Summary
Adult stem cells are often found in specialized niches, where the constituent cells direct self-renewal of their stem cell pool. The niche is therefore crucial for both normal homeostasis and tissue regeneration. In many mammalian tissues, niche cells have classically been difficult to identify, which has hampered any understanding of how tissues first construct niches during development. Fortunately, the Drosophila germline stem cell (GSC) niche is well defined, allowing for unambiguous identification of both niche cells and resident stem cells. The testis niche first forms in the early embryo, during a late stage of gonadogenesis. Here, using live-imaging both in vivo and ex vivo, we follow pro-niche cells as they assemble and assume their final form. We show that after ex vivo culture the niche appears fully functional, as judged by enrichment of adhesion proteins, the ability to activate STAT in adjacent GSCs, and to direct GSCs to divide orthogonally to the niche, just as they would in situ. Collectively, our imaging has generated several novel insights on niche morphogenesis that could not be inferred from fixed images alone. We identify dynamic processes that constitute an assembly phase and a compaction phase during morphogenesis. The compaction phase correlates with cell neighbor exchange among the assembled pro-niche cells, as well as a burst of divisions among newly recruited stem cells. Before compaction, an assembly phase involves the movement of pro-niche cells along the outer periphery of the gonad, using the extracellular matrix (ECM) to assemble at the anterior of the gonad. Finally, live-imaging in integrin mutants allows us to define the role of pro-niche cell-ECM interaction with regard to the new assembly and compaction dynamics revealed here.
Keywords: Niche morphogenesis, Drosophila testis stem cell niche, Gonadogenesis
Introduction
Niches function in many adult organs to direct the self-renewal of tissue-specific stem cells1. For this reason, the proper specification and development of the niche is critical for adult organ function and regeneration. In mammalian tissues, the cells that comprise most niches are still being identified2–7. Additionally, many niches are reorganized during development, either through the introduction of new cell types in adulthood, as is seen in the neurogenic niche, or due to the migration of the stem cells to colonize a new niche as is seen with hematopoietic stem cells8,9. Together, these complexities make it difficult to study how niches first develop. Recent imaging advances have enabled investigators to visualize stem cell organization in organoid models, ex vivo skin explants, and some intravital imaging, but live imaging of mammalian niche formation is still limited10–12.
Instead, our lab is using the Drosophila male gonad, the testis precursor, as a model to study niche morphogenesis. In the testis, both niche cells and stem cells can be identified with well-characterized markers13,14. Also, the most significant events in the assembly of this stem cell niche occur during gonadogenesis in the embryo, and this same niche architecture persists through larval and adult testis development. Therefore, the male Drosophila gonad provides an excellent system for studying the mechanisms required to assemble a functional stem cell niche.
In the mature testis, the niche is composed of a tight aggregate of post-mitotic somatic cells called hub cells15. These cells secrete signals that recruit two neighboring cell populations and specify them to become stem cells, promoting both attachment to the niche and self-renewal behavior16–23. The two populations include a radial array of germline stem cells (GSCs) and of somatic cyst stem cells (CySCs). Both stem cell populations undergo asymmetric divisions producing one daughter cell that is directed to self-renew by the niche and one daughter cell that exits the niche, allowing it to differentiate24–26. The hub is anchored to the apical tip of the testis through interactions involving Integrins and the extracellular matrix (ECM)27,28. This promotes polarity in the testis by forcing differentiating daughters away from the closed end and towards the base of the testis, as cells mature along the spermatogenic pathway15. Importantly, the polarized position of the niche as well as its architecture are established by the end of gonadogenesis in the embryo and are maintained during testis maturation14.
The earliest steps of gonad formation are reasonably well-understood. The gonad first forms in both male and female embryos by coalescence of the primordial germ cells (PGCs) and somatic gonadal precursors (SGPs). The SGPs are derived from a subset of mesodermal cells within parasegments (PS) 10-1229,30 The initially elongate gonad becomes more spherical and compressed over time, settling within abdominal segment 5 (PS 10)31–33. Several factors are known to be involved in these earliest stages of gonadogenesis. Ecadherin (Ecad) mediates interactions among SGPs, as well as between SGPs and germline cells, enabling compression of the gonad into a sphere33. Additionally, live-imaging analysis identified the actin regulator Enabled as a modulator of these interactions31–33. After these initial stages, male and female gonad development diverge. Niche formation in the female gonad occurs well after embryogenesis, in the third larval instar34–39. In contrast, the male niche is set up in the late-stage embryo, and by early first instar this niche is already fully functioning14,27,40–42.
Niche cells in the male accumulate high levels of the adhesion proteins Ecad, N-Cadherin and Fasciclin III (FasIII), enabling detailed analysis in fixed preparations14. Ten to twelve SGPs associate together at the anterior of the gonad, and comprise the forming niche. These pro-niche cells were specified quite a bit earlier in development42,43. Interestingly, the pro-niche cells derive from subsets of cells initially located in separate spatial domains, PS 10 and PS1114,44. PS10 cells are near the anterior of the gonad already, but it is not known how PS11 cells travel to the gonad anterior. Additionally, the initial association of PS 10 and PS11 cells at the anterior is much looser in organization than the final form of the hub, which presents as a tight cluster of cells by the time a first instar larvae hatches14. How the pro-hub cells re-organize to adopt this final compact shape is also unknown. By the end of embryogenesis, the hub expresses the niche signal unpaired (upd) and has recruited the nearby tier of germ cells to act as GSCs14,40, and the male gonad is operating as a testis45.
In this study, we undertook a live imaging approach to investigate niche morphogenesis. We combined in vivo and ex vivo imaging approaches, which allowed us to identify dynamic processes that constitute an assembly phase and a compaction phase during morphogenesis. During niche assembly, pro-niche cells move along the periphery of the gonad, using the extracellular matrix (ECM) to assemble at the gonad anterior. Following assembly, niche compaction is coincident with cell neighbor exchange among the assembled pro-niche cells, as well as a burst of divisions among newly recruited stem cells. Live-imaging niche morphogenesis in integrin mutants revealed a role for pro-niche cell-ECM interactions the new assembly and compaction dynamics revealed here.
Materials and Methods:
Fly stocks
Fly lines used were six4-nls-eGFP46 (D. Finnegan), P-Dsix4-eGFP::Moesin32, nos-moesin::GFP (R. Lehmann), sqh-Sqh::mcherry (A. Martin), tupAME-moeGFP47 (M. Frasch), TypeIV Collagen::GFP (Kyoto DGRC 110692, FBti0153267; viking), His2Av::mRFP1 (FBtp0056035; (II.2, III.1)), Perlecan::GFP (trolZCL170048, Kyoto DGRC 110807, FBti0129820; trol), abdA-Gal449 (S. Merabet, Marseille), PrdGal4 (FBst0001947), tup-Gal4 (FBst0046960), tupAME-Gal4 (gift of J.-L. Frendo), breathless-GAL4 (FBst0078328), UAS-tdTomato (FBst0036328), UAS-Grim (FBtp0009995), mys1 (FBst0000059), trh8 mutants50 (D. Andrew; FBal0050667), tup1 FBst0036503, tupex4 (S. Campuzano; FBal0216723). Heterozygous siblings were used as controls.
For six4-Gal4, an enhancer from the third intron of six4 was amplified as in Clark et al (2006)46 from wildtype genomic DNA, with a BamH1 and an EcoRI restriction site added on the 5’ and 3’, end respectively. The amplicon was cloned into BamH1-EcoRI cut H-GAL4-vector48,51. Transgenic flies were generated by Genetics Services, Inc, using standard P-element transposition.
Live embryo time-lapse imaging
Embryos were collected overnight on agar plates. The following morning, embryos were dechorionated using 50% bleach and stage 15 embryos were selected according to Campus-Ortega and Hartenstein52. Embryos were mounted with the dorsolateral side towards the objective using tape adhesive dissolved in heptane and then covered with s700 halocarbon oil (Sigma H8898). They were imaged for up to 6hrs on either a Leica DM16000 B spinning disk confocal with a 63X 1.2 N.A. water immersion objective or an IX7 Olympus spinning disk confocal microscope with a 40x 1.1 N.A. or 60X 1.2 N.A. water immersion objective. Images were acquired with an EMCCD camera (Andor iXon 3 897E or Hamamatsu photonics, model C9100-13) controlled by Metamorph software. Z-stacks were taken at 5-15 min intervals depending on the experiment, with 35 1μm z-slices through the gonad.
Ex Vivo culture and time-lapse imaging of gonads
The cover slip bottom of a Matek imaging dish (P35G-1.5-14-C) was coated with a 2mg/ml poly-lysine solution overnight (Sigma P-1274), rinsed well with sterile distilled water, covered and allowed to air dry. Embryos expressing fluorescent markers such as six4-nls-eGFP, six4-eGFP::Moesin, or nos-Moesin::GFP were used to enable visualization of the gonad under a stereofluorescent microscope. Embryos were collected for 1-2 hours on agar plates and then aged at room temperature in a humidified chamber for 17 hours (unless otherwise noted). Embryos were brushed into nytex baskets, rinsed of yeast paste with water and dechorionated in a solution of 50% bleach with occasional agitation for about 2 minutes. The bleach was removed by rinsing well with water, and the nytex basket was blotted to remove excess liquid. Embryos were transferred into a small watch glass containing heptane, using a paint brush dipped in heptane, to which the dechorionated embryos adhere. The embryos clustered in the heptane, making it easy to use a Pasteur pipette to collect and pipette them as a small pool on a microscope slide. Once placed, the edge of a tissue was used to wick away excess heptane, thereby gathering the embryos into a single-layered cluster. The embryos were gently picked up by double-sided tape that had been folded over on a coverslip fragment. Using the double-sided tape, the coverslip was attached inside a Matek imaging dish just to the side of the imaging area. The dish was flooded with Ringers solution (128mM NaCl, 4.7 mM KCl, 1.9mM CaCl2, 2.4mM NaH2PO4, 0.05 mM Na2HPO4,pH7.4; or, alternatively, 130mM NaCl, 5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 36 mM Sucrose, 5 mM HEPES, pH7.3).
Embryos were hand-devitellinized by touching a freshly sharpened tungsten needle to the vitelline membrane at the anterior near the micropyle. The needle was gently guided towards the posterior of the embryo. Some embryos floated out from the opened vitelline membrane, others were gently pierced with the needle at their anterior end and removed from the vitelline membrane. The freed embryos were immediately guided onto the poly-lysine coated cover slip bottom of the Matek dish using the needle, either by gently pushing the embryos, or the surrounding Ringers solution to direct embryos onto the coated area. Under a stereofluorescent microscope enabling visualization of the gonad, very sharp tungsten needles were used to slice open the embryo near the gonad. This exposed the gonad with adherent tissue, which is likely the fat body. We pierced the fat body with the needle, and, without touching the gonad, pulled to free it from the carcass. The attached gonad was directed to a clean portion of the coated cover slip by pulling on the fat body, and the fluorescent gonad was guided until it affixed to the poly-lysine-coated coverslip. With practice, 50-75% of dissected gonads can be successfully placed with the gonad rather than the fat body touching the cover slip, which would otherwise interfere with imaging. Excess tissue was not removed.
The Ringers solution was then slowly pipetted off; the surface tension caused by drawing down this buffer likely assisted in affixing the gonad to the coverslip. In 100 uL increments, 500ul of imaging media was carefully added, which consisted of 15% FBS (Gibco 10082) and 0.5X penicillin/streptomycin (Corning 30-002-CI) in Schneider’s Insect Media (Gibco 21720-024)53–55. The stock imaging media was prepared ahead of time, kept at 4°C, and used within two weeks. Bovine insulin (Sigma l0516) was added to an aliquot of imaging media, just prior to use, to final concentration of 0.2mg/ml.
Gonads were imaged as for embryos, except that Z-stacks were acquired with a 0.5μm step size and a time interval of either 2.5, 5 or 10min. Metamorph image files were exported and post-acquisition analyses carried out in Image J.
Immunostaining
For tup and trh mutants, embryos were collected and aged 21-23 hours in a humidified chamber for late stage 17 embryos. Unhatched larvae were dechorionated, hand-devitellinized and dissected in Ringers and the internal organs gently massaged out as above. Dissecting in this way enabled immunostaining of gonads from stage 17 embryos, as the embryonic cuticle inhibits antibody penetration at this stage. Tissue was fixed in 4% formaldehyde, Ringers and 0.1% Triton-X-100 for 20 minutes, washed in PBS plus Triton-X-100 and blocked for 1 hour in 4% normal donkey serum. Primary antibodies were used overnight at 4°C or 4 hours at room temperature. Secondary antibodies were used at 3.75 ug/mL (Alexa488, Cy3 or Cy5; Molecular Probes; Jackson ImmunoResearch) for 1-2 hours at room temperature. DNA was stained with Hoechst 33342 (Sigma) at 0.2 μg/ml for 5 minutes.
