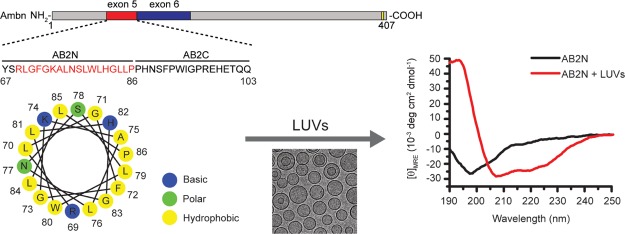
Abstract
Ameloblastin (Ambn), the most abundant non-amelogenin enamel protein, is intrinsically disordered and has the potential to interact with other enamel proteins and with cell membranes. Here, through multiple biophysical methods, we investigated the interactions between Ambn and large unilamellar vesicles (LUVs), whose lipid compositions mimicked cell membranes involved in epithelial cell-extracellular matrix adhesion. Using a series of Ambn Trp/Phe variants and Ambn mutants, we further showed that Ambn binds to LUVs through a highly conserved motif within the sequence encoded by exon 5. Synthetic peptides derived from different regions of Ambn confirmed that the sequence encoded by exon 5 is involved in LUV binding. Sequence analysis of Ambn across different species showed that the N-terminus of this sequence contains a highly conserved motif with a propensity to form an amphipathic helix. Mutations in the helix-forming sequence resulted in a loss of peptide binding to LUVs. Our in vitro data suggest that Ambn binds the lipid membrane directly through a conserved helical motif and have implications for biological events such as Ambn-cell interactions, Ambn signaling, and Ambn secretion via secretory vesicles.
Introduction
Enamel is composed of hydroxyapatite (HAP) crystallites with an architecture that is precisely ordered on several length scales.1 The formation of enamel occurs in the extracellular matrix (ECM),2 which includes proteins that interact with minerals, with other proteins,3−5 and most likely with cells.6 Most ECM proteins in enamel belong to the large secretory calcium-binding phosphoprotein (SCPP) family, which evolved from a common ancestral gene more than 450 million years ago.7,8 In enamel, the ECM consists of a distinct set of macromolecules that are mostly intrinsically disordered, including amelogenin (Amel), ameloblastin (Ambn), enamelin (Enam), and amelotin (Amtn).9−12 These proteins self-assemble or coassemble to create a functional three-dimensional ECM that serves to guide its own replacement by the mineral phase.13−16
Ambn, also known as amelin or sheathlin, is the second most abundant proline-rich enamel matrix protein after Amel.17−19 The teeth of ameloblastin mutant mice that lacked amino acid sequences encoded by exons 5 and 6 were found to have a severely hypoplastic enamel layer, establishing the importance of Ambn for proper enamel formation.6 Ambn is secreted together with Amel and is rapidly processed by matrix metalloproteinase-20 (MMP-20) at its C-terminus after secretion.20 The hydrophobic N-terminal cleavage products accumulate in the “sheath” space throughout the enamel layer, whereas the calcium-binding C-terminal cleavage products accumulate on the enamel rods.21 Because of its localization at the rod–interrod boundary, the N-terminal region of Ambn has been thought to play a role in defining the prismatic structure of enamel. We recently reported that Ambn is not the only protein fragment at the rod–interrod boundaries but colocalizes together with Amel N-terminal fragments.4 In in vitro studies we further demonstrated interactions between the N-termini of Ambn and Amel.4,22
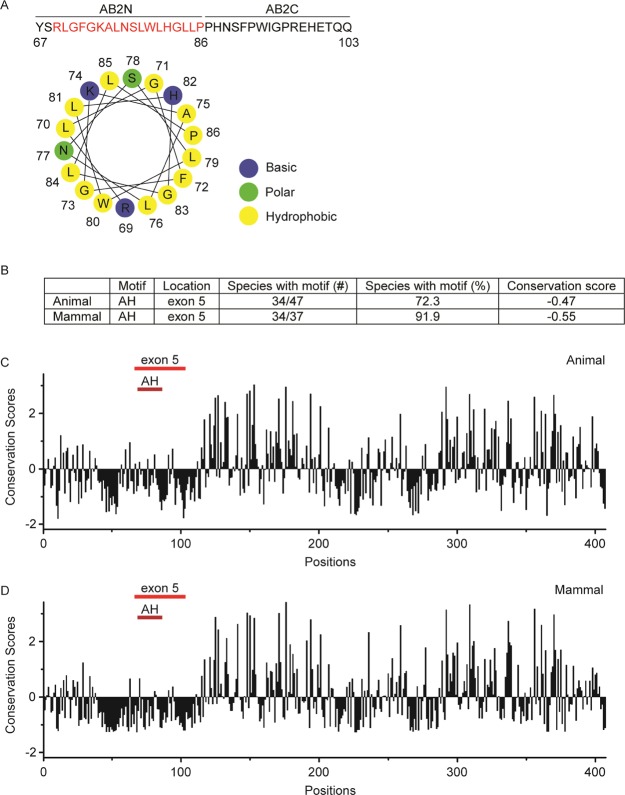
Evolutionary analysis of the Ambn sequences across 47 species shows that this protein has kept its function in tooth enamel formation for more than 200 million years in both prismatic and nonprismatic enamel, which suggests that its functionality extends beyond prismatic structure-building.23 Additional putative functions may include communication between the ECM and ameloblast cells. It has been proposed that Ambn may mediate cell–matrix adhesion through its integrin-binding motif,24 heparin-binding motifs,25 or fibronectin-binding motif.26 However, molecular modeling has not revealed any regions in Ambn with structural similarity to known receptor–ligand systems.27 A systematic analysis of the full-length protein sequences of Ambn from 47 different species in different classes showed that only a few species have these motifs with identical sequences (see average conservation score in Table 1). It is therefore unreasonable to assume that a protein functionality would depend on these poorly conserved motifs. In contrast, the homogeneity in the 37 AA sequences encoded by exon 5 across species is relatively high. This high level of conservation led us to hypothesize that exon 5 motifs are the critical motifs in the Ambn sequence for cell adhesion function (Table 1). Additional support for this hypothesis is based on mutant animal models in which deletion of sequences encoded by exons 5 and 6 resulted in detachment of extracellular enamel matrix from the ameloblasts. The authors suggested that Ambn assists ameloblasts in adhering to the ECM during the secretory stage of enamel formation.6 However, molecular mechanisms underlying such adhesion and interactions have not been fully elucidated. No specific receptors for enamel ECM proteins to adhere to the cells have been identified to date. CD63 and Lamp-1 have been identified as putative receptors for Amel.28,29 Although these receptors are involved in endocytosis,30 they do not support cell–matrix adhesion or construction of a functional matrix. Information on receptors for Enam and Amtn is also limited.
Table 1. Sequence Conservation of Reported Integrin-, Heparin-, and Fibronectin-Binding Motifs and Sequences Encoded by Exons 5 and 6 of Ambn across 47 Different Species from the Classes of Ray-Finned Fishes, Lobe-Finned Fishes, Amphibians, Reptiles, and Mammalsa.
| reported motifs | sequence | location | species with identical motif (#) | species with identical motif (%) | average conservation score |
|---|---|---|---|---|---|
| heparin-binding motifs | HSLSRGPMAHNK | exon 10 | 1/47 | 2.1 | 0.22 |
| VTKG | exon 13 | 4/47 | 8.5 | –0.28 | |
| KVHQPQVHNAWR | exon 13 | 2/47 | 4.3 | 0.30 | |
| fibronectin-binding motif | VPIMDFADPQFPT | exon 7 | 2/47 | 4.3 | 0.94 |
| integrin-binding motif | DGEA | exon 13 | 3/47 | 6.4 | 2.66 |
| N/A | sequence encoded by exon 5 | exon 5 | N/A | N/A | –0.58 |
| N/A | sequence encoded by exon 6 | exon 6 | N/A | N/A | 0.34 |
The conservation scores were calculated by ConSurf. The GenBank accession numbers are provided in Table S1. “N/A” is used in the “species with identical motif” columns for exons 5 and 6 because no defined motifs have been reported for these sequences. Average conservation score and “species with identical motif” are two independent values. See the Experimental Section for the definition of Average conservation score. Note that more negative numbers indicate higher conservation.
In the present in vitro study, we used liposomes or large unilamellar vesicles (LUVs) as a chemical model for cell membrane phospholipid bilayers. These are structures composed of amphiphilic lipids and have been extensively used as cell membrane models.31−33 The lipid composition we used here was based on the membrane domain that is involved in epithelial cell–ECM adhesion.34 Using biophysical and Cryo-TEM methods, we investigated the binding of recombinant mouse Ambn to lipid bilayers. For fluorescence spectroscopy, we designed a series of Ambn Trp variants in which one or two of the three intrinsically fluorescent Trp residues were changed to Phe. Structural changes as a result of LUV addition were then detected using the remaining Trp residues. We synthesized Ambn mutants lacking sequences encoded by exons 5 and 6 to evaluate their function in LUV binding. We further generated a series of Ambn-derived synthetic peptides and peptide variants to identify the liposome-binding motifs in the Ambn sequence. Our in vitro data have biological implications that are relevant not only to Ambn–cell interactions but also to Ambn signaling and to the mechanisms of Ambn secretion via secretory vesicles.
