Abstract
Neutrophils operate at the site of injury or inflammation in the periodontal pocket to ensure periodontal health and clearance of bacterial pathogens. Filifactor alocis is recently identified as a potential periodontal pathogen, and in this study, we assessed the formation of neutrophil extracellular traps (NETs), in response to the presence of the organism. NET formation by human neutrophils was not induced when challenged with F. alocis, independent of opsonization, viability, time, or bacterial dose. F. alocis also failed to induce NETs from TNF-α-primed neutrophils and did not induce the release of extracellular neutrophil elastase. However, significant NET induction was observed when neutrophils were challenged with Streptococcus gordonii or Peptoanaerobacter stomatis, In addition, co-infection studies revealed that the presence of F. alocis with S. gordonii or P. stomatis does not enhance or reduce NETs. Additionally, F. alocis failed to impact pre-formed NETs induced by either S. gordonii or P. stomatis. Pretreatment with F. alocis prior to stimulation with phorbol 12-myristate 13-acetate (PMA), S. gordonii, or P. stomatis revealed that the bacterium is capable of reducing only PMA but not S. gordonii or P. stomatis NET formation. These results indicate that F. alocis manipulates neutrophils, inhibiting the triggering of NET induction.
Keywords: Filifactor alocis, human neutrophils, neutrophil extracellular trap formation, immunomodulation, oral pathogens
Introduction
Periodontitis is a microbially-induced chronic inflammatory disease that affects the gingival tissues supporting the tooth. This multifactorial chronic inflammatory condition of the periodontium represents one of the most common infectious diseases of humans. Indeed, it is estimated that over half the adult population in the USA will experience some form of periodontal disease.1–4 Periodontal disease and the presence of periodontal pathogens have been linked to an increased risk for the development of serious systemic inflammatory conditions such as atherosclerosis, diabetes, and rheumatoid arthritis.2,5 The contribution to periodontal disease of a relatively small group of putative pathogens including Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola is well established.1,6 However, the recent identification of bacterial species directly from nucleic acid sequencing, has led to a reappraisal of the microbial etiology of periodontitis. Several newly recognized organisms, such as Filifactor alocis, have been found to be elevated in periodontal disease sites as compared to healthy sites.5,7,8 F. alocis is a slow-growing, Gram-positive anaerobe and possesses a number of virulence properties consistent with a role in periodontal disease.9–13
Neutrophils are professional phagocytic polymorphonuclear leukocytes that function as the principal innate immune cell abundantly recruited to the periodontal pocket, in response to both bacterial- and host-derived factors. They contribute to the maintenance of periodontal health through employing various killing mechanisms intended to protect the tissue against bacterial infection.14,15 One such mechanism of host-defense described in 2004 by Brinkmann et al. is the formation of neutrophil extracellular traps (NETs).16,17 Although significant progress has been made since the discovery of NETs, questions still remain regarding their composition, role, regulation, and contribution to disease development and progression.18 Additionally, findings from studies of NETs remain controversial, especially whether their effects are beneficial to the host, as they trap and kill microorganisms, or detrimental to the host, as they extrude self-DNA potentially triggering an auto immune response.18 Regulation of NET formation is therefore crucial to ensure production and clearance processes occur when it is most beneficial to the host.19
In NET formation, neutrophils undergo nuclear decondensation, cell membrane disintegration, and eventual extrusion of their DNA extracellularly.20 The DNA structures contain numerous granular proteins, such as myeloperoxidase (MPO), neutrophil elastase (NE), and histones, which give NETs their antimicrobial properties.20,21 This is a dynamic process which can occur in viable or dying neutrophils, and is effective in trapping bacteria due to electrostatic charge interactions and in killing due to localized high concentrations of antimicrobial peptides.20,22–25 Depending on the stimuli used for neutrophil activation, NADPH oxidase activation and production of intracellular reactive oxygen species (ROS) may or may not be precede and be required for NET formation.21,22,25–27
In the oral cavity, it has been reported that NETs are formed in the gingival epithelium and can initiate a first response to periodontal bacteria.28 In the context of periodontal disease, both excessive and ineffective NET production have been associated with development of the disease, highlighting the importance of strict regulation.29 In order to combat NETs produced by the host, several oral bacteria can produce DNases, which degrade DNA, rendering NETs ineffective.29 However, it is also possible that in the environment rich with crevicular exudate outflow, bacterial DNases may not be able to function optimally in degrading NET DNA, and thus NETs may effectively entrap and clear pathogens.29
In this study, we examined the effect of F. alocis on NET formation by human neutrophils. We tested the hypothesis that F. alocis stimulation inhibits NET responses in neutrophils. In addition, we compared NET responses between F. alocis and two other oral bacteria.
Materials and methods
Neutrophil isolation
Blood draws were performed from healthy donors and neutrophils were isolated using plasma-Percoll gradients as previously described.30 Recruitment of donors as well as the blood draws were in accordance with the guidelines approved by the Institutional Review Board of the University of Louisville. Isolated cells showed that ≥90–95% were neutrophils by microscopic evaluation of cytospins and Wright staining. Trypan blue exclusion indicated that >97% of cells were viable.
Bacterial strains, growth conditions, and preparation
F. alocis ATCC 38596 was cultured in brain heart infusion (BHI) broth supplemented with L-cysteine (0.1%) and arginine (20%) for 7 d anaerobically at 37℃ as previously described.10,31 Opsonized F. alocis was prepared in 10% normal human serum at 37℃ for 20 min, and cultures were washed three times with PBS prior to use (Complement Technology, Tyler, TX). For fluorescence microscopy assays, F. alocis was labeled with 5(6)-carboxyfluorescein N-hydroxysuccinimide ester (CFSE; Life Technologies, 4 mg/ml) for 30 min at room temperature (20–25℃) in the dark, and the cultures were washed three times with PBS prior to use. Streptococcus gordonii strain DL1 was cultured in BHI broth overnight (20 h) anaerobically at 37℃. Peptoanaerobacter stomatis, strain CM2,32 was cultured in trypticase soy broth supplemented with 20 g/l yeast extract, 1% hemin, and 1% reducing agent overnight anaerobically at 37℃ as previously described.33 P. stomatis and S. gordonii were labeled with CFSE (4 mg/ml) or hexidium iodide (HI; Life Technologies, 5 mg/ml) for 30 min at room temperature in the dark, and the cultures were washed three times with PBS prior to use.
