton RJ, Eidizadeh A, Vodnala SK, Cam M, Gartner JJ, Jia L, Steinberg SM, Yamamoto TN, et al. (2017). Identification of essential genes for cancer immunotherapy. Nature 548, 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, Xu C, McKenzie JA, Zhang C, Liang X, et al. (2016). Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov. 6, 202–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramilowski JA, Goldberg T, Harshbarger J, Kloppmann E, Lizio M, Satagopam VP, Itoh M, Kawaji H, Carninci P, Rost B, and Forrest AR (2015). A draft network of ligand-receptor-mediated multicellular signalling in human. Nat. Commun 6, 7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, Hodi FS, Martín-Algarra S, Mandal R, Sharfman WH, et al. (2017). Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell 171, 934–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosvall M, and Bergstrom CT (2008). Maps of random walks on complex networks reveal community structure. Proc. Natl. Acad. Sci. USA 105, 1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaer DA, Beckmann RP, Dempsey JA, Huber L, Forest A, Amaladas N, Li Y, Wang YC, Rasmussen ER, Chin D, et al. (2018). The CDK4/6 Inhibitor Abemaciclib Induces a T Cell Inflamed Tumor Microenvironment and Enhances the Efficacy of PD-L1 Checkpoint Blockade. Cell Rep. 22, 2978–2994. [DOI] [PubMed] [Google Scholar]
- Sharma P, and Allison JP (2015). The future of immune checkpoint therapy. Science 348, 56–61. [DOI] [PubMed] [Google Scholar]
- Sharma P, Hu-Lieskovan S, Wargo JA, and Ribas A (2017). Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 168, 707–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, et al. (2017). A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 171, 1437–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. (2016). Dissecting the multicellular ecosystem of metastatic melanoma by singlecell RNA-seq. Science 352, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta JJ, Gennert D, Lu D, Satija R, Shalek AK, and Regev A (2014). Preparation of Single-Cell RNA-Seq Libraries for Next Generation Sequencing. Curr. Protoc. Mol. Biol 107, 4.22.1–4.22.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM, et al. (2015). Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L, and Hinton G (2008). Visualizing Data using t-SNE. J. Mach. Learn Res 9, 2579–2605. [Google Scholar]
- Waltman L, and van Eck NJ (2013). A smart local moving algorithm for large-scale modularity-based community detection. Eur. Phys. J. B 86, 471. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Cell-Type Assignments, Related to Figure 1
(A) Inferred large-scale CNVs distinguish malignant from nonmalignant cells. The inferred CNVs (red, amplification; blue, deletion) of two representative tumors (Mel79 and Mel102) are shown along the chromosomes (x axis) for cells (y axis) that were classified as malignant (left) or non- malignant (right) according CD45 sorting and transcriptional features.
(B-E) Congruence between different assignment methods (STAR Methods).
(B) Each plot shows the distribution of scores (y axis) by one scheme, when assigning the CD45− cells to malignant and nonmalignant cells according to anther scheme. Each scheme evaluates the likelihood that a given cell is malignant in a different way: CNV inference (as in A); the overall expression of melanoma and stroma cell signatures; differential similarity to bulk melanoma tumors compared to normal tissue. Middle line: median; box edges: 25th and 75th percentiles, whiskers: most extreme points that do not exceed ± IQR*1.5; further outliers are marked individually.
(C) Distribution of CNV-R-scores for cells classified as malignant (red) or non-malignant (blue). The CNV-R-score of a cell is the Spearman correlation coefficient (is) between the cell’s CNV profile and its tumor’s inferred CNV profile (STAR Methods).
(D) The distribution of CNV-R-scores across each identified cell subset. Boxplots as in (B).
(E) The CNV-R-score (y axis) versus the overall CNV signal (x axis, STAR Methods) for malignant (red) and non-malignant (blue) cells; Nonmalignant cells with values that exceed the dashed lines were considered unresolved and were omitted from further analyses.
