Abstract
Background
Inflammation plays an important role in the pathogenesis of coronary artery disease (CAD). Studies have reported that inflammatory cytokine interleukin-8 (IL-8) gene −251 A/T (rs4073) polymorphism is correlated with CAD susceptibility, but the result remains controversial. The objective of this study was to clarify the association between IL-8 gene −251 A/T polymorphism and CAD risk.
Material/Methods
A meta-analysis included 8244 patients from 9 individual studies with 10 populations was conducted. Heterogeneity test was conducted, and pooled odds ratio (OR) with 95% confidence interval (CI) was calculated used fixed-effect or random-effects model accordingly. Publication bias was evaluated with the Begg’s funnel plot and Egger’s test. Sensitivity analysis was also conducted.
Results
A significant association between IL-8 gene −251 A/T polymorphism and CAD risk was found in the dominant model (OR 1.42, 95% CI 1.16–1.76, P<0.001), recessive model (OR 1.30, 95% CI 1.12–1.52, P<0.001), allelic model (OR 1.28, 95% CI 1.12–1.47, P<0.001), homozygote model (OR 1.59, 95% CI 1.21–2.08, P<0.001), and heterozygote model (OR 1.35, 95% CI 1.11–1.64, P=0.002). Subgroup analysis by ethnicity found significant associations in the Chinese population in the dominant model(OR 1.43, 95% CI 1.26–1.61, P<0.001), recessive model (OR 1.39, 95% CI 1.21–1.59, P<0.001), allelic model (OR 1.31, 95% CI 1.21–1.42, P<0.001), homozygote model (OR 1.66, 95% CI 1.41–1.95, P<0.001), and heterozygote model (OR 1.34, 95% CI 1.18–1.52, P<0.001), but no significant association was found in the Caucasian population. No significant publication bias was found.
Conclusions
The IL-8 gene −251 A/T polymorphism was significantly associated with CAD risk in the Chinese population but not in the Caucasian population, −251 A allele carrier had an increased risk of CAD in the Chinese population.
MeSH Keywords: Coronary Artery Disease, Genetic Variation, Interleukin-8, Meta-Analysis
Background
Coronary artery disease (CAD), also known as coronary heart disease, is one of the leading causes of death worldwide. Its pathogenesis involves many risk factors including hypertension, diabetes, dyslipidemia. Several hypotheses have been proposed including inflammation hypothesis, immunological theory, etc. The inflammation mechanism in the initiation and development of CAD was once a research hotspot, and CAD was regarded as an inflammatory disease [1,2]. However, the inflammation hypothesis of CAD’s pathogenesis had not been validated until recently with the release of CANTOS (Anti-inflammatory Therapy with Canakinumab for Atherosclerotic Disease) trial. As a landmark study, the CANTOS trial found that anti-inflammation therapy could improve prognosis of high-risk CAD patients significantly, thus validating the inflammation hypothesis of CAD for the first time [3,4].
The inflammatory cytokine interleukin-8 (IL-8), also known as CXCL8, plays an important role in the initiation, progression, and prognosis of atherosclerosis and CAD. Studies have reported high expression levels of IL-8 in human arterial atherosclerotic wall [5]; and IL-8 can rapidly cause rolling monocytes to firmly adhere to endothelial monolayers expressing E-selectin [6]. On the other hand, clinical studies have observed that serum IL-8 concentrations are significantly elevated in CAD patients [7] and can predict cardiovascular events of CAD independent of other cytokines and high sensitivity C-reactive protein [8].
The IL-8 gene is located in chromosome 4q13.3 and belongs to the superfamily of CXC chemokines. One single-nucleotide polymorphism (SNP) of IL-8 gene at position −251 A/T (rs4073) has been well-characterized and was known to influence the expression of IL-8 and susceptibility of several diseases including Alzheimer’s disease [9], cancer[10], and periodontitis [11]. Vogiatzi et al. first reported that IL-8 gene −251 A/T polymorphism was associated with susceptibility to restenosis after PCI [12]. Thereafter, several other studies found that IL-8 gene −251 A/T polymorphism was associated with susceptibility of CAD, and that the −251 A allele may increase CAD risk [13–16]. However, other studies reported no significant correlation between the IL-8 gene −251 A/T polymorphism and the risk of CAD [17–21]. Thus, the association between this SNP of the IL-8 gene and susceptibility of CAD is controversial.
A reliable method to resolve the contradictions between individual studies is meta-analysis. In order to comprehensively evaluate the correlation between the IL-8 gene −251 A/T (rs4073) polymorphism and the risk of CAD, we conducted a meta-analysis that included 4103 CAD patients and 4141 controls from 10 study populations.
Material and Methods
Literature search
All studies about the association between IL-8 gene −251 A/T polymorphism and CAD were identified by comprehensive computer-based searches of PubMed, EMBASE, Web of Science, Wan Fang database, VIP database, and China National Knowledge Infrastructure (CNKI). The keywords used for the literature search were combined: (“interleukin 8” OR “IL-8” OR “CXCL8”) and (“coronary artery disease” OR “coronary heart disease” OR “CAD” OR “CHD” “ OR “myocardial infarction” OR “acute coronary syndrome” OR “angina” OR “ischemic cardiovascular disease”) and (“polymorphism” OR “single nucleotide polymorphism” OR “SNP” OR “mutation” OR “variation” OR “allele” OR “genotype” OR “rs4073” OR “251 A/T”). The literature language was limited to English and Chinese, and the last search was updated on November 20, 2018.
Inclusion criteria
The inclusion criteria for eligible studies were as follow: 1) studies evaluated the association between the IL-8 gene −251 A/T polymorphism and the risk of CAD. 2) The CAD diagnostic criteria were angiographically confirmed CAD or acute myocardial infarction (AMI) diagnosed with general standard criteria. 3) Studies were case-control or officially published cohort studies. 4) Studies had intact original data on genotype distribution and sufficient data to calculate an odds ratio (OR) with 95% confidence interval (CI).
