Summary
S100A7 (psoriasin) and S100A15 (koebnerisin) were first identified in inflamed psoriatic skin. They are of major interest because of their putative functional roles in innate immunity, epidermal cell maturation and epithelial tumorigenesis. Human S100A7 (psoriasin) and S100A15 (koebnerisin) have lately evolved by gene duplications during primate evolution forming a novel S100 subfamily within the Epidermal Differentiation Complex (chromosome 1q21). Therefore, hS100A7 and hS100A15 are almost identical in sequence (>90%) and difficult to discriminate. However, hS100A7 and hS100A15 are distinct in regulation, distribution and function and therefore exemplary for the diversity within the S100 family. Their different properties are compelling reasons to discriminate among these highly homologous proteins in normal tissue, inflammation and cancer.
Keywords: koebnerisin, psoriasin, paralogs, calcium-binding protein, innate immunity, inflammation
Introduction
S100 proteins are small (9–13kDa), calcium-binding proteins constituting the largest, multigenic family of calcium-binding EF-hand proteins (Donato 2003). Many S100 proteins show distinct tissue-and cell-type specific expression patterns indicating local specification.
Members of this protein family are involved in the calcium-/ zink-dependent regulation of various intracellular activities. S100 dependent homeostasis, transcription, protein activation, intracellular trafficking and structural participation in membranes modulate diverse cellular functions such as cell proliferation and maturation. Some S100 members are released into the extracellular space and act as chemoattractants for leukocytes or regulate their activation. To be active, S100 proteins form dimers or multimers in which S100 monomers are assembled by a two-fold axis of rotation. Ion-binding then induces the exposure of an active surface with which the S100 dimers interact with their target proteins (Heizmann et al. 2002; Zimmer et al. 2003).
Many S100 family members are encoded in the epidermal differentiation complex (EDC) located on human chromosome 1q21 (Hardas et al. 1996). This region is of particular interest as it encodes many genes that have been linked to epidermal differentiation and inflammation (Mischke et al. 1996; South et al. 1999; de Cid et al. 2009; Zhang et al. 2009). Two genes in that locus were cloned, hS100A7 (psoriasin) and subsequently hS100A15 (koebnerisin), because of their particularly high expression in inflamed psoriatic lesions, which are characterized by disturbed epidermal differentiation and inflammation (Madsen et al. 1991; Wolf et al. 2003). Despite their small size and conserved functional domains, S100 gene duplications throughout vertebrate evolution led to an increase in number and diversity within the S100 family (Kulski et al. 2003). Minor sequence differences can lead to structural differences that subsequently result in different binding to target proteins and function. The human S100A7/S100A15 subfamily shares over 90% sequence identity. Despite the high homology, hS100A7 (psoriasin) and hS100A15 (koebnerisin) are distinct and therefore exemplary for the functional and expressional diversity within the S100 family.
Identification, genomic organization and protein structure
Human S100A7 (hS100A7) has been identified almost two decades ago and named ‘psoriasin’ because of it is overexpressed by psoriatic keratinocytes (Madsen et al. 1991). Similarly, the human S100A15 (hS100A15) has been discovered by analyzing the differential gene expression in psoriasis (Wolf et al. 2003). Due to overexpression in ‘koeberized’ psoriatic skin, hS100A15 proposed name is ‘koebnerisin’. Both genes map to the S100 gene cluster within the epidermal differentiation complex (EDC, chromosome 1q21), which has been identified as one of the psoriasis candidate loci (PSORS4) (Hardas et al. 1996; Semprini et al. 1999; Semprini et al. 2002).
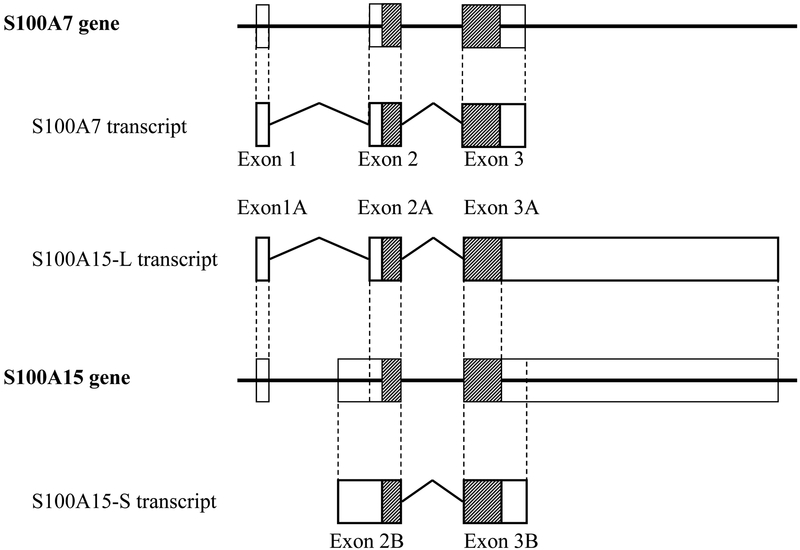
In contrast to other S100 members, hS100A15 reveals an unusual genomic organization. Whereas most S100 genes including hS100A7 encode for a single transcript, two alternative hS100A15 mRNA-isoforms have been discovered. They share the same coding region but show differences in UTR composition and length (0.5 kb vs. 4.4 kb). Both hS100A15 splice variants are differently expressed in psoriatic skin suggesting regulation through alternate promoters (Figure 1).
