Abstract
There is growing public concern surrounding traumatic brain injury (TBI). TBI can cause significant morbidity, and the long-term sequelae are poorly understood. TBI diagnosis and management rely on patient-reported symptoms and subjective clinical assessment. There are no biologic tools to detect mild TBI or to track brain recovery. Emerging evidence suggests that microRNAs (miRNAs) may provide information about the injured brain. These tiny epigenetic molecules are expressed throughout the body. However, they are particularly important in neurons, can cross the blood-brain barrier, and are securely transported from cell to cell, where they regulate gene expression. miRNA levels may identify patients with TBI and predict symptom duration. This review synthesizes miRNA findings from 14 human studies. We distill more than 291 miRNAs to 17 biomarker candidates that overlap across multiple studies and multiple biofluids. The goal of this review is to establish a collective understanding of miRNA biology in TBI and identify clinical priorities for future investigations of this promising biomarker.
Keywords: Concussion, miRNA, biomarkers, traumatic brain injury, diagnosis, prognosis, treatment
Introduction
Head injuries are a common concern in the United States, especially among children and adolescents, whose developing brains may be at an increased risk for long-term sequelae. It is estimated that 20% of adolescents1 have been diagnosed with at least 1 concussion—the sequelae of symptoms induced by mild traumatic brain injury (TBI).2 Despite growing public concern about concussion, there are limited tools available for diagnosis and prognosis.3 Currently, concussion diagnosis is based on subjective assessment.3 There are few biologic measures available to personalize anticipatory guidance or therapeutic decision-making. A lack of sensitive, objective assessment tools hinders the ability of physicians to provide accurate diagnoses and effective treatments.4,5 The resulting one-size-fits-all approach may increase anxiety about brain health for patients and families.6,7 Thus, establishing mild traumatic brain injury (mTBI) biomarkers could have profound clinical impact on the diagnosis and management of concussion.8,9 Emerging evidence suggests that microRNAs (miRNAs) may fill this clinical need.
miRNAs are implicated in the pathophysiology of many diseases, but they are critical for neurodevelopment and brain function.10 Through the regulation of gene activity, miRNAs control cellular processes essential to neuronal injury and repair: differentiation, proliferation, apoptosis, and metabolism.11-13 Disruption in miRNA levels have been reported in numerous disease processes of the central nervous system (CNS).14-16 Here, we review the miRNAs altered in human patients with mild, moderate, and severe TBI, identifying the most consistent findings and proposing avenues for future investigations to validate this promising molecular technology.
Clinical Aspects of TBI
Neuropathology
TBI is defined as an alteration in brain function, or other evidence of brain pathology, caused by an external force.17 The external force can lead to permanent or temporary injuries from the neuronal insult. Primary damage is characterized by the effects of the immediate impact on neuroanatomy and function, whereas secondary damage consists of the biochemical responses to injury that result in neuronal repair or apoptotic cell death.18-21 miRNAs are implicated in both the primary and secondary damage responses to TBI.22 Injured neurons may release miRNAs into the extracellular space, where their small size allows them to navigate the blood-brain barrier (BBB), facilitating peripheral sampling.23,24 In the cellular response to secondary damage,25,26 neurons use miRNA signaling to regulate synaptogenesis and neuroplasticity. As a result, miRNA profiles during the subacute period may also telegraph the trajectory of brain recovery.27
Clinical diagnosis
The Glasgow Coma Scale (GCS) is a scoring system based on verbal, motor, and eye-opening reactions to external stimuli. GCS has been used to classify TBI as mild (GCS ⩾ 13), moderate (GCS 9-12), or severe (GCS ⩽ 8).28 Mild, moderate, and severe TBI involve a spectrum of similar mechanical forces as well as partially overlapping pathophysiology. However, patients with these conditions often display disparate symptomology, and their diagnosis and management can entail distinct approaches. Consideration of the commonalities and differences across TBI severity may help inform biomarker development.
The most common type of TBI is mTBI, accounting for approximately 80% of cases.29 Diagnosis of mTBI is complex, requiring a multi-modal approach. Recent guidelines from the Centers for Disease Control (CDC) recommend neuropsychological tools and age-appropriate symptom scales alongside standard clinical assessment for mTBI diagnosis.3 Although cognition and balance measures may be used, routine use of imaging modalities is discouraged, and objective biomarkers are not recommended outside of a research setting. Recently, the Food and Drug Administration (FDA) approved serum measurements of ubiquitin C terminal hydrolase (UCH-L1) and glial fibrillary acidic protein (GFAP) to identify adult patients at risk for intracranial injury.30,31 The utility of these measures is limited to differentiating patients with moderate-severe TBI who benefit from cranial imaging. This may be because protein biomarkers require moderate-to-severe CNS damage for reliable peripheral detection.32-34 In contrast, miRNAs can be released as signaling vectors by live cells35,36 and do not require cell injury for release. This may make them more amenable to mTBI detection.37-39 This idea is supported by findings that peripheral miRNA levels reflect subtle structural changes in the CNS identified by magnetic resonance imaging.40
Clinical symptoms
Even mTBI can impact cognition, sleep, mood, or physical wellbeing (eg, fatigue, nausea, dizziness, and headache). These sequelae are commonly called “concussion,” a term used herein when stressing the patient-oriented experience of mTBI symptoms.41 Most of the neurologic, cognitive, and behavioral changes associated with mTBI resolve in days to weeks, but for 10% to 30% of those affected, symptoms can persist for months.42-44 Symptom burden at initial presentation45,46 and clinical prediction tools47,48 have demonstrated fair utility for identifying patients at risk for prolonged concussion symptoms, but there are no objective measures currently available. A recent study of saliva miRNA levels in children shows promise for predicting prolonged concussion symptoms.49
Clinical management
Because no single tool strongly predicts outcome in patients with mTBI, physicians are forced to rely on subjective symptom surveys to formulate management plans.3 Such surveys may be manipulated by patients who wish to expedite return-to-play,50 or prolong return-to-work/school activities.51 Although management plans may be partially informed by individual risk factors (eg, previous mTBI, learning difficulties, psychosocial stressors), most patients receive standard anticipatory guidance about sleep, exercise, and headache management. The expectation is that 70% to 80% of patients will recover within 1 month.3 In patients with protracted recovery, referral for multi-disciplinary evaluation is recommended. Thus, patients with persistent headache or dizziness may wait a full month for individualized therapy, during which a critical window for neuroplasticity and intervention may have passed. Currently, there are no specific therapies for mTBI, and many therapeutic trials have provided inconclusive results.52,53 This may result because mTBI is a heterogeneous condition and therapeutic efficacy depends on the recognition of phenotypes and early, personalized intervention.54 The biological diversity and pathologic specificity of miRNAs make them ideal candidates for mTBI phenotyping. Indeed, studies have shown that concussion-related miRNAs measured at the time of injury are associated with chronic symptom characteristics.49
TBI biomarkers
Biomarkers, such as proteins, metabolites, and neuronal imaging, have been extensively explored in patients across the spectrum of TBI severity.9 In particular, studies of protein biomarkers as a means for diagnosing, monitoring, and predicting the course of concussions has increased markedly over the past decade.55 Here we briefly focus on 2 proteins with clinical approval.30,31,46 GFAP and UCH-L1 have demonstrated utility for discerning adult patients with TBI who may benefit from neuroimaging.56-59 Protein biomarkers may require BBB disruption to migrate from cerebral spinal fluid (CSF) into blood.60 As a result, the sensitivity of protein biomarkers for TBI detection may be limited in individuals with mTBI.61 Second, unlike miRNAs, which are packaged in protective exosomes,62 extracellular proteins may be subject to degradation by endogenous proteases.61 This could potentially limit their utility to acute and subacute periods of TBI. Finally, a number of protein biomarkers are released following damage to myocytes and osteocytes, and this could limit their specificity in patients with poly-trauma.63 This is not to say that protein biomarkers have no role in TBI assessment. Rather, we wish to highlight potential limitations that miRNAs may be uniquely suited to address.