(F and G) tSNE plots of all non-malignant cells (dots), colored by (F) overall expression (color bar) of de novo cell type signatures (Table S3B), and (G) detection (red) of T cell markers CD4 or CD8 (CD8A or CD8B), and B cell markers (CD19 and CD22).
Figure S5. The Immune Resistance Program Predicts Survival of TCGA Melanoma Patients, Related to Figure 5
(A-D) Kaplan-Meier (KM) plots stratified by high, intermediate or low Overall Expression of the respective signature in bulk RNA-Seq of TCGA tumors. P: COX regression p value; Pc: COX regression p value that tests if the program further enhances the predictive power of a model with inferred CD8+ T cell infiltration levels as a covariate compared to its prognostic value alone
(E) The number of subjects at risk are indicated for each group at the bottom of the KM curves for five different time points.
Figure S6. The Immune Resistance Program Predicts Response to Anti-PD-1 Therapy in an Independent Cohort, Related to Figure 5
(A-E) KM plots of progression-free survival (PFS) for the 104 of 112 patients in validation cohort 2 with PFS data, with patients stratified by high, intermediate and low Overall Expression values of the respective signature (labeled on top, see Tables S1C and S6B), after controlling for cell cycle as a confounding factor (STAR Methods). Pc p values test if the signature further enhances the predictive power of models with inferred CD8+ T cell infiltration levels as a covariate. The number of subjects at risk are indicated for each group at the bottom of the KM curves for six different time points.
Figure S7. The Immune Resistance Program in Cell Cultures, Mouse Cell Lines, and Various Tumor Types, Related to Figures 6 and 7
(A) The distribution of the immune resistance scores across three human melanoma cell lines, in control conditions (gray) and following abemaciclib treatment (cyan).
(B) The transcriptional state of patient-derived melanoma cells grown in monoculture or in co-culture with autologous ex vivo expanded TILs, with and without abemaciclib; right: tSNE plots; left: the fraction of immune resistant and sensitive cells in each condition.
(C and D) Differences in expression of the resistance program are aligned with the different level of resistance of mouse cell line models to ICI.
(C) Distribution of Overall Expression of the resistance program (x axis) across single cells of three mouse cell lines (B16, CT26 and MC38) grown in in vitro monocultures.
(D) Heatmap (bottom) shows genes (rows) from the immune resistance program that are differentially expressed between MC38 (blue bar), CT26 (green bar) and B16 (red bar) cells (columns) by the comparisons indicated on the left color bar and legend; cells are first sorted by model and then by Overall Expression of the resistance program (top); the corresponding cell cycle score are shown in the middle panel.
(E and F) To test the effect of CD8+ T cell depletion on response to phased combination of CDK4/6i and immunotherapy. B16cellswere implanted in C7BL/6 mice and treated with either vehicle control (n = 5), phased combination of ICI and abemaciclib (n = 10) or phased combination following CD8+ T cell depletion (n = 9, STAR Methods).
(E) The reduction in tumor outgrowth rate seen with phased therapy is lost in animals without CD8+ T cells. (F) KM survival curves of animals in (E).
(G and H) The resistance program across tumor types. Distribution of Overall Expression scores of the resistance program (x axis) in 9,559 tumors from 33 cancer types (y axis) from TCGA, either scored as-is (G) or with a regression-based approach that controls for tumor microenvironment-related signals (H, STAR Methods). Middle line: median; box edges: 25th and 75th percentiles, whiskers: most extreme points that do not exceed ± IQR*1.5; further outliers are marked individually.
Figure S2. The Immune Resistance Program Is Robust to Varying Definitions of T Cell Infiltration and Can Be Decoupled from Cell Cycle, Related to Figures 1 and 2
(A-C) Congruence between the transcriptional programs in malignant that are associated with the exclusion of different T cell subsets.
(A) The inferred abundance of different T cell subsets is highly correlated across TCGA melanoma tumors. Pearson correlation coefficient (color bar) between the inferred proportions of T cells of different subsets.