Exclusion criteria
Studies were excluded as follows: repeated publications, when different studies reported the same or overlapping patients, only the latest or most complete was included; review articles; articles with insufficient information; studies about the association between the IL-8 gene other SNPs and CAD risk; unpublished studies or studies not in English or Chinese language; and studies in which CAD patients were further specially selected with traditional Chinese medicine (TCM) syndrome differentiation.
Data extraction
Two authors (Zhang and Gao) performed the data extraction independently. The following data were collected from all included studies: the first author’s name, publication year, study population (ethnicity, sex, and age), number of genotypes, genotyping methods, allele frequency of cases and controls, diagnostic methods for cases and controls, and P value for Hardy-Weinberg equilibrium (HWE) in controls. The results were compared, and disagreements were settled by consensus. If these 2 authors could not reach a consensus, the results were further reviewed by the third author (Huang).
Study quality assessment
The Newcastle-Ottawa Quality Assessment Scale (NOS) [22] was used to assess the quality of the included studies by 2 authors independently. The NOS uses a “star” rating system ranging between zero (worst) up to 9 stars (best) to judge the quality of observational studies. Any inconsistences about study quality judgement between the 2 authors were solved through discussion.
Statistical analysis
The statistical analysis was performed by using Review Manager 5.30 (Cochrane Collaboration, The Nordic Cochrane Centre, Copenhagen) and Stata 14.0 (STATA Corp., College Station, TX, USA). To measure the strengths of the genetic associations of the IL-8 gene −251 A/T polymorphism with the risk of CAD, the pooled odds ratios (ORs) and 95% confidence interval (CI) were calculated. A χ2 test was used to check whether the frequencies of the genotypes deviated from the HWE [23] in controls of each included study. Heterogeneity between studies was assessed by I2 test, P<0.10 and I2>50% was considered existing significant heterogeneity [24]. If significant heterogeneity was observed, the Mantel-Haenszel test with random-effects model was used to evaluate the pooled ORs (95% CIs), otherwise, the Mantel-Haenszel test with fixed-effect model was used to calculate the pooled ORs (95% CIs) [25]. To further explore the effect of ethnicity on heterogeneity among the studies, subgroup analysis was performed based on different ethnicities (Chinese versus Caucasian). The potential publication bias was checked by using the Begg’s funnel plot and Egger’s test. Sensitivity analysis was conducted to observe the influence of any single study on the pooled ORs. Except for I2 test for assessing heterogeneity, a 2-tailed P<0.05 was considered significant.
Results
Study characteristics
A total of 199 studies were initially identified according to the search criteria described. After screening titles and abstracts, 183 studies were discarded for being irrelevant. After full-text assessment, 7 studies were further excluded for repetitive publications, comparison of different CAD types, improper diagnostic standard for CAD, or about other loci of IL-8 gene. Finally, 9 eligible studies were identified for overall data combination, but 1 of the studies [13] had 2 different populations, so a total of 10 populations with 4103 CAD cases and 4141 controls were included for data analysis in this meta-analysis. The flow diagram of study selection process is shown in Figure 1. HWE of genotype distribution in the controls was tested in all the included studies, and the distribution of genotypes in controls was not in HWE in 3 of the included studies. Of the 9 included studies, polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method was used to determine IL-8 gene −251 A/T genotypes in 5 studies, while the remaining 4 studies used allele-specific polymerase chain reaction (AS-PCR), matrix adsorbed laser desorption-ionization-time of flight (MALDI-TOF), tetra-primer amplification refractory mutation system-polymerase chain reaction (tetra-primer ARMS-PCR), and Sequenom MassARRAY to genotyping respectively. The characteristics and genotype frequencies of the included studies are presented in Tables 1 and 2.
Figure 1.
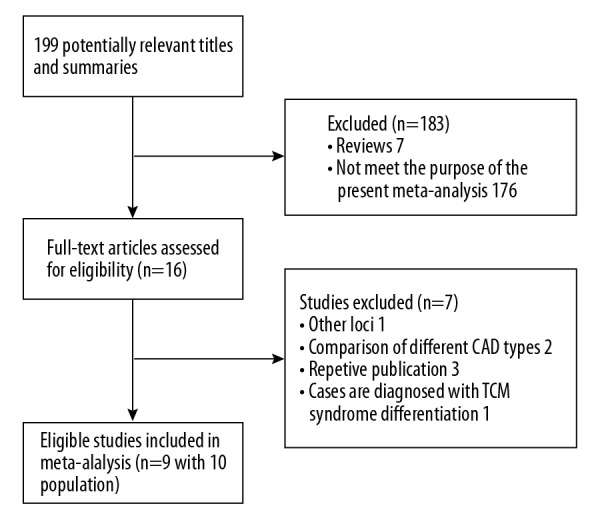
Flow diagram of study selection process. CAD – coronary artery disease; TCM – traditional Chinese medicine.
Table 1.