miRNA Physiology
miRNA processing
miRNAs are short (19-28 nucleotides), endogenous, non-coding RNAs that act at the post-transcriptional level to regulate protein synthesis.64 They are initially transcribed in the nucleus as long (>1000 base pairs) precursor molecules called primary microRNA (pri-miRNA) which fold into a hairpin structure. The pri-miRNA is trimmed by the RNase Drosha into stem-loop structures called precursor microRNA (pre-miRNA) that are 60 to 100 nucleotides long. The pre-miRNA is exported into the cytoplasm where the miRNA processing enzyme Dicer cleaves them into miRNA:miRNA duplexes. One half of this duplex will become a mature miRNA molecule, whereas the other half, known as the minor miRNA, will be degraded.65,66 The mature miRNA strand is bound within the microRNA-induced silencing complex (miRISC) where it suppresses the expression of specific gene targets by inhibiting protein translation or promoting messenger RNA (mRNA) degradation.67 The miRISC complex recognizes target genes through “seed sequences” at the 5′ end of the miRNA that are complementary to the 3′ region of the intended mRNA target. The seed sequence of the miRNA can recognize hundreds of different mRNAs, and there are several miRNAs that share the same seed sequence.68 miRNAs are organized into families based on similarities in the sequence of the hairpin molecules.69
miRNAs in the brain
The CNS contains the highest concentration and highest diversity of miRNAs. It is estimated that 70% of all miRNAs are expressed in the brain, spinal cord, or peripheral nerves.70 Expression of miRNA evolves throughout neurodevelopment and varies across the brain regions.71 Within neurons, miRNAs also display intracellular variation in localization.72 Enrichment of certain miRNAs in the axonal or dendritic compartments suggests that individual miRNAs may have unique functions regulating local protein expression, synapse maturation, or neural circuit formation.73 Expression of miRNAs is critical to the development and function of the CNS. Dysregulated miRNA levels have been linked to impaired learning, memory, and cognition as well as a host of neuropsychiatric disorders.74 These characteristics have led to rising interest in the utility of miRNAs as biomarkers for CNS pathology.75
The role of miRNAs in neuronal injury and repair
Numerous studies have examined physiologic alterations in miRNA expression after TBI. Redell et al22 first described altered levels of hippocampal miRNAs in rats exposed to controlled cortical impacts. Using microarray techniques to interrogate more than 400 miRNAs, they verified changes in 8 miRNAs with quantitative real-time polymerase chain reaction (RT-PCR; miR-107, miR-130a, miR-223, miR-292-5p, miR-433-3p, miR-451, miR-541, and miR-711). Liu et al26 extended miRNA findings to the rat cerebral cortex and established that miRNA levels change dynamically during the first 72 hours after TBI. Notably, the study identified 1 miRNA (miR-21) which remained chronically elevated following the initial injury.
The biochemical cascade that occurs following brain injury consists of mechanical insult, oxidative stress, apoptotic cell death, subacute repair, and chronic remodeling.76 miR-21, a miRNA biomarker identified in both human and animal TBI studies, may inhibit apoptosis and target angiogenesis factors critical to BBB maintenance.77,78 miR-16 is another biomarker candidate identified in animal studies that may regulate apoptosis. In addition, miR-16 has putative targets critical to cell cycle regulation, including Bcl-2 and cyclin dependent kinase 6. Together these molecules may facilitate neurogenesis and acute repair responses following TBI.79 Unlike proteins, which are typically increased after TBI, some miRNAs undergo down-regulation after injury (eg, miR-107, mir-27a). Decreases in miR-107 may be critical for inflammatory processes, allowing transcription of granulin.80 Whereas decreases in miR-27a may facilitate programmed cell death by allowing expression of pro-apoptotic Bcl-2 proteins.81
The exact mechanisms that drive individual miRNA alterations after TBI are not fully understood. As with proteins, endothelial cells and astrocytes forming the BBB may release miRNAs following TBI-induced disruption.82 In addition, some miRNAs may be released from damaged neurons as a result of the shearing forces that occur in axonopathy.83 Although peripheral transport of neuron-derived exosomes containing miRNA has not been firmly established, several studies suggest that nucleic acid content can cross the BBB with the help of vesicular transport.84,85 Glia also express miRNAs, and may alter their expression in response to oxidative, metabolic, and apoptotic demands of traumatic injury.86 Peripheral miRNAs can also undergo concentration shifts in response to sympathetic, hormonal, or neuro-immune mechanisms that regulate the physiologic response to TBI. In this latter scenario, miRNAs could play an important role in neuroplasticity,87 and differential miRNA expression among TBI patients in the subacute period could portend long-term prognosis. Whether miRNA changes are passive artifacts of primary injury or integral players in the secondary injury response, their dynamic expression and versatile role in the human brain provides a distinct opportunity to gauge the CNS response to trauma. Furthermore, the same properties that allow miRNA cross-talk between neurons (small size, vesicle protection) may permit sampling of these brain-related molecules in peripheral biofluids.
Peripheral measurement of brain-related miRNAs
miRNAs can be transported through the extracellular space by exosomes and microvesicles, or bound to protective proteins such as high-density lipoprotein.88 Such carriers facilitate protective transfer and targeted docking of miRNAs at distant cells, where they may traverse the cell membrane and influence gene expression.89 Protection by exosomes and microvesicles also permits ease of miRNA measurement in various biofluids. miRNAs can be quantified in serum, plasma, CSF, urine, and saliva.61,90 Due to their abundance, stability at fluctuating pH levels, resistance to enzymatic degradation, and essential role in transcriptional regulation, miRNAs make ideal biomarker candidates.91 Brain-related miRNAs have been detected in serum less than 1 hour after TBI.92
miRNAs as Biomarkers in Human TBI
Overlapping miRNAs in human TBI
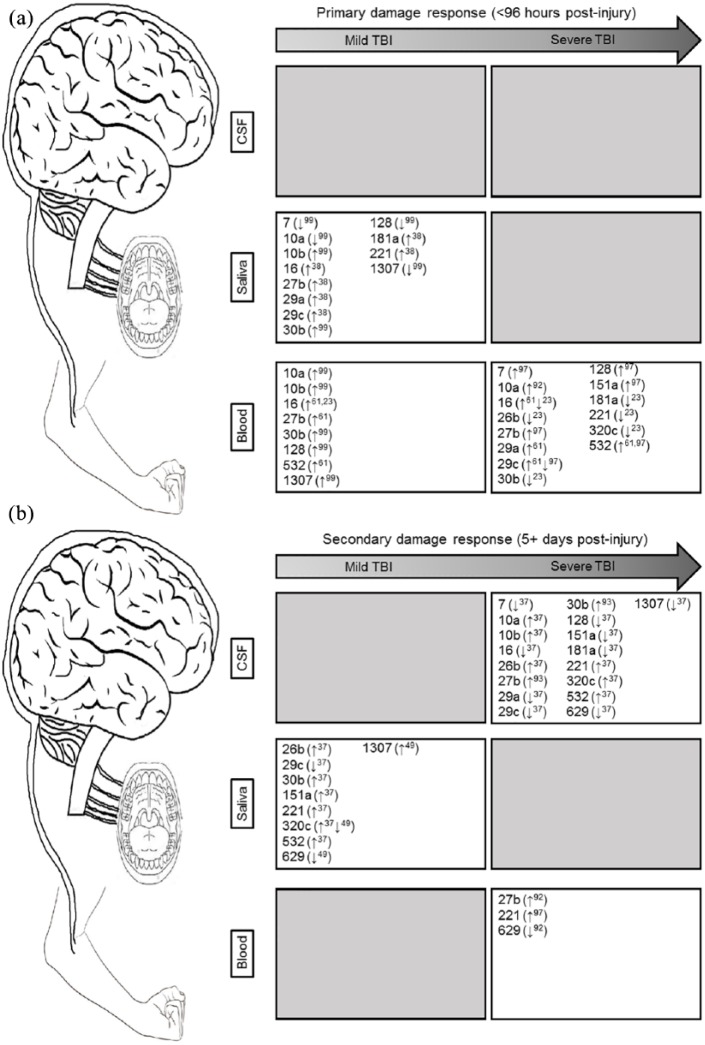
This review summarizes findings from 14 studies that examined miRNA levels in human patients with TBI. Together, the 14 studies have identified a host of miRNA candidates that may herald CNS injury across a spectrum of severity and time (Supplemental Figure 1). Although most of the studies explored miRNA levels in peripheral blood,23,38,39,61,92-99 a handful of recent investigations have also identified saliva as a novel and easily accessible biofluid with potential CNS overlap.37,38,49 Cumulatively, the 14 studies distinguish 291 miRNAs with prospective roles in TBI. Of these, 17 miRNAs are altered in multiple studies, across 3 biofluids (CSF, blood, and saliva; Figure 1A and B). A host of factors may contribute to this heterogeneity, including TBI severity, timing of sample collection, biofluid chosen, method of RNA analysis, and participant ages. Below, we consider such factors and attempt to untangle the web of biomarker complexity pervading miRNA research. The goal of this review is to identify gaps in existing miRNA literature that can be addressed through cohesive methodological approaches to promote valid, reliable results with near-term clinical applications.
Figure 1.
(a) Acute injury response of miRNAs: across 3 biofluids (cerebrospinal fluid, saliva, and blood) in the acute period (<96 hours post injury); (b) secondary injury response of miRNAs: across 3 biofluids (cerebrospinal fluid, saliva, and blood) in the subacute or chronic period (>5 days post injury).
CSF, cerebrospinal fluid; miRNA, microRNA; TBI, traumatic brain injury.
Arrows denote the general direction of change for each miRNA, along with the study in which it was detected (the superscript number corresponds to the reference). Gray boxes denote the time-points or biofluids without existing miRNA TBI studies.