Figure1: Genomic structure of the human S100A7/S10015 subfamily.
Schematic representation of the genomic and exon/intron organization for the S100A7 and the S100A15 genes. Boxes represent exons, hatched regions indicate the coding sequence; intervening lines denote introns. The alternate S100A15 mRNA-splice variants are marked A for hS100A15-L (long) and B for hS100A15-S (short).
Analysis of the hS100A15 deduced amino acid sequence revealed a conserved C-terminal and a variant N-terminal EF-hand typical for S100 proteins (101 amino acids, 11.305 Da, calculated pI of 7.57 kDa). The hS100A15 protein is highly homologous (93% sequence identity) to hS100A7 (psoriasin, 101 amino acids, 11.326 kD, calculated pI of 6.77) (Figure 2). The main differences found at the putative N-terminus led to the prediction of a calcium-binding EF-hand motif for the hS100A15, which is not found to be functional in S100A7. Because of the predicted differences between highly homologous hS100A7 and hS100A15 studies were needed to discriminate their expression and function.
Figure2: Protein structure of the human S100A7/S10015 subfamily.
Alignment of the predicted amino acid sequences of the human S100A7 (hS100A7, NP_002954) with the human S100A15 (hS100A15, NP_79669). Identical amino acid residues are indicated as blue boxes, and chemically similar amino acids are marked as grey boxes. Predicted EF-hand motifs for both proteins are marked above the sequences (variant S100-specific motif: amino acids 12–39, canonical EF-hand: amino acids 54–82).
Epithelial maturation
The gene locus of the S100A7/S100A15 subfamily is linked to epidermal maturation (Epidermal Differentiation Complex; human chromosome 1q21). This chromosomal region encodes additional genes (involucrin, filaggrin, trichoyalin, repetin, etc.) that are sequentially expressed in maturing epidermis (Mischke et al. 1996; South et al. 1999). The epidermis is constantly exposed to a variety of physicochemical and microbial challenges. The upper differentiated layers serve as a physical (cornified envelope) and biological (antimicrobial lipids, antimicrobial proteins) protective hurdle supported by normal microflora on the skin surface (Schroder and Harder 2006). The cornified envelope is a protective structure that is assembled adjacent the inner surface of the cell plasma membrane during the terminal stages of keratinocyte differentiation (Eckert et al. 2004). It is assembled from a pool of precursor proteins that are covalently crosslinked to one another via the action of the membrane-anchored enzyme, type I transglutaminase (Steinert et al. 1996). hS100A7 and other S100 proteins are transglutaminase substrates and several S100 proteins are components of the keratinocyte cornified envelope (Robinson et al. 1997). Accordingly, the hS100A7 transcript shows a calcium and differentiation-dependent regulation in human keratinocytes (Martinsson et al. 2005). During keratinocyte differentiation in epidermis hS100A7 redistributes to the cell periphery (Broome et al. 2003; Ruse et al. 2003) suggesting that hS100A7 is released from differentiated keratinocytes. Studies show that extracellular hS100A7 functions as an antibacterial agent reducing survival of E. coli and other strains (Glaser et al. 2005). However, considering the difficulties distinguishing hS100A7 and hS100A15, both proteins may have contributed to previously reported features. As hS100A7 and hS100A15 are almost identical in sequence (93%), distinguishing their expression has been difficult. Moreover, many of the customized and commercial hS100A7 antibodies are cross-reactive with both S100 proteins (Figure 3A). Thus, hS100A15 specific antibodies were generated that did not cross-react with related recombinant hS100 proteins, particularly hS100A7 (Wolf et al. 2008; Wolf et al. 2009).
Figure3: hS100A7 and hS100A15 function through distinct classes of receptors.
A) Frozen sections of inflamed lesional psoriasis were stained for hS100A7 (green) and hS100A15 (red) showing differential upregulation of both S100 proteins. Nuclei were stained with DAPI (blue). Bar size: 50 μm. B) Recombinant hS100A7, hS100A15 were injected intraperitoneally into C57/BL6 wild-type mice alone or premixed in combination. After four hours, cells attracted into the peritoneal fluid were counted and analyzed by flow cytometry using the granulocyte marker Gr-1. (mean value + s.d. from six mice, * P ≤ 0.05). C) hS100A7 mediates leukocyte chemotaxis through RAGE (receptor of advanced glycated end products). This atypical chemotaxis receptor is pertussis toxin-insensitive, which helps distinguish ligand activity through classical chemokine receptors. hS100A15 chemotactic activity is Gi-protein-dependent, however the receptor has to be specified.