Characteristics of the included studies in the meta-analysis.
| Study | Ethnicity | Genotyping method | Subjects of cases | Matching criteria | Sample size (n) | Quality score | |
|---|---|---|---|---|---|---|---|
| Cases | Controls | ||||||
| Zhang X et al. (a*) [13] | Chinese | PCR-RFLP | CAC patients | Age, gender, LDL-C | 675 | 636 | 6 |
| Zhang X et al. (b*) [13] | Chinese | PCR-RFLP | CAC patients | Age, gender, LDL-C | 360 | 360 | 6 |
| Vogiatzi K et al. [17] | Caucasian | AS-PCR | CAC patients | Age, hypertension | 241 | 157 | 6 |
| Zhang RJ et al. [15] | Chinese | PCR-RFLP | CAC patients | Age, alcohol consumption, FH | 217 | 245 | 6 |
| Velasquez IM et al. [19] | Caucasian | MALDI-TOF | MI patients | Age, gender | 867 | 1035 | 5 |
| Yang HT et al. [21] | Chinese | PCR-RFLP | CAC patients | Age, alcohol consumption | 410 | 410 | 5 |
| Wang S et al. [14] | Chinese | PCR-RFLP | CAC patients | Age, alcohol consumption, FH | 264 | 286 | 6 |
| Hou H et al. [18] | Chinese | PCR-RFLP | CAC patients | Age, gender | 244 | 170 | 6 |
| Ren B et al. [20] | Chinese | Sequenom MassARRAY | CAC patients | Age, gender, alcohol consumption | 325 | 342 | 5 |
| Kaur N et al. [16] | Caucasian | Tetra-primer ARMS-PCR | CAC patients | BMI, HDL-C, TG | 500 | 500 | 6 |
PCR-RFLP – polymerase chain reaction-restriction fragment length polymorphism; AS-PCR – allele specific-polymerase chain reaction; MALDI-TOF – Matrix Adsorbed Laser Desorption-Ionisation-Time of Flight; ARMS-PCR – amplification refractory mutation system-polymerase chain reaction; CAC – coronary angiography-confirmed CAD; MI – myocardial infarction; FH – family history; LDL-C – low-density lipoprotein cholesterol; HDL-C – high-density lipoprotein cholesterol; TG – triglycerides; BMI – body mass index.
This study has two different populations, here we marked as population a and b.
Table 2.
Genotype frequencies of the included studies in the meta-analysis.
| Study | Ethnicity | Case/control (n) | CAD group (−251 A/T) | Control (−251 A/T) | PHWE | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AT | TT | A | T | AA | AT | TT | A | T | ||||
| Zhang X et al. (a*) [13] | Chinese | 675/636 | 123 | 320 | 232 | 566 | 784 | 80 | 292 | 264 | 452 | 820 | 0.96 |
