Abstract
Background
The hemolysin (Hly) secretion system of E. coli allows the one-step translocation of hemolysin A (HlyA) from the bacterial cytoplasm to the extracellular medium, without a periplasmic intermediate. In this work, we investigate whether the Hly secretion system of E. coli is competent to secrete a repertoire of functional single-domain VHH antibodies (nanobodies, Nbs), facilitating direct screening of VHH libraries and the purification of selected Nb from the extracellular medium.
Results
We employed a phagemid library of VHHs obtained by immunization of a dromedary with three protein antigens from enterohemorrhagic E. coli (EHEC), namely, the extracellular secreted protein A (EspA), the extracellular C-terminal region of Intimin (Int280), and the translocated intimin receptor middle domain (TirM). VHH clones binding each antigen were enriched and amplified by biopanning, and subsequently fused to the C-terminal secretion signal of HlyA to be expressed and secreted in a E. coli strain carrying the Hly export machinery (HlyB, HlyD and TolC). Individual E. coli clones were grown and induced in 96-well microtiter plates, and the supernatants of the producing cultures directly used in ELISA for detection of Nbs binding EspA, Int280 and TirM. A set of Nb sequences specifically binding each of these antigens were identified, indicating that the Hly system is able to secrete a diversity of functional Nbs. We performed thiol alkylation assays demonstrating that Nbs are correctly oxidized upon secretion, forming disulphide bonds between cysteine pairs despite the absence of a periplasmic intermediate. In addition, we show that the secreted Nb-HlyA fusions can be directly purified from the supernatant of E. coli cultures, avoiding cell lysis and in a single affinity chromatography step.
Conclusions
Our data demonstrate the Hly secretion system of E. coli can be used as an expression platform for screening and purification of Nb binders from VHH repertories.
Keywords: E. coli/hemolysin, Nanobodies, Protein secretion, Single-domain antibodies
Background
Secretion of proteins to the extracellular medium reduces toxicity for the producing cell and eliminates the need of cell lysis for downstream processes [1, 2]. The hemolysin A (HlyA; 110 kDa) and the protein components required for its secretion were originally isolated from uropathogenic E. coli (UPEC) strains [3–6] and it serves as a paradigm of the bacterial Type I Secretion Systems (T1SS) [7, 8]. The Hly export machinery is composed of only three polypeptides, namely the inner membrane (IM) proteins HlyB and HlyD, and the outer membrane (OM) protein TolC. HlyB and HlyD are encoded, along with HlyA, in the Hly operon found in plasmids or the chromosome of UPEC strains, whereas TolC is encoded in a different location of the chromosome in most E. coli strains [4, 6, 9]. HlyB is a dimeric ABC-transporter anchored to the IM that has a cytosolic domain with ATPase activity [10, 11], while HlyD is a member of trimeric adaptor proteins [12] with a larger portion spanning much of the periplasm [13]. TolC forms homotrimeric OM channel comprising a β-barrel pore with long α-helical region that extends 10 nm toward the periplasm, forming a cylinder that opens to the extracellular medium, but is closed in the periplasmic entrance [14]. Upon engagement of HlyA in the cytosol by HlyB/D, TolC is recruited by this complex and its periplasmic entrance is opened to assemble a continuous channel connecting the IM and OM of E. coli [15–17] through which the HlyA polypeptide is secreted in a one-step mechanism, from the cytosol to the extracellular medium without a periplasmic intermediate (Fig. 1a) [7, 8]. TolC is a multifunctional protein that also participates in the secretion of other toxins and small molecules associated to different IM protein complexes [18].
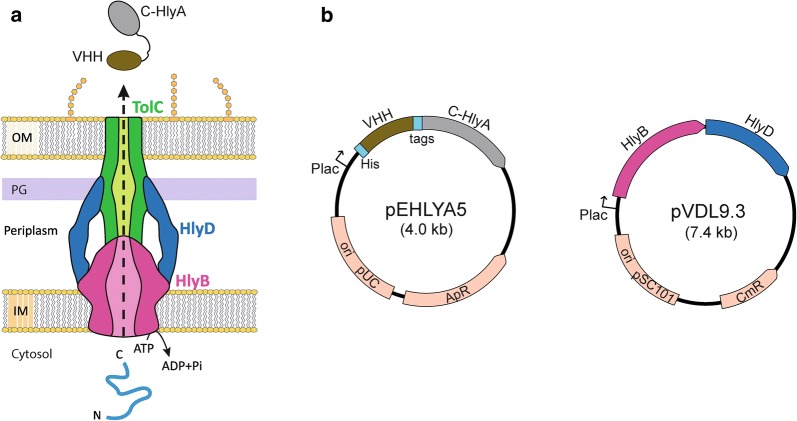
Fig. 1.
The E. coli hemolysin system for secretion of nanobodies. a Schematic representation of the HlyB, HlyD and TolC components of the Hly secretion system that spans the inner membrane (IM), the periplasmic space with the peptidoglycan (PG) layer, and outer membrane (OM) of E. coli. TolC is a trimeric OM protein with a large periplasmic domain; HlyB is an ATPase and forms a dimer in the IM. HlyD is a trimeric adaptor IM protein that interacts with HlyB and with the periplasmic domain of TolC. For simplicity, a longitudinal section of the Hly-protein complex is represented showing only two subunits of HlyD and a continuous open channel for protein export. The HlyBD complex recognises the C-terminal domain of HlyA (C-HlyA) in the bacterial cytosol to export the fusion protein with the nanobody (Nb) VHH domain to the extracellular medium. b Plasmids pEHLYA5 and pVDL9.3 used in this work for the secretion of Nb-HlyA fusions on E. coli bacteria (TolC+). Plasmid pEHLYA5 is used to generate fusion of the VHH sequence with an N-terminal His-tag and C-HlyA secretion signal. The linker region between the VHH and C-HlyA sequences includes tags for immunodetection (HA-tag, E-tag) and a human rhinovirus 3C protease recognition site. Plasmid pVDL9.3 encodes HlyB and HlyD components. Expression of the VHH-HlyA, HlyB and HlyD is controlled under the Plac promoter in both plasmids
Contrary to classical N-terminal signal peptides of proteins secreted to the periplasm by the Sec pathway [19], the signal for secretion of HlyA is located in the C-terminal end and is not removed during transport [20, 21]. The C-terminal location of HlyA signal also implies a post-translational mechanism of secretion. The last 50 amino acids of C-terminal end of HlyA appear to contain the signal recognized by HlyB/D complex [22, 23], but heterologous proteins have been secreted with this system fused to larger C-terminal fragments, comprising the last 60 amino acids [20, 24] and, more frequently, the last 218 residues (C-HlyA; 23 kDa), which includes three glycine- and aspartic-rich repeats, named as repeats in toxins (RTX) [7, 21]. Different heterologous proteins have been secreted in a functional form with C-HlyA, including the maltose binding protein [25], alkaline phosphatase [26, 27], β-lactamase [28], mammalian fatty acid binding protein [29], streptokinase [30], a single-chain Fv antibody [31], and a nanobody [32].
Nanobodies (Nbs) are recombinant single domain antibodies derived from the variable VHH domains from heavy chain-only antibodies (HCAbs) found in camelids (e.g., dromedaries, llamas, alpacas, etc.) [33, 34]. The VHHs have acquired important adaptations to be soluble and functional in the absence of the light chain, having longer and more diverse complementarity-determining regions (CDRs), strict monomeric behaviour, reversible folding properties, and higher resistance to proteolysis and thermal denaturation than VH domains from conventional antibodies [34]. These intrinsic biophysical properties facilitate their expression in bacteria, yeast, and mammalian hosts [35–38]. Their small size (ca. 14 kDa; 2–4 nm diameter) and long CDRs also allow binding of less accessible epitopes than those recognized by conventional antibodies, including active sites of enzymes [39, 40] and inner regions in the surface proteins of pathogens [41]. VHHs are also highly similar to human VH3 sequences, which is important to have a low immunogenicity in therapeutic applications of Nbs [42, 43]. These characteristics have made Nbs very attractive molecules for multiple applications, including cell biology studies [44], protein crystallography [45], therapy and in vivo diagnosis of human diseases [46–49]. Hence, methodologies for Nb selection, characterization and production are of great interest.
