Abstract
Synapse density is reduced in postmortem cortical tissue from schizophrenia patients, which is suggestive of increased synapse elimination. Using a reprogrammed in vitro model of microglia-mediated synapse engulfment, we demonstrate increased synapse elimination in patient-derived neural cultures and isolated synaptosomes. This excessive synaptic pruning reflects abnormalities in both microglia-like cells and synaptic structures. Further, we find that schizophrenia risk-associated variants within the human complement component 4 locus are associated with increased neuronal complement deposition and synapse uptake; however, they do not fully explain the observed increase in synapse uptake. Finally, we demonstrate that the antibiotic minocycline reduces microglia-mediated synapse uptake in vitro and its use is associated with a modest decrease in incident schizophrenia risk compared to other antibiotics in a cohort of young adults drawn from electronic health records. These findings point to excessive pruning as a potential target for delaying or preventing the onset of schizophrenia in high-risk individuals.
Schizophrenia (SZ) is a heritable psychiatric disorder with disabling psychotic and cognitive symptoms. Its pathophysiology remains largely elusive despite the identification of more than 100 regions of the genome associated with SZ liability1, repeated imaging studies displaying reduced gray matter thickness as well as abnormal functional connectivity2–10, and postmortem studies reporting reduced numbers of dendritic spines11–13. Given the extensive elimination of synapses in the human cerebral cortex during late adolescence and early adulthood14, the time when the early symptoms of SZ usually become clinically evident, it has been proposed that excessive synaptic pruning by microglia contributes to the observed reduction in synapse density in SZ patients15,16. In particular, rodent studies suggest a pivotal role for complement signaling in microglia-mediated elimination of synapses in the developing visual system16–18, providing a potential mechanism by which structurally distinct alleles of the complement component 4 (C4) genes may be associated with SZ liability1. However, the inability to model microglial functions in humans using patient-derived primary cells has limited efforts to directly link excessive microglial synapse elimination to SZ pathology. Mouse models cannot fully address this question because human microglia differ markedly from their rodent counterparts, with a higher expression of complement components and multiple genes important in brain development19.
We have recently developed and validated high-throughput methods for modeling synaptic pruning in vitro using patient-derived cells20. Similar to induced pluripotent stem cell (iPSC)-derived microglia-like cells21–23, human monocytes cultured under conditions resembling the CNS microenvironment recapitulate the important traits of primary human microglia, including a ramified morphology, expression of genes highly enriched in acutely isolated microglia (for example, P2RY12, C1QA, GAS6, MERTK, GPR34, and PROS1), and transcriptomic clustering with primary fetal microglia rather than peripheral monocyte-derived macrophages20. Mimicking primary fetal microglia, induced microglia-like (iMG) cells under serum-free in vitro conditions engulf synapses isolated from iPSC-derived neural cultures20. By using live imaging, as well as confocal microscopy, we have been able to develop robust methods to quantify synapse uptake20.
In this study, we employ a high-throughput synaptosome (SYN) system based on patient-derived iMG cells and iPSC-derived neurons to demonstrate increased, and complement-dependent, iMG uptake of patient-derived synaptic structures when deriving these, as well as iMG cells, from SZ patients as distinct to matched healthy controls (HCs), with concordant results in iMG- and iPSC-derived neural cocultures, and determine that these effects may be modulated by, but are not fully attributable to, effects of human C4 risk variants. Finally, we demonstrate that minocycline, a second-generation tetracycline with high brain penetrance, reduces synapse uptake in vitro in a dose-responsive fashion, and detect a modest reduction in incidence of psychosis associated with minocycline exposure in adolescents and young adults as indicated by electronic health records (EHRs).
Results
Patient-derived microglia-like cells.
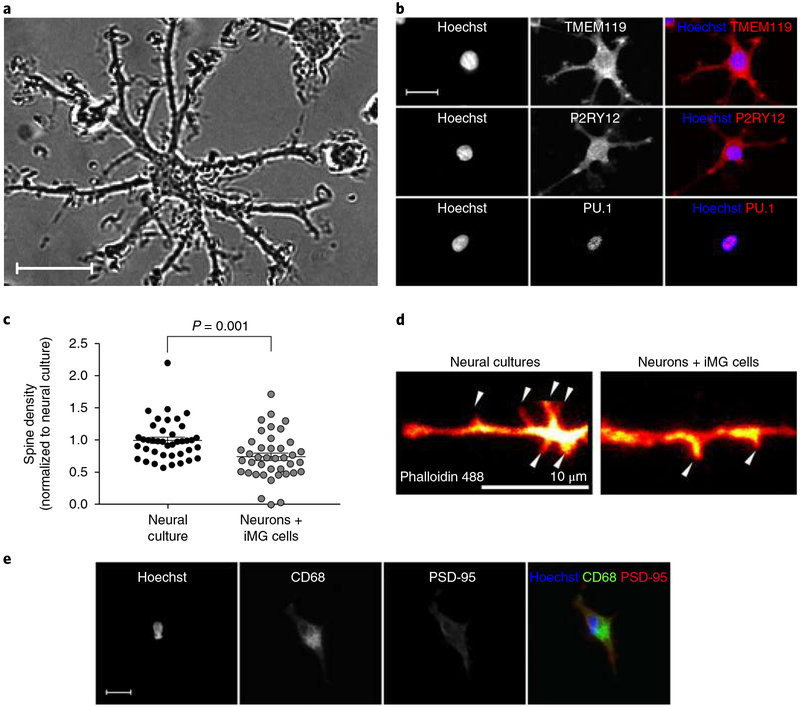
To derive iMG cells from SZ patients and matched HCs, monocytes were isolated from blood drawn from male individuals with SZ and age-matched male HCs. Briefly, microglial induction was achieved by exposure to interleukin-34 (IL-34) and granulocyte-macrophage colony-stimulating factor, under serum-free culture conditions, on an extracellular matrix resembling astrocyte-derived extracellular matrix, containing laminin, collagen, and nidogen-1 (entactin)20. No significant differences in morphological measurements, or yield per preparation, could be observed between patient- and control-derived cultures (Supplementary Fig. 1) with the vast majority of cells displaying a ramified morphology resembling resting state microglia (Fig. 1a), and expressing microglia markers such as transmembrane protein 119 (TMEM119), P2Y purinoceptor 12 (P2RY12), and transcription factor PU.1 (PU.1) (Fig. 1b). To further characterize iMG cells we performed transcriptome analyses comparing mRNA expression levels in iMG cells to matched monocytes (Supplementary Fig. 2), as well as monocyte-derived macrophages (granulocyte-macrophage colony-stimulating factor; 10% fetal bovine serum (FBS); Supplementary Fig. 3). Compared to monocytes, and to ordinary monocyte-derived macrophages, iMG cells displayed a strong enrichment of upregulated (fold change > 20) microglia-specific genes (based on 2 gene sets of published microglia-specific genes in acutely isolated microglia19,24; see Supplementary Tables 1–4), with iMG cells significantly clustering apart from monocytes and monocyte-derived macrophages (Supplementary Fig. 4 (displaying a hierarchical cluster analysis with uncertainty in clustering assessed by multiscale bootstrap resampling); see also Sellgren et al.20 for direct comparison of gene expression in iMG cells and primary human microglia). Corroborating our previous observations with isolated synapses from HCs20, we also noted a clear decrease in spine density after coculturing iMG cells with iPSC-derived neural cultures (Fig. 1c,d), with a concurrent robust uptake of the post-synaptic marker postsynaptic density protein 95 (PSD-95) as well as the presynaptic marker synaptosome-associated protein, 25 kDa (SNAP-25) in iMG cells (Fig. 1e and Supplementary Fig. 5).
Fig. 1 |. Characterizations of iMG cells.
a, Representative phase-contrast image of iMG cells captured during a live imaging session (repeated in 3 independent experiments with 36 images collected per session and similar results). Scale bar, 20 μm. b, Confocal images of iMG cells stained for TMEM119, P2RY12, and PU.1. Images are representative of 3 independent experiments with 20 images collected per experiment. Scale bar, 20 μm. c, Spine density (spines per 10-μm dendrite) in an iPSC-derived neural line with and without coculture with iMG cells (derived from 2 individuals) for 48 h (n = 40 randomly selected dendrites examined per group). Data are normalized to neural culture only and were analyzed using a t test (equal variance); t(78) = 3.40, P = 0.001 (two-sided). Mean ± s.e.m. is indicated for each group. d, Phalloidin 488-stained dendrite of an iPSC-derived neuron in pure culture and cocultured with iMG cells for 48 h.; arrows indicate dendritic spines. Images are representative of the experiments described in c. Scale bar, 10 μm. e, Colocalization and engulfment of PSD-95 (red) by iMG cells (green, CD68) in coculture. Images are representative of 3 independent experiments with 20 images collected per experiment. Scale bar, 30 μm.
Isolation of active synapses from human iPSC-derived neural cultures.
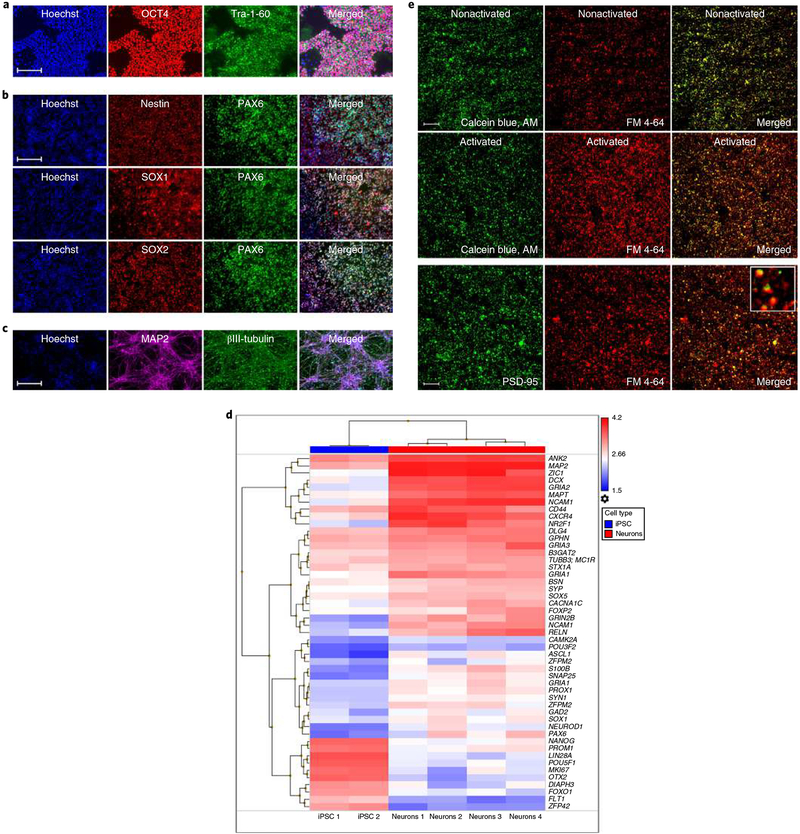
To model the interactions between synapses and microglia in SZ, we collected and reprogrammed iPSCs (Fig. 2a and Supplementary Fig. 6) from the dermal fibroblasts of male patients with SZ and age-matched HC donors, with clinical status confirmed by structured clinical interview, using an integration-free and all-synthetic RNA-based reprogramming system as previously described25. Purified and expandable neural progenitor cells (NPCs) were derived from these iPSCs using a neural induction supplement (NIS) based on defined small molecule induction26, followed by magnetic-activated cell sorting (MACS; polysialylated-neural cell adhesion molecule (PSA-NCAM+), CD271−, CD133+) and verified by immunocytochemical analyses for standard NPC markers (Fig. 2b). We then performed an inclusive neuronal differentiation by mitogen removal, which resulted in mixed populations of neural subtypes, including telencephalic cortical neurons (Fig. 2c,d and Supplementary Table 5)27,28. To be able to model interactions between iMG cells and synapses in patients versus HCs, we proceeded with the isolation of SYNs, that is, synaptic nerve terminals. As we have previously shown, iPSC-derived neural cultures yield a highly purified SYN preparation compared to isolation from postmortem human brain20. Briefly, neural cultures were collected, homogenized, and purified using the Syn-PER Synaptic Protein Extraction Reagent (Thermo Fisher Scientific), which was followed by repeated centrifugal fractionation. In addition to previous validation of iPSC-derived SYNs20, we performed additional analyses confirming that SYNs after freeze-thaw exhibited canonical structural and functional synaptic properties using transmission electron microscopy (Supplementary Fig. 7a), enrichment of the presynaptic marker SNAP-25 (Western blot analyses; Supplementary Fig. 7b), labeling for active synaptic vesicle endocytosis on depolarization (Fig. 2e)29, and fixation followed by staining for the postsynaptic marker PSD-95 (Fig. 2e and Supplementary Fig. 7c). In line with previous reports29,30, these investigations confirmed specificity of the SYN extraction procedure as well as the functionality of isolated and freeze-thawed SYNs, and further enabled us to control for line-specific differences of SYN yield in functional assays.