| Zhang X et al. (b*) [13] | Chinese | 360/360 | 76 | 176 | 108 | 328 | 392 | 58 | 159 | 143 | 275 | 445 | 0.22 |
| Vogiatzi K et al. [17] | Caucasian | 241/157 | 41 | 127 | 73 | 209 | 273 | 28 | 76 | 53 | 132 | 182 | 0.93 |
| Zhang RJ et al. [15] | Chinese | 217/245 | 69 | 101 | 47 | 239 | 195 | 57 | 108 | 80 | 222 | 268 | 0.08 |
| Velasquez IM et al. [19] | Caucasian | 867/1035 | 269 | 416 | 182 | 954 | 780 | 330 | 516 | 189 | 1176 | 894 | 0.61 |
| Yang HT et al. [21] | Chinese | 410/410 | 114 | 178 | 118 | 406 | 414 | 105 | 171 | 134 | 381 | 439 | <0.01 |
| Wang S et al. [14] | Chinese | 264/286 | 78 | 125 | 61 | 281 | 247 | 56 | 128 | 102 | 240 | 332 | 0.17 |
| Hou H et al. [18] | Chinese | 244/170 | 41 | 130 | 73 | 212 | 276 | 21 | 85 | 64 | 127 | 213 | 0.37 |
| Ren B et al. [20] | Chinese | 325/342 | 93 | 147 | 85 | 333 | 317 | 85 | 149 | 108 | 319 | 365 | 0.02 |
| Kaur N et al. [16] | Caucasian | 500/500 | 199 | 225 | 76 | 623 | 377 | 148 | 195 | 157 | 491 | 509 | <0.01 |
HWE – Hardy-Weinberg equilibrium; CAD – coronary artery disease.
This study has two different populations, here we marked as population a and b.
Meta-analysis results
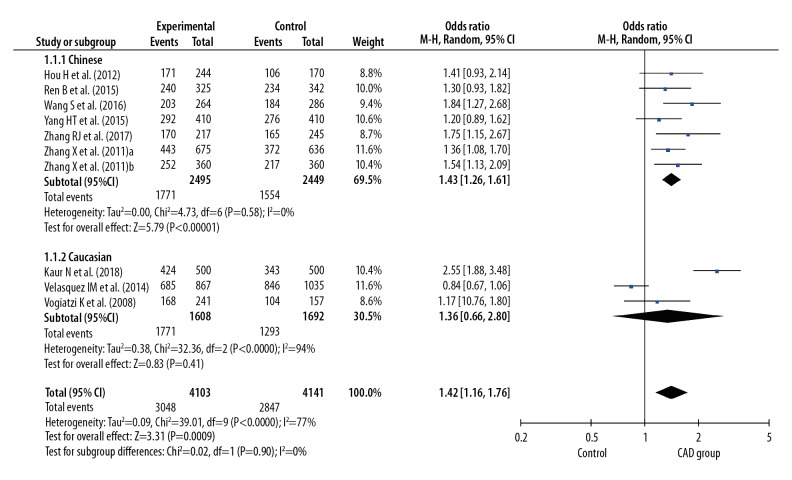
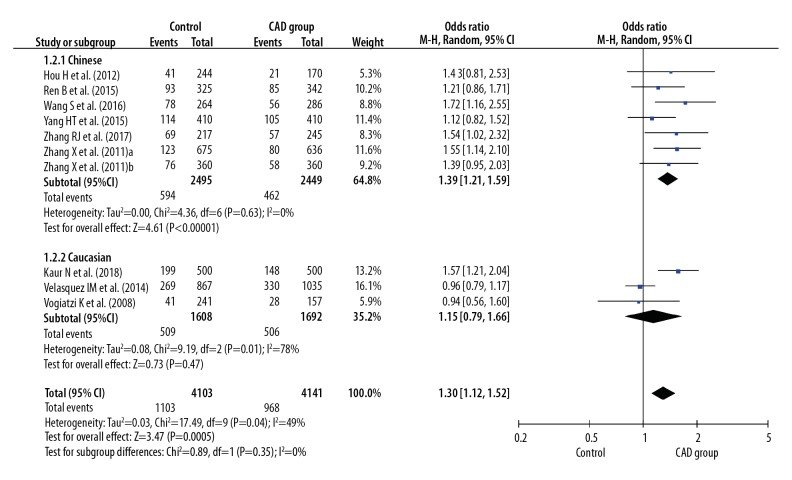
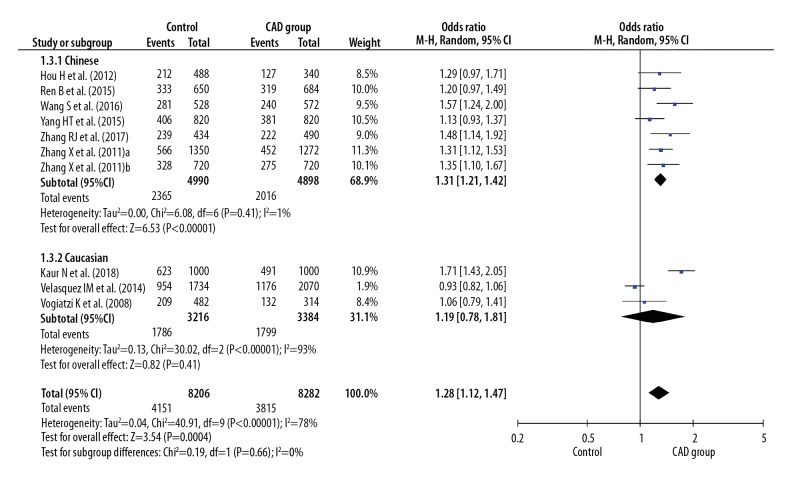
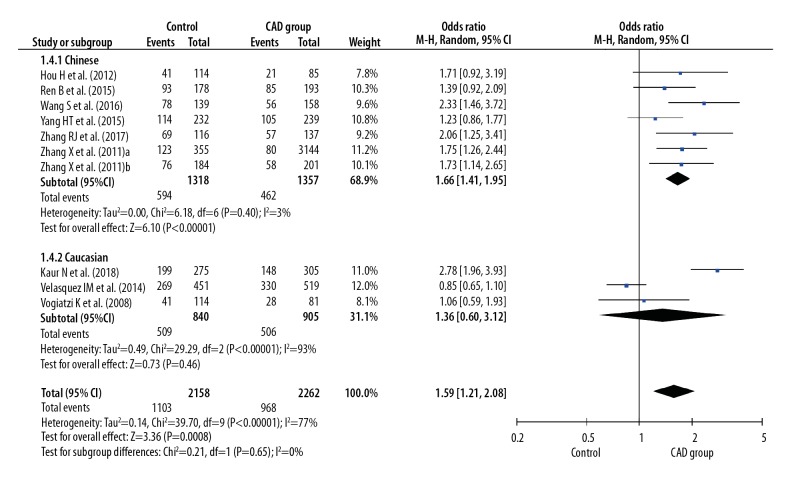
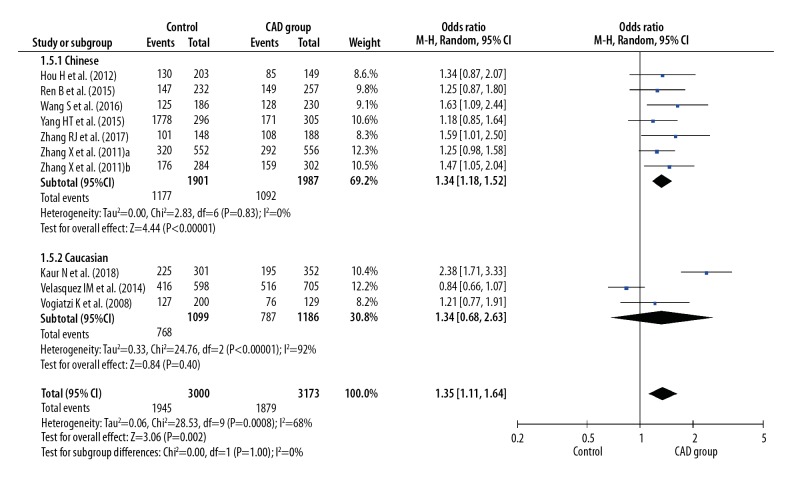
Before calculating pooled ORs, a heterogeneity test was conducted, and heterogeneity was found in the allelic model (I2=78%, P<0.001), dominant model (I2=7 7%, P<0.001), recessive model (I2=49%, P=0.04), homozygote model (I2=77%, P<0.001), and heterozygote model (I2=68%, P<0.001) of the IL-8 gene −251 A/T polymorphism. We used random-effects models to merge ORs for all the comparison models of the IL-8 gene −251 A/T polymorphism. A significant association was found between the IL-8 gene −251 A/T polymorphism and the risk of CAD in all dominant (OR 1.42, 95% CI 1.16–1.76, P<0.001), recessive (OR 1.30, 95% CI 1.12–1.52, P<0.001), allelic (OR 1.28, 95% CI 1.12–1.47, P<0.001), homozygote (OR 1.59, 95% CI 1.21–2.08, P<0.001), and heterozygote (OR 1.35, 95% CI 1.11–1.64, P=0.002) models (Figures 2–6).
Figure 2.