Secretion of Nbs to the extracellular medium of E. coli cultures could simplify the screening of clones binding an antigen of interest and their purification for in vitro characterization. Leakiness of the bacterial OM after overexpression in the periplasm allows, in some cases, the detection of Nbs in culture supernatants of E. coli [50, 51]. However, this is due to a non-specific release of periplasmic and cellular proteins due to bacterial lysis, and not to an actual secretion mechanism. Large accumulation of Nbs in the periplasm also leads to an impaired growth of the producing bacteria. Contrary to this, the Hly system specifically secretes the heterologous protein to the extracellular media, with little effect on the growth of the producing bacteria, given that the secreted protein is not accumulated in the periplasm and the integrity of the OM is not compromised [31]. Considering that a Nb binding α-amylase had been secreted in an active form with Hly-secretion system of E. coli [32], we wondered whether the Hly-secretion system could secrete a large diversity of Nbs that can be found in an immune library. With this aim, we constructed and screened with the Hly-system an immune library of VHHs against three protein antigens from enterohemorrhagic E. coli (EHEC), namely the extracellular secreted protein A (EspA), the extracellular 280 amino acid C-terminal region of Intimin (Int280), and the translocated intimin receptor middle domain (TirM) [52–54]. Our data show that screening of E. coli culture supernatants after secretion of Nb-HlyA fusions allows the identification of a diverse set of Nbs binding these antigens. We also demonstrate that the secreted Nb-HlyA fusions present disulphide bonds, indicating a correct oxidation state of the cysteine pairs. Lastly, secreted Nb-HlyA fusions were directly purified in a single affinity chromatography step from the supernatants of small-scale E. coli cultures (200 ml), with yields ranging between ca. 200–800 μg of purified Nb-HlyA fusion (~ 0.3–1.3 mg/l per OD600).
Results and discussion
Screening of Nbs against EspA, Int280, and TirM antigens of EHEC using the Hly secretion system
His-tagged versions of EspA, Int280, and TirM antigens from EHEC were purified and used for the immunization of a dromedary (Camelus dromedarius) (Fig. 2a). The generation of camel antibodies against these antigens in the blood serum of the immunized animal was confirmed by ELISA (Fig. 2b). The VHH gene segments were amplified by RT-PCR from the mRNA of peripheral blood lymphocytes (~ 2 × 107) and cloned in a phagemid vector (pCANTAB 5Ehis) [31]. Upon transformation into E. coli strain TG1, a VHH phagemid library was constructed with a diversity of ≥ 1 × 107 clones. Phage antibody (Phab) particles displaying the Nbs were produced and subjected to two cycles of biopanning in immunoplates coated with EspA, Int280, and TirM antigens (“Methods”). Phabs bound to each of the antigens were recovered, amplified in E. coli TG1 bacteria, and pool of phagemid DNA isolated for each repertoire.
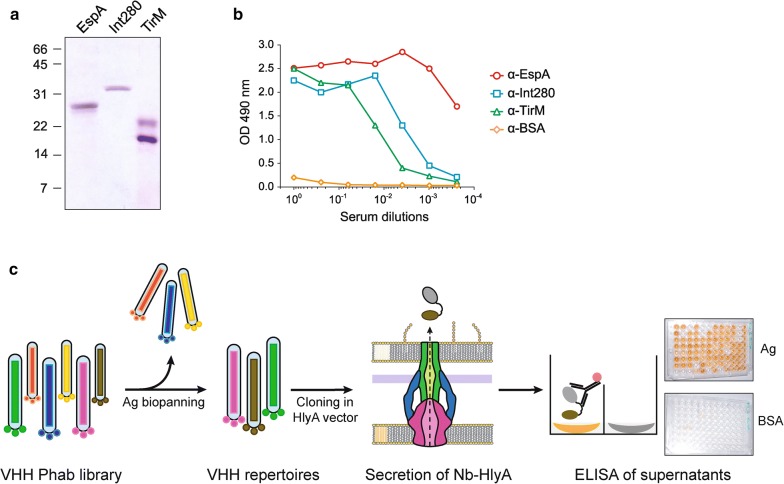
Fig. 2.
Identification of Nb binders secreted with the Hly-system from a VHH immune library against EHEC antigens. a Coomassie staining of purified EHEC protein antigens EspA, Int280 and TirM used for camel immunization. b ELISA of camel serum after immunization to reveal antibody response against EspA, Int280, TirM and BSA (negative control). The camel antibody response against each of the proteins using the indicated serum dilutions was developed with protein-A peroxidase (POD). c The VHH sequences amplified from the immunized animal were used to generate a phage antibody (Phab) library. Phab binders were enriched by panning to obtain VHH repertoires against each antigen (Ag). The VHH repertoires were cloned into pEHLYA5 and the Nb-HlyA fusions were secreted in E. coli bacteria carrying pVDL9.3. The culture supernatants of individual clones from each VHH repertoire were tested by ELISA against their corresponding antigen (EspA, Int280, TirM) and BSA (negative control). Bound Nb-HlyA fusions were developed with anti-E-tag-mAb
To evaluate the capacity of the Hly system to secrete a diversity of functional Nbs, the three repertoires of VHHs were excised from phagemid DNA and fused to the C-HlyA signal in the vector pEHLYA5 (Fig. 1b). This multicopy plasmid (pUC18-derivative; ApR) carries an IPTG-inducible Plac promoter, an N-terminal His-tag, unique SfiI and NotI restriction sites flanking the VHH against α-amylase (Vamy) [32], a linker region having epitopes for immunodetection (HA- and E-tag) and the C-HlyA signal. The VHH repertoires were cloned replacing the Vamy sequence in pEHLYA5 and transformed into E. coli strain HB2151 (TolC+) harbouring pVDL9.3, which expresses HlyB and HylD under the control of Plac (Fig. 1b) [55]. 96 individual transformants from each repertoire were picked, inoculated in liquid media (96-well plates) and induced with IPTG. After centrifugation to remove bacteria, cultures supernatants were used directly in ELISA to determine binding to their respective antigen (EspA, Int280, or TirM) and to BSA, as a negative control antigen. To detect bound Nb-HlyA fusions, the ELISA plates were incubated with anti-E-tag mouse monoclonal antibody (mAb) and secondary anti-mouse IgG conjugated to peroxidase (POD). A scheme of the screening method is shown in Fig. 2c. We found that culture supernatants of ca. 30–70% of the clones, depending on the antigen, showed a positive ELISA signal against their corresponding antigen (OD490 ≥ 0.3), whereas their binding to BSA had signals ≤ 0.10. These data suggested the secretion of functional Nb-HlyA fusions against the three antigens with variable affinity and/or secretion levels. Between 24 and 40 positive clones for each antigen, showing diverse signals in the ELISA ranging between OD490 values 0.3 and 2.5 units, were chosen for plasmid preparation. The sequence of the cloned VHHs was determined and aligned for comparison, which confirmed the isolation of different Nb sequences with distinct CDRs for each antigen (Fig. 3). These clones were named after their recognized antigen (I for Int280, T for TirM and E for EspA). We found four different Nbs against Int280 (IA1, IB7, IB10, IC1), six Nbs against TirM (TD4, TE1, TE4, TF1, TF2, TG10) and three Nbs against EspA (EC1, EC7, EE6). Table 1 summarizes the identified Nbs, their frequency of isolation, and their CDR3 sequence.
Fig. 3.
Amino acid sequence of VHHs clones identified after Hly-screening. Alignment of the amino acid sequences of the VHH domain in the clones selected by Hly-screening. The CDRs are labelled in colours: CDR1 (red), CDR2 (blue) and CDR3 (green). Cysteine residues are highlighted in yellow
Table 1.
Nanobodies identified against EHEC antigens by Hly screening
| Antigen | Representative clone | Frequency | CDR3 sequence |
|---|---|---|---|
| Int280 | IA1 | 14/27 | GIYYSVFSVCAGRIEH |
| IB7 | 6/27 | DSGPYCLDCGYCDRYNY | |
| IC1 | 6/27 | RQGVTSWLRDTEYSY | |
| IB10 | 1/27 | ELGAGSGRCYGYHY | |
| TirM | TF2 | 10/24 | PKYGGTWRWRVEEEKTTI |
| TD4 | 4/24 | SAGHTIRTVTSCPKYGINY | |
| TF1 | 4/24 | PDLSTNCDTVLTNSGALYNY | |
| TG10 | 4/24 | GTAPYWHTPIPTLSEDKYFY | |
| TE1 | 1/24 | DRCHSSTQVAGFGTNPRGRYGYAY | |
| TE4 | 1/24 | DRRVHFCKAPLSTSGHDT | |
| EspA | EC7 | 33/40 | ATDSYLCNPSRGGYNV |
| EC1 | 6/40 | GGGRLGWGAMASFAY | |
| EE6 | 1/40 | GNQYSDGGCRYSGTRGYNN |
Next, we compared the secretion levels and binding activity of representative clones from each group. E. coli bacteria carrying the empty vector (pUC18) were used as a negative control. Nb-HlyA fusions with the expected size (ca. 40–45 kDa) were detected by Western blot in the culture supernatant of all representative clones (Fig. 4a). The levels of secreted Nb-HlyA fusions were similarly high for 8 out of 14 clones, whereas the remaining 6 showed lower secretion levels. Binding of these clones to their respective antigen and BSA was compared by ELISA using serial dilutions of the culture supernatants. Results from these experiments are shown in Fig. 4b, representing the OD490 values obtained for the dilutions of the culture supernatant of each clone against the specific antigen, subtracting background values to BSA. From these data, clones IB10, TD4, and EC7 clearly showed the highest binding activities for their respective antigens, with positive ELISA signals at dilutions ≥ 10−2 (Fig. 4b). These results demonstrate that the Hly-system allows the secretion of different Nb sequences against all antigens tested, with variable expression levels and binding activities, as can be expected in the screening of VHH libraries. Hence, the Hly-system has the capacity to secrete a diversity of VHHs in a functional form, with secretion levels in the culture supernants high enough to allow the detection of positive Nb binders with different affinities for the antigen.