Results and Discussion
Ambn Interacts with LUVs
LUVs were generated using a mixture of several lipids to mimic the primary lipid composition of the membrane domains involved in epithelial cell–ECM adhesion (Figure 1A).34 Mouse ameloblastin (Ambn) was expressed in Escherichia Coli, and the purified Ambn was characterized by SDS-PAGE, mass spectroscopy, and dynamic light scattering (DLS) (Figures S1 and S2). Fluorescence (Figure 1B) and CD spectroscopy (Figure 1C), Cryo-TEM (Figure 2A–D), membrane leakage (Figure 2E), and static light scattering (Figure 2F) were conducted to determine the interaction between recombinant mouse Ambn and LUVs.
Figure 1.
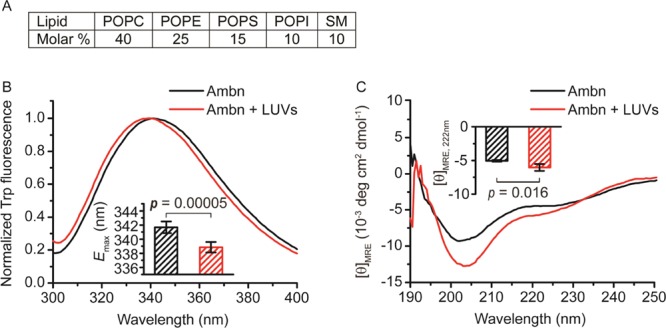
Changes in Ambn secondary structure as the result of LUV addition. (A) Lipid composition used to assemble 100 nm diameter LUVs (see experimental). (B) Fluorescence and (C) CD spectra of Ambn with 300 μM 100 nm LUVs. The inset in (B) shows the emission maxima of the Trp fluorescence spectra (Student’s t-test, n = 6). The inset in (C) shows the changes of averaged mean residue ellipticity values at 222 nm ([θ]MRE,222nm) in the presence of LUVs (Student’s t-test, n = 4).
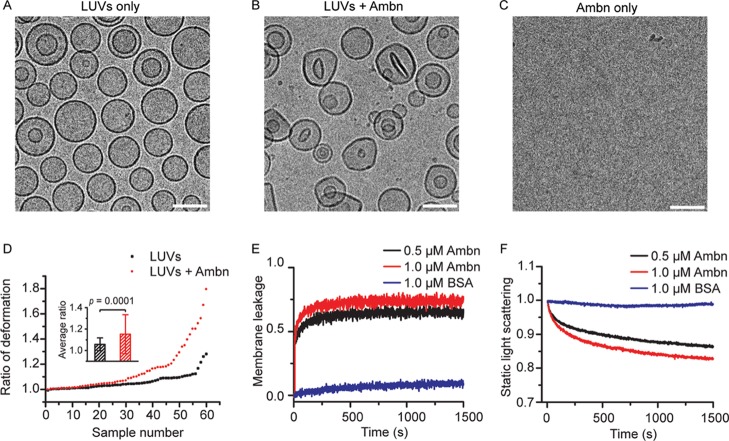
Figure 2.
Changes in LUV morphology and size distribution as the result of Ambn addition. (A,B) Representative Cryo-TEM images of LUVs in the absence (A) and presence (B) of 1.0 μM Ambn. (C) Cryo-TEM image of 1.0 μM Ambn. Scale bar is 100 nm. (D) Quantitative analysis of LUV deformation based on Cryo-TEM images. The extent of LUV deformation was defined by the ratio of the longest dimension to the shortest dimension for each vesicle. The ratios of 60 selected LUVs for each category (LUVs only and LUVs with Ambn) were measured. The inset shows the averaged ratio of deformation (Student’s t-test, n = 60). (E) Normalized membrane leakage of LUVs in the presence of two concentrations of Ambn and BSA, which was used as the control. (F) Normalized static light scattering of 300 μM LUVs with two concentrations of Ambn and BSA, which was used as the control.
Trp fluorescence spectra showed that Ambn had a blue shift in the presence of LUVs (Figure 1B). The emission maximum wavelength (Emax) had a blue shift of 2.8 ± 1.3 nm (Student’s t-test, p = 0.00005, n = 6) in the presence of 100 nm LUVs, suggesting that the presence of LUVs changed the local environment of the Trp residues to a more hydrophobic one, presumably as the result of its interactions with lipids.
We monitored changes in the secondary structure by CD spectroscopy. The small peak at 208 nm and the shoulder at 222 nm in the absence of lipids compared to the deeper, more pronounced peaks upon the addition of LUVs indicate a coil-helix transition and suggest that Ambn is mainly intrinsically disordered in solution (Figure 1C), which is consistent with a previous report.11 This coil–helix transition is also supported by the rightward shift in the main downward peak. When LUVs were added, the mean residue ellipticity at 190 nm (MRE190nm) increased, forming a more pronounced peak, suggesting that Ambn became more structured. The MRE208nm and MRE222nm decreased in the presence of 100 nm LUVs, forming more pronounced negative peaks, suggesting that α-helix content in Ambn increased in the presence of LUVs. Analyses of the CD spectra using DICHROWEB showed that the α-helix content increased from approximately 8.9 ± 1.3% to approximately 15.5 ± 2.7% in the presence of LUVs, whereas the percentage of β-sheet and disordered structure decreased.
Direct visualization by Cryo-TEM revealed that LUVs without Ambn were spherical with a diameter of 103.8 ± 20.3 nm (Figure 2A), whereas LUVs with Ambn lost much of their spherical shape and had an average longest dimension of 106.8 ± 19.3 nm (Figure 2B). Cryo-TEM images of 1.0 μM Ambn did not show any structures in the control or Ambn groups (Figure 2C). The extent of LUV deformation was defined by the ratio of the longest dimension over the shortest dimension for each vesicle. About 30% of the LUVs were deformed significantly following the addition of Ambn. The average ratio of deformation was 1.06 ± 0.06 for 60 selected LUVs in the control group and 1.15 ± 0.18 for 60 selected LUVs in the Ambn group (Figure 2D, inset). This difference was statistically significant (Student’s t-test, p = 0.0001), suggesting that the morphology of LUVs has changed in the presence of Ambn. We interpret these observations to suggest that Ambn interacts with the phospholipid bilayer surface directly causing this deformation. Under our experimental conditions and Ambn concentration used (1.0 μM) to investigate Ambn–LUVs interactions, Ambn neither formed detectable large aggregates nor showed any defined morphology (Figure 2C).
Independent of Cryo-TEM observations, a membrane leakage assay showed that Ambn caused concentration-dependent membrane leakage starting from 0.5 μM and increasing at 1.0 μM (Figure 2E). These observations suggest that Ambn had a high affinity for the lipid bilayer of LUVs and disrupted their membrane integrity in a dose-dependent manner. Disintegration of the LUVs by Ambn was evidenced by decrease in their size distribution when Ambn was added. Changes in the LUV size were monitored by measuring static light scattering at 300 nm over time in a lipid vesicle clearance assay. Static light scattering decreased rapidly in the presence of 0.5 μM Ambn and more rapidly in the presence of 1.0 μM Ambn. BSA which was used as a negative control did not affect the size distribution of LUVs even at 1.0 μM concentration (Figure 2F), implying that Ambn altered the LUVs’ integrity in a specific manner.
Ambn Binds Phospholipids via an N-Terminal Motif
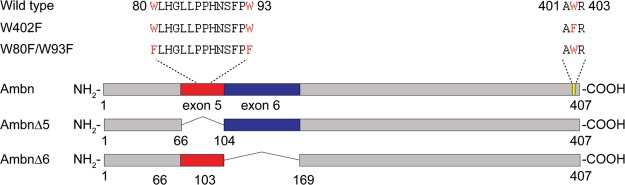
The presence of Trp in the sequences on enamel matrix proteins such as amelogenin has allowed us to identify interacting regions of these proteins with their targets using intrinsic fluorescence techniques.31,35 If a Trp residue is part of a membrane-binding region, a blue shift in fluorescence would be detectable upon LUV addition because of immersion of the Trp residue in the hydrophobic environment of the membrane.31,35 Here, in order to identify the membrane-binding regions of Ambn, two Ambn W/F variants were designed and expressed (Figure S1). W402F (at the C-terminus) and W80F/W93F (at the N-terminus) Ambn variants were created to identify whether the membrane binding occurs at the C- or N-terminal end. W402F Ambn has only two tryptophan residues at the N-terminus, and W80F/W93F Ambn has only one tryptophan residue at the C-terminus (Figure S1), allowing us to distinguish the regions in which LUVs cause conformational changes in Ambn upon interaction. As was previously shown for amelogenin,31,32,35 CD spectra of the variants have confirmed that such variation does not affect protein conformation (Figure S3). The emission maximum wavelength of the W80F/W93F variant did not change significantly (p = 0.19) in the presence of LUVs (Figure 3A), indicating that the environment of the Trp402 at the C-terminus did not change implying that the C-terminus of Ambn is not involved in liposome binding. In contrast, the emission maximum wavelength of the W402F variant shifted from 339.3 ± 0.6 to 336.3 ± 1.2 nm (p = 0.008) in the presence of LUVs (Figure 3B), indicating that the solvent environment around the N-terminal Trp residues at positions 80 and 93 of Ambn has changed implying that the N-terminus is involved in interaction with LUVs. These data together with the CD spectra suggest that a key region possibly encompassing W80 and/or W93 binds membranes because of hydrophobic interactions between its residues and membrane lipids. We thus sought to locate possible key binding regions.