For gonads dissected and cultured ex vivo, immunostaining was performed as above within the imaging dish. Registration marks were placed on the imaging dish to aid in re-locating each gonad after staining. Imaging media was carefully removed by pipetting slowly once the live-imaging session was completed, and replaced with 4% formaldehyde in PBS, again being careful not to disturb the attached tissue. Fixation (20 min) was carried out stationary at room temperature. Following fixation, the solution was removed, and PBS, 0.1% Triton-X-100 was added to wash out residual fixative. Antibody staining was carried out as described above. With care during solution exchanges, greater than 50% of gonads remained attached to the dish, and after immunostaining was complete, each could be could be re-imaged and registered with its pre-fixation time-lapse.
We used rabbit antibody against Vasa 1:5000, (gift from R. Lehmann, NYU), STAT92E 1:1000 (gift from E. Bach, NYU), and RFP 1:500 (Abcam, ab62341); goat antibody against Vasa 1:200 (Santa Cruz, dC-13, now discontinued), mouse antibody against Fasciclin III 1:50 (DSHB, 7G10), Islet 1:100 (DSHB, 40.3A4; Drosophila Tailup) and Crumbs 1:20 (DSHB, cq4); rat antibody against DE-cadherin 1: 20 (DSHB, DCAD2); guinea pig antibody against Traffic jam 1:10,000 (gift from D. Godt); and chick antibody against GFP 1:1000 (Aves Labs, GFP-1020). Images of fixed samples were acquired on a Zeiss Imager with Apotome using a 40x, 1.2 N.A. lens, or one of the Spinning Disk Confocal systems listed above.
Imaging and Statistical Analysis
Time lapse images were analyzed, Z projections were generated, and hub cell area was measured using Fiji software. Mann Whitney U-tests or Kruskal-Wallis tests were used for statistical comparisons, where appropriate.
Sex identification and Genotyping
Embryos were staged according to Campos-Ortega and Hartenstein52. Male embryos and larvae were identified by various criteria depending on the experiment: identifying msSGPs at the posterior of the gonad, which are visible to a trained eye in many stains, robust expression driven by the six4 enhancer in the msSGPs31, and occasionally by the larger size of the male gonad45. Sibling controls were distinguished from homozygous mutants by using fluorescent balancer chromosomes (CyO, P{w[+mC]=ActGFP}JMR1, FBst0004533; TM6B, P{w[+mW.hs]=Ubi-GFP.S65T}PAD2, Tb[1], FBst0004887; or TM3, P{w[+mC]=GAL4-twi.G}2.3, P{UAS-2xEGFP}AH2.3, Sb[1] Ser[1], FBst0006663).
Quantification of the internal tilt of the niche
To quantify the position of the assembled niche on the face of the gonad, we examined our live-imaging series in detail. We first noted the subset of z-slices that covered the full depth of the gonad, from the external-most position to the internal-most. We normalized these values from 0 to 1 (external to internal). A fraction of the z-slices represented the breadth of the assembled niche. We then located that span of niche positions on the 0-1 scale. Niches that equally spanned the midpoint value (0.5) were considered anteriorly-centered. Niches that were shifted toward higher values were considered to have an internal tilt, while niches shifted toward lower values were considered to have an external tilt.
Quantification of niche position relative to other organs
Along the external-internal axis of the gonad, we noted which z-slices included clear visualization of other organs. Often there was a portion of the niche that was visible in sections that included the trachea or the alary muscle. We report the percentage of the overall niche size (measured in microns along the external-internal axis of the gonad) that overlapped with 1) the branch of the trachea that was near the gonad anterior, or 2) the alary muscle region near the gonad anterior (Table S1).
Quantitation of STAT92E accumulation
Embryos expressing nos-moesin::GFP were collected and aged to 15-17hrs AEL. Gonads were processed for mounting as for ex vivo imaging. Mounted embryos were not imaged, but were aged at 25°C for 5 hours, and then fixed for 25 minutes at 4°C in 8% formaldehyde in Buffer B with Phosphatase Inhibitor (Sigma, 04906837001). Tissues were rinsed carefully, washed in PBS plus Triton-X-100 and blocked for 1 hour at room temperature using normal donkey serum with phosphatase inhibitor. Antibody incubations were carried out as above. Z-stack images of gonads were obtained on a Zeiss 880 point-scanning confocal using a 40x, 1.2 N.A. lens. Images were imported into Image J, and antibody fluorescence was measured using a custom plugin. Briefly, the user thresholds nuclei based on the DNA signal, and these ROIs are converted to masks, which are used to return measurements from each slice, through each nucleus. Slices were summed, with an average signal reported from each GSC returned. The plot compares STAT92E signal in GSCs relative to those germ cells several tiers away from the niche. Lines connect the average signal for all GSCs in a gonad to the average signal from all non-GSCs in that same gonad.
Measurement of cell division orientations
Gonads expressing six4-eGFP::Moe and His2Av::mRFP were imaged live from the end of assembly through compaction. six4-eGFP::Moe allowed precise definition of hub-GSC interfaces, while Metaphase, Anaphase and Telophase figures were identified by His::RFP. The graph presents the 'deviation from expected’ in degrees for both GSCs and non-GSCs. GSCs were defined as germ cells in contact with the hub. Germ cells more than 2 tiers away from the hub were scored as non-GSCs. For Metaphase, an idealized division plane of a GSC would generate a Metaphase plate perfectly parallel to the hub-GSC interface. Using Image J, straight lines were drawn along the hub-GSC interface, as well as along the metaphase plate. We then calculated the deviation from parallel, with ‘no deviation’ equaling zero degrees for the graph. For Anaphase and Telophase, an idealized GSC division plane would be orthogonal to the hub-GSC interface. Thus, we drew a line along the division plane, and plotted the absolute value of deviation from ninety degrees; thus, no deviation would equal zero degrees on the graph.
Measurement of niche area and circularity
To measure hub area, we took a Z-stack image through the entire hub and generated a Z-projection. We rotated the Z-projection to view the anterior niche face, and used Image J to determine niche area within the projection borders. To calculate niche circularity we measured the circumference of the region occupied by the niche, and compared it to the circumference of a perfect circle with the same area to generate a ratio. Values approaching 1 are more circular.
Please see the KRT Table.
KEY RESOURCES TABLE
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti Vasa | R. Lehmann | N/A |
| Goat polyclonal anti Vasa | Santa Cruz | Cat#dC-13 (discontinued) |
| Rabbit polyclonal anti STAT92E | E. Bach | N/A |
| Rabbit polyclonal anti RFP | Abcam | Cat#ab62341 |
| Mouse monoclonal anti Fasciclin III | Developmental Studies Hybridoma Bank (DSHB) | Cat#7G10 |
| Mouse monoclonal anti Islet | DSHB | Cat#40.3A4 |
| Mouse monoclonal anti Crumbs | DSHB | Cat#cq4 |
| Rat monoclonal anti DE-cadherin | DSHB | Cat#DCAD2 |
| Guinea pig polyclonal anti Traffic jam | D. Godt | N/A |
| Chick polyclonal anti GFP | Aves Labs | Cat#GFP-1020 |
| Normal Donkey Serum (NDS) | Jackson Immunoresearch Laboratories | 017-000-121 |
| Alexafluor Secondary Antibodies (488, 647) | Molecular Probes | |
| Cy3 Affinipure Secondary Antibodies | Jackson ImmunoResearch | |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| s700 halocarbon oil | Sigma | Cat#H8898 |
| Poly Lysine | Sigma | Cat#P-1274 |
| FBS | GIBCO | Cat#10082 |
| penicillin/streptomycin | Corning | Cat#30-002-Cl |
| Schneider’s Insect Media | GIBCO | Cat#21720-024 |
| Insulin,Bovine | Sigma | Cat#l0516 |
| Formaldehyde, 16% | Electron Microscopy Sciences | Cat#15710 |
| Formaldehyde, 40% | Electron Microscopy Sciences | Cat#15715-S |
| Hoechst | Sigma | Cat#33342 |
| Phosphatase Inhibitor | Sigma | Cat#04906837001 |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: six4-nls-eGFP | D. Finnegan | Clark et al, 2006 |
| D. melanogaster: P-Dsix4-eGFP::Moesin | R. Lehmann | Sano et la 2012 |
| D. melanogaster: nos-moesin::GFP | R. Lehmann | Sano et al 2005; FBtp0040584 |
| D. melanogaster: sqh-Sqh::mCherry | A. Martin | Mason et al, 2013 |
| D. melanogaster: tupAME-moeGFP | M. Frasch | Boukhatmi et al 2014 |
| D. melanogaster: TypeIV Collagen::GFP | Kyoto Stock Center (KSC)110692 | FBti0153267 |
| D. melanogaster: His2Av::mRFP1 | Bloomington Drosophila Stock Center (BDSC) | FBtp0056035 |
| D. melanogaster: Perlecan::GFP | trolZCL1700, KSC 110807 | FBti0129820 |
| D. melanogaster: abdA-Gal4 | Merabet, Marseille | Hudry et al, 2011 |
| D. melanogaster: Prd-Gal4 | BDSC | FBst0001947 |
| D. melanogaster: tup-Gal4 | BDSC | FBst0046960 |
| D. melanogaster: tupAME-Gal4 | J.-L. Frendo | N/A |
| D. melanogaster: breathless-GAL4 | BDSC | FBst0078328 |
| D. melanogaster: UAS-tdTomato | BDSC | FBst0036328 |
| D. melanogaster: P{UAS-grim.Y}2 | H. Bellen | FBtp0009995 |
| D. melanogaster: mys1 / FM4 | BDSC | FBst0000059 |
| D. melanogaster: trh8 / TM3, twi-Gal4, UAS-GFP | D. Andrew | FBal0050667 |
| D. melanogaster: y1 w*; tup1 P{neoFRT}40A / CyO | BDSC | FBst0036503 |
| D. melanogaster: tupex4 | S. Campuzano | FBal0216723 |
| D. melanogaster: CyO, P{w[+mC]=ActGFP}JMR1 | BDSC | FBst0004533 |
| D. melanogaster: TM3, P{w[+mC]=GAL4-twi.G}2.3, P{UAS-2xEGFP}AH2.3, Sb[1] Ser[1] | BDSC | FBst0006663 |
| Oligonucleotides | ||
| GAA GAA TCC CAG CAA AGA CCG TGA GTT G | Clark et al, 2006 | D-six4 third intron, forward, EcooRI site |
| GTT GGA TCC ATT GCC ATC CAG TTG | Clark et al, 2006 | D-six4 third intron, reverse, BamH1 site |
| Recombinant DNA | ||
| H-GAL4 vector | S. Barolo | Castro et al, 2005; 10.1242/dev.01920 |
| Software and Algorithms | ||
| FIJI (Image J) | www.fiji.sc | N/A |
| Image J | www.imagej.nih.gov/ij/ | N/A |
| Metamorph Microscopy Automation and Image Analysis Software | Leica | v7.8.40 |
| Axio-Vision Imaging Software | Zeiss | v4.8.1 |
| Graphpad Prism | Graphpad Software | v7.0 |
| Other | ||
| Matek imaging dish | Thermofisher | P35G-1.5-14-C |
Results
Hub cells move along the periphery during anterior assembly
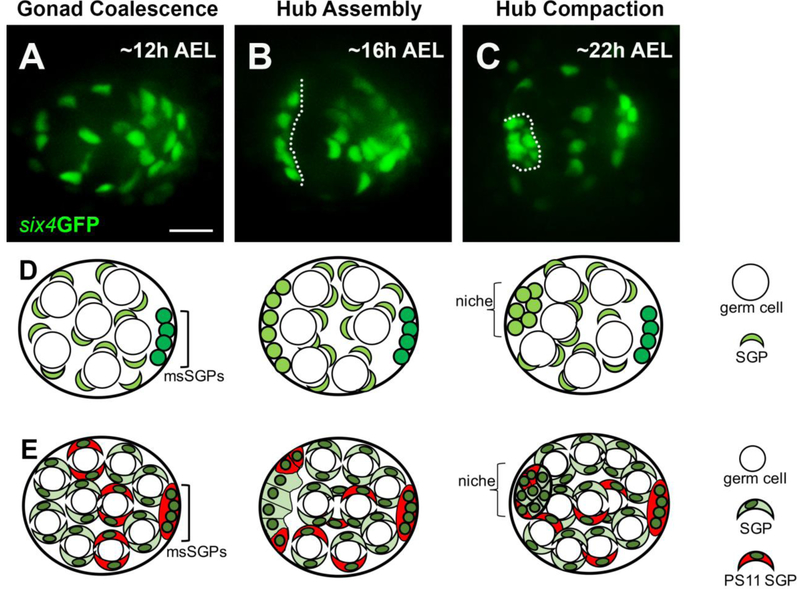
We first sought to visualize niche formation in embryonic gonads using timelapse imaging. Hub markers become visible only after the niche has formed. Therefore, in order to investigate niche morphogenesis, we first needed to determine whether we could image through late gonadogenesis and identify a hub. A construct containing a six4 enhancer driving GFP with a nuclear localization signal (six4-nls-eGFP) marks all SGPs and was previously used to live-image earlier stages of gonadogenesis, which included the coalescence of primordial germ cells and SGPs into a spherical gonad31,32. We used this to image Stage 15 embryos (approximately 12 hours after egg lay; 12h AEL) at five-minute intervals until approximately stage 17 (17h AEL) when the onset of muscle contractions precluded further time-lapse imaging. At the beginning of imaging, SGP nuclei were dispersed and intermingled with germ cells (Fig. 1A,D-E). However, towards the end of imaging, a subset of SGPs had assembled along the periphery, roughly located at the anterior of the gonad (Fig. 1B, outline; Fig. 1D-E, see Movie, S1). Furthermore, there was a tier of negative space between the anteriorly assembled SGPs and the remaining SGPs indicating that germ cells were being recruited to the anterior. Previous analysis of fixed gonads also described an anterior “cap” of E-cadherin-enriched cells marking the prospective hub (pro-hub) visible prior to other markers of hub cell fate14. Our quantification during in vivo imaging revealed that the pro-hub had a mean of about 12 cells (Fig. 2D, n=22 niches). Shortly after these nuclei assembled at the anterior, embryonic muscle contractions began, which precluded further in vivo imaging. Although the hub appeared more compact in gonads dissected from embryos later in embryogenesis (Fig. 1C, outline; 23 hr AEL; Fig. 1D,E), we speculate that the assembled cells at the earlier stage represents the pro-hub based on location and the recruitment of germ cells (see ex vivo imaging, below). Since we could visualize the formation of the pro-hub, we could now interrogate the behavior and requirements for hub assembly.