Fig. 4.
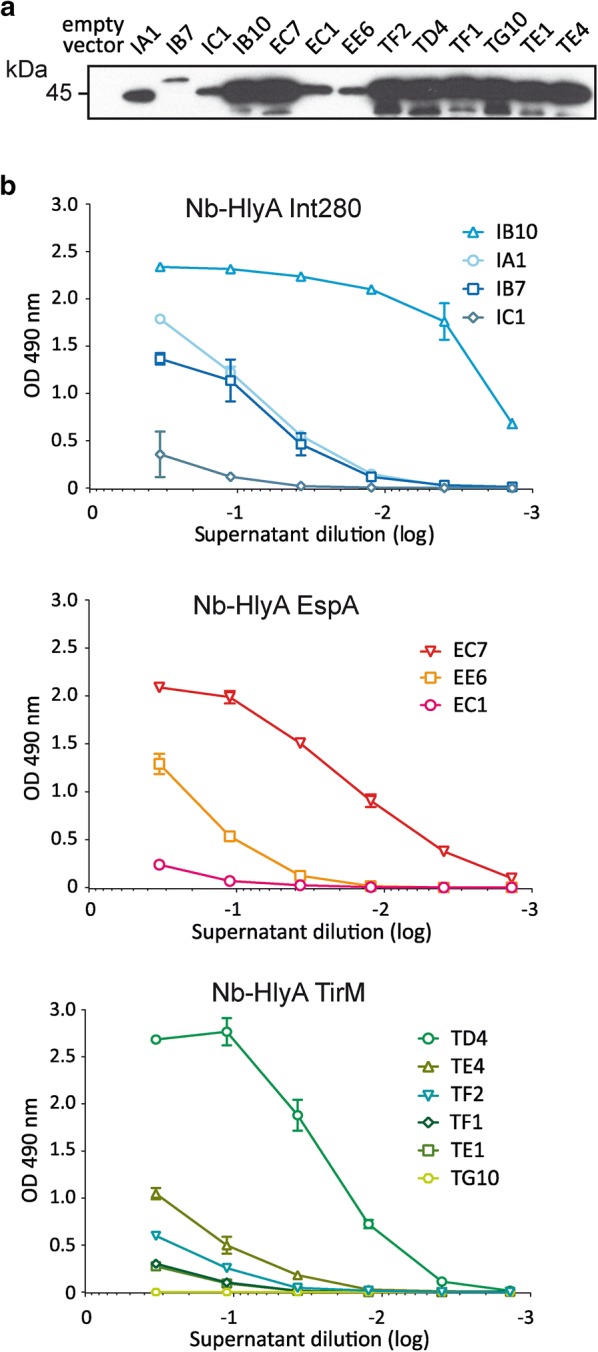
Secretion of Nb-HlyA fusions and antigen binding of representative clones. Representative clones of the identified VHH sequences against Int280, EspA and TirM were secreted as Nb-HlyA fusions. a Western blot developed with anti-E-tag-mAb of supernatants from induced cultures of the indicated clones showing a major protein band of ca. 42–45 kDa corresponding to Nb-HlyA fusions. The image shown is overexposed to visualize clearly protein bands of clones with lower expression levels. b ELISA of culture supernatants containing secreted Nb-HlyA fusions of the indicated clones against Int280 (top), EspA (middle) and TirM (bottom) developed with anti-E-tag mAb-POD. The represented OD490 values are the average from triplicate culture supernatants of each clone. Background signals to BSA are subtracted from the values obtained against the specific antigen (Int280, EspA, TirM)
Disulphide bonds in the secreted Nb-HlyA fusions
In E. coli, the cytosol is a strongly reducing environment, and cytoplasmic proteins usually do not form stable disulphide bonds [56, 57]. In contrast, cysteines in extracellular and periplasmic proteins secreted by the Sec-pathway are frequently oxidized, forming disulphide bonds catalysed by specialized Dsb-enzymes located, or having their catalytic sites, in the periplasm of E. coli [58, 59]. Proteins secreted by the Hly-system do not have access to the periplasm but, despite this, disulphide bonds were formed in a secreted scFv-HlyA fusion independently of Dsb-enzymes [60]. In contrast, when the same scFv was secreted fused to an autotransporter β-domain, which uses a periplasmic-dependent pathway, the presence of disulphide bonds required Dsb-enzymes (e.g. DsbA), which were not formed spontaneously in the extracellular medium [60].
Camelid VHHs frequently have, in addition to the conserved cysteine pair found in most immunoglobulin (Ig) domains, extra cysteine residues in the CDRs that also form disulphide bonds [61]. Out of the Nb sequences selected in the Hly-screening (Fig. 3), three clones have the two cysteines of the canonical disulphide bond of Ig-domains (IC1, TD4, EC1), whereas the rest contain one extra pair of cysteines (IA1, IB10, TF2, TG10, TE4, TF1, TE1, EE6, EE7) or two (IB7). We chose two clones having either two (TD4) or four (TF1) cysteines and investigated the oxidation state of the Nb-HlyA fusions, secreted and in the producing bacteria, by incubation with the thiol alkylating reagent 4-acetamido-4′-maleimidylstilbene-2,2′-disulphonic acid (AMS). AMS has a molecular mass of ca. 500 Da and its covalent binding to free thiol groups increase the protein mass and induce a retardation of its mobility in non-reducing SDS-PAGE. In contrast, proteins having disulphide bonds do not react with AMS and show a faster electrophoretic mobility.
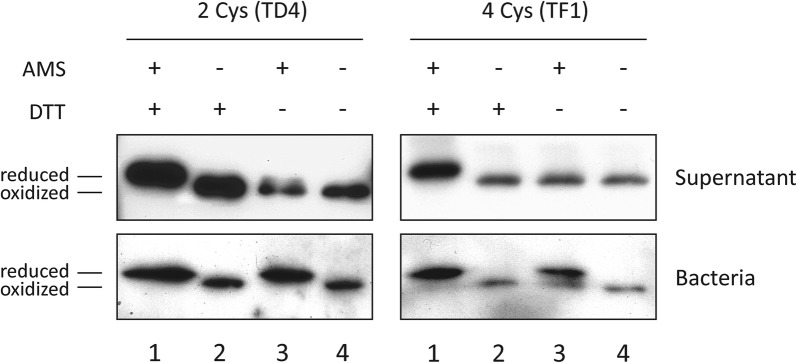
Bacterial cultures of TD4 and TF1 clones were induced and their culture supernatants and bacteria were collected. These samples were either directly precipitated with trichloroacetic acid (TCA), or previously incubated with dithiothreitol (DTT), to obtain positive controls of reduced samples before TCA-precipitation. Precipitated samples were resuspended in buffer having AMS, or lacking this alkylating reagent to obtain negative controls of alkylation (see “Methods”). All samples were subsequently subjected to non-reducing SDS-PAGE and Western blot. As expected, protein bands corresponding to Nb-HlyA fusions migrated differently depending on the treatment (Fig. 5). Secreted Nb-HlyA fusions were retarded in samples treated with DTT and AMS (reduced and alkylated controls; Fig. 5, lanes 1) and showed a faster mobility in untreated samples (Fig. 5, lanes 4), which were indicative of the mobilities of these Nb-HlyA fusions in their fully reduced and oxidized forms, respectively. Importantly, both Nb-HlyA fusions were found to be oxidized in the extracellular media treated with AMS (Fig. 5, lanes 3, Supernatants) and were reduced in the bacterial samples treated with AMS (Fig. 5, lanes 3, Bacteria). Non-alkylated samples had no migration drift. Hence, these data showed that thiol groups of Nb-HlyA fusions having 2 or 4 cysteine pairs, which are the most common among VHHs, have disulphide bonds in the secreted fusions but are reduced in the bacterial cytoplasm prior to their secretion. These results agree with those previously reporting the formation of disulphide bonds in a secreted scFv-HlyA fusion [60].
Fig. 5.