Fig. 2 |. Isolation of active synaptic structures from iPSC-derived neural cultures.
a, iPSCs stained for octamer-binding transcription factor (OCT4; POU domain, class 5, transcription factor 1) or Tra-1–60 as indicated. b, iPSC-derived NPCs stained for nestin, paired box protein Pax-6 (PAX6), transcription factor SOX1, or transcription factor SOX2 as indicated. c, Neurons stained for the neuronal markers tubulin beta-3 chain (βIII-tubulin) and microtubule-associated protein 2 (MAP2). Images in a–c are representative of 8 independent experiments with 20 images collected per well in each experiment. Scale bars, 100 μm. d, Heat map of mRNA expression values determined in iPSCs (n = 2 patients) and iPSC-derived neurons (n = 4 patients); for details see Supplementary Table 5. e, Confocal images of SYNs labeled with calcein blue, AM and FM 4–64 (first and second rows). SYNs were activated by the addition of 40 mM KCl as indicated. The last row displays SYNs labeled with FM 4–64 and the synaptic marker PSD-95. Images are representative of experiments performed in 8 independent patients with 40 images collected in cell culture triplicates per experiment. Scale bar, 60 μm.
Increased engulfment of synaptic structures in patient-derived models of synaptic pruning.
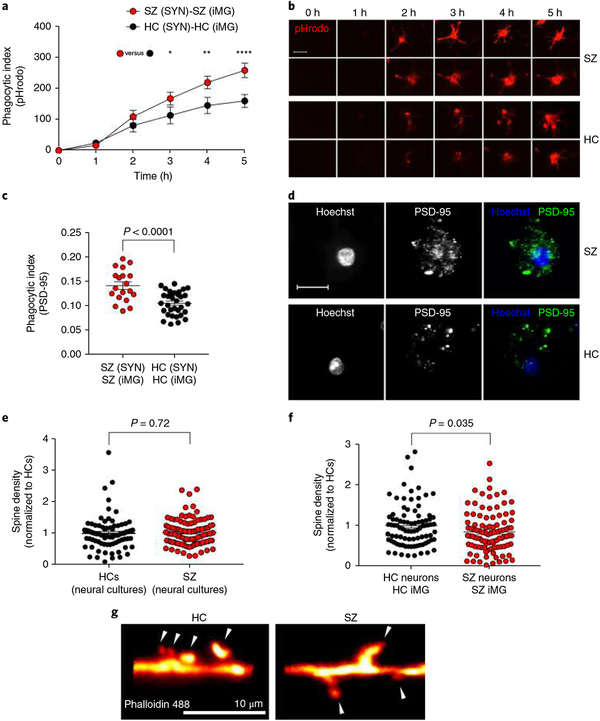
We first compared overall phagocytosis in assays incorporating iMG cells and SYNs derived from males with SZ to those derived from age-matched male HCs in real-time live imaging using SYNs labeled with a pH-sensitive dye (pHrodo) that binds non-specifically to proteins and exclusively fluoresces bright red in the post-phagocytic phagolysosome compartments. Each SYN line (derived from 3 SZ patients and 3 HCs) was assessed with iMG cultures from at least 3 different patients (in total, iMG cells from 13 SZ patients and 9 matched HCs). Quantification of bright red fluorescence (Supplementary Fig. 8), indicating cellular uptake of pHrodo-labeled SYNs31,32, revealed significantly increased phagocytosis in SZ-derived iMG cultures compared to matched HC iMG cultures (Fig. 3a,b; for a detailed description of phagocytic indexes see Methods). Using confocal microscopy on fixed samples with SYNs labeled with the pH-sensitive cyanine dye, CypHer5E, which is more resistant than pHrodo to fluorescence dampening in the acidic lysosome on fixation33, we could also observe that the vast majority of dyed particles inside iMG cells also stained positive for the postsynaptic marker PSD-95 (Supplementary Fig. 9) confirming that we detected specific engulfment of SYNs. Using confocal microscopy, we additionally quantified phagocytic inclusions (PSD-95+ inclusions, 0.5–1.5 μ m; Supplementary Fig. 10) using SYNs derived from 4 SZ patients and 4 HCs (combined with iMG cells from 13 SZ patients and 18 matched HCs). In agreement with real-time live imaging experiments, using confocal microscopy to quantify PSD-95+ phagocytic inclusions confirmed an increased uptake of SZ-derived SYNs (Fig. 3c,d).
Fig. 3 |. Increased engulfment of synaptic structures in schizophrenia-derived models.
a, Quantification of pHrodo (red)-labeled SYN uptake in iMG cells during live imaging. The phagocytic index represents the mean pHrodo+ area per iMG cell. SZ-derived models were designed by using iMG cells derived from 13 patients and SYNs derived from 3 patients (combined to form n = 13 SZ (SYN)-SZ (iMG) models), while HC models were derived from 9 HCs and SYN from 3 HCs (both groups matched with correspondent SZ patients); n = 13 HC (SYN)-HC (iMG) models. Nine images (20×) per well were automatically acquired every hour and the means were then extracted and analyzed using a two-way repeated ANOVA. There was a significant interaction between the effects of time and group on phagocytic index (F(5,60) = 4.84; P = 0.0009). Šidák’s multiple comparison tests gave an adjusted P value of 0.035 at 3 h, 0.002 at 4 h, and < 0.0001 at 5 h. All other comparisons were non-significant. b, The first two rows display representative images from the SZ models in a; the last two rows represent images from the HC models in a. Scale bar, 25 μm. c, Quantification of phagocytic inclusions (PSD-95+ inclusions, 0.5–1.5 μm) using confocal microscopy in a sample containing SYNs derived from a total of 4 SZ patients and 4 matched HCs, combined in different combinations with iMG cells from 13 SZ patients and 18 matched HCs (n = 19 SZ (SYN)-SZ (iMG) models and n = 33 HC (SYN)-HC(iMG) models). The phagocytic index represents the number of PSD-95+ particles (0.5–1.5 μm) per iMG cytoplasm area. Twenty randomly selected confocal images were taken per well and the means were analyzed using a t test (equal variance); t(50) = 4.55, P < 0.0001 (two-sided). d, Representative confocal images for the two groups in c. Scale bar, 30 μm. e, Quantification of spine density (spines per 10-μm dendrite) in neural lines derived from 3 SZ patients versus 2 HCs (n = 80 randomly selected dendrites examined in the SZ group and n = 100 randomly selected dendrites examined in the HC group); Welch-corrected t test; t(148) = 0.35, P = 0.72 (two-sided). f, Quantification of spine density in the same neural lines but cocultured with iMG cells (derived from 3 SZ patients or 2 HCs; n = 88 randomly selected dendrites examined in HC group and n = 99 randomly selected dendrites examined in the SZ group) as indicated; t test (equal variance); t(185) = 2.1, P = 0.035 (two-sided). Data in both graphs are normalized to the HC group. g, Representative images of phalloidin 488-stained high-magnification confocal images of dendritic spines (indicated by the arrowheads) in HC and SZ neural cultures described in f. The mean number of spines per 10-μm dendrite was 4.5 (s.e.m. = 0.25) for HC cultures and 3.8 (s.e.m. = 0.23) in SZ cultures. All reported P values are two-sided; the mean ± s.e.m. is indicated for each group in all graphs. *P < 0.05, **P < 0.01, ****P < 0.0001.
To further validate our findings using isolated SYNs we also performed coculture experiments (combining iMG cells and iPSC-derived neurons) comparing synapse density in SZ-derived cocultures to HC-derived cocultures. While we observed no difference in baseline spine density between SZ and HC neural monocultures (derived from the same set of patients; Fig. 3e), in accordance with our data from the isolated SYN system, SZ-derived cocultures displayed a significant reduction in spine density (Fig. 3f,g).
Microglial factors influence synapse engulfment independent of neuronal phenotype.
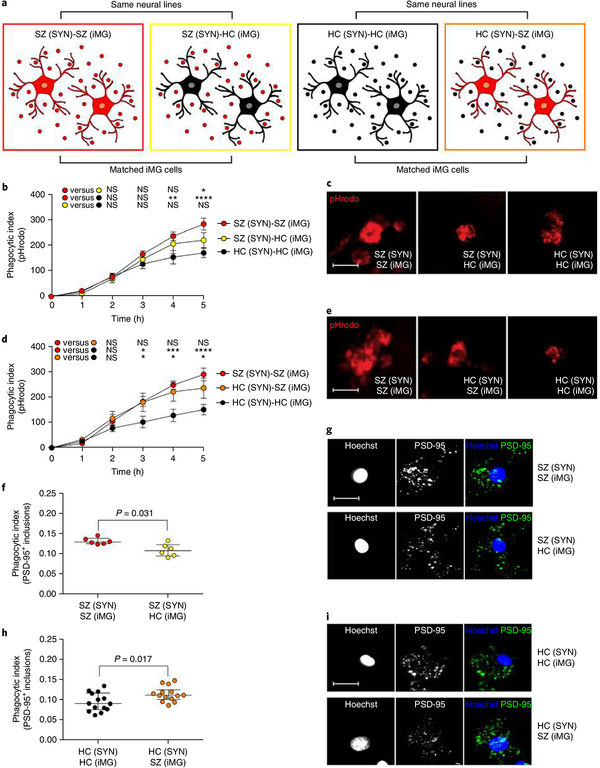
To better dissect the individual contributions of SYNs and iMG cells to the observed increase in phagocytic uptake of patient-derived synaptic particles, we repeated the phagocytosis live imaging experiments by crossing SZ-derived SYNs (as well as HC-derived SYNs) with iMG cells from HCs and SZ patients, respectively (Fig. 4a). Specifically, we first compared synapse elimination in ‘pure’ disease models, that is, iMG cultures from eight patients with SZ and SYNs from two matched SZ patients, with ‘crossed’ or ‘mixed’ models in which the same synaptic structures from these SZ patients were added to iMG cells derived from eight matched HCs. We also performed experiments comparing SZ and matched HC iMG cells (derived from three and five patients, respectively) using SYNs derived from three HCs. These live imaging experiments (Fig. 4b–e), as well as confocal microscopy experiments (Fig. 4f–i) revealed that SZ-derived iMG cells show a greater uptake of SYNs (SZ-derived as well as HC-derived) than HC-derived iMG cells, whereas the uptake of HC-derived SYNs by HC-derived iMG cells was smallest in both experiments. This suggests that both neuronal and microglial factors contribute to the increased uptake of synaptic structures by SZ-derived iMG cells versus HC-derived iMG cells.