NETs immunofluorescence microscopy
To assess NET formation by neutrophils, we used an adaption of a previously described method.34 Neutrophils (1 × 106 cells/condition) were seeded onto sterile 12 mm coverslips in a 24-well plate in NETs assay media (RPMI + 0.5% BSA + 10 mM HEPES) and incubated for 1 h at 37℃ with 5% CO2 to allow cells to attach to coverslips. After 1 h incubation, neutrophils were left unstimulated, or stimulated with phorbol 12-myristate 13-acetate (PMA, Sigma, 50 nM), or challenged with CFSE-labeled non-opsonized or opsonized F. alocis, CFSE-labeled S. gordonii, or CFSE-labeled P. stomatis. Neutrophil challenges with PMA or the different oral bacteria were for 180 min.
For co-infection studies, neutrophils were challenged with HI-labeled S. gordonii + CFSE-labeled non-opsonized or opsonized F. alocis for 180 min, or with HI-labeled P. stomatis + CFSE-labeled non-opsonized or opsonized F. alocis for 180 min.
For degradation of pre-formed NETs, neutrophils were challenged with HI-labeled S. gordonii or P. stomatis for 90 min to induce NETs. Following the initial 90 min of S. gordonii or P. stomatis stimulation, neutrophils were challenged with CFSE-labeled non-opsonized or opsonized F. alocis for an additional 90 min. For the priming assays, neutrophils were stimulated with TNF-α (2 ng/ml) or stimulated with TNF-α for 10 min followed by a challenge with either CFSE-labeled non-opsonized or opsonized F. alocis. For inhibition of NETs, neutrophils were stimulated with PMA (50 nM, 180 min) or challenged with HI-labeled S. gordonii or HI-labeled P. stomatis or pre-treated with CFSE-labeled non-opsonized or opsonized F. alocis (MOI 10, 60 min) followed by the different NET inducers PMA or HI-labeled S. gordonii or HI-labeled P. stomatis.
For bacterial-challenged conditions, phagocytosis was synchronized by centrifugation at 600 g at 14℃ and plates were incubated at 37℃ with 5% CO2 for the different experimental conditions. Followed by 2% paraformaldehyde for 2 h and blocked overnight at 4℃ with 1% BSA. Following day cells were stained with MPO Ab (Biolegend, 667802, 1:1000) at 37℃ with 5% CO2 for 1 h. Followed by washings steps and staining with secondary Ab AlexaFluor 647 (Life Technologies, 1:1000). DAPI (3 µM) was applied for 5 min at room temperature as a nuclear stain and cells were washed in PBS 1 time for 5 min. Confocal images (1 µm thickness for each slice) were obtained using a Fluoview FV1000 confocal microscope with a 63X oil objective to determine NET induction. Ten images taken randomly from different regions of each coverslip in an experiment were taken.
To quantify the NET formation, we used methods previously described.19 The image files were loaded as separate image stacks for each channel in ImageJ/FIJI software. To collect the data of total cell number in the DAPI fluorescence image stack, automatic particle analysis was set to 20 pixels minimum size and summarized the result output. To collect the data of total cell number in the MPO fluorescence image stack, automatic particle analysis was set to 75 pixels minimum size and summarized the result output. The output list results were imported into an Excel spreadsheet for further processing. The percentage of NETs formed was calculated by the following formula: NET-rate (%) = 100 × objects counted (MPO channel)/objects counted (DAPI channel). The percentage of NETs formed was calculated for each of the ten images per condition acquired and then summarized as an average per condition.
NE extracellular release assay
The Cayman Chemical NETosis assay kit, which allowed for the detection of extracellular NE present on NETs and distinguish it from the NE release due to granule exocytosis, was used to quantify NETs induced by F. alocis. Neutrophils (1 × 106 cells/condition) were seeded into a 24-well plate in NETs assay media (RPMI + 0.5% BSA + 1 M CaCl2) and incubated for 30 min at 37℃ with 5% CO2 to allow cells to settle. After 30 min incubation, neutrophils were left unstimulated, or challenged with non-opsonized or opsonized F. alocis for different time points. Phagocytosis was synchronized by centrifugation at 600 g at 14℃. The assay was performed according to the manufacturer’s protocol. The ability of extracellular NE, present in the supernatants, to cleave the synthetic substrate was measured at 405 nm using a SpectraMax Soft Max Pro 5.4 spectrophotometer. NE values are expressed as mU/ml.
Statistical analysis
For all the experimental conditions tested in this study, data were statistically analyzed by one-way ANOVA with the Tukey–Kramer multiple-comparison test or paired two-tailed Student’s t-test (GraphPad Prism software, San Diego, CA, USA). Differences were considered statistically significant at the level of P < 0.05.
Results
F. alocis challenge of human neutrophils resulted in minimal NET induction, independent of opsonization or bacterial dose
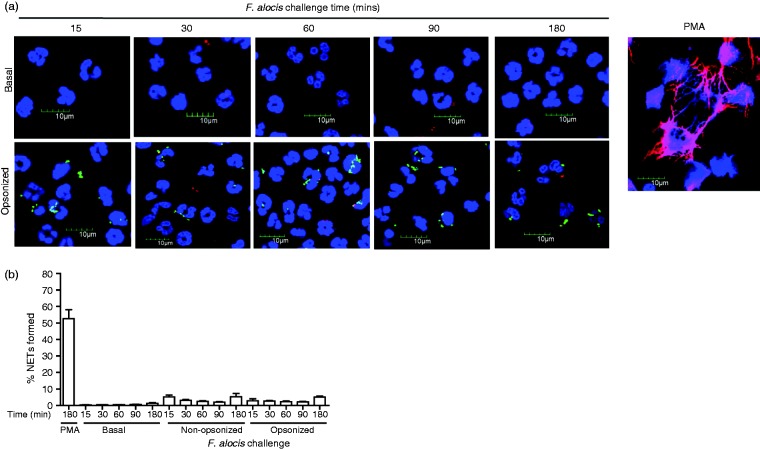
Since NETs have implications in development of periodontal disease and can be found in oral biofilms, we used immunostaining to determine if F. alocis is capable of inducing NET formation in human neutrophils.23,29,35 As it has been demonstrated that NETs can be induced at an early or late time point depending on the stimulus,22,23 we performed a time course of F. alocis challenge. Assessed by confocal microscopy, PMA, a known pharmacological stimulus of NETs at 180 min, induced robust NET formation, as indicated by the presence of colocalization of neutrophil DNA and MPO, an antimicrobial protein present in azurophil granules (Figure 1). However, quantification of NET formation from 10 different fields of view, of neutrophils from three independent donors challenged with opsonized F. alocis induced minimal NET formation over 15–180 min. Less than 5% NETs were produced in response to F. alocis, similar to the level with unchallenged neutrophils (Figure 1). Previous studies determined that the presence of serum and complement could have a negative impact on NET induction,25,36 therefore we challenged neutrophils with non-opsonized F. alocis. Similar to the opsonized bacteria and the unchallenged neutrophils, the non-opsonized F. alocis induced minimal NET formation across the time course (Figure 1b). These data suggest that failure of F. alocis to induce NETs is independent of serum opsonization. Several studies have reported that NET induction by neutrophils requires the extracellular release of NE,37–39 and thus we also measured NE in neutrophil supernatants. High levels of NE were detected from the supernatants collected after 2 h stimulation with the positive control, PMA (46 mU/ml, n = 2 independent donors). However, between 15 and 60 min after F. alocis challenge, NE levels were lower than the levels of unstimulated cells (data not shown). At the 2 h time point, similar low levels of NE were detected in the supernatants from non-opsonized (12 mU/ml, n = 2 independent donors) or opsonized (12.7 mU/ml, n = 2 independent donors) F. alocis. Overall these results indicate that F. alocis does not induce NET formation independent of opsonization or time.