Disulphide bond formation in Nb-HlyA fusions. Thiol alkylation assay to determine the oxidation state of Cysteines (Cys) in secreted and cytoplasmic Nb-HlyA fusions from TD4 and TF1 clones, containing 2 and 4 Cys residues, respectively. Secreted proteins in culture supernatants and bacterial samples are treated (+) or not (−) with AMS (alkylating agent) and DTT (reducing agent), as indicated. Retarded mobility of alkylated polypeptides is indicative of reduced thiol groups. Protein bands of Nb-HlyA fusions developed after non-reducing SDS-PAGE and Western blot with anti-E-tag mAb
Purification of Nb-HlyA fusions from culture supernatants
To test the purification of secreted Nb-HlyA fusions, we chose the clones IB10, EC7 and TD4, which showed the higher binding signals against each antigen in culture supernatants (Fig. 3). E. coli HB2151(pVDL9.3) bacteria with the corresponding pEHLYA5-derivative were grown in 200 ml LB liquid cultures in shake flasks and induced with IPTG (see “Methods”). The culture supernatants were collected after centrifugation, equilibrated to PBS 1X and directly passed through a Cobalt-containing agarose resin for immobilized metal affinity chromatography (IMAC). The secreted Nb-HlyA fusions bound to the IMAC column were eluted by addition of imidazole. SDS-PAGE and Coomassie staining of the purified protein samples revealed major protein bands with the expected size of the Nb-HlyA fusions (ca. 42 kDa; Fig. 6a). Minor protein bands of smaller size are also visible, which likely correspond to proteolytic fragments of the full-length Nb-HlyA detected by Western blot (Fig. 4a). We quantified the amount of Nb-HlyA fusions obtained for each clone, which ranged between ca. 200–800 μg from these 200 ml cultures, indicating a yield equivalent to 0.3–1.3 mg/l per OD600.
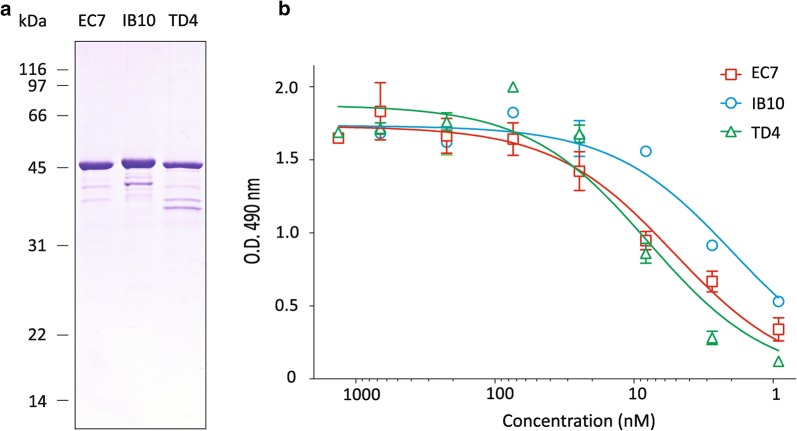
Fig. 6.
Purification and antigen binding activities of secreted Nb-HlyA fusions. a Coomassie staining after SDS-PAGE of purified His-tagged Nb-HlyA fusions from induced culture supernatants of EC7, IB10 and TD4 clones produced in E. coli HB2151(pVDL9.3). b Antigen binding curves determined by ELISA of purified Nb-HlyA fusions from EC7, IB10 and TD4 clones at the indicated concentrations. Bound Nb-HlyA proteins were developed with anti-E-tag mAb. OD490 values indicated are obtained against the specific antigen (EspA for EC7, Int280 for IB10, TirM for TD4) after subtraction of OD490 values against BSA (control antigen). Data are the average of triplicate ELISA experiments
Lastly, we confirmed the binding of the purified Nb-HlyA fusions for the corresponding antigen (i.e., Int280, TirM, EspA) by ELISA, using BSA as a negative control antigen. ELISA were developed with anti-E-tag mAb-POD and OD490 values to BSA (ca. ≤ 0.1) were subtracted from those obtained with the specific antigen at the different concentrations of the Nb-HlyA fusions (from 1 to 1000 nM; Fig. 6b). These data confirmed the activity of the purified Nb-HlyA fusions with curves indicating binding at low concentrations (1–10 nM). Hence, a single IMAC step allowed the purification of active Nb-HlyA fusions secreted in the culture supernatants of E. coli.
Conclusions
The Hly-system had been used previously for the secretion of a model Nb binding α-amylase [32], but its potential to secrete repertoires of Nbs had not been tested. This work has shown that VHH repertoires can be secreted as Nb-HlyA fusions with the Hly-system of E. coli. Culture supernatants can be used directly in ELISA to identify antigen-specific binders among the secreted Nb-HlyA fusions, as we did for an immune library of VHHs against Int280, TirM and EspA antigens of EHEC. In addition, we have shown that active Nb-HlyA fusions can be purified from culture supernatants using a single IMAC step, avoiding bacterial cell lysis. Secretion facilitates purification, reduces toxicity for the producing bacteria and minimizes the action of intracellular proteases [1, 2]. Finally, we have demonstrated that the cysteine pairs found in VHHs form disulphide bonds in the secreted Nb-HlyA fusions. In contrast, these cysteines are reduced in the cytoplasm of bacteria, indicating that oxidation of Nb-HlyA fusions occurs associated to protein secretion. Taken together, these data reveal the Hly-system as a suitable platform for the secretion of Nbs in screenings of VHH repertoires, which could be enriched by phage display or alternative yeast and E. coli display technologies [36]. The amount of Nb produced in this work using small-scale shake-flask cultures is sufficient for in vitro characterization of the antigen-binding properties of the selected Nb clones. Higher yields of production may be attained in fed-batch cultures of E. coli (in bioreactors) in which very high cell densities can be achieved (OD600 > 50) and by optimization of the expression of Hly-export machinery and Nb-HlyA in the producing E. coli strain [1, 62–64].
Methods
Conditions for bacterial growth
Escherichia coli strains used in this work are listed in Table 2. Bacteria were grown at 30 °C or 37 °C, as indicated, in LB-agar plates (1.5% w/v) or in liquid LB medium. Antibiotics for plasmid selection were added at the following concentrations: ampicillin (Ap) 150 µg/ml; chloramphenicol (Cm) 30 µg/ml; kanamycin (Km) 50 µg/ml. Cultures of E. coli strain HB2151 carrying pVDL9.3 (hlyB hlyD) and the indicated pEHLYA5-derivative were grown overnight (o/n) at 30 °C (170 rpm) in liquid LB + Ap + Cm. Next, bacteria were inoculated in fresh medium and grown at 37 °C (170 rpm) until OD600 reached 0.5. At this point, bacteria were induced with 1 mM isopropyl-1-thio-β-d-galactoside (IPTG) and further incubated for 6 h with shaking (100 rpm). Average final OD600 determined for these cultures were ~ 3.0 (± 0.25). For secretion on 96-well plates, 200 μl of liquid media per well were used for growth and induction, as above. Culture supernatants were obtained after centrifugation (400g, 10 min). For secretion in flasks (200 ml of liquid media in 1 l flask), cultures were centrifuged twice (10 min, 10,000g, 4 °C) and the supernatants were collected for IMAC purification. In all cases, culture supernatants were equilibrated to PBS 1× (by adding 1/10th volume of PBS 10×) before downstream use.
Table 2.
Bacterial strains and plasmids used in this work
| Name | Relevant characteristics | Source or reference |
|---|---|---|
| DH10B-T1R | (F− λ−) mcrA Δmrr-hsdRMS-mcrBC φ80lacZΔM15, ΔlacX74, recA1, endA1, Δ(ara, leu)7697 galU galK rpsL(StrR), nupG tonA | Novagen-Merck |
| HB2151 | Δlac-pro, ara, nalR, thi, F’(proAB lacIQ lacZΔM15) | [68] |
| TG1 | Δ(lac-proAB) Δ(mcrB-hsdSM)5 (rK− mK−) thi-1 supE [F´ traD36 proAB lacIqZΔM15] | Stratagene-ThermoFisher |
| BL21 (DE3) | F−; ompT hsdSB(rB−, mB−) gal dcm lon λ(DE3[lacI lacUV5-T7 gene1 ind1 sam7 nin5]) | Novagen-Merck |
| pCANTAB-5Ehis | ApR; phagemid vector for periplasmic production of Nb-pIII fusions with His and E-tag | [31] |
| pEHlyA5 | ApR; pUC ori, lac promoter, N-terminal His tag, VHH, HA and E-tags, C-HlyA | This work |
| pVDL9.3 | CmR; expression of HlyB and HlyD | [55] |
| pET28a | KmR; pBR ori, T7 promoter, N-terminal His-tagged fusions | Novagen-Merck |
| pET28a-TirMEHEC | pET28a derivative; expression of His-tagged TirM of EHEC | This work |
| pET28a-Int280EHEC | pET28a derivative; expression of His-tagged Int280 of EHEC | This work |
| pET28a-EspAEHEC | pET28a derivative; expression His-tagged EspA of EHEC | This work |
Plasmids, DNA constructs, and oligonucleotides
Plasmids used in this study are listed in Table 2. E. coli strain DH10B-T1R was used as host for the cloning and propagation of plasmids. PCRs were performed with the Taq DNA polymerase (Roche, NZyTech) for standard amplifications in screenings and with the proofreading DNA polymerase Herculase II Fusion (Agilent Technologies) for cloning purposes. Int280 (residues 660 to 939 of Intimin), EspA (full-lenght) and TirM (residues 252 to 360 of Tir) sequences of EHEC were amplified by PCR from genomic DNA of EHEC strain EDL933stx- [65] and cloned EcoRI–HindIII into the pET28a plasmid backbone under the T7 promoter and fused to a N-terminal His-tag for purification. Plasmid pEHLY5 was constructed using vector pEHLYA2SD [31], by insertion of a chemically synthesized (GeneArt-ThermoFisher) XbaI–BamHI DNA fragment of 566 bp encoding the His-tag-VHH(amylase)-HA-3Csite-E-tag region of pEHLYA5, which contains unique SfiI/NcoI and NotI sites flanking the VHH. All plasmid constructs were fully sequenced (Secugen, Madrid, Spain). Oligonucleotide used as primers (Sigma) are described in Table 3.