Fig. 4 |. Microglial factors influence synapse engulfment.
a, ‘Pure’ disease models, derived from patients with SZ, were compared to ‘mixed’ models in which the same synaptic structures from SZ patients were added to iMG cells derived from matched HCs, as well as ‘pure’ HC models compared to ‘mixed’ models in which the same synaptic structures from HC patients were added to iMG cells derived from matched SZ patients. For both experimental designs, as a reference, we included matched ‘pure’ HC and SZ models, respectively. b, Quantification of pHrodo (red)-labeled SY) uptake in iMG cells during live imaging sessions of 5 h. SZ-SZ models were based on SYN from 2 SZ patients and iMG cells from 8 SZ patients (n = 8 models), while SZ-HC models were based on the same SYN from SZ patients and 8 models based on iMG cells derived from 7 matched HCs (n = 8 matched models). HC-HC models were based on iMG cells from 8 HCs and SYN from 3 HCs (n = 8 matched models). Nine images (20×) per well were automatically acquired every hour and the means were then extracted and analyzed using a two-way repeated ANOVA. There was a significant interaction between the effects of time and group on the phagocytic index (F(10,105) = 3.07; P = 0.0018). Šidák’s multiple comparison tests (across the three groups at every time point) gave an adjusted significant P value of 0.005 at 4 h for the SZ-SZ group versus the HC-HC group, and at 5 h for comparison of the SZ-SZ group versus the HC-HC group (P < 0.0001), as well as the comparison of the SZ-SZ group and the SZ-HC group (P = 0.042). All other comparisons were non-significant. c, Representative live images (5 h) from SZ-SZ, SZ-HC, and HC-HC models in the experiments described in b. d, Identical live imaging design as in b but in this experiment comparing HC-HC models (based on SYNs from 3 HCs and iMG cells from 5 HC patients; n = 7 models), with HC-SZ models based on the same SYN from HCs but iMG cells derived from 3 matched SZ patients (n = 7 matched models). SZ-SZ models were based on iMG cells from 7 SZ patients and SYNs from 3 SZ patients (n = 7 matched models). Data were analyzed using a two-way repeated ANOVA. There was a significant interaction between the effects of time and group on the phagocytic index (F(10,90) = 3.89; P = 0.0002). Šidák’s multiple comparison tests (across the three groups at every time point) revealed a significant different in phagocytic indexes at 3 h between the SZ-SZ group and the HC-HC group (adjusted P value = 0.031) as well as between the HC-HC group and the HC-SZ group (adjusted P value = 0.041). The mean difference was also stable for these comparisons at 4 h (SZ-SZ versus HC-HC: P = 0.0008; HC-HC group versus HC-SZ: P = 0.011) and at 5 h (SZ-SZ versus HC-HC: P < 0.0001; HC-HC group versus HC-SZ: P = 0.023). All other group comparisons were non-significant. e, Representative live images from the different combinations in d. Scale bar, 25 μ m. f, Quantification of phagocytic inclusions (PSD-95+ inclusions, 0.5–1.5 μ m) using confocal microscopy and SYNs derived from 2 neural SZ lines matched with iMG cells from 6 SZ patients or 6 matched HCs (n = 6 models per group) with representative confocal images in g; scale bar, 30 μ m. Wilcoxon signed-rank test: P = 0.031. h, Quantification of PSD-95+ inclusions using confocal microscopy and SYNs derived from 3 neural HC lines matched with iMG cells from 6 HCs or 6 matched SZ patients (n = 14 models per group), with representative confocal images in i; Scale bar, 30 μ m. Wilcoxon signed-rank test: P = 0.017. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. All reported P values are two-sided. Mean ± s.e.m. is indicated in b and d; the median with the interquartile range is shown in f and h.
C4 SZ risk variants increase C3 deposition on neurons and synapse engulfment.
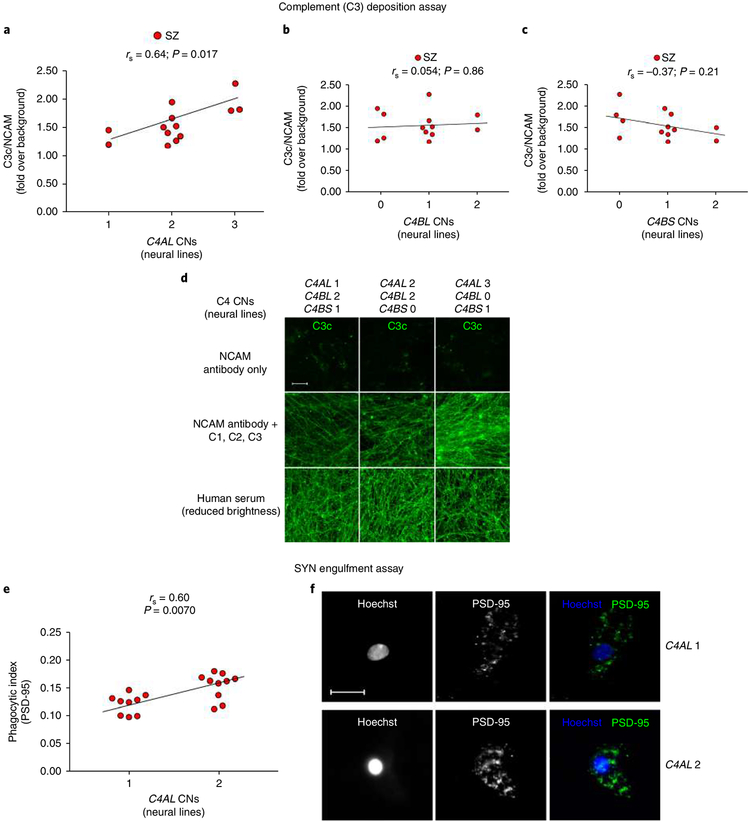
To examine whether the increased engulfment of synaptic structures by microglia in the SZ models could be partly explained by SZ risk variants at the C4 locus (previously suggested to increase C4A expression in postmortem brain)16, we first determined the C4 genotypes; then, we developed and applied a complement activation assay (Supplementary Fig. 11) to assess neuronal cell-specific C4 expression. For initial validation, we used a sample of iPSC-derived neuronal cultures from 13 individuals with childhood-onset SZ. We used droplet digital PCR to determine total copy numbers (CNs) of C4A and C4B, as well as those C4 alleles with human endogenous retroviral insertion creating the long form (C4-HERV (C4L)) and those without this insertion (short form (C4S)). C4A expression (increasing risk of SZ) is influenced by the CN of C4AL, the rare C4AS haplotype, and to a lesser extent by C4BL CN, while C4B expression is influenced by the CN of both C4BL and C4BS16. Therefore, we imputed the C4AL, C4AS, C4BL, and C4BS CNs from these patients, hypothesizing that C4A has a greater tendency to induce complement activation on neuronal structures than C4B (explaining the association between C4A expression and SZ through an effect on microglial synapse engulfment). Accordingly, we expected to observe a larger effect on complement deposition with increasing CN of C4AL and C4AS than C4BL and C4BS. To determine the effect of C4 haplotypes on complement deposition in these iPSC-derived neuronal cultures, we first treated them with an immunoglobulin M (IgM) anti-NCAM antibody to sensitize complement activation. Following antibody treatment, the upstream complement components C1, C2, and C3 were then supplemented. Because C4 is necessary for complement cascade activation, these conditions test the direct synthesis of C4 from the neuronal cultures. As predicted, we observed a strong and significant positive correlation between C4AL CN and C3 complement deposition (Fig. 5a), while no significant positive correlation was observed for C4BL CN (Fig. 5b) or C4BS CN (Fig. 5c). Because only one individual in the sample carried the rare C4AS haplotype according to our imputation, we could not directly assess the effect of C4BS CN. Sensitivity analyses, using directly genotyped C4A and C4B CN, yielded similar results (Supplementary Fig. 12). Further, to control for the tendency of complement components to deposit on neurons, we tested C3 deposition after treatment with 5% serum (excluding C5). We observed substantially greater overall C3 deposition on serum exposure, but the amount of C3 deposition did not correlate with C4A or imputed C4AL CN (Fig. 5d, and Supplementary Fig. 13). Control experiments without IgM anti-NCAM antibody pretreatment further confirmed specific C3 deposition without sensitization, with strong correlation with C3 deposition after IgM anti-NCAM (Supplementary Fig. 14), and a similar effect of C4AL CNs on C3 deposition (Spearman’s rank correlation coefficient rs = 0.78; P = 0.0025). Finally, we also analyzed a sample of iPSC-derived neural cultures from 11 healthy parents of the probands used in the analysis of SZ patients. In contrast to our analysis of SZ patients we did not observe a significant positive correlation between C4AL CN and C3 deposition in HCs (Supplementary Fig. 15a), although the effect was in the same direction (rs = 0.50; P = 0.11). This may indicate a more pronounced effect of C4AL CN on C3 complement deposition in SZ. Also, in line with the results from probands, we did not observe a significant positive effect of C4BL or C4BS CN on complement deposition in first-degree relatives (rs = − 0.064; P = 0.85, rs = − 0.57; P = 0.069, respectively, Supplementary Fig. 15b,c).
Fig. 5 |. C4 SZ risk variants increase complement deposition on neurons and increase synapse engulfment in in vitro models derived from SZ patients.
a, Significant correlation between neural C4AL CNs and neuronal C3 complement deposition; no effect of C4BL (b) or C4BS (c) CN on C3 deposition was observed. n = 13 SZ patients (means from 36 fields per well and 8 wells per line for each experimental condition. d, Representative confocal images froma,b of C3 deposition after IgM anti-NCAM antibody alone (negative control, top row), with the addition of C1, C2, and C3 (second row) across lines with increasing C4AL CNs and decreasing C4B CNs, and with the addition of C5-depleted serum (positive control, bottom row). Scale bar, 100 μ m. e, Similar correlation as in a but between C4AL CNs (SZ patients also described in Fig. 3c; n = 19 patients) and SYN uptake as measured by PSD-95+ phagocytic inclusions. f, Representative confocal images for the two groups. Scale bar, 30 μ m. All reported P values are two-sided; correlation coefficients are Spearman’s.
If C4AL CN increases neuronal complement deposition through increased neuronal C4A expression, it should be possible to observe an effect of C4AL CN on complement-dependent iMG uptake of synaptic structures. Therefore, we determined the C4AL, C4AS, C4BL, and C4BS CN status in SZ patients used for iPSC-derived neural cultures in our SYN assays. Paralleling our observation in the complement deposition assays, this revealed a strong correlation between C4AL CN and iMG synapse uptake in our functional assay (PSD-95+ inclusions; Fig. 5e,f). To test the relevance of neuronal C3 deposition for synapse pruning by iMG cells in SZ we pretreated neuron-iMG cocultures from two SZ patients with an anti-α M (clone M1/70) antibody to inhibit the microglia-specific C3 receptor. This confirmed that iMG uptake of synaptic structures in SZ largely depends on C3-C3 receptor interaction, and explains the similar effects of C4AL CNs on neuronal C3 complement deposition and engulfment of synaptic structures by iMG cells (Supplementary Fig. 16).
Since all SZ patients carried the same C4BL and C4BS CN (1 and 0 CN, respectively) we could not study the effect of C4B variants in this sample. The HCs examined in the iMG engulfment assays were likewise genotyped; consistent with our results from the complement deposition assay we observed no significant association between C4BL and C4BS CN on the phagocytic index (rs = − 0.067, P = 0.71;rs = 0.16, P = 0.36, respectively; Supplementary Fig. 14c,d). In contrast to the SZ sample, HCs displayed no variability over C4AL CN. Thus, it was not possible to directly compare the effect of neuronal C4 variants on synapse engulfment between patients and controls.
We also examined whether the C4AL risk variants in iMG cells contributed to increased synapse engulfment in our assay (PSD-95+ inclusions). The C4AL CN status in the lines used to generate iMG cells was analyzed against the phagocytic index, but without evidence of a significant correlation (rs (SZ) = 0.32, P = 0.19; rs (HC) = − 0. 067, P = 0.71; Supplementary Fig. 17). The correlation between neural C4AL CN and phagocytic index also remained significant after adjusting for C4AL CN in iMG cells used in same experiments (P = 0.034), and the difference between SZ patients and HCs remained similar and significant when compared across the same neural C4AL CN (mean difference 0.05; 95% confidence interval (CI) 0.03–0.07). These results further support our findings from the experiments with mixed combinations of patient- and HC-derived iMG cells and neuronal SYNs or neural cultures, suggesting that patient neuronal and microglial factors contribute to the observed increased uptake of synaptic structures, while indicating that variability at the C4 locus may have influence on synaptic pruning through increased C4A expression in neurons.
As previously described20, SYNs prepared from iPSC-derived neural cultures treated with astrocyte-conditioned medium contain C4 protein (Supplementary Fig. 18) and C4+ puncta within iMG cells (post-assay) strongly colocalize with PSD-95+ puncta (Costes P value for colocalization = 1; Supplementary Fig. 19). To further support the hypothesis that increased C4A expression is involved in SZ overpruning through increased complement deposition on synaptic structures, we then confirmed PSD-95+ SYN uptake by quantifying colocalized C4+ inclusions (Supplementary Fig. 20). While we cannot exclude the possibility that iMG cells upregulated C4AL expression while exposed to SYNs (in a manner not detected in our analysis of iMG C4AL CN), it is important to note that such upregulation could not explain the effect of neuronal C4AL CN on iMG uptake of PSD-95+ inclusions.
Minocycline decreases synapse engulfment in vitro.