Figure 1.
F. alocis challenge fails to induce NET formation by human neutrophils independent of time or serum opsonization. Neutrophils were unchallenged (Basal), exposed to PMA (50 nM, 180 min) or challenged with CFSE-labeled viable opsonized or non-opsonized F. alocis (MOI 10) for 15, 30, 60, 90, and 180 min. Following infection, cells were fixed and immunostained using Abs directed against MPO (AlexaFluor647), DNA stained with DAPI, and imaged for NET immunofluorescence by confocal microscopy. (a) Representative confocal images (from 4 independent experiments of 100 quantified cells per experiment) of basal neutrophils, neutrophils challenged with CFSE-labeled opsonized F. alocis or neutrophils exposed to PMA (180 min). CFSE-F. alocis (shown in green); Neutrophil nucleus/DNA-DAPI (shown in blue); neutrophil MPO (AlexaFluor647 shown in red), Merge image: NET formation. (b) Quantification of percentage of NETs formed using ImageJ analysis. Data are expressed as means of % NETs formed ± SEM from four independent experiments.
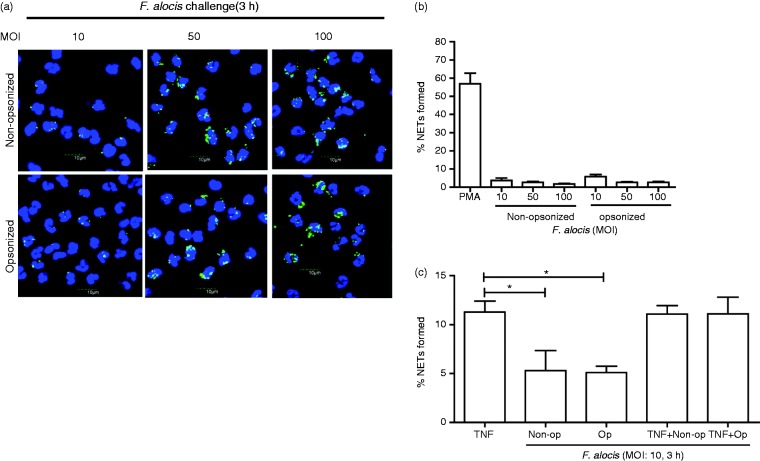
Previous studies have reported an association between increasing amounts of bacteria and the induction of NETs,40,41 therefore we examined whether higher MOIs of F. alocis from MOI 10 to 100, would induce significant NET formation. Minimal NET formation by neutrophils challenged with MOI 10, 50, and 100 of non-opsonized or opsonized F. alocis was observed (Figure 2a and b).
Figure 2.
F. alocis challenge at increasing MOIs fails to induce NET formation by human neutrophils. Neutrophils were challenged with CFSE-labeled viable non-opsonized F. alocis (MOI 10, 50, and 100) or with opsonized F. alocis (MOI 10, 50, and 100) for 180 min. Following infection, cells were fixed and immunostained using Abs directed against MPO (AlexaFluor647), DNA was stained with DAPI, and imaged for NET immunofluorescence by confocal microscopy. (a) Representative confocal images (from 3 independent experiments of 100 quantified cells per experiment) of neutrophils challenged with CFSE-labeled non-opsonized F. alocis (MOI 10-50-100) for 180 min or neutrophils challenged with CFSE-labeled opsonized F. alocis (MOI 10-50-100) for 180 min. CFSE-F. alocis (shown in green); neutrophil nucleus/DNA-DAPI (shown in blue); neutrophil MPO (AlexaFluor647 shown in red), merge image: NET formation. (b) Quantification of percentage of NETs formed using ImageJ analysis. Data are expressed as means of % NETs formed +/- SEM from 3 independent experiments. (c) Neutrophils were stimulated with TNF-α (10 min) or challenged with CFSE-labeled non-opsonized F. alocis (MOI 10), or CFSE-labeled opsonized F. alocis (MOI 10) for 180 min, or pretreated with TNF-α (10 min) followed by CFSE-labeled non-opsonized F. alocis (MOI 10) challenge, or CFSE-labeled opsonized F. alocis (MOI 10) challenge for 180 min. Following infection, cells were fixed and immunostained using Abs directed against MPO (AlexaFluor647), DNA stained with DAPI, and imaged for NET immunofluorescence by confocal microscopy. Quantification, using ImageJ analysis, of percentage of NETs formed from neutrophils at the different experimental conditions detailed above was performed. Data are means ± SEM from four independent experiments. *P < 0.05
Primed neutrophils operate in a pre-activated state, where upon encountering a secondary stimulus, they will induce a robust response,42 and it has been reported that neutrophils from patients with periodontal disease display a primed phenotype.43 As it has been determined that priming of neutrophils can enhance NET production,17 we sought to determine if primed neutrophils that were subsequently challenged with F. alocis would induce significant NET production. Neutrophils were pretreated with TNF-α, a known priming agent, and then challenged with non-opsonized or opsonized F. alocis for 180 min. The results showed that exposing neutrophils to TNF-α alone for 3 h induced significantly higher NET formation compared to cells challenged with either non-opsonized or opsonized F. alocis (Figure 2c). No increase in NET formation was observed when TNF-α-primed neutrophils were challenged with non-opsonized or opsonized F. alocis compared to the TNF-α alone condition (Figure 2c). Overall these data show that F. alocis does not induce NET formation when exposed to naïve or primed neutrophils.