Figure 1: Live-imaging reveals the stages of hub formation. A – C).
Three stages of niche formation: Gonad Coalescence, Hub Assembly, and Hub Compaction are shown using a selected still from live-imaging of six4GFPnls (green), which marks all SGPs. A) A view of a coalesced gonad, imaged in vivo at 12h AEL, showing that SGPs were relatively dispersed, intermingled with germ cells (large negatively-marked spaces). B) A view of a gonad at Hub Assembly, imaged in vivo at 16h AEL, showing that a subset of SGPs had assembled at the anterior forming the prospective hub (left of dotted line), and had recruited a tier of prospecive germline stem cells (negative space; GSCs). C) A view of a gonad after Hub Compaction, imaged ex vivo at 22 h AEL. D, E) For each of the three stages, two schematics, each at an increasing level of detail, are drawn directly below their respective live image view. The schematics are labeled using the same terms as in the text, and a key is shown along the right hand side. All SGPs, green; msSGPs, bright green. In E), in addition to all SGPs (green), this series marks PS 11 cells and msSGPs (red). Some PS11 cells will assemble with PS10 cells as a pro-niche. Scale bar is 10 microns.
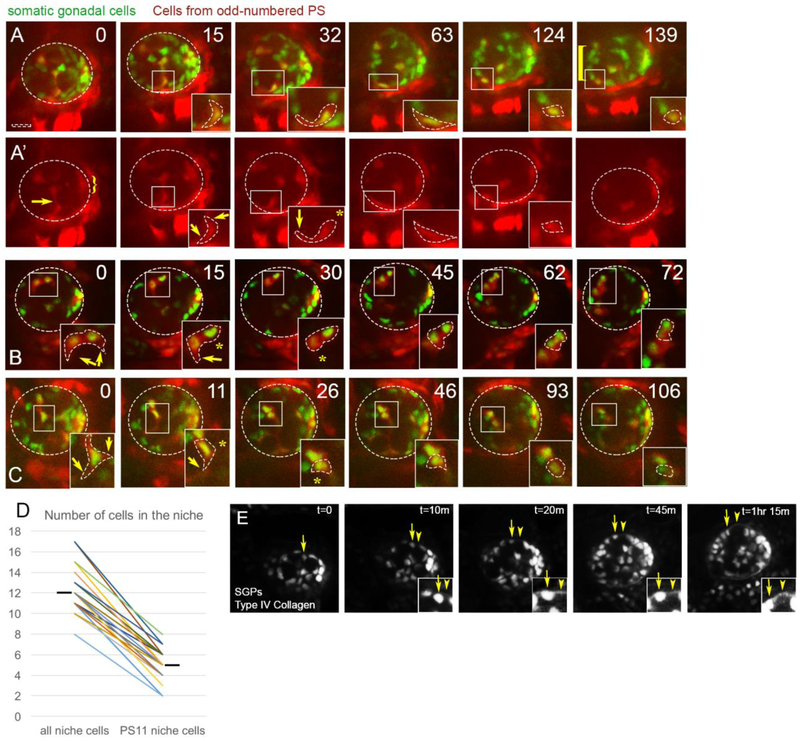
Figure 2: PS11 hub cells migrate anteriorly along the periphery.
Representative stills (time in minutes) from live imaging of six4-nlsGFP marking all SGPs (green) and Prd-GAL4>tdTomato to mark SGPs originating from PS11 (red). Scalebar is 10 microns. A) Merge, and A’) Prd>tdTomato channel alone. At Time 0 before anterior movement of pro-niche cells, tdTomato is expressed only in a central column of SGPs (originating from PS11) and the posterior msSGPs (from PS13, bracket), and not SGPs from even-numbered parasegments. The stills in this time series were from Z-slices at mid-depth within the gonad (13-16 um in from the periphery of the gonad (26.5 um represents full depth in this gonad)). At Time 0 a PS11 hub cell can be seen extending a protrusion towards the gonad periphery, and moving on to it. This same cell also has a trailing extension that is likely wrapped around a germ cell (arrow). As this cell moves to the lower edge of the gonad, it is moving from within the internal milieu of the gonad to the periphery. This cell is followed from time 0 through 139 min, and the boxed region is magnified in the inset, where the cell body is outlined. A trailing extension visible at Time 0 and 15 min, is retracted by 32 min (asterisk), while the anterior extension persists as the cell moves along the gonad periphery (arrow). By 124min, this PS11 cell has joined the anterior assembly of pro-niche cells, and retracted its anterior extension.B) Two PS11 pro-niche cells (inset) that were already located on the gonad periphery at the start of imaging. Both cells migrated anteriorly and ended in assembly (72 min) with a third PS11 pro-niche cell, and two nonPS11 SGPs (expressing six4-nlsGFP, but not Prd>tdTomato). Both PS11 cells intially had cytoplasmic protrusions surrounding a germ cell (0min; arrows) that were subsequently retracted (asterisks, 15 and 30min). (C) The inset initially focuses on one peripheraly located PS11 pro-niche cell, with cytoplasmic protrusions surrounding a germ cell (arrows, 0min) that were retracted (asterisk, 11min) or remodeled (0 – 26min, asterisk) as the cell moved anteriorly. Pro-niche cells tracked in (B) and (C) were visible in the first z-slice containing cells from their region of the gonad; i.e., within 1 micron of the gonad periphery. (D) Quantification of total niche cells and PS11 niche cells in each gonad imaged (n = 22; hashmark, mean). Connectors pair data collected from each gonad. A larger contribution from PS 11 did not always correlate with a larger niche. E) Basement membrane is desposited during hub assembly. Live-imaging of SGPs (six4-nlsGFP) simultaneously with Collagen::GFP to monitor depositon of ECM. A PS11 pro-hub cell moves out to and then along the periphery arrow). ECM was detectable by the time the PS11 cell reached the periphery (t=10m and onward, arrowhead; and insets).
While the niche forms at the anterior, it includes hub cells that are specified more centrally in the gonad. Previous lineage tracing has demonstrated that a fraction of SGPs derived from parasegment PS11 become hub cells, suggesting that they must move or sort anteriorly to assemble with PS10-derived hub cells14,44,56 (Fig. 1E). To visualize the PS11 niche cells, we expressed cytoplasmic tdTomato in cells derived from odd-numbered PS (prd-GAL4), while simultaneously marking all SGPs with nuclear GFP (Fig. 2). At the beginning of imaging, tdTomato was visible in centrally-located PS11 SGPs and posterior PS13 male-specific SGPs (Fig. 2A’, arrow and bracket, respectively). After niche assembly, quantification revealed that between 1/3 to 1/2 of the cells in each niche derived from PS11 (n = 22 gonads, Fig. 2D) demonstrating that we could track their behavior as they migrate to join the hub.
After each time-lapse series, we identified the tdTomato+ cells that joined the pro-hub. These cells were in an aggregate with other six4-nls-eGFP+ nuclei at the anterior of the gonad (Fig. 2A, 139min, square bracket). We then traced the path that PS11 cells took to reach the pro-hub by stepping back earlier in the series. These cells travelled short distances, from 15 −20 μm, in order to assemble with anterior, PS10-derived pro-hub cells (Fig 2, arrows). Prior to assembly at the anterior, PS11 pro-niche cells had cellular extensions that encysted nearby germ cells, as has been described generally for SGPs at this stage of gonadogenesis33 (Fig. 2A’, arrow, 0 min; see Movie S2). However, the PS11 pro-hub cells retracted their trailing extension as they moved anteriorly (Fig. 2A', compare 15 to 32 min panel, arrows to arrow and asterisk, respectively). Other examples of PS 11 cellular extensions that were remodeled during cell movement are shown (Fig. 2B, 0 to 30 min; 2C, 0 to 30 min; arrows and asterisks).
By the assembly stage, the pro-niche cells were located as a cap on the periphery of the gonad. In principle, PS11 pro-niche cells could have traveled through the internal milieu of the gonad to a peripheral point of assembly, or these cells might first move out to the gonad periphery before traveling toward the anterior. The precise path taken by migrating pro-niche cells would give insight into the mechanism that directs this movement. Analyzing the time lapse movies in detail revealed that anterior movement of the pro-niche cells always occurred along the periphery of the gonad and never within the internal milieu (n = 22 gonads). In one example, prior to anterior movement, the cytoplasmic arm of a PS11 pro-niche cell was encysting an unlabeled germ cell (Fig. 2A, 0 min). The pro-niche cell was located in a z-slice representing a central cross section of the gonad. This cell extended a cytoplasmic protrusion toward the lower edge of the cross section, which is at the periphery of the gonadal sphere (Fig. 2A, 15min inset, lower arrow). Once this extension contacted the periphery, the pro-niche cell retracted the trailing cytoplasmic arm that had been in contact with the germ cell it encysted (Fig. 2A, 32min inset, asterisk). This behavior resulted in a PS11 pro-niche cell that was located on the periphery of the gonad (Fig. 2A, 63 min). PS11 cells appeared to begin anterior movement after contacting the periphery, since moving cells were always visible less than 1 micron from the closest peripheral edge of the gonad (Fig. 2A, 124 min; 2B, C). Even though a reasonable fraction of PS11 cells end up in the pro-hub (Fig. 2D), we never observed a large, tight cluster of migrating cells. Instead we observed moving PS11 pro-niche cells strung out along the periphery (Fig. 2B,C). Once all of the PS11 pro-niche cells reached the anterior of the gonad, they aggregated into an elongated cluster with resident PS10 (tdTomato-negative) pro-niche cells. The aggregated pro-niche cells sorted closer to one another than to other SGP nuclei and were separated from the remainder of the SGP nuclei by a tier of negatively marked germ cells, which will likely become GSCs (Fig. 2B,C).
The movement of PS11 cells to and then along the periphery suggested the presence of an extracellular matrix (ECM) at the gonad periphery. It was previously demonstrated that ECM proteins were enriched around late stage embryonic gonads27. Live-imaging now afforded the opportunity to investigate the timing and location of ECM deposition. We used timelapse imaging of a GFP protein trap for Type IV-Collagen (Col::GFP), along with six4-nls-eGFP to determine when ECM is deposited relative to the pro-hub cell movements described above. As the pro-hub cells moved outwards, Col::GFP was detected at the gonad periphery, and the PS11 pro-hub cells were in close association with the ECM throughout their anterior migration (Fig. 2E, t=20m through 1hr 15min). Similar results were obtained imaging Perlecan::GFP (data not shown). These data indicate that ECM is present during the assembly phase, and suggest a mechanism whereby pro-niche cells travel anteriorly (see below and Discussion).
The gonadal niche assembles near a branch of the trachea, and an alary muscle.
The positional bias of niche placement at the gonad anterior might suggest an extra-gonadal cue to direct niche assembly. Therefore, we reasoned that the tissues nearest to where the hub assembles would be good candidates for directing hub assembly. We used live imaging to examine the position of the niche relative to various tissues.
Imaging six4-nls-eGFP in combination with a luminal marker for the trachea, revealed that the niche is located in proximity to the dorsal trunk and a transverse connective of the trachea (Fig S1A). To test whether any signals from the trachea might play a functional role in niche assembly, we examined final niche architecture in fixed tissue from trachealess (trh) mutants50. However, in all cases, niches appeared just as in sibling controls (Fig. S1, D-E), suggesting that while the trachea is in close proximity to the niche, it is not required for its assembly.
