Abstract
Studying nuclear positioning in developing tissues of the model nematode Caenorhabditis elegans greatly contributed to the discovery of SUN and KASH proteins and the formation of the LINC model. Such studies continue to make important contributions into both how LINC complexes are regulated and how defects in LINC components disrupt normal development. The methods described explain how to observe and quantify the following: nuclear migration in embryonic dorsal hypodermal cells, nuclear migration through constricted spaces in larval P cells, nuclear positioning in the embryonic intestinal primordia, and nuclear anchorage in syncytial hypodermal cells. These methods will allow others to employ nuclear positioning in C. elegans as a model to further explore LINC complex regulation and function.
Keywords: LINC, KASH, SUN, C. elegans, Nuclear migration, Nuclear anchorage, Nuclear envelope
1. Introduction
The normal development of most eukaryotes depends on actively moving and anchoring the nucleus to a specific location within the cell. Failures in nuclear positioning lead to a wide variety of defects and diseases [1, 2]. The connection between the cytoskeleton and the nuclear envelope is essential for these processes and is often mediated by a nuclear envelope bridge of Sad1p/UNC-84 (SUN) proteins at the inner nuclear membrane and Klarsicht/ANC-1/Syne homology (KASH) proteins in the outer nuclear membrane. SUN and KASH proteins interact with each other in the perinu-clear space, and together, they form LINC complexes to connect the nucleoskeleton to the cytoskeleton [3–5]. LINC complexes are found throughout eukaryotes and have been shown in a variety of systems to mediate nuclear migration and anchorage [1, 2].
Caenorhabditis elegans played a central role in the discovery of SUN and KASH proteins and the development of the LINC complex model [3]. C. elegans is a particularly well-suited system for studying LINC components and nuclear positioning because it combines powerful genetics with the ability to study nuclear positioning events by real-time imaging in the context of a developing organism. In the early 1980s, mutations in unc-83 and unc-84 that disrupted nuclear migration were isolated by Horvitz and Sulston [6], while mutations in anc-1 that disrupted nuclear anchorage were found by Hedgecock and Thomson [7]. Malone, Starr, and Han molecularly characterized UNC-84 (SUN), UNC-83 (KASH), and ANC-1 (KASH), which led to the discovery of SUN and KASH domains at the nuclear envelope and the creation of the LINC model [8–11].
Here we discuss three tissues in C. elegans that serve as excellent models for studying nuclear migration events and focus on the adult hypodermis to study nuclear anchorage. Together, these invariant and developmentally regulated nuclear positioning events have played huge roles in characterizing the mechanisms of LINC complexes. The goal of this methods paper is to make these experimental models accessible to others interested in studying LINC complexes in C. elegans.
The first nuclear migration event described here occurs during embryogenesis, when left and right groups of dorsal epithelial cells intercalate, and their nuclei migrate contralaterally across the length of hyp7 precursor cells (Fig. 1A). These cells subsequently fuse, forming the dorsal hypodermal syncytium with laterally positioned nuclei [12–14]. Mutations in unc-83 or unc-84 completely disrupt hyp7 cell nuclear migration (Fig. 1B), resulting in nuclei that are mispositioned to the dorsal cord (Fig. 1C, D) [8, 10, 15]. Subheading 3.1 describes how nuclear migration is studied in hyp7 precursors by scoring finalized nuclear positioning defects in larvae, and Subheading 3.2 describes how to view the migration defect by live imaging in the embryo.
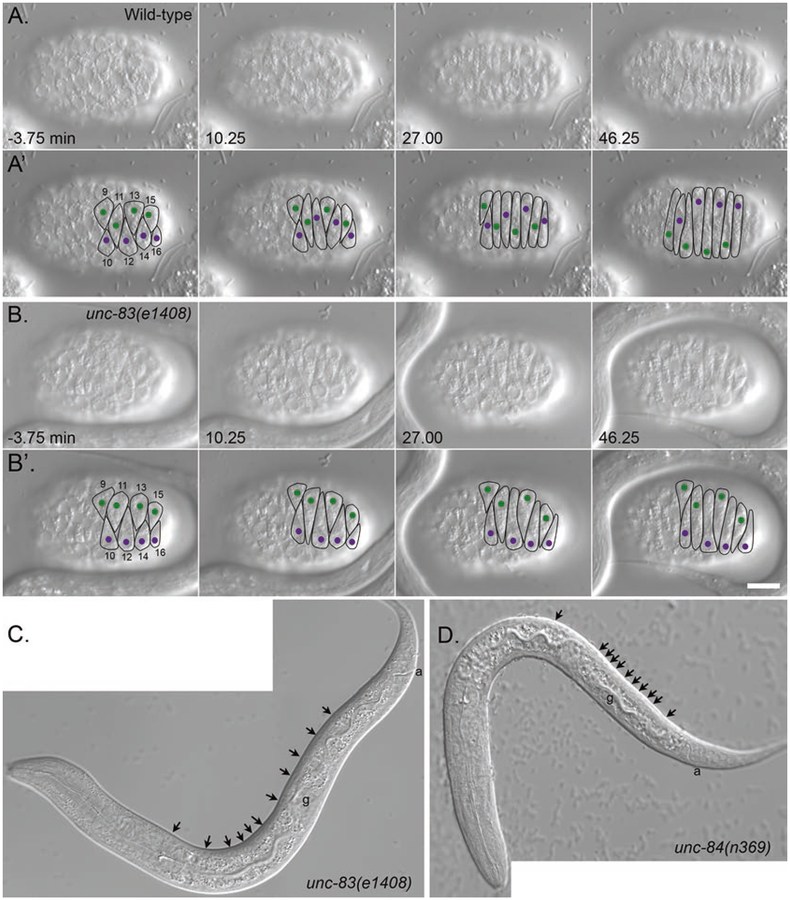
Fig. 1.