Forest plot for overall comparison in dominant model (AA+AT versus TT).
Figure 3.
Forest plot for overall comparison in recessive model (AA versus AT+TT).
Figure 4.
Forest plot for overall comparison in allelic model (A versus T).
Figure 5.
Forest plot for overall comparison in homozygote model (AA versus TT).
Figure 6.
Forest plot for overall comparison in heterozygote model (AT versus TT).
In a subgroup analysis stratified by ethnicity (Chinese versus Caucasian), 7 original studies with 4944 patients (2495 cases and 2449 controls) were allocated into a Chinese population subgroup, and 3 original studies with 3300 patients (1608 cases and 1692 controls) were allocated into a Caucasian subgroup population. No significant heterogeneity was found in the Chinese population subgroup in the recessive model (I2=0%, P=0.63), dominant model (I2=0%, P=0.58), allelic model (I2=1%, P=0.41), homozygote model (I2=3%, P=0.40), and heterozygote model (I2=0%, P=0.83), but significant heterogeneity was found in the Caucasian population subgroup in the recessive model (I2=78%, P=0.01), dominant model (I2=94%, P<0.001), allelic model (I2=93%, P<0.001), homozygote model (I2=93%, P<0.001), and heterozygote model (I2=92%, P<0.001) (Figures 2–6). Significant associations between the IL-8 gene −251 A/T polymorphism and the risk of CAD were found in the Chinese population in the dominant model (OR 1.43, 95% CI 1.26–1.61, P<0.001), recessive model (OR 1.39, 95% CI 1.21–1.59, P<0.001), allelic model (OR 1.31, 95% CI 1.21–1.42, P<0.001), homozygote model (OR 1.66, 95% CI 1.41–1.95, P<0.001), and heterozygote model (OR 1.34, 95% CI 1.18–1.52, P < 0.001), but no significant association was found in the Caucasian population in all dominant (OR 1.36, 95% CI 0.66–2.80, P=0.41), recessive (OR 1.15, 95% CI 0.79–1.66, P=0.47), allelic (OR 1.19, 95% CI 0.78–1.81, P=0.41), homozygote (OR 1.36, 95% CI 0.60–3.12, P=0.46), and heterozygote (OR 1.34, 95% CI 0.68–2.63, P=0.40) models (Figures 2–6).
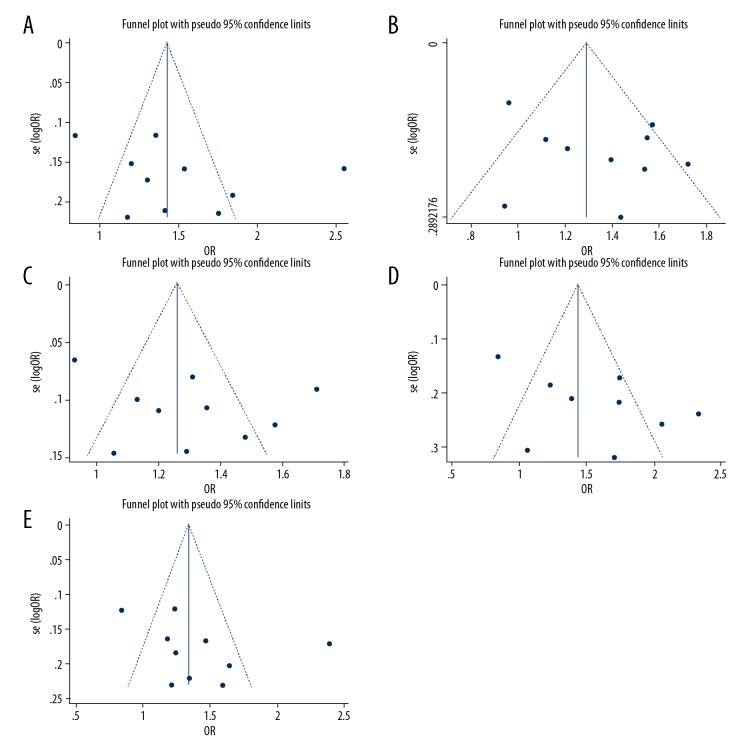
Publication bias
The Begg’s funnel plot and Egger’s test [26] were used to evaluate publication bias of the included studies. No obvious asymmetry was found in funnel plots for all the genetic models (Figure 7). The Begg’s test suggested no significant publication bias in the recessive model (Z=0.09, P=0.929), dominant model (Z=0.63, P=0.531), allelic model (Z=0.09, P=0.929), homozygote model (Z=0.45, P=0.655), and heterozygote model (Z=1.16, P=0.245). In comparison to the Begg’s test, the Egger’s test is more sensitive, so we further conducted the Egger’s test to rule out possible publication bias. The Egger’s test results also showed no significant publication bias in the recessive model (t=1.27, P=0.239), dominant model (t=1.12, P=0.296), allelic model (t=1.20, P=0.264), homozygote model (t=0.93, P=0.382), and heterozygote model (t=1.25, P=0.248). These results demonstrated that there was no significant publication bias in the included studies, indicating the reliability of this meta-analysis.
Figure 7.
Funnel plots of the association between interlukein-8 gene −251 A/T polymorphism and coronary artery disease risk: (A) dominant model; (B) recessive model; (C) allelic model; (D) homozygote model; and (E) heterozygote model.
Sensitivity analysis
To evaluate the influence of each study on the pooled ORs and to ensure no single study was completely responsible for the combined results, sensitivity analysis was conducted. The pooled ORs in all models were not significantly altered by omitting each of the individual studies (Table 3), which indicated that the results of this meta-analysis were robust [27].
Table 3.