Figure 3.
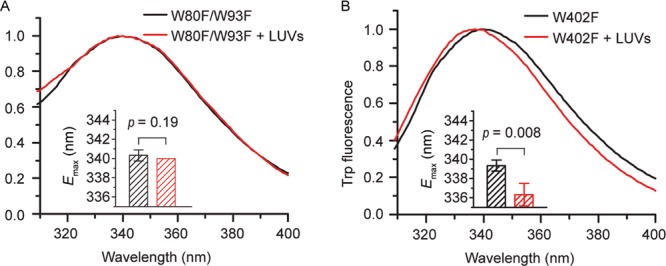
Identification of the liposome-binding region using Ambn Trp variants. Trp fluorescence spectra of (A) 2.0 μM AmbnW80F/W93F and (B) AmbnW402F variants titrated with 300 μM LUVs. The insets show the emission maxima of the Trp fluorescence spectra. Student’s t-test, n = 3.
In a mouse model (originally described as Ambn KO6 but later demonstrated to be a mutant AmbnΔ5-636 in which exons 5 and 6 of Ambn were deleted), a truncated form of ameloblastin that included the C-terminus was expressed, resulting in severe disturbance of enamel formation and detachment of ameloblasts from the ECM.6 The authors then concluded that the cell binding capacity of Ambn resides in the deleted portion, namely, sequences encoded by exons 5 and 6. To examine whether sequences encoded by exons 5 or 6 are involved in membrane binding, we created recombinant mutants; the sequence encoded by exon 5 was deleted to create Ambn p.Y67_Q103del (AmbnΔ5), and the sequence encoded by exon 6 was deleted to create Ambn p.P104_V168del (AmbnΔ6) (Figure S1). Membrane leakage assays showed that lipid vesicles leaked more slowly in the presence of the AmbnΔ5 mutant (Figure 4A, red line). In contrast, vesicles became leaky faster in the presence of wild-type Ambn and AmbnΔ6 (Figure 4A, blue and black lines). This difference suggests that the ability of Ambn to disrupt membrane integrity was lost when the sequence encoded by exon 5 was deleted. Lipid vesicle clearance assays showed that the LUVs disintegrated into small entities more slowly in the presence of AmbnΔ5, whereas they disintegrated more quickly in the presence of wild-type Ambn and the AmbnΔ6 mutant (Figure 4B), suggesting that Ambn lost its ability to interact with vesicles without the sequence encoded by exon 5.
Figure 4.
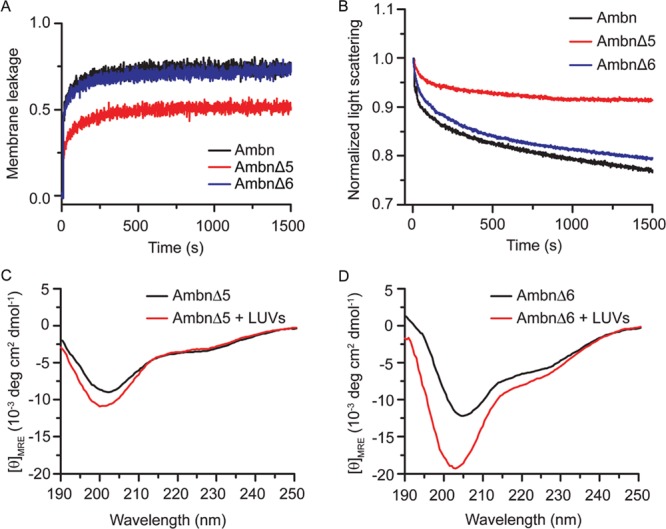
Ambn mutants lose their liposome binding ability. (A) Normalized membrane leakage of LUVs in the presence of 1.0 μM WT Ambn, AmbnΔ5, and AmbnΔ6. (B) Normalized static light scattering of 300 μM LUVs with 1.0 μM WT Ambn, AmbnΔ5, and AmbnΔ6. (C) CD spectra of AmbnΔ5 and (D) AmbnΔ6 in the absence and presence of LUVs.
CD spectra of the mutants in solution are shown in Figure S4, and CD spectra of mutants with and without LUVs are shown in Figure 4C,D. The spectrum of AmbnΔ5 in solution without LUVs differs from AmbnΔ6 and WT Ambn spectra, possibly indicating changes in the secondary structure that may lead to misfolding or aggregation. Upon addition of LUVs (Figure 4C), AmbnΔ5 showed little to no change in CD spectra, whereas AmbnΔ6 showed a slight change in CD spectra, as the peak at 208 nm and the shoulder at 222 nm were augmented in the presence of lipids (Figure 4D). Therefore, both of these regions of Ambn are important for the helix–coil transition induced by the presence of a membrane. The differences between WT Ambn and AmbnΔ5 CD spectra, both in solution and in the presence of LUVs, as well as the lack of membrane interaction observed in membrane leakage and static light scattering experiments on AmbnΔ5 (Figure 4A,B) altogether suggest that exon 5 is required for both membrane binding and maintenance of the ordered structure of the protein. Although AmbnΔ6 disrupted LUV membranes nearly as efficiently as WT Ambn (Figure 4A,B) and showed a slight increase in α-helicity in the presence of LUVs (Figure 4D), its CD spectra upon addition of LUVs lacked the significant rightward shift at 208 nm observed in the case of WT Ambn (Figure 1C). This nuanced result suggests that AmbnΔ6 has slightly diminished binding to liposomes possibly because of folding or conformational changes. We therefore suggest that although membrane binding occurs via the sequence encoded by exon 5, the exon 6 sequence has an important structural role to achieve the desired folding of the protein during membrane binding. A family with amelogenesis imperfecta (AI) was found to carry a genomic deletion of Ambn exon 637 and for the most part had thin, aprismatic enamel, underscoring the functional importance of this region.
Synthetic Peptide Encoded by Exon 5 Interacts with Phospholipids
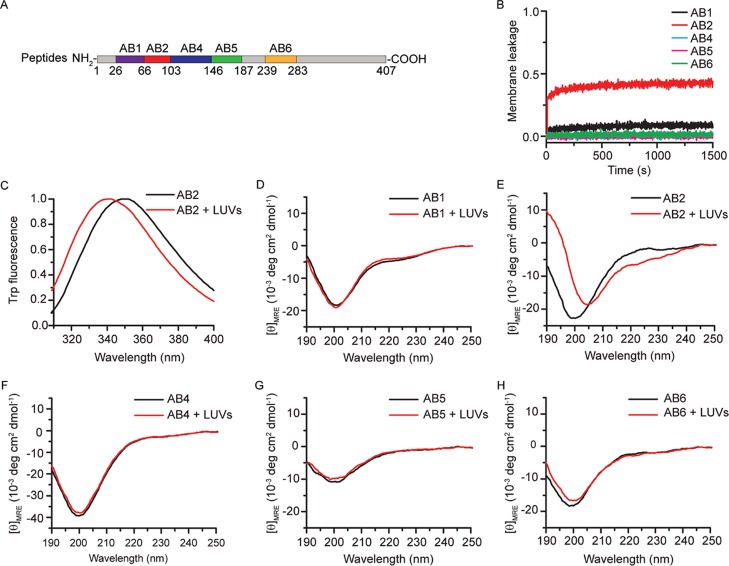
To further pinpoint the cell membrane-binding motif of Ambn and confirm that the lack of AmbnΔ5 binding to liposomes was not merely the result of protein misfolding, we examined the binding affinity of a series of Ambn-derived peptides (AB1, AB2, AB4, AB5, and AB6) to LUVs (Figure 5A, Table 2). AB1 included 40 amino acid residues encoded by exons 3 and 4, AB2 included 37 amino acid residues encoded by exon 5, AB4 included 43 amino acid residues encoded by exon 6, AB5 included 41 amino acid residues encoded by exons 6 and 7, and AB6 included 44 amino acid residues encoded by exon 13. AB5 was the linker sequence between two domains.
Figure 5.
Ambn-derived peptides confirmed that the sequence encoded by exon 5 is involved in liposome-binding. (A) Schematic of Ambn-derived peptides (see Table 2). (B) Normalized membrane leakage of LUVs in the presence of 1.0 μM peptides AB1, AB2, AB4, AB5, and AB6. (C) Trp fluorescence spectra of 10.0 μM peptide AB2 with 300 μM LUVs. (D–H) CD spectra of 10.0 μM peptides AB1, AB2, AB4, AB5, and AB6 with LUVs.