Table 3.
Oligonucleotides used in this study
| Name | Sequence (5′→3′) |
|---|---|
| CALL001 | GTCCTGGCTCTCTTCTACAAGG |
| CALL002 | GGTACGTGCTGTTGAACTGTTCC |
| VHH-SfiI2 | GTCCTCGCAACTGCGGCCCAGCCGGCCATGGCTCAGGTGCAGCTGGTGGA |
| VHH-NotI2 | GGACTAGTGCGGCCGCTGAGGAGACGGTGACCTGGGT |
| BamEcoTirM-EHEC | CGCGGATCCGAATTCCAGGCGCTTGCATTGACGCCGGAG |
| XhoHindInt280-EHEC | CCGCTCGAGAAGCTTTTACGATGAAACTTTCAGCTCCTCCTG |
| BamEcoInt280-EHEC | CGCGGATCCGAATTCATTACTGAGATTAAGGCTGATAAG |
| XhoHindInt280-EHEC | CCGCTCGAGAAGCTTTTATTCTACACAAACCGCATAGAC |
| BamEcoEspA-EHEC | CGCGGATCCGAATTCATGGATACATCAAATGCAACATCC |
| XhoHindEspA-EHEC | CCGCTCGAGAAGCTTTTATTTACCAAGGGATATTGCTGA |
Immunization and generation of the VHH library
Purified His-tagged Int280, TirM and EspA from EHEC were diluted in sterile water (2.5 ml) and mixed at 0.2 mg/ml to the same volume of adjuvant (Veterinary Vaccine Adjuvant, GERBU) reaching a total volume of 5 ml. This solution was injected subcutaneously to one male dromedary camel (Camelus dromedarius) corresponding to 0.5 mg of each antigen. After the initial immunization, four boosting immunizations were performed in the same manner once per week. Pre-immune serum was prepared from a small blood sample (5 ml) before the first injection. The immune serum was collected 7 days after the last immunization. Serial dilutions of pre-immune and immune sera were used in ELISA to confirm the immune response against the respective antigens with protein A-POD as secondary. Additional 50 ml of blood of the immunized animal were collected from the jugular vein in tubes containing EDTA as anticoagulant, using Venoject system. For lymphocyte isolation, 50 ml of RPMI-1640 medium (Sigma) were added to the 50 ml blood sample and the final mixture was divided in 4 aliquots of 25 ml. Each aliquot was added on top of a 25 ml of Ficoll-Paque Plus (StemCell Technologies) in sterile capped 50 ml Falcon tubes. After centrifugation (800g, 30 min, RT), lymphocytes were taken from the interphase, washed twice in RPMI-1640 by centrifugation (800g, 10 min), resuspended in 5 ml of RPMI-1640, and the number of cells determined in a Neubauer hematocytometer (Hausser Scientific). About 2 × 107 cells were lysed with 2 ml of Trizol (Invitrogen) for RNA extraction following manufacturer instructions. The poly-A+ mRNA was purified from total RNA using an oligo-dT resin (Oligotex mRNA Minikit, Qiagen) and directly employed as template for first-strand cDNA synthesis reactions with random hexamers and oligo-dT primers (iScript cDNA Synthesis, Bio-Rad). About 1 μl of each cDNA synthesis was used as template in 50 μl PCR reactions with oligonucleotides CALL001 and CALL002. The amplified fragments of ≈ 0.6 kb, corresponding to VHH-CH2 domains, and ≈ 0.9 kb, corresponding to conventional VH-CH1-CH2 domains, were separated in 1.2% (w/v) low melting agarose gel and the ~ 0.6 kb band was purified (Qiaex II Gel Extraction kit, Qiagen). This fragment was used as template in a second PCR reaction with oligonucleotides VHH-Sfi2 and VHH-Not2 (Table 3) to finally obtain the amplified fragments of ~ 0.4 kb, corresponding to the VHH domains. The amplified VHH fragments were cloned SfiI-NotI into the phagemid pCANTAB 5Ehis-backbone [66]. Ligations were electroporated in E. coli TG1 cells (Stratagene) and a library size of approx. 2 × 107 clones were determined by plating dilutions on LB + Ap agar plates with 2% w/v glucose at 30 °C.
Protein purification
Cultures of E. coli BL21(DE3) carrying the corresponding pET28a-derivative were grown at 30 °C in 500 ml of LB + Km and induced at OD600 ~ 0.5 with 1 mM IPTG during 2 h. Cells were harvested by centrifugation (10 min,10,000g, 4 °C), resuspended in 20 ml of Solution A—NaPO4 pH 7, 300 mM NaCl, DNase (0.1 mg/ml; Roche) and protease inhibitor cocktail (Roche)—, and lysed by passing three times through a French-Press at 1200 psi. The resultant lysate was ultracentrifuged (60 min, 40,000g, 4 °C) to obtain a cleared lysate supernatant. For purification of the His-tagged Int280EHEC, EspAEHEC, TirMEHEC, the lysates were passed through 2 ml of pre-equilibrated Cobalt-containing resin (Clontech) in a chromatography column and were washed with HEPES buffer (20 mM HEPES pH 7.4, 200 mM NaCl). The bound His-tagged proteins were eluted adding the same buffer complemented with 100 mM imidazole and were collected in 0.5 ml aliquots. For the purification of the secreted His-tagged Nb-HlyA fusions, supernatants from induced cultures were loaded at ca. 1 ml/min onto chromatography columns with pre-equilibrated Cobalt-containing resin (Clontech). Columns were washed with PBS and eluted with the same buffer containing 150 mM imidazole. Fractions eluted from metal-affinity chromatography were concentrated to a final ~ 1 ml in PBS using a 3 kDa centrifugal filter unit (Amicon Ultra-15, Millipore). Protein concentration was estimated using the Bicinchoninic acid protein assay kit (Thermo Scientific).
Packaging of Phabs into M13 particles
For preparation of phage antibodies (Phabs) a mixture of E. coli TG1 cells representing the library or a subpopulation after panning, was incubated in 25 ml of LB-Ap supplemented with 2% (w/v) glucose at 30 °C until OD600 ~ 0.2. At this point, 1010 plaque forming units (PFU) of VCS-M13 helper phage (KmR; Stratagene) were added for 1 h incubation at 37 °C with gentle agitation. Then, E. coli cells were harvested by centrifugation (5 min, 4000g, RT) and resuspended in 250 ml of fresh LB + Ap + Km. After 16 h of incubation at 30 °C, the cultures were chilled on ice and centrifuged (10 min, 8000g, 4 °C). To recover the M13-particles from the supernatant, 50 ml of PEG-NaCl solution (20% w/v polyethylene glycol 8000; 2.5 M NaCl) were added and the resulting mixture was kept on ice for additional 45 min. Phage pellets obtained after centrifugation (20 min, 10,000g, 4 °C) were resuspended in 2 ml of TE (10 mM Tris–HCl, 1.0 mM EDTA, pH 8.0) and stored at − 80 °C.
Library enrichment by Phab panning
All steps were performed at room temperature (RT). Each antigen (10 µg/ml, in PBS) was adsorbed for 2 h to 4 wells (50 µl/well) of a microtiter immunoplate (Maxisorb, Nunc). These solutions were discarded, and the wells were blocked by adding 200 µl/well of PBS with 3% w/v skimmed milk and 1% w/v BSA (Sigma). After 2 h, the blocking solution was replaced by a total of 2 × 1011 PFU of Phabs in 50 µl of PBS with 3% w/v skimmed milk. Phabs were allowed to bind for 1 h, and the unbound Phabs were removed from the plates by 20 washes of 1 min employing 200 µl/well of PBS with Tween20 0.05% and another 20 washes with PBS. To collect the bound Phabs, wells were incubated during 5 min with 0.1 M glycine pH 2.5 (50 µl/well). The solution from the wells was pooled together and immediately equilibrated by addition of one volume (400 µl) of 1 M Tris–HCl, pH 7.5. Phabs in this solution were later used to infect TG1, which were plated on LB + Ap agar plates. After 24 h incubation at 30 °C, colonies grown on these plates were harvested as a pool and used for phagemid packaging.