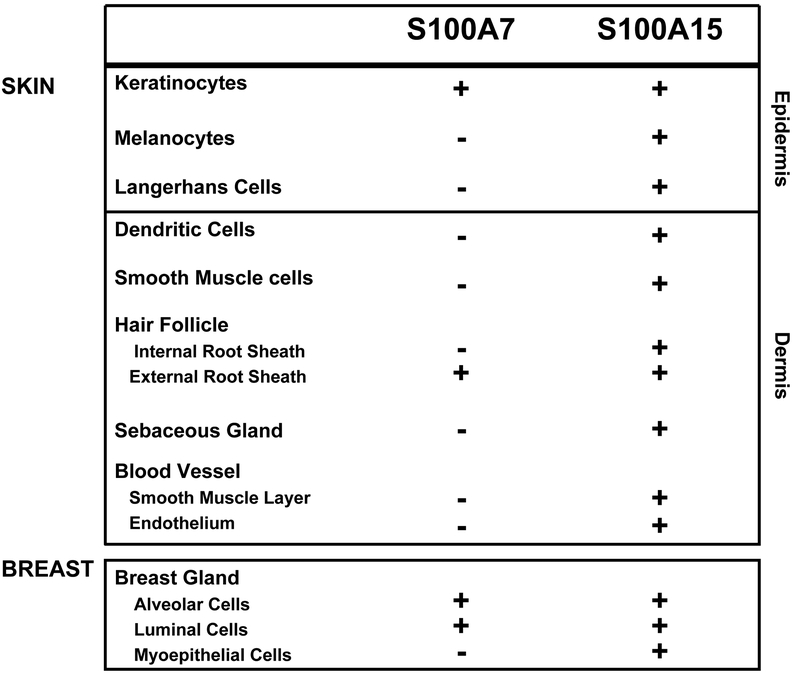
In normal skin, hS100A7 expression is confined to the granular/ cornified layers of the interfollicular epidermis and hair follicles. hS100A15 co-localized there but unlike hS100A7, it is expressed by epidermal basal cells and dendritic-shaped cells. The latter hS100A15 expressing cells were identified as melanocytes (MART-1 positive) and epidermal Langerhans cells (MHCII positive). In the dermis, hS100A7 was not detected; however hS100A15 was further expressed by dendritic cells, endothelial cells and vascular smooth muscle cells and peripheral nerves (Figure 3B, C). Similar to skin, a differential staining pattern has been shown in normal breast tissue (Figure 3D) (Wolf et al. 2009).
As hS100A15 is expressed within the differentiated epidermal layers of normal skin, the regulation of S100A15 during calcium-induced keratinocyte differentiation was investigated. While both S100A15 mRNA variants were induced by calcium, the hS100A15-L response was more pronounced. Induction was more rapid at higher calcium concentrations concordant with expression of late differentiation markers similar to hS100A7 (Martinsson et al. 2005).
Consistent with upregulation of hS100A7 and hS100A15 with epidermal differentiation, data indicate that the hS100A15 also participates in antimicrobial defence. Similar to hS100A7, hS100A15 functions as an antibacterial agent reducing survival of E. coli and other strains (Buchau et al. 2007). Together with hS100A7, both hS100A15 transcripts were regulated by bacterial components of several strains, where hS100A15-L followed hS100A7 induction pattern. Further, E.coli induced expression of hS100A7 and hS100A15 were dependent on Toll-like-receptor (TLR) 4. Although hS100A15 show an expression pattern distinct from hS100A7, both proteins are co-expressed by differentiated keratinocytes. They are co-regulated through inducers of differentiation and antimicrobial agents and may therefore co-participate in both physical and antimicrobial defence (Glaser et al. 2005; Buchau et al. 2007; Abtin et al. 2008). MECHANICAL IMPLIES MOVING PARTS
hS100A7 and hS100A15 exhibit distinct expression pattern in human skin, and both proteins can be specifically detected in cellular subsets of alveolar and small duct luminal cells within normal breast (Wolf et al. 2009) suggesting both proteins might participate in the microbial homeostasis within the host breast as well as in the digestive tract of nursing newborns. That hS100A15 is expressed by epithelial-derived myoepithelial cells around acini and in surrounding blood vessels may reflect its biological diversity from hS100A7 with additional, distinct functions for hS100A15. This study emphasizes the importance to discriminate the highly related hS100A7 and hS100A15 paralogs and opens the opportunity to further dissect their differential roles in normal tissue and their use as distinct markers in epithelial pathology in skin and beyond.
Epithelial tumorigenesis
Disruption of the calcium signalling pathway has been implicated as a central mechanism in tumorigenesis, specifically tumor invasion and metastasis (Kohn and Liotta 1995).