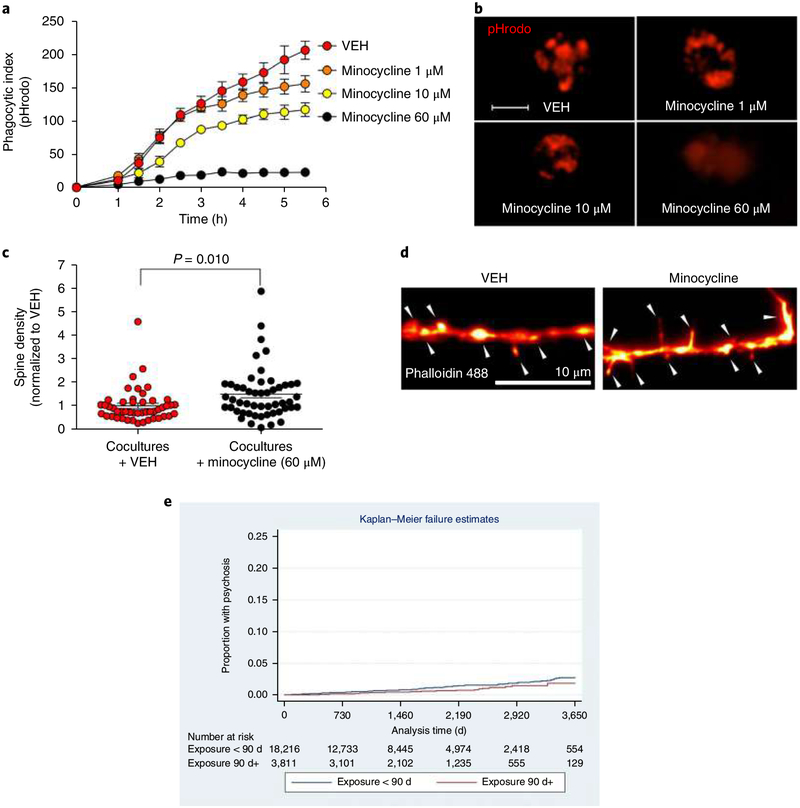
Minocycline, a semisynthetic brain-penetrant tetracycline antibiotic, has been suggested to be beneficial in neurodevelopmental and neurodegenerative diseases because of putative anti-inflammatory effects34. While its mechanism of action is unknown, it has been postulated that minocycline works by targeting synaptic remodeling35. Consistent with this hypothesis, repeated high-dose intraperitoneal administrations of minocycline in postnatal mice cause a decrease in engulfment of retinal ganglion cell inputs, a commonly used rodent model for synaptic pruning18. Clinically, however, minocycline is typically administered in lower doses, yielding significantly lower brain concentrations36,37, and human cellular data is lacking. Guided by previous rodent studies38,39 we pretreated iMG cell cultures (HCs, n = 8 patients) with minocycline in a series of clinically relevant doses (1 μM, 10 μM, and 60 μM). Real-time live imaging revealed a dose-dependent decrease in engulfment of SYNs (HCs; derived from 2 patients) with nearly undetectable SYN uptake at 60 μM (P < 0.0001; Fig. 6a,b and Supplementary Fig. 21). The effect of minocycline (60 μM) appeared to be similar in SZ patients and HCs as measured by PSD-95 uptake (mean ± s.e.m. reduction in phagocytic index in HCs (2 patients) = 0.6 ± 0.009, and in SZ (2 patients) = 0.5 ± 0.005). For validation, we also applied minocycline (60 μM) to our coculture system and confirmed a decrease in spine density in vehicle (VEH)-treated compared to minocycline-treated cocultures (Fig. 6c,d). We note that these experiments were not designed to detect more modest differences in effects nor kinetic effects.
Fig. 6 |. Minocycline inhibits synapse engulfment in vitro and decreases SZ risk in EHRs.
a, Quantification of pHrodo (red)-labeled SYN uptake in iMG cells during live imaging sessions of 5.5 h. iMG cultures (n = 8 models with iMG derived from 8 patients and SYNs from 2 patients) were pretreated for 30 min with VEH or minocycline at a concentration of 1 μ M, 10 μ M, or 60 μ M. Nine images (20×) per well were automatically acquired every hour; the means were then extracted and analyzed using a two-way repeated ANOVA. There was a significant interaction between the effects of time and group on the phagocytic index (F(30, 270) = 17.0; P < 0.0001). Šidák’s multiple comparison tests gave significant adjusted P values for all comparisons at the last time point (5.5 h); P values ranged from < 0.0001 to 0.008. b, Representative images from a displaying pHrodo+ uptake at 5.5 h in different conditions. Scale bar, 25 μ m. c, Quantification of spine density (spines per 10-μm dendrite) in a neural line derived from an HC and treated with 60 μm minocycline as indicated (n = 80 randomly selected dendrites examined in the SZ group and n = 100 randomly selected dendrites examined in the HC group). Welch-corrected t test: t(148) = 0.35, P = 0.72 (two-sided). Quantification of spine density in co-cultures (neurons and iMG cells) derived from an HC line and treated with 60 μm minocycline or VEH as indicated (n = 48 randomly selected dendrites examines in the VEH group and n = 51 in the minocycline group). Welch-corrected t test): t(87.2) = 2.66, P = 0.010. Data are normalized to the VEH group in the graph; the mean number of spines per 10 μm was 3.2 (s.e.m. = 0.34) among untreated cultures and 4.7 (s.e.m. = 0.48) in minocycline-treated cultures. d, Representative images of phalloidin-stained high-magnification confocal images of dendritic spines (indicated by the arrowheads) in VEH- and minocycline-treated cocultures in the experiments described in c. e, Time to diagnosis in individuals exposed to monocycline or doxycycline for at least 90 d or exposed for fewer than 90 d (log-rank χ2(1 d.f.) = 5.66; P = 0.017). Cumulative hazard at 10 years was 0.0185 (95% CI 0.0110–0.0312) in the exposed group, and 0.0279 (95% CI 0.0225–0.0346) in the unexposed group. Likewise, in a Cox (proportional hazards) regression model, monocycline or doxycycline exposure for at least 90 d was associated with decreased risk of incident psychosis (HR 0.58, 95% CI 0.39–0.88). Results were unchanged after excluding 672 individuals exposed to isotretinoin. The error bars in panels a and c represent s.e.m. All P values are two-sided; mean ±s.e.m. values are indicated in a and c.
Minocycline exposure during adolescence and risk of incident psychotic disorder.
These results indicate that minocycline dose-dependently decreases microglial engulfment of synaptic structures in human in vitro models of synaptic pruning at clinically relevant doses. Therefore, we hypothesized that chronic exposure to minocycline, or the chemically and mechanistically similar brain-penetrant drug doxycycline, during adolescence, that is, at a time of intense synaptic pruning14, might decrease SZ risk. These two medications are among several antibiotics commonly prescribed for the treatment of acne vulgaris40, allowing an investigation using the EHRs of two large academic medical centers with up to 10 years of follow-up. Given the challenges in terms of feasibility of prospective randomized studies, we sought to identify proof of concept that might motivate such next-step investigation, rather than definitive evidence of efficacy.
Specifically, we identified individuals who received at least one electronically prescribed antibiotic from among erythromycin, clarithromycin, minocycline, doxycycline, and trimethoprim/sulfa-methoxazole between the ages of 10 and 18. Primary analysis utilized survival methodology, with participants entering the risk pool at first antibiotic prescription and exiting at last observed ‘fact’ in the EHR, or first diagnosis of primary psychotic disorder, whichever came first. We used a Kaplan–Meier log-rank test to compare individuals who did or did not receive minocycline or doxycycline for at least 90 d in total during the risk period, a threshold selected a priori to represent chronic exposure.
In the cohort as a whole, 12,498 out of 22,027 (56.7%) individuals were female and 13,977 out of 22,027 (63.5%) were white. Mean age at entry was 14.7 years (s.d. 2.2). Of this cohort, 3,811 (17.3%) received either minocycline (2,151, 9.8%) or doxycycline (2,056, 9.3%) for at least 90 d (proportions exceed 17.3% because some individuals received both).
Among the 22,027 individuals, 203 (0.9%) received a subsequent diagnosis of a primary psychotic disorder (10th revision of the International Statistical Classification of Diseases and Related Health Problems (ICD-10) F20–F29). In Kaplan–Meier survival analysis, total follow-up (at-risk) time was 92,354 patient years, median follow-up duration was 3.8 years, and the mean was 4.2 years. Figure 6e illustrates time to diagnosis in individuals exposed to minocycline or doxycycline for at least 90 d, or exposed for fewer than 90 d (log-rank χ2 (1 d.f.) = 5.66; P = 0.017). Cumulative hazard at 10 years was 0.0185 (95% CI 0.0110–0.0312) in the exposed group, and 0.0279 (95% CI 0.0225–0.0346) in the unexposed group. Likewise, in a Cox (proportional hazards) regression model, minocycline or doxycycline exposure for at least 90 d was associated with a significantly decreased risk of incident psychosis (hazard ratio 0.58, 95% CI 0.39–0.88) after adjustment for age, sex, self-reported ethnicity, and year of entry. In sensitivity analysis, results were unchanged after excluding 672 individuals exposed to isotretinoin, another acne treatment associated with psychiatric symptoms.
Discussion
By applying cellular reprogramming methods to create patient-specific in vitro models of synaptic pruning in individuals with SZ and carefully matched HCs, we demonstrate that complement-dependent elimination of isolated synaptic structures in SZ-derived iMG cultures is increased, attributable to the effects of synapses and iMG cells. Notably, these effects are not fully attributable to human SZ risk variants within the C4 locus, although such variants are associated with increased complement deposition on neurons, as well as increased engulfment of synaptic structures by iMG cells. In a dose-dependent fashion, minocycline then inhibited the elimination of synaptic structures. Further, minocycline use during adolescence was associated with a modest but significant reduction in incidence of psychosis in a preliminary, proof-of-concept EHR cohort study.
Our experimental data using patient-derived cells offer a potential mechanism explaining structural and functional disturbances observed in SZ patients. Loss of cortical gray matter volume in SZ patients has been repeatedly observed41. Decreased synaptic density has been reported from postmortem material11–13 and functional magnetic resonance imaging studies display aberrant functional brain connectivity8–10,41. Studies of ‘at-risk’ adolescence or early adulthood populations further reveal strong evidence for an accelerated decrease in gray matter thickness in ‘converters’ to psychosis2–7, suggesting that the observed structural aberrations are already present at transition to illness and are likely to be related to disturbance in cortical maturation in the given age interval. Rodent studies demonstrate that microglia, dependent on complement signaling, play a major role in synaptic pruning17,18,42, a prominent feature of human neurodevelopment during adolescence14, while microglial activity, as measured by [11C]PBR28 positron-emission tomography scans, correlate to cortical gray matter loss in patients at ultra-high-risk of psychosis43.
Our results are consistent with, but not fully explained by, previous reports that the most strongly associated locus for SZ in meta-analysis of genome-wide association data involves structurally distinct alleles of the human C4A and C4B genes, with the risk associated at least in part with increased C4A expression (foremost C4AL CN)1,16. We found that these variants are associated with increased complement deposition on synaptic structures; in particular, by examining structural C4 alleles in human iPSC-derived neural lines, we identified an association between C4AL CN and neuronal complement deposition (C3), with no significant effect observed for C4BL (predominately affecting C4B expression) or C4BS (only affecting C4B expression). Likewise, in our engulfment assays, greater CNs of C4AL in neural lines was associated with increased uptake of synaptic structures by iMG cells, while blocking C3-C3 receptor interaction in SZ neuronal cultures drastically reduced the decrease in spine density caused by iMG cells. However, we found differences in phagocytosis that are not fully accounted for by the genetic variation at the C4 locus. Thus, our results suggest a model where SZ risk variants within the C4 locus cause an excessive neuronal complement deposition mediated by C4A, which is one contributor, but not the only contributor, to increased microglial synapse engulfment.
While we could not observe a clear effect of C4 risk variants in iMG lines, we detected SZ-associated differences in phagocytosis by iMG cells, and phagocytosis of SYNs, via our patient-derived models. In line with the demonstrated polygenicity of SZ risk1, this observation suggests that several independent factors contribute to aberrations in synaptic pruning in SZ patients. Furthermore, by pretreating iMG cells with the antibiotic minocycline, we observed a dose-dependent decrease in SYN uptake. This first observation in our human model system accords with a previous animal study reporting decreased engulfment of retinal ganglion cell inputs in mice treated with minocycline17. Engulfment of viable NPCs, a microglial function that we have previously shown can be mimicked in vitro by iMG cells20, is also inhibited by minocycline using a rodent model where an induced status epilepticus increases the numbers of progenitors in the subgranular zone of the dentate gyrus44. In contrast, minocycline does not decrease the phagocytosis of amyloid-β fibrils in precursor protein transgenic mice45, although it prevents secretion of proinflammatory cytokines45, and even rescues decreased engulfment of latex beads by adult hippocampal microglia isolated from polyinosinic:polycytidylic acid mice46. Taken together, this work demonstrates that minocycline influences microglial phagocytic function in a highly complex but dissectible manner, where factors such as activation state and given substrate may be of crucial importance.