Results of sensitivity analysis.
| Study omitted | Cases/controls (n) | Crude OR (95%CI) | ||||
|---|---|---|---|---|---|---|
| Recessive (AA vs. AT+TT) | Dominant (AA+AT vs. TT) | Allele (A vs. T) | Homozygote (AA vs. TT) | Heterozygote (AT vs. TT) | ||
| Zhang X et al. (a*) [13] | 675/636 | 1.28 (1.09–1.50) | 1.44 (1.12–1.84) | 1.28 (1.09–1.49) | 1.57 (1.16–2.13) | 1.37 (1.09–1.71) |
| Zhang X et al. (b*) [13] | 360/360 | 1.30 (1.10–1.53) | 1.41 (1.12–1.78) | 1.27 (1.09–1.48) | 1.57 (1.17–2.12) | 1.34 (1.08–1.66) |
| Vogiatzi K et al. [17] | 241/157 | 1.33 (1.14–1.56) | 1.45 (1.16–1.82) | 1.30 (1.13–1.51) | 1.64 (1.24–2.19) | 1.36 (1.11–1.68) |
| Zhang RJ et al. [15] | 217/245 | 1.29 (1.10–1.51) | 1.40 (1.12–1.75) | 1.26 (1.09–1.46) | 1.55 (1.06–2.06) | 1.33 (1.08–1.63) |
| Velasquez IM et al. [19] | 867/1035 | 1.39 (1.24–1.57) | 1.53 (1.29–1.80) | 1.34 (1.21–1.49) | 1.74 (1.42–2.14) | 1.44 (1.23–1.68) |
| Yang HT et al. [21] | 410/410 | 1.33 (1.13–1.58) | 1.45 (1.15–1.84) | 1.30 (1.12–1.51) | 1.64 (1.21–2.21) | 1.37 (1.11–1.70) |
| Wang S et al. [14] | 264/286 | 1.27 (1.09–1.48) | 1.39 (1.11–1.73) | 1.25 (1.09–1.44) | 1.52 (1.15–2.02) | 1.32 (1.08–1.63) |
| Hou H et al. [18] | 244/170 | 1.30 (1.11–1.53) | 1.43 (1.14–1.79) | 1.28 (1.10–1.48) | 1.58 (1.18–2.11) | 1.35 (1.10–1.67) |
| Ren B et al. [20] | 325/342 | 1.32 (1.11–1.56) | 1.44 (1.14–1.82) | 1.29 (1.11–1.50) | 1.61 (1.19–2.18) | 1.36 (1.10–1.69) |
| Kaur N et al. [16] | 500/500 | 1.27 (1.08–1.48) | 1.32 (1.11–1.57) | 1.23 (1.09–1.40) | 1.47 (1.15–1.88) | 1.24 (1.07–1.45) |
OR – odds ratio; CI – confidence interval.
This study has two different populations, here we marked as population a and b.
As shown in Table 2, the genotype distribution in controls is not in HWE in 3 of the included studies [16,20,21], this may influence the reliability of the study results. So to overcome this shortcoming, we recalculated the pooled ORs by excluding these 3 studies, and found that a significant association was still present in all dominant (OR 1.35, 95% CI 1.08–1.70, P=0.01), recessive (OR 1.32, 95% CI 1.07–1.63, P=0.01), allelic (OR 1.26, 95% CI 1.07–1.48, P=0.007), homozygote (OR 1.54, 95% CI 1.11–2.14, P=0.01), and heterozygote (OR 1.27, 95% CI 1.04–1.55, P=0.02) models of the IL-8 gene −251 A/T polymorphism. The results further demonstrated the steadiness and reliability of this meta-analysis.
Discussion
In the present meta-analysis, we included 9 studies (10 study populations) with a total of 4103 CAD patients and 4141 controls to assess the correlation between the IL-8 gene −251 A/T polymorphism and the risk of CAD. A significant association between −251 A/T polymorphism and CAD risk was found in dominant, recessive, allelic, homozygote, and heterozygote models, −251 A allele carrier has an increased risk of CAD. After subgrouping by ethnicity, the association between −251 A/T polymorphism and CAD risk was significant in all the genetic models in the Chinese population, but not in all the genetic models in the Caucasian population.
For publication bias evaluation, we first conducted funnel plot analysis, and found the funnel plots presented symmetrical shape in all the genetic models, indicating no obvious publication bias in this meta-analysis [28]. However, observing the symmetry of the funnel plot shape visually is subjective to some extent [29], so we conducted the Begg’s test [30] and the Egger’s test [26] respectively to evaluate publication bias. Both the Egger’s and the Begg’s tests identified no significant publication bias in all recessive, dominant, allelic, homozygote, and heterozygote models. These results indicated that there is no significant publication bias in the included studies, suggesting our meta-analysis was reliable. Sensitivity analysis showed no single study was completely responsible for the combined results, and significant associations between this SNP and CAD risk were still present in all the genetic models after omitting the 3 studies in which the genotype distribution was not in HWE in controls, further demonstrating the stability and reliability of our meta-analysis.
CAD threatens people’s health seriously worldwide, its pathogenesis involves many risk factors, such as genetic predisposition, dyslipidemia, hypertension, diabetes mellitus, but its exact mechanism has not been completely elucidated. The major pathological feature of CAD is atherosclerosis of coronary arteries. Over the past 10 years, the important role of inflammation in the pathogenesis of atherosclerotic cardiovascular diseases (ASCVD) aroused people’s attention, atherosclerosis was once regarded as an inflammatory disease [1]. The newly released CANTOS trial found that canakinumab, a therapeutic monoclonal antibody targeting the inflammatory cytokine interleukin-1β (IL-1β), can significantly reduce recurrent cardiovascular events of CAD, validated the inflammatory hypothesis of CAD pathogenesis for the first time [2,3].
