Abstract
The dendritic cell vaccine DC-Ad-GM∙CAIX is an active, specific immunotherapy with the potential of providing a safe and effective therapy against renal cell carcinoma (RCC). Using immunocompetent Balb/c mouse models we tested the efficacy and mechanism of the vaccine to prevent and treat the growth of a syngeneic RCC (RENCA) engineered to overexpress the human TAA carbonic anhydrase IX (NPR-IX). In a prevention model, NPR-IX tumor development was specifically and significantly delayed by 13 days in DC-Ad-GM∙CAIX-treated mice (P<0.001), tumor volumes were 79% smaller (day 24, P<0.007) and body weight was maintained at study termination compared to the controls. Six of these mice remained tumor-free for >1 year. In a treatment model, NPR-IX tumors remained smaller in DC-Ad-GM∙CAIX-treated mice for 8 days (P<0.002), achieving a 60% growth inhibition at termination. No vaccine-related organ toxicity was observed in either model. The critical mechanistic parameter separating responsive from non-responsive tumors was hCAIX protein expression, demonstrated by aggressive growth of tumors that did not express hCAIX protein and in sham-treated mice (DC-Ad-Null). No murine serum anti-hCAIX antibodies were detected. Moreover, altered mechanisms of immunoediting as a means for immune evasion were suggested by differential gene expression (Ccl1, Hmgb1, Fgl2, Cd209a, Klra2) and therapy evasion miRNAs (miR-1186, miR-98, miR-5097, miR-1942, miR-708) in tumors that evaded DC-Ad-GM∙CAIX immunotherapy. This is the first study in immunocompetent mice that provides a proof of concept for the specificity, efficacy, safety, and activity of the DC-Ad-GM∙CAIX immunotherapy, forming the basis for a first-in-human phase I trial in RCC patients.
Keywords: Dendritic cell, GM∙CAIX, renal cell carcinoma, immunotherapy, immunoediting
INTRODUCTION
Despite the FDA approval of six targeted therapies for advanced renal cell carcinoma (RCC) in the past six years, complete and durable remissions of metastatic disease are rarely if ever achieved. Moreover, these agents are mainly cytostatic, are associated with significant side effects, require chronic administration, and treatment resistance typically develops within one year. On the other hand, RCC belongs with melanoma to a class of solid tumors that are significantly responsive to and are, in a small percentage of cases, curable by systemic treatment with high-dose interleukin-2 (1). The identification of tumor-associated antigens (TAAs) overexpressed in RCC, such as carbonic anhydrase IX (CAIX), has led to the development of humoral and cell-mediated immunotherapies (2, 3).
CAIX regulates proton exchange to buffer intracellular pH, a function crucial for cell survival (4). CAIX up-regultaion has been described in many cancerous tissues (4, 5) and is directly regulated by the hypoxia-inducible factor 1α-von Hippel-Lindau (VHL) pathway. Upon loss of pVHL function in clear cell RCC, stabilization of HIF-1α occurs, leading to the overexpression of CAIX and rendering it useful as a diagnostic and therapeutic TAA (6–10).
The recent FDA approval of sipuleucel-T and its associated body of research have clearly shown the immunotherapeutic benefit of fusing a potent cytokine such as granulocyte-macrophage colony stimulating factor (GM-CSF) with a TAA. Transduction of such a fusion protein ex vivo into antigen presenting cells is capable of leading to an effective immune stimulation leading to the improved survival of patients with advanced solid cancer (11, 12). We have previously shown the in vitro ability of the fusion protein GM-CSF-CAIX (GM∙CAIX) to induce a CAIX-targeted, T-cell mediated, and MHC-restricted anti-tumor activity (13–15). To model the role of T-cell anti-tumor activity against clear cell RCC in mice with an intact immune system, we constitutively and stably overexpressed human CAIX (hCAIX) in a CAIX-negative parental RENCA cell line (16). The cells were then sorted for hCAIX expression (Newly Purified RENCA-CAIX; NPR-IX) and transplanted in syngeneic Balb/c mice. An adenoviral vector was used to transduce the GM∙CAIX fusion protein in syngeneic DCs that were then transplanted in the same mice. Our study shows for the first time that murine DCs expressing GM∙CAIX can generate a potent T-cell mediated hCAIX-specific inhibition of tumor growth in immunocompetent mice. Loss of hCAIX expression was essential for tumor evasion from the DC-Ad-GM∙CAIX immunotherapy. Furthermore, differential gene and miRNA expression characterization revealed candidate markers including for tumor cell proliferation and immune evasion mechanisms. Collectively, these encouraging pre-clinical results provide the basis for the initiation of clinical trials using DC-Ad-GM∙CAIX immunotherapy.
MATERIALS and METHODS
Cells and cell culture.
RENCA cells were a gift from Dr. R. Wiltrout (NCI/NIH; ref. 17). NPR-IX cells are FACS-purified RENCA cells overexpressing hCAIX (elaborated in Supplementary Text).
In vitro DC generation and gene transduction.
Bone marrow cells of 8-week-old female Balb/c mice were induced to differentiate into DCs as described previously by Koya et al (18). The differentiated DCs were transduced with Ad-GM∙CAIX (15) or with Ad-Null (Welgen, MA; elaborated in Supplementary Text).
Animals.
Eight-week-old Balb/cAnNCr female mice were obtained from the Radiation Oncology Defined Flora Animal Research Facility, UCLA. All animals had free access to water and food. All procedures were approved by the UCLA Animal Research Committee and followed the UCLA Institutional Animal Care and Use Committee guidelines according to the American Association for Laboratory Animal Science.
RCC in vivo models.
The prevention model had 6 groups of 8 immunocompetent Balb/c mice each. Groups A-D were immunized by subcutaneous transplantation of 2×10^6 DCs in 150 μl of PBS 1x into the right flank. Two groups (A, B) were transplanted with DC-Ad-GM∙CAIX and two groups (C, D) with DC-Ad-Null. Groups E and F had no treatment. The DC transplantation was repeated after 6 days. Twelve days (day 0) after the first DC transplantation, 5×10^5 RCC cells in 100 μl of PBS 1x were subcutaneously transplanted into the left flank. Three groups (A, C, E) were transplanted with NPR-IX, and three groups (B, D, F) with RENCA cells. The treatment model had three groups (G, H, I) of 16 immunocompetent Balb/c mice each, which were subcutaneously transplanted in the left flank with 5×10^5 NPR-IX. On days 8 and 14, group G was subcutaneously transplanted in the right flank with 2×10^6 DC-Ad-GM∙CAIX and group H with 2×10^6 DC-Ad-Null, while group I had no treatment. Mouse performance status was assessed daily, body weight and tumors measured twice a week. Tumor volume “V” was calculated according to the formula V=ab2×Pi/6, where “a” is the longest diameter of the tumor and “b” is the longest diameter perpendicular to “a”.
Immunohistochemistry.
In three mice of each group of the prevention model, pathology-relevant organs, blood, and tumors were collected. Paraffin-embedded sections were cut at 4 μm thickness and rehydrated through graded ethanol. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide in methanol for 10 min. Heat-induced antigen retrieval and proteolytic induced epitope retrieval were used. The slides were then stained with antibodies as described the Supplementary Text. All sections were visualized with the diaminobenzidine reaction and counterstained with hematoxylin.
