Summary
DNA replication errors generate complex chromosomal rearrangements and thereby contribute to tumorigenesis and other human diseases. One mechanism that triggers these errors is mitotic entry before the completion of DNA replication. To address how mitosis might impact DNA replication, we used Xenopus egg extracts. When mitotic CDK (Cyclin B1-CDK1) is used to drive interphase egg extracts into a mitotic state, the replicative CMG (CDC45/MCM2–7/GINS) helicase undergoes ubiquitylation on its MCM7 subunit, dependent on the E3 ubiquitin ligase TRAIP. Whether replisomes have stalled or undergone termination, CMG ubiquitylation is followed by its extraction from chromatin by the CDC48/p97 ATPase. TRAIP-dependent CMG unloading during mitosis is also seen in C. elegans early embryos. At stalled forks, CMG removal results in fork breakage and end joining events involving deletions and templated insertions. Our results identify a mitotic pathway of global replisome disassembly that can trigger replication fork collapse and DNA rearrangements.
eTOC
Mitotic entry before completion of DNA replication causes genome instability via an unknown mechanism. Using Xenopus egg extracts, Deng et al. find that mitotic cyclin-dependent kinase triggers replication fork breakage and DNA rearrangements. The mechanism requires TRAIP-dependent ubiquitylation of the replicative helicase followed by p97 ATPase-dependent helicase removal from chromatin.
INTRODUCTION
Genome evolution occurs through the gradual accrual of genetic changes or in a saltatory manner, with bursts of chromosomal alterations originating from single catastrophic events (Holland and Cleveland, 2012; Leibowitz et al., 2015; Liu et al., 2011; Stephens et al., 2011). Many chromosomal alterations can be traced to DNA breaks that arise during DNA replication (Hills and Diffley, 2014; Mankouri et al., 2013; Techer et al., 2017). However, when and how replication fork breakage is triggered remains incompletely understood (Toledo et al., 2017).
In normal cells, multiple cell cycle regulatory controls and error correction mechanisms prevent DNA replication errors (Hills and Diffley, 2014). Cells prepare for DNA replication in the G1 phase of the cell cycle, when pairs of MCM2–7 ATPases are recruited to each origin (“licensing”). In S phase, cyclin-dependent kinase (CDK) promotes the association of CDC45 and GINS with MCM2–7, leading to formation of the replicative CMG helicase complex (CDC45-MCM2–7-GINS) (“initiation”). CMG unwinding of the origin nucleates the assembly of two DNA replication forks that travel away from the origin, copying DNA as they go (“elongation”). When converging forks from adjacent origins meet, the replisome is disassembled (“termination”). Replisome disassembly in metazoans requires the E3 ubiquitin ligase, CRL2Lrr1, which ubiquitylates the MCM7 subunit of CMG, leading to CMG’s extraction from chromatin by the p97 ATPase (Dewar et al., 2017; Sonneville et al., 2017). In the absence of CRL2Lrr1, CMGs persist on chromatin until mitosis, but are then removed by a secondary, p97-dependent pathway that is controlled by an unknown E3 ubiquitin ligase (Sonneville et al., 2017). Re-replication is inhibited because de novo licensing of origins is suppressed in the S and G2 phases of the cell cycle. Thus, faithful DNA replication requires the seamless integration of replication licensing, initiation, elongation, and termination. Errors in the process are detected by the DNA damage response, which activates repair mechanisms and prevents entry into mitosis in the setting of incomplete or abnormal replication.
DNA replication forks become stressed in a variety of circumstances, including the activation of oncogenes, collision with DNA lesions and other obstacles, and nucleotide starvation (Cortez, 2015; Hills and Diffley, 2014; Saldivar et al., 2017). Replication stress can cause replication fork “collapse”, an irreversible state from which replication does not restart (Cortez, 2015; Hills and Diffley, 2014; Pasero and Vindigni, 2017; Saldivar et al., 2017; Toledo et al., 2017). Numerous experiments suggested that fork collapse involves replisome disassembly (Cortez, 2015). However, these studies did not establish a causal relationship between replisome disassembly and collapse, and other studies have concluded that fork collapse may not involve replisome disassembly (De Piccoli et al., 2012; Dungrawala et al., 2015). Fork collapse is frequently associated with MUS81-dependent breakage of DNA strands at the fork (Pasero and Vindigni, 2017), but whether collapse and breakage are mechanistically distinct phenomena remains unclear.
Although replication fork breakage is generally viewed as a source of gross chromosomal rearrangements, breakage may sometimes preserve genome integrity (Bhowmick and Hickson, 2017). A well-studied example involves common fragile sites (CFS), which are among the most frequently rearranged genomic loci in cancer genomes (Glover et al., 2017). Common fragile site “expression”, the appearance of cytologically visible breaks and gaps, is promoted by low doses of aphidicolin, which delays duplication of these already late-replicating loci. Unreplicated DNA at CFS forms ultrafine DNA bridges between anaphase chromosomes (Baumann et al., 2007; Chan et al., 2007), and severance of these bridges by MUS81 is thought to allow chromosome segregation. However, random breakage of the fork would yield deleterious outcomes such as the generation of acentric or iso-chromosomes. So far, no mechanism has emerged that explains if and how such outcomes can be avoided.
Although breakage of a few stressed forks may be be beneficial, concurrent breakage of many forks generates catastrophic chromosomal rearrangements. Several lines of evidence implicate mitotic entry with unreplicated DNA as one potential cause of extensive fork breakage. Cell fusion experiments (Johnson and Rao, 1970) and experiments on cells with micronuclei (Kato and Sandberg, 1968) showed that S phase chromosomes undergo “pulverization” upon exposure to mitotic cytoplasm. Although there was early disagreement about whether chromosome pulverization reflects discontinuous condensation or DNA breakage (Rao et al., 1982), current work indicates that chromosome pulverization usually reflects DNA fragmentation. First, premature mitotic entry triggered by inhibition of the WEE1 kinase causes extensive fork breakage that depends on the formation of an active MUS81 complex (Dominguez-Kelly et al., 2011; Duda et al., 2016). Second, chromothripsis, a mutational process involving extensive chromosome fragmentation and rearrangement, may involve entry into mitosis of micronuclei undergoing DNA replication (Crasta et al., 2012; Leibowitz et al., 2015). Extensive fork breakage during mitosis is especially problematic as both homologous recombination (HR) and classical non-homologous end joining (NHEJ) are suppressed at this stage of the cell cycle (Hustedt and Durocher, 2016). In summary, it has become apparent that genome instability in a variety of contexts is linked to mitotic replication fork breakage. However, why forks are so fragile in mitosis is unclear.
Here, we use Xenopus egg extracts to explore the relationship between DNA replication and mitosis. We find that in extracts supplemented with the mitotic kinase Cyclin B1-CDK1, the RING E3 ubiquitin ligase TRAIP is required for the ubiquitylation of CMG helicase at stalled and terminated forks. Ubiquitylated CMG is extracted from chromatin by the CDC48/p97 ATPase. At stalled forks, CMG unloading leads to fork breakage and end joining events that appear to depend on DNA polymerase θ (Polθ). Unlike Cyclin B1-CDK1 treatment, ATR inhibition does not lead to breakage of stalled forks. Together, our results identify TRAIP-dependent replisome disassembly as a crucial step in mitotic replication fork collapse and breakage. We propose that the effect of TRAIP-induced fork breakage on genome stability depends on how many forks are present when cells enter mitosis.
RESULTS
Mitotic CDK triggers aberrant processing of stressed DNA replication forks
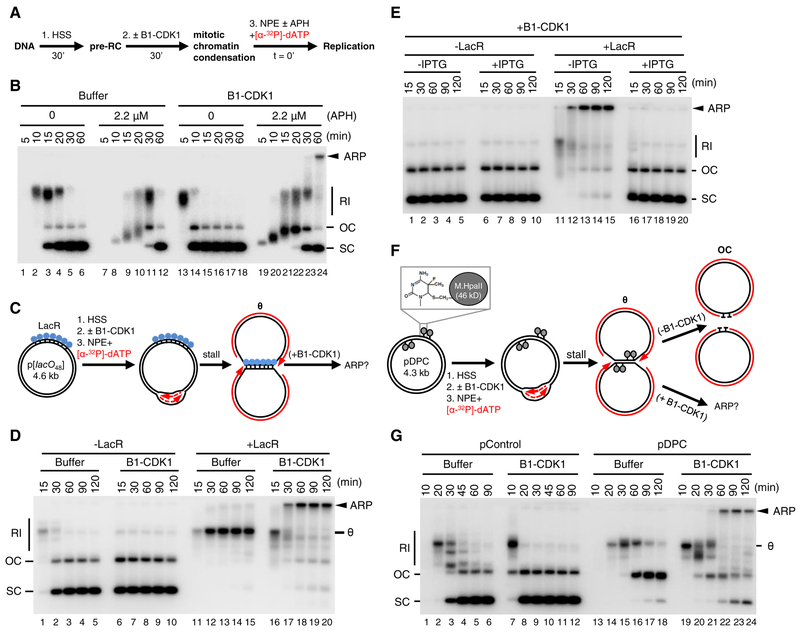
To examine the effect of mitotic CDK on DNA replication and fork stability, we used Xenopus egg extracts, which can recapitulate S phase or mitosis. For S phase, plasmid DNA was first incubated in a high-speed supernatant (HSS) of Xenopus egg extract. HSS promotes the assembly onto DNA of pre-replication complexes (pre-RCs) containing double hexamers of the MCM2–7 ATPase (Figure 1A). The subsequent addition of a nucleoplasmic extract (NPE) leads to the association of CDC45 and GINS with each MCM2–7 hexamer to form two active CMG DNA helicases, which unwind DNA, promoting a single, complete round of DNA replication, manifested as the appearance of supercoiled (SC) daughter molecules (Figure 1B, lanes 1–6) (Walter et al., 1998). To achieve replication in a mitotic state, we added Cyclin B1-CDK1 (B1-CDK1) after licensing and before NPE addition because this kinase inhibits licensing (Hendrickson et al., 1996; Prokhorova et al., 2003) (Figure 1A). We confirmed that in this sequence, B1-CDK1 induced chromosome condensation (Figures S1A–S1C) and condensin recruitment (Figures S1D–E). As we showed previously (Prokhorova et al., 2003), B1-CDK1 increased the rate of DNA replication (Figure 1B, compare lanes 1–6 and 13–18), due in part to increased origin firing (Figure S1F). However, in the absence of other perturbations, all replication products comprised open circular or supercoiled species (Figure 1B, lanes 13–18), indicating that B1-CDK1-induced chromatin condensation does not cause aberrant DNA replication.
Figure 1. Mitotic CDK triggers aberrant processing of stalled DNA replication forks in Xenopus egg extracts.
(A) Schematic of experimental approach to test effect of B1-CDK1 on DNA replication. APH, DNA polymerase inhibitor aphidicolin.
(B) A 3 kb pBlueScript plasmid was replicated according to (A) and products were separated on a native agarose gel followed by autoradiography. Unless stated otherwise, the ‘0 minute’ time point refers to NPE addition.
(C) Schematic of DNA replication for LacR-bound p[lacO48] plasmid.
(D) p[lacO48] was replicated according to (C) under the indicated conditions.
(E) p[lacO48] was replicated according to (C) in the absence or presence of LacR and IPTG (10 mM, 15 min incubation in NPE before mixing with “licensing” mixture), as indicated.
(F) Schematic of replication for pDPC, containing four 46 kDa M.HpaII DNA methyltransferases at the indicated positions. Products formed in the presence and absence of B1-CDK1 are indicated.
(G) pControl or pDPC was replicated according to (F) using the indicated conditions. From (A) to (G), B1-CDK1 was added to “licensing” mixture at a concentration of 50 ng/μL and its final concentration in the overall reaction is 16.7 ng/μL (see method). RI, replication intermediate; OC, open circle; SC: supercoil; θ, theta structure; ARP, aberrant replication product.
See also Figure S1.