Nuclear migration in embryonic hyp7 precursors. (A, B) DIC images from a time-lapse series of images of nuclear migration in dorsal hyp7 precursors in wild-type (A) and unc-83(null) (B) embryos. The time that cell 12 completed intercalation was defined as t = 0. Dorsal view, anterior is to the left. (A’, B’) Cell borders are outlined in black, nuclei migrating left to right are purple, and nuclei migrating right to left are green. Bar, 10 μm. Reproduced from [15] with permission from The Journal of Cell Biology. (C, D) L1 larva showing hyp7 nuclei mislocalized in the dorsal cord (arrowheads) in unc-83(null) (C) and unc-84(null) (D). In wild type, there would be no nuclei in the dorsal cord. Not all nuclei are seen in this focal plane. Lateral view; anterior is to the left, and dorsal is up. The four-cell germ line (g) and the anus (a) are marked to help with dorsal-ventral orientation. Note that the anchorage phenotype shown in (D) is the most severe phenotype seen in unc-84(null) larvae
In the second discussed tissue, larval hypodermal cells, P-cell nuclei migrate from a lateral to a ventral position (Fig. 2A). These nuclear migration events are particularly interesting because nuclei must flatten to ~5% of their width to squeeze through a constricted space between muscles and the cuticle [16] (Fig. 2B, C). Thus, P cells are an excellent model for how nuclei squeeze through constricted spaces in humans, such as leukocyte extravasation and some cancer metastases [17, 18]. After the completion of nuclear migration to the ventral cord, P cells divide and give rise to the vulva, hypodermal cells, and motor neurons. Failure in P-cell nuclear migration results in P-cell death and, in turn, Egl (egg-laying deficient) and Unc (uncoordinated) animals due to the lack of vulval cells and motor neurons, respectively. These phenotypes in unc-83 and unc-84 null animals are temperature sensitive; at the restrictive temperature of 25 °C, about 50% of P-cell nuclei fail to migrate to the ventral cord, resulting in Egl and Unc phenotypes. However, at the permissive temperature of 15 °C, 90% of P-cell nuclei migrate normally to the ventral cord, and the animals have no obvious phenotype [6, 8, 10, 19]. In Subheading 3.3, we describe how to score P-cell nuclear migration by counting missing P-cell progeny using GABA neuron (Fig. 2D–F) or P-cell nuclear markers (Fig. 2C).
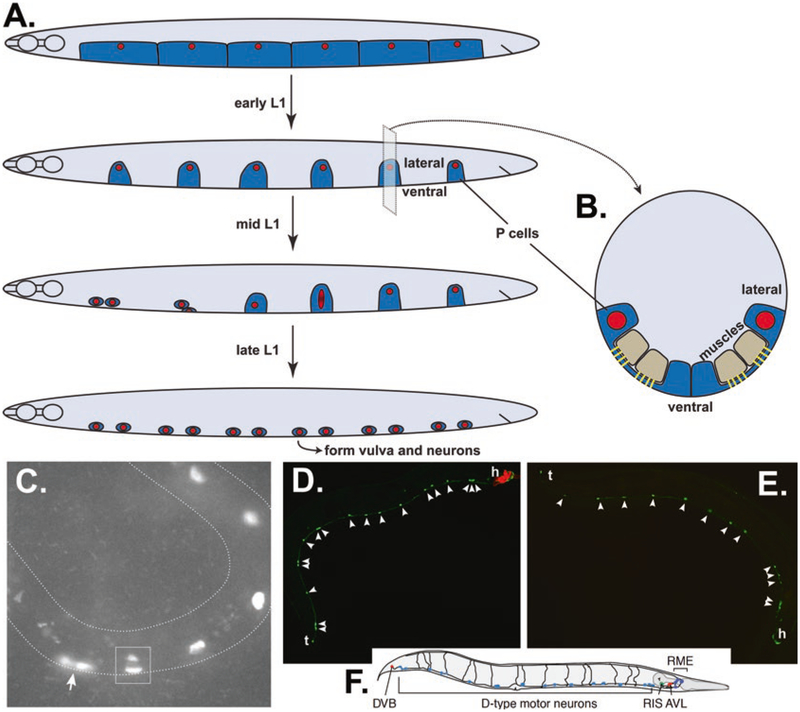
Fig. 2.
Nuclear migration through constricted spaces in larval P cells. (A) Cartoon of P cells throughout L1 larval development; lateral view, ventral is down, anterior to the left. Shortly after hatching, P-cell cytoplasm (dark blue) covers the ventral surface of the larva, and there are six P-cell nuclei (red) on each lateral side. P cells narrow in early L1 and migrate during mid L1 to form a row of 12 P cells in the ventral cord by late L1. Migration usually initiates with the anterior-most pair of P cells. During nuclear migration, P-cell nuclei stretch from lateral to ventral. (B) A cross section of an L1 larva just before P-cell nuclear migration. In order for the P-cell nucleus (red) to migrate from the lateral to the ventral compartments of the P-cell cytoplasm (dark blue), it must squeeze through a narrow constriction between body wall muscles (tan) and the cuticle. Fibrous organ-elles (yellow) form posts in this constricted space to attach muscles to the cuticle. (C) A mid-L1 larva expressing an RFP P-cell nuclear marker in an otherwise wild-type animal. Anterior is left, and ventral is down and on the outside of the curve. The anterior-most pair of P cells has completed migration to the ventral cord (arrow). The next P-cell nucleus is stretched between lateral and ventral compartments; the dark space within the nucleus is nucleoplasm within the constricted space under body wall muscles. P-cell nuclei more posterior have yet to enter the constricted space. Image kindly provided by Courtney Bone (BioMarin). (D–F) Adult animals expressing unc-47::gfp to mark GABA neurons. Tails (t) are left, and heads (h) are right. Lateral view; ventral is down. (D) An unc-84(null) animal expressing an unc-84(+) transgene (transgenic animals express a red marker in the head) with 18 GABA-positive neurons (arrowheads). (E) An unc-84(null) animal with only 13 GABA-positive neurons (arrowheads) between the head and tail. (F) A cartoon showing all the GFP-marked GABA neurons in wild type. Reproduced from [34] with permission from Cell Press. We only count the 19 D-type motor neurons in the ventral cord (light blue), 12 of which are derived from P cells. The DVB in the tail and neurons in the head is derived from other lineages
The third set of nuclear migration events occur in the embryonic intestinal primordium. These nuclei move toward the future apical surface where the intestinal lumen forms [20] (Fig. 3). un-83 and unc-84 null animals have strong nuclear migration defects in the developing embryonic intestine as indicated by nuclei failing to position at the midline of the primordium at both 25 °C and 15 °C [10] (Fig. 3). However, mutant embryos raised at 25 °C have slightly less severe defects than embryos raised at 15 °C, opposite the effect of temperature on P-cell nuclear migrations. In Subheading 3.4 we describe how to assay nuclear positioning in the intestinal primordium.