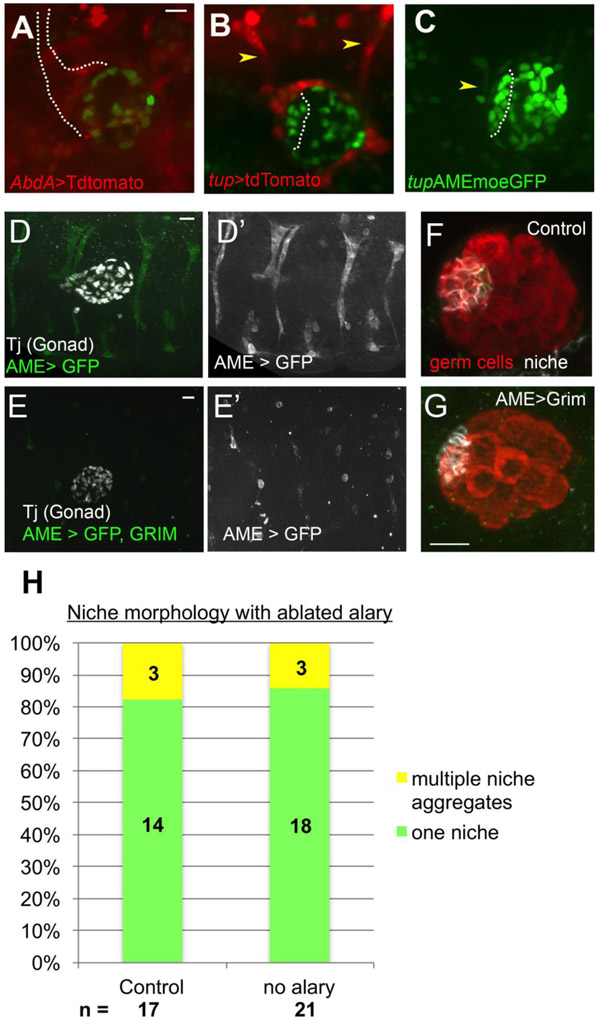
To identify other tissues near the anterior of the gonad, we imaged six4-nls-eGFP with AbdA-GAL4> tdTomato. This highlighted a branched structure just anterior to the gonad that appeared to be an Alary muscle (Fig. 3A). Alary muscles (AMs) are present in each segment, extending from the dorsally located heart, and projecting ventrally along the boundary of the segment57. We confirmed the structure to be an alary muscle by expressing tdTomato using tailup (tup) regulatory sequences, since tup is expressed in Alary muscles47. Using six4-nls-eGFP to mark the gonad, the segment 4/5 Alary muscle (tup>tdTomato) contacted the anterior gonad, near the niche (dotted line), while the segment 5/6 Alary muscle was at the gonad posterior (Fig. 3B). Furthermore, live imaging using an AM-specific enhancer from tup to express the actin-binding domain of Moesin (tupAME-moeGFP47) along with six4-nls-eGFP revealed the juxtaposition of the segment 4/5 alary muscle prior to and throughout hub assembly (Fig. 3C).
Figure 3: An alary muscle is located near the niche.
A) Live-imaging AbdA>tdTomato revealed a Y-shaped structure anterior to assembled hub (dotted line, six4-nlsGFP). B) Tup> tdTomato identified the structure as an alary muscle (AM); one located at the anterior and one at the posterior of the gonad (arrowheads). C) tupAME-moe::GFP specifically marked the AM (arrowhead), which was present as pro-hub cells assembled (outline, six4-nlsGFP). (D, D’) Control embryo carrying tupAME-GAL4>GFP, stained with Traffic jam to mark the gonad. D) A control embryo with GFP under control of the AME driver, revealing the position of the alary muscles. E, E’) Embryo with Alary muscles ablated by tupAME-GAL4>Grim, GFP. Lone GFP+ cells are likely pycnotic, unfused alary muscle precursors. Dissected gonads from control (F) and alary ablated (G) embryos, stained for germ cells (Vasa, red) and hub cells (FIII, white). Ablation did not disrupt niche formation, which was consistently at the gonad anterior, located opposite the msSGPs (not shown in the image). (H) Quantification of niche morphology from control and ablated embryos. Scale bars are 10 microns.
To test for a functional role for AMs in guiding hub cell assembly, we first analyzed tup mutants, which disrupt AM formation47. There were defects in hub organization (Fig. S2), however, tup is expressed in and necessary for development of several other tissues, including cardiac and dorsal muscles47,58,59. To determine more specifically whether the AM was necessary for proper niche assembly, we ablated only the alary muscle by driving expression of the cell death gene, grim60, under control of an alary muscle-specific driver tupAME-Gal4. We validated the expression of this driver in the AMs, and confirmed their successful ablation (Fig. 3D,E). We isolated gonads from embryos expressing tupAME>Grim, or controls without Grim, and immunostained for FasIII to assess niche assembly. Alary-specific ablation did not appear to affect the niche (Fig. 3F,G). In more than 82% of alary muscle ablations, just as in the controls, we observed one niche aggregate at the anterior of the gonad, relative to msSGP position (Control n = 17, Alary ablation n = 21) (Fig. 3H). Thus, the AM is not required for proper niche assembly. Further work is required to identify other tissues potentially responsible for niche formation (see Discussion).
Hub cells assemble with a positional bias
Niche cells assemble as a peripheral cap at the anterior of the gonad14 (Fig. 1B). We were curious as to whether this cap exhibited any consistent positional bias further than this that could indicate a tissue near the gonad that might be directing assembly. Indeed, our live imaging suggested a reproducible, internal “tilt” to the position of the niche on the anterior gonad periphery. To assess the tilt more rigorously, we analyzed multiple time series, comparing the approximate position of the assembled niche to the full arc of the gonad (see Materials and Methods). At the time of assembly, the gonad is a coalesced sphere14,33. While stepping through z-slices from the external face of the sphere to the internal, we noted which z-slices included the assembled niche, and normalized those slices onto a scale from 0 (external face) to 1 (internal face) (see Movie S3). Niches that occupied an equivalent number of z-slices on either side of 0.5 are directly centered. Niches that occupied more space between 0.5 and 1, than between 0 and 0.5, were tilted internally.
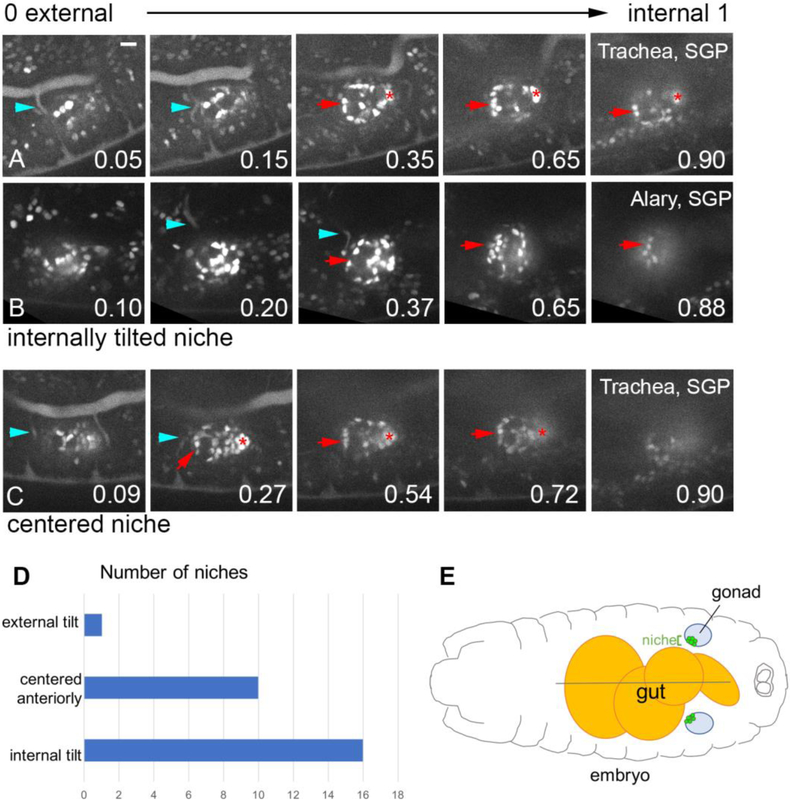
Such quantitation confirmed that the niche assembled with a slight internal tilt relative the embryo exterior-interior axis. For example, Fig. 4A shows 5 z-slices through a gonad with the first, most external slice at position 0.05, and z-slices representing fractional positions 0.15, 0.35, 0.65 and 0.90 as one steps internally. In these panels, niche cells appeared from position 0.35, continuing to remain in view even at position 0.90. While we show only select slices that included this niche, in this instance the niche occupied fractional positions 0.27-1 (Fig. 4A). Because the majority of the slices occupied by the niche occupied more internal-facing regions of the gonad, this niche was scored as internally tilted. An additional example of an internally tilted niche, and an anteriorly centered niche, is also shown (Fig. 4B,C). Our quantification revealed that 59% of niches assembled with an internal tilt, 37% of niches were directly centered at the anterior of the gonad, and only 4% tilted externally (Fig. 4D, n = 27). These results indicate a positional bias of niche assembly, with a location facing anterior and frequently toward internal regions of the embryo (Fig. 4D).
Figure 4: The hub often adopts an internal tilt, relative to A-P axis of embryo.
A, B, C)The final time point from a movie visualizing niche assembly (six4-nlsGFP). Each sequence shows Z slices starting from the external region of the gonad, and moving more internally, with the fractional position along the external-internal gonad axis indicated (0, most external; 1, most internal). Niche cells (arrow), msSGPs (asterisk), and either trachea (A, C; breathless>GFP, blue arrowhead), or the Alary (C; tupAME-GFP, blue arrowhead). Scale bar is 10 microns. D) Frequency distribution of the tilt of the niche. E) Schematic of an embryo showing a dorsal view of the gut, the gonads, and an internally tilted niche in each gonad.
Once we recognized the internal tilt of the assembled niche, we mapped the trachea and 4/5 Alary muscle along this external-internal axis of the gonad. Given the internal tilt of the niche, neither the trachea nor 4/5 Alary muscle overlapped substantially with it (Table S1), consistent with the finding of no functional relationship between either tissue and the position of the niche.
We developed a protocol to live image late-stage niche morphogenesis ex vivo
Our in vivo imaging of gonadogenesis could proceed only up until the onset of muscle contractions. By this stage, the prospective hub cells had assembled to a peripheral, anterior and internal position on the gonad, but the niche had not compacted into a tighter aggregate (Fig1, compare B and C). Given that pro-niche cells had already assembled at the anterior and a basement membrane had been deposited the gonad, we hypothesized that gonads could be cultured at late stages ex vivo. Therefore, we developed a protocol for dissecting out gonads from early stage 17 embryos, then culturing and imaging niche morphogenesis (see Materials and Methods). To verify the physiological relevance of hub formation ex vivo, we compared resultant niches to those formed in vivo using markers of hub cell fate, hub cell number and hub-GSC signaling.
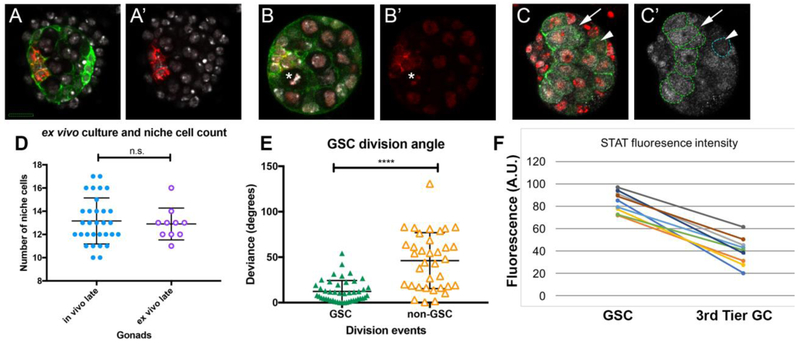
As described previously, hub cells accumulate the adhesion proteins FasIII and ECad. Gonads at the assembly stage were dissected from embryos, cultured for 5 hours and then fixed and immunostained. Following ex vivo culture, the niche cells accumulated FasIII, which is hub cell-specific (Fig. 5A), and ECad, which is enriched on hub cells relative to other SGPs (Fig. 5B). We next wished to test whether the number of niche cells in ex vivo cultured gonads was comparable to the number of niche cells in gonads that developed in vivo. To do so, we cultured gonads ex vivo and then fixed and immunostained to count the number of hub cells. In parallel, we counted hub cell number in gonads that were allowed to develop in vivo up until an age comparable to the end of our ex vivo culture. We found no difference in the number of FasIII+ niche cells (Fig. 5D mean hub cell number = 12; n=30 gonads in vivo, n = 10 ex vivo).
Figure 5: ex vivo culture enables analysis of niche compaction.
A, A’) Gonad expressing six4-Moe-GFP to visualze F-actin enrichment at SGP cell cortices (green), cultured ex vivo for ~5 hours, then post-fixed and stained for the niche (FasIII, red), and nuclei (Hoechst, white). FasIII accumulated properly at niche cell boundaries (not all boundaries are visible in this section). Hub cells and, especially, msSGPs at posterior of gonad accumulate higher levels of F-actin than other SGPs. B, B’) Gonad from an embryo expressing six4-Moe-GFP (green) and HistoneRFP (red), cultured ex vivo for ~5 hours, then post-fixed and stained for ECad (red), and Hoechst (white), also revealing nuclei. E-cad accumulated as expected among niche boundaries (red). Note the metaphase plate of a GSC (asterisk); a division plane perpendicular to the hub-GSC interface, as expected. C, C’) Gonad expressing nos-Moe-GFP to visualize F-actin in germ cells (green), cultured ex vivo for ~5 hours, then post-fixed and stained for STAT92E (white), and nuclei (Hoechst, white), nos-Moe-GFP outlines all germ cells. A GSC (arrow) and a non-GSC (arrowhead, blue dotted line) are marked. Germ cells in contact with the niche in this slice are outlineed with a green dotted line, and represent GSCs. D) Niche cell number (FasIII) per gonad (circles), comparing cases where the niche developed fully in vivo (dissected from late-stage embryos, ~23h AEL, blue closed circles), to gonads cultured ex vivo until the compact architecture was achieved (purple open circles). E) For GSCs (n=46, green closed triangles) and non GSCs (n=35, orange open triangles), the graph plots the deviation of each division from an idealized axis perpendicular to the niche-GSC boundary. F) Average STAT accumualtion (arbirtrary units) in ex vivo culture, comparing the GSC tier to germ cells removed from the hub/GSC boundary (3rd tier germ cells). Connectors report values from the same gonad. In D,E bars represent mean and standard deviation; n.s., not significant; ****, p < 0.0001. For A-C, scale bar is 10 microns.