Selection of individual specific binders
For rapid screening of individual Phab binders, a small-scale rescue of phagemids was performed in 150 µl cultures of TG1 grown in 96-well microtitre plates using 109 PFU of helper-M13 phages. After production of the phagemids the plate was centrifuged (10 min, 600g, RT) and the supernatants (containing the Phabs) were used in ELISA to determine their specific binding to the antigen. At least 2 × 106 independent colonies from each enriched Phab library were harvested from agar plates, and 50 units of OD600 were used for plasmid isolation (NucleoBond Xtra Midi, Macherey–Nagel). The VHH fragments were cloned SfiI-NotI into the pEHLYA5 backbone vector under the Plac promoter. The size of each library was 2–3 × 106 clones, as determined by plating dilutions on LB + Ap agar plates with 2% (w/v) glucose incubated at 30 °C. Isolated plasmids from the pools were transformed in strain HB2151 harbouring plasmid pVDL9.3 and individual colonies were isolated in 96-well microtitre plates with 200 µl of liquid LB-media with appropriated antibiotics and 1 mM IPTG for the secretion of the Nb-HlyA fusions.
Alkylation of thiol groups in Nb-HlyA fusions
Protein samples from extracellular media and whole-cell bacteria were treated with the alkylating reagent 4-acetamido-4′-maleimidylstilbene-2,2′-disulphonic acid (AMS; ThermoFisher Scientific) as reported previously [60]. Briefly, for Nb-HlyA in supernatants, four aliquots of 800 µl from each induced culture supernatant were taken. Two of these aliquots were incubated with dithiothreitol (DTT, 25 mM) to obtain positive controls of reduced proteins. After incubation for 10 min at 37 °C, all four aliquots were precipitated with trichloroacetic acid (TCA, 10% w/v) for 1 h at 4 °C, and precipitated proteins were recovered by centrifugation (14,000g, 15 min). The protein pellets were washed twice with 1 ml of ice-cold acetone followed by centrifugation (14,000g, 15 min). Two of these protein pellets (AMS positive samples) were resuspended in 75 µl of a freshly prepared alkylation buffer (150 mM Tris HCl pH 7.5, 1% w/v SDS, 5% v/v glycerol, 15 mM AMS). The other two samples were resuspended in 75 µl of the same buffer without AMS. After 1 h incubation at 22 °C, 75 µl of non-reducing SDS-PAGE sample buffer was added (see below).
For intracelullar Nb-HlyA, bacteria from 1 ml of induced cultures were harvested, washed once, and resuspended in the same volume of LB. Samples were divided in four 200 µl aliquots and two of them were adjusted to 0.1 M DTT (DTT+ samples). Following 20 min incubation on ice, TCA was added (10% w/v). After 1 h incubation at 4 °C, the precipitated proteins collected by centrifugation (14,000g, 15 min, 4 °C). Pellets were washed once with 1 ml of ice-cold acetone, air-dried, and resuspended in 20 µl of alkylation buffer (AMS positive samples), or 20 µl of the same buffer without AMS. After incubation for 1 h at 22 °C, 10 µl of non-reducing SDS-PAGE sample buffer were added. All samples were boiled at 100 °C for 10 min before loading for SDS-PAGE (5–10 µl/well).
SDS-PAGE and Western blot
Sodium Dodecyl Sulfate–Polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot were performed following standard methods [67] using the Miniprotean III system (Bio-Rad). The sample buffer for reducing SDS-PAGE is 60 mM Tris–HCl pH 6.8, 1% w/v SDS, 5% v/v glycerol, 0.005% w/v bromophenol blue and 1% v/v 2-mercapto-ethanol (2-ME). In non-reducing SDS-PAGE, the sample buffer was devoid of 2-ME. Proteins separated by SDS-PAGE were either stained with Coomassie Blue R-250 (Bio-Rad) or subjected to Western blot. For the latter, the proteins were transferred to a polyvinylidene difluoride membrane (PVDF, Immobilon-P, Millipore) using semi-dry electrophoresis (Bio-Rad), as previously described [67]. Nb-HlyA fusions were detected with primary mAb anti-E-tag (Phadia, 1:5000) and secondary polyclonal rabbit anti-mouse IgG antibodies fused to POD (1:5000, Sigma). Membranes were developed using a mixture of 100 mM Tris–HCl (pH 8.0) containing 1.25 mM luminol (Sigma), 0.22 mM cumaric acid (Sigma), and 0.0075% (v/v) H2O2 (Sigma). The membranes were then developed by exposure to X-ray films (AGFA).
Enzyme-linked immunosorbent assays (ELISAs)
ELISA conditions were based on those described previously [57]. Briefly, 96-well immunoplates (Maxisorp, Nunc) were coated for 120 min at RT with purified antigens (as indicated) diluted in PBS at a concentration of 5 µg/ml. Bovine serum albumin (BSA, Roche) was used as a negative control for detection. Phabs, culture supernatants or purified Nb-HlyA fusions were added to the wells at the indicated dilutions. For detection of bound Phabs, an anti-M13-HRP mAb (GE Healthcare) was added (1:5000). For detection of Nb-HlyA fusions, anti-E-tag mAb (1:2000; Phadia) and anti-mouse IgG-POD (1:2000; Sigma), as secondary antibody, were added. The reaction was developed with o-phenylenediamine (OPD, Sigma) and H2O2 (Sigma) and the OD490 was determined using a microplate reader (iMark ELISA plate reader, Bio-Rad).
Authors’ contributions
DRG and SF performed the experiments; CG performed camel immunization and collect camel blood samples; DR, SF, CG and LAF designed the experiments and analysed data; LAF conceived this study and secured funding. DRG and LAF wrote the manuscript and prepared the Figures and Tables. All authors read and approved the final manuscript.
Acknowledgements
We thank Prof. Víctor de Lorenzo (CNB-CSIC) and Prof. Gad Frankel (Imperial College London) for helpful discussions and support. We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated and/or analysed during this study are included in this published article.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The camel immunization protocol followed animal experimentation guidelines published by the regional government of the Canary Islands (Spain) and was approved by the Ethics Committee, Veterinary Medicine Service, Las Palmas de Gran Canaria University Foundation (Ref.: 0014/2011).
Funding
Work in the lab of LAF is partially supported by the Grants BIO2017-89081-R (Agencia Española de Investigación AEI/MICIU/FEDER, EU), ERC-2012-ADG_20120314 (European Research Council, EU) and LCCM_P59474 (Wellcome Trust, UK).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- Ab
antibody
- ABC
ATP-binding cassette
- AMS
4-acetamido-4′-maleimidylstilbene-2,2′-disulphonic acid
- Amy
α-amylase
- Ap
ampicillin
- BSA
bovine serum albumin
- C-HlyA
C-terminal domain of 218 amino acids of HlyA
- CDR
complementary determining region
- Cm
chloramphenicol
- Cys
cysteine
- Dsb
disulphide bond
- DTT
dithiothreitol
- EHEC
enterohemorrhagic E. coli
- ELISA
enzyme-linked immunosorbent assay
- EspA
extracellular secreted protein A
- HCAb
heavy-chain only antibodies
- Hly
hemolysin
- HlyA
hemolysin A
- Ig
immunoglobulin
- IM
inner membrane
- IMAC
immobilized metal affinity chromatography
- Int280
extracellular 280 amino acid C-terminal region of Intimin
- IPTG
isopropyl-1-thio-β-galactoside
- Km
kanamycin
- mAb
monoclonal antibody
- Nb-HlyA
nanobody fused to C-HlyA
- Nb
nanobody
- o/n
overnight
- OD490
optical density at 490 nm
- OD600
optical density at 600 nm
- OM
outer membrane
- OMP
outer membrane protein
- OPD
o-phenylenediamine
- PBS
phosphate-buffered saline
- PFU
plaque forming units
- PG
peptidoglycan
- Phab
phage antibody
- POD
peroxidase
- PVDF
polyvinylidene difluoride membrane
- RT-PCR
reverse transcription polymerase chain reaction
- RT
room temperature
- RTX
repeats in toxins
- TCA
trichloroacetic acid
- TirM
translocated intimin receptor middle domain
- UPEC
uropathogenic E. coli
- VHH
variable heavy chain domain from heavy-chain only antibodies
Contributor Information
David Ruano-Gallego, Email: d.ruano-gallego@imperial.ac.uk.