Flow cytometry.
FITC-conjugated anti-hCAIX (U.S. Biologicals, MA) and PE-conjugated anti-mouse Cd86 (BD, MA) were used. Fixation and permeabilization were performed using BD Fix/Wash and BD Perm/Wash (BD, NJ). To detect anti-hCAIX antibodies, 1:10 diluted mouse sera from groups A and E and FITC-conjugated goat anti-mouse IgG (Abcam, MA) were used as primary and secondary antibodies, respectively.
Quantitative real-time reverse transcription-PCR.
RNA was isolated using Trizol Reagent (Invitrogen, CA). Total RNA (1 μg) was reverse transcribed in a volume of 50 μl. Five μl of the resulting solution was then used for PCR according to the manufacturer’s instructions (Applied Biosystems, Inc., Foster City, CA). Gene expression signatures were quantified relative to the expression level of ribosomal 18s, and for miRNA relative to snoRNA 234 or snoRNA202. All probes were purchased from Applied Biosystems or Integrated DNA Technologies (IDT, CA). Normalized data are presented as fold difference in log2 gene expression. Primers are listed in Supplementary Table S3.
Gene expression analysis.
The gene expression was investigated by SurePrint G3 Mouse GE 8×60K, Agilent, CA. Background corrected signal values determined by Agilent feature extraction software were normalized by median centering of the ratios to a reference, which is the median of all control arrays. Global expression profiles of samples were examined by multidimensional scaling of about 36,000 features that were found at least in 50% of the arrays. Euclidean distance was used as the dissimilarity measure. The features having presence calls in <80% of samples and a geometric mean signal <50 in both groups A and C were eliminated before further analyses resulting in mRNA features differentially expressed by 1.5-fold (two tailed p-value < 0.015 by T-test). Ingenuity pathway analyses were done on the datasets of differentially expressed genes (P<0.05). Publicly available literature from 1966 to 2012 was surveyed using CoreMine and PubMed (elaborated in Supplementary Text).
miRNA expression analysis.
Exiqon miRNA arrays (miRCURY LNA™ microRNA array, v6, Exiqon, Denmark) having more than 1,891 capture probes covering all human, mouse, rat, and viral miRNAs and 66 proprietary miRNAs spotted in quadruplicate were used for measuring genome miRNA expression. Intensities were determined by GenePix Pro 6.0 software (Molecular Devices, Sunnyvale, CA) and were normalized by median centering of the ratios to a reference control array. Global miRNA expression profiles of samples were examined by multidimensional scaling of about 4,200 features that were flagged as found in ≥50% of the samples. The features found in <60% of samples in both groups A and C were eliminated from further analysis resulting in miRNAs differentially expressed by 1.5-fold (two tailed p-value < 0.015 by T-test), and 11 mouse miRNA replicates appeared 50–100% of the time at signal levels ≥200 (elaborated in Supplementary Text).
Statistical analysis of tumor volume, weights and survival data.
Kruskal-Wallis one-way ANOVA were used to examine mouse weight and tumor volume. Wilcoxon’s rank-sum tests were used to compare differences in weight and tumor volume between pairs of the six groups. The rates of weight change and tumor growth were analyzed using a linear mixed effects model with a random intercept and a random slope (19). Seven primary pairwise comparisons and three primary pairwise comparisons were performed in the prevention and the treatment model, respectively, with Bonferroni’s adjustment. Time to tumor development (prevention model: tumor volume >0mm3; treatment model: >600 mm3) and to 15% weight loss from baseline were compared using the Kaplan-Meier curves and the log-rank test (20). Missing values of weight and tumor volume up to day 24 were imputed using the Last Observation Carried Forward method. Berger and Boos’ method was used to estimate the time intervals of treatment effect in weight loss and tumor volume of the groups A and G in their respective experiments (21). P-values <0.05 were considered statistically significant. SAS Version 9.2 (SAS Institute Inc., Cary, NC) was used.
RESULTS
Prevention model
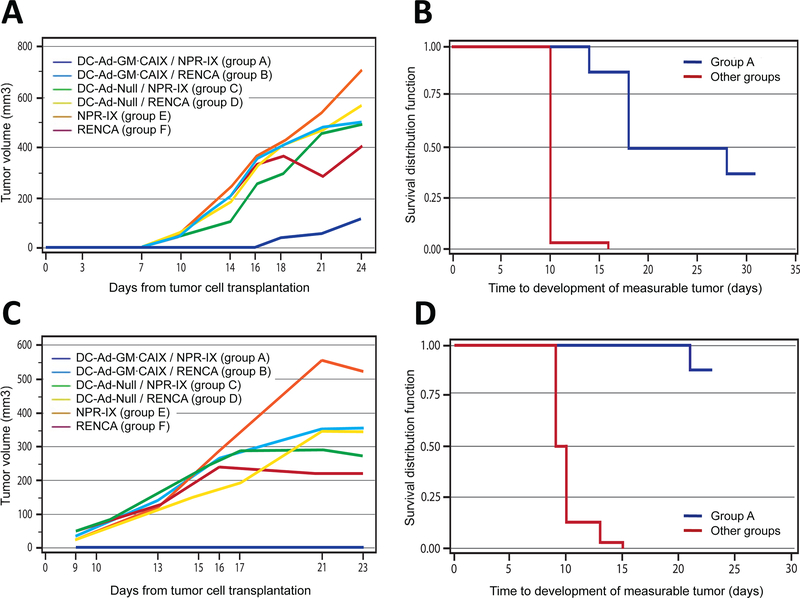
DC-Ad-GM∙CAIX immunotherapy significantly and specifically prevented the growth of the NPR-IX tumors in syngeneic Blab/c mice (group A in Fig. 1A). In this group the mean time to development of a palpable tumor (~50 mm3) was 13 days longer than in the control groups (23 days in group A vs. 10 days in the sham and RENCA tumors control groups B-F, P<0.001). At study termination, median tumor volume in group A was 79% smaller than in the control groups B-F (day 24, all P<0.007; Fig. 2A), and 50% of the mice were tumor-free (Fig. 2B). Repetition of this study yielded similar results, with 7 of 8 (88%) mice in group A remaining tumor-free at termination (Fig. 2C-D). Tumor growth was prevented for >1 year in 6 of 16 mice.