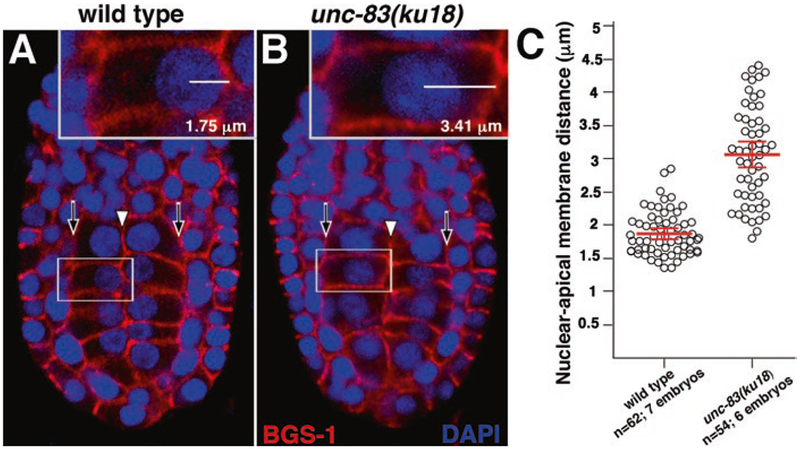
Fig. 3.
Nuclear migration in the C. elegans intestinal primordium. Wild-type and unc-83(ku18) E16 stage embryos were stained with a cortical marker (anti-BGS-1, red) and a nuclear dye (DAPI, blue). (A, B) Representative single optical sections from a ventral view (anterior up) are shown with white arrowheads denoting the apical surfaces and black arrows denoting the basal surfaces of intestinal cells. The nuclear apical membrane distance was determined by measuring the distance between the center of the nucleus and the apical membrane (insets). (C) Each circle represents the nuclear apical membrane distance of a single intestinal cell. The mean distance is shown in red, and the error bars denote the 95% confidence interval
There are at least two other C. elegans tissues with interesting nuclear migration events mediated by LINC complexes. Pronuclear migration after fertilization, which is mediated by the SUN protein SUN-1 and the KASH protein ZYG-12, can be easily filmed in the one-cell embryo [21, 22]. The syncytial germ line of the C. elegans hermaphrodite also relies on SUN-1 and ZYG-12 to organize nuclei [23]. As they are both well-established systems used by many different labs, we will not discuss the methods for studying them here.
After nuclei migrate to a specific location, they are anchored in place. Defects in nuclear anchorage can be seen in many of the syncytia in C. elegans including binucleated intestinal cells, pharynx isthmus muscle cells, seam cells, and the adult hyp7 syncytium [7, 9]. The phenotype is most dramatic in the adult hyp7 syncytium, which contains 139 nuclei that are normally evenly spaced apart [24]. Mutations in either unc-84 or anc-1 have a nuclear anchorage defect that can be observed with DIC optics (Fig. 1D) but is best quantified when the hyp7 nuclei are marked with a nuclear GFP (Fig. 4). Subheading 3.5 describes how we quantify nuclear anchorage in the adult hyp7.
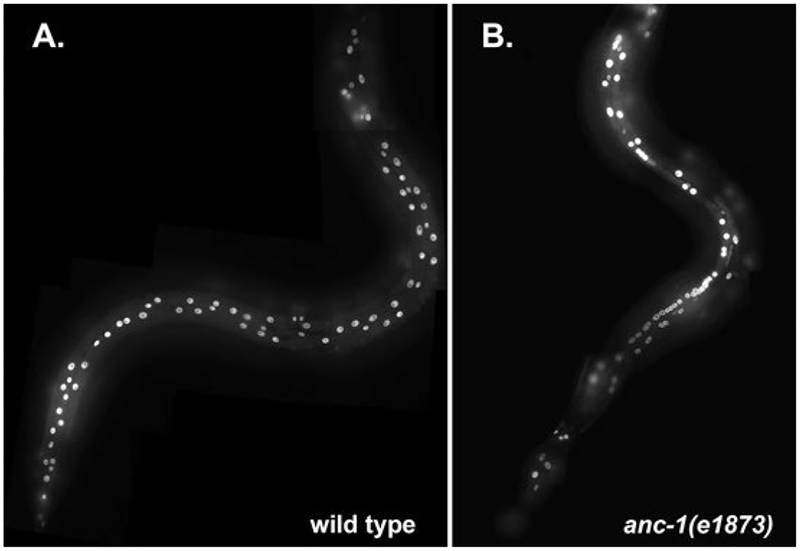
Fig. 4.
Nuclear anchorage in the adult hyp7 and seam cell syncytia. Adult animals expressing nuclear-localized GFP in hypodermal cells under control of the col-19 promoter are shown. (A) Wild-type animal with well spaced-out hypodermal nuclei. (B) anc-1(e1873) null animal with many clustered hypodermal nuclei. Heads are up
In order to enhance the study of nuclear positioning, we have developed fluorescent fusion proteins for live imaging in transgenic animals. We have taken advantage of three tissue-specific markers: the promoter of lbp-1 for expression of fluorescent proteins in embryonic hyp7 precursors [15, 25], the hlh-3 promoter for expression in larval P cells [19, 26], and the col-19 promoter for expression in the adult hyp7 syncytium [27]. Expression of fluorescent fusion proteins only in the desired tissue improves imaging quality. Refer to the referenced papers to see how these plasmids were cloned and how they could be of use for you to clone your gene of interest to be expressed at the time of nuclear migration or anchorage.
2. Materials
2.1. Microscopy
Dissecting microscope with epifluorescence.
Wide field fluorescence (or confocal) microscope with 10× and 63× or 100× objectives with DIC optics and epifluorescence (filters for GFP and RFP).