In normal skin, hS100A7 and hS100A15 are coexpressed in the differentiated layers indicating a specific association with squamous cell differentiation (Moubayed et al. 2007). In epithelial tumors, expression is altered in association with early stages of skin and breast tumorigenesis with highest levels of expression within preinvasive squamous cell carcinoma in-situ (unpublished and Wolf 2009). However, while factors related to cellular differentiation clearly comprise an important aspect of the regulation of the S100A7/S100A15 subfamily, the downregulation that is frequently seen in squamous and breast ductal carcinoma cells within invasive compared to in-situ components(Alowami et al. 2003; Emberley et al. 2004b) suggests regulation by additional factors that may also be associated with the invasive process in these tumors. Since much of the previous work on hS100A7 precedes the discovery of the highly homologous hS100A15 and hS100A7 antibodies, thetools used might have detected both proteins in association with tumor progression.
The ability to distinguish the closely related hS100A7 and hS100A15 at the RNA and protein level reveals significant distinctions in their regulation. Breast cancer specimens studied link high expression levels of both hS100A7 and hS100A15 transcripts with ER negativity and imply a correlation to clinical outcome as previously indicated for only hS100A7(Emberley et al. 2003; Emberley et al. 2004a). While hS100A7 protein expression closely follows corresponding RNA levels, hS100A15 protein is ubiquitous in invasive carcinomas and appears to be preferentially modified/ cross-linked, and thus providing a more stable and potentially longer lived protein even when transcript levels are low (Wolf et al. 2009). The coincident but differential expression and intracellular localization of these almost identical S100 paralogs could have significant biological implications for normal breast and breast cancer. Co-expression of transcripts for both hS100A7 and hS100A15 proteins in ER/PR negative tumors suggests a joint regulation related to tumor progression. While the secreted proteins have distinct roles as chemoattractants (Wolf et al. 2008), they also act synergistically to enhance inflammation and thus could influence breast tumors.
Beyond skin, this study emphasizes the importance to discriminate the highly related hS100A7 and hS100A15 paralogs and opens the opportunity to further dissect their differential roles and their use as distinct markers in breast cancer pathogenesis and other epithelial tumors.
Skin inflammation
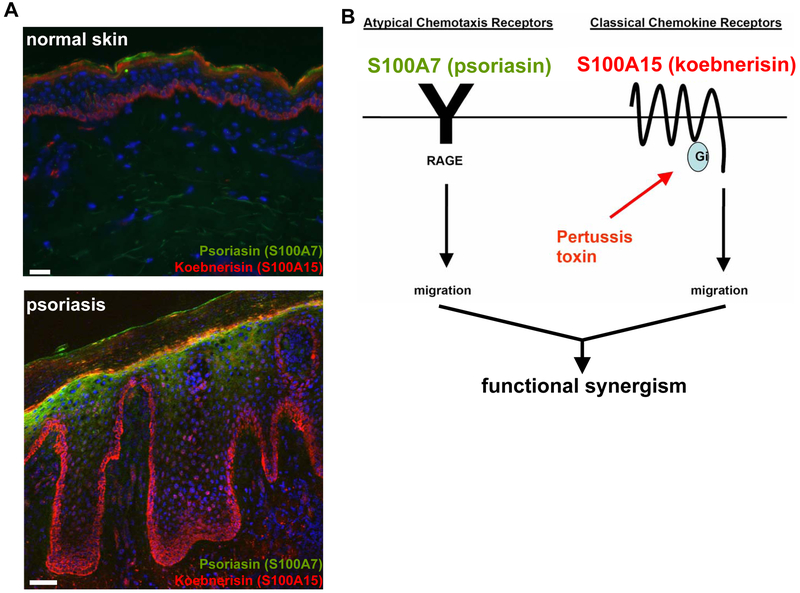
The human calcium-binding protein S100A15 was first identified in inflamed hyperplastic psoriatic skin, where the hS100A15 gene is transcribed into two mRNA splice variants, hS100A15-S (short isoform) and hS100A15-L (long isoform) (Wolf et al. 2003). Both isoforms showed elevated levels in lesional psoriatic skin, where hS100A15-L was more pronounced compared with hS100A15-S. There, the detection of the hS100A15-L transcript was pronounced in the basal and granular layer of non-lesional psoriatic skin and further extended throughout the hyperplastic epidermis of lesional psoriatic skin. Similar to lesional psoriasis, increased hS100A15 expression was observed pan-epidermal in the skin of chronic atopic eczema (Wolf et al. 2007). Sporadic staining of hS100A15 in single cells and cell clusters was detected in the dermis of inflammatory psoriatic and atopic skin. .The hS100A7 transcript is also upregulated atopic and psoriatic skin and distributed in upper epidermis of both atopic and psoriatic skin (Madsen et al. 1991; Glaser et al. 2009). Using specific antibodies, both hS100A7 and hS100A15 proteins are upregulated in inflamed lesional psoriatic skin and co-expressed by the epidermal suprabasal compartments (Figure 4A). In addition, hS100A15 is highly expressed by basal psoriatic keratinocytes at the epidermal-dermaljunction, as well as dendritic and stromal cells in the dermis. Similarly, both hS100A7 and hS100A15 proteins are expressed and secreted at a higher rate in cultured psoriatic keratinocytes, which is important for their function as chemoattractants (Wolf et al. 2008).