The evolution of miRNA exploration in human TBI
Initial investigations of miRNA expression in TBI patients predominantly focused on serum miRNA differences between small numbers of injured participants and controls. A 2012 study by Pasinetti and colleagues used microarray to detect 13 small RNAs in blood mononuclear cells from 9 veterans with mTBI compared with 9 veterans without mTBI (73% male, mean age: 30 years). The authors reported 1 miRNA change (miR-671-5p) that was validated with PCR.96
Another study compared vesicular RNA in the CSF of 11 severe TBI patients and 17 controls to detect differential expression (53% male, mean age: 46 years). The authors determined that most of the RNA packaged in CSF microparticles was non-coding RNA, and that 2 of these non-coding RNAs (miR-9 and miR-451) were differentially expressed in severe TBI patients. They also found that the transfer of microparticles in vitro resulted in the regulation of specific neuronal genes.95
A study by Yang et al94 explored serum miRNA levels in an expanded cohort of adult patients with severe TBI (n = 76), compared with healthy (n = 38) controls (mean age: 47 years). They found increased levels of miR-93, miR-191, and miR-499 in TBI patients, which peaked on days 2 to 7 post injury, and was correlated with injury severity.94
You and colleagues described CSF miRNA levels in 26 comatose patients with severe TBI compared with 21 controls (55% male, mean age: 50 years). They detected 14 miRNAs with distinct expression levels in severe TBI patients, and 1 of these (miR-431-3p) had an associated single-nucleotide polymorphism in its promoter region.93
Studies have also incorporated multiple biofluids to compare miRNA levels. A study by Bhomia and colleagues is notable for its use of both CSF and serum from severe TBI patients, as well as its use of controls with orthopedic injury. The authors used RT-PCR to measure miRNA from 8 patients with mTBI, 8 patients with severe TBI, 7 orthopedic controls, and 8 healthy controls. The severe TBI serum was collected on average 33.8 hours after injury, and the CSF was collected 26.3 hours after injury, on average. Those with mTBI and orthopedic controls had their serum collected on average 3.1 hours after injury. They identified 10 miRNAs up-regulated in the serum of both mild and severe TBI groups relative to the orthopedic controls: miR-151-5p, miR-328, miR-362-3p, miR-486, miR-505, miR-451, miR-30d, miR-20a, miR-195, and miR-92a. Changes of 4 of these miRNAs were validated in the CSF of severe TBI patients.61
In 2018, Hicks and colleagues used a similar approach to identify overlapping CSF and saliva miRNA profiles among children with severe (n = 8) and mTBI (n = 60; mean age: 14 years). The authors used RNA sequencing to identify 6 salivary miRNAs with overlapping CSF alterations (miR-182-5p, miR-221-3p, miR-26b-5p, miR-320c, miR-29c-3p, and miR-30e-5p) <24 hours after injury. Furthermore, they showed that these miRNAs could predict mTBI status relative to healthy controls and that saliva miRNA levels were correlated with the severity of subjective symptoms.37
However, this was not the first study to demonstrate the diagnostic utility of peripheral miRNA levels. A study by Redell et al23 used RT-PCR to explore 108 miRNAs in the plasma of 10 adults with severe TBI, and 10 adults with mTBI (<10 after injury). There were 33 miRNAs increased and 19 miRNAs decreased in the plasma of TBI patients relative to healthy controls (8 were unique to severe TBI patients). Two miRNAs (miR-92a and miR-16) demonstrated diagnostic potential in both severe and mTBI relative to healthy and orthopedic injury controls.23
Recently, a study of saliva miRNA levels from 32 rugby players (mean age: 24 years) detected 5 miRNAs (miR-27b-3p, miR-142-3p, let-7i, miR-107, and miR-135b-5p) with differential expression 48 to 72 hours after sports-related concussion. Patterns for these 5 miRNAs correlated with reaction time on ImPACT testing and were able to predict a TBI diagnosis better than 92 protein biomarker candidates.38
Several studies have also focused on the longitudinal response of miRNAs to TBI. A study by Taheri and colleagues used RT-PCR to analyze 740 miRNAs in the serum of 38 patients (71% male, mean age: 43 years) receiving intensive care for severe TBI at 1, 7, and 28 days post injury, as well as 25 patients 5 years post injury. In patients with TBI-induced hypopituitarism, 2 miRNAs (miR-126-3p and miR-3610) demonstrated alterations during both the acute and chronic phases.97
Di Pietro and colleagues screened 754 miRNAs in the serum of 5 mTBI, 5 severe TBI, and 5 healthy controls at 1 and 15 days post injury. They identified 10 miRNA candidates that changed over time. In 30 additional patients with TBI, 2 of the miRNA candidates (miR-425-5p and miR-502) were validated at earlier time-points (0-1 hour and 4-12 hours post injury) and 2 (miR-21 and miR-335) were validated relative to healthy and orthopedic controls.92
Zhangjie et al98 also examined the time course of miRNA levels following sports-related concussion in 15 athletes (relative to 8 healthy peers). At both 1 and 2 weeks post injury, serum levels of miR-425-5p were down-regulated in concussed patients and correlated with the time since injury.98
The ability to measure dynamic miRNA responses after TBI has led some groups to posit that miRNA levels may be used to predict medical outcomes. One study measured plasma miRNA levels on arrival to the emergency department, and again 5 and 30 days post injury in adults with TBI (mean age: 43 years). The authors reported that plasma levels of miR-142-3p and miR-423-3p differentiated patients with mild head injury from those with higher post-concussive syndrome (PCS) risk, based on standardized post-traumatic amnesia testing.39
A study published in JAMA Pediatrics by Johnson and colleagues used RNA sequencing to identify 5 saliva miRNAs (miR-320c, miR-133a-5p, miR-769-5p, let-7a-3c, and miR-1307-3p) that differentiated risk for prolonged concussion symptoms among 52 children with mTBI (58% male, mean age: 14 years). Levels of 3 miRNAs at the time of injury were also associated with the presence of specific symptoms (eg, memory difficulty, headaches, fatigue) 1 month later.49
Recently, a large-scale study of 50 adult mixed martial arts fighters (mean age: 27 years) compared the utility of serum and saliva miRNAs for detecting hits-to-the-head, relative to protein markers. Serum and saliva were collected immediately before fight and again after fight (15 minutes, 2-3 days, 1 week, and 3-4 weeks). Saliva and serum miRNA levels showed an equivalent ability to detect the number of hits-to-the-head a fighter experienced. This ability exceeded the accuracy of serum protein markers. Intriguingly, saliva miRNA levels demonstrated a more acute response to head trauma than serum miRNA, and acute saliva miRNA levels were correlated with functional measures of cognition and balance.99
A Sampling of Top miRNA Candidates and Their Putative Functions in TBI
miR-320c represents one intriguing biomarker candidate in patients with TBI. miR-320c is altered in the blood of adults with severe TBI and the saliva of children with mTBI.23,49,97 Furthermore, this miRNA demonstrates longitudinal variation in CSF after severe TBI and predicts the duration of TBI symptoms. Previous studies in depressed adults have found that miR-320c levels change in the cerebral cortex following suicide.100 Such findings are intriguing given the association of prolonged concussive symptoms with depressed mood. Such an association might be facilitated by the role of miR-320c in neuroplasticity.37 For example, an investigation of miRNA expression in neuronal precursor cells has demonstrated that miR-320c levels are critical for neuronal differentiation and axonal outgrowth.101
Another compelling TBI biomarker candidate is miR-92a. miR-92a was up-regulated in the blood of patients with mTBI and the blood and CSF of patients with severe TBI.23,61 This microRNA was up-regulated in the acute (hours) and subacute (days) period after injury. Studies have shown that up-regulated miR-92a blocks angiogenesis after an ischemic event.102,103 Thus, acute up-regulation of miR-92a in patients with severe TBI may be critical in those with intracranial bleeding or neurovascular compromise.
Changes in the miR-30 family have been detected in several human TBI studies. Changes in miR-30 expression were noted in the blood of patients with severe TBI, both hours and days after injury. miR-30 changes have also been detected in the CSF weeks after brain injury. In patients with mTBI, subacute changes in miR-30 expression have been reported in both saliva and blood.23,38,61,93 The miR-30 family plays a role in neuroinflammation and is negatively regulated by pro-inflammatory cytokines. A decreased expression of miR-30 has been associated with changes in cell morphology, loss of cell adhesion, and increased cell migration due to tight junction disruption.104-106 Therefore, miR-30 may play a critical role in the maintenance of the BBB.
The biologic rationale for peripheral miRNA biomarkers in TBI can be inferred from the roles of their putative mRNA targets in brain-related processes. Here, we use DIANA mirPath bioinformatics along with the microT-CDS database to interrogate the 17 miRNAs implicated in the primary (hours-to-days) and secondary (days-to-weeks) damage responses to TBI (Figures 1A and B). The 17 miRNAs target 2014 mRNA transcripts with high confidence (microT-CDS ⩾ 0.975). These mRNAs demonstrate enrichment for 22 KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways (false discovery rate [FDR] < 0.05), including 8 implicated in brain-related mechanisms (Table 1). The pathway targets included those specific to neuronal outgrowth (neurotrophin signaling, FDR = 0.021) and brain function (amphetamine addiction, FDR = 2.6E-05; cocaine addiction, FDR = 0.020), as well as those with broad-ranging implications in cell signaling (ErbB signaling, FDR = 0.0056). Among the 17 miRNAs, miR-27b-3p targets the largest number of transcripts from brain-related pathways (43 mRNAs, accounting for 14% of its 313 total targets). Five genes (COL27A, KMT2C, ACVR1C, ACVR2A, and ITGA5) were targeted by at least 2 of the 17 miRNAs. It is worth noting that these targets and functions are putatively based on predicted miRNA-mRNA interactions, or observed in vitro targeting experiments. Additional mechanistic studies defining the specific physiologic contributions of individual miRNAs in TBI will be required to enhance our understanding of the exact role peripheral miRNAs play in CNS injury.