A properly functioning hub should activate the STAT signaling pathway in the adjacent tier of germline cells16,18,61,62. Indeed, after ex vivo culturing, fixation and immunostaining for STAT, there was a two- to three-fold higher level of pathway activation in germ cells contacting the hub, compared to more distal germ cells (Fig. 5C,E).
As a final test of a properly constructed hub, we assayed for oriented GSC divisions, as these cells should divide along a plane perpendicular to the interface with the hub24,63. We quantified the division axis during ex vivo imaging by measuring the division angle relative to the hub interface (Fig. 5E). For comparison, we also quantified the axis for germ cells farther away from the developing niche. These data showed that GSC divisions were oriented appropriately in ex vivo culture, as the average deviation from perpendicular was minimal (12 degrees), with the bulk of divisions occurring within 30 degrees (n = 46). In contrast, non-GSC germline cells divided at angles that deviated from perpendicular much more substantially than GSCs. There was also a much greater spread of division angles for non-GSC germline cells, with standard deviation of GSC division angles at 12 degrees and that of non-GSCs at 31 degrees (Fig. 5E).
Collectively, the characteristics of adhesion, STAT activation and oriented GSC divisions suggest strongly that development of the niche in explant culture proceeds normally. Therefore ex vivo imaging should allow insight into the changes that take place as the loosely assembled pro-hub transitions to its final, more compact state14 (Fig. 1B, C).
Hub cells arrange into a smaller aggregate with smooth boundaries after assembling at the anterior
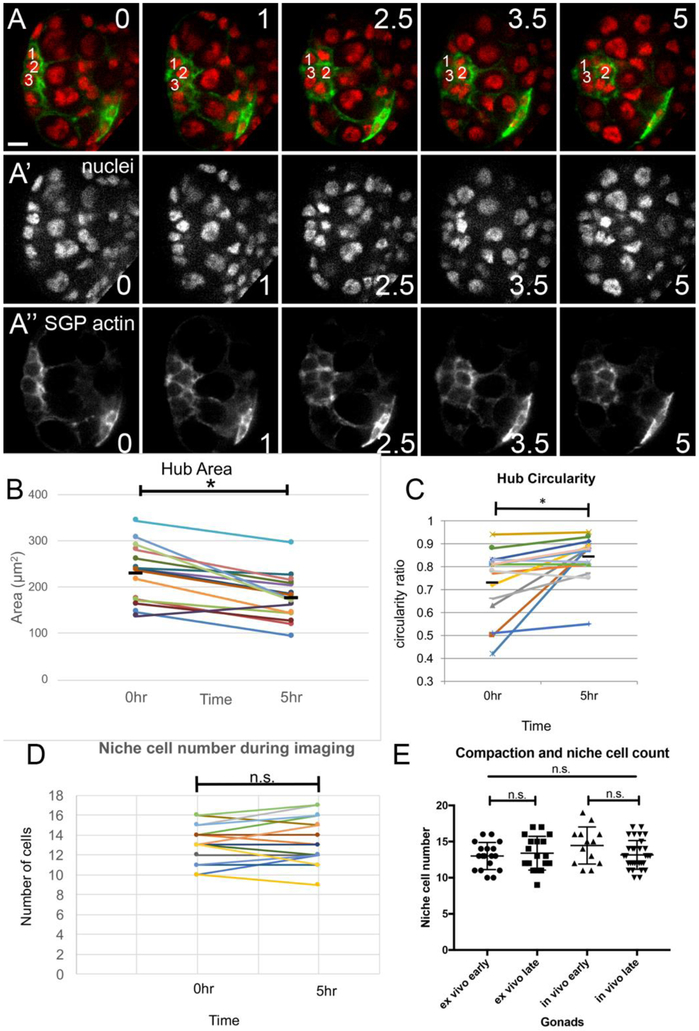
After ex vivo imaging it appeared that the hub covered less area, and that a pronounced smoothening of the boundary between the hub and its recruited GSCs had occurred (Fig. 6A; see Movie S4). To quantify these parameters, we first measured the area occupied by the hub (see Materials and Methods) and found that compaction reduced area to on average about 78% of its initial value (n = 15, Fig. 6B). Comparing the initial and final values for each gonad showed that virtually all gonads exhibited an area reduction (Fig. 6B, connection lines). Next, to quantify the smoothening of the niche boundary during compaction, we calculated a ‘circularity parameter’ (see Materials & Methods) from measurements of the boundary between the niche and the surrounding tier of GSCs, in gonads both before and after compaction. We found that on average the boundary became 11.2% more circular after compaction (Fig. 6C, n=15), with about half of the gonads exhibiting fairly substantial smoothening (Fig. 6C, connection lines). In principle, compaction of the hub could mean some pro-hub cells are lost between the assembly stage and the niche assuming its final shape. Alternatively, compaction could reflect cell biological changes among the pro-hub cells such that the same number of cells covered a smaller outward-facing area of the gonadal sphere. To discriminate between these possibilities, we imaged gonads ex vivo, using Histone-RFP (HisRFP) to mark all nuclei and six4 to drive the actin-binding domain of Moesin fused to GFP (six4-eGFP::Moesin) to outline all SGPs. We counted the number of niche cells at assembly before compaction was underway, and then in these same gonads once compaction was complete. The number of niche cells before and after compaction was virtually identical, with a mean of 13 in both cases (Fig. 6D, E). To verify that the number of niche cells also remained unchanged when compaction occurred in vivo, we performed an additional experiment in which we dissected gonads prior to niche compaction (~16h AEL), and after compaction (~23h AEL) and counted the number of niche cells in each case. Again, there was no significant difference in the number of niche cells counted in gonads prior to compaction versus after compaction was completed (Fig. 6E). We noted in tracking niche cell number ex vivo that there were small fluctuations up and down (Fig. 6D). Given that there was no consistent decrease in number, the fluctuations cannot mechanistically account for the observed decrease in hub area during compaction. Instead, the fluctuations likely reflect dynamic cell behavior that we observe during this process (see next).
Figure 6: After compaction the hub occupies a smaller area and forms smoother boundaries facing the GSC tier.
A) ex vivo culture during compaction, showing all nuclei (HistoneRFP, red; single channel, A’), and F-actin in SGPs (six4-moesin-GFP; green, single channel, A”). The prospective hub is outlined, with three, cells labeled that were tracked. These cells exchanged neighbor relationships during compaction (t=0 through t=2.5hr). msSGPs, which exhibit higher F-actin, are labled (asterisk). Scale bar is 10 microns. B - D) Gonads were cultured ex vivo, and measured before Compaction (time =0) and after Compaction was completed (5hr), with connectors reporting values from the same gonad. B) Hub area showed a significant decrease (p < 0.05). C) The hub-GSC boundary became significantly smoother, reflected in increased circularity. Several niches exhibited fairly substantial changes. D) Niche cell number remained relatively constant during compaction, while area was decreasing and circularity was increasing. E) A direct comparison revealed no differences to niche cell number between ex vivo culture and in vivo development. Ex vivo early and late are the 0 and 5hr time points from (D). The in vivo samples are from gonads that were dissected and immediately fixed and stained, either early before compaction (~16h AEL), or late, after compaction was completed (~23h AEL).
Cells exchange neighbors and there is a burst of GSC divisions during hub compaction
Compaction was evident by the decrease in negative space between hub cell nuclei and tightening of encircling germ cells (Fig. 6A). Our timelapse imaging revealed cell behaviors that play a role in hub compaction. First, cells exchange neighbors during compaction. At assembly, the pro-hub cells were in a more elongated configuration, but by compaction they had converged into a disc-shaped (Fig. 6A, compare time 0 to 5hrs). In the series shown, one pro-niche cell shifted to the right, while its neighbor cells above and below, which were initially not in contact, now converged (Fig. 6A, numbered cells, compare 0hr to 2.5hr). By the end of this series, the pro-niche cell that had shifted to the right had also ingressed, such that it was directly internal to pro-niche cells on the gonad periphery (data not shown). A complete analysis of these neighbor exchanges is underway, as the movements are complex (see Discussion).
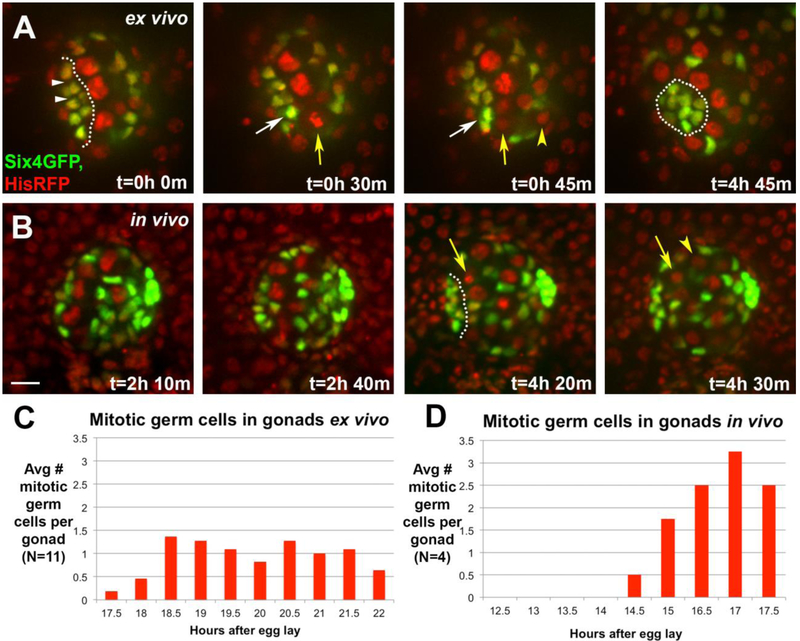
The second cell behavior associated with compaction was a burst of GSC divisions. The division axis was oriented perpendicular to the GSC-niche cell interface (see Fig. 5E, and below). Such an orientation could direct force on adjacent niche cells. To quantify the GSC divisions more rigorously, we imaged six4-nls-eGFP to mark all SGP nuclei along with HisRFP to mark all nuclei, including germ cells (Fig. 7). Germ cells in mitosis were identified by chromosome condensation and segregation revealed by HisRFP. GSCs contacting the assembled hub divided orthogonal to the GSC-hub interface as expected (Fig. 7A, t=30min, 45min, yellow arrow and arrowhead). Monitoring six4-nls-eGFP revealed that these GSC divisions often resulted in displacement of the nearest hub cell away from the GSC and towards the other hub cells (Fig. 7A, t=30min, 45min, white arrow). At the end of compaction, hub cell nuclei were tightly aggregated and germ cells were found in a radial array around the aggregate (Fig. 7A, t=4h 45min). We quantified GSC mitoses during ex vivo imaging of compaction and found an average of 1 mitotic GSC for every 30 minutes of imaging (Fig. 7C).
Figure 7: There is a burst in germline divisions during hub compaction.
A) An ex vivo timelapse, monitoring SGPs (six4-nlsGFP) and all nuclei (HistoneRFP). At t=0, the pro-hub is just passed initial assembly, and covers an extended region (outline). Its nuclei are separated by negative space (white arrowheads). A GSC enters metaphase (t=30min, yellow arrow) and then anaphase (t=45min, arrow and arrowhead mark daughters), with division orthogonal to the hub-GSC interface. Coincident with anaphase, the hub cell nucleus (white arrow) nearest the dividing GSC, moves closer to its neighbor (compare internuclear distance at t=30 and 45min). By the end of the timelapse, the hub has compacted with less negative space among hub cell nuclei (t=4h 45min, outline). B) An in vivo timelapse of a gonad monitoring SGPs (six4-nlsGFP) and all nuclei (HistoneRFP). The initial panels show a stage just prior to and at assembly (t=2h 10min – 2h 40min). After assembly, a GSC divided orthogonal to the hub (t=4h 20min – 4h 30min, arrow; daughter cell marked by arrowhead). For A-B, scale bar is 10 microns. C, D) Quantification of mitotic germ cells by stage, comparing gonads imaged ex vivo (C) and in vivo (D). Compaction begins ~16h AEL, and is completed by ~23h AEL.