Figure 4: S100A7 and S100A15 function through distinct classes of receptors.
a Frozen skin sections were stained for S100A7 (green) and S100A15 (red) showing their upregulation and differential distribution in normal skin compared to psoriasis. Nuclei were stained with DAPI (blue). Bar 50 μm. b S100A7 mediates leukocyte chemotaxis through RAGE (receptor of advanced glycated end products). This atypical chemotaxis receptor is pertussis toxin-insensitive, which helps to distinguish ligand activity through classical chemokine receptors. S100A15 chemotactic activity is Gi protein-dependent, but the receptor has yet to be specified. The distinct mechanisms of actions within the S100A7 (psoriasin)/S100A15 (koebnerisin) subfamily contribute to their synergistic effect in inflammation
Psoriasis and chronic atopic eczema are chronic inflammatory skin diseases characterized by skin-infiltrating immune cells. These cells are known to secrete proinflammatory cytokines mainly produced by granulocytes and macrophages and Th1/Th17-differentiated lymphocytes (Numerof and Asadullah 2006). hS100A15 expression is induced in cultured human keratinocytes upon treatment with TNF-α and IFN-γ as well as IL-1β, suggesting that the proinflammatory environment in diseased skin contributes to hS100A15 expression in the epidermis. The alternate hS100A15-transcripts (S100A15-L and hS100A15-S) are differentially regulated by these cytokines. hS100A15-L predominantly responded to proinflammatory cytokines in cultured keratinocytes compared with hS100A15-S. Whereas hS100A15-S weakly followed the induction of hS100A15-L by Th1 cytokines, IL-1β solely induced hS100A15-L, which further indicates specific S100A15 isoform regulation by alternate promoters. The unresponsiveness of keratinocytes to regulate hS100A15 by the Th2 derived cytokines IL-4 and IL-13 suggests that hS100A15 is preferentially induced by Th1 driven psoriasis and late chronic atopic eczema inflammation rather than in Th2 dominated diseases (Grewe et al. 1998). A similar regulation pattern through Th1 cytokines has been shown for hS100A7 concordant with co-regulation of both hS100A7 and hS100A15 in inflammation (Glaser et al. 2005; Glaser et al. 2009). Also, the S100A7/S100A15 subfamily in regulated by Th17 cytokines important in the pathogenesis of psoriasis and other inflammatory skin diseases (Sabat et al. 2007; Eyerich et al. 2009); however a specific discrimination of this subfamily members is needed in future investigations.
That hS100A7 and hS100A15 are co-upregulated in similar pathophysiological conditions through similar epidermotropic proinflammatory mediators suggests that both proteins functionally cooperate in inflammation.
The upregulation and secretion of both human S100A7 and S100A15 in chronic inflammatory diseases suggeststhey contribute to the inflammatory phenotype. When extracellular, either hS100 protein induced an inflammatory response as shown by intraperitoneal injection into mice (Wolf et al. 2008). When injected together, the inflammatory response was amplified resembling the increased expression and release of both hS100A7 and hS100A15 by psoriatic keratinocytes with implications for their pathogenetic role in the disease (Figure 4B). Both proteins are chemoattractants but differ in their chemotactic activity towards specific leukocyte subtypes. With the discovery of hS100A7 (Madsen et al. 1991), structural and functional data had stimulated the quest for mechanistic clarification of its extracellular action. The multiligand receptor RAGE is implicated in inflammatory processes including leukocyte migration (Zen et al. 2007; Ramasamy et al. 2008). RAGE is expressed at low levels in normal tissues and becomes upregulated wherever its ligands accumulate. Ligands initiate a sustained cellular activation through MAP-kinases culminating in the activation of NFkB. Through recognition of β-sheet fibrillar structures, proinflammatory cytokine-like mediators of the S100/ calgranulin family or high mobility group box-1 (HMGB-1), RAGE participates in the phenotype of inflammatory skin diseases, diabetes, amyloidosis and promotes tumor progression (Taguchi et al. 2000; Santilli et al. 2009; Sourris and Forbes 2009; Srikanth et al. 2009). Mechanistically, hS100A7 and hS100A15 stimulate chemotaxis through activation of different classes of receptors. hS100A15-mediated chemotaxis is blocked by pertussis toxin suggesting a signalling through a classical Gi protein coupled receptor. In contrast, hS100A7-mediated chemotaxis is pertussis toxin-insensitive and is mediated through the pattern recognition receptor RAGE (receptor of advanced glycated end products) (Figure 4C). Further, hS100A7 but not hS100A15 binds and directly mediates chemotaxis through the RAGE in both in vitro chemotaxis assays and in vivo in mouse models (Wolf 2008. ALREADY SHOWN ABOVE
hS100A7-RAGE binding, signaling and chemotaxis are zinc-dependent, reflecting the zinc-mediated changes in the hS100A7 dimer structure. This finding identifies zinc as an important mediator of hS100A7 chemotactic activity similar to S100A7 zinc-dependent antimicrobial action (Glaser et al. 2005; Lee and Eckert 2007). That RAGE is not the receptor for both S100 paralogs is likely due to the structural disparity between hS100A7 and hS100A15. Whereas most S100 proteins including hS100A7 and hS100A15, bind calcium at their conserved C-terminal EF-hand, the calcium binding at the variant N-terminal EF hand is impaired in hS100A7 due to lack of glutamate residues (Donato 2003; Zimmer et al. 2003). In contrast to hS100A7, the hS100A15 protein inherits those glutamate residues suggesting that hS100A15 binds calcium at its N-terminal EF-hand which may contribute to a quaternary structure distinct to hS100A7 (Boeshans et al. 2006).