Agarose pad made of 2% agarose in H2O on a glass microscope slide (see Note 1).
Worm pick (see Note 2).
25 × 75 × 1 mm glass microscope slides.
M9 buffer: 3 g KH2PO4, 6 g Na2HPO4, 5 g NaCl, 1 mL 1 M MgSO4, and H2O up to 1 L. Autoclave to sterilize [28].
1 mM tetramisole, 25 mM sodium azide, or other paralytic in M9 buffer.
25 × 25 × 1 mm coverslips.
2.2. Staining Nuclei and the Cell Cortex of C. elegans Embryos
0.1% (w/v) poly-l-lysine.
Custom epoxy-coated slides with three square 14 mm wells (30–2066A-Brown. Cell-Line/Thermo Fisher Scientific).
25 × 25 × 1 mm and 25 × 40 × 1 mm coverslips.
Methanol or 4% buffered paraformaldehyde: 60 mM Pipes pH 6.8, 25 mM Hepes pH 6.8, 10 mM EGTA pH 6.8, 2 mM MgCl2, 0.1 mg/mL l-α-lysolecithin.
1× PBS-T: 1× PBS with 0.1% Tween-20.
PBS-BSA: 1× PBS-T, 1% bovine serum albumin, 0.1% sodium azide.
Rabbit anti-BGS-1 (diluted 1:900 in PBS-BSA) [29] and Donkey anti-Rabbit IgG-Rhodamine Red (Jackson ImmunoResearch; diluted 1:400 in PBS-BSA).
0.33 μM Alexa Fluor 488 phalloidin (Thermo Fisher Scientific; diluted in 1× PBS + 0.2% TX-100).
PBS-DAPI: 1× PBS, 0.1 μg/mL DAPI.
Mounting media: 80% glycerol, 0.223 M DABCO.
Clear nail polish.
ZEN software (Zeiss) or equivalent.
3. Methods
3.1. Counting Mispositioned hyp7 Nuclei in the Dorsal Cord
Culture C. elegans on NGM agar plates spotted with OP50 E. coli.
Prepare microscope slides with 2% agarose pad (see Note 1).
Pipette ~5 μL of M9 buffer to the agarose pad. 1 mM tetramisole or other paralytic can be optionally added to the M9.
Using a dissecting microscope and worm pick (see Note 2), transfer a large number (at least 20) of L1 and L2 worms to the M9 buffer.
Cover the agarose pad with a 25 × 25 × 1 mm coverslip.
Use DIC optics and a 10× objective on a compound microscope to locate an L1 or L2 worm.
Switch to a higher objective (63× or 100×) using DIC optics to zoom in on the selected worm (see Note 3).
Ensure that the larva is on its lateral side by identifying the four-cell germ line and anus on the ventral side (Fig. 1C, D). Identify the dorsal cord, which is opposite the developing germ line (see Note 4).
Score “fried egg”-shaped nuclei in the dorsal cord posterior of the pharynx to the anus.
Repeat steps 5–8 on additional L1 and L2 worms. Wild-type animals should have no hypodermal nuclei in the dorsal cord, whereas unc-83 or unc-84 null mutants have about 14 nuclei in the dorsal cord per L1 or L2 animal.
3.2. Filming and Analysis of hyp7 Nuclear Migration
Culture C. elegans on NGM agar plates spotted with OP50 E. coli.
Prepare microscope slides with 2% agarose pad (see Note 1).
Pipette ~5 μL of M9 buffer to the agarose pad.
Using a dissecting microscope and worm pick (see Note 2), transfer a large number of gravid adults and/or embryos to the M9 buffer. It is best to use a plate that has just starved, so the adults hold their embryos. These embryos will be at a later stage than normally found in the adult worm and will be close to the pre-comma stage needed for imaging. A newly starved plate will also have a lot of laid embryos close to the correct stage.
Cover the agarose pad with a 25 × 25 × 1 mm coverslip. When the coverslip is placed on top of the gravid adults, apply gentle pressure, and embryos will squeeze out of the vulva.
Use DIC optics and a 10× objective on a compound microscope to locate a pre-comma stage embryo (see Note 5).
Switch to a higher objective (63× or 100×) using DIC optics to zoom in on the selected embryo (see Note 3).
Capture images at one frame every 15 s, for at least 40 min (see Note 6).
Export the images as an AVI at 15 frames per second without compression.
Import the video into ImageJ [30], and crop embryo to 640 × 480 pixels for analysis.
Analyze nuclear movement in each cell individually by designating the center of the nucleus and the forward border of the hyp7 precursor using the Manual Tracking plugin for ImageJ (see Note 7).
Track the center of a single nucleus at each time point of the movie and then repeat with the forward border of the same hyp7 precursor (see Note 8).
Import the x/y coordinates for both the center of the nucleus and the forward border of the cell into Microsoft Excel to calculate the distance and time of migration (see Note 9).
Migration is completed when the nucleus reaches the opposite side of the cell (or when the nucleus/opposite border cannot be seen anymore due to the embryo rolling).
Make measurement for distance traveled from center of nucleus to the hyp7 boundary that the nucleus migrates toward (see Note 10).
Repeat steps 12–15 to analyze additional hyp7 nuclear migrations in the embryo. Only analyze nuclei 11–16 as designated in [14].
3.3. Counting Missing P-Cell Progeny Using GABA Neuron or P-Cell Nuclear Markers
Culture C. elegans on NGM agar plates spotted with OP50 E. coli at the required temperature (see Note 11 about temperature and Note 12 about the strain with the GFP marker for the assay).
Using a dissecting microscope with an RFP filter (see Note 12) and a worm pick (see Note 2), transfer an equal number of young adult animals with and without the red rescuing construct onto a fresh NGM plate.
Prepare microscope slides with 2% agarose pad (see Note 1).
Pipette ~5 μL of M9 with 1 mM tetramisole or other paralytic on the center of the agarose pad.
With a worm pick, transfer all of the young adult animals from step 2 into the drop of paralytic and gently swirl pick tip without damaging the agarose pad.
Cover the agarose pad with a 25 × 25 × 1 mm coverslip.
Place slide on the stage of an epifluorescence compound microscope equipped with GFP and RFP fluorescence filters.