To determine if GSC divisions also correlated with compaction in vivo, we returned to imaging gonads in intact embryos. We quantified germ cell divisions using HisRFP, and related this to hub assembly using six4-nls-eGFP. Primordial germ cells (PGCs) are arrested in the cell cycle during their migration to the gonadal soma, but they reenter the cell cycle during gonadogenesis due to Jak/STAT signaling45,64. As expected, prior to assembly mitotic germ cells were not observed (Fig. 7B, t=2h 10min – 2h 40min panels). However, 3-4 hours into imaging (the approximate equivalent of 16-17h AEL), mitotic germ cells were visible at every timepoint (10 min intervals) and germ cells contacting the assembled hub divided orthogonal to it (Fig. 7B, t=4h 20min – 4h 30min panels). Quantification of mitotic germ cells in vivo demonstrated that germ cells resumed dividing concurrently with the end of hub assembly and beginning of hub compaction (Fig. 7D).Taken together, these data suggest a possible role for oriented GSC divisions in hub compaction (see Discussion). Due to muscle contractions we could not image longer in vivo, but ex vivo imaging showed that divisions continued for up to five hours, through the compaction phase (Fig. 7C). Although the apparent rate of divisions appeared higher during in vivo compared to ex vivo imaging, whether the differential might be caused by the ex vivo conditions is unclear as, by necessity, there was an overlap of only one, half-hour time point between the two types of imaging.
Integrins are required to assemble the stem cell niche
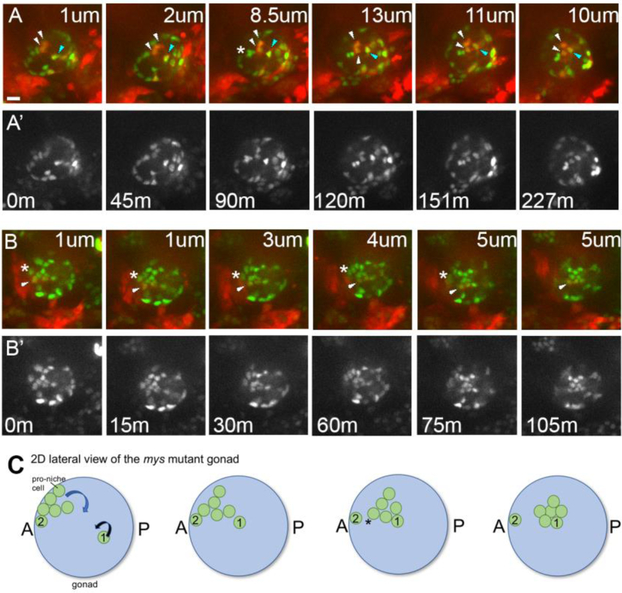
Previous work showed that Integrin-matrix interactions were essential in anchoring the formed niche to the anterior of the gonad and the testis27,28. We wished to determine if live-imaging could more precisely define the role of Integrins during niche morphogenesis, and specifically ask if there might be a requirement at the earlier steps that live-imaging had now identified. In previous work with embryos mutant for myospheroid (mys), which lacks the βPS integrin subunit essential to integrin function65, the niche was located internally, at the center of the gonad sphere27. That work had to rely on fixed samples, and a sensible conclusion was that a compact niche formed normally at the anterior of the gonad, but due to lack of stable ECM attachment, it then sorted internally, to the center of the gonad27. We imaged hub assembly in vivo in mys1 mutants using six4-nls-eGFP, and prd>tdTomato. As expected from the previous endpoint analysis27, at the end of imaging, mys mutants contained a tight aggregate of internalized hub cells rather than one anchored at the periphery (Fig. 8A; t=227min panel). However, by tracking the behavior of 18 cells in six different mys mutant gonads, it was evident that defects became apparent well before compaction, as pro-niche cells began to leave the periphery of the gonad before being fully compacted (Figure 8A, B show representative examples, as described next).
Figure 8: Integrin mutants exhibit defects in niche morphogenesis.
A-B) In intergrin mutants niche cells begin assembly, with most cells relocating to the anterior periphery of the gonad, but then dropping internally. A, B) Selected stills from two different in vivo time series of myospheroid mutants, imaged to follow all SGPs (six4nlsGFP), and PS11 SGPs (prd>tdTomato). A, B, merge; A’, B’ single six4nlsGFP channel. Each frame is taken at a different depth into the gonadal sphere, with the distance in from the periphery marked for each panel in microns (um; minutes, m).Scale bar is 10 microns. A) t=0, an assembly phase gonad, with the pro-hub consisting of a mix of PS10 cells (green only, 5 cells visible) and PS11 cells (red+green, 2 visible). Within 45 minutes, the assembled niche began to internalize before completing compaction. Over the next 2 hours, two PS11 SGPs (white arrowheads) dropped inwards; note the section depth changing from 1 to 13 um. Simultaneously, another PS11 SGP (blue arrowhead), which had not assembled at the anterior, migrated in and joined the internalizing niche. Lastly, between 45min and 90 min three orignially assembled PS10 SGPs separated (asterisk), with two internalizing and one remaining peripheral. B) Shows a partial, face-on view of the niche, just after initial assembly. A PS11 SGP was initally on the periphery of the gonad, but was internalized as the niche dropped inwards (arrowhead). Similar to the case shown in ‘A’, internalization began before compaction was completed. Note also that an adjacent PS10 SGP that remained on the gonad periphery (asterisk) and did not internalize with the rest of the pro-niche cells. C) A schematic of a mys mutant gonad depicting various defects observed in live imaging of panels A and B. In this lateral, 2D view, only relevant proniche cells are shown. Of seven pro-niche cells (green), 6 have asembled at the anterior. These cells are all on the gonad periphery (left panel), which is difficult to convey in 2D. Five of these cells leave the periphery and move internally over time (curved blue arrow), similar to the case shown in panels A-B. The cluster adopts a more compact form only after it internalizes (i.e., by the fourth frame). Note also that one proniche cell, Cell #1, never migrated to the anterior. Instead, it migrates into the milieu of the gonad (curved black arrow) from its initially more posterior position, and joins the five internalizing niche cells (frames 2 -4). This behavior is similar to the the cell marked by the blue arrowhead in ‘A’. Finally, of the six cells that assembled at the anteior only five leave the gonad periphery. Cell #2 is left behind at the gonad periphery during internalization of the niche. This behavior is similar to the cell marked by an asterisk in ‘B’.
In Figure 8A, the pro-niche cells have dropped internally by t=90m. Comparing the architecture of the pro-niche cell aggregates at the early time point to the cluster at the end of the time series suggests that compaction of the niche was not complete when internalization of the niche began. In fact, it appeared that pro-niche cells were only loosely associated at the time of internalization. Fig. 8A, t=45m shows that two PS11 pro-niche cells dropped inwards first (white arrowheads), and were only later followed by two PS10 pro-niche cells (asterisk, t=90m). Furthermore, in some instances, cells that were initially part of the pro-niche assembly did not internalize with the rest of the niche (Fig. 8B, follow the asterisk). Thus, pro-niche cells internalized before compaction was completed in mys mutants (Fig. 8B).
To determine whether earlier steps of niche formation were disrupted in mys mutants, we asked if pro-niche cells from PS11 were able to reach the anterior periphery of the gonad. Indeed, an anterior cap of niche cells began to assemble in mys mutants (Fig. 8A,B t=0min), including some PS11 cells as judged by prd>tdTomato (Fig. 8A,B, white arrowheads). Thus, in the absence of βPS Integrin, some PS11 cells could still arrive at the gonad anterior. However, in other cases, PS11 cells had not completed movement towards the anterior of the gonad before the niche dropped internally. For instance, a PS11 pro-niche cell that had not assembled at the anterior with other proniche cells (Figure 8A, t=0min, blue arrowhead) later joined an internalizing niche, but entered from its more posterior, PS11 location (Figure 8A, t= 45m through 227min, blue arrowhead). Together, these imaging data show that that Integrin function is not simply necessary to anchor the compacted hub at the anterior, but rather is also important for proper migration of some pro-niche cells during assembly.
Discussion
This work is the first to characterize the dynamics of testis niche morphogenesis. Through live-imaging both in vivo and ex vivo, we are able to follow pro-niche cells as they assemble and assume the final compact form of this niche. After ex vivo culture, the niche appears fully functional, as judged by enrichment of adhesion proteins, the ability to activate STAT in adjacent germline cells, and to direct the now GSCs to divide orthogonally to the niche, just as they would in situ. Collectively, our imaging has generated several novel insights on niche morphogenesis that could not be inferred from fixed images alone. The compaction phase correlates with cell neighbor exchange among the assembled pro-niche cells, as well as a burst of divisions among newly recruited stem cells. Before compaction occurs, an assembly phase involves the movement of PS11 pro-niche cells along the outer periphery of the gonad, using the ECM to assemble at the anterior of the gonad. There they join PS10 pro-niche cells, forming an anterior and internally-facing cap. Finally, live-imaging in integrin mutants allows us to define the role of pro-niche cell-ECM interaction more precisely with regard to the new assembly and compaction dynamics revealed here.
Hub compaction
Previously, analysis of fixed samples nicely inferred a change from an initial cap of pro-hub cells to a compact niche14. Our ex vivo imaging now visualizes compaction directly, and by comparing cell numbers at assembly and after compaction, shows that the initial assemblage of the elongate cap of cells indeed constitutes the pro-hub. Scoring the same gonad before and after compaction showed little, if any, shuffling in the number of pro-niche cells. This finding is consistent with the fact that specification of niche cells occurs much earlier in development, before the stages imaged here42,43 Lastly, our work suggests two cell biological phenomena that correlate with the process of compaction.
Firstly, while compaction is underway, there is a burst of GSC divisions orthogonal to the hub. Interestingly, at late anaphase, the niche cells adjacent to a dividing GSC occasionally appeared to become compressed against their niche cell neighbors, subsequently occupying less space. It is known that activation of the STAT pathway is necessary to induce germline divisions in the (male) gonad45. Since the ligand for pathway activation is secreted by hub cells, this suggests a possible scenario where niche cells orient GSC divisions, which subsequently generate some of the force necessary for niche compaction, pushing hub cells closer together. In principle, this scenario can now be tested by attempting to block GSC divisions during ex vivo culture.
Secondly, hub compaction occurs concurrently with cell neighbor exchanges. These exchanges are reminiscent of convergence and extension66, where directed junctional re-organization changes the overall shape of the sheet of cells67,68. For example, at assembly, an individual cell within the elongate cap will shrink one of its boundaries, allowing cells that were previously on either side of this cell to form new contacts adhering to one another. Such events may impart an increasingly more compact overall shape to the incipient niche. In addition to lateral neighbor exchange, we observed ingression of some pro-niche cells during compaction. This results in stratification of the niche, where a few internal pro-hub cells now lie more internal to the superficial cap of peripheral hub cells. These lateral and stratifying exchanges are complex, but their comprehensive cataloging, now enabled by ex vivo imaging, may well reveal patterns of cell movements. In turn, those patterns will likely suggest specific circuits to test as regulators of compaction.
The neighbor exchange behavior associated with compaction is reminiscent of cell sorting, where “like” cells will maximize contacts among themselves, while minimizing those with different neighbor types69. Indeed, our imaging describes a significant smoothening of the boundary between the pro-niche and the tier of stem cells, an outcome expected from sorting phenomena. Increased accumulation of adhesion proteins such as ECadherin, NCadherin and Fasciclin III, has been noted at the Assembly stage, although manipulation of these did not appear to grossly affect compaction14. While unidentified factors or significant redundancy among these might drive sorting, the live-imaging parameters established here now enable analysis of the dynamics of the process. Future work might reveal changes in, for example, timing of compaction and smoothening, rather than relying on endpoint analysis alone.
An extra-gonadal cue for hub assembly
The positioning of the niche at the gonad anterior could, in principle, be driven by a gonad-intrinsic mechanism. For example, SGPs across the gonad have different antero-posterior identities, as they originate from three separate parasegments. Thus, they accumulate Hox proteins to differing levels, and AbdB mutant embryos can exhibit defects in hub position14. However, it remains possible that these defects could be caused by broader patterning changes in tissues outside the gonad, rather than reflecting an intrinsic requirement for AbdB. Still, the fact that pro-hub cells are enriched for adhesion proteins from the assembly stage onward is consistent with the idea that sorting driven by homophilic adhesions could be a main driver for niche formation14,27. Nevertheless, two novel observations from our live-imaging strongly suggest the existence of an extra-gonadal influence.
First, while the niche assembles at the anterior, we find that it exhibits a consistent tilt towards the interior of the embryo. This biased placement of the hub towards internal organs suggests that an extra-gonadal cue is influencing niche position. Our live-imaging did reveal tissues located near cells of the assembling proniche: a branch of the trachea, and an alary muscle. The ablations we conducted rule out any major impact from these two tissues on niche formation. However, other tissues, such as the nearby gut and its surrounding musculature, remain to be tested.