RAGE is thought to recognize spatial structures rather than amino acid sequences (Bierhaus et al. 2005). Since sequences and secondary structures of the hS100A7 and hS100A15 monomers are alike, their distinct quaternary structures may determine if they are either perceptible by RAGE (hS100A7) or not (hS100A15). Similar results have been reported for hS100A12 binding to RAGE in a multimeric form (Moroz et al. 2009).
The proposed structural differences and distinct functional mechanisms between hS100A7 and hS100A15 provides evidence for the disparity within the S100A7/S100A15 subfamily beyond expression differences. Their independent actions through distinct receptors regulate physiological functions and potentiate their proinflammatory activities in disease (Figure 4).
Perspective
hS100A7 (psoriasin) and hS100A15 (koebnerisin) were cloned because of their particularly high expression in psoriatic lesions. They are encoded within the S100 protein complex on chromosome 1q21 (PSOR4) that has been genetically linked to disturbed differentiation and inflammation. Although both proteins are highly homologous, they are differentially expressed and regulated in normal and diseased tissues and have distinct functions and mechanisms of action. It is therefore important to discriminate hS100A7 (psoriasin) and hS100A15 (koebnerisin) and to learn more about their distinct functional roles and synergistic action in immunity and tumorigenesis. This understanding is crucial for developing therapeutic interventions for pathological conditions mediated by the S100A7/ S100A15 subfamily.
Acknowledgements
This work was supported by grants from the the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and the German Research Foundation (DFG).
Abbreviations:
- UTR
untranslated region
- EDC
epidermal differentiation complex
- hS100A15-L
long human S100A15 transcript
- hS100A15-S
short human S100A15 transcript
- TLR
Toll-like receptor
References
- Abtin A, Eckhart L, Mildner M, Gruber F, Schroder JM et al. (2008) Flagellin is the principal inducer of the antimicrobial peptide S100A7c (psoriasin) in human epidermal keratinocytes exposed to Escherichia coli. Faseb J 22(7): 2168–2176. [DOI] [PubMed] [Google Scholar]
- Alowami S, Qing G, Emberley E, Snell L, Watson PH (2003) Psoriasin (S100A7) expression is altered during skin tumorigenesis. BMC Dermatol 3: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierhaus A, Humpert PM, Morcos M, Wendt T, Chavakis T et al. (2005) Understanding RAGE, the receptor for advanced glycation end products. J Mol Med 83(11): 876–886. [DOI] [PubMed] [Google Scholar]
- Boeshans KM, Wolf R, Voscopoulos C, Gillette W, Esposito D et al. (2006) Purification, crystallization and preliminary X-ray diffraction of human S100A15. Acta Crystallogr Sect F Struct Biol Cryst Commun 62(Pt 5): 467–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broome AM, Ryan D, Eckert RL (2003) S100 protein subcellular localization during epidermal differentiation and psoriasis. J Histochem Cytochem 51(5): 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchau AS, Hassan M, Kukova G, Lewerenz V, Kellermann S et al. (2007) S100A15, an antimicrobial protein of the skin: regulation by E. coli through Toll-like receptor 4. J Invest Dermatol 127(11): 2596–2604. [DOI] [PubMed] [Google Scholar]
- de Cid R, Riveira-Munoz E, Zeeuwen PL, Robarge J, Liao W et al. (2009) Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat Genet 41(2): 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato R (2003) Intracellular and extracellular roles of S100 proteins. Microsc Res Tech 60(6): 540–551. [DOI] [PubMed] [Google Scholar]
- Eckert RL, Broome AM, Ruse M, Robinson N, Ryan D et al. (2004) S100 proteins in the epidermis. J Invest Dermatol 123(1): 23–33. [DOI] [PubMed] [Google Scholar]
- Emberley ED, Murphy LC, Watson PH (2004a) S100A7 and the progression of breast cancer. Breast Cancer Res 6(4): 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emberley ED, Alowami S, Snell L, Murphy LC, Watson PH (2004b) S100A7 (psoriasin) expression is associated with aggressive features and alteration of Jab1 in ductal carcinoma in situ of the breast. Breast Cancer Res 6(4): R308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emberley ED, Niu Y, Njue C, Kliewer EV, Murphy LC et al. (2003) Psoriasin (S100A7) expression is associated with poor outcome in estrogen receptor-negative invasive breast cancer. Clin Cancer Res 9(7): 2627–2631. [PubMed] [Google Scholar]