Table 1.
Putative targets of the 17 miRNAs with the highest TBI biomarker potential.
| MicroRNA Targets | miR-181a-5p ↑ (4) | miR-128-3p ↓ (4) | miR-16-5p ↑ (4) | miR-221-3p ↓ (4) | miR-26b-5p ↑ (3) | miR-27b-3p ↑ (6) | miR-29a/c-3p ↓ (4) | miR-30a-5p ↑ (5) | miR-320c ↑ (4) | miR-532-5p ↑ (3) | miR-1307-3p ↑ (3) | miR-151a-3p ↑ (2) | miR-7-5p ↓ (3) | miR-629-5p ↓ (3) | miR-10a/b-5p ↑ (3) | mRNA targets |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total no. of transcripts | 49 | 197 | 203 | 20 | 246 | 313 | 168 | 380 | 72 | 7 | 124 | 40 | 105 | 8 | 82 | 2014 |
| ECM-receptor interaction (FDR = 2.3E-39) | 1 | 4 | 0 | 0 | 1 | 4 | 13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | ITGA8, COL4A5, COL27A1, ITGA5, COL3A1, SV2A, COL2A1, RELN, COL5A1, COL4A4, COL1A2, LAMC1, COL11A1, COL6A3, LAMA2, COL5A3, COL5A2, SPP1, COL4A1, VEGFA |
| Pluripotency of stem cells (FDR = 1.5E-06) | 1 | 5 | 11 | 0 | 6 | 7 | 2 | 3 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | BMI1, JARID2, GSK3B, WNT7A, INHBB, APC, WNT10B, REST, MAPK14, RAF1, INHBA, WNT4, ZFHX3, FZD3, ACVR1, ACVR2B, FZD10, LIFR, PIK3R1, ACVR2A, FGF2, ACVR1C, IGF1, AKT3, BMPR1A, WNT3A, ISL1, GRB2, SMAD1, PIK3R2 |
| Amphetamine addiction (FDR = 2.6E-05) | 0 | 4 | 0 | 0 | 1 | 5 | 1 | 5 | 1 | 0 | 2 | 0 | 1 | 0 | 2 | AFT2, CAMK4, CREB5, PPP1CC, PPP3R1, DRD1, GRIA1, CREB1, PPP3CA, SLC6A3, PRKX, PRKCB, CAMK2B, GRIA4, GRIN2D |
| Cocaine addiction (FDR = 0.020) | 0 | 2 | 1 | 0 | 2 | 2 | 0 | 4 | 1 | 0 | 1 | 0 | 0 | 0 | 2 | ATF2, CREB5, DRD1, GPSM1, GRM3, CDK5R1, BDNF, CREB1, SLC6A3, PRKX, GRIN2D |
| Neurotrophin signaling (FDR = 0.021) | 0 | 6 | 4 | 0 | 3 | 6 | 3 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | GSK3B, SH2B3, CAMK4, MAP2K7, RAP1A, MAPK14, RAF1, BCL2, PRKCD, BDNF, KIDINS220, RPS6KA6, PIK3R1, SOS1, IRS1, GAB1, AKT3, CAMK2B, PRDM4, GRB2, RAP1B, NGFR, PIK3R2 |
| Glioma (FDR = 0.0022) | 0 | 1 | 3 | 0 | 2 | 2 | 3 | 1 | 1 | 0 | 3 | 0 | 1 | 0 | 1 | REF1, EGFR, CDK6, E2F3, PIK3R1, SOS1, PRKCB, IGF1, AKT3, CAMK2B, PTEN, GRB2, PIK3R2 |
| ErbB signaling (FDR = 0.0056) | 0 | 4 | 1 | 1 | 2 | 7 | 1 | 3 | 0 | 0 | 1 | 0 | 3 | 0 | 1 | GSK3B, HBEGF, MAP2K7, RAF1, CDKN1B, EGFR, CBLB, NRG3, PIK3R1, SOS1, PRKCB, GAB1, AKT3, CAMK2B, MAP2K4, ABL2, GRB2, PIK3R2 |
| Long-term potentiation (FDR = 0.016) | 0 | 4 | 3 | 0 | 3 | 7 | 0 | 7 | 1 | 0 | 0 | 0 | 2 | 0 | 1 | CAMK4, PPP1CC, RAP1A, GRM5, PPP3R1, RAF1, GRIA1, PPP3CA, PLCB1, RPS6KA6, PRKX, PRKCB, CAMK2B, GRIN2D, RAP1B |
ECM, extracellular matrix; FDR, false discovery rate; KEGG, Kyoto Encyclopedia of Genes and Genomes; TBI, traumatic brain injury.
The 17 miRNAs identified in ⩾2 human TBI studies, across 3 biofluids (cerebrospinal fluid, saliva, and blood), were interrogated for putative mRNA targets. Together, they targeted 2014 coding transcripts with high confidence (microT-CDS > 0.975) which demonstrated enrichment (FDR < 0.05) for 22 KEGG signaling pathways. Of the 22 pathways, 8 implicated in brain-related processes are shown here (FDR P-values in parentheses). The number of mRNAs targeted by each miRNA in the respective pathway is displayed. Arrows denote the general direction of change for each miRNA, along with the number of studies in which it was detected (in parentheses).
A close examination of miRNA gene targets reveals numerous transcripts with potential biologic significance in TBI. For example, vascular endothelial growth factor A (VEGFA) induces proliferation and migration of vascular endothelial cells and is essential for angiogenesis.107 VEGFA is targeted by miR-29a-3p (microT-CDS = 0.989) which rises in the acute TBI response (when the brain might wish to block VEGFA protein expression and prevent further bleeding), but drops in the subacute/chronic periods (when allowing VEGFA expression might be beneficial). The reelin transcript (RELN) encodes a secretory matrix protein that is imperative for the cell-cell positioning involved in neuronal migration.108 Expression of RELN is disrupted in temporal lobe epilepsy and major depressive disorder.109 RELN is targeted by miR-27b-3p (microT-CDS = 0.999), which is elevated across CSF, blood, and saliva in TBI patients.24,38,39,93,95,98 In addition, miR-27b-3p levels have been shown to correlate with reaction time on ImPACT testing.38 Putative miRNA-mRNA interactions provide a simplistic biologic construct for the potential function of miRNA expression following TBI. Additional work to elucidate the exact role of individual miRNAs in the CNS injury response will help guide the selection of ideal biomarker candidates in various clinical circumstances (eg, severe-versus-mild injury; prognostic-versus-diagnostic utility).
Future Directions: Providing Clarity and Clinical Applicability Through miRNA Research
Methodology
Pilot studies have now established a putative role for miRNAs in TBI pathophysiology.22,77 Moreover, it is clear that the mechanistic findings in animal models extend to human patients with TBI,95 allowing measurement of brain-related miRNAs in peripheral biofluids.23,39,61,92 Studies of children and adults with severe TBI have identified the miRNAs that are altered in the CNS and deserve primary consideration as TBI biomarkers.37,93,95 The miRNAs identified in this review (that overlap across multiple studies, or demonstrate dynamic longitudinal expression patterns across multiple biofluids) may provide high yield targets for future hypothesis-driven investigations. In animal models, such investigations might manipulate miRNA expression to define cell- or tissue-specific actions, or explore TBI resilience, as Ge et al77 have done with miR-21.
It will be critical that future investigations eschew the small sample sizes employed in pilot research. Hundreds of samples from TBI patients will likely be required to control for the inter-individual variation that exists in baseline miRNA expression110 and produce reliable results. Alternatively, as LaRocca et al99 recently demonstrated, pre-injury miRNA baseline levels can be employed, although this approach is costly and time-consuming. Starting from hypothesis-driven miRNA targets, future studies should consider building diagnostic/prognostic miRNA algorithms in a training cohort, then validating the findings in large, external hold-out sets. This rigorous approach was recently demonstrated with success by Di Pietro et al38 in their study of salivary miRNA among concussed athletes. To ensure the reliability and reproducibility of such findings, investigators should provide explicit details for sample collection, sample storage, miRNA isolation, and miRNA quantification whenever possible. Selecting reagents and instruments from companies that have established miRNA protocols with test-retest reliability will allow others to reproduce findings and validate the clinical promise of miRNA biomarkers.111-113
Considering context-specific miRNAs
Severity of injury, timing of sample collection, biofluid selection, participant age, and medical comorbidities can all impact miRNA patterns following TBI. Careful consideration of these factors when designing future miRNA trials and interpreting results will be necessary to discern reliable biomarkers from statistical artifact. For example, a number of miRNAs demonstrate overlapping expression patterns in both severe and mTBI,23,37,92,94 but it is likely that distinct miRNA panels will be necessary to provide the clinical specificity and sensitivity required for these disparate conditions. After all, mTBI and severe TBI involve different pathophysiologic mechanisms and unique medical management considerations. Among patients with severe TBI, GCS and clinical endpoints have been used with some success to delineate clinical severity, whereas in mTBI GCS does not provide sufficient granularity, so employing sensitive measures of balance and cognition38,99 will provide objective measures of functional deficits while aligning with clinical guidelines for mTBI care.