Given that stressed DNA replication forks appear to undergo breakage during mitosis, we added a low concentration of the replicative DNA polymerase inhibitor aphidicolin (APH) to slow fork progression (Figure 1B, lanes 7–12). The combination of B1-CDK1 and APH led to the appearance of a new replication product that migrated at the very top of the gel (Figure 1B, lane 24). This aberrant replication product (ARP) comprised ~6% of total replication for a 3 kb plasmid and up to 30% for a 9 kb plasmid (data not shown), presumably because the larger plasmid hosts more replication forks. ARPs recovered from extract were not resolved by Topoisomerase I or Topoisomerase II, indicating they are not plasmid topoisomers (data not shown). Thus, in the presence of replication stress, mitotic CDK induces aberrant DNA replication.
To examine the effect of B1-CDK1 on replication forks that have stalled at a defined location, we replicated a plasmid containing an array of 48 lacO sites (p[lacO48]) bound by the lac repressor (LacR) (Figure 1C). As expected (Dewar et al., 2015), replication forks accumulated at the outer edges of the LacR array, generating a “theta” (θ) structure (Figures 1C and 1D, lanes 11–15). In the presence of B1-CDK1, the theta molecules disappeared and ARPs accumulated (Figure 1D, lanes 16–20). ARPs were not generated when LacR-mediated fork stalling was prevented with IPTG (Figure 1E), or in the presence of the CDK1 inhibitor (CDK1-i) RO-3306 (Figure S1G). Furthermore, addition of Cyclin E-CDK2 or Cyclin A2 (which preferentially associates with endogenous CDK1; (Strausfeld et al., 1996)), did not induce ARPs, although their addition accelerated DNA replication as expected (Figure S1H). Second, we replicated a plasmid substrate (pDPC), which contains two site-specific DNA-protein crosslinks (DPCs) on each leading strand template (Figure 1F). As expected (Duxin et al., 2014), in the absence of B1-CDK1, replication of pDPC first yielded theta structures when forks transiently paused at the DPC. Plasmids then resolved into open circular (OC) species that persisted due to slow translesion synthesis past the peptide adduct generated by DPC proteolysis (Figure 1F, upper arrow and Figure 1G, lanes 13–18). In the presence of B1-CDK1, we again observed abundant ARPs (Figure 1G, lanes 19–24). In summary, mitotic CDK caused aberrant processing of replication forks stalled by aphidicolin, non-covalent nucleoprotein complexes, and DPCs.
Mitotic processing of stalled replication forks leads to complex chromosomal rearrangements
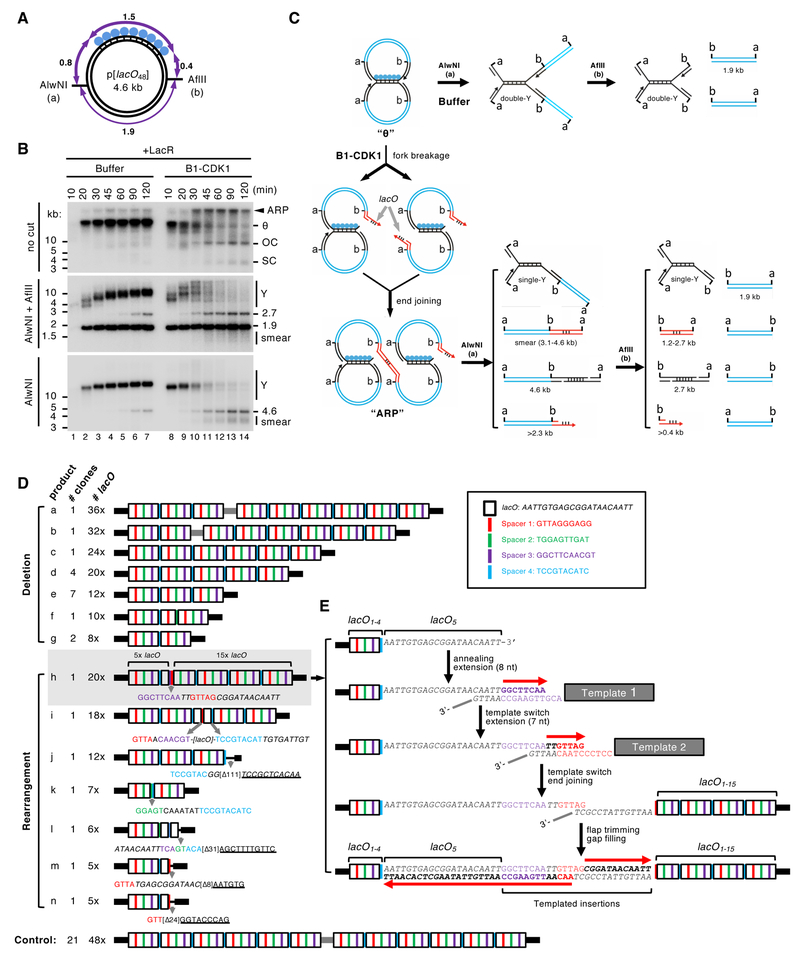
To determine the structure of mitotic ARPs, we replicated the 4.6 kb LacR plasmid in the presence and absence of B1-CDK1 and digested the replication products with AlwNI and AflII, which cuts the plasmid into a 1.9 kb fragment and a 2.7 kb fragment encompassing the lacO repeats (Figure 2A). In the absence of B1-CDK1, fully replicated 1.9 kb fragments quickly accumulated, whereas the rest of the plasmid migrated as a double-Y structure that gradually increased in size due to slow progression of forks through the LacR array (Figure 2B, middle panel, lanes 1–7 and Figure 2C, “Buffer”; (Dewar et al., 2015)). In the presence of B1-CDK1, the 1.9 kb fragment again accumulated quickly and persisted, demonstrating that this lacO-free region was replicated efficiently (Figure 2B, middle panel, lanes 8–14). However, the double-Y structure containing the lacO array rapidly disappeared. Thus, in the presence of B1-CDK1, aberrant DNA processing occurs specifically on molecules containing stalled forks.
Figure 2. Mitotic processing of stalled replication forks leads to complex DNA rearrangements.
(A) Structure of the 4.6 kb p[lacO48] plasmid. Numbers mark the length of the indicated DNA segments in kilo-basepairs (kb).
(B) p[lacO48] was replicated in the presence of Buffer or B1-CDK1. At the indicated time points, replication products were isolated and digested with AlwNI and AflII, or AlwNI, as indicated. Numbers label the size of linear fragments in kb; Y, double-Y or single-Y structure (see panel C).
(C) Model explaining the restriction products observed in (B). Although the model favors fork breakage on the leading strand, the possibility of fork breakage on the lagging strand has not been excluded. A more detailed model is presented in Figure S2A.
(D) The smear of ~3–4 kb mitotic DNA replication products generated after AlwNI digestion in (B) was self-ligated, cloned and sequenced. The controls are replication products of the same plasmid from a mitotic reaction lacking LacR. The lacO repeats, shown as white boxes, are separated by four unique spacers shown in different colors. Inset, DNA sequences of the lacO repeat and four spacers. The detailed structure of the entire lacO array is shown in Figure S2C.
(E) A model for the generation of product h in (D) from multiple template-switching events.
See also Figure S2.
When the replication products were digested only with AlwNI, we observed B1-CDK1-dependent disappearance of the now larger double-Y structure (Figure 2B, bottom panel, lanes 8–14). In addition, we detected a new series of species migrating between ~3 and ~4 kb (Figure 2B, bottom panel; smear). We hypothesized that when replication forks enter the array and slow down or stall, B1-CDK1 promotes their collapse and breakage. The resulting double-strand breaks (DSBs) subsequently undergo joining with DSBs from broken forks on other plasmids, generating ARPs (Figures 2C, “B1-CDK1” and S2A). If replication forks collapse at the outer edges of the array, the size of the end joining product after AlwNI digestion is close to 3.1 kb because most of the 1.5 kb lacO array is lost; collapse further into to the array generates larger products, accounting for the 3–4 kb range of products observed (Figure S2B). To test this hypothesis, the 3–4 kb species were cloned and sequenced using primers immediately flanking the lacO array (Figure S2C). In contrast to control clones (generated from replication in the absence of LacR), all of which contained 48 lacO repeats, the 24 clones from the 3–4 kb smear contained fewer than 48 lacO repeats (Figure 2D, products a-n). This result confirms that replication forks collapsed within the lacO array and then underwent end joining with loss of lacO repeats. Seventeen of these products (a-g) involved only deletions of the lacO repeats. This suggests that the deletions might occur via single strand annealing (SSA) (Bhargava et al., 2016), which generates deletions between homologous sequences. The remaining 7 clones contained complex rearrangements, with microhomogy at the junction or insertions that likely arose from replication template-switching events (Figure 2D; product h-n). For example, product h appears to have arisen from fork collapse at the 5th repeat, followed by two successive microhomology-mediated strand invasion and copying events, followed by joining to a second fork that broke at the 15th repeat (Figure 2E). The sequencing data strongly suggests that stressed replication forks collapse in the presence of B1-CDK1, generating DSBs that subsequently undergo end joining, sometimes after repeated template-switching.
Immunodepletion of DNA Polθ reduces mitotic ARPs
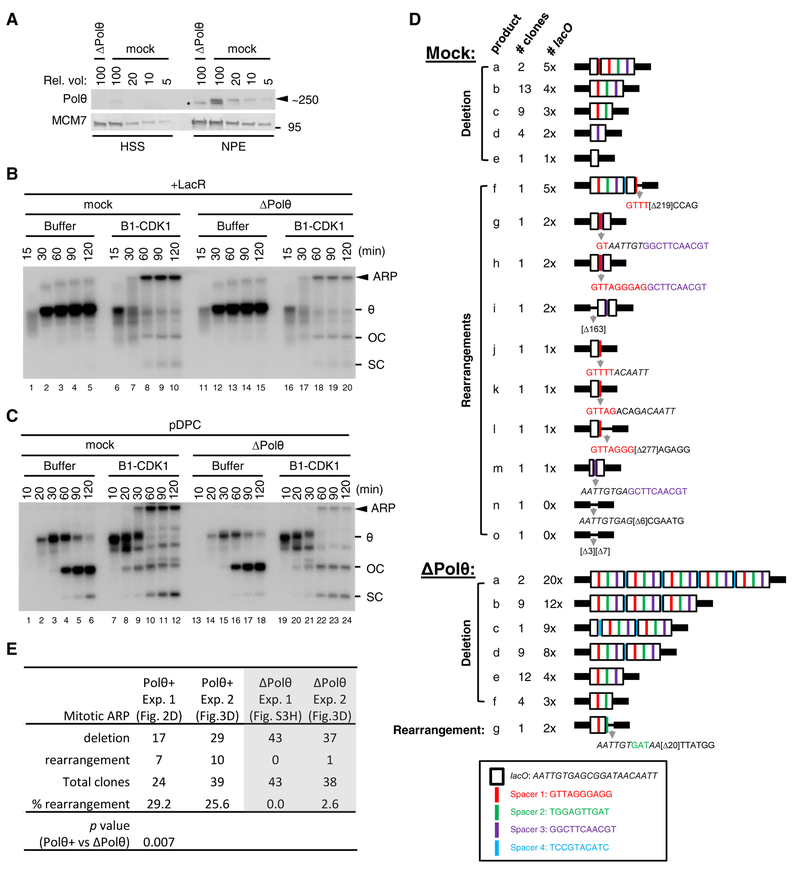
We next addressed the mechanism of end joining after B1-CDK1-induced fork collapse. As expected (Peterson et al., 2011), RAD51, which is essential for homologous recombination (HR), did not bind chromatin in the presence of B1-CDK1 (Figure S3A), and inhibition of RAD51 function in extracts had no effect on B1-CDK1-induced ARP formation (Figures S3B–D). Further, classical non-homologous end joining (NHEJ) was not required for ARP formation (Figure S3E). The structures of the mitotic ARPs (Figures 2C–E) suggested that MMEJ (microhomology-mediated end joining, also called alternative end joining) is responsible for some mitotic DSB repair. Indeed, immunodepletion of DNA polymerase Polθ (Figure 3A), a major mediator of MMEJ known to undergo template-switching (Wyatt et al., 2016), decreased ARPs during replication of LacR plasmid (Figures 3B and S3F) and pDPC (Figures 3C and S3G). Additionally, Polθ depletion resulted in overall lower amounts of replication products (Figures S3F–G), probably due to resection of unligated nascent strands. Finally, Polθ depletion virtually eliminated ARPs containing complex rearrangements (Figures 3D–3E). Thus, in mitotic extracts where HR and NHEJ are inactive, MMEJ appears to play an important role in joining DNA ends after fork breakage.