Second, the individual movements of PS11 cells to join the PS10 cohort suggests that homotypic adhesions and sorting alone cannot explain the position of assembly. During the stages leading up to those shown here, SGPs exhibit robust cytoplasmic extensions that probe the internal milieu of the newly formed gonad, tightly embracing each other, and germline neighbors32,33. Indeed, our imaging revealed PS11 cellular extensions around germ cells at the onset of assembly. If sorting were the main driver for assembly, these extensions should be sufficient to cause PS11 cells to move through the internal gonad milieu, seek out and coalesce with anteriorly located PS10 SGPs. Instead, PS11 cells move out to the gonad periphery, and retract their trailing extensions. They then move along the periphery towards the anterior. In addition, if PS 11 cells were sorting together one might have expected these pro-niche cells to move as one tight collective, reminiscent of how the somatic border cell cluster migrates through the internal milieu of the Drosophila egg chamber70,71. In contrast, the PS11 cells often move individually or as a string of cells along the periphery. Taken together, PS 11 cell behavior is much more in line with individual cell migration directed by a cue.
Previous work described an ECM visible at the gonad periphery at the end of embryogenesis27 Pigment cells ensheath the gonad36, and are the likely source of this ECM. Indeed, our live-imaging revealed that this ECM is present while PS11 cells are moving anteriorly. Thus, we suspect that the matrix provides traction for movement in response to the posited external guidance cue.
In fact, previous work has suggested a role for ECM interaction, since in the absence of Integrin function, the newly formed hub was located inside the gonadal sphere27. Since that work had to rely on endpoint analysis, it was reasonable to assume that the hub had fully formed at the gonad anterior and then was internalized whole due to lack of Integrin-based adhesion. Instead, our live-imaging of Integrin mutants now reveals that Integrin-ECM interactions are necessary well before compaction, as pro-niche cells can leave the periphery of the gonad before the niche compacts. This is the case for PS10 cells, which can be observed internalizing from the anterior of the gonad before compaction, and for PS11 cells, either after they arrive at the anterior or directly internalizing from their more posterior, PS11 location. Hub compaction occurs in Integrin mutants27, but our imaging reveals this to occur mostly after the niche internalizes in the mutants. The observation that PS11 cells can directly join an internalized cluster suggests that, left to their own, pro-hub cells might sort to inappropriate locations due to preferential adhesion. This implies that Integrin-ECM interactions at the periphery antagonize the sorting tendency of pro-niche cells. If Integrin-ECM interactions during migration are compromised, the resulting testis would exhibit aberrant polarity, as the niche would not be localized at the closed end of the tubule. Although we and others27 have characterized some of the components making up the ECM surrounding the gonad, which of these engage Integrins on the pro-niche cells is not clear.
We also do not yet know the nature of the directional cue read by PS11 cells. Our imaging shows that some but not all PS11 cells successfully arrive at the anterior in Integrin mutants. This suggests that pro-niche cells have the capability to respond to the cue, at least partially, even if they cannot interact with peripheral ECM. Normally, however, since all the PS11 cells that we tracked in wild type gonads migrated to the anterior along the ECM at the periphery of the gonad, these Integrin-ECM interactions serve to ensure reproducible positioning of the niche.
The combination of in vivo and ex vivo imaging presented here overcome previous challenges to understanding how a stem cell niche is formed. Visualizing the dynamics of niche formation has proven powerful in suggesting unanticipated mechanisms for niche morphogenesis in this work. This imaging should now allow for in-depth study of both the process of assembly and compaction into a fully functioning stem cell niche.
Supplementary Material
Figure S1: The Trachea is located near the assembling niche. A) six4-nlsGFP (SGPs) and GFP in the tracheal lumen reveal the assembled hub (dotted outline) located near the dorsal branch (arrowheads) and a transverse connective (arrow) of the trachea. B-C) Wild type, and trh8 mutant embryos are labeled with Crumbs immunostaining, which marks the trachea (arrowheads mark the dorsal trunk, and an arrow marks a transverse connective), as well as other epithelial tissues. C) trh8 mutants lack the trachea. D-F) Gonads dissected from control and trh8 mutant embryos. All gonads exhibited the niche located opposite to the posterior msSGPs (arrows show niche; dotted outline marks msSGPs, which in our hands, accumulated signal in Vasa stains, but remained quite distinguishable from nearby germ cells, as msSGP cells were much smaller; n =10 gonads). Scale bars are 10 microns.
Figure S2. tup mutants have niche morphogenesis defects. (A-B) Gonads (outlined) dissected from control and heteroallelic tup mutant embryos (stage 17, 21-23h AEL) stained with anti-GFP (six4-nlsGFP), anti-Tj to mark SGPs and anti-FasIII to mark hub cells. Scale bar is 10 microns. A-A”) Control gonad exhibiting a single aggregate of hub cells enriched for FasIII (asterisk). Routinely, the niche was located opposite the posteriorly-located msSGPs, which were identified by bright six4-nlsGFP (arrowhead). B-B”) tup mutant gonad exhibiting two separate aggregates of FasIII-enriched hub cells (arrows). Additionally, aggregates were often not located at the anterior relative to posterior msSGPs (arrowhead). C) Quantification of the niche morphology in control and tup mutant gonads. We counted the number of gonads that had one anterior niche aggregate, and the number of gonads with multiple niche aggregates for each genotype.
Movie S1. Niche assembly in vivo imaging of niche assembly. The gonad is visible as an oval-shaped collection of SGPs marked with six4-nls-eGFP. Dotted line denotes gonad boundaries. AbdA>RFP labels nearby tissues in the embryo. The movie begins with SGPs dispersed throughout the gonad; germ cells are negatively marked. As the movie progresses, pro-niche cells collect at the gonad anterior (left). The movie ends with an elongate column of assembled pro-niche cells. Bright six4-nls-eGFP is visible in msSGPs at the gonad posterior (right).
Movie S2. Migration along the gonad periphery in vivo imaging of niche assembly. six4-nls-eGFP marks all SGPs (green), while PS11 and PS 13 SGPs are labelled with prd>tdTomato. Sections presented in the time lapse focus on one PS11 cell (arrow, first frame) at the gonad periphery. The cell initially exhibited cellular extensions along the periphery and also one back into the gonad, the latter stretching over a germ cell (arrowhead). Over time, this SGP moves anteriorly (left) along the periphery of the gonad, by the end of the timelapse the PS11 cell (arrow) has joined PS10 pro-niche cells (green only) forming an elongate, assembled column at the anterior. Note that during movement of the PS11 cell, it retracted the cellular extension that had embarced a germ cell.
Movie S3. Internal tilt of the niche in vivo imaging of an assembled niche. A z-stack taken from the last time point I shown, progressing from the external surface of the embryo, towards internal regions. Z positions are labelled in microns. six4-nls-eGFP labels SGP nuclei in the gonad, as well as some neighboring mesodermal nuclei to lower levels. As the sections progress further into the embryo, the gonad becomes visible in the lower left. The assembly of pro-niche cells only becomes visible in the deeper sections (note section depth). A branch of the trachea (labeled by breathless>GFP) can be observed in more external sectionos the series relative to those in which the pro-niche niche cells were visible.
Movie S4. Niche compaction ex vivo imaging of a gonad undergoing niche compaction. F-actin in SGPs is visualized with six4-eGFP::Moesin. All nuclei are labelled with His2Av::mRFP1. The gonad (dotted outline) is shown in an approximately face-on view; the small-profile cells (dotted outline within the gonad) comprise the compacting niche. The larger cells and nuclei represent prospective GSCs arranged in an arc to the right around the niche (other GSCs are in deeper sections, and not shown). Throughout the time-lapse, the pro-niche cell cluster tightens, occupying less surface area and their nuclei becoming closer to one another. Note also that the boundary between the niche and the surrounding GSCs becomes smoother and more circular (see dotted outline, final panel). Finally, note the reorganization over time of cortical f-actin in pro-niche cells. Early in the time series, f-actin is present along all interfaces of pro-niche cells, but adopts a more asymmetric appearance, exhibiting a relative increase on hub cell-GSC interfaces, and relative decrease on niche-niche cell interfaces.
Table S1. The Niche overlaps with other organs along the external-internal arc of the gonad. We examined the external to internal axis of the gonad, relative to the anterior-posterior axis of the embryo (see Materials and Methods). Along the external-internal gonad axis, we measured the size of the gonad (in microns), and the space occupied by the niche (in microns). We also evaluated where, along this axis, the trachea and the alary muscle overlapped with the niche. We indicate the percentage of the niche space that overlaps with either tissue as the Portion of niche overlap.
Highlights.
3 to 5 bullet points (maximum 85 characters, including spaces, per bullet point).
Assembling pro-niche cells move along the gonad peripheral extra-cellular matrix
The niche assembles with a biased tilt towards the interior of the embryo
We developed a physiologically relevant ex vivo method to analyze compaction
Niche compaction is accompanied by cell rearrangement and a burst of GSC divisions
ECM is required to anchor pro-niche cells during anterior migration and assembly
Acknowledgements
We thank the fly community for their generosity with reagents, especially Dorothea Godt, Ruth Lehmann, Manfred Frasch, Erika Bach, Samir Merabet, Jean-Louis Frendo, and Deborah Andrew. We also thank the Bloomington Stock Center and the Developmental Studies Hybridoma Bank, and especially Andrea Stout, director of the CDB Microscopy Core, for expert advice on live imaging. We are grateful to Mark Van Doren, the DiNardo, Ghabrial, Little and Capelson laboratories for insightful discussions. We also would like to thank an anonymous reviewer for helpful suggestions that improved the manuscript. This work was supported by the NIH training grant in Developmental Biology T32HD007516 to L.W., GM125123 to L.A., and NIH R01 GM60804 and R33 AG047915 to S.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Morrison SJ & Spradling AC Stem Cells and Niches : Mechanisms That Promote Stem Cell Maintenance throughout Life. Cell 132, 598–611 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan KS et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc. from Natl. Acad. Sci. 109, 466–471 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeda N et al. Interconversion between intestinal stem cell populations in distinct niches. Science 334, 1420–4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calvi LM et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 425, 841–846 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Sasaki N, Sachs N, Wiebrands K, Ellenbroek SIJ & Fumagalli A Reg4 + deep crypt secretory cells function as epithelial niche for Lgr5 + stem cells in colon. Proc. from Natl. Acad. Sci E5399–E5407 (2016). doi: 10.1073/pnas.1607327113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasaki N, Sato T & Clevers H in Biology and Engineering of Stem Cell Niches (eds. Vishwakarma A & Karp JM) 111–125 (Academic Press, 2017). [Google Scholar]
- 7.Mitroulis I et al. Secreted protein Del-1 regulates myelopoiesis in the hematopoietic stem cell niche. J. Clin. Invest. 127, 3624–3639 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjornsson CS, Apostolopoulou M, Tian Y & Temple S It Takes a Village : Constructing the Neurogenic Niche. Dev. Cell 32, 435–446 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dzierzak E & Speck NA Of lineage and legacy : the development of mammalian hematopoietic stem cells. Nat. Immunol. 9, 129–136 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ritsma L et al. Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature 507, 362–365 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cetera M, Leybova L, Joyce B & Devenport D Counter-rotational cell flows drive morphological and cell fate asymmetries in mammalian hair follicles. Nat. Cell Biol. 20, 541–552 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heitman N, Saxena N & Rendl M Advancing insights into stem cell niche complexities with next-generation technologies. Curr. Opin. Cell Biol. 55, 87–95 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonczy P, Viswanathan S & Dinardo S Probing spermatogenesis in Drosophila with P-element enhancer detectors. Development 98, 89–98 (1992). [DOI] [PubMed] [Google Scholar]
- 14.Le Bras S & Van Doren M Development of the male germline stem cell niche in Drosophila. Dev. Biol. 294, 92–103 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Fuller MT in The Development of Drosophila melanogaster (eds. Bate M & Martinez Arias A) (Cold Spring Harbor Labs Press, 1993). [Google Scholar]
- 16.Kiger AA, Jones DL, Schulz C, Rogers MB & Fuller MT Stem cell selfrenewal specified by JAK-STAT activation in response to a support cell cue. Science 294, 2542–5 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Tulina N & Matunis E Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science 294, 2546–9 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Leatherman JL & Dinardo S Germline self-renewal requires cyst stem cells and stat regulates niche adhesion in Drosophila testes. Nat. Cell Biol. 12, 806–811 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawase E, Wong MD, Ding BC & Xie T Gbb / Bmp signaling is essential for maintaining germline stem cells and for repressing bam transcription in the Drosophila testis. Development 131, 1365–1375 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Michel M, Raabe I, Kupinski AP, Pérez-Palencia R & Bökel C Local BMP receptor activation at adherens junctions in the Drosophila germline stem cell niche. Nat. Commun. 2, 415 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Shivdasani AA & Ingham PW Regulation of Stem Cell Maintenance and Transit Amplifying Cell Proliferation by TGF-Beta Signaling in Drosophila Spermatogenesis. Curr. Biol. 13, 2065–2072 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Schulz C et al. A missexpression screen reveals effects of bag-of-marbles and TGFBeta class signaling on the Drosophila male germ-line stem cell lineage. Genetics 167, 707–723 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voog J, D’Alterio C & Jones DL Multipotent somatic stem cells contribute to the stem cell niche in the Drosophila testis. Nature 454, 1132–1136 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardy RW, Tokuyasu KT, Lindsley DL & Garavito M The germinal proliferation center in the testis of Drosophila melanogaster. J. Ultrastruct. Res. 69, 180–190 (1979). [DOI] [PubMed] [Google Scholar]