Mounting evidence demonstrates that saliva miRNAs may provide a novel window into the brain,114-117 and that salivary miRNA levels may have utility in TBI.37,38,49,99 Although saliva and blood share some common miRNA biomarkers following TBI (Figures 1A and B), the 2 biofluids also demonstrate distinct signatures after injury.99 This may result from the origin of miRNAs in these 2 fluids. Saliva, which can receive exosomal miRNAs directly from cranial nerves in the oropharynx, demonstrates a rapid miRNA response after TBI. In comparison, blood-based miRNAs must traverse the BBB, which may account for their relative delay in temporal patterns.99 In the secondary response to TBI, it is likely that serum miRNAs carry information about the inflammatory mechanisms underlying neuronal injury (although this response may be muted without disruption of the BBB). Salivary miRNAs, on the other hand, appear to reflect the secondary neuroplasticity response. This may explain why saliva miRNA levels measured 1 week after injury predict prolonged concentration/memory difficulties with such accuracy.49 Saliva collection allows immediate sideline assessment by coaches and athletic trainers without venipuncture training.118 Non-invasive saliva collection also has the added benefit of room-temperature miRNA stabilization (when using RNA-specific devices).119,120 Ultimately, serum and saliva miRNA profiles are likely to provide complementary information following TBI. Investigations that harness both biofluids in parallel may help clarify the clinical role for each approach.
Clinical utility must also be considered when selecting time-points for miRNA collection. Numerous studies have demonstrated that CNS37 and peripheral38,39,94,97-99 miRNA profiles fluctuate across the primary and secondary phases of TBI. The miRNAs responding within hours of injury are likely to be most suitable as a diagnostic tool. Studies with this clinical focus should employ large control groups, matched by age, sex, and ethnicity. For translation to a clinical setting, control groups must include individuals with non-cranial orthopedic injury,23,61 as well as post-exercise participants (for field-side applicability in concussed athletes).121,122 Conversely, miRNAs involved in the subacute response (days to weeks after injury) may provide more information for prognosis and clinical outcomes. Investigations geared toward prognostic biomarkers should consider timing of typical follow-up visits in outpatient TBI management (7-14 days) and use controls whose medical comorbidities may mirror prolonged TBI symptoms (eg, patients with depression, attention deficits, or disordered sleep).96 Finally, with the rising concern over traumatic encephalopathy and the long-term implication of TBI, measuring miRNA levels at chronic time-points alongside sensitive functional outcomes (eg, memory, balance) and imaging may help fill critical gaps in the current miRNA TBI literature (see gray boxes in Figure 1).
Clinical applications
The primary emphasis of most miRNA studies has been TBI diagnosis. Certainly, an objective marker of mTBI would provide clinical benefit in emergency department and outpatient settings. Currently, clinicians in these settings rely on subjective symptom reports because neurologic examination and imaging are typically normal. Recent epidemiologic research suggests that health care providers may have previously under-diagnosed mTBI.123 As such, addition of an objective diagnostic biomarker could improve mTBI recognition. In cases of negative testing, it would also provide valuable reassurance to patients and families that head trauma did not result in biologically detectable brain injury.
This sort of reassurance will require development of markers sensitive enough to detect even subconcussive hits to the head. It will also require that we understand how such blows impact long-term brain health. The interplay between repeated minor head trauma and chronic traumatic encephalopathy necessitates examining athletes in contact sports to identify miRNAs that change from minor head impacts. The study by LaRocca and colleagues begins this critical work, by identifying saliva and blood-based miRNA signatures in mixed martial arts fighters with subconcussive hits to the head. By treating the miRNA response as a function of the number of blows to the head, and not an “all-or-none” concussion experience, the study defines miRNAs that portend minor brain disruption.
What happens after TBI diagnosis? Surveys of health care providers demonstrate a critical need for objective toolsets that guide prognosis and clinical management.4,124,125 This need extends from mTBI through severe TBI. Clinical risk scores using information about injury mechanisms and patient factors are currently used to predict outcomes,126,127 but these tools can be time-consuming and inaccurate. In intensive care settings, separate sets of biomarkers will likely be required to predict outcomes involving morbidity and mortality. It would be naïve to expect such biomarkers to completely overlap with those used for mTBI diagnosis.
In mTBI, there is a unique opportunity to harness miRNA profiles to divide patients into phenotypic subgroups and guide individualized treatment plans. For example, recent findings that salivary miRNAs identify children at risk for PCS and that specific miRNAs may be associated with individual symptoms could be used for initiation of early individualized therapy.49In patients with miRNA profiles that portend PCS characterized by chronic headaches, this might mean early initiation of aggressive triptan therapy. In patients with miRNA levels implicated in prolonged concentration difficulties, this might inform individualized return-to-learn decisions or trigger prescription of stimulant medication.
Accumulating research demonstrates that graded exercise routines can accelerate TBI recovery in a subset of patients identified through treadmill testing.128,129 Interestingly, some of the same miRNAs that demonstrate alterations in patients with TBI show contrary patterns with endurance exercise.121 This concept has also been validated in mouse studies, where voluntary running mitigates the miRNA response to TBI,130 and post-injury running triggers miR-21 alterations associated with cognitive improvements.131 In this context, miRNAs represent a means for phenotypic assessment, as well as potential therapeutic targets in and of themselves.
Conclusions
Despite extensive research involving TBI biomarkers, significant gaps remain. There are limited tools for diagnosis, prognosis, and therapeutic decisions after TBI. Emerging research suggests that miRNAs are ideally suited to fill this need. miRNAs are highly expressed in the brain, can cross the BBB, and are stable in peripheral biofluids. Moreover, the unique function of miRNAs in neuronal communication may provide a distinct window into the injured brain. The miRNA response to TBI is complex, and at times messy. However, with careful consideration of the physiologic factors involved, it is possible to discern commonalities across studies. Focusing on the miRNA candidates with reproducible findings across multiple biofluids and time-points may yield clinical utility in the near future. It will be critical that future miRNA investigations employ large sample sizes, while considering confounding factors such as patient ages, or injury severity. If these factors are addressed, miRNAs may one day guide diagnostic, prognostic, and therapeutic decision making in patients with TBI.
Supplemental Material
Supplemental material, Supplemental_Figure_1A for A Review of MicroRNA Biomarkers in Traumatic Brain Injury by Hamna Atif and Steven D Hicks in Journal of Experimental Neuroscience
Supplemental Material
Supplemental material, Supplemental_Figure_1B for A Review of MicroRNA Biomarkers in Traumatic Brain Injury by Hamna Atif and Steven D Hicks in Journal of Experimental Neuroscience
Supplemental Material
Supplemental material, Supplemental_Figure_1C for A Review of MicroRNA Biomarkers in Traumatic Brain Injury by Hamna Atif and Steven D Hicks in Journal of Experimental Neuroscience
Footnotes
Funding:The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of conflicting interests:The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: SDH is a paid consultant for Quadrant Biosciences Inc. He is named as an inventor on intellectual property using miRNA technology in patients with traumatic brain injury. AF has no conflicts of interest to disclose.
Author Contributions: SDH conceived of the study. AH and SDH contributed equally to the literature review and drafting the manuscript.
Supplemental Material: Supplemental material for this article is available online.