Figure 3. Depletion of DNA polymerase θ disrupts the generation of aberrant replication product in the presence of mitotic CDK.
(A) Mock-depleted and Polθ-depleted Xenopus egg extracts were blotted for Polθ and MCM7, alongside a serial dilution of mock-depleted extracts. Asterisk, background band.
(B) LacR-bound p[lacO48] was replicated in mock-depleted or Polθ-depleted extracts with or without B1-CDK1 treatment. Total DNA replication and ARP were quantified in Figure S3F.
(C) pDPC was replicated in mock-depleted or Polθ-depleted egg extracts with or without B1-CDK1 treatment. Total DNA replication and ARP were quantified in Figure S3G.
(D) Structure of clones derived from mitotic ARPs in mock- or Polθ-depleted extracts. LacR-bound p[lacO48] was replicated in mock- or Polθ-depleted extracts with B1-CDK1 treatment. The smear of ~3–4 kb mitotic DNA replication products generated after AlwNI digestion was self-ligated, cloned and sequenced. Elements in the box at the bottom show the sequences of lacO and spacers.
(E) Comparison of mitotic ARP-derived clones in the presence or absence of Polθ. Mock depletion (Figure 2D) and Polθ depletion (Figure S3H) in experiment 1 (Exp. 1) were performed independently whereas they were performed side by side (Figure 3D) in experiment 2 (Exp. 2). The shown p-value was from unpaired two-tailed Student’s t-test. In (B) and (C), OC, open circle; SC, supercoil; θ, theta structure; ARP, aberrant replication product.
See also Figure S3.
Condensin is dispensible for mitotic CDK-induced fork instability
Mitotic chromatin condensation has long been proposed to cause DNA damage in under-replicated regions (El Achkar et al., 2005; Lukas et al., 2011). We therefore investigated the role of chromatin condensation on fork collapse in mitotic egg extracts. Although immunodepletion of the condensin subunit SMC2 inhibited B1-CDK1-induced chromosome condensation (Figures S4A–B), it did not affect the formation of ARPs (Figures S4C–D). Therefore, chromatin condensation, per se, is neither necessary nor sufficient for fork instability in mitotic egg extracts.
CMG unloading at stalled forks initiates mitotic fork breakage
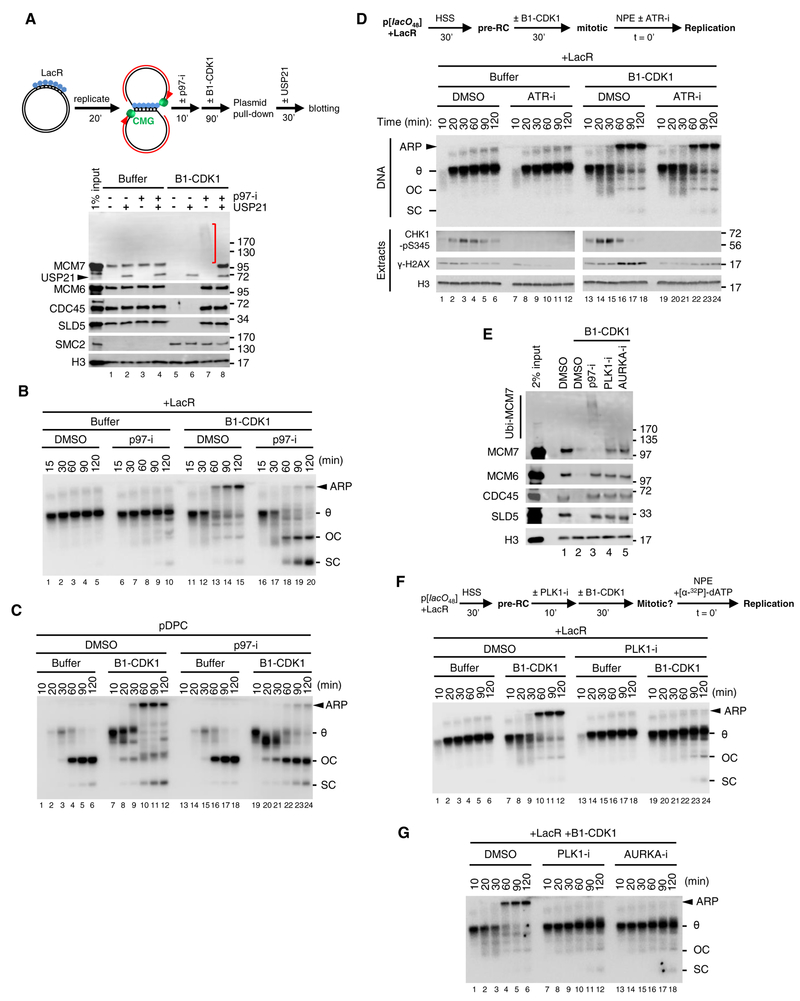
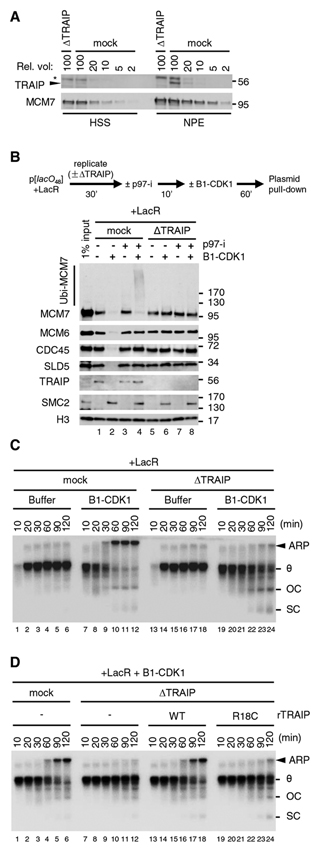
When two replication forks converge on a DNA inter-strand crosslink (ICL) in interphase egg extracts, CMG ubiquitylation and unloading from chromatin by the CDC48/p97 ATPase leads to DNA incisions that generate a double-stranded DNA break (Amunugama et al., 2018; Fullbright et al., 2016; Klein Douwel et al., 2014; Semlow et al., 2016). We therefore asked whether B1-CDK1-induced fork breakage at single stalled forks is caused by CMG unloading. As shown previously (Dewar et al., 2015), CMGs that stalled at a LacR array did not dissociate from chromatin in interphase extracts (Figure 4A, lane 1). In contrast, in the presence of B1-CDK1, CMGs were unloaded (Figure 4A, lane 5). Addition of the p97 inhibitor NMS-873 (p97-i) prevented B1-CDK1-triggered CMG unloading and revealed a ladder of MCM7 species (Figure 4A, lane 7, red bracket) that was collapsed by USP21, a non-specific deubiquitylating enzyme (Figure 4A, lane 8). Therefore, B1-CDK1 induces MCM7 ubiquitylation and p97-dependent CMG unloading at single stalled forks, demonstrating that in mitotic conditions, fork convergence is not required for CMG unloading. Strikingly, p97-i suppressed the formation of ARPs on the LacR plasmid (Figure 4B), suggesting that fork breakage requires CMG unloading. Consistent with this interpretation, CMG unloading normally preceded replication fork breakage (Figure S4E). Interestingly, in the presence of p97-i, theta structures were converted to mature replication products (OC and SC) more efficiently in the presence of B1-CDK1 than in its absence (Figure 4B, compare lanes 16–20 and 6–10), suggesting that B1-CDK1 may promote fork progression through the array when CMG unloading is prevented. Treatment with p97-i also reduced the mitotic CDK-induced H2AX phosphorylation (γ-H2AX) signal, consistent with inhibition of DSB formation (Figure S4F, compare lanes 13–18 and 19–24). As seen for LacR plasmid, p97-i also prevented ARP formation on pDPC (Figures 4C and S4G). Our data demonstrate that breakage of stalled forks in the presence of mitotic CDK requires p97 activity.
Figure 4. Mitotic CDK-induced fork collapse requires p97-dependent CMG unloading.
(A) LacR-bound p[lacO48] plasmid was replicated and treated as schemed. Chromatin-bound proteins were recovered and blotted with the indicated antibodies. Red bracket, ubiquitylated MCM7. Histone H3 served as loading control. Note that the MCM7 antibody cross-reacts with USP21.
(B) LacR-bound p[lacO48] was replicated in the presence or absence of p97-i and B1-CDK1, as indicated.
(C) pDPC was replicated in the presence or absence of p97-i and B1-CDK1, as indicated. ARP, OC+SC and overall DNA replication were quantified in Figure S4G.
(D) Effect of ATR inhibition on stalled replication forks. LacR-bound p[lacO48] plasmid was replicated as schemed. Final concentration of ATR inhibitor (ATR-i; ETP-46464) in the reactions was 200 μM. Extracts with [α−32P]-dATP were sampled to track DNA replication while extracts without [α−32P]-dATP were sampled in parallel to track CHK1-S345 phosphorylation (CHK1-pS345), γ-H2AX. Histon H3 was included as loading control.
(E) LacR-bound p[lacO48] plasmid was replicated and treated as in (A). The final concentrations of PLK1 inhibitor (PLK1-i, BI-2536) and Aurora kinase A inhibitor (AURKA-i, MLN-8237) were 50 μM and 10 μM, respectively. DMSO and p97-i treatments were included as controls. Chromatin-bound proteins were recovered and blotted with the indicated antibodies.
(F) LacR-bound p[lacO48] was replicated as schemed. PLK1-i was added 10 minutes before B1-CDK1 treatment, with a final concentration of 50 μM in the overall reaction.
(G) LacR-bound p[lacO48] was replicated in the presence of PLK1-i or AURKA-i, as schemed in (F). The final concentrations of PLK1-i and AURKA-i were 50 μM and 10 μM, respectively.
In (B), (C), (D), (F) and (G), OC, open circle; SC: supercoil; θ, theta structure; ARP, aberrant replication product.
See also Figure S4.
B1-CDK1-induced fork breakage requires PLK1 and AURKA, but not inhibition of ATR signaling
In mammalian cells, inhibition of ATR signalling leads to fork breakage and chromosomal fragmentation that depends on the protein kinases CDK1, PLK1, and AURKA (Aurora kinase A) (Brown and Baltimore, 2000; Ragland et al., 2013). We therefore examined whether these kinases affect fork breakage in egg extracts. As shown in Figure 4D, a potent ATR inhibitor (ATR-i, ETP-46464) did not induce breakage of forks stalled at a LacR array in interphase egg extract lacking B1-CDK1 (measured by ARP formation), even though ATR-i abolished CHK1-pS345 and γ-H2AX (lanes 7–12). Conversely, the fork breakage observed in the presence of B1-CDK1 occurred even though ATR signaling was active, as seen from CHK1-pS345 (Figure 4D, lanes 13–18), and ATR-i did not further enhance breakage in this setting (Figure 4D, lanes 19–24). Therefore, in interphase egg extract, ATR inhibition is insufficient to cause fork breakage, and in mitotic extract, B1-CDK1 induces fork breakage in the presence of ATR activity. Strikingly, B1-CDK1-induced CMG ubiquitylation, CMG unloading, and fork breakage were all suppressed by selective inhibitors of PLK1 or AURKA (Figures 4E–G). We conclude that in egg extracts that are arrested in a mitotic state, ATR is unable to suppress fork breakage, whereas breakage depends on PLK1 and AURKA, consistent with findings in mammalian cells (Ragland et al., 2013).