Use DIC optics and a 10× objective on a compound microscope to locate a young adult animal.
Switch to a higher objective (63× or 100×) to zoom in on the selected young adult (see Note 3). Once an animal is in focus, make sure the anus and pharynx are in the same focal plane. If not, the specimen should not be counted. If so, switch to epi-fluorescence in the GFP channel.
Score the number of GABA neurons. Count green fluorescent cell bodies along the ventral side of the worm. Take care not to count autofluorescent gut granules, which are less bright and a slightly more yellow shade of green and in a different plane of focus. Record the number of GABA neuron cell bodies identified in the ventral cord of the animal (see Note 13).
Switch to the RFP fluorescence, and check for the presence of the ycEx60 rescuing array as marked by bright red neurons in the head of the animal. Record the presence or absence of the ycEx60 array with the respective GABA neuron cell body count.
Alternatively, score P-cell nuclear migration in late L1 larvae using a P-cell-specific nuclear red fluorescent marker (see Note 14).
3.4. Scoring Nuclear Migration in the Embryonic Intestinal Primordium
Culture C. elegans on NGM agar plates spotted with OP50 E. coli at the required temperature (see Note 11).
Wash hatched animals off of NGM plates by gently adding and removing successive rounds of H2O being careful not to detach embryos from the agar surface.
Aspirate embryos by briskly pipetting H2O until the force of the water releases the embryos from the agar surface. Pipette the dislodged embryos into a microfuge tube.
Pellet the embryos by centrifuging for 10 s at maximum speed in a benchtop microfuge.
Remove the bacteria containing supernatant, and resuspend the embryos in 1 mL H2O.
Repeat steps 4 and 5 until the supernatant is clear and free of bacterial contamination.
Remove the supernatant, leaving approximately 100 μL above the embryo pellet.
Resuspend the embryos in the remaining supernatant, and pipette onto the center well of a poly-l-lysine-coated slide (see Note 15).
After 1–2 min, remove excess H2O by dabbing the corner of the embryo-containing well with a Kimwipe. Move briskly to step 10 to avoid embryos drying out.
To stain the intestinal cell cortex with (1) antibodies, such as anti-BGS-1, permeabilize, and fix with −20 °C MeOH or (2) fluorescent phalloidin, permeabilize and fix with 3–4% paraformaldehyde as described [20].
After blocking and staining, rinse one time with PBS-DAPI and one time with PBS, 10 min each.
Remove excess PBS from the slide as described in step 9. Then place 5 μL of mounting media and a coverslip on the embryos. Seal the slide with nail polish. Place slides at 4 °C or view immediately.
Use wide-field or confocal microscopy to capture two-channel fluorescence Z stacks through E16 bean stage embryos of the correct orientation (see Note 16).
Open the stack with ZEN or other image analysis software. Identify a focal plane where the nucleus, apical, and basal surfaces of a single intestinal cell are in focus (Fig. 3A, B).
For each nucleus in the intestinal primordium, measure the distance from the center of the nucleus to the apical membrane.
3.5. Quantifying Nuclear Anchorage Defects in the Adult hyp7 Syncytium
Create a line of C. elegans with the genetic mutation of interest in a background expressing a GFP nuclear marker in the hypodermis (see Note 17).
Culture C. elegans to young adults on NGM agar plates spotted with OP50 E. coli.
Prepare microscope slides with 2% agarose pad (see Note 1).
Pipette ~5 μL of M9 with 1 mM tetramisole or other paralytic on the center of the agarose pad.
Using a dissecting microscope and worm pick (see Note 2), transfer a large number (~20) of young adults to the M9 buffer.
Cover the agarose pad with a 25 × 25 × 1 mm coverslip.
Place slide on the stage of an epifluorescence compound microscope equipped with GFP fluorescence filters.
Use DIC optics and a 10× objective on a compound microscope to locate an adult animal.
Switch to a higher objective (63× or 100×) to zoom in on the selected adult (see Note 3). Once an animal is in focus, make sure the anus and pharynx are in close to the same focal plane. If not, the specimen should not be counted. If so, switch to epifluorescence in the GFP channel.
Assay nuclear anchorage by counting the number of GFP-positive hypodermal nuclei that are clustered (see Note 18).
Fig. 5.
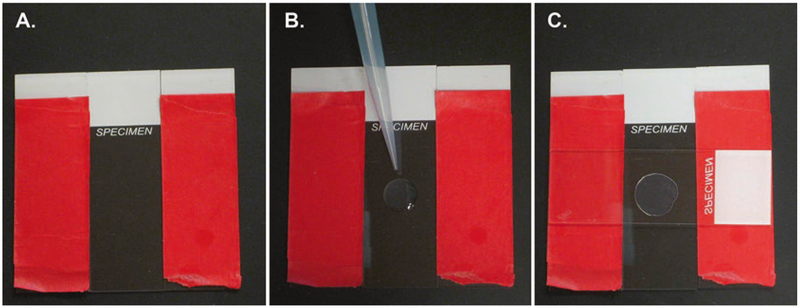
Making agarose pads. (A) The base slide is placed between two other slides with two layers of tape (red) to act as spacers for the pad. (B) A drop of about 100 μL of molten 2% agarose is placed on the slide. (C) Immediately, another slide is placed on top of the molten agarose to create a pad of uniform thickness. See Note 1 for details
Acknowledgments
We thank past and present members of the Starr and Hermann labs for the continual development of these assays over the past 15 years. We thank Courtney Bone for the image in Fig. 2C and Venecia Valdez, Hongyan Hao, and Linda Ma for commenting on the manuscript. Studies in the Starr Lab are supported by the National Institutes of Health grant R01 GM073874. Studies in the Hermann lab are supported by the National Science Foundation grant MCB1613804 and the National Institutes of Health grant R15 GM120639.