Our investigations using naturalistic data from EHRs revealed a decrease in the risk of incident psychosis after chronic exposure to minocycline or doxycycline, but not non-tetracycline antibiotics, during adolescence. In light of the challenges in extrapolating from non-randomized, naturalistic cohorts47,48, this preliminary finding requires confirmation in prospective randomized trials. Previous clinical studies have yielded mixed effects for minocycline in small-scale clinical investigation, but importantly these studies only examined acute efficacy in chronic SZ patients. More generally, the model we describe should facilitate further drug discovery studies aiming at identifying potential targets for decreasing synaptic pruning, which could represent the first demonstrated means of preventing this chronic and disabling disorder.
Several limitations of the current study should be highlighted. First, the transcriptomic signature of human ex vivo microglia changes rather rapidly in vitro19. iMG cells, similar to recent iPSC-derived microglia-like cells and primary cultured microglia, do not fully recapitulate the molecular signature of human immediate ex vivo microglia despite adjusting conditions to mimic the brain microenvironment. Second, spine composition in iPSC-derived neural cultures may not fully recapitulate in vivo conditions49,50. However, despite these limitations, our in vitro assays using iMG cells were still able to recapitulate mechanisms observed in vivo using rodent models20. It is also worth noting that the current study lacks direct evidence supporting that the observed effects of minocycline in vitro are per se causing the decreased incidence of SZ observed in our register-based analyses. Even if this effect can be confirmed, while the timing of minocycline exposure in our health records study overlaps with cortical maturation due to extensive synaptic pruning, it is fully possible that minocycline modestly reduces the risk of transition to psychosis (in this study, ~40% reduction in risk, based on the hazard ratio) through another as-yet-unidentified mechanism.
Nonetheless, taken in aggregate, our work supports increased synapse elimination in SZ, offering a mechanism linking known genetic risk variants to observed structural brain changes in early disease. Specific pharmacological interventions targeting microglial elimination of synapses merit further study for their capacity to delay or prevent the onset of SZ in high-risk individuals.
Methods
Study design.
For the patient versus control comparison of SYN uptake by iMG cells, using live imaging, we assumed a between-individual effect size of 0.5 (repeated analysis of variance (ANOVA), α = 0.05; correlation among repeated measurements, 0.5). With a power of 0.9 this indicated a total sample size of at least 26 (n = 13 per group)51,52. Using 3 iPSC-derived neural lines from SZ patients and 3 iPSC-derived neural lines from HCs, we created 13 SZ models and 13 HCs models by combining the derived SYNs with iMG cells from 13 male SZ patients and 9 male HCs. Patients versus controls, as well as each SYN-iMG pair, were matched on age. To reduce technical variance 9 images per well were taken (fixed location per software) and averaged. The patient versus control experiments using confocal microscopy were designed to at least exceed the power in our live imaging experiments (SYNs derived from 4 patients per group and iMG cells derived from 18 HCs and 13 SZ patients; combined into a total of 33 HC models and 19 SZ models). Throughout the experiments all available samples were used. Because iMG derivation is destructive, that is, we cannot expand peripheral blood mononuclear cell-derived iMG cells, the number of samples available for any given experiment varied but still matched internally per experiment, depending on the available remaining peripheral blood mononuclear cells. If more than one buffy coat was obtained, or an individual already contributed with cells, we matched additional iMG cultures with SYNs from another neural line to maximize the numbers of unique replicates in terms of SZ and HC models. (The number of patients for each main analysis is stated in Supplementary Table 6 as well as in the figure legends.) Twenty randomly selected confocal images (per software) were taken per well and, as predefined, only images displaying > 20 cells and < 100 cells were included in further analyses. (Previous experiments in our laboratory suggest that the risk of outliers increases unacceptably outside this range.) Technical variance between wells for a given HC iMG line has then been shown to be very low20. To examine the consistency of within-individual microglial assays in a patient versus control setting and using different buffy coats, for eight randomly selected individuals (four SZ patients and four HCs) microglia samples were now derived from two different buffy coat samples. The correlation between pairs (R2) of samples was 0.92 (Supplementary Fig. 22). Investigators performing the experiments were blinded to well conditions. To analyze spine density in the coculture experiments, fields lacking identifiable neurons or spines were excluded from the analysis. For further details regarding design, see also the Life Sciences Reporting Summary.
Study cohort.
Study participants aged 18–75 years old were recruited into a tissue bank, the Massachusetts General Hospital (MGH) NeuroBANK, from outpatient clinics at MGH, as well as via advertisements and study volunteer lists. All individuals signed a written informed consent before participating in the study, as approved by the Institutional Review Board of Partners HealthCare. All relevant ethical regulations were followed when performing the study. A structured psychiatric evaluation was performed for all individuals (17 SZ patients and 18 HCs) using the Structured Clinical Interview for DSM-IV and the Mini-International Neuropsychiatric Interview by a single psychiatrist with at least 5 years of clinical experience53. Participants included in the study were randomly selected and matched by age and sex. The fibroblasts of participants for the iPSCs used in the C3 deposition assays were derived from the fibroblasts obtained from the laboratory of J. Rapoport (National Institute of Mental Health under NIH-2514–2 (10–09)). This sample contained 13 probands with childhood-onset SZ and 11 healthy first-degree relatives (parents).
Generation of iMG cells.
iMG cells were derived from monocytes using established methods previously described in detail20, but with some modifications. Briefly, the peripheral blood mononuclear cells generated from whole blood were rapidly thawed at 37 °C and diluted into prewarmed Roswell Park Memorial Institute (RPMI) 1640 medium (500 ml; Sigma-Aldrich) with 10% heat-inactivated, low-endotoxin FBS (Rockland Immunochemicals, catalog no. FBS-01–0100) and spun at 300g for 5 min (break off). The supernatant was aspirated completely and the pellet was resuspended in RPMI 1640 medium with 10% heat-inactivated, low-endotoxin FBS and 1% penicillin-streptomycin. Cells were plated onto Geltrex-coated (Thermo Fisher Scientific, catalog. no. A1048002) 24-well plates at a density of 1 × 106 cells per well and incubated overnight at 37 °C. Media was then changed to RPMI 1640 medium with GlutaMAX (Thermo Fisher Scientific) with 1% penicillin-streptomycin, 0.1 μg ml−1 of IL-34 (R&D Systems, catalog no. 5265-IL-010) and 0.01 μg ml−1 of granulocyte-macrophage colony-stimulating factor (R&D Systems, catalog no. 215-GM-010/CF). After 10 d the plates were washed thoroughly to remove unbound cells and fresh media was added. Cells were collected or used for functional assays the following day.
Collecting dermal biopsies and establishing fibroblast cultures.
Following subcutaneous injection of lidocaine (1%), a physician investigator obtained biopsies from the non-dominant upper arm (using a 3.0 or 4.0 mm punch tool). A scalpel blade was used to cut the biopsy into small pieces. The biopsy pieces were then transferred to a 60-mm tissue culture-treated dish (Corning) on which the pieces were plated dermis side down. The dish was placed in an incubator at 37 °C for 15 min for the dermal sample to adhere to the dish. Media that contained DMEM (Thermo Fisher Scientific, catalog no. 11995–065) with 10% heat-inactivated FBS (Thermo Fisher Scientific, catalog no. 10082147) and 1% penicillin-streptomycin-L-glutamine (Corning, catalog no. 30–009-CI) was gently added over the pieces of biopsy. The resulting fibroblasts were passaged, confirmed as Mycoplasma negative (MycoAlert Mycoplasma Detection Kit; Lonza), frozen down, and then thawed as needed for iPSC reprogramming.
iPSC reprogramming.
Human fibroblasts were reprogrammed, stabilized, and expanded under xeno-free conditions by Cellular Reprogramming, Inc. (www.cellular-reprogramming.com), as described previously25. Briefly, iPSC colonies were obtained using mRNA reprogramming in a feeder-free culture system. Stable iPSCs were expanded in NutriStem XF medium (Biological Industries) and on biolaminin 521 LN-coated (BioLamina) plates to at least passage 3. iPSCs were then purified using MACS with anti-TRA-1–60 MicroBeads (Miltenyi Biotec) on LS columns according to the manufacturer’s instructions. All fibroblasts and iPSCs were screened and found negative for Mycoplasma; they stained positive for octamer-binding transcription factor 4 (POU domain, class 5, transcription factor 1) and TRA-1–60.
Neural differentiation.
The induction of the NPCs used to derive neural cultures for SYN isolations was initiated by media replacement to neurobasal medium (NBM; Thermo Fisher Scientific, catalog no. 21103049) supplemented with 1× NIS (Thermo Fisher Scientific). After confluence, cells were passaged with Accutase (100 ml; Sigma-Aldrich) and purified using a PSA-NCAM MACS sorting procedure (Miltenyi Biotec). The medium was changed to a neural expansion medium (50% NBM, 50% advanced DMEM/F-12 with 1× NIS; Thermo Fisher Scientific) for further expansion. When confluent, putative NPCs were further purified by double sorting using MACS columns and CD271 and CD133 MACS kits (Miltenyi Biotec). CD271−/CD133+ NPCs were analyzed for nestin, transcription factor SOX1, transcription factor SOX2, and paired box protein Pax-6 expression by immunocytochemistry. NPCs were further differentiated into neurons for SYN isolation by plating the expanded cells at a seeding density of 40,000 cells per cm2 on poly-L-ornithine/laminin-coated 6-well tissue culture plates in neuronal differentiation medium (70% DMEM, 30% Ham’s F-12 medium (Corning)) supplemented with N-2 Supplement (Thermo Fisher Scientific, catalog no. 17502048) and B-27 supplement (Thermo Fisher Scientific, catalog no. 17504044) with medium replacement every 3–5 d for 6 weeks.
For neural culture used in the coculture experiments, we generated inducible neurogenin 2 (NGN2) expressing stable NPC lines using TALEN-based plasmids we designed to insert a doxycycline-inducible NGN2 construct into the adeno-associated virus integration site 1 (AAVS1) safe-harbor locus54,55. The doxycyclineinducible NGN2 AAVS1 knock-in plasmid, based on an AAVS1 SA-2A-puro-pA donor (plasmid no. 22075; Addgene), was generated by replacing the puromycin resistance gene with the neomycin resistance gene and cloning a cassette containing the Tet-On 3G promoter driving the human NGN2 complementary DNA followed by P2A, a zeocin resistance gene, the BGH polyA, CMV early enhancer/chicken β -actin (CAG) promoter driving Tet-On 3G and BGH polyA. The AAVS1-targeting TALENs are based on hAAVS1 1R TALEN and hAAVS1 1L TALEN (plasmid nos. 35432 and 35431, respectively; Addgene) and were generated by golden gate assembly. Plasmid maps can be provided upon request. NPCs were collected with Accutase and counted. For each transfection, 4 × 106 cells were pelleted at 300g and resuspended in 100 μl prewarmed nucleofector solution (Human Stem Cell Kit 1, Lonza, catalog no. VPH-5012); 4 μ gof NGN2 plasmid and 1.5 μg each of 1R and 1L plasmids were added directly to the resuspended cells, or negative control without plasmid, followed by nucleofection (Amaxa Nucleofector II; Lonza) according to the manufacturer’s protocol using program B-016. After nucleofection, cells were plated onto Geltrex-coated 6-well plates in neural expansion medium (50% NBM, 50% advanced DMEM/F-12 with 1× NIS). Stable lines were expanded by selection with the addition of 125 μl ml−1 G418 (Geneticin; Thermo Fisher Scientific) over 8–10 d or until the negative transfection control cells completely died. Characterization of induced NGN2 neurons shows that a majority of the neurons are glutamatergic (vesicular glutamate transporter 1-positive) and express the postsynaptic marker PSD-95 (Supplementary Fig. 23). These cells were used in iMG neural cocultures as described later.