ORCID iD: Steven D Hicks  https://orcid.org/0000-0002-9579-6798
https://orcid.org/0000-0002-9579-6798
References
- 1. Veliz P, McCabe SE, Eckner JT, Schulenberg JE. Prevalence of concussion among US adolescents and correlated factors. JAMA. 2017;318:1180-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Raftery M, Kemp S, Patricios J, Makdissi M, Decq P. It is time to give concussion an operational definition: a 3-step process to diagnose (or rule out) concussion within 48 h of injury: World Rugby guideline. Br J Sports Med. 2016;50:642-643. [DOI] [PubMed] [Google Scholar]
- 3. Lumba-Brown A, Yeates KO, Sarmiento K, et al. Centers for Disease Control and Prevention guideline on the diagnosis and management of mild traumatic brain injury among children. JAMA Pediatr. 2018;172:e182853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zonfrillo MR, Master CL, Grady MF, Winston FK, Callahan JM, Arbogast KB. Pediatric providers’ self-reported knowledge, practices, and attitudes about concussion. Pediatrics. 2012;130:1120-1125. [DOI] [PubMed] [Google Scholar]
- 5. Finch CF, McCrory P, Ewing MT, Sullivan SJ. Concussion guidelines need to move from only expert content to also include implementation and dissemination strategies. Br J Sports Med. 2013;47:12-14. [DOI] [PubMed] [Google Scholar]
- 6. Covassin T, Crutcher B, Bleecker A, Heiden EO, Dailey A, Yang J. Postinjury anxiety and social support among collegiate athletes: a comparison between orthopaedic injuries and concussions. J Athl Train. 2014;49:462-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zemek R, Clarkin C, Farion KJ, et al. Parental anxiety at initial acute presentation is not associated with prolonged symptoms following pediatric concussion. Acad Emerg Med. 2013;20:1041-1049. [DOI] [PubMed] [Google Scholar]
- 8. Zetterberg H, Smith DH, Blennow K. Biomarkers of mild traumatic brain injury in cerebrospinal fluid and blood. Nat Rev Neurol. 2013;9:201-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jeter CB, Hergenroeder GW, Hylin MJ, Redell JB, Moore AN, Dash PK. Biomarkers for the diagnosis and prognosis of mild traumatic brain injury/concussion. J Neurotrauma. 2013;30:657-670. [DOI] [PubMed] [Google Scholar]
- 10. Follert P, Cremer H, Béclin C. MicroRNAs in brain development and function: a matter of flexibility and stability. Front Mol Neurosci. 2014;7:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Karp X, Ambros V. Encountering microRNAs in cell fate signaling. Science. 2005;310:1288-1289. [DOI] [PubMed] [Google Scholar]
- 12. Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen C-Z, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83-86. [DOI] [PubMed] [Google Scholar]
- 14. Lynam Lennon N, Maher SG, Reynolds JV. The roles of microRNA in cancer and apoptosis. Biol Rev. 2009;84:55-71. [DOI] [PubMed] [Google Scholar]
- 15. Latronico MV, Catalucci D, Condorelli G. Emerging role of microRNAs in cardiovascular biology. Circ Res. 2007;101:1225-1236. [DOI] [PubMed] [Google Scholar]
- 16. Williams MD, Mitchell GM. MicroRNAs in insulin resistance and obesity. Exp Diabetes Res. 2012;2012:484696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Menon DK, Schwab K, Wright DW, Maas AI. Position statement: definition of traumatic brain injury. Arch Phys Med Rehabil. 2010;91:1637-1640. [DOI] [PubMed] [Google Scholar]
- 18. Amorini AM, Lazzarino G, Di Pietro V, et al. Metabolic, enzymatic and gene involvement in cerebral glucose dysmetabolism after traumatic brain injury. Biochim Biophys Acta. 2016;1862:679-687. [DOI] [PubMed] [Google Scholar]
- 19. Di Pietro V, Amorini AM, Tavazzi B, et al. Potentially neuroprotective gene modulation in an in vitro model of mild traumatic brain injury. Mol Cell Biochem. 2013;375:185-198. [DOI] [PubMed] [Google Scholar]
- 20. Tavazzi B, Vagnozzi R, Signoretti S, et al. Temporal window of metabolic brain vulnerability to concussions: oxidative and nitrosative stresses—part II. Neurosurgery. 2007;61:390-396. [DOI] [PubMed] [Google Scholar]
- 21. Vagnozzi R, Tavazzi B, Signoretti S, et al. Temporal window of metabolic brain vulnerability to concussions: mitochondrial-related impairment—part I. Neurosurgery. 2007;61:379-389. [DOI] [PubMed] [Google Scholar]
- 22. Redell JB, Liu Y, Dash PK. Traumatic brain injury alters expression of hippocampal microRNAs: potential regulators of multiple pathophysiological processes. J Neurosci Res. 2009;87:1435-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Redell JB, Moore AN, Ward NH, III, Hergenroeder GW, Dash PK. Human traumatic brain injury alters plasma microRNA levels. J Neurotrauma. 2010;27:2147-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu D-Z, Tian Y, Ander BP, et al. Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J Cereb Blood Flow Metab. 2010;30:92-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu Z, Yu D, Almeida-Suhett C, et al. Expression of miRNAs and their cooperative regulation of the pathophysiology in traumatic brain injury. PLoS ONE. 2012;7:e39357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu L, Sun T, Liu Z, et al. Traumatic brain injury dysregulates microRNAs to modulate cell signaling in rat hippocampus. PLoS ONE. 2014;9:e103948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Truettner JS, Alonso OF, Bramlett HM, Dietrich WD. Therapeutic hypothermia alters microRNA responses to traumatic brain injury in rats. J Cereb Blood Flow Metab. 2011;31:1897-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Teasdale G, Maas A, Lecky F, Manley G, Stocchetti N, Murray G. The Glasgow Coma Scale at 40 years: standing the test of time. Lancet Neurol. 2014;13:844-854. [DOI] [PubMed] [Google Scholar]
- 29. Fischer H. U.S. military casualty statistics: operation new dawn, operation Iraqi freedom, and operation enduring freedom; 2013. https://apps.dtic.mil/docs/citations/ADA590694.
- 30. Scheers I, Palermo J, Freedman S, et al. Recommendations for diagnosis and management of autoimmune pancreatitis in childhood: consensus from INSPPIRE [published online ahead of print May 9, 2018]. J Pediatr Gastroenterol Nutr. doi: 10.1097/MPG.00000000000020282018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bazarian JJ, Biberthaler P, Welch RD, et al. Serum GFAP and UCH-L1 for prediction of absence of intracranial injuries on head CT (ALERT-TBI): a multicentre observational study. Lancet Neurol. 2018;17:782-789. [DOI] [PubMed] [Google Scholar]
- 32. Honda M, Tsuruta R, Kaneko T, et al. Serum glial fibrillary acidic protein is a highly specific biomarker for traumatic brain injury in humans compared with S-100B and neuron-specific enolase. J Trauma. 2010;69:104-109. [DOI] [PubMed] [Google Scholar]
- 33. Papa L, Akinyi L, Liu MC, et al. Ubiquitin C-terminal hydrolase is a novel biomarker in humans for severe traumatic brain injury. Crit Care Med. 2010;38:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bloomfield SM, McKinney J, Smith L, Brisman J. Reliability of S100B in predicting severity of central nervous system injury. Neurocrit Care. 2007;6:121-138. [DOI] [PubMed] [Google Scholar]
- 35. Morel L, Regan M, Higashimori H, et al. Neuronal exosomal miRNA-dependent translational regulation of astroglial glutamate transporter GLT1. J Biol Chem. 2013;288:7105-7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Goldie BJ, Dun MD, Lin M, et al. Activity-associated miRNA are packaged in Map1b-enriched exosomes released from depolarized neurons. Nucleic Acids Res. 2014;42:9195-9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hicks SD, Johnson J, Carney MC, et al. Overlapping microRNA expression in saliva and cerebrospinal fluid accurately identifies pediatric traumatic brain injury. J Neurotrauma. 2018;35:64-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Di Pietro V, Porto E, Ragusa M, et al. Salivary microRNAs: diagnostic markers of mild traumatic brain injury in contact-sport. Front Mol Neurosci. 2018;11:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mitra B, Rau TF, Surendran N, et al. Plasma micro-RNA biomarkers for diagnosis and prognosis after traumatic brain injury: a pilot study. J Clin Neurosci. 2017;38:37-42. [DOI] [PubMed] [Google Scholar]
- 40. Ignacio C, Hicks SD, Burke P, Lewis L, Szombathyne-Meszaros Z, Middleton FA. Alterations in serum microRNA in humans with alcohol use disorders impact cell proliferation and cell death pathways and predict structural and functional changes in brain. BMC Neurosci. 2015;16:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Quarrie KL, Murphy IR. Towards an operational definition of sports concussion: identifying a limitation in the 2012 Zurich consensus statement and suggesting solutions. Br J Sports Med. 2014;48:1589-1591. [DOI] [PubMed] [Google Scholar]
- 42. McCrea M, Guskiewicz K, Randolph C, et al. Incidence, clinical course, and predictors of prolonged recovery time following sport-related concussion in high school and college athletes. J Int Neuropsychol Soc. 2013;19:22-33. [DOI] [PubMed] [Google Scholar]
- 43. Dean PJ, O’Neill D, Sterr A. Post-concussion syndrome: prevalence after mild traumatic brain injury in comparison with a sample without head injury. Brain Inj. 2012;26:14-26. [DOI] [PubMed] [Google Scholar]