B1-CDK1 induces p97-dependent replication fork collapse
Replication fork collapse is defined as a state from which replication cannot restart, and we wanted to determine whether B1-CDK1 induces such a state in egg extracts. As we showed previously (Dewar et al., 2015), replication forks stalled at a LacR array are able to resume synthesis upon addition of IPTG, leading to mature, supercoiled replication products (Figure S4H, lanes 7–12). In the presence of B1-CDK1, IPTG treatment did not generate mature replication products (Figure S4H, lanes 19–24), presumably because forks broke and underwent end-joining. However, when p97i was included with B1-CDK1, mature replication products were fully recovered after IPTG addition (Figure S4H, lanes 31–36). Thus, B1-CDK1 induces collapse of stalled replication forks and inhibition of p97 is sufficient to prevent this collapse.
TRAIP promotes B1-CDK1-induced CMG unloading at stalled forks
We next sought the E3 ubiquitin ligase responsible for B1-CDK1-dependent CMG unloading. While the Cullin inhibitor MLN-4924 (Cul-i) blocked CMG unloading during replication termination in interphase (Figure S5A, compare lanes 1 and 4) (Dewar et al., 2017), it had almost no effect on mitotic CMG unloading from stalled forks (Figure S5A, compare lanes 3 and 6), indicating the latter process does not involve CRL2Lrr1 or any other Cullin-ring ligase.
The E3 ubiquitin ligase TRAIP counteracts replication stress to maintain genome integrity (Feng et al., 2016; Harley et al., 2016; Hoffmann et al., 2016; Soo Lee et al., 2016), and we recently found that it is bound to replication forks that have stalled at a LacR array (Dewar et al., 2017). Strikingly, immunodepletion of TRAIP from egg extract (Figure 5A) prevented B1-CDK1-induced CMG unloading at stalled forks (Figure 5B, compare lanes 2 and 6), and it eliminated polyubiquitylation of MCM7 in the presence of p97-i (Figure 5B, compare lanes 4 and 8). Furthermore, TRAIP depletion abolished the formation of ARPs during replication of LacR plasmid (Figure 5C, compare lanes 7–12 and 19–24) and pDPC (Figure S5B). Re-addition of recombinant wild TRAIP (rTRAIPWT) purified from bacteria to TRAIP-depleted egg extracts rescued the formation of mitotic ARPs (Figure 5D; and Figures S5C–S5E). We also added back rTRAIPR18C, a point mutant of TRAIP that was identified in a human patient with primordial dwarfism (Harley et al., 2016). Unlike rTRAIPWT, rTRAIPR18C supported only low levels of ARP formation on LacR plasmid (Figure 5D, compare lanes 19–24 and 13–18). rTRAIPΔPIP lacking its C-terminal PCNA interaction motif (PIP box, amino acid 460–469) induced mitotic ARPs as efficiently as rTRAIPWT (Figure S5F), consistent with TRAIP’s PIP box being dispensible for the suppression of genome instability in mammalian cells (Hoffmann et al., 2016). We conclude that in the context of stalled forks, TRAIP is essential for mitotic CDK-induced CMG unloading and fork collapse.
Figure 5. E3 ubiquitin ligase TRAIP promotes mitotic CMG unloading from a stalled replication fork.
(A) Mock-depleted and TRAIP-depleted egg extracts were blotted for TRAIP and MCM7 alongside a serial dilution of mock-depleted extracts. Asterisk, background band.
(B) LacR-bound p[lacO48] plasmid was replicated in mock-depleted or TRAIP-depleted egg extracts and treated as schemed. Chromatin-bound proteins were recovered and blotted with the indicated antibodies.
(C) LacR-bound p[lacO48] was replicated in mock-depleted or TRAIP-depleted extracts with or without B1-CDK1 treatment.
(D) LacR-bound p[lacO48] was replicated in mitotic mock-depleted or TRAIP-depleted egg extracts with or without recombinant wildtype TRAIP (rTRAIPWT) or R18C mutant (rTRAIPR18C), as indicated. rTRAIPWT and rTRAIPR18C were added to NPE at a concentration of 21 ng/μL (~7-fold over endogenous TRAIP, see quantification in Figure S5C). Matched buffer without recombinant protein was added to control reactions. Addition of rTRAIPWT at endogenous level (Figure S5C) into TRAIP-depleted extracts also led to substantial rescue of mitotic ARPs (Figures S5D and S5E).
In (C) and (D), open circle; SC: supercoil; θ, theta structure; ARP, aberrant replication product.
See also Figure S5.
Chromatin recruitment of TRAIP is not regulated by B1-CDK1
To understand how TRAIP is regulated, we monitored its binding to chromatin. As we showed previously (Dewar et al., 2017), in interphase egg extract TRAIP is associated with replisomes that have stalled at a LacR array (Figure 5B, lane 1). Therefore, TRAIP is present at forks before they are exposed to B1-CDK1. Upon addition of B1-CDK1, TRAIP was lost from the chromatin, but not when CMG unloading was inhibited with p97-i (Figure 5B, compare lanes 2 and 4). Interestingly, chromatin-bound TRAIP did not increase in the presence of B1-CDK1 and p97-i compared to the level observed before B1-CDK1 addition (Figure 5B, compare lanes 1 and 4). These data suggest that mitotic CDK activates TRAIP in a manner that does not involve its de novo recruitment to the fork.
Fork breakage in mitotic extracts is distinct from programmed incisions during ICL repair
The breakage of single stalled forks in mitotic egg extracts shown here is remeniscent of breakage at forks that have converged on cisplatin ICLs in interphase egg extracts (Figure 5). We therefore asked whether B1-CDK-induced breakage at single forks requires FANCI-FANCD2, XPF-ERCC1, or SLX1-SLX4, which promote DNA incisions during ICL repair (Klein Douwel et al., 2014; Knipscheer et al., 2009). Immunodepletion of FANCI-FANCD2 did not prevent mitotic ARP formation on LacR plasmid (Figures S5G–H), nor did depletion of SLX4, XPF, or MUS81 (data not shown). We speculate that there might be redundancy among SLX1, XPF, and MUS81 for mitotic fork breakage, or that other nucleases are involved. Our results indicate that while ICL incisions and B1-CDK1-dependent replication fork collapse both appear to require TRAIP-dependent CMG unloading, these processes are otherwise mechanistically distinct.
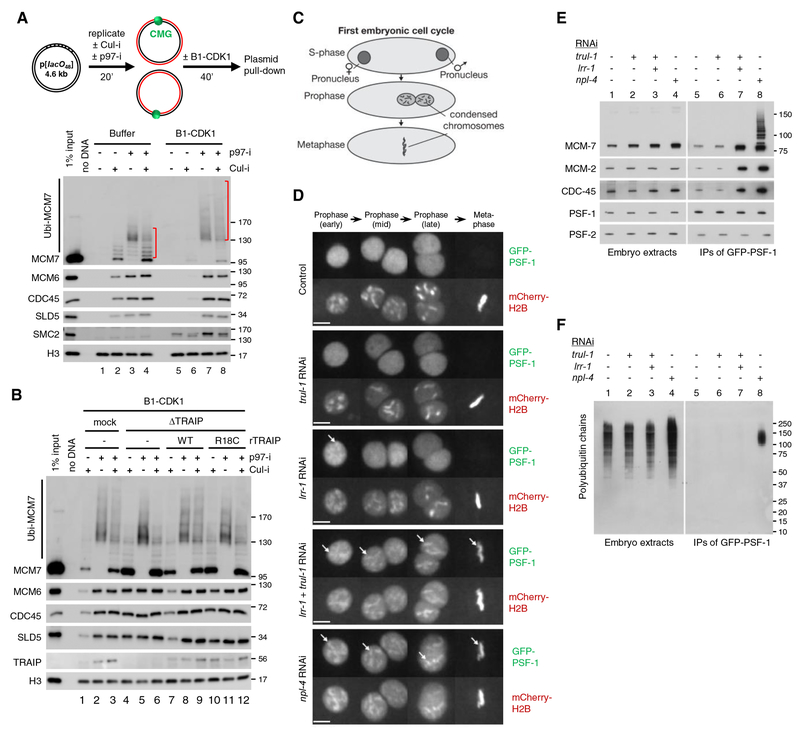
TRAIP promotes CMG unloading from terminated replisomes in mitosis
In C. elegans early embryos lacking CUL2LRR−1, CMGs persist on chromatin until late prophase, when they are unloaded from chromatin by p97 (Sonneville et al., 2017). This observation indicated that an alternative ubiquitylation pathway acts to unload terminated CMGs in mitosis, but the relevant E3 ubiquitin ligase has not been identified. To determine whether TRAIP is involved in this pathway, we first addressed whether Xenopus egg extracts recapitulate mitotic unloading of CMGs that have undergone replication termination. To this end, we replicated a plasmid in interphase egg extracts in the presence of Cul-i. DNA synthesis went to completion (Figure S6A), but CMG unloading was blocked due to inhibition of CRL2Lrr1 (Figure 6A, compare lanes 1 and 2; (Dewar et al., 2017)). Importantly, upon addition of B1-CDK1, CMG was unloaded despite the presence of Cul-i (Figure 6A, lane 6), and this unloading was blocked by p97-i (Figure 6A, lane 8). Therefore, as seen in worms, mitotic frog egg extracts support CRL2Lrr1-independent unloading of terminated CMGs. Interestingly, in the presence of p97-i, MCM7 was ubiquitylated even more extensively than in interphase extracts (Figure 6A, compare lanes 7–8 and 3–4 and Figure S6B, compare lanes 5–6 and 1–2). This hyper-ubiquitylation was unaffected by Cul-i (Figure 6A, lane 8), indicating that it is CRL2Lrr1-independent. Importantly, TRAIP depletion inhibited B1-CDK1-induced CMG unloading from terminated forks (Figure 6B, compare lanes 1 and 4, and Figure S6C, compare lanes 1 and 4) and MCM7 hyper-ubiquitylation in the presence of p97-i (Figure 6B, compare lanes 2 and 5 as well as lanes 3 and 6). These defects were reversed by rTRAIPWT but not rTRAIPR18C (Figures 6B and S6C). Therefore, in the absence of CRL2Lrr1 activity, TRAIP promotes an alternative pathway to unload terminated CMGs in mitotic egg extract.
Figure 6. TRAIP mediates unloading of terminated CMGs in mitosis.
(A) p[lacO48] plasmid, in the absence of LacR, was replicated and treated as schemed. Chromatin-bound proteins were recovered and blotted with the indicated antibodies. Red brackets indicate the levels of MCM7 ubiquitylation.
(B) p[lacO48] plasmid, in the absence of LacR, was replicated in mock-depleted or TRAIP-depleted egg extracts supplemented with or without rTRAIPWT (~4-fold of endogenous TRAIP), or rTRAIPR18C (~9-fold of endogenous TRAIP), followed by indicated treatments. Chromatin-bound proteins were recovered and blotted with the indicated antibodies.
(C) Illustration of the first cell cycle of the C. elegans embryo. Following S-phase, the female and male pronuclei migrate towards each other and chromosomes condense during prophase. Subsequently, the two sets of chromosomes intermingle during metaphase.
(D) Timelapse video microscopy of the first embryonic mitosis, in embryos exposed to the indicated RNAi and expressing GFP-PSF-1 and mCherry-Histone H2B. The female pronucleus is shown during early prophase, before convergence with the male pronucleus (mid and late prophase). The arrows indicate examples of persistence of GFP-PSF-1 on condensed chromatin during mitosis. Scale bar, 5 μm.
(E-F) Worms in which the PSF-1 subunit of the CMG helicase was tagged with GFP were subjected to the indicated RNAi treatment. GFP-PSF-1 was recovered by immunoprecipitation, and the association of the indicated proteins was then monitored by immunoblotting against the indicated proteins (E) or ubiquitin (F).
See also Figure S6.