Table 2. The Amino Acid Sequences and Masses of Ambn (AB1, AB2, AB2N, AB2C, AB4, AB5, and AB6) Peptidesa.
| peptides | exons | sequence | mass (Da) |
|---|---|---|---|
| AB1 | exon 3 and 4 | 27VPAFPQQPGAQGMAPPGMASLSLETMRQLGSLQGLNALSQ66 | 4080.75 |
| AB2 | exon 5 | 67YSRLGFGKALNSLWLHGLLPPHNSFPWIGPREHETQQ103 | 4284.89 |
| AB4 | N-terminus of exon 6 | 104PSLQPHQPGLKPFLQPTAATGVQVTPQKPGPQPPMHPGQLPLQ146 | 4542.33 |
| AB5 | exon 7 and C-terminus of exon 6 | 147EGELIAPDEPQVAPSENPPTPEVPIMDFADPQFPTVFQIAR187 | 4491.05 |
| AB6 | exon 13 | 240YGTLFPRFGGFRQTLRRLNQNSPKGGDFTVEVDSPVSVTKGPEK283 | 4884.53 |
| AB2N | N-terminus of exon 5 | 67YSRLGFGKALNSLWLHGLLP86 | 2242.67 |
| AB2C | C-terminus of exon 5 | 87PHNSFPWIGPREHETQQ103 | 2060.23 |
AB2 is the sequence encoded by exon 5.
Comparative membrane leakage assays using these peptides showed that lipid vesicles became leaky in the presence of AB2 (a sequence encoded by exon 5) but only slightly so in the presence of AB1 and still less in the presence of AB4, AB5, or AB6 (Figure 5B), suggesting that the sequence encoded by exon 5 could disrupt LUV membrane integrity. Trp fluorescence spectra of AB2 showed a significant blue shift in the presence of LUVs (Figure 5C). The emission maximum wavelength shifted significantly from 349.3 ± 0.4 to 339.5 ± 0.7 nm, suggesting that the microenvironment of the Trp of AB2 became much more hydrophobic in the presence of LUVs. CD spectra of peptides (AB1, AB2, AB4, AB5, and AB6) showed that only the secondary structure of AB2 significantly changed in the presence of LUVs (Figure 5D–H), confirming that AB2 is involved in membrane binding. When LUVs were added to AB2, the MRE190nm of AB2 increased from to −7.2 ± 0.3 to 9.4 ± 0.2 deg cm2 dmol–1 and the MRE222nm decreased from −2.2 ± 0.2 to −6.0 ± 0.8 deg cm2 dmol–1. Analyses of the CD spectra using DICHROWEB showed that the α-helix content of AB2 increased from approximately 7.9 ± 1.6 to 25.0 ± 3.1% in the presence of LUVs, whereas the β-sheet content and disordered structure content decreased, suggesting that LUVs induced AB2 to form an α-helix.
Using synthetic peptides, we show that the sequence encoded by exon 5 and not exon 6 interacts with phospholipids. We found that the lack of sequence encoded by exon 6 resulted in slightly diminished binding of Ambn to liposomes (Figure 4D). This may well be related to misfolding or premature aggregation of Ambn which may lead to blockage of the interacting domain (i.e., sequence encoded by exon 5) or interruption of ordered Ambn self-assembly.15
The Liposome-Binding Motif Is at the N-Terminus of the Sequence Encoded by Exon 5
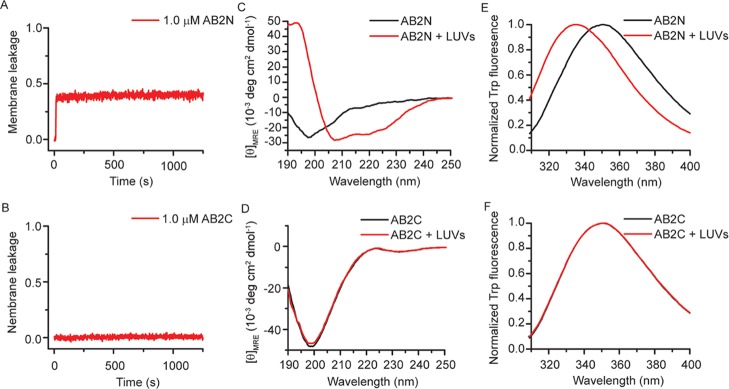
In order to further identify and narrow down the lipid binding domain within the sequence encoded by exon 5, we synthesized AB2N peptide, representing the N-terminus of AB2, and AB2C, representing the C-terminus of AB2 (Table 2). Membrane leakage assays showed that AB2N made LUVs leaky, whereas AB2C did not (Figure 6A,B). CD spectra showed that AB2N was intrinsically disordered, and its α-helix content increased from 7.0 ± 0.4 to 61.8 ± 6.6% in the presence of LUVs (Figure 6C). In contrast, AB2C had no random coil to α-helix transition (Figure 6D). Trp fluorescence showed that the emission maximum wavelength of AB2N shifted from about 350.0 to 336 nm (Figure 6E), suggesting that the microenvironment of Trp became more hydrophobic in the presence of LUVs. In contrast, the fluorescence spectra of AB2C did not change significantly (Figure 6F).
Figure 6.
The liposome-binding motif of Ambn is at the N-terminal end of the sequence encoded by exon 5. (A,B) Normalized membrane leakage of 300 μM LUVs with 1.0 μM AB2N or AB2C, representing the N- or C-terminal region of AB2, respectively. (C,D) CD and (E,F) Trp fluorescence spectra of 10.0 μM AB2N or AB2C with 300 μM LUVs.
Among full-length Ambn, AB2, and AB2N, AB2N shows the purest coil-helix transition, as evidenced by the isosbestic point of its CD spectra at 203 nm (Figure 6C), the point at which many peptides’ CD spectra in random coil and α-helical states intersect.38 In contrast, Ambn has an isosbestic point at ∼196 nm (Figure 1C) and AB2 at ∼205 nm (Figure 5E), indicating the presence of regions with other structures or lacking in structure alongside the region that turns helical in the presence of liposomes. AB2N appears to consist only of the amino acids that undergo the helix–coil transition observed in all three forms.
N-Terminus of the Sequence Encoded by Exon 5 Contains an Amphipathic Helix
An amphipathic helix (AH) is a common motif encountered in various proteins and peptides, and it can act as a membrane anchor, as a tool to deform lipid membranes, or as a sensor of membrane curvature.39,40 We analyzed the entire sequence of mouse Ambn for the presence of amphipathic helices using Heliquest (Table S2). There are four overlapping regions at the N-terminus of Ambn between residues 58 and 86 that have a tendency to form AH. We noted that the N-terminus of the sequence encoded by exon 5 (AB2N) contains a highly conserved sequence (91.9%) among 37 mammalian species tested with a propensity to form an AH (69RLGFGKALNSLWLHGLLP86). The projection of AB2N on a helical wheel showed that AB2N has a proper hydrophobic moment and net charge (Figure 7A and Table 3) and a putative hydrophobic face at its N-terminus (Table S2), suggesting that AB2N has the sequence requirements to form an AH upon its binding to LUVs (Figure 7A). The highly negative conservation scores (−0.55 among mammals and −0.47 across 47 species) of the residues corresponding to AB2N (AH motif) (Figure 7B–D) indicate that evolution has preserved this AH-forming motif and reflect on its probable functional importance. The high AH motif sequence conservation among mammalian species may suggest a function in conservation of prismatic structures in mammals, and it is the subject of future studies.41,42
Figure 7.
AH in Ambn. (A) The amino acid sequence of the segment encoded by exon 5 of mouse Ambn and a schematic presentation of the AH wheel formed by AB2N. (B) Analysis of the AH motif (AB2N) homogeneity within exon 5 among 47 species and 37 mammals. (C,D) Position-specific conservation scores of full-length mouse Ambn in 47 animal species (C) and 37 mammals (D). The amino acids sequences of 47 species were aligned by using ClustalX2 (Table S1).43 Then, the conservation scores of mouse Ambn were computed by Consurf44 using the multiple sequence alignment of these sequences and the empirical Bayesian algorithms. The locations of exons 5 and the amphipathic sequence (AH) within exon 5 are shown by red lines. Note that highly conserved regions are below zero.
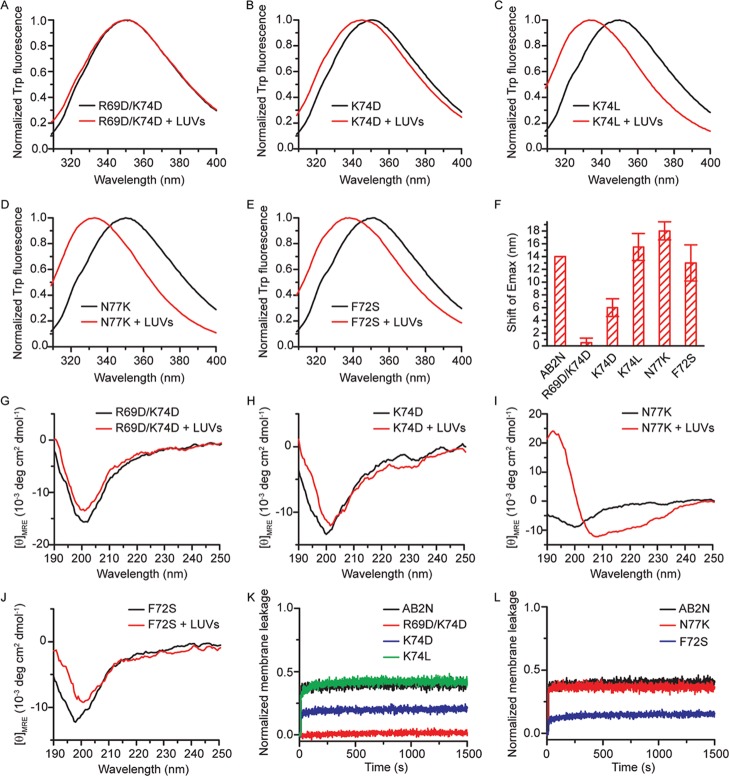
Table 3. Sequences and CD Structural Characteristics of Peptide Variants R69D/K74D, K74D, K74L, N77K, and F72S Derived from the Identified Membrane-Binding Motif (AB2N)a.