Sofía Fraile, Email: sfraile@cnb.csic.es.
Carlos Gutierrez, Email: cgutierrez@dpat.ulpgc.es.
Luis Ángel Fernández, Phone: +34 91 585 48 54, Email: lafdez@cnb.csic.es.
References
- 1.Kleiner-Grote GRM, Risse JM, Friehs K. Secretion of recombinant proteins from E. coli. Eng Life Sci. 2018;18:532–550. doi: 10.1002/elsc.201700200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mergulhao FJ, Summers DK, Monteiro GA. Recombinant protein secretion in Escherichia coli. Biotechnol Adv. 2005;23:177–202. doi: 10.1016/j.biotechadv.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Welch RA, Dellinger EP, Minshew B, Falkow S. Haemolysin contributes to virulence of extra-intestinal E. coli infections. Nature. 1981;294:665–667. doi: 10.1038/294665a0. [DOI] [PubMed] [Google Scholar]
- 4.Mackman N, Nicaud JM, Gray L, Holland IB. Identification of polypeptides required for the export of haemolysin 2001 from E. coli. Mol Gen Genet. 1985;201:529–536. doi: 10.1007/BF00331351. [DOI] [PubMed] [Google Scholar]
- 5.Bhakdi S, Mackman N, Menestrina G, Gray L, Hugo F, Seeger W, Holland IB. The hemolysin of Escherichia coli. Eur J Epidemiol. 1988;4:135–143. doi: 10.1007/BF00144740. [DOI] [PubMed] [Google Scholar]
- 6.Wandersman C, Delepelaire P. TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc Natl Acad Sci USA. 1990;87:4776–4780. doi: 10.1073/pnas.87.12.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holland IB, Peherstorfer S, Kanonenberg K, Lenders M, Reimann S, Schmitt L. Type I protein secretion-deceptively simple yet with a wide range of mechanistic variability across the family. EcoSal Plus. 2016 doi: 10.1128/ecosalplus.ESP-0019-2015. [DOI] [PubMed] [Google Scholar]
- 8.Thomas S, Holland IB, Schmitt L. The type 1 secretion pathway—the hemolysin system and beyond. Biochim Biophys Acta. 2014;1843:1629–1641. doi: 10.1016/j.bbamcr.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 9.Nicaud JM, Mackman N, Gray L, Holland IB. The C-terminal, 23 kDa peptide of E. coli haemolysin 2001 contains all the information necessary for its secretion by the haemolysin (Hly) export machinery. FEBS. 2001;1986(204):331–335. doi: 10.1016/0014-5793(86)80838-9. [DOI] [PubMed] [Google Scholar]
- 10.Holland IB, Schmitt L, Young J. Type 1 protein secretion in bacteria, the ABC-transporter dependent pathway (review) Mol Membr Biol. 2005;22:29–39. doi: 10.1080/09687860500042013. [DOI] [PubMed] [Google Scholar]
- 11.Pimenta AL, Racher K, Jamieson L, Blight MA, Holland IB. Mutations in HlyD, part of the type 1 translocator for hemolysin secretion, affect the folding of the secreted toxin. J Bacteriol. 2005;187:7471–7480. doi: 10.1128/JB.187.21.7471-7480.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Symmons MF, Marshall RL, Bavro VN. Architecture and roles of periplasmic adaptor proteins in tripartite efflux assemblies. Front Microbiol. 2015;6:513. doi: 10.3389/fmicb.2015.00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim JS, Song S, Lee M, Lee S, Lee K, Ha NC. Crystal structure of a soluble fragment of the membrane fusion protein HlyD in a type I secretion system of gram-negative bacteria. Structure. 2016;24:477–485. doi: 10.1016/j.str.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 15.Thanabalu T, Koronakis E, Hughes C, Koronakis V. Substrate-induced assembly of a contiguous channel for protein export from E. coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 1998;17:6487–6496. doi: 10.1093/emboj/17.22.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen C, Hughes C, Koronakis V. Chunnel vision. Export and efflux through bacterial channel-tunnels. EMBO Rep. 2000;1:313–318. doi: 10.1093/embo-reports/kvd075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balakrishnan L, Hughes C, Koronakis V. Substrate-triggered recruitment of the TolC channel-tunnel during type I export of hemolysin by Escherichia coli. J Mol Biol. 2001;313:501–510. doi: 10.1006/jmbi.2001.5038. [DOI] [PubMed] [Google Scholar]
- 18.Koronakis V. TolC—the bacterial exit duct for proteins and drugs. FEBS Lett. 2003;555:66–71. doi: 10.1016/S0014-5793(03)01125-6. [DOI] [PubMed] [Google Scholar]
- 19.Tsirigotaki A, De Geyter J, Sostaric N, Economou A, Karamanou S. Protein export through the bacterial Sec pathway. Nat Rev Microbiol. 2017;15:21–36. doi: 10.1038/nrmicro.2016.161. [DOI] [PubMed] [Google Scholar]
- 20.Hess J, Gentschev I, Goebel W, Jarchau T. Analysis of the haemolysin secretion system by PhoA-HlyA fusion proteins. Mol Gen Genet. 1990;224:201–208. doi: 10.1007/BF00271553. [DOI] [PubMed] [Google Scholar]
- 21.Mackman N, Baker K, Gray L, Haigh R, Nicaud JM, Holland IB. Release of a chimeric protein into the medium from Escherichia coli using the C-terminal secretion signal of haemolysin. EMBO J. 1987;6:2835–2841. doi: 10.1002/j.1460-2075.1987.tb02580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chervaux C, Holland IB. Random and directed mutagenesis to elucidate the functional importance of helix II and F-989 in the C-terminal secretion signal of Escherichia coli hemolysin. J Bacteriol. 1996;178:1232–1236. doi: 10.1128/jb.178.4.1232-1236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koronakis V, Koronakis E, Hughes C. Isolation and analysis of the C-terminal signal directing export of Escherichia coli hemolysin protein across both bacterial membranes. EMBO J. 1989;8:595–605. doi: 10.1002/j.1460-2075.1989.tb03414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarchau T, Chakraborty T, Garcia F, Goebel W. Selection for transport competence of C-terminal polypeptides derived from Escherichia coli hemolysin: the shortest peptide capable of autonomous HlyB/HlyD-dependent secretion comprises the C-terminal 62 amino acids of HlyA. Mol Gen Genet. 1994;245:53–60. doi: 10.1007/BF00279750. [DOI] [PubMed] [Google Scholar]
- 25.Bakkes PJ, Jenewein S, Smits SH, Holland IB, Schmitt L. The rate of folding dictates substrate secretion by the Escherichia coli hemolysin type 1 secretion system. J Biol Chem. 2010;285:40573–40580. doi: 10.1074/jbc.M110.173658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Angkawidjaja C, Kuwahara K, Omori K, Koga Y, Takano K, Kanaya S. Extracellular secretion of Escherichia coli alkaline phosphatase with a C-terminal tag by type I secretion system: purification and biochemical characterization. Protein Eng Des Sel. 2006;19:337–343. doi: 10.1093/protein/gzl017. [DOI] [PubMed] [Google Scholar]
- 27.Erb K, Vogel M, Wagner W, Goebel W. Alkaline phosphatase which lacks its own signal sequence becomes enzymatically active when fused to N-terminal sequences of Escherichia coli haemolysin (HlyA) Mol Gen Genet. 1987;208:88–93. doi: 10.1007/BF00330427. [DOI] [PubMed] [Google Scholar]
- 28.Chervaux C, Sauvonnet N, Clainche AL, Kenny B, Hunt AL, Broome-Smith JK, Holland IB. Secretion of active b-lactamase to the medium mediated by the Escherichia coli haemolysin transport pathway. Mol Gen Genet. 1995;249:237–245. doi: 10.1007/BF00290371. [DOI] [PubMed] [Google Scholar]
- 29.Schwarz CKW, Landsberg CD, Lenders MHH, Smits SHJ, Schmitt L. Using an E. coli Type 1 secretion system to secrete the mammalian, intracellular protein IFABP in its active form. J Biotechnol. 2012;159:155–161. doi: 10.1016/j.jbiotec.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Kern I, Ceglowski P. Secretion of streptokinase fusion proteins from Escherichia coli cells through the hemolysin transporter. Gene. 1995;163:53–57. doi: 10.1016/0378-1119(95)00395-M. [DOI] [PubMed] [Google Scholar]
- 31.Fernández LA, Sola I, Enjuanes L, de Lorenzo V. Specific secretion of active single-chain Fv antibodies into the supernatants of Escherichia coli cultures by use of the hemolysin system. Appl Environ Microbiol. 2000;66:5024–5029. doi: 10.1128/AEM.66.11.5024-5029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fraile S, Muñoz A, de Lorenzo V, Fernández LA. Secretion of proteins with dimerization capacity by the haemolysin type I transport system of Escherichia coli. Mol Microbiol. 2004;53:1109–1121. doi: 10.1111/j.1365-2958.2004.04205.x. [DOI] [PubMed] [Google Scholar]