For cortical excitatory neuronal cultures used in the C4 deposition assays, iPSCs were passaged in TeSR-E8 medium (Stem Cell Technologies) on vitronectin recombinant human protein (Thermo Fisher Scientific, catalog no. A14700) using standard conditions. Cortical excitatory neuronal differentiation is based on a modified dual SMAD inhibition protocol52 with slight modifications. Briefly, the protocol was adapted to run as an adherent culture with automated liquid handling on a Microlab STAR Liquid Handling System (Hamilton Co.) and modified to reduce the number of neural crest cells. To generate NPCs, iPSCs were plated onto vitronectin-coated 6-well plates in TesR-E8 medium. Two days before the start of differentiation the cells were switched to mTeSR1 (Stem Cell Technologies). When cells reached 80–90% confluence (day 0), ‘phase I’ medium was added: advanced DMEM/F-12 (Thermo Fisher Scientific, catalog no. 12634010), 1% GlutaMAX, 1% penicillin-streptomycin, 500 μM N-acetyl-L-cysteine (Sigma-Aldrich, catalog no. A7250), 2 μg ml−1 heparin (Sigma-Aldrich, catalog no. H3393), 10 μM SB431542 (Selleck Chemicals, catalog no. S1067), 10 μM LDN193189 (Stem Cell Technologies, catalog no. 72144), 2 μM XAV939 (Stem Cell Technologies, catalog no. 72674), and 1% N-2 Supplement (Thermo Fisher Scientific, catalog no. 17502048). The medium was exchanged 50:50 every other day until day 7, when cells were replated onto 6-well plates at 5 × 104 cells per well in ‘phase II’ medium: advanced DMEM/F-12, 1% GlutaMAX, 1% penicillin-streptomycin, 500 μM N-acetyl-L-cysteine, 2 μg ml−1 heparin, 1% B-27 Supplement (Thermo Fisher Scientific, catalog no. 17504044), 2.5 ng ml−1 bFGF recombinant human protein (Thermo Fisher Scientific, catalog no. 13256029), 20 nM CHIR99021 (Stem Cell Technologies, catalog no. 72054), 100 nM LDN193189, 5 nM all-trans retinoic acid (Stem Cell Technologies, catalog no. 72262), and 0.5% N-2 Supplement. After 28 d the medium was exchanged to induce the production of early born cortical neurons for 28 d (56 in total), with DMEM/F-12, 2% GlutaMAX, 1% MEM, NEAA (Thermo Fisher Scientific), 2% B-27, and 1% N-2 Supplement, with 10 μM forskolin (Stem Cell Technologies, catalog no. 72114) added from days 28–49 and 5 ng ml−1 brain-derived neurotrophic factor (PeproTech, catalog no. AF-450–02) and 5 ng ml−1 glial-derived neurotrophic factor (PeproTech, catalog no. AF-450–10) added from days 49–56.
Synaptic pruning in iMG neural coculture.
Twenty-four well plates were coated with 20 μg ml−1 poly-L-ornithine hydrobromide (Sigma-Aldrich, catalog no. P3655) for 1 h followed by overnight coating of 5 μg ml−1 laminin from Engelbreth-Holm-Swarm murine sarcoma basement membrane (Sigma-Aldrich, catalog no. L2020). Stable inducible NGN2 cells expressing NPC lines were collected with Accutase, pelleted, and seeded at a density of 150,000 cells per well in neuronal differentiation medium (50% DMEM/F-12, 50% NBM supplemented with 1× NEAA, 1× N-2 Supplement, 10 ng ml−1 brain-derived neurotrophic factor (Peprotech, catalog no. 450–02), 10 ng ml−1 recombinant human NT-3 (PeproTech, catalog no. 450–03). After 2 d, the medium was replaced with fresh differentiation medium including 1× B-27 and the addition of 2 μg ml−1 doxycycline (Sigma-Aldrich). The medium was changed after 2 d with the same medium and the addition of 5 μg ml−1 zeocin (Thermo Fisher Scientific); this medium was continued for 5 d, and replaced every other day. For neuronal maturation, the medium was replaced without zeocin and neural cultures were cocultured with rat primary astrocytes (a gift from the Greenberg Laboratory, Harvard Medical School) indirectly using 1.0 μM PET Millicell 24-well hanging cell culture inserts (MilliporeSigma); inserts were seeded with 6,000 astrocytes per insert in neuronal differentiation medium (NED) and placed in each neural culture well for indirect media conditioning. After21 d of neural differentiation as described, neurons were used for the coculture experiments. Mature iMG cells were collected from plates using Accutase and pelleted; 1,000 iMG cells were added to each neural culture well. Neural cultures displayed a variety of spine subtypes including thin, stubby, and mushroom (Supplementary Fig. 24). The coculture plates were then imaged using an IncuCyte ZOOM live imaging system (Essen BioScience) with images collected every 4 h for a period of 48 h. After the assay, cells were washed and fixed with 4% paraformaldehyde (PFA) for 15 min and stained with Alexa Fluor 488 phalloidin (Thermo Fisher Scientific, catalog no. A12379) according to the manufacturer’s instructions. Cells were then imaged on the IN Cell Analyzer 6000 (GE Healthcare Life Sciences) as described earlier. Randomly selected raw 16-bit TIFF files containing image data were converted to RGB images using ImageJ (National Institutes of Health (NIH)). From these images, 10 μm sections of phalloidin-stained dendritic spines were identified, cropped, and opened in SynPAnal for spine analysis by an operator (B.W.) blinded to well conditions56. The 10 μm dendrite regions were then highlighted, and the spine regions were identified and counted manually. Lastly, the numerical data were exported to a Calc spreadsheet (LibreOffice, The Document Foundation) for analysis. Further inferential statistics were performed using Prism 6.0 (GraphPad Software); 10–15 fields of view per well were analyzed, with four 10 μm process sections selected per field.
C3 functional blocking in iMG neural coculture.
NGN2 neurons were generated as described earlier. After 21 d of differentiation, mature NGN2 neurons were used for the coculture experiments with iMG cells as described earlier. For the C3 blocking experiments, neurons were pretreated for 30 min before iMG cells were added with either 6 μg ml−1 of anti-CD11b functional antibody (ab212505; Abcam) or 6 μg ml−1 of low-endotoxin, azide-free isotype control (ab18428; Abcam). The coculture was then placed in the incubator for 48 h at 37 °C and 5% CO2. Cells were fixed with 4% PFA for 15 min at room temperature, followed by staining with Alexa Fluor 488 phalloidin (catalog no. A12379) according to the manufacturer’s instructions. Images were taken using an automated high-content analysis system (ImageXpress; Molecular Devices), which took 36 random images per well. Raw TIFF files containing phalloidin 488 fluorescence data were processed using customized ImageJ software (NIH), available upon request, which performed 3 functions: (1) generated a binary mask image containing objects representing areas of high actin fluorescence in the cell and a separate set of masks representing total neuronal material per field; (2) generated an object image file that identified and numbered individual actin objects within the field of view, excluding objects based on unwanted size and circularity characteristics; and (3) measured the parameters from wanted objects, including dimensionality and frequency, and saved the results to a CSV text file. The data in the CSV files were read in RStudio (RStudio Team). A custom R routine was applied (available from https://gitlab.com/bradwatmuff/microglia-szd-paper-2018); this normalized the number of actin objects per field of view against total neuronal fluorescence. Data collection and analysis were performed with blinding to the conditions of the experiments.
Global gene expression microarray hybridization and data analyses.
Cells were collected using Accutase and pelleted by centrifugation at 300g. The supernatant was removed and the pellets were stored at − 80 °C. Total RNA was extracted using the RNeasy Mini kit (QIAGEN) according to the manufacturer’s instructions and concentrations; 260/280 nm readings were determined using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific). RNA quality was further assessed on an 2100 Bioanalyzer Instrument (Agilent Technologies) and samples with RNA integrity numbers > 6 were used for analysis. Whole-genome gene expression profiling was performed using the Clariom S Pico human gene expression assay (Thermo Fisher Scientific) with 4.5 ng total RNA per sample at the Boston Children’s Hospital IDDRC Molecular Genetics Core Facility (supported by NIH-P30-HD 18655). A GeneChip Fluidics Station 450 (Thermo Fisher Scientific) performed all the liquid handling steps and chips were scanned with the GeneChip Scanner 3000 7G (Thermo Fisher Scientific). Quality control was performed using the manufacturer’s software Transcriptome Analysis Console version 4.0 (Thermo Fisher Scientific), according to the manufacturer’s instructions (principal component analysis, labeling controls, hybridization controls, positive versus negative area under the curve, and signal box plot) with no indication of outliers. Normalization was performed according to a robust multichip analysis approach. Differentially expressed genes were identified using a one-way ANOVA. Uncorrected as well as false discovery rate-adjusted (Benjamini–Hochberg) P values are reported. To assess the uncertainty in the hierarchical cluster analysis of iMG cells versus monocytes and monocyte-derived macrophages (Supplementary Fig. 4) we applied bootstrap resampling (10,000 iterations) using the R package pvclust. For each suggested cluster pvclust provides two types of P values: the bootstrap probability value from the ordinary bootstrap resampling and the approximately unbiased probability value from multiscale bootstrap resampling. The multiscale bootstrap resampling method was introduced to develop an approximately unbiased test; therefore, it provides better estimations of the probability values36. The s.e.m. of the approximately unbiased P values were also plotted against the approximately unbiased P values with no indication of sampling error.
Isolating and labeling nerve terminals.
Twenty-four hours before collecting the neural cultures by scraping, the medium was changed to astrocyte-conditioned medium (ScienCell Research Laboratories). SYNs were then isolated as described previously using a synaptic protein extraction reagent (Syn-PER; Thermo Fisher Scientific)20. The resulting SYNs were collected and freeze-thawed (identically prepared and handled), and then pre-assay-labeled with an amine-reactive and pH-sensitive dye (pHrodo Red, succinimidyl ester; Thermo Fisher Scientific, catalog no. P36600) for real-time (live) assays according to the manufacturer’s instructions. For fixed experiments, to show colocalization with synaptic markers (Supplementary Fig. 9), SYNs were labeled with the fixable pH-sensitive dye CypHer5E NHS Ester (GE Healthcare Life Sciences, catalog no. PA15401) as described previously33.
Synaptic vesicle endocytosis and synapse markers in isolated nerve terminals.
With some modifications, we adopted the methods of Daniel et al.29 for high-throughput quantification of synaptic vesicle endocytosis to validate functionality of SYNs isolated from iPSC-derived neuronal cultures. First, 96-well plates (ibidi GmbH, catalog no. 89626) were coated with poly(ethyleneimine) (1:15,000 diluted in water; Sigma-Aldrich, P3143) at 37 °C overnight. The next day, plates were flicked to remove excess poly(ethyleneimine), further dried at 37 °C for 30 min and stored at 4 °C for a maximum of 30 d. Frozen SYNs were thawed and sonicated for 1 h at room temperature; 200 μl of Tris-HCl/EDTA/sucrose (SET) buffer (5 mM Tris-HCl, 1 mM EDTA, and 0.32 M sucrose buffer dissolved in water and adjusted to pH 7.4) was added to the SYNs and mixed thoroughly. A 1:1,000 dithiothreitol solution (Sigma-Aldrich, catalog no. D5545; 250 mM stock solution dissolved in SET buffer) was added to the SYN solution, and 100 μl of the final (300 μl) solution was added to each well to give triplicate data points. The plate was centrifuged at 1,500g for 30 min at 4 °C. HEPES-buffered Krebs-like (HBK) buffer (143 mM NaCl, 4.7 mM KCl, 1.3 mM MgSO4× 7H2O, 1.2 M CaCl2, 0.1 mM Na2HPO4, and 10 mM d-glucose dissolved in water and adjusted to pH 7.4) was prewarmed at 37 °C and infused with 5% CO2 for 30 min. After SYN centrifugation, the SET buffer was carefully removed using a multichannel pipette and 60 μ l of HBK buffer was added. The SYNs were then incubated at 37 °C for 15 min to acclimate to HBK. Simultaneously, a solution of 1.5 μ M calcein blue, AM (Thermo Fisher Scientific, catalog no. C1429) was prepared in the HBK buffer. After incubation, the HBK buffer was carefully removed and calcein blue, AM (60 μl per well) was added followed by incubation for 30 min at 37 °C in the dark. FM 4–64 dye (Thermo Fisher Scientific, catalog no. T13320) was diluted 1:100 in water for unstimulated control wells, and diluted 1:100 in 400 mM KCl for stimulated wells. These solutions were protected from light. After incubation with calcein blue, AM, SYNs were removed from the incubator and the solutions were carefully replaced with 81 μl of HBK buffer. Starting with the wells for the unstimulated controls, 9 μl of the unstimulated solution was added to the unstimulated wells, followed by 9 μl of stimulated solution for the stimulated wells, using a multichannel pipette. The plate was incubated for 2 min at 37 °C. The medium was aspirated carefully with a pipette and replaced by 81 μl of HBK buffer and 9 μl ADVASEP-7 (Sigma-Aldrich, catalog no. A3723) was added to stop depolarization and wash off the excess FM 4–64 from the plasma membranes. The plate was incubated for another 2 min at 37 °C, after which the ADVASEP-7 solution was replaced by 90 μl of HBK buffer using a multichannel pipette. The plate was then immediately imaged with an IN Cell Analyzer 6000. Forty randomly selected images were acquired per well with all settings kept constant between conditions. After imaging, plates were fixed with 4% PFA for 15 min at room temperature. The SYNs were washed carefully with PBS and incubated with anti-PSD-95 antibody conjugated to Alexa Fluor 488 (ab195004; Abcam) for 20 min at room temperature. Wells were then washed once with PBS and imaged with the IN Cell Analyzer 6000 using the same settings (including the same areas of the well) as FM 4–64. Quantification was performed using the IN Cell Analyzer 1000 software (GE Healthcare Life Sciences). The number of PSD-95+ puncta (0.5–1.5 μm) colocalized with FM 4–64 was quantified on each of the 40 randomly selected images per well and used to create a mean count (over 3 wells per line) representative of the concentration of PSD-95+ SYNs for a specific line. To ensure that our main analyses were not substantially influenced by selective bias due to differences in collected SYN counts, we also performed all main analyses adding these counts as covariates (regression modeling). Given limited line-to-line variability with a random distribution between cases and controls, all adjusted analyses displayed similar effect sizes and significances for the different phagocytic indexes (data not shown).