- Eyerich K, Pennino D, Scarponi C, Foerster S, Nasorri F et al. (2009) IL-17 in atopic eczema: linking allergen-specific adaptive and microbial-triggered innate immune response. J Allergy Clin Immunol 123(1): 59–66 e54. [DOI] [PubMed] [Google Scholar]
- Glaser R, Harder J, Lange H, Bartels J, Christophers E et al. (2005) Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nat Immunol 6(1): 57–64. [DOI] [PubMed] [Google Scholar]
- Glaser R, Meyer-Hoffert U, Harder J, Cordes J, Wittersheim M et al. (2009) The antimicrobial protein psoriasin (S100A7) is upregulated in atopic dermatitis and after experimental skin barrier disruption. J Invest Dermatol 129(3): 641–649. [DOI] [PubMed] [Google Scholar]
- Grewe M, Bruijnzeel-Koomen CA, Schopf E, Thepen T, Langeveld-Wildschut AG et al. (1998) A role for Th1 and Th2 cells in the immunopathogenesis of atopic dermatitis. Immunol Today 19(8): 359–361. [DOI] [PubMed] [Google Scholar]
- Hardas BD, Zhao X, Zhang J, Longqing X, Stoll S et al. (1996) Assignment of psoriasin to human chromosomal band 1q21: coordinate overexpression of clustered genes in psoriasis. J Invest Dermatol 106(4): 753–758. [DOI] [PubMed] [Google Scholar]
- Heizmann CW, Fritz G, Schafer BW (2002) S100 proteins: structure, functions and pathology. Front Biosci 7: d1356–1368. [DOI] [PubMed] [Google Scholar]
- Kohn EC, Liotta LA (1995) Molecular insights into cancer invasion: strategies for prevention and intervention. Cancer Res 55(9): 1856–1862. [PubMed] [Google Scholar]
- Kulski JK, Lim CP, Dunn DS, Bellgard M (2003) Genomic and phylogenetic analysis of the S100A7 (Psoriasin) gene duplications within the region of the S100 gene cluster on human chromosome 1q21. J Mol Evol 56(4): 397–406. [DOI] [PubMed] [Google Scholar]
- Lee KC, Eckert RL (2007) S100A7 (Psoriasin)--mechanism of antibacterial action in wounds. J Invest Dermatol 127(4): 945–957. [DOI] [PubMed] [Google Scholar]
- Madsen P, Rasmussen HH, Leffers H, Honore B, Dejgaard K et al. (1991) Molecular cloning, occurrence, and expression of a novel partially secreted protein “psoriasin” that is highly up-regulated in psoriatic skin. J Invest Dermatol 97(4): 701–712. [DOI] [PubMed] [Google Scholar]
- Martinsson H, Yhr M, Enerback C (2005) Expression patterns of S100A7 (psoriasin) and S100A9 (calgranulin-B) in keratinocyte differentiation. Exp Dermatol 14(3): 161–168. [DOI] [PubMed] [Google Scholar]
- Mischke D, Korge BP, Marenholz I, Volz A, Ziegler A (1996) Genes encoding structural proteins of epidermal cornification and S100 calcium-binding proteins form a gene complex (“epidermal differentiation complex”) on human chromosome 1q21. J Invest Dermatol 106(5): 989–992. [DOI] [PubMed] [Google Scholar]
- Moroz OV, Burkitt W, Wittkowski H, He W, Ianoul A et al. (2009) Both Ca2+ and Zn2+ are essential for S100A12 protein oligomerization and function. BMC Biochem 10: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayed N, Weichenthal M, Harder J, Wandel E, Sticherling M et al. (2007) Psoriasin (S100A7) is significantly up-regulated in human epithelial skin tumours. J Cancer Res Clin Oncol 133(4): 253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numerof RP, Asadullah K (2006) Cytokine and anti-cytokine therapies for psoriasis and atopic dermatitis. BioDrugs 20(2): 93–103. [DOI] [PubMed] [Google Scholar]
- Ramasamy R, Yan SF, Herold K, Clynes R, Schmidt AM (2008) Receptor for advanced glycation end products: fundamental roles in the inflammatory response: winding the way to the pathogenesis of endothelial dysfunction and atherosclerosis. Ann N Y Acad Sci 1126: 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NA, Lapic S, Welter JF, Eckert RL (1997) S100A11, S100A10, annexin I, desmosomal proteins, small proline-rich proteins, plasminogen activator inhibitor-2, and involucrin are components of the cornified envelope of cultured human epidermal keratinocytes. J Biol Chem 272(18): 12035–12046. [DOI] [PubMed] [Google Scholar]