F. alocis cannot significantly enhance or inhibit NETs in a co-infection setting and cannot degrade pre-formed NETs
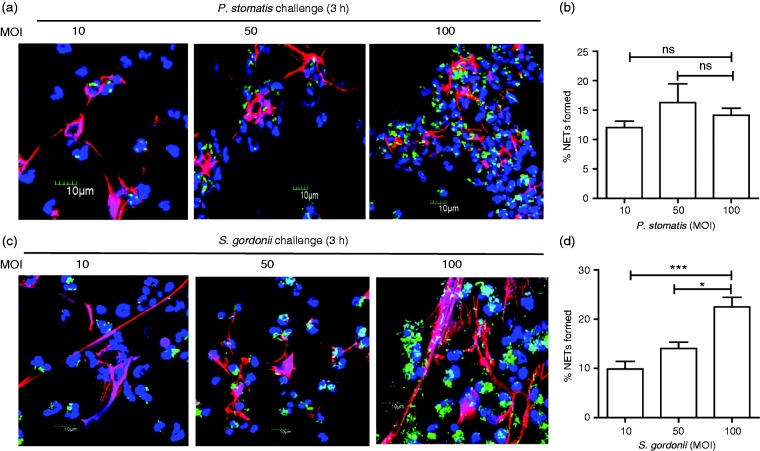
Our results thus far indicate that NETs were not formed when neutrophils were infected with F. alocis. Next, we wanted to determine if other Gram-positive oral bacteria would trigger a similar neutrophil response. Neutrophils were challenged with the emerging oral pathogen P. stomatis, which strongly activates human neutrophils by inducing high ROS production and degranulation;33 however, the ability of this organism to trigger NET formation has not been described. In addition, the ability of the oral commensal S. gordonii to induce NET formation was previously reported,44 and thus served as a good positive control for this functional response. After challenging neutrophils with P. stomatis or S. gordonii at MOI 10 for 15–180 min, NET formation was detected for both organisms only at the 180 min (data not shown). Next, neutrophils were challenged with increasing MOIs from 10 to 100 with either P. stomatis or S. gordonii and NET formation visualized by confocal microscopy after 180 min (Figure 3a and c). P. stomatis induced NET formation which was similar across MOI 10, 50 and 100, while S. gordonii elicited a dose-dependent increase in NET formation (Figure 3b and d).
Figure 3.
Both P. stomatis and S. gordonii induce NET formation by human neutrophils. Neutrophils were unchallenged (Basal), or challenged with CFSE-labeled P. stomatis (MOI 10, 50, and 100), or CFSE-labeled S. gordonii (MOI 10, 50, and 100) for 180 min. Following infection, cells were fixed and immunostained using Abs directed against MPO (AlexaFluor647), DNA stained with DAPI, and imaged for NET immunofluorescence by confocal microscopy. (a, c) Representative confocal images (from 3 independent experiment of 100 quantified cells per experiment) of CFSE-labeled P. stomatis or S. gordonii challenged neutrophils at 180 min at MOI 10, 50 and 100, respectively. CFSE-P. stomatis or S. gordonii (shown in green); neutrophil nucleus/DNA-DAPI (shown in blue); neutrophil MPO (AlexaFluor647 shown in red); merge image: NET formation. (b, d) Quantification of percentage of NETs formed, by P. stomatis or S. gordonii, using ImageJ. Data are expressed as means of % NETs formed ± SEM from three independent experiments. *P < 0.05, ***P < 0.0001. ns, non-significant.
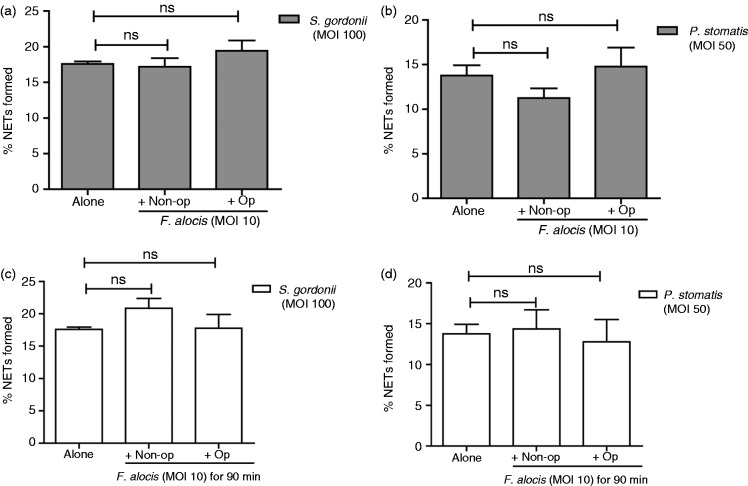
To investigate the impact of F. alocis on NETs in the context of an oral community, we performed co-infection studies with F. alocis and S. gordonii or P. stomatis. In comparison with S. gordonii (MOI 100) alone at 180 min, there was no significant change in NET formation when neutrophils were co-infected with non-opsonized or opsonized F. alocis (MOI 10) and S. gordonii (MOI 100) for 180 min (Figure 4a). Similarly, no significant change in NET formation was observed in co-infected neutrophils with P. stomatis (MOI 50) with either non-opsonized or opsonized F. alocis (MOI 10) compared to P. stomatis alone (Figure 4b). These results show that in co-infected neutrophils, F. alocis could not inhibit or prevent S. gordonii and P. stomatis induction of NETs. We next sought to determine if F. alocis could actively degrade NETs that were pre-formed by S. gordonii and P. stomatis. Neutrophils were challenged with S. gordonii (MOI 100) for 90 min followed by challenge with either non-opsonized or opsonized F. alocis (MOI 10) for additional 90 min. No degradation of NET formation induced by S. gordonii was observed in the presence of F. alocis (Figure 4c). Similarly, when neutrophils were challenged with P. stomatis (MOI 50, for 90 min), no degradation of NETs were observed with the subsequent presence of F. alocis (Figure 4d). These results indicate that F. alocis lacks the ability to degrade NETs that were formed by S. gordonii or P. stomatis challenge.