4 Notes
Preparing 2% agarose pads. Layer two pieces of lab tape along the length of a 25 × 75 × 1 mm slide. Repeat this on an additional slide. Flank each long side of a 25 × 75 × 1 mm slide with a taped slide (Fig. 5A). The layered tape on the flanking slides ensures proper thickness of the agarose pad. Melt 2% agarose in water in the microwave. Drop a spot of molten aga-rose (~100 μL) in the center of the middle slide (Fig. 5B). Immediately drop another slide, face down, on the molten drop perpendicular to the middle slide. Gently press the top slide where it is overlapping the tape, and apply gentle pressure until the agarose solidifies (Fig. 5C). Finally, gently separate the top slide to expose the agarose pad to be used for microscopy. Try to limit the number of bubbles in the agarose pad, as this will disrupt the DIC visualization of the animals. Move quickly to prevent desiccation of your pad and sample.
Worm picks. We typically use 30 gauge platinum wire to pick up and transfer worms between plates and to an agarose pad for imaging. About an inch of wire is attached by melting to the end of a glass pipette. The worm-picking end of the wire is flattened with a blunt instrument like the back of a scissor blade. Alternatively (recommended) we buy premade worm picks from www.wormstuff.com that are made of a stiffer alloy of 90% platinum/10% iridium and nicely flattened and feature an ergonomic design that is more comfortable than a glass pipette. Scrape a small amount of bacteria from a plate onto the tip of the worm pick to move worms/embryos to a new plate or agarose pad. Try to limit the amount of bacteria transferred to an agarose pad because it will hinder imaging.
We use a Leica DM6000 compound microscope with DIC optics and a 63× Plan Apo 1.40 NA objective for imaging, but any high-quality setup with a 63× or 100× objective and DIC optics should suffice.
Identifying dorsal vs. ventral in an L1 larva. Worms of all stages usually lay or crawl on either their left or right side. The easiest way to identify the ventral side is to spot the anus or the developing germ line (four large cells in an elliptical organ in L1), both of which reside on the ventral side (Fig. 1C, D). One trick to see nuclei in the dorsal cord is to get nuclei on the ventral side in focus. Hypodermal nuclei abnormally in the dorsal cord are more circular and slightly larger than body wall muscle nuclei, which are right next to the dorsal cord and sometimes mistaken for hypodermal nuclei.
To image nuclear migration in hyp7 precursors, embryos must be properly staged and oriented with the dorsal surface on top. A pre-comma stage embryo is rounded and fills up the eggshell. The intercalation of hyp7 precursors occurs between 250 and 390 min after first cleavage and has a well-established order of intercalation [14]. As a general rule, hyp7 precursors intercalate in a posterior to anterior fashion [12]. The exception to this pattern is a pair of hyp7 precursors called pointer cells, which do not intercalate until the ventral hypodermal cells have migrated to the nascent ventral midline at 370–385 min post-cleavage [12, 13]. At the posterior end, hyp7 precursors appear as wedge-shaped cells with the cellular boundaries creating depressions in the hypodermis. The nuclei are large and can be seen by the exclusion of cytoplasmic granules (for example of pointer cells, see nuclei 9 and 10 in Fig. 1A, B).
We capture images with a Leica DC350 camera and Leica LAS AF software. Any imaging software that is able to take time-lapse images and export the images in the AVI format should suffice. The embryo is continuously moving during filming, and it is necessary to check the focal plane before each image is captured to ensure that the hyp7 nuclei are in focus. Small adjustments to the focus are required before almost every image. To image another embryo, a new slide will need to be prepared due to desiccation of the agarose pad and the lack of embryos at pre-comma stage.
For the Manual Tracking plugin, set the x/y calibration to 0.102 μm (for 640 × 480 image) and then 10 pixels = 1 μm for a Leica DC350 camera. These numbers will need adjustments for different cameras.
When tracking a nucleus, align the marker with the center of the nucleus at each time point. To make measurements of distance traveled, also track the hyp7 cell boundary opposite of where the nucleus being analyzed begins (not the edge of the cell itself) as a reference point. This will be approximately where the tip of the hyp7 cell touches when intercalation is complete.
Intercalation is considered complete in the first frame that the tip of the cell can be seen touching the opposite hyp7 cell border. Define the completion of intercalation as time = 0 for each cell individually; this marks the beginning of migration for that nucleus. The distance between the nucleus and the forward border is calculated using Pythagorean theorem and the x/y coordinates determined at each time point.
Two quantitative traits have proven most useful for analyzing nuclear migration in hyp7 embryonic precursors [15, 31]. First is the distance a nucleus travels in the 10 min after its cell completes intercalation, and second is the time it takes for a nucleus to cross the dorsal midline of the embryo. A wild-type nucleus will migrate an average of 3.3 μm in 10 min and cross the midline at an average of 11.2 min after intercalation. An unc-83 or unc-84 null mutant nucleus will move an average of less than 0.1 μm in 10 min. More than half the mutant nuclei will never cross the midline, and those that do so, cross at an average of 36.4 min after intercalations are complete [15].
Null mutations in unc-83 or unc-84 are temperature sensitive. About 50% of P-cell nuclei fail to migrate at 25 °C, while almost all P-cell nuclei complete their migrations when raised at 15 °C [8, 10]. The temperature-sensitive period is only during P-cell nuclear migration when the animal is in the mid-L1 larval stage. For embryonic intestinal cells, the temperature sensitivity is opposite. Nuclear localization in unc-83 or unc-84 mutant backgrounds is slightly more severe at 15 °C than at 25 °C, and the temperature-sensitive period is in the pre-comma stage embryo [10].
We score P-cell nuclear migration defects blindly. We score a mixture of unc-84(null) and unc-84(null); ycEx60[odr-1::rfp; unc-84 (+)] rescued animals. The rescued animals express a red fluorescent marker in a few chemosensory neurons in the head of the animal off of an extrachromosomal array [19]. It is important to count the GFP GABA neurons before determining if the animal you are counting is a null mutant or a rescued animal in order to reduce experimental bias.