| name | sequence | net charge | μH | H face | Δ[θ]MRE,222nm | AH |
|---|---|---|---|---|---|---|
| AB2N | YSRLGFGKALNSLWLHGLLP | +2.25 | 0.129 | Y | –17.3 ± 1.2 | Y |
| R69D/K74D | YSDLGFGDALNSLWLHGLLP | –1.75 | 0.133 | Y | –0.1 ± 1.0 | N |
| K74D | YSRLGFGDALNSLWLHGLLP | +0.25 | 0.119 | Y | –0.7 ± 0.5 | N |
| K74L | YSRLGFGLALNSLWLHGLLP | +1.25 | 0.090 | Y | ND | N |
| N77K | YSRLGFGKALKSLWLHGLLP | +3.35 | 0.128 | Y | –7.6 ± 0.6 | Y |
| F72S | YSRLGSGKALNSLWLHGLLP | +2.25 | 0.112 | N | –0.8 ± 0.5 | N |
The amino acids that were the targets for variations are highlighted by style. The positively charged residues R and K are bold, the negatively charged residue D is bold-italic, the polar residue S is underlined, and the hydrophobic residues F and L are italic. μH: hydrophobic moment calculated by HeliQuest, AH: amphipathic Helix, ND: not determined.
The Positive Charge and Hydrophobic Surface in the Amphipathic Helix of Ambn Are Vital for Its Liposome-Binding Ability
To examine whether the amphipathic character of AB2N is vital for membrane-binding ability, a series of peptide variants of AB2N with disruption to the hydrophobic face and charges (R69D/K74D, K74D, K74L, N77K, and F72S) were synthesized (Table 3). In R69D/K74D, K74D, K74L, and N77K, the positive charge on the polar side of the AH was changed to different extents, but the hydrophobic face was not affected. These changes were expected to affect the long-range electrostatic interaction between the peptide and LUVs. In F72S, the hydrophobic face was disrupted but the positive charge on the polar side of the AH was unchanged. We expected that this change to the hydrophobic face would disrupt the hydrophobic effect that transfers the peptide to the membrane. We predicted that N77K might form an AH, whereas all other peptide variants would not because of their altered net charge, insufficient hydrophobic moment, or lack of a hydrophobic face. Fluorescence spectra of R69D/K74D with LUVs showed no significant change upon LUV addition (Figure 8A,F), suggesting that R69D/K74D did not interact with LUVs. Fluorescence spectra of K74D with LUVs had a slight blue shift (Figure 8B,F), suggesting that K74D interacted with LUVs weakly. Fluorescence spectra of K74L, N77K, and F72S had similar, significant blue shifts (Figure 8C–F), implying that the LUVs interacted with these peptide variants. CD spectra of R69D/K74D and K74D showed no significant change upon LUV addition (Figure 8G–H), suggesting that the mutated AB2N did not undergo a random coil to α-helix transition because of a lack of sufficient positive charge. The MRE190nm and MRE222nm of N77K significantly increased and decreased, respectively (Figure 8I), suggesting that N77K became more structured, with 37.1.0 ± 3.0% α-helical content (calculated by DICHROWEB) in the presence of LUVs. This peptide also shows a typical isosbestic point near 203 nm between its CD spectra. In contrast to the fluorescence spectra suggesting that F72S accumulated in the vicinity of negatively charged membranes of LUVs through long-range electrostatic interactions (Figure 8E), CD spectra of F72S showed no significant change (Figure 8J), indicating that F72S did not undergo the random coil to α-helix transition (see Δθ @ θ = 222 in Table 3). Membrane leakage assays showed that R69D/K74D did not disrupt membrane integrity and K74D disrupted it only weakly, whereas K74L and N77K disrupted membrane integrity strongly (Figure 8K,L). These observations suggest that the positive charge of the AH is important for membrane-binding affinity. This outcome is as expected because the membrane is 25% negatively charged and therefore should attract positively charged motifs via electrostatic interactions. F72S with a mutation from a nonpolar to a polar amino acid residue on the hydrophobic face (Figure 8L) only weakly disrupted membrane integrity. This observation suggests that the hydrophobic face of the AH is also important for membrane binding. Mutations on either charged or hydrophobic residues showed attenuation of membrane binding, supporting the notion that AB2N forms an AH in the presence of LUVs and its amphipathic character is vital for liposome binding.
Figure 8.
Formation of AH is vital for liposome-binding ability. (A–E) Trp fluorescence spectra of 10.0 μM peptide variants titrated with 300 μM LUVs. (F) Blue shifts of the emission maxima of the Trp fluorescence spectra of the peptide variants shown in A to E (n = 2). (G–J) CD spectra of 10.0 μM peptide variants titrated with 300 μM LUVs. (K,L) Normalized membrane leakage of LUVs in the presence of 1.0 μM peptide variants R69D/K74D, K74D, K74L, N77K, and F72S.
Conclusions
In order to explore the potential of Ambn to directly bind cell membranes, we investigated interactions between Ambn and large unilamellar vesicles (LUVs) as in vitro models for cell membrane lipid bilayers. By applying intrinsic fluorescence spectroscopy of Ambn variants in which Trp residues were mutated to Phe, we identified the phospholipid-binding region to be at the N-terminus. The finding that the recombinant Ambn mutant AmbnΔ5 did not bind to phospholipids supported the notion that interactions occur within the sequence encoded by exon 5. An isolated synthetic peptide representing the sequence encoded by exon 5 interacted with the phospholipids, as we observed the peptide to adopt an increased helical conformation as a result of LUV addition. Other Ambn-derived peptides encoded by exons 3, 4, 6, 7, and 13 did not show any conformational changes upon LUV addition suggesting a lack in the ability to interact with phospholipids.
The current in vitro study has provided important basic information on the structure of Ambn and its properties and sets a foundation for future mechanistic in vivo studies. Ambn is an intrinsically disordered protein that gains ordered structure, specifically an α-helical structure, in the presence of partially negatively charged lipids. This happens as the N-terminal part of the sequence encoded by exon 5, which consists of a highly conserved AH motif, undergoes a coil-helix transition and binds the membrane. We found that both hydrophobic and positively charged residues in the helical motif are crucial for this Ambn–membrane interaction.
Ameloblastin self-assembles into ribbon-like structures via a three Y motif (Y/F-x-x-Y/L/F-x-Y/F motif)45 located within the sequence encoded by exon 5. It was recently shown that triple mutations in this motif affected its self-assembly in vitro. As reported by Wald et al. in a mouse model with mutations in these three Y motifs, enamel appeared with disordered hydroxyapatite crystallites.45 Although these observations might highlight the importance of Ambn self-assembly, evidence for Ambn self-assembly in the mutant mice in vivo was limited. The defect in enamel formation in those animals may well be the result of perturbation in Ambn–Amel or Ambn–cell interactions.
In vitro studies together with present data collectively show that the exon 5 of Ambn is involved not only in its self-assembly and cell membrane binding but also in its interactions with amelogenin.3,4,22,45 Ambn may play a role in tethering the enamel matrix to the cell surfaces of ameloblasts, possibly through its interaction and coassembly with Amel.4,46 Like Ambn, Amel interacts with multiple targets via the same interacting domains in vitro and in vivo.4,22 We previously reported that recombinant Amel interacts with negatively charged small unilamellar vesicles via its N-terminal motif.32 Notably, the N-terminus of Amel is also involved in its interactions with Ambn, as well as in its self-assembly.22,47
Much study is needed to demonstrate such multitarget interactions for both Ambn and Amel in vitro and in vivo and gain further insight into the biological significance of Amel/Ambn–membrane interaction. However, the findings that Ambn is intrinsically disordered and has the potential to interact with different targets support such molecular mechanisms in vivo.4,46,48,49 In addition the biological significance of Ambn–cell interactions has been highlighted in recent mutant mouse models.6 Cell culture studies will be needed to confirm and explore Ambn interactions with ameloblast plasma membranes. Finally, the outcomes of these studies have biological implications that are relevant not only to Ambn–cell interactions but also to Ambn signaling and to the mechanisms of Ambn secretion via secretory vesicles.
Experimental Section
Sequence Analysis of Ambn from Different Species Using Consurf
The amino acid sequences were aligned using ClustalW. Then, the multiple sequence alignment was uploaded to Consurf server (http://consurf.tau.ac.il/2016/), and the amino sequence of Mus musculus was used as the query sequence. The conservation score for each amino acid of M. musculus Ambn was calculated by the Consurf server using the Bayesian method. For each motif of interest, the conservation scores of all the included amino acids were averaged. As described by the Consurf server: “The conservation scores calculated by ConSurf are normalized, so that the average score for all residues is zero, and the standard deviation is one. The conservation scores calculated by ConSurf are a relative measure of evolutionary conservation at each sequence site of the target chain. The lowest score represents the most conserved position in a protein. It does not necessarily indicate 100% conservation (e.g., no mutations at all) but rather indicates that this position is the most conserved in this specific protein calculated using a specific MSA.”