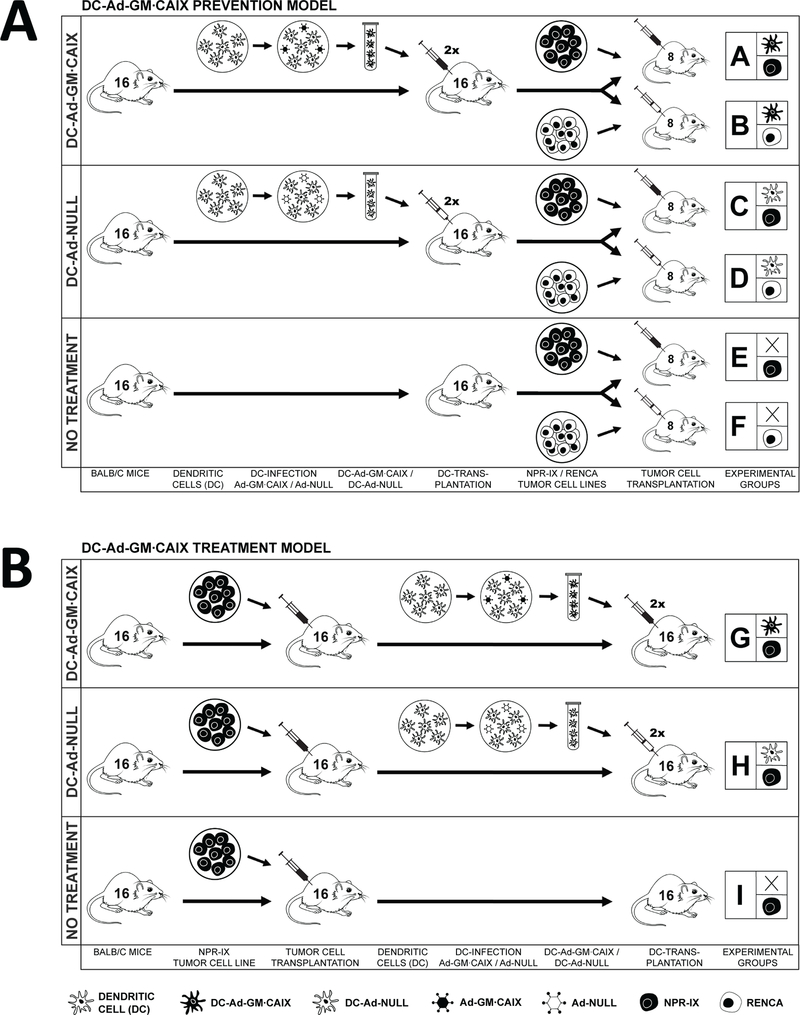
Fig. 1.
Experimental outlines.
(A) Prevention Model: Bone marrow cells are harvested from syngeneic Balb/c mice, ex vivo differentiated to DCs and transduced with either Ad-GM∙CAIX (DC-Ad-GM∙CAIX) or with sham Ad-Null (DC-Ad-Null). First, mice are vaccinated by two rounds of subcutaneous transplantation of DC-Ad-GM∙CAIX (groups A, B) or DC-Ad-Null (groups C, D) or not vaccinated (groups E, F). Twelve days after the second vaccination, mice are challenged by subcutaneous transplantation with either NPR-IX (groups A, C, E) or RENCA (groups B, D, F) tumor cells.
(B) Treatment Model: NPR-IX tumor cells are first subcutaneously transplanted, and only after tumors are established the mice are vaccinated with DC-Ad-GM∙CAIX (group G), DC-Null (group H) or not vaccinated (group I).
Fig. 2.
Tumor volume, survival, and body weight.
Prevention model: (A) Tumor growth over time, (B) time to observable tumor growth (Kaplan-Meier curve), and (C, D) repeated study. (G, I) Body weight over time and (H, J) time to 15% weight loss (Kaplan-Meier curve).
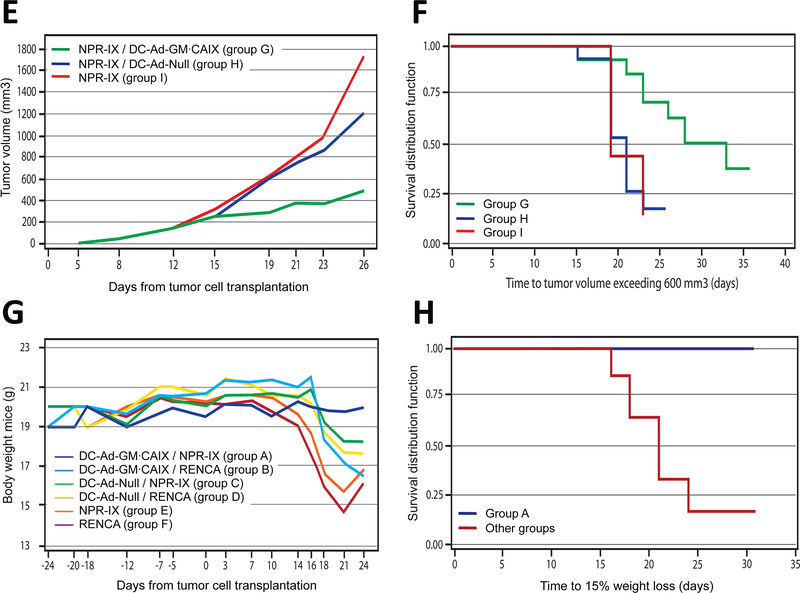
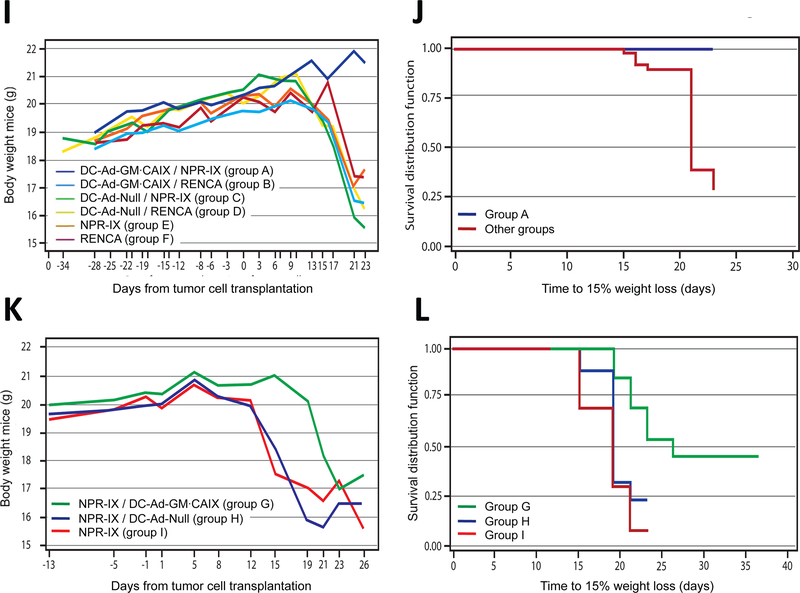
Treatment model: (E) Tumor growth over time and (F) time to tumor growth >600 mm3 (Kaplan-Meier curve). (K) Body weight over time and (L) time to 15% weight loss (Kaplan-Meier curve).
Although the growth rate of NPR-IX tumors (group E; 45.3 mm3/day) was significantly faster than that of RENCA tumors (group F; 27.6 mm3/day; P<0.001), the growth rate of tumors that eventually developed in group A showed no significant difference compared to groups B-F (all P>0.03).