UD87 (unc-84(n369), oxIs[punc-47::gfp]) is an excellent control strain for counting GABA neurons; these transgenic animals are homozygous for the GABA neuron marker that is integrated on the X chromosome at genetic position 2.8 [32, 33]. The oxIs12[punc-47::gfp] marker is expressed in 19 D-type motor GABA neurons in the ventral cord (Fig. 2D–F), a single neuron in the tail (DVB) and 6 neurons in the head (RMEL, RMED, RMER, RMEV, AVL, RIS) of the adult [32]. Make sure not to count larvae, as it takes until early adulthood for all 19 GABA neurons to express the GFP marker. Twelve of these 19 D-type GABA neurons, named VD2 to VD13 [24], are derived from P cells. If there are no P-cell nuclear migration defects, 19 GABA neurons are present in the ventral cord. Animals with P-cell nuclear migration defects have less than 19 GABA neurons in the ventral cord. The single most posterior neuron and the six neurons around the head are excluded when counting because they are not derived from P cells and are also in the interior of the animal, out of the ventral cord [34].
The transgene phlh-3::nls::tdTomato specifically labels P-cell nucleoplasm in L1 animals. This marker is integrated in strains UD381 (ycIs11[phlh-3::nls::tdTomato]) and UD59 (unc-84(n369); ycIs11[phlh-3::nls::tdTomato]) [19]. This marker is used to score P-cell nuclear migration in L1 larvae [19]. Transgenic animals can be synchronized by bleaching as described [35] and hatched into M9 without food. Arrested L1 larvae are then fed standard OP50 to release the arrest for 16, 13, or 10 h at 15 °C, 20 °C, or 25 °C, respectively [19]. This will enrich for L1 larvae around the time of P-cell nuclear migration. Migrating P-cell nuclei flatten to move through the constricted space formed between the muscle and cuticle (Fig. 2). It takes 15–25 min for one nucleus to completely move through this constriction [16]. These migrations can be filmed live using protocols described in [16, 36]. tdTomato-labeled nuclei unable to complete the migration through this constriction remain on the lateral side of the late L1 animal and are classified as failed migration events [16, 19].
Prepare poly-l-lysine slides. Hold a microscope slide so that the epoxy-coated side is facing up, and place a small drop of 0.1% poly-l-lysine to the glass surface in the center well. Use the barrel of a long Pasteur pipette to quickly spread a thin layer of the solution over the entire well. Quickly flame the bottom of the slide until the liquid evaporates. Embryos can be placed on the slide once it has cooled.
To measure the distance between the nucleus and intestinal cell apical surface, the embryos must be properly staged and oriented in a precise dorsal- or ventral-up position. The location of the dorsally positioned hyp7 precursor cells can be used to easily determine if the embryo is properly oriented. At the early E16 stage, intestinal nuclei are centrally positioned and are not polarized along the apical-basal axis [20]. We therefore score nuclear position at the mid/late E16 stage when the nuclei have migrated and are apically polarized. To identify this stage, we observe hyp7 precursor intercalation as described in Note 5. The intestinal primordium is at the E16 (16 E descendants) stage between 262 and 408 min after first cleavage [13, 20]. Therefore, an embryo displaying intercalation of all hyp7 precursors except for the pointer cells indicates that the embryo is at the mid/late E16 stage of intestinal development and properly staged to score the location of intestinal nuclei.
A fluorescent marker to follow hypodermal nuclei was constructed by insertion of a 1 kb fragment upstream of the col-19 gene, which is expressed in the hypodermal cells (including the dorsal and ventral hyp7 and lateral seam cells) of late larvae and adults [27], into pPD96.04 (from Andrew Fire, Addgene plasmid #1502) to create pcol-19::gfp-nls::lacZ (pSL779). N2 animals were injected with pSL779 at 40 ng/μL + 100 ng/μL of odr-1::rfp to make strain UD522 [ycEx249]. This marker can be crossed into other strains using standard protocols [37].
Nuclei were scored as clustered if within 10% nuclear diameter proximity to another nucleus in the same focal plane along the longitudinal axis of the worm, as determined by DIC microscopy. Contacts between nuclei on the perpendicular axis were not counted, as the marker could not distinguish seam cell nuclei in proximity to hyp7 nuclei from clusters of hyp7 nuclei [24]. Only nuclei situated between the pharynx and the anus were counted, as nuclei near the mouth and at the very end of the tail were observed to cluster in wild-type animals. For most strains, resolution of nuclei on the opposite lateral side of the worm is too poor to allow accurate counting, and therefore, clustering should only be counted on the top-facing lateral side. In wild type, an average of 0.8 ± 0.4 (average ± 95% CI) GFP-positive nuclei per animal were clustered, while in an anc-1(e1873) null background, 52.5 ± 3.7 nuclei per animal were clustered. Thus, this assay is much more quantitative than previously published nuclear anchorage assays [8, 38].