Figure 4.
In co-infection studies, F. alocis has no impact on either inhibition of NET formation or degradataion of pre-formed NETs induced by S. gordonii or P. stomatis. (a) Neutrophils were challenged with HI-labeled S. gordonii (MOI 100, Alone), or co-infected with HI-labeled S. gordonii (MOI 100) + CFSE-labeled non-opsonized F. alocis (MOI 10, Non-op) or CFSE-labeled opsonized F. alocis (MOI 10, Op) for 180 min; or (b) challenged with HI-labeled P. stomatis (MOI 50, Alone), or co-infected with HI-labeled P. stomatis (MOI 50) + CFSE-labeled non-opsonized F. alocis (MOI 10, Non-op) or CFSE-labeled opsonized F. alocis (MOI 10, Op) for 180 min. Following infection, cells were fixed and immunostained using Abs directed against MPO (AlexaFluor647), DNA stained with DAPI, and imaged for NET immunofluorescence by confocal microscopy. (a, b) Quantification of percentage of NETs formed using ImageJ analysis. In (a), data are expressed as means of % NETs ± SEM from four independent experiments. In (b), data are means ± SEM from three independent experiments. ns, non-significant. (c) Neutrophils were challenged with HI-labeled S. gordonii (MOI 100, Alone) for 180 min, or HI-labeled S. gordonii (MOI 100) for 90 min and then infected with CFSE-labeled non-opsonized F. alocis (MOI 10, Non-op) or CFSE-labeled opsonized F. alocis (MOI 10, Op) for additional 90 min or (d) challenged with HI-labeled P. stomatis (MOI 50, Alone) for 180 min, or HI-labeled P. stomatis (MOI 50) for 90 min and then infected with CFSE-labeled non-opsonized F. alocis (MOI 10, Non-op) or CFSE-labeled opsonized F. alocis (MOI 10, Op) for additional 90 min. Following infection, cells were fixed and immunostained using Abs directed against MPO (AlexaFluor647), DNA stained with DAPI, and imaged for NET immunofluorescence by confocal microscopy. (c, d) Quantification of percentage of NETs formed using ImageJ analysis. Data are expressed as means of % NETs ± SEM from three independent experiments. ns, non-significant.
F. alocis can manipulate neutrophils ability to induce NETs
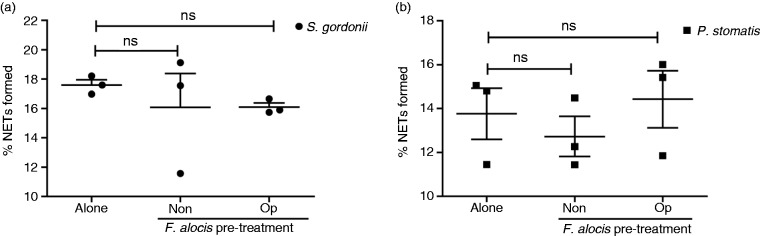
Thus far, our data showed that in co-infection studies F. alocis was not able to prevent NETs induced by the other two Gram-positive oral bacteria; next, we wanted to determine if pre-exposure of neutrophils to F. alocis could inhibit bacteria-induced NETs. To test this hypothesis, neutrophils were pre-treated with non-opsonized or opsonized F. alocis for 60 min before S. gordonii challenge. Quantification of NET formation from three independent donors, showed no significant change when neutrophils were pre-treated with either non-opsonized or opsonized F. alocis compared to S. gordonii alone (Figure 5a). Similarly, pre-treatment of neutrophils with either non-opsonized or opsonized F. alocis prior to P. stomatis challenge resulted in no significant change when compared to the percentage of NETs formed by P. stomatis alone (Figure 5b).
Figure 5.
Pretreatment with F. alocis, in the presence or absence of serum, had no effect on either S. gordonii or P. stomatis-induced NETs. (a) Neutrophils were challenged with HI-labeled S. gordonii (MOI 100, Alone) for 180 min or pre-treated with non-opsonized (Non-op) or opsonized (Op) CFSE-labeled F. alocis for 60 min and then challenged with HI-labeled S. gordonii (MOI 100) for 180 min. (b) Neutrophils were challenged with HI-labeled P. stomatis (MOI 50, Alone) for 180 min or pre-treated with non-opsonized (Non-op) or opsonized (Op) CFSE-labeled F. alocis for 60 min and then challenged with HI-labeled P. stomatis (MOI 50) for 180 min. In both (a) and (b), following infection, cells were fixed, exposed to Abs directed against MPO (Red- AlexaFluor647), stained with DAPI, and then imaged for NET immunofluorescence by confocal microscopy. Quantification of percentage of NETs formed using ImageJ analysis. Data are expressed as means of % NETs ± SEM from three independent experiments. ns, non-significant.
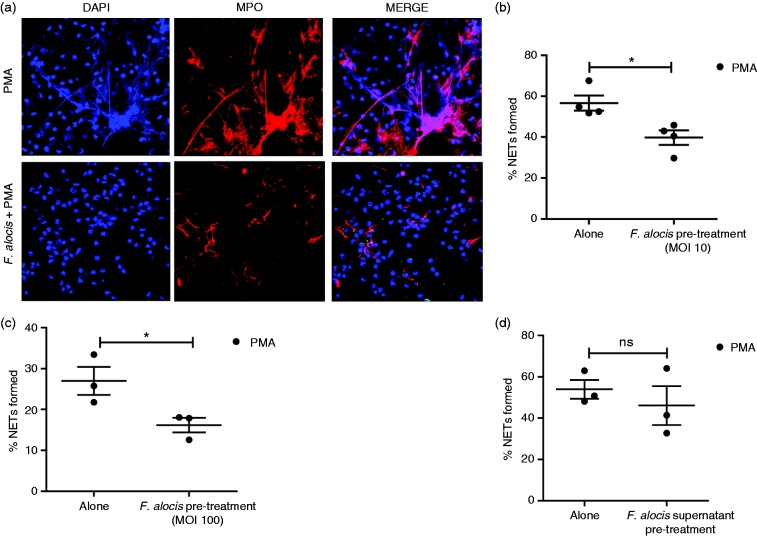
Next, we wanted to determine if F. alocis pre-treatment of neutrophils could impact PMA induction of NETs (Figure 6a). There was a significant 30% reduction in NET formation when neutrophils were pretreated with opsonized F. alocis (MOI 10) followed by PMA stimulation when compared to PMA alone (Figure 6b). Pre-treatment of neutrophils with opsonized F. alocis at a higher MOI of 100, showed a significant 40% inhibition of PMA-induced NETs (Figure 6c). A similar inhibitory effect was observed when neutrophils were pre-treated with non-opsonized F. alocis followed by PMA (data not shown). Next, we wanted to determine if F. alocis inhibition of PMA-induced NETs was mediated by secreted F. alocis products. To test this possibility, neutrophils were pre-treated with F. alocis culture supernatants followed by PMA stimulation. The results showed that pre-treatment with the F. alocis culture supernatant had no inhibitory effect on PMA induced NET formation compared to PMA alone (Figure 6d). Overall, these results indicate that pre-treatment with F. alocis, but not its secreted products, could inhibit PMA-induced NET formation.