Treatment model
To test whether the DC-Ad-GM∙CAIX immunotherapy would have an inhibitory effect on established RCC tumors (Fig. 1B), three groups of mice bearing syngeneic NPR-IX tumors were treated with DC-Ad-GM∙CAIX, DC-Ad-Null, or no DCs (groups G, H, and I, respectively). Tumor growth was inhibited in group G relative to the control groups H-I, achieving a 60% median growth inhibition at day 26 (Fig. 2E-F). Group G had a smaller median tumor volume than the control groups from day 19 to 26 (P<0.002). Additionally, median tumor growth rate was significantly lower in group G (27.7 mm3/day) compared to the control groups (group H 62.3 mm3/day; group I 66.0 mm3/day; both P<0.001). Tumor growth rates between group H and I were statistically not different (P=0.602).
Histopathology, toxicity, and in vitro immune-monitoring
In the prevention model, mice in group A maintained their body weight throughout the experiment, while mice in each of the control groups B-F suffered significant weight loss (P<0.001; Fig. 2G-J). In the treatment model, group G showed a significantly smaller median weight loss compared to the control groups H-I combined from day 15 to 21 (P<0.011; Fig. 2K-L). At study termination, none of the organs in groups A-I showed evidence of metastases or systemic toxicity caused by the immunotherapy, such as histopathologic inflammatory lesions or atrophy.
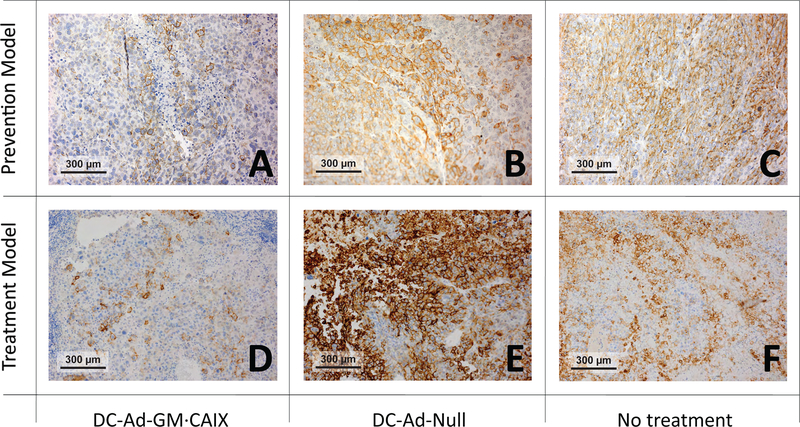
Immunohistological analyses showed absent or minimal hCAIX expression in tumors that evaded DC-Ad-GM∙CAIX immunotherapy (groups A and G), but strong expression in the control groups C, E, H-I (Fig. 3A-F), suggesting that DC-Ad-GM∙CAIX immunotherapy is specifically targeting and killing hCAIX-expressing tumor cells. Murine CAIX (mCAIX) staining, however, was minimal in all groups (Table 1). In addition, tumor necrosis levels and staining for Ki-67, murine macrophages (F4/80), and murine granulocytes (Cd11b) were similar in groups A, C and E (Table 1).
Fig. 3.
Immunohistochemical analysis of NPR-IX tumors with human CAIX (hCAIX).
The panels show staining of membranous hCAIX in brown. Original magnification, x200. Prevention model: (A) DC-Ad-GM∙CAIX-treated mouse (group A, 10% hCAIX positive); (B) DC-Ad-Null-treated mouse (group C, 70% hCAIX positive); (C) Non-treated mouse (group E, 70% hCAIX positive). Treatment model: (D) DC-Ad-GM∙CAIX-treated mouse (group G, 10% hCAIX positive); (E) DC-Ad-Null-treated mouse (group H, 80% hCAIX positive); (F) Non-treated mouse (group I, 50% hCAIX positive).
Table 1.
Prevention Model: Histopathologic analysis
| Group A | Group C | Group E | |
|---|---|---|---|
| CAIX | |||
| hCAIX | −/+ | +++ | ++/+++ |
| mCAIX | − | + | −/+ |
| Tumor necrosis | |||
| H&E | + | + | +/++ |
| Tunel | + | + | +/++ |
| Ki-67 | +++ | +++ | +++ |
| F4/80 | Tumor cells: − macrophages: +++ | Tumor cells: − macrophages: +++ | Tumor cells: − macrophages: +++ |
| Cd11b | +++ | +++ | +++ |
− negative + 1–33% pos. / low ++ 34–66% pos. / medium +++ 67–100% pos. / high
Next we tested the sera for the presence of murine antibodies against the hCAIX protein expressed in NPR-IX and DC-Ad-GM∙CAIX. FACS analysis of sera from groups A and E did not show evidence for the presence of murine anti-hCAIX antibodies (Supplementary Fig. S1)
Differential gene and miRNA expression in tumors that evaded DC-Ad-GM∙CAIX therapy and control
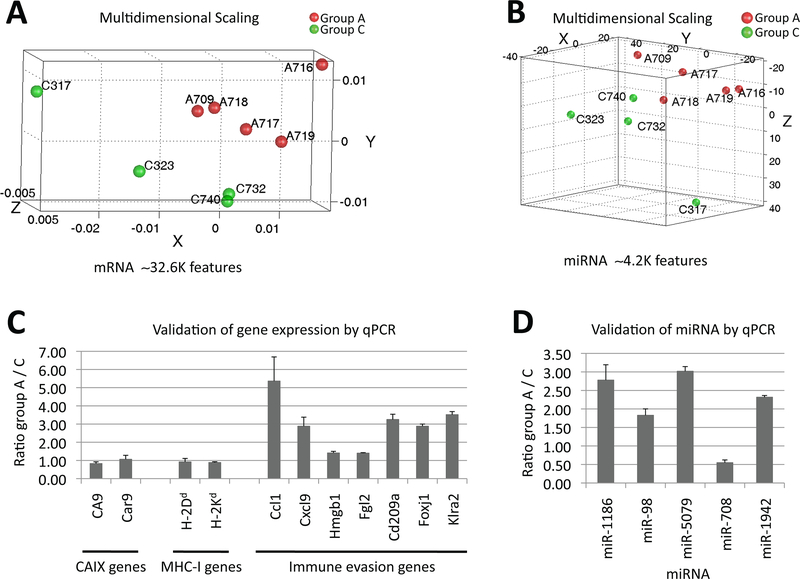
To test if the down-regulation in hCAIX protein expression originated at the transcription or protein level (Fig. 3), qPCR analysis was performed and revealed similar hCAIX expression levels in tumors that evaded DC-Ad-GM∙CAIX therapy (group A) and in tumors of mice treated with DC-Ad-Null (groups C), suggesting hCAIX protein regulation as the cause of its reduced expression. A similar transcription signature was obtained for the endogenous mouse CAIX gene (Car9; Table 2, Fig. 4C).