Protein Expression and Purification
Recombinant mouse ameloblastin (Ambn) and mutants were expressed and purified with histidine (His), thioredoxin (Trx), and S-tags in BL21 E. coli using pET-32a plasmid (Novagen) into which mouse ameloblastin gene (GenBank no. AAB93765.1) was inserted. The method is described in detail in our recent publication.50 In brief, Ambn and its mutants expressed in E. coli were purified using nickel affinity chromatography (Ni-NTA Agarose, Qiagen), followed by dialysis through a 10k MWCO dialysis membrane (SnakeSkin Dialysis Tubing, ThermoFisher). The protein concentration after dialysis was determined by BCA assay (Pierce BCA Protein Assay Kit, ThermoFisher) and His-, Trx-, and S-tags were cleaved by Enterokinase (light chain, New England Biolabs) at the designed cleavage site (Asp–Asp–Asp–Asp–Lys) using 0.8 μL enzyme per milligram of protein (pH 7.5, in the presence of 1 M urea, at 37 °C for 6 h with gentle mixing). The cleaved fragments were separated from the protein with a reversed-phase high performance liquid chromatography (HPLC) system (Varian Prostar system with Agilent OpenLab CDS software). Phenomenex C4 column (10 × 250 mm, 5 μm) was used and proteins were eluted with an increasing gradient of 32–72% acetonitrile over 80 min, at a flow rate of 1.5 mL/min. The collected proteins were lyophilized and characterized with SDS-PAGE (Figure S1). Mutagenesis was performed using Q5 Site-Directed Mutagenesis Kit (NEB). The primers were designed according to the NE Base Changer. The Ambn variants Ambn W80F/W93F, Ambn W402F, Ambn Y67_Q103del (exon 5 deleted, AmbnΔ5), and Ambn P104_V168del (exon 6 deleted, AmbnΔ6) were expressed and purified as described for the wild type. A schematic presentation of the Ambn variants and mutants is shown in Figures 9 and 5A. Recombinant WT Ambn was characterized using mass spectroscopy (Figure S1). DLS analysis of WT Ambn and AmbnΔ6 revealed the presence of large assemblies at concentrations above 1 μM but monomeric entities were also detected (Figure S2A,B,E,F,G,H,K,L). AmbnΔ5, on the other hand, had a much more heterogeneous size distribution with lower level assemblies at lower concentration when compared to the WT and AmbnΔ6 (Figure S2C,D,I,J). This behavior was consistent with what has been previously reported regarding assembly characteristics of human recombinant Ambn lacking the sequence encoded by exon 5.15
Figure 9.
Schematic drawing of Ambn variants (AmbnW80F/W93F and AmbnW402F) and mutants (AmbnΔ5 and AmbnΔ6) derived from WT mouse Ambn.
Peptide Synthesis
All peptides were synthesized by Chempeptide Limited (Shanghai, China). Seven peptides (AB1, AB2, AB2N, AB2C, AB4, AB5, and AB6) (Table 2) were designed based on the amino acid sequence of mouse Ambn. Five peptide variants (R69D/K74D, K74D, K74L, N77K, and F72S) were derived from the identified membrane-binding motif (AB2N) (Table 3). The lyophilized peptides were dissolved in distilled water at a concentration of 10 mg/mL and stored at −20 °C before use. Purity of the peptides was determined through HPLC equipped with a Kromasil-C18 column (4.6 × 250 mm, 5 μm), and their concentrations were determined using a Pierce BCA protein assay kit (Thermofisher).
Unilamellar Lipid Vesicle Preparation
Large unilamellar vesicles (LUVs) were prepared as previously described.51 The lipid composition, which was based on the membrane domain involved in epithelial cell-ECM adhesion, is shown in Figure 1A.34 Commercial lipids (Avanti) were dissolved in chloroform or methanol, and then were mixed to obtain a desirable lipid molar ratio of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC)/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE)/1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (POPS)/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoinositol (POPI)/sphingomyelin (SM) = 40:25:15:10:10. Note that POPC, POPE, and SM have neutral charge, whereas POPS and POPI are negatively charged. Therefore, the ECM-adhesion-imitating LUVs used here are about 25% anionic. The chloroform and methanol in the mixture were evaporated under a stream of nitrogen, and the remaining lipid mixture was dried in a vacuum desiccator overnight. The size of the LUVs used in all experiments was 100 nm.
LUV–Ambn Interactions
The interactions between LUVs and Ambn were analyzed by Cryo-transmission electron microscopy, membrane leakage assay, and lipid vesicle clearance assay (static light scattering), as previously reported51 and as described below in detail. CD spectroscopy and tryptophan fluorescence spectroscopy were used to analyze the conformational changes in proteins and peptides as the result of their interactions with LUVs.
Membrane Leakage Assay
For membrane leakage assays, the dried lipid mixtures were rehydrated in pH 7.4 buffer containing 10.0 mM HEPES, 50.0 mM KCl, 1.0 mM EDTA, 3.0 mM NaN3, 9.0 mM ANTS (8-aminonaphthalene-1,3,6-trisulfonic acid), and 25.0 mM DPX (p-xylene-bispyridinium bromide). The lipid suspension was subjected to 10 freeze/thaw cycles, followed by 21 extrusions through 400 nm-cutoff and 100 nm-cutoff polycarbonate filters (Avanti). Unencapsulated ANTS and DPX were removed by size exclusion chromatography using the Sephadex G-100 medium (GE Healthcare). The LUVs were collected in pH 7.4 buffer containing 10.0 mM HEPES, 50.0 mM KCl, 1.0 mM EDTA, and 3.0 mM NaN3.
The 300 μM 100 nm LUVs encapsulating 9.0 mM ANTS and 25.0 mM DPX were mixed separately with 0.5 μM Ambn, 1.0 μM Ambn, 1.0 μM AmbnΔ5, 1.0 μM AmbnΔ6, or 1.0 μM peptides, as previously described.51 One μM BSA and 0.05% Triton X-100 were used as the controls. The release of fluorophore ANTS and its quencher DPX was monitored by measuring the increase of ANTS fluorescence intensity using a Quanta Master 4 fluorescence spectro-fluorometer. The excitation and emission wavelengths were 380 and 520 nm, respectively. All of the split widths were 5 nm. All data were normalized to the fluorescence intensity when all vesicles were disrupted by Triton X-100.
Lipid Vesicle Clearance Assay (Static Light Scattering)
For lipid vesicle clearance and electron microscopy assays, the dried lipid mixtures were rehydrated in pH 7.4 buffer containing 10.0 mM HEPES and 50.0 mM KCl, then shaken in a glass tube to form large, multilamellar vesicles (LMVs). LUVs with a diameter of 100 nm were prepared by forcing the lipid suspension through polycarbonate filters with an extruder.
Clearance of LUVs was monitored by measuring the change of static scattering light intensity as a function of time using a QuantaMaster 4 fluorescence spectro-fluorometer as previously described.51 The excitation wavelength was 300 nm, and the split width was 1 nm. The emission wavelength was 300 nm, and the split width was 5 nm. LUVs with a diameter of 100 nm were diluted in pH 7.4 buffer containing 10.0 mM HEPES, 50.0 mM KCl, 1.0 mM EDTA, and 3.0 mM NaN3 to a molar concentration of 300 μM. After monitoring this mixture for 500 s, 0.5 μM Ambn, 1.0 μM Ambn, 1.0 μM BSA, 1.0 μM AmbnΔ5, or 1.0 μM AmbnΔ6 was added. The data were then collected for another 2000 s. Then, 0.05% Triton X-100 was added to disrupt all vesicles, and the data were normalized to the intensity after adding Triton X-100.
Cryo-Transmission Electron Microscopy
LUVs (10 μM, 100 nm) were mixed with 1.0 μM Ambn for 1 h at room temperature. To prepare Cryo-EM grids, a 2.5 μl sample was applied to a Quantifoil grid, blotted for 3.0 seconds with a filter paper in 100% humidity, and then plunged into liquid ethane with a manual plunger. The image was collected on a FEI Tecnai TF20 at an accelerating voltage of 200 kV using the TVIPS EM-Menu program. The instrument was equipped with a 16-megapixel CCD camera. The nominal magnification used was 29 000 with 2 × 2 binning to increase the contrast. The images were collected at the Electron Imaging Center for Nanomachines, UCLA.
Circular Dichroism Spectroscopy
Ambn or Ambn variant (2 μM) was mixed with 300 μM 100 nm LUVs in pH 7.4, 10.0 mM Tris-HCl buffer and kept at room temperature for 5.0 min before measurements. Far-UV circular dichroism spectra were recorded on a J-815 circular dichroism spectrometer (JASCO) over a wavelength range of 190–260 nm using a 1 mm path length quartz cell. The circular dichroism spectra were analyzed using DICHROWEB online with the CDSSTR algorithm.52
Intrinsic Tryptophan Fluorescence Spectroscopy
Samples were prepared as described for CD. Tryptophan fluorescence spectra were recorded on a Quanta Master 4 fluorescence spectro-fluorometer. The excitation wavelength was 295 nm, the emission wavelength range was 300–400 nm, and all split widths were 5 nm. The changes of emission maximum were analyzed to reveal the tertiary structural changes of Ambn.