- Ruse M, Broome AM, Eckert RL (2003) S100A7 (psoriasin) interacts with epidermal fatty acid binding protein and localizes in focal adhesion-like structures in cultured keratinocytes. J Invest Dermatol 121(1): 132–141. [DOI] [PubMed] [Google Scholar]
- Sabat R, Philipp S, Hoflich C, Kreutzer S, Wallace E et al. (2007) Immunopathogenesis of psoriasis. Exp Dermatol 16(10): 779–798. [DOI] [PubMed] [Google Scholar]
- Santilli F, Vazzana N, Bucciarelli LG, Davi G (2009) Soluble forms of RAGE in human diseases: clinical and therapeutical implications. Curr Med Chem 16(8): 940–952. [DOI] [PubMed] [Google Scholar]
- Schroder JM, Harder J (2006) Antimicrobial skin peptides and proteins. Cell Mol Life Sci 63(4): 469–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semprini S, Capon F, Bovolenta S, Bruscia E, Pizzuti A et al. (1999) Genomic structure, promoter characterisation and mutational analysis of the S100A7 gene: exclusion of a candidate for familial psoriasis susceptibility. Hum Genet 104(2): 130–134. [DOI] [PubMed] [Google Scholar]
- Semprini S, Capon F, Tacconelli A, Giardina E, Orecchia A et al. (2002) Evidence for differential S100 gene over-expression in psoriatic patients from genetically heterogeneous pedigrees. Hum Genet 111(4–5): 310–313. [DOI] [PubMed] [Google Scholar]
- Sourris KC, Forbes JM (2009) Interactions between advanced glycation end-products (AGE) and their receptors in the development and progression of diabetic nephropathy - are these receptors valid therapeutic targets. Curr Drug Targets 10(1): 42–50. [DOI] [PubMed] [Google Scholar]
- South AP, Cabral A, Ives JH, James CH, Mirza G et al. (1999) Human epidermal differentiation complex in a single 2.5 Mbp long continuum of overlapping DNA cloned in bacteria integrating physical and transcript maps. J Invest Dermatol 112(6): 910–918. [DOI] [PubMed] [Google Scholar]
- Srikanth V, Maczurek A, Phan T, Steele M, Westcott B et al. (2009) Advanced glycation endproducts and their receptor RAGE in Alzheimer’s disease. Neurobiol Aging. [DOI] [PubMed] [Google Scholar]
- Steinert PM, Kim SY, Chung SI, Marekov LN (1996) The transglutaminase 1 enzyme is variably acylated by myristate and palmitate during differentiation in epidermal keratinocytes. J Biol Chem 271(42): 26242–26250. [DOI] [PubMed] [Google Scholar]
- Taguchi A, Blood DC, del Toro G, Canet A, Lee DC et al. (2000) Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature 405(6784): 354–360. [DOI] [PubMed] [Google Scholar]
- Wolf R, Lewerenz V, Buchau AS, Walz M, Ruzicka T (2007) Human S100A15 splice variants are differentially expressed in inflammatory skin diseases and regulated through Th1 cytokines and calcium. Exp Dermatol 16(8): 685–691. [DOI] [PubMed] [Google Scholar]
- Wolf R, Mirmohammadsadegh A, Walz M, Lysa B, Tartler U et al. (2003) Molecular cloning and characterization of alternatively spliced mRNA isoforms from psoriatic skin encoding a novel member of the S100 family. Faseb J 17(13): 1969–1971. [DOI] [PubMed] [Google Scholar]
- Wolf R, Voscopoulos C, Winston J, Dharamsi A, Goldsmith P et al. (2009) Highly homologous hS100A15 and hS100A7 proteins are distinctly expressed in normal breast tissue and breast cancer. Cancer Lett 277(1): 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf R, Howard OM, Dong HF, Voscopoulos C, Boeshans K et al. (2008) Chemotactic activity of S100A7 (Psoriasin) is mediated by the receptor for advanced glycation end products and potentiates inflammation with highly homologous but functionally distinct S100A15. J Immunol 181(2): 1499–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zen K, Chen CX, Chen YT, Wilton R, Liu Y (2007) Receptor for advanced glycation endproducts mediates neutrophil migration across intestinal epithelium. J Immunol 178(4): 2483–2490. [DOI] [PubMed] [Google Scholar]
- Zhang XJ, Huang W, Yang S, Sun LD, Zhang FY et al. (2009) Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat Genet 41(2): 205–210. [DOI] [PubMed] [Google Scholar]
- Zimmer DB, Wright Sadosky P, Weber DJ (2003) Molecular mechanisms of S100-target protein interactions. Microsc Res Tech 60(6): 552–559. [DOI] [PubMed] [Google Scholar]