Table 2.
qPCR validated MHC-I, CAIX, and differential expression of genes and miRNA in tumors that evaded DC-Ad-GM∙CAIX immunotherapy (group A) relative to DC-Ad-Null (group C).
| Marker | Expression group A vs. C | Interest | Ref. |
|---|---|---|---|
| CAIX | |||
| CA9 | no change | Subject TAA (hCAIX) in the current study | |
| Car9 | no change | Mouse ortholog of subject TAA in the current study | |
| MHC-I | |||
| H-2Kd | no change | Balb/c MHC-I | (49) |
| H-2Dd | no change | Balb/c MHC-I | (49) |
| Immune evasion | |||
| Ccl1 | up | Induction of Treg immunosuppressive function | (28, 29) |
| Cxcl9 | up | In melanoma prompts evasion of immune surveillance; in RCC correlates with T-cell infiltration and favorable prognosis | (30, 31) |
| Hmgb1 | up | Late in tumor progression or in response to therapies promotes recruitment and sustenance of immunosuppressive myeloid-derived suppressor cells and Treg | (32–34) |
| Fgl2 | up | Inhibitor of DC maturation and T-cell proliferation | (37) |
| Cd209a (DC-SIGN) | up | Initiates innate immunity; in advanced RCC may also lead to evasion from the immune surveillance; Important target in DC immunotherapy | (35, 36) |
| Foxj1 | up | Promotes T-cell tolerance and inhibition of spontaneous autoimmunity | (50) |
| Klra2 | up | Negative regulator of murine myeloid cells and NK cells | (38–40) |
| miRNA | |||
| miR-1186 | up | Short term expression: promotion of tumor growth through Cyclin B1 (Ccnb1) activation | (42–44) |
| miR-98 | up | Promotion and suppression of tumor growth | (45–47) |
| miR-5097 | up | Novel description | |
| miR-1942 | up | Novel description in RCC / immunoediting | |
| miR-708 | down | Tumor suppressor of RCC | (48) |
Fig. 4.
Clusters of differential (A) mRNA and (B) miRNA expression between tumors that evaded the DC-Ad-GM∙CAIX prevention therapy of group A and sham treated of group C (DC-Ad-Null). Multidimensional scaling using 1-correlation as distance metric. Validation of (C) gene expression and (D) miRNA by qPCR. Error bars shown are standard error of the mean. Tumor expression levels relative to control are shown. Gene and miRNA expressions were normalized to 18S rRNA and to snoRNA234 (except for mir-708, which was normalized to snoRNA202), respectively.
Since MHC class I (MHC-I) expression is crucial for CTL targeting of tumor cells, we next evaluated whether tumors that evaded the DC-Ad-GM∙CAIX therapy exhibited differential expression of MHC-I. The qPCR analysis suggested that the Balb/c MHC-I markers H-2Kd and H-2Dd were expressed and at similar levels in tumors that evaded the DC-Ad-GM∙CAIX therapy and in tumors of mice treated with DC-Ad-Null (Table 2, Fig. 4C).
To characterize and evaluate the molecular divergence between NPR-IX tumors that evaded DC-Ad-GM∙CAIX therapy (group A; 5 tumors) and NPR-IX tumors of mice treated with DC-Ad-Null (group C; 4 tumors), we preformed genome wide mRNA and miRNA expressions analysis. We found 367 differentially expressed gene features between groups A and C altered at least by 1.5-fold at P<0.015 representing 314 coding genes that are listed at NCBI EntrezGene database (Supplementary Table S1). Hierarchical clustering of mean centered logarithmic expressions of these genes by average linkage algorithm using 1-correlation as distance metric clustered the tumor samples into two groups (groups A and C; Fig. 4A). Similar clustering was observed for the miRNA expression microarray spots (n=28; P<0.015) of which 11 miRNAs clustered the samples into groups A and C (Fig. 4B). The differentially expressed genes were associated with significantly enriched gene ontology categories (IPA, Ingenuity, CA; P<0.05) including immune response, proliferation, cell growth, cell movement, and cell-to-cell signaling (Supplementary Table S2A-B). Then, using an extensive literature survey (CoreMine and PubMed) we identified which of the differentially expressed genes and miRNAs are known to play a predominant role in mechanisms of tumor evasion. Of the evasion genes we observed and validated by qPCR were the differential expression of chemokines (Ccl1, Cxcl9), regulators and markers of the myeloid cells, DCs, natural killer (NK) cells and T-cells (Hmgb1, Cd209a, Fgl2, Klra2, Foxj1), and of 5 miRNAs (miR-1186, miR-98, miR-5097, miR-1942, miR-708; Table 2, Fig. 4C-D).
DISCUSSION
Designing targeted and effective immunotherapy strategies for the treatment of cancer requires not only identifying specific TAAs, but also utilizing an innovative approach which re-engages the immune system to specifically recognize and kill tumor cells (22). Our current in vivo results, coupled with our previously published in vitro data, support the hypothesis that DC-Ad-GM∙CAIX can generate an in vivo T-cell mediated anti-hCAIX-specific immune response resulting in both prevention and treatment of established RCC tumors. Moreover, this therapy appears to be capable of inducing significant inhibition of RCC tumor growth in immunocompetent mice without obvious systemic toxicity.
As previously established, murine and human GM-CSF act in a species-specific manner (23, 24), and therefore the therapeutic efficacy demonstrated here can likely be attributed to only the hCAIX part of the fusion molecule. Indeed, overexpression of hCAIX was the critical mechanistic determinant of tumor response to DC-Ad-GM∙CAIX immunotherapy. Only tumor cells overexpressing hCAIX protein (NPR-IX) were inhibited and only in groups immunized with DC-Ad-GM∙CAIX (groups A, G). Moreover, in both the prevention and the treatment models, tumor tissue that subsequently evaded the therapy appeared to have done so only in tumors with little or no hCAIX protein expression. A significant finding is that, while both mouse and human CAIX proteins were absent in the immunotherapy-evading tumors, the CAIX transcripts were expressed without significant change from the controls, suggesting that CAIX expression may also be regulated at the protein level (Table 2; Fig. 3). Similarly, although HLA class I gene down-regulation or loss often occurs in RCC, presumably enabling the tumor to evade cytotoxic T-cells (25), in our system both the evading tumors and controls expressed the same transcription levels of the Balb/c MHC class I markers H-2Kd and H-2Dd (Table 2). Our in vivo pharmacodynamics data, the lack of murine serum antibodies against hCAIX, the uncontrolled growth of NPR-IX tumors in the control groups C and E, and our previous and reconfirmed in vitro detection of hCAIX-targeted cytotoxic T-cell activity (IFN-γ; ref. 13), all suggest a hCAIX-specific cytotoxic T-cell response and not a humoral one.