- 33.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 34.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 35.Fleetwood F, Devoogdt N, Pellis M, Wernery U, Muyldermans S, Ståhl S, Löfblom J. Surface display of a single-domain antibody library on gram-positive bacteria. Cell Mol Life Sci. 2013;70:1081–1093. doi: 10.1007/s00018-012-1179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salema V, Fernandez LA. Escherichia coli surface display for the selection of nanobodies. Microb Biotechnol. 2017;10:1468–1484. doi: 10.1111/1751-7915.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMahon C, Baier AS, Pascolutti R, Wegrecki M, Zheng S, Ong JX, Erlandson SC, Hilger D, Rasmussen SGF, Ring AM, et al. Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat Struct Mol Biol. 2018;25(3):289–296. doi: 10.1038/s41594-018-0028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Munter S, Ingels J, Goetgeluk G, Bonte S, Pille M, Weening K, Kerre T, Abken H, Vandekerckhove B. Nanobody based dual specific CARs. Int J Mol Sci. 2018;19:403. doi: 10.3390/ijms19020403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lauwereys M, Arbabi Ghahroudi M, Desmyter A, Kinne J, Holzer W, De Genst E, Wyns L, Muyldermans S. Potent enzyme inhibitors derived from dromedary heavy-chain antibodies. EMBO J. 1998;17:3512–3520. doi: 10.1093/emboj/17.13.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conrath KE, Lauwereys M, Galleni M, Matagne A, Frere JM, Kinne J, Wyns L, Muyldermans S. Beta-lactamase inhibitors derived from single-domain antibody fragments elicited in the camelidae. Antimicrob Agents Chemother. 2001;45:2807–2812. doi: 10.1128/AAC.45.10.2807-2812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stijlemans B, Conrath K, Cortez-Retamozo V, Van Xong H, Wyns L, Senter P, Revets H, De Baetselier P, Muyldermans S, Magez S. Efficient targeting of conserved cryptic epitopes of infectious agents by single domain antibodies. African trypanosomes as paradigm. J Biol Chem. 2004;279:1256–1261. doi: 10.1074/jbc.M307341200. [DOI] [PubMed] [Google Scholar]
- 42.Saerens D, Ghassabeh GH, Muyldermans S. Single-domain antibodies as building blocks for novel therapeutics. Curr Opin Pharmacol. 2008;8:600–608. doi: 10.1016/j.coph.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 43.Steeland S, Vandenbroucke RE, Libert C. Nanobodies as therapeutics: big opportunities for small antibodies. Drug Discov Today. 2016;21:1076–1113. doi: 10.1016/j.drudis.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 44.Helma J, Cardoso MC, Muyldermans S, Leonhardt H. Nanobodies and recombinant binders in cell biology. J Cell Biol. 2015;209:633–644. doi: 10.1083/jcb.201409074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pardon E, Laeremans T, Triest S, Rasmussen SG, Wohlkonig A, Ruf A, Muyldermans S, Hol WG, Kobilka BK, Steyaert J. A general protocol for the generation of nanobodies for structural biology. Nat Protoc. 2014;9:674–693. doi: 10.1038/nprot.2014.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu Y, Liu C, Muyldermans S. Nanobody-based delivery systems for diagnosis and targeted tumor therapy. Front Immunol. 2017;8:1442. doi: 10.3389/fimmu.2017.01442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chakravarty R, Goel S, Cai W. Nanobody: the “magic bullet” for molecular imaging? Theranostics. 2014;4:386–398. doi: 10.7150/thno.8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oliveira S, Heukers R, Sornkom J, Kok RJ, En Henegouwen PMVB. Targeting tumors with nanobodies for cancer imaging and therapy. J Control Release. 2013;172:607–617. doi: 10.1016/j.jconrel.2013.08.298. [DOI] [PubMed] [Google Scholar]
- 49.Wesolowski J, Alzogaray V, Reyelt J, Unger M, Juarez K, Urrutia M, Cauerhff A, Danquah W, Rissiek B, Scheuplein F, et al. Single domain antibodies: promising experimental and therapeutic tools in infection and immunity. Med Microbiol Immunol. 2009;198:157–174. doi: 10.1007/s00430-009-0116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harmsen MM, De Haard HJ. Properties, production, and applications of camelid single-domain antibody fragments. Appl Microbiol Biotechnol. 2007;77:13–22. doi: 10.1007/s00253-007-1142-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Georgiou G, Segatori L. Preparative expression of secreted proteins in bacteria: status report and future prospects. Curr Opin Biotechnol. 2005;16:538–545. doi: 10.1016/j.copbio.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 52.Clements A, Young J, Constantinou N, Frankel G. Infection strategies of enteric pathogenic E coli. Gut Microbes. 2012;3(2):71–87. doi: 10.4161/gmic.19182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong AR, Pearson JS, Bright MD, Munera D, Robinson KS, Lee SF, Frankel G, Hartland EL. Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol Microbiol. 2011;80:1420–1438. doi: 10.1111/j.1365-2958.2011.07661.x. [DOI] [PubMed] [Google Scholar]
- 54.Frankel G, Phillips AD. Attaching effacing Escherichia coli and paradigms of Tir-triggered actin polymerization: getting off the pedestal. Cell Microbiol. 2008;10:549–556. doi: 10.1111/j.1462-5822.2007.01103.x. [DOI] [PubMed] [Google Scholar]
- 55.Tzschaschel BD, Guzman CA, Timmis KN, de Lorenzo V. An Escherichia coli hemolysin transport system-based vector for the export of polypeptides: export of Shiga-like toxin IIeB subunit by Salmonella typhimurium aroA. Nat Biotechnol. 1996;14:765–769. doi: 10.1038/nbt0696-765. [DOI] [PubMed] [Google Scholar]
- 56.Bessette PH, Åslund F, Beckwith J, Georgiou G. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc Natl Acad Sci USA. 1999;96:13703–13708. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jurado P, Ritz D, Beckwith J, de Lorenzo V, Fernández LA. Production of functional single-chain Fv antibodies in the cytoplasm of Escherichia coli. J Mol Biol. 2002;320:1–10. doi: 10.1016/S0022-2836(02)00405-9. [DOI] [PubMed] [Google Scholar]
- 58.Nakamoto H, Bardwell JC. Catalysis of disulfide bond formation and isomerization in the Escherichia coli periplasm. Biochim Biophys Acta. 2004;1694:111–119. doi: 10.1016/j.bbamcr.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 59.Kadokura H, Katzen F, Beckwith J. Protein disulfide bond formation in prokaryotes. Annu Rev Biochem. 2003;72:111–135. doi: 10.1146/annurev.biochem.72.121801.161459. [DOI] [PubMed] [Google Scholar]
- 60.Fernández LA, De Lorenzo V. Formation of disulphide bonds during secretion of proteins through the periplasmic-independent type I pathway. Mol Microbiol. 2001;40:332–346. doi: 10.1046/j.1365-2958.2001.02410.x. [DOI] [PubMed] [Google Scholar]
- 61.Govaert J, Pellis M, Deschacht N, Vincke C, Conrath K, Muyldermans S, Saerens D. Dual beneficial effect of interloop disulfide bond for single domain antibody fragments. J Biol Chem. 2012;287:1970–1979. doi: 10.1074/jbc.M111.242818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pfleger BF, Pitera DJ, Smolke CD, Keasling JD. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat Biotechnol. 2006;24:1027–1032. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- 63.Sharma SS, Blattner FR, Harcum SW. Recombinant protein production in an Escherichia coli reduced genome strain. Metab Eng. 2007;9:133–141. doi: 10.1016/j.ymben.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Welch M, Govindarajan S, Ness JE, Villalobos A, Gurney A, Minshull J, Gustafsson C. Design parameters to control synthetic gene expression in Escherichia coli. PLoS ONE. 2009;4:e7002. doi: 10.1371/journal.pone.0007002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perna NT, Plunkett G, 3rd, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 66.McCafferty J, Fitzgerald KJ, Earnshaw J, Chiswell DJ, Link J, Smith R, Kenten J. Selection and rapid purification of murine antibody fragments that bind a transition-state analog by phage display. Appl Biochem Biotechnol. 1994;47:157–173. doi: 10.1007/BF02787932. [DOI] [PubMed] [Google Scholar]
- 67.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short protocols in molecular biology. 5. New York: Wiley; 2002. [Google Scholar]
- 68.Carter P, Bedouelle H, Winter G. Improved oligonucleotide site-directed mutagenesis using M13 vectors. Nucleic Acids Res. 1985;13:4431–4443. doi: 10.1093/nar/13.12.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated and/or analysed during this study are included in this published article.