Figure 6.
F. alocis pre-treatment causes a significant decrease in NET formation induced by PMA. Neutrophils were challenged with PMA for 180 min, or pre-treated with opsonized CFSE-labeled F. alocis (MOI 10 or 100) for 60 min and then exposed to PMA for 180 min. Following infection, cells were fixed and immunostained using Abs directed against MPO (AlexaFluor647), DNA stained with DAPI, and imaged for NET immunofluorescence by confocal microscopy. (a) Representative confocal images (from 3 independent experiment of 100 quantified cells per experiment) of neutrophils stimulated with PMA only (Alone), or pretreated for 60 min with op-CFSE-labeled F. alocis followed by PMA stimulation for 180 min. Neutrophil nucleus/DNA-DAPI (shown in blue); neutrophil MPO (AlexaFluor647 shown in red), Merge image: NET formation and CFSE-F. alocis (shown in green). (b, c) Quantification of percentage of NETs formed using ImageJ analysis. Data are expressed as means of % NETs ± SEM from four independent experiments in (b) and three independent experiments in (c) * P < 0.05. (d) Neutrophils were challenged with PMA only (Alone) for 180 min, or pre-treated with F. alocis culture supernatants for 60 min and then exposed to PMA for 180 min. Quantification of percentage of NETs formed used ImageJ analysis. Data are expressed as means of % NETs ± SEM from three independent experiments. ns, non-significant.
Discussion
Given that neutrophils represent the most abundant and highly recruited innate immune cell to the periodontal pocket, processes such as NET production are an important component of the host-microbe interface. Previous literature has linked NET formation to other chronic inflammatory conditions,29 and in vivo studies have detected NETs in the oral biofilm, the saliva, and the crevicular exudate.44 NETs could serve as a more efficient mechanism of control and clearance by neutrophils in the oral cavity, as in this environment, bacteria are widely dispersed and phagocytosis is not always effective.27 In the present study, we demonstrated that F. alocis fails to trigger NET formation by human neutrophils, independent of time of challenge, opsonization and MOI. However, F. alocis can manipulate neutrophils in order to inhibit their ability to effectively produce NETs in response to a known pharmacological inducer, PMA.
Different factors can impact and influence NET production, one of these being the MOI of the bacterial challenge.20 For example, studies performed with a Gram-negative oral pathogen, Aggregatibacter actinomycetemcomitans, showed that NET production was dependent on bacterial load.40 Studies performed with Burkholderia pseudomallei showed that with increasing MOI and later time intervals, more NETs were formed.41 However, in our study, increasing F. alocis dose, in the presence or absence of serum, did not result in NET production. On the other hand, our findings showed that release of NETs induced by S. gordonii was dose dependent. Similarly to the observations obtained in this study where S. gordonii challenge at MOI 10 produced ∼ 10% NET formation, Hirschfeld et al. reported NET formation was observed by a variety of oral bacteria, including S. gordonii, taken from supragingival biofilm and whole saliva samples in healthy donors.44 Furthermore, a recent study compared the ability of several oral bacteria to induce ROS production and NET formation in human neutrophils.45 Although the oral bacteria tested were heat-killed and at much higher MOI (1000), S. gordonii was one of the organisms, together with Veillonella parvula, to induce higher percentages of NETs as well as high ROS production compared to the keystone pathogen P. gingivalis.45 The present study, to the best of our knowledge, is the first to report that F. alocis does not induce NET formation; in contrast to another emerging oral pathogen, P. stomatis, that is able to induce NET formation.
Previous work demonstrated that priming neutrophils with TNF-α can induce NET formation.17 Neutrophils arriving at the gingival tissue are primed and hence could promote more release of NETs. However, based on our in vitro findings, F. alocis will not induce release of NETs even if exposed to TNF-α-primed neutrophils. These results would suggest that F. alocis interaction with neutrophils in the inflamed gingival tissue will not contribute to NET release. In the gingival tissue there are several priming agents besides TNF-α that could activate neutrophils. Hence future studies may determine if these results are specific to TNF-α priming or if this result would change when neutrophils are exposed to different priming agents such as IL-8 or C5a.
NET formation is a dynamic process and depending on the stimuli it can occur within 10 min upon S. aureus challenge or take more time to occur between 90 and 180 min upon PMA stimulation.23,46 The time course studies performed with F. alocis show that there was no significant induction of NETs at any time point tested up to 180 min. In co-infection conditions, a potential in vivo scenario at the gingival tissue, F. alocis did not inhibit or exacerbate NETs formed by either S. gordonii or P. stomatis. However, if F. alocis challenge preceded challenge with the pharmacological inducer, PMA, there was a significant reduction in NET formation. Similarly, Bordetella parapertussis utilizes adenylate cyclase toxin (CyaA) to inhibit ROS production, which further inhibits PMA-induced NET formation.47 Additionally, probiotic Lactobacillus rhamnosus was determined to be effective in inhibiting both PMA- and S. aureus-induced NETs in murine neutrophils and HL-60 neutrophil-like cell lines.48 Our results lead us to conclude that F. alocis is most likely manipulating neutrophil signaling mechanisms that lead to NET formation, because when NETs are pre-formed, they cannot be degraded by the organism. However, F. alocis is capable of preventing NET formation even after exposure to known inducers like PMA.