While the absence of the hCAIX protein appears to have been the critical determinant of evasion, the evasion phase may depend also on other genes and can be regarded as part of the concept of “cancer immunoediting” (26). Clinically and biologically, cancer immunoediting is a complex but orderly continuum that can be separated into a series of three overlapping phases: elimination (tumor destruction), equilibrium (dormancy), and evasion (escape; ref. 27). To gain further molecular insight into the immunoediting processes active in our models, we compared the gene and miRNA expression in tumors that evaded the DC-Ad-GM∙CAIX immunotherapy with the expression in tumors subject to DC-Ad-Null therapy (groups A and C, respectively). The genes and miRNA expression datasets of tumors that evaded the prevention immunotherapy significantly clustered the tumors apart from the tumors in group C. Our gene ontology analysis of the differential gene expression suggests a significant enrichment of categories such as activation of leukocytes, inflammatory response, and chemotaxis of myeloid cells (Supplementary Table S2). Consistent with this idea is the differential up-regulation we observed for the chemokines Ccl1 and Cxcl9 in tumors that evaded immunotherapy. Ccl1 has been suggested to induce Treg mediated immunosuppressive functions (28, 29), and Cxcl9 has been correlated with both T-cell infiltration and promotion of immune surveillance evasion (30, 31). There is also compelling evidence for the role of myeloid cells in tumor evasion. Hmgb1, Cd209a (DC-SIGN) and Fgl2, all of which were found to be up-regulated in the evading tumors (group A), have been suggested to function as recruiters of immunosuppressive myeloid-derived suppressor cells and Treg (32–34), leading to evasion from immune surveillance (35, 36) and to inhibition of DC maturation and T-cell proliferation (Table 2, Fig. 4C; ref. 37). Another characteristic of RCC are the presence and role of NK cells (38). One of the negative regulators of murine myeloid cells is Klra2 (Ly49B). While Klra2 is not normally expressed at detectable levels on NK cells, it appears to be up-regulated on myeloid cells and NK cells following encounter with inflammatory cytokines or bacterial products, decreasing their reactivity and preventing non-specific inflammatory responses (39, 40). Now for the first time we report that Klra2 is also up-regulated in tumors that have evaded the DC-Ad-GM∙CAIX immunotherapy, suggesting an intriguing new role in inhibiting NK cell-mediated target RCC lysis (Table 2, Fig. 4C, Supplementary Table S1).
Tumors that evaded the prevention immunotherapy up-regulated four miRNAs (miR-1186, miR-98, miR-5097, miR-1942) and down-regulated one (miR-708; Table 2, Fig. 4D, Supplementary Table S1). Our discovery of the up-regulation of miR-5097 in tumors that evaded immunotherapy is, to the best of our knowledge, the first publication on this miRNA. The function of miR-1942 in tumors that evaded therapy is yet to be identified (41). The short-term up-regulation of miR-1186 was recently suggested to promote tumor growth through Cyclin B1 (Ccnb1) activation, which is substantially involved in RCC carcinogenesis and progression (42–44). miR-98 is suggested to have tumor suppressive function in breast tumors, tumor promoting function in other types of tumors (45–47). miR-708 expression, which we found to be down-regulated in tumors that evaded immunotherapy, was recently described to be attenuated in RCC and was suggested to have a role as a pro-apoptoic tumor suppressor (48).
Collectively, the identification and molecular characterization of tumors that evaded immunotherapy may provide information on additional immune evasion processes and may ultimately be useful for the development of tumor evasion diagnostic markers and for the rational development of next generation immunotherapies.
In summary, we have developed, tested, and characterized a novel immunotherapy targeting hCAIX in immunocompetent mouse models, which proved to be highly responsive to both prevention and treatment strategies. These encouraging pre-clinical in vivo data, coupled with our previously reported in vitro and severe combined immunodeficiency (SCID) mouse model studies, provide the basis for initiating a first–in-human phase I trial using clinical-grade Ad-GM∙CAIX-based immunotherapy in patients with advanced RCC.
Supplementary Material
Supplementary Fig. S1. Dot plots of indirect FACS for anti-hCAIX antibodies using (A) NPR-IX and (B) RENCA stained by mouse serum from group A, and (C) NPR-IX and (D) RENCA stained by mouse serum from group E, followed by staining with FITC-conjugated anti-mouse IgG. Comparable rates of positive staining for mouse IgG within group A and group E suggest that sera of mice of group A and E do not contain measurable amounts of mouse anti-hCAIX antibodies.
Supplementary Fig. S2. Dot plots of FACS for hCAIX in NPR-IX and RENCA cells. (A) Unstained NPR-IX; (B) NPR-IX stained with anti-hCAIX-FITC, showing 95.30% of cells positive for hCAIX expression; (C) Unstained RENCA; (D) RENCA stained with anti h CAIX-FITC, showing 2.15% of cells positive for hCAIX expression, which is assumed to be an artifact of nonspecific binding.
Supplementary Fig. S3. Western blot of the fusion protein hGMCAIX. The 70 kDa weight seen in both blots shows that the hGM·CAIX fusion gene is primarily expressed as a fused protein. Lane 1: 293T control; lane 2: 293T infected with Ad-Null; lane 3: 293T infected with Ad-GM·CAIX; (A) Gel exposed for actin loading control and hCAIX; (B) Gel exposed for actin loading control and hGM-CSF.
Supplementary Fig. S4. Dot plots of FACS for hCAIX (representative of hGM·CAIX expression) and mCd86 in DC-Ad-GM·CAIX; (A) Unstained DC-Ad-GM·CAIX; (B) DC-Ad-GM·CAIX stained with anti-hCAIX-FITC, showing 70.1% of cells positive for hCAIX expression, indicating successful infection of DCs with Ad-GM·CAIX; (C) DC-Ad-GM·CAIX stained with anti-mCd86-PE, showing 96.3% of cells positive for mCd86 expression, indicating successful differentiation from BMCs into DCs; (D) DC-Ad-GM·CAIX double stained with anti-hCAIX-FITC and with anti-mCd86-PE, showing 28.4% of cells positive for both hCAIX and mCd86 expression, representing the percentage of hCAIX-expressing DCs.
Supplementary Fig. S5. Dot plots of FACS for hCAIX and hGM-CSF expression in DC-Ad-GM·CAIX. (A) Unstained DC-Ad-GM·CAIX; (B) DC-Ad-GM·CAIX stained with anti hCAIX-FITC, showing 49.51% of cells positive for hCAIX expression; (C) DC-Ad-GM·CAIX stained with anti-hGM-CSF-PerCP, showing 30.35% of cells positive for hGM-CSF expression; (D) DC-Ad-GM·CAIX double-stained with anti-hCAIX-FITC and anti-hGMCSF-PerCP, showing 12.70% of cells positive for both hCAIX and hGM-CSF expression.
ACKNOWLEDGMENTS
This project was supported in part by the Swiss National Science Foundation (SNSF) individual fellowship PBBSP3–133403 (F.B.), the Swiss Board of Urology (SGU) annual fellowship (F.B.), an individual fellowship of the Gottfried und Julia Bangerter-Rhyner Foundation (F.B.), and the Wissenschaftliche Urologische Gesellschaft e.V. (N.K.). The project was further supported in part by NCI RAID Initiative number NSC 740833 from the National Cancer Institute. We thank Mr. Randy Caliliw for his excellent technical help. We are grateful to Dr. Melissa Stauffer for editing the manuscript. We thank all the co-workers of Dr. Antoni Ribas’ laboratory for their generous support. We also thank Drs. Aya Jakobovits and Adrian Bot for fruitful discussions and evaluation, Drs. Xinmin Li, Jian Zhou, and Wan Haolei for microarray and qPCR support. Flow cytometry was performed in the UCLA Jonsson Comprehensive Cancer Center and Center for AIDS Research Flow Cytometry Core Facility.