We next asked whether the C. elegans orthologue of TRAIP, which we called TRUL-1 (Traip Ubiquitin Ligase 1, encoded by the previously uncharacterised C. elegans gene B0432.13), controls mitotic removal of CMG from chromatin in the first embryonic cell cycle (Figure 6C). On its own, RNAi depletion of TRUL-1 had no impact on CMG disassembly. However, simultaneous depletion of TRUL-1 and LRR-1 led to the persistence on mitotic chromatin of the PSF-1 subunit of GINS (Figure 6D, four of five embryos examined) and CDC-45 (Figures S6D, five of five embryos examined), indicating that C. elegans TRAIP is required for the removal of CMG from mitotic chromatin in animals. Moreover, compared to single depletion of TRUL-1 or LRR-1, double depletion led to the accumulation of CMG complexes containing unmodified MCM7 (Figure 6E, lane 7). This contrasts with the persistence of ubiquitylated CMGs observed upon depletion of p97’s cofactor NPL-4 (Figures 6E and 6F, lane 8) (Sonneville et al., 2017). Thus, unloading of terminated CMGs in mitosis is a conserved function of TRAIP in metazoans.
DISCUSSION
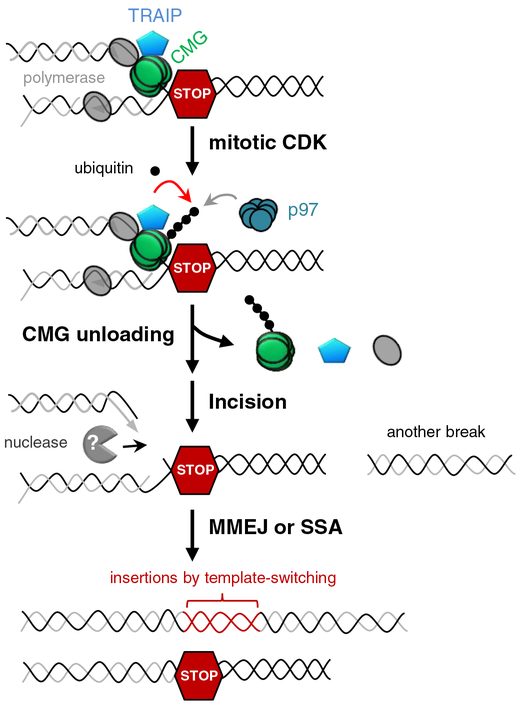
The molecular events underlying mitotic replication fork breakage and how such breakage affects genome stability remain unclear. Here, we show that in mitotic egg extracts, the E3 ubiquitin ligase TRAIP promotes p97-dependent replisome disassembly, followed by replication fork breakage and end joining events involving SSA and MMEJ (Figure 7). As discussed below, we propose that TRAIP-dependent fork breakage can be beneficial or detrimental, depending primarily on the burden of stressed forks at mitotic entry.
Figure 7. Model of CMG unloading, fork breakage and complex DNA rearrangements upon premature mitotic entry.
When a replication fork encounters a replication barrier (indicated as a red hexagonal STOP sign), the replisome containing CMG and TRAIP is stably stalled during interphase. With the increase of mitotic CDK activity, E3 ubiquitin ligase TRAIP is activated (directly or indirectly) to cause CMG ubiquitylation on MCM7 subunit, which in turn triggers CMG unloading from chromatin by CDC48/p97 ATPase. Loss of CMG leads to incision by so far unknown DNA nuclease(s), followed by error-prone double-strand repair by MMEJ and/or SSA, which results in DNA rearrangements such as deletions and insertions from template-switching events.
See also Figure S7.
TRAIP’s regulation of CMG ubiquitylation is critically dependent on cell-cycle status. In the presence of B1-CDK1, TRAIP targets stalled CMGs, which encircle ssDNA, and terminated CMGs, which probably encircle dsDNA (Figure S6Ei and ii; (Dewar et al., 2015)). In contrast, TRAIP’s action in interphase extracts is more selective. In this setting, TRAIP does not target terminated CMGs, a function performed by CRL2Lrr1 in S phase [Figure S6Eiii; (Dewar et al., 2017; Sonneville et al., 2017)], nor does TRAIP appear to target CMG at single moving or stalled forks, which would cause premature fork collapse. Future work will be required to understand how TRAIP’s selectivity is modulated by B1-CDK1.
It has been widely proposed that replisome disassembly causes fork collapse (Cortez, 2015; Toledo et al., 2017), but without knowing the mechanism of disassembly, testing this idea has been difficult. Here, we show that B1-CDK1 induces CMG ubiquitylation and unloading, fork breakage, and fork collapse. This cascade is inhibited via multiple independent manipulations (PLK1-i, p97-i, TRAIP depletion) that all target the CMG unloading step. Thus, our data establish a firm relationship between replisome disassembly, fork breakage, and collapse in a vertebrate cell-free system that exhibits physiological complexity. Whether the inability to restart the fork (collapse) results from replisome disassembly per se or a downstream event such as fork breakage is presently unclear. Moreover, without active recombinant MCM2–7, we cannot make point mutations to directly test whether CMG ubiquitylation is responsible for fork breakage. Nevertheless, multiple lines of evidence point to CMG as the most likely target. First, TRAIP associates with stalled and terminated replication forks (Dewar et al., 2017; Hoffmann et al., 2016), ideally positioning TRAIP for CMG ubiquitylation. Second, prior to fork collapse, B1-CDK1 induces rapid and quantitative ubiquitylation of MCM7, the same protein that is ubiquitylated when CMG is unloaded during replication termination. Finally, CMG is unique among replisome components in that it cannot be reloaded de novo in S phase (Deegan and Diffley, 2016). Thus, loss of CMG provides a simple explanation for the irreversibility of fork collapse. It will be interesting to determine how this pathway relates to the depletion of RPA at the fork, which has also been proposed to trigger fork collapse and breakage (Toledo et al., 2013).
After fork breakage in mitotic extracts, the newly formed DNA ends undergo two classes of joining events. The first class involves deletions the highly repetitive lacO repeats and is thus best explained by single-strand annealing. SSA, which is usually RAD52 dependent (Bhargava et al., 2016), has recently been shown to mediate DNA repair synthesis during mitosis (Bhowmick et al., 2016). However, we have not been able to test the involvement of RAD52 due to an inability to raise antibodies against Xenopus RAD52. The second class of end joining products is mediated by micro-homology, sometimes with multiple template-switching events, indicative of DNA Polθ-mediated end joining (MMEJ, (Wyatt et al., 2016)). Consistent with this idea, aberrant replication products were reduced and complex rearrangements were eliminated in Polθ-depleted extracts. Notably, we detected only short-tract template switches typical of MMEJ. If template-switching events mediated by Polθ or other factors were followed by more processive DNA synthesis near the break, duplications could result that resemble copy number alterations in human cancer and congenital disease (Carvalho and Lupski, 2016; Leibowitz et al., 2015).
We envision at least two beneficial effects of TRAIP-dependent replisome disassembly in mitosis. One arises when converging forks are unable to complete DNA replication by anaphase, as seen at common fragile sites (CFS) (West and Chan, 2018). We propose that TRAIP-dependent CMG unloading leads to preferential breakage on the two leading strand templates because these are normally protected by CMG (Fu et al., 2011) and therefore exposed after CMG dissociation (Figure S7). In this scenario, one intact daughter chromosome would immediately be restored by gap filling, and the other could be regenerated via joining of the two broken ends, albeit with sister chromatid exchange and at the cost of a small deletion (Figure S7, left branch). Importantly, this mechanism avoids the formation of acentric and dicentric chromosomes that would result if the forks underwent random breakage (Figure S7, right branch), thus biasing breakage at CFS towards more beneficial outcomes. Strikingly, CFS expression induces chromosomal alterations that exhibit key features predicted by our model: submicroscopic deletions covering the CFS locus, microhomologies at the breakpoint junctions, and a very high frequency of sister chromatid exchanges (Glover et al., 2017) (Figure S7, left branch). In contrast, break-induced replication models of CFS expression (Bhowmick et al., 2016; Minocherhomji et al., 2015) do not account for the sister chromatid exchanges at CFS, and they would not be beneficial at CFS located distant from chromosome ends. A second possible benefit of TRAIP activity in mitosis is to disassemble terminated CMGs that evaded the action of CRL2Lrr1 in the previous S phase. In principle, such CMGs might interfere with transcription, replication, or other processes in the next cell cycle. Future studies will address whether the dwarfism phenotype observed in patients with TRAIP mutations (Harley et al., 2016) is caused by defective CMG unloading in mitosis or other TRAIP-dependent processes.
In addition to its beneficial effects, we propose that TRAIP-dependent CMG unloading contributes to genome instability phenomena that were previously linked to mitotic DNA replication. These include chromosome breakage after S and M phase cells are fused (Duelli et al., 2007; Johnson and Rao, 1970; Rao et al., 1982) or when mitotic CDK is prematurely activated in S phase by WEE1 inhibition (Dominguez-Kelly et al., 2011; Duda et al., 2016), and chromothripsis in micronuclei that are still engaged in replication when they enter mitosis (Crasta et al., 2012; Leibowitz et al., 2015; Ly et al., 2017). In these cases, massive chromosomal breakage leads to genome instability or cell death. Notably, chromosome fragmentation in the presence of WEE1 inhibitor and common fragile site expression are both MUS81-dependent (Dominguez-Kelly et al., 2011; Duda et al., 2016; Naim et al., 2013; Ying et al., 2013). In contrast, fork breakage in our experiments was not inhibited by MUS81 depletion. Whether this reflects incomplete MUS81 depletion in extracts, greater redundancy with other nucleases, or a difference in experimental systems remains to be determined.
Many studies of fork collapse focus on its regulation by ATR. While ATR-dependent phosphorylation of SMARCAL1 and WRN regulates fork stability, these ATR substrates do not appear to account for ATR’s essential role in preventing fork collapse (Cortez, 2015; Pasero and Vindigni, 2017; Saldivar et al., 2017). Instead, a growing body of evidence suggests that ATR affects fork stability indirectly (Toledo et al., 2017). For example, ATR inhibition of late origin firing prevents exhaustion of the nuclear RPA pool, causing fork deprotection and breakage (Toledo et al., 2013). However, given the concentration of RPA in egg extracts (~10 μM; (Walter et al., 1998; Wuhr et al., 2015)), and the concentration of DNA in our experiments, RPA exhaustion cannot account for our results. Another hypothesis to explain the effect of ATR on fork stability involves the suppression of mitotic kinases. In cells treated with Aphidicolin and ATRi, replication fork collapse depends on B1-CDK1, AURKA, and PLK1 (Eykelenboom et al., 2013; Ragland et al., 2013). Even in the absence of exogenous replication stress, ATR prevents the premature accumulation of Cyclin B and PLK1 in S phase, which is critical to suppress replication fork collapse and genome instability (Ruiz et al., 2016; Saldivar et al., 2018). Thus, replication fork collapse in interphase can be due to premature activation of mitotic kinases. Consistent with the central importance of ATR in restraining B-CDK1, ATR is not required to stabilize stalled DNA replication forks in egg extracts that are permanently arrested in interphase (Figure 4D; (Luciani et al., 2004)). Conversely, when stressed forks are exposed to B1-CDK1, forks break, even in the presence of ATR activity. Based on these considerations, in many studies where ATR inhibition induces replication fork collapse, this collapse may be due to premature activation of B1-CDK1 and TRAIP-dependent replisome disassembly.
In summary, our data suggest that when TRAIP is activated by mitotic CDK, a short temporal window opens in which replication forks can finish replication and terminate. The window closes when CMGs are ubiquitylated and extracted from chromatin. In the presence of a few unreplicated loci (e.g. fragile sites), CMG unloading and fork breakage promotes chromosome segregation and genome integrity, but when many forks are present (e.g. micronuclei, premature CDK1 activation in S phase), massive DNA fragmentation results, leading to cell death or transformation.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Johannes Walter (Johannes_Walter@hms.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Xenopus laevis
Egg extracts were prepared using Xenopus laevis (Nasco Cat #LM0053MX). All experiments involving animals were approved by the Harvard Medical School Institutional Animal Care and use Committee (IACUC) and conform to relevant regulatory standards.