While the potential signaling pathways leading to NET formation have been outlined, few studies have been able to identify signaling mechanism involved in NET inhibition. A recent study found that TLR signaling could be linked to NET formation, as observed with anti-inflammatory drug, dexamethasone treatment on S. aureus and PMA-induced NETs.49 Another recent study, looking at the ROS production and NET formation by several oral bacteria, showed that inhibition of TLR3,7, and 9, through the use of chloroquine, or TLR2 and 4, through the use of oxidized 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine, had no significant impact on bacteria-induced ROS or NET formation.45 F. alocis promotes neutrophil granule exocytosis which is dependent on TLR2 and phosphorylation of both p38MAPK and ERK1/2;31 however, which mechanism(s) F. alocis may use to inhibit PMA-induced NETs are currently under investigation in our laboratory.
It has been reported that numerous Gram-positive bacteria, such as S. pneumoniae, express DNases, which can aid in NET degradation.50 The production of extracellular nucleases is well established for bacterial pathogens, including anaerobes, however their role in virulence is only recently appreciated.16,28 Recently, it was determined that Neisseria gonorrhoeae produces a heat-stable thermonuclease (Nuc), which provides the bacterium an effective virulence factor against NETs, as it is capable of DNA degradation, and results in bacterial survival.51 A thermonuclease was found in a proteomic analysis of F. alocis culture supernatants performed in our laboratory (unpublished observation); however, no significant reduction in PMA-induced NET formation was found in the presence of F. alocis culture supernatants. These results would suggest that although F. alocis possesses a thermonuclease, it may be present at a low concentration, therefore ineffective in degradation of NETs in our in vitro studies. Future studies in our laboratory are aimed at generating a F. alocis recombinant thermonuclease and titrating its potential effect on degradation of NETs.
In conclusion, the findings reported in this study show that F. alocis does not trigger the formation of NETs. Moreover, F. alocis is effective in manipulating neutrophils to impair their ability to form NETs, through a yet-to-be-determined mechanism. For F. alocis, this could ensure its survival as well as provide benefits to the entire oral community, as it can manipulate neutrophil deployment of NET formation, helping other bacteria that would normally be recognized and effectively killed by NETs to go undetected, further leading to their survival and persistence in the oral cavity. This could be especially important for those bacteria that are not effectively internalized, and neutrophils relying on extracellular killing mechanisms to clear the organism. In addition, this study provides novel findings about the different neutrophil responses provoked by the interaction with two emerging oral pathogens, F. alocis and P. stomatis. In the disease setting, it is tempting to speculate that neutrophils infected with F. alocis would be less effective in the release of NETs upon later encounter by P. stomatis; a scenario that might benefit the bacterial survival, and hinder neutrophils in their ability to control the dysbiotic community and resolve inflammation.
Acknowledgments
The authors want to thank Terri Manning for neutrophil isolation and for her expert technical help.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by NIH/National Institute of Dental and Craniofacial Research (NIDCR) (grant numbers DE024509, DE011111, DE012505, and DE017921).
References
- 1.Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol 2012; 27: 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol 2010; 8: 481–490. [DOI] [PubMed] [Google Scholar]
- 3.Eke PI, Dye BA, Wei L, et al. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res 2012; 91: 914–920. [DOI] [PubMed] [Google Scholar]
- 4.Eke PI, Dye BA, Wei L, et al. Update on prevalence of periodontitis in adults in the United States: NHANES 2009 to 2012. J Periodontol 2015; 86: 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dewhirst FE, Chen T, Izard J, et al. The human oral microbiome. J Bacteriol 2010; 192: 5002–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berezow AB, Darveau RP. Microbial shift and periodontitis. Periodontol 2000 2011; 55: 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wade WG. Has the use of molecular methods for the characterization of the human oral microbiome changed our understanding of the role of bacteria in the pathogenesis of periodontal disease? J Clin Periodontol 2011; 38(Suppl 11): 7–16. [DOI] [PubMed] [Google Scholar]
- 8.Griffen AL, Beall CJ, Firestone ND, et al. CORE: a phylogenetically-curated 16S rDNA database of the core oral microbiome. PLoS One 2011; 6: e19051–e19051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahlen G, Leonhardt A. A new checkerboard panel for testing bacterial markers in periodontal disease. Oral Microbiol Immunol 2006; 21: 6–11. [DOI] [PubMed] [Google Scholar]
- 10.Moffatt CE, Whitmore SE, Griffen AL, et al. Filifactor alocis interactions with gingival epithelial cells. Mol Oral Microbiol 2011; 26: 365–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aruni AW, Roy F, Fletcher HM. Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by Porphyromonas gingivalis. Infect Immun 2011; 79: 3872–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aruni AW, Zhang K, Dou Y, et al. Proteome analysis of coinfection of epithelial cells with Filifactor alocis and Porphyromonas gingivalis shows modulation of pathogen and host regulatory pathways. Infect Immun 2014; 82: 3261–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aruni W, Chioma O, Fletcher HM. Filifactor alocis: the newly discovered kid on the block with special talents. J Dent Res 2014; 93: 725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hart TC, Atkinson JC. Mendelian forms of periodontitis. Periodontol 2000 2007; 45: 95–112. [DOI] [PubMed] [Google Scholar]
- 15.Uriarte SM, Edmisson JS, Jimenez-Flores E. Human neutrophils and oral microbiota: a constant tug-of-war between a harmonious and a discordant coexistence. Immunol Rev 2016; 273: 282–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brinkmann V, Reichard U, Goosmann C, et al. Neutrophil extracellular traps kill bacteria. Science 2004; 303: 1532–1535. [DOI] [PubMed] [Google Scholar]
- 17.Amulic B, Cazalet C, Hayes GL, et al. Neutrophil function: from mechanisms to disease. Annu Rev Immunol 2012; 30: 459–489. [DOI] [PubMed] [Google Scholar]