Analysis of AH
The projection of sequences on a helical wheel was conducted by Heliquest.53 The factors determining the presence of the AH are the hydrophobic moment (μH), number of proline residues (Pro), net charge, and whether there is a putative hydrophobic face (H face).
Statistical Analysis
Statistical analysis was carried out using one-tailed Student’s t-tests. P-values were determined in Microsoft Excel. The differences were considered significant if P < 0.05. The P-values are described in figure legends. All data were representative of at least three independent experiments using different batches of purified proteins.
Acknowledgments
We thank Wong Hoi (Electron Imaging Center for Nanomachines, UCLA) for collecting Cryo-TEM images, Gayathri Visakan for technical assistance with protein purification, and Dr. Shuxing Li (NanoBiophysics Center, USC) for assistance with CD and fluorescence spectra.
Glossary
Abbreviations
- ANTS
8-aminonaphthalene-1,3,6-trisulfonic acid
- Ambn
ameloblastin
- AmbnΔ;5
Ambn p.Y67_Q103del
- AmbnΔ;6
Ambn p.P104_V168del
- Amel
amelogenin
- Amtn
amelotin
- AH
amphipathic helix
- CD
circular dichroism
- DLS
dynamic light scattering
- DPX
p-xylene-bispyridinium bromide
- Enam
enamelin
- ECM
extracellular matrix
- HAP
hydroxyapatite
- LUVs
large unilamellar vesicles
- MMP-20
matrix metalloproteinase-20
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPE
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- POPS
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine
- POPI
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoinositol
- SM
sphingomyelin
- TEM
transmission electron microscopy
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b03582.
GenBank accession numbers of Ambn protein sequences for bioinformatics analysis, helical regions of mouse Ambn as calculated by Heliquest, characterization of WT mouse Ambn and its mutants AmbnΔ5 and AmbnΔ6, and DLS and CD spectra of Ambn and its mutants (PDF)
Author Contributions
J.S. designed and performed the experiments, analyzed the data, and wrote the manuscript; N.C.K. designed the leakage assay experiments, performed DLS experiments, and critically revised the manuscript; R.A.B. expressed and purified the recombinant proteins; J.M.-O. designed the experiments, analyzed data, and wrote and critically revised the manuscript.
This project was funded by NIH-NIDCR grants R01DE013414 and DE020099 to J.M.-O.
The authors declare no competing financial interest.
Supplementary Material
References
- Cui F.-Z.; Ge J. New observations of the hierarchical structure of human enamel, from nanoscale to microscale. J. Tissue Eng. Regen. Med. 2007, 1, 185–191. 10.1002/term.21. [DOI] [PubMed] [Google Scholar]
- Margolis H. C.; Beniash E.; Fowler C. E. Role of macromolecular assembly of enamel matrix proteins in enamel formation. J. Dent. Res. 2016, 85, 775–793. 10.1177/154405910608500902. [DOI] [PubMed] [Google Scholar]
- Gallon V.; Chen L.; Yang X.; Moradian-Oldak J. Localization and quantitative co-localization of enamelin with amelogenin. J. Struct. Biol. 2013, 183, 239–249. 10.1016/j.jsb.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder P.; Prajapati S.; Bapat R.; Moradian-Oldak J., Amelogenin-Ameloblastin Spatial Interaction around Maturing Enamel Rods. J. Dent. Res. 2016, 95, . 10.1177/0022034516645389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradian-Oldak J. Protein-mediated enamel mineralization. Front. Biosci.: J. Virt. Libr. 2012, 17, 1996. 10.2741/4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto S.; Kiba T.; Hall B.; Iehara N.; Nakamura T.; Longenecker G.; Krebsbach P. H.; Nanci A.; Kulkarni A. B.; Yamada Y. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J. Cell Biol. 2004, 167, 973–983. 10.1083/jcb.200409077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki K.; Weiss K. M. Mineralized tissue and vertebrate evolution: the secretory calcium-binding phosphoprotein gene cluster. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 4060–4065. 10.1073/pnas.0638023100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sire J.-Y.; Davit-Béal T.; Delgado S.; Gu X. The origin and evolution of enamel mineralization genes. Cells Tissues Organs 2007, 186, 25–48. 10.1159/000102679. [DOI] [PubMed] [Google Scholar]
- Lakshminarayanan R.; Bromley K. M.; Lei Y.-P.; Snead M. L.; Moradian-Oldak J. Perturbed amelogenin secondary structure leads to uncontrolled aggregation in amelogenesis imperfecta mutant proteins. J. Biol. Chem. 2010, 285, 40593–40603. 10.1074/jbc.m110.131136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delak K.; Harcup C.; Lakshminarayanan R.; Sun Z.; Fan Y.; Moradian-Oldak J.; Evans J. S. The Tooth Enamel Protein, Porcine Amelogenin, Is an Intrinsically Disordered Protein with an Extended Molecular Configuration in the Monomeric Form. Biochemistry 2009, 48, 2272–2281. 10.1021/bi802175a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald T.; Bednárová L.; Osička R.; Pachl P.; Šulc M.; Lyngstadaas S. P.; Slaby I.; Vondrášek J. Biophysical characterization of recombinant human ameloblastin. Eur. J. Oral Sci. 2012, 119, 261–269. 10.1111/j.1600-0722.2011.00913.x. [DOI] [PubMed] [Google Scholar]
- Iwasaki K.; Bajenova E.; Somogyi-Ganss E.; Miller M.; Nguyen V.; Nourkeyhani H.; Gao Y.; Wendel M.; Ganss B. Amelotin-a Novel Secreted, Ameloblast-specific Protein. J. Dent. Res. 2016, 84, 1127–1132. 10.1177/154405910508401207. [DOI] [PubMed] [Google Scholar]
- Fang P.-A.; Margolis H. C.; Conway J. F.; Simmer J. P.; Beniash E. CryoTEM study of effects of phosphorylation on the hierarchical assembly of porcine amelogenin and its regulation of mineralization in vitro. J. Struct. Biol. 2013, 183, 250–257. 10.1016/j.jsb.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincham A. G.; Moradian-Oldak J.; Diekwisch T. G. H.; Lyaruu D. M.; Wright J. T.; Bringas P. Jr.; Slavkin H. C. Evidence for Amelogenin ″Nanospheres″ as Functional Components of Secretory-Stage Enamel Matrix. J Struct Biol 1995, 115, 50–59. 10.1006/jsbi.1995.1029. [DOI] [PubMed] [Google Scholar]
- Wald T.; Osickova A.; Sulc M.; Benada O.; Semeradtova A.; Rezabkova L.; Veverka V.; Bednarova L.; Maly J.; Macek P. Intrinsically disordered enamel matrix protein ameloblastin forms ribbon-like supramolecular structures via an N-terminal segment encoded by exon 5. J. Biol. Chem. 2013, 288, 22333–22345. 10.1074/jbc.m113.456012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberth S. A.; Bacino M. S.; Sandhu S.; Li W.; Bonde J.; Habelitz S. Progression of Self-Assembly of Amelogenin Protein Supramolecular Structures in Simulated Enamel Fluid. Biomacromolecules 2018, 19, 3917–3924. 10.1021/acs.biomac.8b00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebsbach P. H.; Lee S. K.; Matsuki Y.; Kozak C. A.; Yamada K. M.; Yamada Y. Full-length Sequence, Localization, and Chromosomal Mapping of Ameloblastin. J. Biol. Chem. 1996, 271, 4431–4435. 10.1074/jbc.271.8.4431. [DOI] [PubMed] [Google Scholar]
- Hu C.-C.; Fukae M.; Uchida T.; Qian Q.; Zhang C. H.; Ryu O. H.; Tanabe T.; Yamakoshi Y.; Murakami C.; Dohi N.; Shimizu M.; Simmer J. P. Sheathlin: cloning, cDNA/polypeptide sequences, and immunolocalization of porcine enamel sheath proteins. J. Dent. Res. 1997, 76, 648–657. 10.1177/00220345970760020501. [DOI] [PubMed] [Google Scholar]
- Fong C. D.; Hammarström L.; Lundmark C.; Wurtz T.; Slaby I. Expression patterns of RNAs for amelin and amelogenin in developing rat molars and incisors. Adv. Dent. Res. 1996, 10, 195–200. 10.1177/08959374960100021301. [DOI] [PubMed] [Google Scholar]
- Iwata T.; Yamakoshi Y.; Hu J. C.-C.; Ishikawa I.; Bartlett J. D.; Krebsbach P. H.; Simmer J. P. Processing of ameloblastin by MMP-20. J. Dent. Res. 2007, 86, 153–157. 10.1177/154405910708600209. [DOI] [PubMed] [Google Scholar]
- Uchida T.; Murakami C.; Wakida K.; Satoda T.; Dohi N.; Takahashi O. Synthesis, secretion, degradation, and fate of ameloblastin during the matrix formation stage of the rat incisor as shown by immunocytochemistry and immunochemistry using region-specific antibodies. J. Histochem. Cytochem. 2016, 45, 1329–1340. 10.1177/002215549704501002. [DOI] [PubMed] [Google Scholar]