IL-8 as another important inflammatory mediator, its role in the pathogenesis of CAD has been extensively researched [5,6,8]. Several polymorphisms have been detected in the IL-8 gene, and a common SNP of IL-8 gene at −251 position (−251 A/T, or rs4073) of the promoter region has been well-characterized. Studies proved that the −251 A/T polymorphism of IL-8 can affect susceptibility of a large number of inflammatory diseases, as well as disease severity and clinical prognosis, including CAD. Several studies reported that −251 A allele of the IL-8 gene can increase CAD risk [13–15], but negative results were also observed in other studies [17,19,21]. In order to overcome the result contradictories between different studies, we conducted this meta-analysis and found that the −251 A/T polymorphism of the IL-8 gene was significantly associated with CAD risk in the Chinese population, but not in the Caucasian population.
There are several possible metabolic and molecular mechanisms to explain the conclusion. This polymorphism has been associated with transcriptional activity of IL-8 gene, the −251 A allele was associated with its increased expression [31,32]. Hull et al. reported that the −251 A allele increases IL-8 levels after stimulation with lipopolysaccharide (LPS) [33]. IL-8 is a powerful pro-inflammatory cytokine, and as aforementioned, CAD is an inflammatory disease, so it is reasonable to conclude that an elevated level of IL-8 in −251 A allele carriers will increase the risk of CAD. This is in line with the results of our meta-analysis that −251 A allele increases the risk of CAD significantly.
This is the first meta-analysis assessing the association between the IL-8 gene −251 A/T polymorphism and the susceptibility of CAD. Inevitably, there are some limitations in our meta-analysis. First, the number of qualified studies was not large. Second, inclusion criteria were limited to articles published only in English and Chinese, this may have missed related non-English or non-Chinese literatures, resulting in the risk of bias [34]. Third, the genotype distribution in controls in 3 of the included studies was not in HWE, which may influence the reliability of the study results.
On the other hand, this meta-analysis has its distinct merits. First, a total number of 8244 subjects were recruited into the final pooled data analysis under our strict inclusion criteria. Second, the cases in all the included studies were angiographically confirmed CAD or MI patients, guaranteeing the diagnostic reliability of the cases. And the NOS quality scores of all the included studies were relatively high (6 or 5 score), indicating the original studies of this meta-analysis have a good quality [22]. Third, no significant publication bias was found in the included studies by both the Begg’s and Egger’s tests. Fourth, sensitivity analysis found no single study significantly influences the pooled ORs in all the genetic models, and, although as aforementioned, 3 of the included studies were not in HWE, we recalculated the pooled data after excluding these 3 studies and found significant associations were still present in all the genetic models, further confirming the reliability and stability of our results [27]. In addition, a subgroup analysis by ethnicity of study population was conducted, and the results showed a significant association between the IL-8 gene −251 A/T polymorphism and the risk of CAD in the Chinese population, but no significant association was found in the Caucasian population.
Conclusions
Our meta-analysis found that the IL-8 gene −251 A/T polymorphism was significantly associated with the risk of CAD in the Chinese population, individuals with the A allele of the IL-8 gene −251 A/T polymorphism had an increased risk of suffering from CAD. But this association was not found in the Caucasian population. This conclusion may be helpful for formulating individualized prevention and treatment strategies for CAD in the Chinese population in the future. However, in the light of the limitations of this meta-analysis, additional well-designed studies are needed to further verify the association of the IL-8 gene −251 A/T polymorphism with CAD risk.
Footnotes
Conflict of interests
None.
Source of support: This study was financially supported by Zhejiang Provincial Natural Science Foundation of China under Grant No. Y15H020003, Zhejiang Medical and Health Science and Technology Project under Grant No. 2016ZDB010, and Zhejiang Science and Technology Plan Project under Grant No. 2016C33207
References
- 1.Taleb S. Inflammation in atherosclerosis. Arch Cardiovasc Dis. 2016;109:708–15. doi: 10.1016/j.acvd.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Harrington RA. Targeting inflammation in coronary artery disease. N Engl J Med. 2017;377:1197–98. doi: 10.1056/NEJMe1709904. [DOI] [PubMed] [Google Scholar]
- 3.Ridker PM, Everett BM, Thuren T, et al. Anti-inflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med. 2017;377:1119–31. doi: 10.1056/NEJMoa1707914. [DOI] [PubMed] [Google Scholar]
- 4.Weber C, von Hundelshausen P. CANTOS trial validates the inflammatory pathogenesis of atherosclerosis: Setting the stage for a new chapter in therapeutic targeting. Circ Res. 2017;121:1119–21. doi: 10.1161/CIRCRESAHA.117.311984. [DOI] [PubMed] [Google Scholar]
- 5.Rus HG, Vlaicu R, Niculescu F. Interleukin-6 and interleukin-8 protein and gene expression in human arterial atherosclerotic wall. Atherosclerosis. 1996;127:263–71. doi: 10.1016/s0021-9150(96)05968-0. [DOI] [PubMed] [Google Scholar]
- 6.Gerszten RE, Garcia-Zepeda EA, Lim YC, et al. MCP-1 and IL-8 trigger firm adhesion of monocytes to vascular endothelium under flow conditions. Nature. 1999;398:718–23. doi: 10.1038/19546. [DOI] [PubMed] [Google Scholar]