Sources of support:
This work was supported in part by the Swiss National Science Foundation (SNSF) individual fellowship PBBSP3–133403 (F.B.), the Swiss Board of Urology (SGU) annual fellowship (F.B.), an individual fellowship of the Gottfried und Julia Bangerter-Rhyner Foundation (F.B.), the Wissenschaftliche Urologische Gesellschaft e.V. (N.K.). The project was further in part supported by NCI RAID Initiative number NSC 740833 from the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosure: Kite Pharma Inc., executive chairman, founder and stock holder (ASB), co-founder and stock holder (AJP), scientific advisory board and stock holder (AR); patents, Ad-GM∙CAIX (ASB), biomarkers for the immunoediting-escape phase (JR, ASB, FDB, AJP, AR); all other authors have declared there are no financial interests in regards to this work.
REFERENCES
- 1.Rosenberg SA. Cancer immunotherapy comes of age. Nat Clin Pract Oncol. 2005;2:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shablak A, Hawkins RE, Rothwell DG, et al. T cell-based immunotherapy of metastatic renal cell carcinoma: modest success and future perspective. Clin Cancer Res. 2009;15:6503–10. [DOI] [PubMed] [Google Scholar]
- 3.Gouttefangeas C, Stenzl A, Stevanovic S, et al. Immunotherapy of renal cell carcinoma. Cancer Immunol Immunother. 2007;56:117–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swietach P, Hulikova A, Vaughan-Jones RD, et al. New insights into the physiological role of carbonic anhydrase IX in tumour pH regulation. Oncogene. 2010;29:6509–21. [DOI] [PubMed] [Google Scholar]
- 5.Stillebroer AB, Mulders PF, Boerman OC, et al. Carbonic anhydrase IX in renal cell carcinoma: implications for prognosis, diagnosis, and therapy. Eur Urol. 2010;58:75–83. [DOI] [PubMed] [Google Scholar]
- 6.Finley DS, Pantuck AJ, Belldegrun AS. Tumor biology and prognostic factors in renal cell carcinoma. Oncologist. 2011;16 Suppl 2:4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanov S, Liao SY, Ivanova A, et al. Expression of hypoxia-inducible cell-surface transmembrane carbonic anhydrases in human cancer. Am J Pathol. 2001;158:905–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uemura H, Fujimoto K, Tanaka M, et al. A phase I trial of vaccination of CA9-derived peptides for HLA-A24-positive patients with cytokine-refractory metastatic renal cell carcinoma. Clin Cancer Res. 2006;12:1768–75. [DOI] [PubMed] [Google Scholar]
- 9.Lamers CH, Sleijfer S, Vulto AG, et al. Treatment of metastatic renal cell carcinoma with autologous T-lymphocytes genetically retargeted against carbonic anhydrase IX: first clinical experience. J Clin Oncol. 2006;24:e20–2. [DOI] [PubMed] [Google Scholar]
- 10.Siebels M, Rohrmann K, Oberneder R, et al. A clinical phase I/II trial with the monoclonal antibody cG250 (RENCAREX(R)) and interferon-alpha-2a in metastatic renal cell carcinoma patients. World J Urol. 2011;29:121–6. [DOI] [PubMed] [Google Scholar]
- 11.Kantoff PW, Higano CS, Shore ND, et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med. 2010;363:411–22. [DOI] [PubMed] [Google Scholar]
- 12.Borrello I, Pardoll D. GM-CSF-based cellular vaccines: a review of the clinical experience. Cytokine Growth Factor Rev. 2002;13:185–93. [DOI] [PubMed] [Google Scholar]
- 13.Tso CL, Zisman A, Pantuck A, et al. Induction of G250-targeted and T-cell-mediated antitumor activity against renal cell carcinoma using a chimeric fusion protein consisting of G250 and granulocyte/monocyte-colony stimulating factor. Cancer Res. 2001;61:7925–33. [PubMed] [Google Scholar]
- 14.Mukouyama H, Janzen NK, Hernandez JM, et al. Generation of kidney cancer-specific antitumor immune responses using peripheral blood monocytes transduced with a recombinant adenovirus encoding carbonic anhydrase 9. Clin Cancer Res. 2004;10:1421–9. [DOI] [PubMed] [Google Scholar]
- 15.Hernandez JM, Bui MH, Han KR, et al. Novel kidney cancer immunotherapy based on the granulocyte-macrophage colony-stimulating factor and carbonic anhydrase IX fusion gene. Clin Cancer Res. 2003;9:1906–16. [PubMed] [Google Scholar]
- 16.Shvarts O, Janzen N, Lam JS, et al. RENCA/carbonic anhydrase-IX: a murine model of a carbonic anhydrase-IX-expressing renal cell carcinoma. Urology. 2006;68:1132–8. [DOI] [PubMed] [Google Scholar]
- 17.Wiltrout RH, Gregorio TA, Fenton RG, et al. Cellular and molecular studies in the treatment of murine renal cancer. Semin Oncol. 1995;22:9–16. [PubMed] [Google Scholar]
- 18.Koya RC, Kimura T, Ribas A, et al. Lentiviral vector-mediated autonomous differentiation of mouse bone marrow cells into immunologically potent dendritic cell vaccines. Mol Ther. 2007;15:971–80. [DOI] [PubMed] [Google Scholar]
- 19.Littell RC, Milliken GA, Stroup WW, et al. SAS System for Mixed Models, Cary, NC: SAS Institute Inc; 1996. [Google Scholar]
- 20.Klein J, Moeschberger M. Survival Analysis: Techniques for Censored and Truncated Data, 2nd ed. Springer-Verlag; ISBN: 038795399X. 2003. [Google Scholar]
- 21.Berger RL, Boos DD. Confidence Limits for the Onset and Duration of Treatment Effect. Biometrical Journal. 1999;41:517–31. [Google Scholar]
- 22.Sharma P, Wagner K, Wolchok JD, et al. Novel cancer immunotherapy agents with survival benefit: recent successes and next steps. Nat Rev Cancer. 2011;11:805–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitamura T, Hayashida K, Sakamaki K, et al. Reconstitution of functional receptors for human granulocyte/macrophage colony-stimulating factor (GM-CSF): evidence that the protein encoded by the AIC2B cDNA is a subunit of the murine GM-CSF receptor. Proc Natl Acad Sci U S A. 1991;88:5082–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe Y, Matsumoto Y, Yamaguchi M, et al. Absorption of recombinant human granulocyte colony-stimulating factor (rhG-CSF) and blood leukocyte dynamics following intranasal administration in rabbits. Biol Pharm Bull. 1993;16:93–5. [DOI] [PubMed] [Google Scholar]
- 25.Atkins D, Ferrone S, Schmahl GE, et al. Down-regulation of HLA class I antigen processing molecules: an immune escape mechanism of renal cell carcinoma? J Urol. 2004;171:885–9. [DOI] [PubMed] [Google Scholar]
- 26.Chow MT, Moller A, Smyth MJ. Inflammation and immune surveillance in cancer. Semin Cancer Biol. 2012;22:23–32. [DOI] [PubMed] [Google Scholar]
- 27.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–70. [DOI] [PubMed] [Google Scholar]
- 28.Daurkin I, Eruslanov E, Stoffs T, et al. Tumor-associated macrophages mediate immunosuppression in the renal cancer microenvironment by activating the 15-lipoxygenase-2 pathway. Cancer Res. 2011;71:6400–9. [DOI] [PubMed] [Google Scholar]