- 44. Bazarian JJ, Wong T, Harris M, Leahey N, Mookerjee S, Dombovy M. Epidemiology and predictors of post-concussive syndrome after minor head injury in an emergency population. Brain Inj. 1999;13:173-189. [DOI] [PubMed] [Google Scholar]
- 45. Meehan WP, Mannix R, Monuteaux MC, Stein CJ, Bachur RG. Early symptom burden predicts recovery after sport-related concussion. Neurology. 2014;83:2204-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Grubenhoff JA, Deakyne SJ, Brou L, Bajaj L, Comstock RD, Kirkwood MW. Acute concussion symptom severity and delayed symptom resolution. Pediatrics. 2014;134:54-62. [DOI] [PubMed] [Google Scholar]
- 47. Drake AI, McDonald EC, Magnus NE, Gray N, Gottshall K. Utility of Glasgow Coma Scale-Extended in symptom prediction following mild traumatic brain injury. Brain Inj. 2006;20:469-475. [DOI] [PubMed] [Google Scholar]
- 48. Zemek R, Barrowman N, Freedman SB, et al. Clinical risk score for persistent postconcussion symptoms among children with acute concussion in the ED. JAMA. 2016;315:1014-1025. [DOI] [PubMed] [Google Scholar]
- 49. Johnson JJ, Loeffert AC, Stokes J, Olympia RP, Bramley H, Hicks SD. Association of salivary microRNA changes with prolonged concussion symptoms. JAMA Pediatr. 2018;172:65-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Erdal K. Neuropsychological testing for sports-related concussion: how athletes can sandbag their baseline testing without detection. Arch Clin Neuropsychol. 2012;27:473-479. [DOI] [PubMed] [Google Scholar]
- 51. Silver JM. Effort, exaggeration and malingering after concussion. J Neurol Neurosurg Psychiatry. 2012;83:836-841. [DOI] [PubMed] [Google Scholar]
- 52. Burke MJ, Fralick M, Nejatbakhsh N, Tartaglia MC, Tator CH. In search of evidence-based treatment for concussion: characteristics of current clinical trials. Brain Inj. 2015;29:300-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jensen RH. Pharmacological treatment of acute and chronic post-traumatic headache. In: Mitsikostas D, Paemeleire K. (eds) Pharmacological Management of Headaches. Cham: Springer; 2016:179-188. [Google Scholar]
- 54. Ellis MJ, Leddy JJ, Willer B. Physiological, vestibulo-ocular and cervicogenic post-concussion disorders: an evidence-based classification system with directions for treatment. Brain Inj. 2015;29:238-248. [DOI] [PubMed] [Google Scholar]
- 55. Papa L, Ramia MM, Kelly JM, Burks SS, Pawlowicz A, Berger RP. Systematic review of clinical research on biomarkers for pediatric traumatic brain injury. J Neurotrauma. 2013;30:324-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Papa L, Brophy GM, Welch RD, et al. Time course and diagnostic accuracy of glial and neuronal blood biomarkers GFAP and UCH-L1 in a large cohort of trauma patients with and without mild traumatic brain injury. JAMA Neurol. 2016;73:551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Posti JP, Takala RS, Runtti H, et al. The levels of glial fibrillary acidic protein and ubiquitin C-terminal hydrolase-L1 during the first week after a traumatic brain injury: correlations with clinical and imaging findings. Neurosurgery. 2016;79:456-464. [DOI] [PubMed] [Google Scholar]
- 58. Takala RS, Posti JP, Runtti H, et al. Glial fibrillary acidic protein and ubiquitin C-terminal hydrolase-L1 as outcome predictors in traumatic brain injury. World Neurosurg. 2016;87:8-20. [DOI] [PubMed] [Google Scholar]
- 59. Meier TB, Nelson LD, Huber DL, Bazarian JJ, Hayes RL, McCrea MA. Prospective assessment of acute blood markers of brain injury in sport-related concussion. J Neurotrauma. 2017;34:3134-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Middeldorp J, Hol E. GFAP in health and disease. Prog Neurobiol. 2011;93:421-443. [DOI] [PubMed] [Google Scholar]
- 61. Bhomia M, Balakathiresan NS, Wang KK, Papa L, Maheshwari RK. A panel of serum MiRNA biomarkers for the diagnosis of severe to mild traumatic brain injury in humans. Sci Rep. 2016;6:28148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lv L-L, Cao Y, Liu D, et al. Isolation and quantification of microRNAs from urinary exosomes/microvesicles for biomarker discovery. Int J Biol Sci. 2013;9:1021-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hergenroeder GW, Redell JB, Moore AN, Dash PK. Biomarkers in the clinical diagnosis and management of traumatic brain injury. Mol Diagn Ther. 2008;12:345-358. [DOI] [PubMed] [Google Scholar]
- 64. Nam J-W, Rissland OS, Koppstein D, et al. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol Cell. 2014;53:1031-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nelson P, Kiriakidou M, Sharma A, Maniataki E, Mourelatos Z. The microRNA world: small is mighty. Trends Biochem Sci. 2003;28:534-540. [DOI] [PubMed] [Google Scholar]
- 66. Suzuki HI, Miyazono K. Emerging complexity of microRNA generation cascades. J Biochem. 2010;149:15-25. [DOI] [PubMed] [Google Scholar]
- 67. Tritschler F, Huntzinger E, Izaurralde E. Role of GW182 proteins and PABPC1 in the miRNA pathway: a sense of deja vu. Nat Rev Mol Cell Biol. 2010;11:379-384. [DOI] [PubMed] [Google Scholar]
- 68. Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kaczkowski B, Torarinsson E, Reiche K, Havgaard JH, Stadler PF, Gorodkin J. Structural profiles of human miRNA families from pairwise clustering. Bioinformatics. 2008;25:291-294. [DOI] [PubMed] [Google Scholar]
- 70. Adlakha YK, Saini N. Brain microRNAs and insights into biological functions and therapeutic potential of brain enriched miRNA-128. Mol Cancer. 2014;13:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ziats MN, Rennert OM. Identification of differentially expressed microRNAs across the developing human brain. Mol Psychiatry. 2014;19:848-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bak M, Silahtaroglu A, Møller M, et al. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14:432-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Corbin R, Olsson-Carter K, Slack F. The role of microRNAs in synaptic development and function. BMB Rep. 2009;42:131-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang W, Kwon EJ, Tsai L-H. MicroRNAs in learning, memory, and neurological diseases. Learn Mem. 2012;19:359-368. [DOI] [PubMed] [Google Scholar]
- 75. Rao V, McCann U, Bergey A, Han D, Brandt J, Schretlen DJ. Correlates of apathy during the first year after traumatic brain injury. Psychosomatics. 2013;54:403-404. [DOI] [PubMed] [Google Scholar]
- 76. Werner C, Engelhard K. Pathophysiology of traumatic brain injury. Br J Anaesth. 2007;99:4-9. [DOI] [PubMed] [Google Scholar]
- 77. Ge X, Han Z, Chen F, et al. MiR-21 alleviates secondary blood-brain barrier damage after traumatic brain injury in rats. Brain Res. 2015;1603:150-157. [DOI] [PubMed] [Google Scholar]
- 78. Harrison EB, Hochfelder CG, Lamberty BG, et al. Traumatic brain injury increases levels of miR-21 in extracellular vesicles: implications for neuroinflammation. FEBS Open Bio. 2016;6:835-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944-13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang W-X, Wilfred BR, Madathil SK, et al. miR-107 regulates granulin/progranulin with implications for traumatic brain injury and neurodegenerative disease. Am J Pathol. 2010;177:334-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sabirzhanov B, Zhao Z, Stoica BA, et al. Downregulation of miR-23a and miR-27a following experimental traumatic brain injury induces neuronal cell death through activation of proapoptotic Bcl-2 proteins. J Neurosci. 2014;34:10055-10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lau LT, Yu AC. Astrocytes produce and release interleukin-1, interleukin-6, tumor necrosis factor alpha and interferon-gamma following traumatic and metabolic injury. J Neurotrauma. 2001;18:351-359. [DOI] [PubMed] [Google Scholar]
- 83. Laterza OF, Lim L, Garrett-Engele PW, et al. Plasma microRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem. 2009;55:1977-1983. [DOI] [PubMed] [Google Scholar]
- 84. Tyler BM, Jansen K, McCormick DJ, et al. Peptide nucleic acids targeted to the neurotensin receptor and administered i.p. cross the blood-brain barrier and specifically reduce gene expression. Proc Natl Acad Sci U S A. 1999;96:7053-7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. García-Romero N, Carrión-Navarro J, Esteban-Rubio S, et al. DNA sequences within glioma-derived extracellular vesicles can cross the intact blood-brain barrier and be detected in peripheral blood of patients. Oncotarget. 2017;8:1416-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ponomarev ED, Veremeyko T, Weiner HL. MicroRNAs are universal regulators of differentiation, activation, and polarization of microglia and macrophages in normal and diseased CNS. Glia. 2013;61:91-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kalsotra A, Zhao J, Anakk S, Dash PK, Strobel HW. Brain trauma leads to enhanced lung inflammation and injury: evidence for role of P4504Fs in resolution. J Cereb Blood Flow Metab. 2007;27:963-974. [DOI] [PubMed] [Google Scholar]
- 88. Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Shivdasani RA. MicroRNAs: regulators of gene expression and cell differentiation. Blood. 2006;108:3646-3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-659. [DOI] [PubMed] [Google Scholar]