Transmission electron microscopy.
Isolated SYN suspensions were fixed with 2.0% glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.4; Electron Microscopy Sciences) for 2 h at room temperature, then centrifuged at 14,000 r.p.m. for 10 min to obtain a working pellet. Pellets were infiltrated in 1.0% osmium tetroxide in cacodylate buffer for 1 h at room temperature and gently rinsed several times in cacodylate buffer. Pellets were stabilized in 2% agarose; agarose blocks (containing pelleted material) were dehydrated through a graded series of ethanol to 100%, then dehydrated briefly in 100% propylene oxide. Specimens were preinfiltrated for 2 h in a 2:1 mix of propylene oxide and Eponate resin (Ted Pella), then transferred into a 1:1 mix of propylene oxide and Eponate resin and allowed to infiltrate overnight on a gentle rotator. The following day, specimens were allowed to infiltrate in fresh 100% Eponate resin for several hours, embedded in flat molds with 100% Eponate, and allowed to polymerize 24–48 h at 60 °C. Thin (70 nm) sections were cut with a Leica EM UC7 ultramicrotome (Leica Microsystems), collected onto formvar-coated grids, stained with uranyl acetate and Reynold’s lead citrate, and examined with a JEM 1011 transmission electron microscope (JEOL, Inc.) at 80 kV. Images were collected using an AMT digital imaging system (Advanced Microscopy Techniques). Electron microscopy was performed in the Microscopy Core of the Center for Systems Biology/Program in Membrane Biology at MGH, which was partially supported by an Inflammatory Bowel Disease grant (no. DK043351) and a Boston Area Diabetes and Endocrinology Research Center award (no. DK057521).
Western blot analyses.
SYNs were centrifuged at 12,000g for 15 min and pellets were resuspended in radioimmunoprecipitation assay buffer with 0.2% Triton X-100 containing protease inhibitors (Roche, catalog no. 05892791001), kept on ice for 15 min and centrifuged again at 20,000g for 10 min. The supernatant was then loaded onto a 4–20% Mini-PROTEAN TGX Precast Protein Gel (Bio-Rad, catalog no. 4561096) and run at 120 V. The gel was then transferred onto an Immobilon-FL PVDF membrane (EMD Millipore) at 120 V for 1 h. After transfer, the membrane was blocked for 1 h with Odyssey blocking buffer (LI-COR Biosciences, catalog no. 927 40100). The Western blot was then cut and incubated with primary antibodies (all Abcam) overnight at 4 °C on a shaker: anti-synaptophysin (1:7,500, ab32127); anti-SNAP25 (1:1,000, ab53723); anti-C4 (1:1,000, ab173577); or anti-beta actin loading control (1:1,000, ab8226). The Western blot was washed three times with PBS containing 0.1% Tween and incubated for 1 h at room temperature with secondary antibodies (LI-COR IRDye 800CW anti-rabbit, NC0809364; LI-COR Biosciences) diluted 1:10,000 in a buffer solution containing 50% Odyssey blocking buffer, 50% PBS, 0.1% Tween, and 0.02% SDS. After incubation, Western blots were washed three times with PBS containing 0.1% Tween and imaged with an Odyssey CLx system (LI-COR) to confirm that the SYNs were positive for typical synaptic markers.
Real-time live imaging.
We used a live cell imaging system, the IncuCyte ZOOM live imaging system (Essen Biosciences), incubated at 37 °C and 5% CO2, as described previously20, but with some modifications regarding image analyses. Briefly, sonicated and pHrodo-labeled SYNs were added to iMG cell cultures on 24-well plates and imaged at a resolution of 0.61 μm per pixel. Images (1,392 × 1,040 pixels) were further preprocessed with the IncuCyte ZOOM software (version 2016A) and exported as 16 bit grayscale PNG files. For the minocycline experiments, iMG cells were pretreated with 1 μM, 10 μM, 60 μM, or VEH for 30 min before initiation of the phagocytosis assay. Data were further analyzed in MATLAB (version R2014b; MathWorks) to quantify phagocytized particles. Briefly, unsharp masking using a 35-pixel s.d. Gaussian filter was used to remove background from the images, followed by distribution-based thresholding. For each image, the threshold was calculated as 10 s.d. above the average value for the local maxima. A threshold was applied to the resulting segmented regions to remove particles smaller than 10 pixels in area. The area of the remaining particles was summed to produce the total amount of phagocytized particles. This sum was divided by the number of iMG cells per image and represents the phagocytic index. Investigators were blinded to the conditions of the experiments during data collection and analyses.
Immunocytochemistry.
Cells were fixed in 4% PFA for 15 min at 4 °C. After fixation, cells were washed two times with PBS and then blocked for 1 h with a solution containing 5% FBS and 0.3% Triton X-100 in PBS. Cells were then washed three times for 5 min each with 1% FBS in PBS and incubated with primary antibodies (details in Supplementary Table 7) for 2 h at room temperature. After incubation, the cells were washed three times for 5 min each with 1% FBS in PBS and incubated for 1 h at room temperature in a mixture of appropriate secondary antibodies (1:500) and Hoechst dye (1:5,000) diluted in 5% FBS in PBS. Cells were then washed again three times for 5 min each with 1% FBS in PBS and imaged. Antibody specificity was confirmed by performing ‘secondary only’ experiments (data not shown). Investigators were blinded to the conditions of the experiments during data collection and analyses.
Confocal microscopy.
Images were acquired using an IN Cell Analyzer 6000. Twenty randomly selected images were acquired per well (24-well plates) with all settings kept constant between different conditions. Quantification was performed using the IN Cell Analyzer 1000 software. Nuclei were identified using a tophat transform, while cells were identified using region growing segmentation. PSD-95+ or C4+ particles were identified using a multiscale top-hat transform (10 transformations), a 0.5–1.5 μm filter, minimum sensitivity for detection of inclusions, and restriction of detection of inclusions to the cytoplasm. To measure outcome we counted the number of identified particles per iMG cell divided by the area of the cytoplasm to create a phagocytic index. The mean of the phagocytic indexes for all images per well was then used in further analyses. Investigators were blinded to the conditions of the experiments during data collection and analyses.
Molecular analyses of C4 alleles.
Similar to Sekar et al.16, we measured the CNs of each individual C4 structural allele (C4A, C4B, C4L, and C4S) using droplet digital PCR, on genomic DNA isolated from blood samples from a total of 130 controls and 54 SZ patients from the MGH Neuropsychiatric Biobank. In summary, 50 ng of genomic DNA was fragmented by digestion with the restriction enzyme AluI16. The CNs of C4A, C4B, C4L, and C4S were then quantified from genomic DNA using droplet digital PCR on a QX200 Droplet Digital PCR System (Bio-Rad Laboratories) using specific and reference primers and probes as described by Wu et al.57. Data were analyzed with the QuantaSoft software (v.1.4.0.99; Bio-Rad Laboratories), which estimated the absolute copies of C4 allele-specific DNA templates by comparison to the reference RPP30 template present as two copies per genome. C4AL, C4AS, C4BL, and C4BS CNs were determined based on the assumption of C4A CN + C4B CN = C4L CN + C4S CN, and given the low prevalence of C4S CN (around 1% in the normal population)16 C4L = C4AL CN if C4L CN ≥ C4A CN. In total, according to this imputation, we observed two patients with C4AS in the sample used for the C3 deposition assay; no patients displayed C4AS in the sample used contributing to our engulfment assays.
Complement deposition assay.
Anti-NCAM antibody (MA5324; EMD Millipore) was added to the neuronal cultures at a final dilution of 1:400 and incubated for 1 h at 37 °C. After this, complement factors C1, C2, and C3 (A098, A112, and A113 respectively; Complement Technology), at 2, 5, and 50 μ g ml−1, respectively, or 5% C5-depleted human serum (A320; Complement Technology) was added. The concentrations and durations of complement factor treatment were tested in initial pilot experiments (not shown). Plates were incubated for 1 h at 37 °C, then fixed with 4% PFA with 4% sucrose in Dulbecco’s PBS. After washing in PBS, plates were blocked in PBS + 5% goat serum + 0.1% BSA + 1 mm EDTA for 1 h at room temperature, then stained with antibodies at room temperature in the same blocking buffer for 1 h and 45 min. C3 and NCAM were visualized with fluorescein isothiocyanate-coupled rabbit anti-C3c antibody (PA1–29718, 1:100 dilution; Thermo Fisher Scientific), goat anti-mouse IgM (heavy chain) cross-adsorbed secondary antibody, Alexa Fluor 568 (A21043, 1:500; Thermo Fisher Scientific). In pilot studies, we found this C3c antibody to produce substantial signal on rodent neurons exposed to human serum but not rodent neurons exposed to heat-inactivated human serum; in rodent neurons exposed to anti-NCAM antibody plus a mixture of C1, C2, C3, and C4 complement, deposited antibody signal was detected and was increased in neurons treated with small interfering RNA (siRNA) directed against the complement regulator CD55 versus control siRNA-treated neurons (data not shown). After this incubation, the staining solution was supplemented for 15 min with CellMask Deep Red Plasma Membrane Stain (1:10,000 final dilution; Thermo Fisher Scientific) and Hoechst dye (1:5,000; Thermo Fisher Scientific) to visualize cell bodies and nuclei, respectively. After washing, wells were imaged (36 fields per well, 8 wells per line for each experimental condition) on an IN Cell Analyzer 6000. Considerable clumping and fasciculation of neurons is observed in these cultures, and there is variability in cell density from line to line. Due to the challenges of segmentation and quantification of clumped cells, we excluded clumped cells from our analysis. To do this, we first identified these clumps using size-restricted Otsu thresholding on the nuclei image. Size restriction was determined empirically. We then used these identified objects as a mask for both nuclei and CellMask channels. Using the masked nuclei image, nuclei bodies were identified. From the identified nuclei, neuronal bodies were segmented by outward propagation from the location of the nuclei objects on the masked CellMask image. The anti-C3c immunofluorescence signal and anti-IgM NCAM signal intensity were calculated in this region.
Risk of SZ in EHRs.
We utilized de-identified data from the EHRs of the Partners HealthCare system, which spans multiple academic medical centers and affiliated outpatient facilities in Massachusetts, under a protocol reviewed by the institution’s Partners Human Research Committee. The reliability of psychiatric phenotypes and medication exposure has previously been demonstrated for genetic and longitudinal outcome studies58–61, including pharmacovigilance studies of similar design62–64. Medication data reflected electronic prescribing by any clinician and included duration of prescription and number of refills. We have previously demonstrated that electronic prescribing is strongly associated with presence of a detectable blood level65. Of note, in the present analysis, diagnostic misclassification and medication non-adherence would both be expected to bias results toward the null hypothesis, that is, no effect of treatment with minocycline on psychosis risk. Follow-up continued until diagnosis of primary psychotic disorder (ICD-10 F20–F29), last available fact in the EHR, or 1 June 2018, whichever came first. A priori, we considered exposure to require a minimum of 90 d exposure to minocycline or doxycycline for two reasons. First, this exposure requires documentation of at least two prescriptions, increasing confidence in adherence. Second, very short durations could still reflect other indications for antibiotic use, while treatment for this duration is more typical in acne patients66, minimizing but not eliminating risk of confounding by indication, for example, if another chronic infection was associated with SZ risk or non-antibiotics were preferred in a particular patient subgroup.