(B and C) Overlap (B, Jaccard similarity coefficient) and correlation in expression across melanoma cells (C, left) or bulk tumors (C, middle and right) between malignant cell programs associated with the exclusion of different T cell subsets (rows, columns).
(D) Congruence between exclusion programs derived with and without controlling for the cell cycle. Venn diagrams show the number of genes shared in up- regulated (left) and downregulated (right) portions of indicated programs when they are derived with (top) and without (bottom) regressing out cell cycle from the scRNA-seq and bulk RNA-Seq data.
(E) Resistance program is stronger in cycling cells even when filtering cell cycle effects. The distribution of Overall Expression scores of the resistance program (y axis) derived with (right) and without (left) filtering out cell cycle effects (STAR Methods). Solid line: mean of the respective distribution; dashed line: mean across all malignant cells.
(F and G) Regression of cell cycle effects masks differences between cycling and non-cycling cells, but preserves differences in the resistance program.
(F) Expression of cell cycle genes (columns) across the malignant cells (rows), with (left) and without (right) regressing out cell cycle. Cells are sorted as cycling or non-cycling (red/black, respectively; leftmost color bar).
(G) Expression of genes in the exclusion (right) and post- treatment (left) programs (columns), after regressing out cell cycle from the data. Cells (rows) are sorted by untreated and post-treatment tumors (right color bar) and clustered within each set; rightmost color bar: cycling status.
Figure S3. Co-variation of the Immune Resistance Program Genes across Single Cells within Each Tumor, Related to Figure 3
Gene-gene Pearson correlation coefficients (color bar) between the genes in the resistance program, across individual malignant cells from each specific tumor (as labeled). Genes are sorted in the same order in all heatmaps (and in Figure 3D). The consistent intra-tumor correlation suggests shared regulation.
Figure S4. Integrative Analysis of scRNA-Seq and In Situ Immunofluorescence Data, Related to Figure 4
(A-D) Integrative analysis of scRNA-seq and CyCIF multiplex protein data (STAR Methods) from each of four representative tumors: (A) Mel79, (B) Mel80, (C) Mel74, and (D) Mel89. Left: tSNE plots of co-embedding of cells from scRNA-seq and images of each tumors, with cells colored by (from left): clusters, data source, or combined source and cell type. Right: Log-odds ratio (color bar, STAR Methods) assessing for each pair of cell types (rows, columns) if they are assigned to the same cluster significantly more (> 0, red) or less (< 0, blue) than expected by chance.
Data Availability Statement
Data availability
Processed scRNA-seq data generated for this study, and the Overall Expression of the different signatures that were used in the analysis of the two clinical validation cohorts, are provided through the Single Cell Portal along with interactive plots at the following URL: https://portals.broadinstitute.org/single_cell/study/melanoma-immunotherapy-resistance. The expression of the immune resistance program genes in Validation Cohort 2 is provided in Table S6B. The processed scRNA-seq data is provided via the Gene Expression Omnibus (GEO), accession number GEO: GSE115978. The raw scRNA-seq data is being deposited in dbGAP.
Code availability
All the relevant code, with instructions is provided via GitHub https://github.com/livnatje/ImmuneResistance. The code enables one to repeat the analyses that were performed, reproduce the results, figures, and tables, and apply the computational approaches to other datasets.
Processed scRNA-seq data generated for this study, and the Overall Expression of the different signatures that were used in the analysis of the two clinical validation cohorts, are provided through the Single Cell Portal along with interactive plots at the following URL: https://portals.broadinstitute.org/single_cell/study/melanoma-immunotherapy-resistance. The expression of the immune resistance program genes in Validation Cohort 2 is provided in Table S6B. The processed scRNA-seq data is provided via the Gene Expression Omnibus (GEO), accession number GEO: GSE115978. The raw scRNA-seq data is being deposited in dbGAP.