- 91. Gilad S, Meiri E, Yogev Y, et al. Serum microRNAs are promising novel biomarkers. PLoS ONE. 2008;3:e3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Di Pietro V, Ragusa M, Davies D, et al. MicroRNAs as novel biomarkers for the diagnosis and prognosis of mild and severe traumatic brain injury. J Neurotrauma. 2017;34:1948-1956. [DOI] [PubMed] [Google Scholar]
- 93. You W-D, Tang Q-L, Wang L, et al. Alteration of microRNA expression in cerebrospinal fluid of unconscious patients after traumatic brain injury and a bioinformatic analysis of related single nucleotide polymorphisms. Chin J Traumatol. 2016;19:11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yang T, Song J, Bu X, et al. Elevated serum miR-93, miR-191, and miR-499 are noninvasive biomarkers for the presence and progression of traumatic brain injury. J Neurochem. 2016;137:122-129. [DOI] [PubMed] [Google Scholar]
- 95. Patz S, Trattnig C, Grünbacher G, et al. More than cell dust: microparticles isolated from cerebrospinal fluid of brain injured patients are messengers carrying mRNAs, miRNAs, and proteins. J Neurotrauma. 2013;30:1232-1242. [DOI] [PubMed] [Google Scholar]
- 96. Pasinetti GM, Ho L, Dooley C, Abbi B, Lange G. Select non-coding RNA in blood components provide novel clinically accessible biological surrogates for improved identification of traumatic brain injury in OEF/OIF veterans. Am J Neurodegener Dis. 2012;1:88. [PMC free article] [PubMed] [Google Scholar]
- 97. Taheri S, Tanriverdi F, Zararsiz G, et al. Circulating microRNAs as potential biomarkers for traumatic brain injury-induced hypopituitarism. J Neurotrauma. 2016;33:1818-1825. [DOI] [PubMed] [Google Scholar]
- 98. Zhangjie S, Davies D, Wang M, et al. Circulating microRNA as novel early biomarker of concussion in elite athletes. Br J Sports Med. 2017;51:A2-A3. [Google Scholar]
- 99. LaRocca D, Barns S, Hicks S, et al. Comparison of serum and saliva miRNAs for identification and characterization of mTBI in adult mixed martial arts fighters. PLoS ONE. 2019;14:e0207785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lopez JP, Fiori LM, Gross JA, et al. Regulatory role of miRNAs in polyamine gene expression in the prefrontal cortex of depressed suicide completers. Int J Neuropsychopharmacol. 2014;17:23-32. [DOI] [PubMed] [Google Scholar]
- 101. Savaskan NE, Bräuer AU, Nitsch R. Molecular cloning and expression regulation of PRG-3, a new member of the plasticity related gene family. Eur J Neurosci. 2004;19:212-220. [DOI] [PubMed] [Google Scholar]
- 102. Bonauer A, Carmona G, Iwasaki M, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710-1713. [DOI] [PubMed] [Google Scholar]
- 103. Doebele C, Bonauer A, Fischer A, et al. Members of the microRNA-17-92 cluster exhibit a cell-intrinsic anti-angiogenic function in endothelial cells. Blood. 2010;115:4944-4950. [DOI] [PubMed] [Google Scholar]
- 104. Joglekar MV, Patil D, Joglekar VM, et al. The miR-30 family microRNAs confer epithelial phenotype to human pancreatic cells. Islets 2009;1:137-147. [DOI] [PubMed] [Google Scholar]
- 105. Wu Y-D, Zhou B. TNF-α/NF-κB/Snail pathway in cancer cell migration and invasion. Br J Cancer. 2010;102:639-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Cano A, Pérez-Moreno MA, Rodrigo I, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76-83. [DOI] [PubMed] [Google Scholar]
- 107. Nagy JA, Dvorak AM, Dvorak HF. VEGF-A and the induction of pathological angiogenesis. Annu Rev Pathol. 2007;2:251-275. [DOI] [PubMed] [Google Scholar]
- 108. Howell BW, Herrick TM, Cooper JA. Reelin-induced tyrosine phosphorylation of disabled 1 during neuronal positioning. Genes Dev. 1999;13:643-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Fatemi S, Earle J, McMenomy T. Reduction in Reelin immunoreactivity in hippocampus of subjects with schizophrenia, bipolar disorder and major depression. Mol Psychiatry. 2000;5:654-663. [DOI] [PubMed] [Google Scholar]
- 110. Witwer KW. Circulating microRNA biomarker studies: pitfalls and potential solutions. Clin Chem. 2015;61:56-63. [DOI] [PubMed] [Google Scholar]
- 111. Doleshal M, Magotra AA, Choudhury B, Cannon BD, Labourier E, Szafranska AE. Evaluation and validation of total RNA extraction methods for microRNA expression analyses in formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2008;10:203-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. McAlexander MA, Phillips MJ, Witwer KW. Comparison of methods for miRNA extraction from plasma and quantitative recovery of RNA from cerebrospinal fluid. Front Genet. 2013;4:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Mestdagh P, Hartmann N, Baeriswyl L, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014;11:809-815. [DOI] [PubMed] [Google Scholar]
- 114. Hicks SD, Ignacio C, Gentile K, Middleton FA. Salivary miRNA profiles identify children with autism spectrum disorder, correlate with adaptive behavior, and implicate ASD candidate genes involved in neurodevelopment. BMC Pediatr. 2016;16:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hicks SD, Khurana N, Williams J, Greene CD, Uhlig R, Middleton FA. Diurnal oscillations in human salivary microRNA and microbial transcription: implications for human health and disease. PLoS ONE. 2018;13:e0198288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Walton EL. Saliva biomarkers in neurological disorders: a “spitting image” of brain health? Biomed J. 2018;41:59-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Farah R, Haraty H, Salame Z, Fares Y, Ojcius DM, Sadier NS. Salivary biomarkers for the diagnosis and monitoring of neurological diseases. Biomed J. 2018;41:63-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Sterling J. Tracking microRNAs to study brain dysfunction: quadrant biosciences believes human saliva holds key to better neuro disorder therapies. Gen Eng Biotech. 2018;38:6. [Google Scholar]
- 119. Slowey PD. Saliva collection devices and diagnostic platforms. In: Streckfus C. (ed.) Advances in Salivary Diagnostics. Berlin: Springer; 2015:33-61. [Google Scholar]
- 120. Park NJ, Yu T, Nabili V, et al. RNAprotect saliva: an optimal room-temperature stabilization reagent for the salivary transcriptome. Clin Chem. 2006;52:2303-2304. [DOI] [PubMed] [Google Scholar]
- 121. Hicks SD, Jacob P, Middleton FA, Perez O, Gagnon Z. Distance running alters peripheral microRNAs implicated in metabolism, fluid balance, and myosin regulation in a sex-specific manner. Physiol Genomics. 2018;50:658-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Uhlemann M, Möbius-Winkler S, Fikenzer S, et al. Circulating microRNA-126 increases after different forms of endurance exercise in healthy adults. Eur J Prevent Cardiol. 2014;21:484-491. [DOI] [PubMed] [Google Scholar]
- 123. Prien A, Grafe A, Rössler R, Junge A, Verhagen E. Epidemiology of head injuries focusing on concussions in team contact sports: a systematic review. Sports Med. 2018;48:953-969. [DOI] [PubMed] [Google Scholar]
- 124. Carl RL, Kinsella SB. Pediatricians’ knowledge of current sports concussion legislation and guidelines and comfort with sports concussion management: a cross-sectional study. Clin Pediatr. 2014;53:689-697. [DOI] [PubMed] [Google Scholar]
- 125. Zuckerbraun NS, Atabaki S, Collins MW, Thomas D, Gioia GA. Use of modified acute concussion evaluation tools in the emergency department. Pediatrics. 2014;133:635-642. [DOI] [PubMed] [Google Scholar]
- 126. Bazarian JJ, Atabaki S. Predicting postconcussion syndrome after minor traumatic brain injury. Acad Emerg Med. 2001;8:788-795. [DOI] [PubMed] [Google Scholar]
- 127. De Kruijk J, Leffers P, Menheere P, Meerhoff S, Rutten J, Twijnstra A. Prediction of post-traumatic complaints after mild traumatic brain injury: early symptoms and biochemical markers. J Neurol Neurosurg Psychiatry. 2002;73:727-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Baker JG, Freitas MS, Leddy JJ, Kozlowski KF, Willer BS. Return to full functioning after graded exercise assessment and progressive exercise treatment of postconcussion syndrome. Rehabil Res Pract. 2012;2012:705309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Leddy JJ, Willer B. Use of graded exercise testing in concussion and return-to-activity management. Curr Sports Med Rep. 2013;12:370-376. [DOI] [PubMed] [Google Scholar]
- 130. Miao W, Bao T, Han J, et al. Voluntary exercise prior to traumatic brain injury alters miRNA expression in the injured mouse cerebral cortex. Braz J Med Biol Res. 2015;48:433-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Hu T, Zhou F-J, Chang Y-F, et al. miR21 is associated with the cognitive improvement following voluntary running wheel exercise in TBI mice. J Mol Neurosci. 2015;57:114-122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplemental_Figure_1A for A Review of MicroRNA Biomarkers in Traumatic Brain Injury by Hamna Atif and Steven D Hicks in Journal of Experimental Neuroscience
Supplemental material, Supplemental_Figure_1B for A Review of MicroRNA Biomarkers in Traumatic Brain Injury by Hamna Atif and Steven D Hicks in Journal of Experimental Neuroscience
Supplemental material, Supplemental_Figure_1C for A Review of MicroRNA Biomarkers in Traumatic Brain Injury by Hamna Atif and Steven D Hicks in Journal of Experimental Neuroscience