- 18.Barrientos L, Marin-Esteban V, de Chaisemartin L, et al. An improved strategy to recover large fragments of functional human neutrophil extracellular traps. Front Immunol 2013; 4: 166–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brinkmann V, Goosmann C, Kuhn LI, et al. Automatic quantification of in vitro NET formation. Front Immunol 2012; 3: 413–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brinkmann V, Zychlinsky A. Neutrophil extracellular traps: is immunity the second function of chromatin? J Cell Biol 2012; 198: 773–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Remijsen Q, Kuijpers TW, Wirawan E, et al. Dying for a cause: NETosis, mechanisms behind an antimicrobial cell death modality. Cell Death Differ 2011; 18: 581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker H, Winterbourn CC. Reactive oxidants and myeloperoxidase and their involvement in neutrophil extracellular traps. Front Immunol 2012; 3: 424–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White PC, Chicca IJ, Cooper PR, et al. Neutrophil extracellular traps in periodontitis: a web of intrigue. J Dent Res 2016; 95: 26–34. [DOI] [PubMed] [Google Scholar]
- 24.Yipp BG, Petri B, Salina D, et al. Infection-induced NETosis is a dynamic process involving neutrophil multitasking in vivo. Nat Med 2012; 18: 1386–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuchs TA, Abed U, Goosmann C, et al. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol 2007; 176: 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pilsczek FH, Salina D, Poon KK, et al. A novel mechanism of rapid nuclear neutrophil extracellular trap formation in response to Staphylococcus aureus. J Immunol 2010; 185: 7413–7425. [DOI] [PubMed] [Google Scholar]
- 27.Vitkov L, Klappacher M, Hannig M, et al. Extracellular neutrophil traps in periodontitis. J Periodont Res 2009; 44: 664–672. [DOI] [PubMed] [Google Scholar]
- 28.Doke M, Fukamachi H, Morisaki H, et al. Nucleases from Prevotella intermedia can degrade neutrophil extracellular traps. Mol Oral Microbiol 2017; 32: 288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooper PR, Palmer LJ, Chapple IL. Neutrophil extracellular traps as a new paradigm in innate immunity: friend or foe? Periodontol 2000 2013; 63: 165–197. [DOI] [PubMed] [Google Scholar]
- 30.Uriarte SM, Rane MJ, Luerman GC, et al. Granule exocytosis contributes to priming and activation of the human neutrophil respiratory burst. J Immunol 2011; 187: 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Armstrong CL, Miralda I, Neff AC, et al. Filifactor alocis promotes neutrophil degranulation and chemotactic activity. Infect Immun 2016; 84: 3423–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sizova MV, Chilaka A, Earl AM, et al. High-quality draft genome sequences of five anaerobic oral bacteria and description of Peptoanaerobacter stomatis gen. nov., sp. nov., a new member of the family Peptostreptococcaceae. Stand Genomic Sci 2015; 10: 37–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jimenez Flores E, Tian S, Sizova M, et al. Peptoanaerobacter stomatis primes human neutrophils and induces granule exocytosis. Infect Immun 2017; 85: e01043–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brinkmann V, Laube B, Abu Abed U, et al. Neutrophil extracellular traps: how to generate and visualize them. J Visualized Exp 2010, pp. 36–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White P, Sakellari D, Roberts H, et al. Peripheral blood neutrophil extracellular trap production and degradation in chronic periodontitis. J Clin Periodontol 2016; 43: 1041–1049. [DOI] [PubMed] [Google Scholar]
- 36.Palmer LJ, Damgaard C, Holmstrup P, et al. Influence of complement on neutrophil extracellular trap release induced by bacteria. J Periodont Res 2016; 51: 70–76. [DOI] [PubMed] [Google Scholar]
- 37.Papayannopoulos V, Metzler KD, Hakkim A, et al. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol 2010; 191: 677–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metzler KD, Goosmann C, Lubojemska A, et al. A myeloperoxidase-containing complex regulates neutrophil elastase release and actin dynamics during NETosis. Cell Rep 2014; 8: 883–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolaczkowska E, Jenne CN, Surewaard BG, et al. Molecular mechanisms of NET formation and degradation revealed by intravital imaging in the liver vasculature. Nat Commun 2015; 6: 6673–6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirschfeld J, Roberts HM, Chapple IL, et al. Effects of Aggregatibacter actinomycetemcomitans leukotoxin on neutrophil migration and extracellular trap formation. J Oral Microbiol 2016; 8: 33070–33070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riyapa D, Buddhisa S, Korbsrisate S, et al. Neutrophil extracellular traps exhibit antibacterial activity against Burkholderia pseudomallei and are influenced by bacterial and host factors. Infect Immun 2012; 80: 3921–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El-Benna J, Hurtado-Nedelec M, Marzaioli V, et al. Priming of the neutrophil respiratory burst: role in host defense and inflammation. Immunol Rev 2016; 273: 180–193. [DOI] [PubMed] [Google Scholar]
- 43.Matthews JB, Wright HJ, Roberts A, et al. Hyperactivity and reactivity of peripheral blood neutrophils in chronic periodontitis. Clin Exp Immunol 2007; 147: 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirschfeld J, Dommisch H, Skora P, et al. Neutrophil extracellular trap formation in supragingival biofilms. Int J Med Microbiol 2015; 305: 453–463. [DOI] [PubMed] [Google Scholar]
- 45.Hirschfeld J, White PC, Milward MR, et al. Modulation of neutrophil extracellular trap and reactive oxygen species release by periodontal bacteria. Infect Immun 2017; 85: e00297–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parker H, Dragunow M, Hampton MB, et al. Requirements for NADPH oxidase and myeloperoxidase in neutrophil extracellular trap formation differ depending on the stimulus. J Leukocyte Biol 2012; 92: 841–849. [DOI] [PubMed] [Google Scholar]
- 47.Gorgojo J, Scharrig E, Gomez RM, et al. Bordetella parapertussis circumvents neutrophil extracellular bactericidal mechanisms. PLoS One 2017; 12: e0169936–e0169936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vong L, Lorentz RJ, Assa A, et al. Probiotic Lactobacillus rhamnosus inhibits the formation of neutrophil extracellular traps. J Immunol 2014; 192: 1870–1877. [DOI] [PubMed] [Google Scholar]
- 49.Wan T, Zhao Y, Fan F, et al. Dexamethasone inhibits S. aureus-induced neutrophil extracellular pathogen-killing mechanism, possibly through toll-like receptor regulation. Front Immunol 2017; 8: 60–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beiter K, Wartha F, Albiger B, et al. An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr Biol 2006; 16: 401–407. [DOI] [PubMed] [Google Scholar]
- 51.Juneau RA, Stevens JS, Apicella MA, et al. A thermonuclease of Neisseria gonorrhoeae enhances bacterial escape from killing by neutrophil extracellular traps. J Infect Dis 2015; 212: 316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]