- Su J.; Chandrababu K. B.; Moradian-Oldak J. Ameloblastin peptide encoded by exon 5 interacts with amelogenin N-terminus. Biochem. Biophys. Rep. 2016, 7, 26–32. 10.1016/j.bbrep.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F.; Gasse B.; Sire J. Y. Evolutionary analysis of selective constraints identifies ameloblastin (AMBN) as a potential candidate for amelogenesis imperfecta. BMC Evol. Biol. 2015, 15, 148. 10.1186/s12862-015-0431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerný R.; Slaby I.; Hammarström L.; Wurtz T. A novel gene expressed in rat ameloblasts codes for proteins with cell binding domains. J. Bone Miner. Res. 1996, 11, 883–91. 10.1002/jbmr.5650110703. [DOI] [PubMed] [Google Scholar]
- Sonoda A.; Iwamoto T.; Nakamura T.; Fukumoto E.; Yoshizaki K.; Yamada A.; Arakaki M.; Harada H.; Nonaka K.; Nakamura S.; Yamada Y.; Fukumoto S. Critical role of heparin binding domains of ameloblastin for dental epithelium cell adhesion and ameloblastoma proliferation. J. Biol. Chem. 2009, 284, 27176–27184. 10.1074/jbc.m109.033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyeler M.; Schild C.; Lutz R.; Chiquet M.; Trueb B. Identification of a fibronectin interaction site in the extracellular matrix protein ameloblastin. Exp. Cell Res. 2010, 316, 1202–1212. 10.1016/j.yexcr.2009.12.019. [DOI] [PubMed] [Google Scholar]
- Vymětal J.; Slabý I.; Spahr A.; Vondrášek J.; Lyngstadaas S. P. Bioinformatic analysis and molecular modelling of human ameloblastin suggest a two-domain intrinsically unstructured calcium-binding protein. Eur. J. Oral Sci. 2008, 116, 124–134. 10.1111/j.1600-0722.2008.00526.x. [DOI] [PubMed] [Google Scholar]
- Zou Y.; Wang H.; Shapiro J. L.; Okamoto C. T.; Brookes S. J.; Lyngstadaas S. P.; Snead M. L.; Paine M. L. Determination of protein regions responsible for interactions of amelogenin with CD63 and LAMP1. Biochem. J. 2007, 408, 347–354. 10.1042/bj20070881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompkins K.; George A.; Veis A. Characterization of a mouse amelogenin [A–4]/M59 cell surface receptor. Bone 2006, 38, 172–180. 10.1016/j.bone.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Shapiro J. L.; Wen X.; Okamoto C. T.; Wang H. J.; Lyngstadaas S. P.; Goldberg M.; Snead M. L.; Paine M. L. Cellular uptake of amelogenin, and its localization to CD63, and Lamp1-positive vesicles. Cell. Mol. Life Sci. 2007, 64, 244–256. 10.1007/s00018-006-6429-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokappa S. B.; Chandrababu K. B.; Moradian-Oldak J. Tooth enamel protein amelogenin binds to ameloblast cell membrane-mimicking vesicles via its N-terminus. Biochem. Biophys. Res. Commun. 2015, 464, 956–961. 10.1016/j.bbrc.2015.07.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekshe Lokappa S.; Balakrishna Chandrababu K.; Dutta K.; Perovic I.; Spencer Evans J.; Moradian-Oldak J. Interactions of amelogenin with phospholipids. Biopolymers 2014, 103, 96–108. 10.1002/bip.22573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peetla C.; Stine A.; Labhasetwar V. Biophysical interactions with model lipid membranes: applications in drug discovery and drug delivery. Mol. Pharm. 2009, 6, 1264–1276. 10.1021/mp9000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez M. G.; Leocata Nieto F.; Fernández-Tome M. C.; Favale N. O.; Sterin-Speziale N. Membrane lipid composition plays a central role in the maintenance of epithelial cell adhesion to the extracellular matrix. Lipids 2008, 43, 343–352. 10.1007/s11745-008-3152-y. [DOI] [PubMed] [Google Scholar]
- Bromley K. M.; Kiss A. S.; Lokappa S. B.; Lakshminarayanan R.; Fan D.; Ndao M.; Evans J. S.; Moradian-Oldak J. Dissecting Amelogenin Protein Nanospheres. J. Biol. Chem. 2011, 286, 34643–34653. 10.1074/jbc.m111.250928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wazen R. M.; Moffatt P.; Zalzal S. F.; Yamada Y.; Nanci A. A mouse model expressing a truncated form of ameloblastin exhibits dental and junctional epithelium defects. Matrix Biol. 2009, 28, 292–303. 10.1016/j.matbio.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter J. A.; Murillo G.; Brookes S. J.; Smith C. E. L.; Parry D. A.; Silva S.; Kirkham J.; Inglehearn C. F.; Mighell A. J. Deletion of ameloblastin exon 6 is associated with amelogenesis imperfecta. Human Mol. Gen. 2014, 23, 5317–5324. 10.1093/hmg/ddu247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzer M. E.; Holtzer A. ?-helix to random coil transitions: Determination of peptide concentration from the CD at the isodichroic point. Biopolymers 1992, 32, 1675–1677. 10.1002/bip.360321209. [DOI] [PubMed] [Google Scholar]
- Bornholdt Z. A.; Noda T.; Abelson D. M.; Halfmann P.; Wood M. R.; Kawaoka Y.; Saphire E. O. Structural rearrangement of ebola virus VP40 begets multiple functions in the virus life cycle. Cell 2013, 154, 763–774. 10.1016/j.cell.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G.; Antonny B. Amphipathic helices and membrane curvature. FEBS Lett. 2010, 584, 1840–1847. 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- Wood C. B.; Dumont E. R.; Crompton A. W. New Studies of Enamel Microstructure in Mesozoic Mammals: A Review of Enamel Prisms as a Mammalian Synapomorphy. J. Mammal. Evol. 1999, 6, 177–213. 10.1023/a:1020624222324. [DOI] [Google Scholar]
- Sander P. M.Non-mammalian synapsid enamel and the origin of mammalian enamel prisms: the bottom-up perspective. In Tooth Enamel Microstructure, 1st ed.; Koenigswald W. v.; Sander P. M., Eds.; Taylor & Francis: Rotterdam, 1997, pp 41−62.
- Thompson J.; Gibson T. J.; Plewniak F.; Jeanmougin F.; Higgins D. G. The CLUSTAL_X Windows Interface: Flexible Strategies for Multiple Sequence Alignment Aided by Quality Analysis Tools. Nucleic Acids Res. 1997, 25, 4876–4882. 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H.; Erez E.; Martz E.; Pupko T.; Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010, 38, W529–W533. 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald T.; Spoutil F.; Osickova A.; Prochazkova M.; Benada O.; Kasparek P.; Bumba L.; Klein O. D.; Sedlacek R.; Sebo P.; Prochazka J.; Osicka R. Intrinsically disordered proteins drive enamel formation via an evolutionarily conserved self-assembly motif. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, E1641–E1650. 10.1073/pnas.1615334114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder P.; Prajapati S.; Lokappa S. B.; Gallon V.; Moradian-Oldak J. Analysis of co-assembly and co-localization of ameloblastin and amelogenin. Front. Physiol. 2014, 5, 274. 10.3389/fphys.2014.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine M. L.; Luo W.; Zhu D.-H.; Bringas P. Jr.; Snead M. L. Functional domains for amelogenin revealed by compound genetic defects. J. Bone Miner. Res. 2003, 18, 466–472. 10.1359/jbmr.2003.18.3.466. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Zhang X.; Lu X.; Atsawasuwan P.; Luan X. Ameloblastin regulates cell attachment and proliferation through RhoA and p27. Eur. J. Oral Sci. 2012, 119, 280–285. 10.1111/j.1600-0722.2011.00887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Diekwisch T. G. H.; Luan X. Structure and function of ameloblastin as an extracellular matrix protein: adhesion, calcium binding, and CD63 interaction in human and mouse. Eur. J. Oral Sci. 2012, 119, 270–279. 10.1111/j.1600-0722.2011.00889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J.; Bapat R. A.; Moradian-Oldak J.. The expression and purification of recombinant mouse ameloblastin in E. coli. In Odontogenesis: Methods and Protocols, 1st ed.; Papagerakis P., Ed.; Humana Press Inc., 2019; Vol. 1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegulian N. C.; Sankhagowit S.; Apostolidou M.; Jayasinghe S. A.; Malmstadt N.; Butler P. C.; Langen R. Membrane Curvature-sensing and Curvature-inducing Activity of Islet Amyloid Polypeptide and Its Implications for Membrane Disruption. J. Biol. Chem. 2015, 290, 25782–25793. 10.1074/jbc.m115.659797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore L.; Wallace B. A. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 2008, 89, 392–400. 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- Gautier R.; Douguet D.; Antonny B.; Drin G. HELIQUEST: a web server to screen sequences with specific -helical properties. Bioinformatics 2008, 24, 2101–2102. 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.