C. elegans maintenance
The C. elegans strains were maintained according to standard procedures (Brenner, 1974) and were grown on ‘Nematode Growth Medium’ (NGM: 3 g/L NaCl; 2.5 g/L peptone; 20 g/L agar; 5 mg/L cholesterol; 1 mM CaCl2; 1 mM MgSO4; 2.7 g/L KH2PO4; 0.89 g/L K2HPO4). The following worm strains were used:
KAL1: psf-1(lab1[gfp::TEV::S-tag::psf-1 + loxP unc-119(+) loxP])
KAL3: psf-1(lab1); ltIs37[pie-1p::mCherry::his-58 + unc-119(+)]
TG1754: unc-119(ed3) III; gtIs65[pie-1p::gfp::cdc-45 + unc-119(+)]; ltIs37
METHOD DETAILS
No statistical methods were used to predetermine sample size. All experiments were performed at least twice independently using separate preparations of Xenopus egg extracts. A representative result is shown.
Protein purification
To purify biotinylated LacR, the LacR-Avi expressing plasmid pET11a[LacR-Avi] (Avidity, Denver, CO) and biotin ligase expressing plasmid pBirAcm (Avidity, Denver, CO) were co-transformed into T7 Express cells (New England Biolabs). Cultures were supplemented with 50 mM biotin (Research Organics, Cleveland, OH). Expression of LacR-Avi and the biotin ligase was induced by addition of IPTG (Isopropyl β-D-thiogalactoside, Sigma, St. Louis, MO) to a final concentration of 1 mM. Biotinylated LacR-Avi was then purified as described (Dewar et al., 2015). BRC (a ~35 amino acid peptide derived from BRCA2 that binds RAD51) and BRC*** (BRC peptide with mutations at RAD51 binding sites), a gift of K. Vrtis, were purified as reported (Long et al., 2011).
Recombinant TRAIP was expressed in Rosetta 2 (DE3) pLysS (Novagen). Bacteria transformed with His6-SUMO-X. laevis TRAIP wild-type (WT), R18C and PIP box mutant (ΔPIP) were grown in media containing 100 μg/mL ampicillin and 27 μg/mL chloramphenicol at 37°C to OD600 0.6, transferred to 16°C for 30 min, and induced overnight with the addition of 0.1 mM IPTG and 50 μM ZnSO4. The bacteria were collected by centrifugation, resuspended in Lysis Buffer (20 mM HEPES-NaOH pH 7.5, 400 mM sodium acetate, 10% glycerol, 20 mM imidazole, 10 μM ZnSO4, 0.1% NP-40, 1 mM DTT, and 1x Roche cOmplete protease inhibitor cocktail), and sonicated.
Ammonium sulfate was added to a final concentration of 300 mM, followed by addition of polyethyleneimine to a final concentration of 0.45%. The lysate was rotated for 15 min at 4°C, centrifuged at 40,000 × g for 45 min, a nd the soluble fraction recovered and precipitated with saturating ammonium sulfate. The precipitated fraction was collected by centrifugation at 40,000 × g for 45 min, resuspended in Lysis Buffer, and rotated for 30 min with NiNTA resin at room temperature. The resin was washed three times with Wash Buffer (20 mM HEPES-NaOH pH 7.5, 400 mM sodium acetate, 10% glycerol, 20 mM imidazole, 10 μM ZnSO4, 0.01% NP-40, 1 mM DTT) and the protein was eluted with Elution Buffer (20 mM HEPES-NaOH pH 7.5, 400 mM sodium acetate, 10% glycerol, 120 mM imidazole, 10 μM ZnSO4, 0.01% NP-40, 1 mM DTT). The SUMO protease Ulp1 was added to the eluate to a final concentration of 0.03 mg/mL and dialyzed against Dialysis Buffer (20 mM HEPES-NaOH pH 7.5, 400 mM sodium acetate, 120 mM imidazole, 10% glycerol) overnight at 4°C. Aliquots were flash frozen and stored at −80°C.
Other proteins used in this study were Cyclin B1-CDK1 (Life Technologies Cat #PR4768C and EMD Millipore Cat #14–450M), Cyclin A2 (Creative Biomart, Cat #CCNA2–6798H) and Cyclin E-CDK2 (EMD Millipore Cat #14–475). USP21 was a gift from D. Finley.
DNA constructs
The 4.6 kb p[lacO48] plasmid (a generous gift of K. Vrtis) contains an array of 48 lacO sites which can be bound by the lac repressor (LacR) to form replication barriers. The pDPC plasmid (4.3 kb), a generous gift of J. Sparks, was constructed based on a previous protocol (Duxin et al., 2014). Control plasmid (pControl) used in Figure 1G has the same DNA sequence as pDPC, but lacks crosslinks.
Xenopus egg extracts and DNA replication
Xenopus egg extracts including Low Speed Supernatant (LSS), High Speed Supernatant (HSS), and Nucleoplasmic egg extract (NPE) were prepared as described (Blow and Laskey, 1986; Lebofsky et al., 2009).
To assess the effects of mitotic cyclins, demembranated sperm chromatin from Xenopus laevis males was incubated in LSS (4,000 sperms/μL LSS) for 40 minutes at room temperature to form nuclei. The reactions were subsequently incubated with a range of concentrations of mitotic B1-CDK1. Nuclear envelope integrity and chromatin condensation were monitored by microscopy after Hoechst staining (see below). The concentration (50 ng/μL) that triggered nuclear envelopment breakdown and chromosome condensation was chosen to trigger mitotic entry in subsequent experiments.
For interphase DNA replication, sperm chromatin or plasmid DNA was first incubated in HSS (final concentration of 7.5–15.0 ng DNA/μL HSS) for 30 minutes at room temperature to license the DNA for replication (“licensing”), followed by the addition of 2 volumes of NPE to initiate CDK2-dependent replication. To radiolabel the nascent strands during replication, NPE was supplemented with trace amounts of [α−32P]-dATP. Mitotic DNA replication was performed essentially as described (Prokhorova et al., 2003). Briefly, after 30 minutes, 0.9 volumes of licensing reaction was incubated with 0.1 volumes of mitotic B1-CDK1 for 30 minutes at room temperature, followed by addition of 2 volumes of NPE. In the “licensing” mixture, the concentration of B1-CDK1 was 50 ng/μL, and its concentration in the final replication reaction was 16.7 ng/μL. Unless stated otherwise, the ‘0 minute’ time point refers to the moment of NPE addition. 2 μL aliquots of replication reaction were stopped with 5 μl of stop solution A (5% SDS, 80 mM Tris pH8.0, 0.13% phosphoric acid, 10% Ficoll) supplemented with 1 μl 20 mg/ml Proteinase K (Roche, Nutley, NJ). Samples were incubated for 1 hour at 37°C prior to electrophoresis on a 0.9% native agarose gel. Gels were dried and radioactivity was detected using a phosphorimager (Lebofsky et al., 2009).
To induce replication fork stalling using LacR, one volume of p[lacO48] (200 ng/μL) was incubated with one volume of recombinant LacR (36 μM) for 45–60 minutes at room temperature. Next, 0.1 volumes of the mixture was combined with 0.9 volumes of HSS for licensing, followed by addition of 2 volumes of NPE for initiation of replication. To inhibit the binding of LacR to the lacO array, IPTG was added to NPE to a final concentration of 10 mM and incubated for 15 minutes prior to use in replication (Figure 1E) or added into replication reactions after fork stalling (Figures S4H and S6C) at the indicated time.
For replication assays with inhibitors, NPE was supplemented with inhibitors for 15 minutes at room temperature before addition to the licensing mixture. Inhibitors were used at the following final concentrations in replication reaction: Aphidicolin (Sigma Cat #A0781–5MG), 2.2 μM or 0.97 μM, as indicated; CDC7 inhibitor PHA-767491 (Sigma Cat #PZ0178), 266 μM; p97 inhibitor NMS-873 (Sigma Cat #SML1128–5MG), 266 μM; DNA-PKcs inhibitor NU-7441, 133 μM; BRC or BRC***, 1 μg/μL; Cullin inhibitor MLN-4924 (Active Biochem Cat #A-1139), 266 μM; PLK1 kinase inhibitor BI-2536 (Adooq Cat # A10134, 50 μM; Aurora A kinase inhibitor MK-5108 (Selleck Cat #S2770), 10 μM and ATR inhibitor (ETP-46464; Sigma Cat #SML1321), 200 μM. For the CDK1 inhibition assay in Figure S1G, CDK1 inhibitor RO-3306 (EMD Millipore Cat #217699–5MG) was incubated with the replication reaction containing stalled replication forks for 5 minutes before the addition of B1-CDK1.
Immunodepletion and Western blotting
Immunodepletions using antibodies against Xenopus laevis FANCD2 (Knipscheer et al., 2009), FANCI (Duxin et al., 2014), SMC2 (antigen: Ac-CSKTKERRNRMEVDK-OH, New England Peptide), TRAIP (antigen: Ac-CTSSLANQPRLEDFLK-OH, New England Peptide), Polθ (antigen: residues 1212 to 1506, Abgent), and RAD51 (Long et al., 2011) were performed as described previously (Budzowska et al., 2015). Briefly, Protein A Sepharose Fast Flow beads (GE Healthcare) were incubated with antibodies at 4°C overnight. For mock depletion, an equivalent quantity of nonspecific rabbit IgGs was used. Five volumes of pre-cleared HSS or NPE were then mixed with one volume of the antibody-bound sepharose beads. For FANCI-D2 depletion of HSS and NPE, two rounds of depletion using both FANCI and FANCD2 antibodies were performed at room temperature for 20 minutes each. Depletions for other proteins were performed at 4°C, with two rounds for HSS and three rounds for NPE. For each round, a mixture of antibody-bound beads and egg extract was rotated on a wheel for 40 minutes. Immunodepleted extracts were collected and used immediately for DNA replication. Depletion efficiency was assessed by Western blotting. Western blots from depletion or plasmid/sperm chromatin pull-downs were probed using antibodies against SMC2, TRAIP, FANCI (Duxin et al., 2014), FANCD2 (Knipscheer et al., 2009), MCM7 (Dewar et al., 2017), MCM6 (Dewar et al., 2017), RAD51 (Long et al., 2011), ORC2 (Dewar et al., 2017), CDC45 (Walter and Newport, 2000), SLD5 (Dewar et al., 2017), CHK1-pS345 (Cell Signaling Technology Cat #2348S), γ-H2AX (Cell Signaling Technology Cat #2577S) and Histone H3 (Cell Signaling Technology Cat #9715S).
Sperm chromatin spin-down assay
Sperm chromatin spin-down was performed as previously described (Raschle et al., 2015). Briefly, chromatin and associated proteins were isolated by centrifugation through a sucrose cushion, washed three times, resuspended in 2x SDS sample buffer (100 mM Tris pH 6.8, 4% SDS, 0.2% bromophenol blue, 20% glycerol, 10% β-mercaptoethanol) and boiled at 95°C for 3–5 minutes. In Figure S3A, chromatin was spun down 20 minutes after NPE addition for the Buffer control and at 9 minutes after NPE addition for the B1-CDK1 treatment (final concentration, 16.7 ng/μL), at which point replication was ~50% complete for both reactions. In Figure S1D, chromatin and associated proteins were isolated from HSS.
Plasmid pull-down assay
Plasmid pull-down assays were performed as described (Budzowska et al., 2015). Briefly, streptavidin-coupled magnetic beads (Dynabeads M-280, Invitrogen; 6 μl beads slurry per pull-down) were washed three times with wash buffer 1 (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA pH 8, 0.02% Tween-20). Biotinylated LacR was incubated with the beads (12 pmol per 6 μL beads) at room temperature for 40 min. The beads were then washed four times with pull-down buffer 1 (10 mM Hepes pH 7.7, 50 mM KCl, 2.5 mM MgCl2, 250 mM sucrose, 0.25 mg/mL BSA, 0.02% Tween-20) and resuspended in 40 μL of the same buffer. At the indicated times, 4 μL samples of the replication reaction were withdrawn and gently mixed with Biotin-LacR-coated beads. The suspension was immediately placed on a rotating wheel and incubated for 30–60 minutes at 4°C. The beads were washed three times with wash buffer 2 (10 mM Hepes pH 7.7, 50 mM KCl, 2.5 mM MgCl2, 0.25 mg/mL BSA, 0.03% Tween-20). The beads were resuspended in 40 μL of 2× SDS sample buffer and boiled at 95°C for 3– 5 minutes. Chromatin-bound proteins were separated by SDS-PAGE and analyzed by Western blotting.