References
- 1.Bone CR, Starr DA (2016) Nuclear migration events throughout development. J Cell Sci 129:1951–1961. 10.1242/jcs.179788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gundersen GG, Worman HJ (2013) Nuclear positioning. Cell 152:1376–1389. 10.1016/j.cell.2013.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Starr DA, Fridolfsson HN (2010) Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol 26:421–444. 10.1146/annurev-cellbio-100109-104037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tapley EC, Starr DA (2013) Connecting the nucleus to the cytoskeleton by SUN-KASH bridges across the nuclear envelope. Curr Opin Cell Biol 25:57–62. 10.1016/j.ceb.2012.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luxton GG, Starr DA (2014) KASHing up with the nucleus: novel functional roles of KASH proteins at the cytoplasmic surface of the nucleus. Curr Opin Cell Biol 28:69–75. 10.1016/j.ceb.2014.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horvitz HR, Sulston J (1980) Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics 96:435–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hedgecock EM, Thomson JN (1982) A gene required for nuclear and mitochondrial attachment in the nematode Caenorhabditis elegans. Cell 30:321–330 [DOI] [PubMed] [Google Scholar]
- 8.Malone CJ, Fixsen WD, Horvitz HR et al. (1999) UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development 126:3171–3181 [DOI] [PubMed] [Google Scholar]
- 9.Starr DA, Han M (2002) Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science 298:406–409. 10.1126/science.1075119 [DOI] [PubMed] [Google Scholar]
- 10.Starr DA, Hermann GJ, Malone CJ et al. (2001) unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development 128:5039–5050 [DOI] [PubMed] [Google Scholar]
- 11.McGee MD, Rillo R, Anderson AS et al. (2006) UNC-83 IS a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol Biol Cell 17:1790–1801. 10.1091/mbc.E05-09-0894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams-Masson EM, Heid PJ, Lavin CA et al. (1998) The cellular mechanism of epithelial rearrangement during morphogenesis of the Caenorhabditis elegans dorsal hypodermis. Dev Biol 204:263–276. 10.1006/dbio.1998.9048 [DOI] [PubMed] [Google Scholar]
- 13.Chisholm AD, Hsiao TI (2012) The Caenorhabditis elegansepidermis as a model skin. I: development, patterning, and growth. Wiley Interdiscip Rev Dev Biol 1:861–878. 10.1002/wdev.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sulston JE, Schierenberg E, White JG et al. (1983) The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 100:64–119 [DOI] [PubMed] [Google Scholar]
- 15.Fridolfsson HN, Starr DA (2010) Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J Cell Biol 191:115–128. 10.1083/jcb.201004118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bone CR, Chang Y-T, Cain NE et al. (2016) Nuclei migrate through constricted spaces using microtubule motors and actin networks in C. elegans hypodermal cells. Development 143:4193–4202. 10.1242/dev.141192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah P, Wolf K, Lammerding J (2017) Bursting the Bubble—Nuclear Envelope Rupture as a Path to Genomic Instability? Trends Cell Biol. 10.1016/j.tcb.2017.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ungricht R, Kutay U (2017) Mechanisms and functions of nuclear envelope remodelling. Nat Rev Mol Cell Biol 18:229–245. 10.1038/nrm.2016.153 [DOI] [PubMed] [Google Scholar]
- 19.Chang Y-T, Dranow D, Kuhn J et al. (2013) toca-1 is in a novel pathway that functions in parallel with a SUN-KASH nuclear envelope bridge to move nuclei in Caenorhabditis elegans. Genetics 193:187–200. 10.1534/genetics.112.146589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leung B, Hermann GJ, Priess JR (1999) Organogenesis of the Caenorhabditis elegans intestine. Dev Biol 216:114–134. 10.1006/dbio.1999.9471 [DOI] [PubMed] [Google Scholar]
- 21.Malone CJ, Misner L, Le Bot N et al. (2003) The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell 115:825–836 [DOI] [PubMed] [Google Scholar]
- 22.Meyerzon M, Gao Z, Liu J et al. (2009) Centrosome attachment to the C. elegans male pronucleus is dependent on the surface area of the nuclear envelope. Dev Biol 327:433–446. 10.1016/j.ydbio.2008.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou K, Rolls MM, Hall DH et al. (2009) A ZYG-12-dynein interaction at the nuclear envelope defines cytoskeletal architecture in the C. elegans gonad. J Cell Biol 186:229–241. 10.1083/jcb.200902101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall DH, Altun ZF (2008) C. Elegans Atlas, 1st edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 25.Plenefisch J, Xiao H, Mei B et al. (2000) Secretion of a novel class of iFABPs in nematodes: coordinate use of the Ascaris/Caenorhabditis model systems. Mol Biochem Parasitol 105:223–236. 10.1016/S0166-6851(99)00179-6 [DOI] [PubMed] [Google Scholar]
- 26.Doonan R, Hatzold J, Raut S et al. (2008) HLH-3 is a C. elegans Achaete/Scute protein required for differentiation of the hermaphrodite-specific motor neurons. Mech Dev 125:883–893. 10.1016/j.mod.2008.06.002 [DOI] [PubMed] [Google Scholar]
- 27.Liu Z, Kirch S, Ambros V (1995) The Caenorhabditis elegans heterochronic gene pathway controls stage-specific transcription of collagen genes. Development 121:2471–2478. 10.1126/science.6494891 [DOI] [PubMed] [Google Scholar]
- 28.Stiernagle T (2006) Maintenance of C. elegans (February 11, 2006), WormBook, ed. The C. elegans Research Community, WormBook; 10.1895/wormbook.1.101.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moorthy S, Chen L, Bennett V (2000) Caenorhabditis elegans β-G spectrin is dispensable for establishment of epithelial polarity, but essential for muscular and neuronal function. J Cell Biol 149:915–930. 10.1083/jcb.149.4.915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schindelin J, Rueden CT, Hiner MC et al. (2015) The ImageJ ecosystem: an open platform for biomedical image analysis. Mol Reprod Dev 82:518–529. 10.1002/mrd.22489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bone CR, Tapley EC, Gorjanacz M et al. (2014) The Caenorhabditis elegans SUN protein UNC-84 interacts with lamin to transfer forces from the cytoplasm to the nucleoskeleton during nuclear migration. Mol Biol Cell 25:2853–2865. 10.1091/mbc.E14-05-0971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McIntire SL, Reimer RJ, Schuske K et al. (1997) Identification and characterization of the vesicular GABA transporter. Nature 389:870–876. 10.1038/39908 [DOI] [PubMed] [Google Scholar]
- 33.Gysi S, Rhiner C, Flibotte S et al. (2013) A network of HSPG core proteins and HS modifying enzymes regulates netrin-dependent guidance of D-type motor neurons in Caenorhabditis elegans. PLoS One 8:e74908 10.1371/journal.pone.0074908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuske K, Beg AA, Jorgensen EM (2004) The GABA nervous system in C. elegans. Trends Neurosci 27:407–414 [DOI] [PubMed] [Google Scholar]
- 35.Porta-de-la-Riva M, Fontrodona L, Villanueva A et al. (2012) Basic Caenorhabditis elegans methods: synchronization and observation. J Vis Exp. 10.3791/4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chai Y, Li W, Feng G, Yang Y et al. (2012) Live imaging of cellular dynamics during Caenorhabditis elegans postembryonic development. Nat Protoc 7:2090–2102. 10.1038/nprot.2012.128 [DOI] [PubMed] [Google Scholar]
- 37.Fay DS (2013) Classical genetic methods. WormBook 30:1–58. 10.1895/wormbook.1.165.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D’Alessandro M, Hnia K, Gache V et al. (2015) Amphiphysin 2 orchestrates nucleus positioning and shape by linking the nuclear envelope to the actin and microtubule cytoskeleton. Dev Cell 35:186–198. 10.1016/j.devcel.2015.09.018 [DOI] [PubMed] [Google Scholar]