- 7.Herder C, Baumert J, Thorand B, et al. Chemokines and incident coronary heart disease: Results from the MONICA/KORA Augsburg case-cohort study, 1984–2002. Arterioscler Thromb Vasc Biol. 2006;26:2147–52. doi: 10.1161/01.ATV.0000235691.84430.86. [DOI] [PubMed] [Google Scholar]
- 8.Inoue T, Komoda H, Nonaka M, et al. Interleukin-8 as an independent predictor of long-term clinical outcome in patients with coronary artery disease. Int J Cardiol. 2008;124:319–25. doi: 10.1016/j.ijcard.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 9.Qin B, Li L, Wang S, et al. Interleukin-8 gene polymorphism −251T>A contributes to Alzheimer’s disease susceptibility. Medicine (Baltimore) 2016;95:e5039. doi: 10.1097/MD.0000000000005039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Liu Y, Yang L, et al. The polymorphism interleukin-8 −251 A/T is associated with a significantly increased risk of cancers from a meta-analysis. Tumour Biol. 2014;35:7115–23. doi: 10.1007/s13277-014-1881-5. [DOI] [PubMed] [Google Scholar]
- 11.Yang ZJ, Tang XP, Lai QG, et al. Interleukin-8 −251 A/T polymorphism and periodontitis susceptibility: A meta-analysis. Genet Mol Res. 2016:15. doi: 10.4238/gmr15047379. [DOI] [PubMed] [Google Scholar]
- 12.Vogiatzi K, Apostolakis S, Voudris V, et al. Interleukin 8 gene polymorphisms and susceptibility to restenosis after percutaneous coronary intervention. J Thromb Thrombolysis. 2010;29:134–40. doi: 10.1007/s11239-009-0338-y. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, Zhang B, Zhang M, et al. Interleukin-8 gene polymorphism is associated with acute coronary syndrome in the Chinese Han population. Cytokine. 2011;56:188–91. doi: 10.1016/j.cyto.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 14.Wang S, Chen L, Dai Y, et al. Association of IL-8 −251 A/T and +781C/T polymorphisms with the susceptibility to coronary artery disease in a population of China. Int J Clin Exp Pathol. 2016;9:8471–77. [Google Scholar]
- 15.Zhang RJ, Li XD, Zhang SW, et al. IL-8 −251 A/T polymorphism contributes to coronary artery disease susceptibility in a Chinese population. Genet Mol Res. 2017:16. doi: 10.4238/gmr16018224. [DOI] [PubMed] [Google Scholar]
- 16.Kaur N, Singh J, Reddy S. Association of IL-8 −251 A/T rs4073 and IL-10 rs1800872 −592C/A polymorphisms and coronary artery disease in north Indian population. Biochem Genet. 2018 doi: 10.1007/s10528-018-9880-7. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 17.Vogiatzi K, Apostolakis S, Voudris V, et al. Interleukin 8 and susceptibility to coronary artery disease: A population genetics perspective. J Clin Immunol. 2008;28:329–35. doi: 10.1007/s10875-008-9194-3. [DOI] [PubMed] [Google Scholar]
- 18.Hou H, Gong H, Zhao H, et al. Relationship between IL-8 −251 A/T single nucleotide polymorphism and plasma level with coronary heart disease susceptibility. Chin J Arterioscler. 2012;20:261–64. [Google Scholar]
- 19.Velasquez IM, Frumento P, Johansson K, et al. Association of interleukin 8 with myocardial infarction: Results from the Stockholm Heart Epidemiology Program. Int J Cardiol. 2014;172:173–78. doi: 10.1016/j.ijcard.2013.12.170. [DOI] [PubMed] [Google Scholar]
- 20.Ren B, She Q. Study on the association between IL-1β, IL-8 and IL-10 gene polymorphisms and risk of coronary artery disease. Int J Clin Exp Med. 2015;8:7937–43. [PMC free article] [PubMed] [Google Scholar]
- 21.Yang HT, Wang SL, Yan LJ, et al. Association of interleukin gene polymorphisms with the risk of coronary artery disease. Genet Mol Res. 2015;14:12489–96. doi: 10.4238/2015.October.16.16. [DOI] [PubMed] [Google Scholar]
- 22.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–5. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 23.Wakefield J. Bayesian methods for examining Hardy-Weinberg equilibrium. Biometrics. 2010;66:257–65. doi: 10.1111/j.1541-0420.2009.01267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–58. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 25.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48. [PubMed] [Google Scholar]
- 26.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riley RD, Sutton AJ, Abrams KR, Lambert PC. Sensitivity analyses allowed more appropriate and reliable meta-analysis conclusions for multiple outcomes when missing data was present. J Clin Epidemiol. 2004;57:911–24. doi: 10.1016/j.jclinepi.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 28.Sedgwick P, Marston L. How to read a funnel plot in a meta-analysis. BMJ. 2015;351:h4718. doi: 10.1136/bmj.h4718. [DOI] [PubMed] [Google Scholar]
- 29.Terrin N, Schmid CH, Lau J. In an empirical evaluation of the funnel plot, researchers could not visually identify publication bias. J Clin Epidemiol. 2005;58:894–901. doi: 10.1016/j.jclinepi.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–101. [PubMed] [Google Scholar]
- 31.Hacking D, Knight JC, Rockett K, et al. Increased in vivo transcription of an IL-8 haplotype associated with respiratory syncytial virus disease-susceptibility. Genes Immun. 2004;5:274–82. doi: 10.1038/sj.gene.6364067. [DOI] [PubMed] [Google Scholar]
- 32.Wacharasint P, Nakada TA, Boyd JH, et al. AA genotype of IL-8 −251 A/T is associated with low PaO(2)/FiO(2) in critically ill patients and with increased IL-8 expression. Respirology. 2012;17:1253–60. doi: 10.1111/j.1440-1843.2012.02244.x. [DOI] [PubMed] [Google Scholar]
- 33.Hull J, Ackerman H, Isles K, et al. Unusual haplotypic structure of IL8, a susceptibility locus for a common respiratory virus. Am J Hum Genet. 2001;69:413–19. doi: 10.1086/321291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyons G. Language: Another cause of publication bias. Eur J Anaesthesiol. 2016;33:620–21. doi: 10.1097/EJA.0000000000000469. [DOI] [PubMed] [Google Scholar]