In our primary analysis we used survival methodology (via a Kaplan–Meier log-rank test) to compare individuals who did or did not receive minocycline or doxycycline for at least 90 d in total during the proposed risk period of increased microglial synaptic pruning, an exposure threshold selected a priori to represent chronic exposure, with results stratified by age at index antibiotic, sex, self-reported ethnicity (white versus non-white), and calendar year of study entry. We further calculated hazard ratios using Cox regression adjusted for these features, after confirming the assumption of proportionality of hazards by visual inspection of Schoenfeld residuals and formal testing. Sensitivity analyses examined the effects of excluding individuals exposed to isotretinoin. We note important caveats in interpreting non-randomized data. First, recent work suggests that incorporating context and pattern of EHR data—for example, examining site effects—may improve discrimination47. In the present study, all individuals were drawn from a unified EHR precluding the examination of such effects. Second, while we have previously shown that coded data for SZ and schizoaffective disorder is reliable to distinguish serious mental illness, it remains challenging to fully exclude selection bias in EHR analyses48.
Statistics.
All analyses were performed using R and Prism 6.0, with the exception of EHR data that were extracted and transformed with the i2b2 software (version 1.6; i2b2 tranSMART Foundation) and analyzed with Stata 13 (StataCorp). mRNA expression data quality control was performed using the Transcriptome Analysis Console version 4.0, as described earlier. Graphs were produced in Prism 6.0, R, or Transcriptome Analysis Console. The assumptions of each test were checked, including normality and equal variance as applicable. The type of statistical test used is reported in the figure legends or main text. All reported P values are two-sided.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
We thank the study participants. We are also grateful to J. Ruiliera (MGH) for technical assistance with isolating buffy coats, D. Fletcher and S. Kommineni (Novartis) for stem cell reprogramming and automated neural differentiation assistance. This work was supported by grant no. P50-MH106933 (National Institute of Mental Health and National Human Genome Research Institute) and an anonymous donor to R.H.P., grant nos. 2017–02559 (Swedish Research Council) and MMW 2017.0118 (Marianne and Marcus Wallenberg Foundation) to C.M.S., and a National Institute of Mental Health Biobehavioral Research Award for Innovative New Scientists (BRAINS) no. R01MH113858 to R.K.
Footnotes
online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of data availability and associated accession codes are available at https://doi.org/10.1038/s41593-018-0334-7.
Competing interests
C.P.G., A.K., K.W., P.B.W., and J.D.B. are employees of Novartis. R.H.P. has served on the scientific advisory boards of Genomind and Psy Therapeutics, and was a consultant to RID Ventures and Takeda (none related to the present work). C.M.S. discloses lecture and consulting fees from Otsuka Pharmaceutical and H. Lundbeck A/S (none related to the present work). None of the other authors declare any competing interests.
Supplementary information is available for this paper at https://doi.org/10.1038/s41593-018-0334-7.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Data availability
Gene expression data is available for download from the NCBI Gene Expression Omnibus (GEO) at SuperSeries (NCBI GEO no. GSE123349). Additional data supporting the findings of this study are available from the corresponding authors upon reasonable request.
References
- 1.Ripke S et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cannon TD et al. Progressive reduction in cortical thickness as psychosis develops: a multisite longitudinal neuroimaging study of youth at elevated clinical risk. Biol. Psychiatry 77, 147–157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pantelis C et al. Neuroanatomical abnormalities before and after onset of psychosis: a cross-sectional and longitudinal MRI comparison. Lancet 361, 281–288 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Ziermans TB et al. Progressive structural brain changes during development of psychosis. Schizophr. Bull 38, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun D et al. Progressive brain structural changes mapped as psychosis develops in ‘at risk’ individuals. Schizophr. Res 108, 85–92 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borgwardt SJ et al. Reductions in frontal, temporal and parietal volume associated with the onset of psychosis. Schizophr. Res 106, 108–114 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Takahashi T et al. Progressive gray matter reduction of the superior temporal gyrus during transition to psychosis. Arch. Gen. Psychiatry 66, 366–376 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Meyer-Lindenberg AS et al. Regionally specific disturbance of dorsolateral prefrontal-hippocampal functional connectivity in schizophrenia. Arch. Gen. Psychiatry 62, 379–386 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Lawrie SM et al. Reduced frontotemporal functional connectivity in SZ associated with auditory hallucinations. Biol. Psychiatry 51, 1008–1011 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Stephan KE, Friston KJ & Frith CD Dysconnection in schizophrenia: from abnormal synaptic plasticity to failures of self-monitoring. Schizophr. Bull 35, 509–527 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glausier JR & Lewis DA Dendritic spine pathology in schizophrenia. Neuroscience 251, 90–107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glantz LA & Lewis DA Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch. Gen. Psychiatry 57, 65–73 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Konopaske GT, Lange N, Coyle JT & Benes FM Prefrontal cortical dendritic spine pathology in SZ and bipolar disorder. JAMA Psychiatry 71, 1323–1331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petanjek Z et al. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc. Natl Acad. Sci. USA 108, 13281–13286 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cannon TD How schizophrenia develops: cognitive and brain mechanisms underlying onset of psychosis. Trends Cogn. Sci. (Regul. Ed.) 19, 744–756 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekar A et al. Schizophrenia risk from complex variation of complement component 4. Nature 530, 177–183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevens B et al. The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Schafer DP et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gosselin D et al. An environment-dependent transcriptional network specifies human microglia identity. Science 356, eaal3222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sellgren CM et al. Patient-specific models of microglia-mediated engulfment of synapses and neural progenitors. Mol. Psychiatry 22, 170–177 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muffat J et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat. Med 22, 1358–1367 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandya H et al. Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat. Neurosci 20, 753–759 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abud EM et al. iPSC-derived human microglia-like cells to study neurological diseases. Neuron 94, 278–293.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett ML et al. New tools for studying microglia in the mouse and human CNS. Proc. Natl Acad. Sci. USA 113, E1738–E1746 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warren L et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7, 618–630 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W et al. Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc. Natl Acad. Sci. USA 108, 8299–8304 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheridan SD et al. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One 6, e26203 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dhara SK et al. Human neural progenitor cells derived from embryonic stem cells in feeder-free cultures. Differentiation 76, 454–464 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Daniel JA, Malladi CS, Kettle E, McCluskey A & Robinson PJ Analysis of synaptic vesicle endocytosis in SYNs by high-content screening. Nat. Protoc 7, 1439–1455 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Dodd PR et al. Optimization of freezing, storage, and thawing conditions for the preparation of metabolically active synaptosomes from frozen rat and human brain. Neurochem. Pathol 4, 177–198 (1986). [DOI] [PubMed] [Google Scholar]
- 31.Chung WS et al. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature 504, 394–400 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miksa M, Komura H, Wu R, Shah KG & Wang P A novel method to determine the engulfment of apoptotic cells by macrophages using pHrodo succinimidyl ester. J. Immunol. Methods 342, 71–77 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beletskii A et al. High-throughput phagocytosis assay utilizing a pH-sensitive fluorescent dye. BioTechniques 39, 894–897 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Solmi M et al. Systematic review and meta-analysis of the efficacy and safety of minocycline in schizophrenia. CNS Spectr. 22, 415–426 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Inta D, Lang UE, Borgwardt S, Meyer-Lindenberg A & Gass P Microglia activation and schizophrenia: lessons from the effects of minocycline on postnatal neurogenesis, neuronal survival and synaptic pruning. Schizophr. Bull 43, 493–496 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hersch S, Fink K, Vonsattel JP & Friedlander RM Minocycline is protective in a mouse model of Huntington’s disease. Ann. Neurol 54, 841 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Fagan SC et al. Optimal delivery of minocycline to the brain: implication for human studies of acute neuroprotection. Exp. Neurol 186, 248–251 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Raghavendra V, Tanga F & DeLeo JA Inhibition of microglial activation attenuates the development but not existing hypersensitivity in a rat model of neuropathy. J. Pharmacol. Exp. Ther 306, 624–630 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Nutile-McMenemy N, Elfenbein A & Deleo JA Minocycline decreases in vitro microglial motility, β1-integrin, and Kv1.3 channel expression.J. Neurochem 103, 2035–2046 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Del Rosso JQ Oral doxycycline in the management of acne vulgaris: current perspectives on clinical use and recent findings with a new double-scored small tablet formulation. J. Clin. Aesthet. Dermatol 8, 19–26 (2015). [PMC free article] [PubMed] [Google Scholar]
- 41.Birur B, Kraguljac NV, Shelton RC & Lahti AC Brain structure, function, and neurochemistry in schizophrenia and bipolar disorder: a systematic review of the magnetic resonance neuroimaging literature. NPJ Schizophr. 3, 15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paolicelli RC et al. Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Selvaraj S et al. Brain TSPO imaging and gray matter volume in schizophrenia patients and in people at ultra high risk of psychosis: an [11C]PBR28 study. Schizophr. Res 195, 206–214 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo C, Koyama R & Ikegaya Y Microglia engulf viable newborn cells in the epileptic dentate gyrus. Glia 64, 1508–1517 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Familian A, Eikelenboom P & Veerhuis R Minocycline does not affect amyloid β phagocytosis by human microglial cells. Neurosci. Lett 416, 87–91 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Giovanoli S et al. Preventive effects of minocycline in a neurodevelopmental two-hit model with relevance to schizophrenia. Transl. Psychiatry 6, e772 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pivovarov R, Albers DJ, Sepulveda JL & Elhadad N Identifying and mitigating biases in EHR laboratory tests. J. Biomed. Inform 51, 24–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haneuse S & Daniels M A general framework for considering selection bias in EHR-based studies: what data are observed and why? EGEMS (Wash DC) 4, 1203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kirwan P et al. Development and function of human cerebral cortex neural networks from pluripotent stem cells in vitro. Development 142, 3178–3187 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Habela CW, Song H & Ming GL Modeling synaptogenesis in schizophrenia and autism using human iPSC derived neurons. Mol. Cell. Neurosci 73, 52–62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Faul F, Erdfelder E, Lang AG & Buchner A G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 39, 175–191 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Faul F, Erdfelder E, Buchner A & Lang AG Statistical power analyses using G*Power 3.1: tests for correlation and regression analyses. Behav. Res. Methods 41, 1149–1160 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Sheehan DV et al. The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J. Clin. Psychiatry 59(Suppl 20), 22–33 (1998). [PubMed] [Google Scholar]
- 54.Oceguera-Yanez F et al. Engineering the AAVS1 locus for consistent and scalable transgene expression in human iPSCs and their differentiated derivatives. Methods 101, 43–55 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Wang C et al. Scalable production of iPSC-derived human neurons to identify tau-lowering compounds by high-content screening. Stem Cell Rep. 9, 1221–1233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Danielson E & Lee SH SynPAnal: software for rapid quantification of the density and intensity of protein puncta from fluorescence microscopy images of neurons. PLoS One 9, e115298 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu YL et al. Sensitive and specific real-time polymerase chain reaction assays to accurately determine copy number variations (CNVs) of human complement C4A, C4B, C4-long, C4-short, and RCCX modules: elucidation of C4 CNVs in 50 consanguineous subjects with defined HLA genotypes. J. Immunol 179, 3012–3025 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Castro VM et al. Validation of electronic health record phenotyping of bipolar disorder cases and controls. Am. J. Psychiatry 172, 363–372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perlis RH et al. Using electronic medical records to enable large-scale studies in psychiatry: treatment resistant depression as a model. Psychol. Med 42, 41–50 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gallagher PJ et al. Antidepressant response in patients with major depression exposed to NSAIDs: a pharmacovigilance study. Am. J. Psychiatry 169, 1065–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gainer VS et al. The Biobank Portal for partners personalized medicine: a query tool for working with consented Biobank samples, genotypes, and phenotypes using i2b2. J. Pers. Med 6, E11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Castro VM et al. Stratifying risk for renal insufficiency among lithium-treated patients: an electronic health record study. Neuropsychopharmacology 41, 1138–1143 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Castro VM et al. Absence of evidence for increase in risk for autism or attention-deficit hyperactivity disorder following antidepressant exposure during pregnancy: a replication study. Transl. Psychiatry 6, e708 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Balekian DS et al. Pre-birth cohort study of atopic dermatitis and severe bronchiolitis during infancy. Pediatr. Allergy Immunol. 27, 413–418 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roberson AM, Castro VM, Cagan A & Perlis RH Antidepressant nonadherence in routine clinical settings determined from discarded blood samples. J. Clin. Psychiatry 77, 359–362 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Bienenfeld A, Nagler AR & Orlow SJ Oral antibacterial therapy for acne vulgaris: an evidence-based review. Am. J. Clin. Dermatol 18, 469–490 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.