- 29.Hoelzinger DB, Smith SE, Mirza N, et al. Blockade of CCL1 inhibits T regulatory cell suppressive function enhancing tumor immunity without affecting T effector responses. J Immunol. 2010;184:6833–42. [DOI] [PubMed] [Google Scholar]
- 30.Amatschek S, Lucas R, Eger A, et al. CXCL9 induces chemotaxis, chemorepulsion and endothelial barrier disruption through CXCR3-mediated activation of melanoma cells. Br J Cancer. 2011;104:469–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oldham KA, Parsonage G, Bhatt RI, et al. T lymphocyte recruitment into renal cell carcinoma tissue: a role for chemokine receptors CXCR3, CXCR6, CCR5, and CCR6. Eur Urol. 2012;61:385–94. [DOI] [PubMed] [Google Scholar]
- 32.Weiner LM, Lotze MT. Tumor-cell death, autophagy, and immunity. N Engl J Med. 2012;366:1156–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palucka K, Banchereau J. Cancer immunotherapy via dendritic cells. Nat Rev Cancer. 2012;12:265–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin L, Zhong K, Sun Z, et al. Receptor for advanced glycation end products (RAGE) partially mediates HMGB1-ERKs activation in clear cell renal cell carcinoma. J Cancer Res Clin Oncol. 2012;138:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou T, Chen Y, Hao L, et al. DC-SIGN and immunoregulation. Cell Mol Immunol. 2006;3:279–83. [PubMed] [Google Scholar]
- 36.Yang L, Yang H, Rideout K, et al. Engineered lentivector targeting of dendritic cells for in vivo immunization. Nat Biotechnol. 2008;26:326–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu H, Shalev I, Manuel J, et al. The FGL2-FcgammaRIIB pathway: a novel mechanism leading to immunosuppression. Eur J Immunol. 2008;38:3114–26. [DOI] [PubMed] [Google Scholar]
- 38.Geiger C, Nossner E, Frankenberger B, et al. Harnessing innate and adaptive immunity for adoptive cell therapy of renal cell carcinoma. J Mol Med (Berl). 2009;87:595–612. [DOI] [PubMed] [Google Scholar]
- 39.Gays F, Aust JG, Reid DM, et al. Ly49B is expressed on multiple subpopulations of myeloid cells. J Immunol. 2006;177:5840–51. [DOI] [PubMed] [Google Scholar]
- 40.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hino K, Fukao T, Watanabe M. Regulatory interaction of HNF1-alpha to microRNA-194 gene during intestinal epithelial cell differentiation. Nucleic Acids Symp Ser (Oxf). 2007:415–6. [DOI] [PubMed] [Google Scholar]
- 42.Huang V, Place RF, Portnoy V, et al. Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res. 2012;40:1695–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ikuerowo SO, Kuczyk MA, Mengel M, et al. Alteration of subcellular and cellular expression patterns of cyclin B1 in renal cell carcinoma is significantly related to clinical progression and survival of patients. Int J Cancer. 2006;119:867–74. [DOI] [PubMed] [Google Scholar]
- 44.Egloff AM, Vella LA, Finn OJ. Cyclin B1 and other cyclins as tumor antigens in immunosurveillance and immunotherapy of cancer. Cancer Res. 2006;66:6–9. [DOI] [PubMed] [Google Scholar]
- 45.Farazi TA, Horlings HM, Ten Hoeve JJ, et al. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71:4443–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du L, Schageman JJ, Subauste MC, et al. miR-93, miR-98, and miR-197 regulate expression of tumor suppressor gene FUS1. Mol Cancer Res. 2009;7:1234–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu G, Zhou R, Liu J, et al. MicroRNA-98 and let-7 confer cholangiocyte expression of cytokine-inducible Src homology 2-containing protein in response to microbial challenge. J Immunol. 2009;183:1617–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saini S, Yamamura S, Majid S, et al. MicroRNA-708 induces apoptosis and suppresses tumorigenicity in renal cancer cells. Cancer Res. 2011;71:6208–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koch KS, Son KH, Maehr R, et al. Immune-privileged embryonic Swiss mouse STO and STO cell-derived progenitor cells: major histocompatibility complex and cell differentiation antigen expression patterns resemble those of human embryonic stem cell lines. Immunology. 2006;119:98–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Srivatsan S, Peng SL. Cutting edge: Foxj1 protects against autoimmunity and inhibits thymocyte egress. J Immunol. 2005;175:7805–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. Dot plots of indirect FACS for anti-hCAIX antibodies using (A) NPR-IX and (B) RENCA stained by mouse serum from group A, and (C) NPR-IX and (D) RENCA stained by mouse serum from group E, followed by staining with FITC-conjugated anti-mouse IgG. Comparable rates of positive staining for mouse IgG within group A and group E suggest that sera of mice of group A and E do not contain measurable amounts of mouse anti-hCAIX antibodies.
Supplementary Fig. S2. Dot plots of FACS for hCAIX in NPR-IX and RENCA cells. (A) Unstained NPR-IX; (B) NPR-IX stained with anti-hCAIX-FITC, showing 95.30% of cells positive for hCAIX expression; (C) Unstained RENCA; (D) RENCA stained with anti h CAIX-FITC, showing 2.15% of cells positive for hCAIX expression, which is assumed to be an artifact of nonspecific binding.
Supplementary Fig. S3. Western blot of the fusion protein hGMCAIX. The 70 kDa weight seen in both blots shows that the hGM·CAIX fusion gene is primarily expressed as a fused protein. Lane 1: 293T control; lane 2: 293T infected with Ad-Null; lane 3: 293T infected with Ad-GM·CAIX; (A) Gel exposed for actin loading control and hCAIX; (B) Gel exposed for actin loading control and hGM-CSF.
Supplementary Fig. S4. Dot plots of FACS for hCAIX (representative of hGM·CAIX expression) and mCd86 in DC-Ad-GM·CAIX; (A) Unstained DC-Ad-GM·CAIX; (B) DC-Ad-GM·CAIX stained with anti-hCAIX-FITC, showing 70.1% of cells positive for hCAIX expression, indicating successful infection of DCs with Ad-GM·CAIX; (C) DC-Ad-GM·CAIX stained with anti-mCd86-PE, showing 96.3% of cells positive for mCd86 expression, indicating successful differentiation from BMCs into DCs; (D) DC-Ad-GM·CAIX double stained with anti-hCAIX-FITC and with anti-mCd86-PE, showing 28.4% of cells positive for both hCAIX and mCd86 expression, representing the percentage of hCAIX-expressing DCs.
Supplementary Fig. S5. Dot plots of FACS for hCAIX and hGM-CSF expression in DC-Ad-GM·CAIX. (A) Unstained DC-Ad-GM·CAIX; (B) DC-Ad-GM·CAIX stained with anti hCAIX-FITC, showing 49.51% of cells positive for hCAIX expression; (C) DC-Ad-GM·CAIX stained with anti-hGM-CSF-PerCP, showing 30.35% of cells positive for hGM-CSF expression; (D) DC-Ad-GM·CAIX double-stained with anti-hCAIX-FITC and anti-hGMCSF-PerCP, showing 12.70% of cells positive for both hCAIX and hGM-CSF expression.