De-ubiquitination assay
Plasmid pull-downs were performed as described above, except that after the wash steps with wash buffer 2, chromatin-bound proteins were resuspended in 20 μL of USP21 buffer (150 mM NaCl, 10 mM DTT, 50mM Tris pH 7.5) and split into two 10 μL aliquots. Each aliquot was incubated with the non-specific deubiquitinase USP21 or buffer at 37°C for 60 minutes. The reactions were stopped by addition of 2x SDS sample buffer and boiled at 95°C for 3–5 minutes.
Restriction digestion
Two-microliter aliquots of replication reactions were stopped in 20 μL of stop solution B (50 mM Tris pH 7.5, 0.5% SDS, 25 mM EDTA), and replication products were purified as previously described (Raschle et al., 2008). Purified products were digested with restriction enzymes as per the manufacturer’s instructions. Digestion reactions were stopped in 0.5 volumes of stop solution C (5% SDS, 4 mg/mL Proteinase K) and incubated for 60 minutes at room temperature. Digested products were separated on a 1% native agarose gel and visualized by autoradiography.
Sequencing
LacR-bound p[lacO48] plasmid was replicated in the presence of mitotic B1-CDK1 for 120 minutes. Replication products were purified and digested with AlwNI (single cut on the parental DNA) for 60 minutes at 37°C, as described above. After separation on a 0.9% native agarose gel, bands smaller than the 4.6 kb full-length linear fragment were extracted and self-ligated with T4 DNA ligase. The ligation products were transformed into E.coli DH5α or XL1-Gold. As a control, p[lacO48] was replicated without LacR for 120 minutes in the presence of B1-CDK1. Replication products (containing only open circular and supercoiled species) were processed as above, and the only band (4.6 kb) after AlwNI restriction was purified for cloning. Clones from both treatments were sequenced by Sanger method with Forward primer: 5’-AAGGCGATTAAGTTGGGTAA-3’ and Reverse primer: 5’-CATGTTCTTTCCTGCGTTATCCCCTGA-3’.
RNA interference
RNAi was performed by feeding worms with RNAse III-deficient HT115 bacteria transformed with an L4440-derived plasmid that express double-stranded RNA (Timmons and Fire, 1998). For microscopy experiments, bacterial culture grown to OD600=1 was supplemented with 1 mM IPTG to express dsRNA. 400 μL of bacteria were loaded onto a 6 cm RNAi plates (3 g/L NaCl, 20 g/L agarose, 5 mg/L cholesterol, 1 mM CaCl2, 1 mM MgSO4, 2.7 g/L KH2PO4, 0.89 g/L K2HPO4, 1 mM IPTG and 100 mg/L Ampicillin) and the plate was incubated overnight at room temperature. For each immunoprecipitation, 0.5 mL of bacterial pre-culture grown overnight was used to inoculate a 400 mL culture in ‘Terrific Broth’ (12 g/L tryptone, 24 g/L yeast extract, 9.4 g/L K2HPO4, 2.2 g/L KH2PO4, adjusted to pH 7). After 7 h of growth in a baffled flask at 37°C with agitation, expression of dsRNA was induced overnight at 20°C by addition of 3 mM IPTG and the bacteria were pelleted. 8 g of bacterial pellet was resuspended with 2 mL buffer (M9 medium supplemented with 75 mg/L cholesterol; 100 mg/L ampicillin; 50 mg/L tetracycline; 12.5 mg/L amphotericin B; 3 mM IPTG) and spread on a 15 cm plate containing NGM supplemented with 1 mM IPTG and 100 mg/L ampicillin.
The plasmids expressing dsRNA were made by cloning PCR products amplified from cDNA into the vector L4440. lrr-1 fragment was obtained with the primers ATGCGATTACCATGTGAAGTGG and CCTCGTGTGTGTATTCGATATTATC; npl-4 fragment with GTCCAAAAGGGCCCAACTGTC and CCAGCAGGAACATCCACCAGC; B0432.13 (trul-1) with ATGACGTCACAGCCCACGTCATC and CGTATTCCGTAAGATTCGACGTA. To target lrr-1 and B0432.13 simultaneously, DNA fragments from each gene were cloned contigously into a single L4440 plasmid. The empty L4440 plasmid was used as control.
Microscopy
Worms at the larval L4 stage were incubated on 6 cm RNAi feeding plates for 30–34 hours at 25°C. Embryos were dissected in M9 medium (6 g/L Na2HPO4, 3 g/L KH2PO4, 5 g/L NaCl, 0.25 g/L MgSO4) and mounted on a 2% agarose pad. Time lapse images were then recorded at 23–24°C using an Olympus IX81 microscope (MAG Biosystems) with a CSU-X1 spinning-disk confocal imager (Yokogawa Electric Corporation), a Cascade II camera (Photometrics) and a 60X/1.40 Plan Apochromat oil immersion lens (Olympus). Images were captured every 10 seconds using MetaMorph software (Molecular Devices) as previously described (Sonneville et al., 2017).
To image sperm chromatin in egg extracts, 1 μL of nuclear assembly reactions containing LSS egg extract and sperm chromatin was mixed with 1 μL of Hoechst 3300 (2.5 μg/mL) for 5 minutes before imaging. Images in Figures S1A and S1C were single focal planes acquired by a wide field Nikon Eclipse E600 microscope equipped with a Nikon 40x Plan Apo NA 1.0 oil objective. Images in Figure S4B were maximum projections from stacks of z-series acquired with a 0.5 μm step size. Images were collected using a 60x Plan Apo NA 1.4 oil objective with a CoolSnapHQ2 CCD camera (Photometrics) on a Yokogawa CSU-22 spinning disk confocal system (Nikon Instruments, Melville, NY). Fluorophores were excited by a 405 nm laser.
Extracts of worm embryos and immunoprecipitation of protein complexes
Preparation of worm extracts and immunoprecipitation of GFP-PSF-1 was performed as previously described (Sonneville et al., 2017). Briefly, 1 mL of a synchronized population of L4 worms expressing GFP-PSF-1 were fed for 50 h at 20 °C on a 15 cm RNAi plate, supplemented with 8 g of bacterial pellet (see above). After feeding, the worms were washed in M9 medium and then disrupted in ‘bleaching solution’ (for 100 mL: 36.5 mL H2O, 45.5 mL 2 N NaOH and 18 mL ClNaO 4%), before washing of the resulting embryo preparation in M9 medium.
At 4°C, embryos were washed twice with lysis buffer (100 mM HEPES-KOH pH 7.9, 50 mM potassium acetate, 10 mM magnesium acetate, 2 mM EDTA), and then resuspended in three volumes of lysis buffer that was supplemented with 2 mM sodium fluoride, 2 mM sodium β-glycerophosphate pentahydrate, 1 mM dithiothreitol (DTT), 1% Protease Inhibitor Cocktail (P8215, Sigma-Aldrich), and 1× ‘Complete Protease Inhibitor Cocktail’ (05056489001, Roche). The washed embryo suspension was then snap frozen drop-wise in liquid nitrogen and stored at −80°C. Subsequently, ~2.5 g of frozen embryos was ground in a SPEX SamplePrep 6780 Freezer/Mill. After thawing, we added a one-quarter volume of ‘glycerol-mix’ buffer (lysis buffer supplemented with 50% glycerol, 300 mM potassium acetate, 0.5% detergent IGEPAL CA-630, protease inhibitors, and DTT at the concentrations mentioned above). De-ubiquitylase enzymes were inhibited by addition of 5 μM propargylated ubiquitin (Ubi-PrG; MRC PPU, Dundee), and chromosomal DNA was digested with 1,600 U of Pierce Universal Nuclease (123991963, Fisher) for 30 min at 4°C. Extracts were centrifuged at 25,000 × g for 30 min and then for 100,000 × g for 1 h, before pre-incubation with agarose beads (0.4 mL slurry) for 45 min. Samples of each extract were taken and combined with Laemmli buffer, before storage at −80°C. The remainder was then incubated for 90 min with 40 μL of GFP-Trap_A beads (Chromotek). The beads were washed four times with 1 mL of wash buffer (100 mM HEPES-KOH pH 7.9, 100 mM potassium acetate, 10 mM magnesium acetate, 2 mM EDTA, 0.1% IGEPAL CA-630, 2 mM sodium fluoride, 2 mM sodium β-glycerophosphate pentahydrate, plus protease inhibitors as above). Finally, the bound proteins were eluted at 95 °C for 5 min in 100 μl of 1× Laemmli buffer and stored at −80°C.
Data quantification
Autoradiographs and Western blots were quantified using ImageJ 1.48v (National Institute of Health). The quantification methods for individual results are described in the figure legends.
Supplementary Material
HIGHLIGHTS.
Replication fork collapse is triggered by mitotic CDK-dependent CMG unloading
Mitotic CMG unloading requires the E3 ubiquitin ligase TRAIP and the p97 ATPase
Mitotic processing of stalled forks enables high-fidelity chromosome segregation
New model for the generation of complex chromosome rearrangements
ACKNOWLEDGMENTS
We thank James Dewar, Emily Low, Justin Sparks, Kyle Vrtis, Daniel Finley, Puck Knipscheer, Jan-Michael Peters and MRC PPU Reagents and Services (https://mrcppureagents.dundee.ac.uk) for experimental protocols or reagents. We thank Alan D’Andrea, Randy King, Ralph Scully, and members of the Pellman and Walter laboratories for helpful discussion and critical reading of the manuscript. R.A.W. was supported by postdoctoral fellowship 131415-PF-17-168-01-DMC from the American Cancer Society. K.L. was supported by the Medical Research Council (core grant MC_UU_12016/13), the Wellcome Trust (Ref. 102943/Z/13/Z for an investigator award) and Cancer Research U.K. (Ref. C578/A24558). D.P. was supported by NIH grant CA213404. J.C.W. was supported by NIH grants GM080676 and HL098316. D.P. and J.C.W. are investigators of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Amunugama R, Willcox S, Wu RA, Abdullah UB, El-Sagheer AH, Brown T, McHugh PJ, Griffith JD, and Walter JC (2018). Replication Fork Reversal during DNA Interstrand Crosslink Repair Requires CMG Unloading. Cell Rep 23, 3419–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann C, Korner R, Hofmann K, and Nigg EA (2007). PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell 128, 101–114. [DOI] [PubMed] [Google Scholar]
- Bhargava R, Onyango DO, and Stark JM (2016). Regulation of Single-Strand Annealing and its Role in Genome Maintenance. Trends Genet 32, 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick R, and Hickson ID (2017). The “enemies within”: regions of the genome that are inherently difficult to replicate. F1000Res 6, 666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick R, Minocherhomji S, and Hickson ID (2016). RAD52 Facilitates Mitotic DNA Synthesis Following Replication Stress. Mol Cell 64, 1117–1126. [DOI] [PubMed] [Google Scholar]
- Blow JJ, and Laskey RA (1986). Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell 47, 577–587. [DOI] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, and Baltimore D (2000). ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev 14, 397–402. [PMC free article] [PubMed] [Google Scholar]
- Budzowska M, Graham TG, Sobeck A, Waga S, and Walter JC (2015). Regulation of the Rev1-pol zeta complex during bypass of a DNA interstrand cross-link. EMBO J 34, 1971–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, and Lupski JR (2016). Mechanisms underlying structural variant formation in genomic disorders. Nat Rev Genet 17, 224–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, North PS, and Hickson ID (2007). BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J 26, 3397–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D (2015). Preventing replication fork collapse to maintain genome integrity. DNA Repair (Amst) 32, 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, and Pellman D (2012). DNA breaks and chromosome pulverization from errors in mitosis. Nature 482, 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Piccoli G, Katou Y, Itoh T, Nakato R, Shirahige K, and Labib K (2012). Replisome stability at defective DNA replication forks is independent of S phase checkpoint kinases. Mol Cell 45, 696–704. [DOI] [PubMed] [Google Scholar]
- Deegan TD, and Diffley JF (2016). MCM: one ring to rule them all. Curr Opin Struct Biol 37, 145–151. [DOI] [PubMed] [Google Scholar]
- Dewar JM, Budzowska M, and Walter JC (2015). The mechanism of DNA replication termination in vertebrates. Nature 525, 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar JM, Low E, Mann M, Raschle M, and Walter JC (2017). CRL2Lrr1 promotes unloading of the vertebrate replisome from chromatin during replication termination. Genes Dev 31, 275–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Kelly R, Martin Y, Koundrioukoff S, Tanenbaum ME, Smits VA, Medema RH, Debatisse M, and Freire R (2011). Wee1 controls genomic stability during replication by regulating the Mus81-Eme1 endonuclease. J Cell Biol 194, 567–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda H, Arter M, Gloggnitzer J, Teloni F, Wild P, Blanco MG, Altmeyer M, and Matos J (2016). A Mechanism for Controlled Breakage of Under-replicated Chromosomes during Mitosis. Dev Cell 39, 740–755. [DOI] [PubMed] [Google Scholar]
- Duelli DM, Padilla-Nash HM, Berman D, Murphy KM, Ried T, and Lazebnik Y (2007). A virus causes cancer by inducing massive chromosomal instability through cell fusion. Curr Biol 17, 431–437. [DOI] [PubMed] [Google Scholar]
- Dungrawala H, Rose KL, Bhat KP, Mohni KN, Glick GG, Couch FB, and Cortez D (2015). The Replication Checkpoint Prevents Two Types of Fork Collapse without Regulating Replisome Stability. Mol Cell 59, 998–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxin JP, Dewar JM, Yardimci H, and Walter JC (2014). Repair of a DNA-protein crosslink by replication-coupled proteolysis. Cell 159, 346–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Achkar E, Gerbault-Seureau M, Muleris M, Dutrillaux B, and Debatisse M (2005). Premature condensation induces breaks at the interface of early and late replicating chromosome bands bearing common fragile sites. Proc Natl Acad Sci U S A 102, 18069–18074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eykelenboom JK, Harte EC, Canavan L, Pastor-Peidro A, Calvo-Asensio I, Llorens-Agost M, and Lowndes NF (2013). ATR activates the S-M checkpoint during unperturbed growth to ensure sufficient replication prior to mitotic onset. Cell Rep 5, 1095–1107. [DOI] [PubMed] [Google Scholar]
- Feng W, Guo Y, Huang J, Deng Y, Zang J, and Huen MS (2016). TRAIP regulates replication fork recovery and progression via PCNA. Cell Discov 2, 16016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YV, Yardimci H, Long DT, Ho TV, Guainazzi A, Bermudez VP, Hurwitz J, van Oijen A, Scharer OD, and Walter JC (2011). Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 146, 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullbright G, Rycenga HB, Gruber JD, and Long DT (2016). p97 Promotes a Conserved Mechanism of Helicase Unloading during DNA Cross-Link Repair. Mol Cell Biol 36, 2983–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover TW, Wilson TE, and Arlt MF (2017). Fragile sites in cancer: more than meets the eye. Nat Rev Cancer 17, 489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley ME, Murina O, Leitch A, Higgs MR, Bicknell LS, Yigit G, Blackford AN, Zlatanou A, Mackenzie KJ, Reddy K, et al. (2016). TRAIP promotes DNA damage response during genome replication and is mutated in primordial dwarfism. Nat Genet 48, 36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson M, Madine M, Dalton S, and Gautier J (1996). Phosphorylation of MCM4 by cdc2 protein kinase inhibits the activity of the minichromosome maintenance complex. Proc Natl Acad Sci U S A 93, 12223–12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hills SA, and Diffley JF (2014). DNA replication and oncogene-induced replicative stress. Curr Biol 24, R435–444. [DOI] [PubMed] [Google Scholar]
- Hoffmann S, Smedegaard S, Nakamura K, Mortuza GB, Raschle M, Ibanez de Opakua A, Oka Y, Feng Y, Blanco FJ, Mann M, et al. (2016). TRAIP is a PCNA-binding ubiquitin ligase that protects genome stability after replication stress. J Cell Biol 212, 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, and Cleveland DW (2012). Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements. Nat Med 18, 1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hustedt N, and Durocher D (2016). The control of DNA repair by the cell cycle. Nat Cell Biol 19, 1–9. [DOI] [PubMed] [Google Scholar]
- Johnson RT, and Rao PN (1970). Mammalian cell fusion: induction of premature chromosome condensation in interphase nuclei. Nature 226, 717–722. [DOI] [PubMed] [Google Scholar]
- Kato H, and Sandberg AA (1968). Chromosome pulverization in human cells with micronuclei. J Natl Cancer Inst 40, 165–179. [PubMed] [Google Scholar]
- Klein Douwel D, Boonen RA, Long DT, Szypowska AA, Raschle M, Walter JC, and Knipscheer P (2014). XPF-ERCC1 acts in Unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol Cell 54, 460–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, and Walter JC (2009). The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326, 1698–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebofsky R, Takahashi T, and Walter JC (2009). DNA replication in nucleus-free Xenopus egg extracts. Methods Mol Biol 521, 229–252. [DOI] [PubMed] [Google Scholar]
- Leibowitz ML, Zhang CZ, and Pellman D (2015). Chromothripsis: A New Mechanism for Rapid Karyotype Evolution. Annu Rev Genet 49, 183–211. [DOI] [PubMed] [Google Scholar]
- Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, et al. (2011). Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 146, 889–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long DT, Raschle M, Joukov V, and Walter JC (2011). Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science 333, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciani MG, Oehlmann M, and Blow JJ (2004). Characterization of a novel ATR-dependent, Chk1-independent, intra-S-phase checkpoint that suppresses initiation of replication in Xenopus. J Cell Sci 117, 6019–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, Grofte M, Chan KL, Hickson ID, Bartek J, et al. (2011). 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol 13, 243–253. [DOI] [PubMed] [Google Scholar]
- Ly P, Teitz LS, Kim DH, Shoshani O, Skaletsky H, Fachinetti D, Page DC, and Cleveland DW (2017). Selective Y centromere inactivation triggers chromosome shattering in micronuclei and repair by non-homologous end joining. Nat Cell Biol 19, 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri HW, Huttner D, and Hickson ID (2013). How unfinished business from S-phase affects mitosis and beyond. EMBO J 32, 2661–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minocherhomji S, Ying S, Bjerregaard VA, Bursomanno S, Aleliunaite A, Wu W, Mankouri HW, Shen H, Liu Y, and Hickson ID (2015). Replication stress activates DNA repair synthesis in mitosis. Nature. [DOI] [PubMed] [Google Scholar]
- Naim V, Wilhelm T, Debatisse M, and Rosselli F (2013). ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat Cell Biol 15, 1008–1015. [DOI] [PubMed] [Google Scholar]
- Pasero P, and Vindigni A (2017). Nucleases Acting at Stalled Forks: How to Reboot the Replication Program with a Few Shortcuts. Annu Rev Genet 51, 477–499. [DOI] [PubMed] [Google Scholar]
- Peterson SE, Li Y, Chait BT, Gottesman ME, Baer R, and Gautier J (2011). Cdk1 uncouples CtIP-dependent resection and Rad51 filament formation during M-phase double-strand break repair. J Cell Biol 194, 705–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhorova TA, Mowrer K, Gilbert CH, and Walter JC (2003). DNA replication of mitotic chromatin in Xenopus egg extracts. Proc Natl Acad Sci U S A 100, 13241–13246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragland RL, Patel S, Rivard RS, Smith K, Peters AA, Bielinsky AK, and Brown EJ (2013). RNF4 and PLK1 are required for replication fork collapse in ATR-deficient cells. Genes Dev 27, 2259–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao PN, Johnson RT, and Sperling K (1982). Premature Chromosome Condensation: Application in Basic,Clinical, and Mutation Research (New York, Academic Press; ). [Google Scholar]
- Raschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, and Walter JC (2008). Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134, 969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle M, Smeenk G, Hansen RK, Temu T, Oka Y, Hein MY, Nagaraj N, Long DT, Walter JC, Hofmann K, et al. (2015). DNA repair. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science 348, 1253671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz S, Mayor-Ruiz C, Lafarga V, Murga M, Vega-Sendino M, Ortega S, and Fernandez-Capetillo O (2016). A Genome-wide CRISPR Screen Identifies CDC25A as a Determinant of Sensitivity to ATR Inhibitors. Mol Cell 62, 307–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldivar JC, Cortez D, and Cimprich KA (2017). The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat Rev Mol Cell Biol 18, 622–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldivar JC, Hamperl S, Bocek MJ, Chung M, Bass TE, Cisneros-Soberanis F, Samejima K, Xie L, Paulson JR, Earnshaw WC, et al. (2018). An intrinsic S/G2 checkpoint enforced by ATR. Science 361, 806–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semlow DR, Zhang J, Budzowska M, Drohat AC, and Walter JC (2016). Replication-Dependent Unhooking of DNA Interstrand Cross-Links by the NEIL3 Glycosylase. Cell 167, 498–511 e414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneville R, Moreno SP, Knebel A, Johnson C, Hastie CJ, Gartner A, Gambus A, and Labib K (2017). CUL-2LRR-1 and UBXN-3 drive replisome disassembly during DNA replication termination and mitosis. Nat Cell Biol 19, 468–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo Lee N, Jin Chung H, Kim HJ, Yun Lee S, Ji JH, Seo Y, Hun Han S, Choi M, Yun M, Lee SG, et al. (2016). TRAIP/RNF206 is required for recruitment of RAP80 to sites of DNA damage. Nat Commun 7, 10463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. (2011). Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144, 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausfeld UP, Howell M, Descombes P, Chevalier S, Rempel RE, Adamczewski J, Maller JL, Hunt T, and Blow JJ (1996). Both cyclin A and cyclin E have S-phase promoting (SPF) activity in Xenopus egg extracts. J Cell Sci 109, 1555–1563. [DOI] [PubMed] [Google Scholar]
- Techer H, Koundrioukoff S, Nicolas A, and Debatisse M (2017). The impact of replication stress on replication dynamics and DNA damage in vertebrate cells. Nat Rev Genet 18, 535–550. [DOI] [PubMed] [Google Scholar]
- Timmons L, and Fire A (1998). Specific interference by ingested dsRNA. Nature 395, 854. [DOI] [PubMed] [Google Scholar]
- Toledo L, Neelsen KJ, and Lukas J (2017). Replication Catastrophe: When a Checkpoint Fails because of Exhaustion. Mol Cell 66, 735–749. [DOI] [PubMed] [Google Scholar]
- Toledo LI, Altmeyer M, Rask MB, Lukas C, Larsen DH, Povlsen LK, Bekker-Jensen S, Mailand N, Bartek J, and Lukas J (2013). ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103. [DOI] [PubMed] [Google Scholar]
- Walter J, and Newport J (2000). Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell 5, 617–627. [DOI] [PubMed] [Google Scholar]
- Walter J, Sun L, and Newport J (1998). Regulated chromosomal DNA replication in the absence of a nucleus. Mol Cell 1, 519–529. [DOI] [PubMed] [Google Scholar]
- West SC, and Chan YW (2018). Genome Instability as a Consequence of Defects in the Resolution of Recombination Intermediates. Cold Spring Harb Symp Quant Biol. [DOI] [PubMed] [Google Scholar]
- Wuhr M, Guttler T, Peshkin L, McAlister GC, Sonnett M, Ishihara K, Groen AC, Presler M, Erickson BK, Mitchison TJ, et al. (2015). The Nuclear Proteome of a Vertebrate. Curr Biol 25, 2663–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt DW, Feng W, Conlin MP, Yousefzadeh MJ, Roberts SA, Mieczkowski P, Wood RD, Gupta GP, and Ramsden DA (2016). Essential Roles for Polymerase theta-Mediated End Joining in the Repair of Chromosome Breaks. Mol Cell 63, 662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying S, Minocherhomji S, Chan KL, Palmai-Pallag T, Chu WK, Wass T, Mankouri HW, Liu Y, and Hickson ID (2013). MUS81 promotes common fragile site expression. Nat Cell Biol 15, 1001–1007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.