- 25.Yamashita YM, Jones DL & Fuller MT Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science (80-.). 301, 1547–1550 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Cheng J, Tiyaboonchai A, Yamashita YM & Hunt AJ Asymmetric division of cyst stem cells in Drosophila testis is ensured by anaphase spindle repositioning. Development 138, 831–837 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanentzapf G, Devenport D, Godt D & Brown NH Integrin-dependent anchoring of a stem-cell niche. Nat. Cell Biol. 9, 1413–1418 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee S, Zhou L, Kim J, Kalbfleisch S & Scho F Lasp anchors the Drosophila male stem cell niche and mediates spermatid individualization. Mech. Dev. 125, 768–776 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Boyle M & DiNardo S Specification, migration and assembly of the somatic cells of the Drosophila gonad. Development 121, 1815–25 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Brookman JJ, Toosy AT & White RAH The 412 retrotransposon and the development of gonadal mesoderm in Drosophila. Development 116, 1185–1192 (1992). [DOI] [PubMed] [Google Scholar]
- 31.Clark IBN, Jarman AP & Finnegan DJ Live imaging of Drosophila gonad formation reveals roles for Six4 in regulating germline and somatic cell migration. BMC Dev. Biol. 9, 1–9 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sano H et al. The Drosophila Actin Regulator ENABLED Regulates Cell Shape and Orientation during Gonad Morphogenesis. PLoS One 7, e52649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jenkins AB, Mccaffery JM & Van Doren M Drosophila E-cadherin is essential for proper germ cell-soma interaction during gonad morphogenesis. Development 130, 4417–4426 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Maimon I, Popliker M & Gilboa L Without children is required for Stat-mediated zfh1 transcription and for germline stem cell differentiation. Development 141, 2602–2610 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gancz D & Gilboa L Insulin and Target of rapamycin signaling orchestrate the development of ovarian niche-stem cell units in Drosophila. Development 140, 4145–4154 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Panchal T et al. Specification and spatial arrangement of cells in the germline stem cell niche of the Drosophila ovary depend on the Maf transcription factor Traffic jam. PLoS Genet. 2, 1–32 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarikaya DP & Extavour CG The Hippo Pathway Regulates Homeostatic Growth of Stem Cell Niche Precursors in the Drosophila Ovary. PLoS Genet. 11, 1–28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vlachos S et al. A Pak-regulated cell intercalation event leading to a novel radial cell polarity is involved in positioning of the follicle stem cell niche in the Drosophila ovary. Development 142, 82–91 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song X, Call GB, Kirilly D & Xie T Notch signaling controls germline stem cell niche formation in the Drosophila ovary. Development 134, 1071–1080 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Sheng XR et al. Jak-STAT regulation of male germline stem cell establishment during Drosophila embryogenesis. Dev. Biol. 334, 335–344 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinden D et al. Jak-STAT regulation of cyst stem cell development in the Drosophila testis. Dev. Biol. 372, 5–16 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okegbe TC & DiNardo S The endoderm specifies the mesodermal niche for the germline in Drosophila via Delta-Notch signaling. Development 138, 1259–1267 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kitadate Y & Kobayashi S Notch and Egfr signaling act antagonistically to regulate germ-line stem cell niche formation in Drosophila male embryonic gonads. Proc. from Natl. Acad. Sci. 107, 14241–14246 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeFalco T, Camara N, Le Bras S & Van Doren M Nonautonomous Sex Determination Controls Sexually Dimorphic Development of the Drosophila Gonad. Dev. Cell 14, 275–286 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wawersik M et al. Somatic control of germline sexual development is mediated by the JAK / STAT pathway. Nature 436, 563–567 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clark IBN, Boyd J, Hamilton G, Finnegan DJ & Jarman AP D-six4 plays a key role in patterning cell identities deriving from the Drosophila mesoderm. Dev. Biol. 294, 220–231 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Boukhatmi H et al. An Org-1-Tup transcriptional cascade reveals different types of alary muscles connecting internal organs in Drosophila. Development 141, 3761–71 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Morin X, Daneman R, Zavortink M & Chia W A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. from Natl. Acad. Sci. 98, 15050–15055 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hudry B, Viala S, Graba Y & Merabet S Visualization of protein interactions in living Drosophila embryos by the bimolecular fluorescence complementation assay. BMC Biol 9, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Isaac DD & Andrew DJ Tubulogenesis in Drosophila : a requirement for the trachealess gene product. Genes Dev. 10, 103–117 (1996). [DOI] [PubMed] [Google Scholar]
- 51.Castro B, Barolo S, Bailey AM & Posakony JW Lateral inhibition in proneural clusters: cis-regulatory logic and default repression by Suppressor of Hairless. Development 132, 3333–3344 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Campos-Ortega JA & Hartenstein V The Embryonic Development of Drosophila melanogaster. (Springer-Verlag, 1985). [Google Scholar]
- 53.Lenhart KF & DiNardo S Somatic Cell Encystment Promotes Abscission in Germline Stem Cells following a Regulated Block in Cytokinesis. Dev. Cell 34, 192–205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prasad M, Jang ACC, Starz-Gaiano M, Melani M & Montell DJ A protocol for culturing drosophila melanogaster stage 9 egg chambers for live imaging. Nat. Protoc. 2, 2467–2473 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Sheng XR & Matunis E Live imaging of the Drosophila spermatogonial stem cell niche reveals novel mechanisms regulating germline stem cell output. Development 138, 3367–3376 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dinardo S, Okegbe T, Wingert L, Freilich S & Terry N lines and bowl affect the specification of niche cells in the Drosophila testis. Development 138, 1687–96 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Labeau EM, Trujillo DL & Cripps RM Bithorax Complex genes control alary muscle patterning along the cardiac tube of Drosophila. Mech. Dev. 126, 478–486 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boukhatmi H et al. Tup / Islet1 integrates time and position to specify muscle identity in Drosophila. Development 139, 3572–3582 (2012). [DOI] [PubMed] [Google Scholar]
- 59.Mann T, Bodmer R & Pandur P The Drosophila homolog of vertebrate Islet1 is a key component in early cardiogenesis. Development 136, 317–326 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen P, Nordstrom W, Gish B & Abrams JM grim, a novel cell death gene in Drosophila. Genes Dev. 10, 1773–1782 (1996). [DOI] [PubMed] [Google Scholar]
- 61.Leatherman JL & DiNardo S Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell 3, 44–54 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tulina N & Matunis E Control of Stem Cell Self-Renewal in Drosophila Spermatogenesis by JAK-STAT Signaling. Science (80-.). 294, 2546–2550 (2001). [DOI] [PubMed] [Google Scholar]
- 63.Yamashita YM, Jones DL & Fuller MT Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 301, 1547–1550 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Asaoka-Taguchi M, Yamada M, Nakamura a, Hanyu K & Kobayashi S Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat. Cell Biol. 1, 431–437 (1999). [DOI] [PubMed] [Google Scholar]
- 65.Brown NH, Gregory SL & Martin-Bermudo MD Integrins as mediators of morphogenesis in Drosophila. Dev. Biol. 223, 1–16 (2000). [DOI] [PubMed] [Google Scholar]
- 66.Keller R et al. Mechanisms of convergence and extension by cell intercalation. Philos. Trans. R. Soc. B Biol. Sci. 355, 897–922 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bertet C, Sulak L & Lecuit T Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature 429, 667–671 (2004). [DOI] [PubMed] [Google Scholar]
- 68.Zallen JA & Wieschaus E Patterned gene expression directs bipolar planar polarity in Drosophila. Dev. Cell 6, 343–355 (2004). [DOI] [PubMed] [Google Scholar]
- 69.Steinberg MS Reconstruction of tissues by dissociated cells. Science (80-.). 141, 401–408 (1963). [DOI] [PubMed] [Google Scholar]
- 70.Montell DJ, Rorth P & Spradling AC slow border cells, a locus required for developmentally regulated cell migration during oogenesis, encodes Drosophila C/EBP. Cell 71, 51–62 (1992). [DOI] [PubMed] [Google Scholar]
- 71.Montell DJ Border-cell migration: the race is on. Nat. Rev. Mol. Cell Biol. 4, 13–24 (2003). [DOI] [PubMed] [Google Scholar]
- 72.Kadam S, Ghosh S & Stathopoulos A Synchronous and symmetric migration of Drosophila caudal visceral mesoderm cells requires dual input by two FGF ligands. Development 139, 699–708 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: The Trachea is located near the assembling niche. A) six4-nlsGFP (SGPs) and GFP in the tracheal lumen reveal the assembled hub (dotted outline) located near the dorsal branch (arrowheads) and a transverse connective (arrow) of the trachea. B-C) Wild type, and trh8 mutant embryos are labeled with Crumbs immunostaining, which marks the trachea (arrowheads mark the dorsal trunk, and an arrow marks a transverse connective), as well as other epithelial tissues. C) trh8 mutants lack the trachea. D-F) Gonads dissected from control and trh8 mutant embryos. All gonads exhibited the niche located opposite to the posterior msSGPs (arrows show niche; dotted outline marks msSGPs, which in our hands, accumulated signal in Vasa stains, but remained quite distinguishable from nearby germ cells, as msSGP cells were much smaller; n =10 gonads). Scale bars are 10 microns.
Figure S2. tup mutants have niche morphogenesis defects. (A-B) Gonads (outlined) dissected from control and heteroallelic tup mutant embryos (stage 17, 21-23h AEL) stained with anti-GFP (six4-nlsGFP), anti-Tj to mark SGPs and anti-FasIII to mark hub cells. Scale bar is 10 microns. A-A”) Control gonad exhibiting a single aggregate of hub cells enriched for FasIII (asterisk). Routinely, the niche was located opposite the posteriorly-located msSGPs, which were identified by bright six4-nlsGFP (arrowhead). B-B”) tup mutant gonad exhibiting two separate aggregates of FasIII-enriched hub cells (arrows). Additionally, aggregates were often not located at the anterior relative to posterior msSGPs (arrowhead). C) Quantification of the niche morphology in control and tup mutant gonads. We counted the number of gonads that had one anterior niche aggregate, and the number of gonads with multiple niche aggregates for each genotype.
Movie S1. Niche assembly in vivo imaging of niche assembly. The gonad is visible as an oval-shaped collection of SGPs marked with six4-nls-eGFP. Dotted line denotes gonad boundaries. AbdA>RFP labels nearby tissues in the embryo. The movie begins with SGPs dispersed throughout the gonad; germ cells are negatively marked. As the movie progresses, pro-niche cells collect at the gonad anterior (left). The movie ends with an elongate column of assembled pro-niche cells. Bright six4-nls-eGFP is visible in msSGPs at the gonad posterior (right).
Movie S2. Migration along the gonad periphery in vivo imaging of niche assembly. six4-nls-eGFP marks all SGPs (green), while PS11 and PS 13 SGPs are labelled with prd>tdTomato. Sections presented in the time lapse focus on one PS11 cell (arrow, first frame) at the gonad periphery. The cell initially exhibited cellular extensions along the periphery and also one back into the gonad, the latter stretching over a germ cell (arrowhead). Over time, this SGP moves anteriorly (left) along the periphery of the gonad, by the end of the timelapse the PS11 cell (arrow) has joined PS10 pro-niche cells (green only) forming an elongate, assembled column at the anterior. Note that during movement of the PS11 cell, it retracted the cellular extension that had embarced a germ cell.
Movie S3. Internal tilt of the niche in vivo imaging of an assembled niche. A z-stack taken from the last time point I shown, progressing from the external surface of the embryo, towards internal regions. Z positions are labelled in microns. six4-nls-eGFP labels SGP nuclei in the gonad, as well as some neighboring mesodermal nuclei to lower levels. As the sections progress further into the embryo, the gonad becomes visible in the lower left. The assembly of pro-niche cells only becomes visible in the deeper sections (note section depth). A branch of the trachea (labeled by breathless>GFP) can be observed in more external sectionos the series relative to those in which the pro-niche niche cells were visible.
Movie S4. Niche compaction ex vivo imaging of a gonad undergoing niche compaction. F-actin in SGPs is visualized with six4-eGFP::Moesin. All nuclei are labelled with His2Av::mRFP1. The gonad (dotted outline) is shown in an approximately face-on view; the small-profile cells (dotted outline within the gonad) comprise the compacting niche. The larger cells and nuclei represent prospective GSCs arranged in an arc to the right around the niche (other GSCs are in deeper sections, and not shown). Throughout the time-lapse, the pro-niche cell cluster tightens, occupying less surface area and their nuclei becoming closer to one another. Note also that the boundary between the niche and the surrounding GSCs becomes smoother and more circular (see dotted outline, final panel). Finally, note the reorganization over time of cortical f-actin in pro-niche cells. Early in the time series, f-actin is present along all interfaces of pro-niche cells, but adopts a more asymmetric appearance, exhibiting a relative increase on hub cell-GSC interfaces, and relative decrease on niche-niche cell interfaces.
Table S1. The Niche overlaps with other organs along the external-internal arc of the gonad. We examined the external to internal axis of the gonad, relative to the anterior-posterior axis of the embryo (see Materials and Methods). Along the external-internal gonad axis, we measured the size of the gonad (in microns), and the space occupied by the niche (in microns). We also evaluated where, along this axis, the trachea and the alary muscle overlapped with the niche. We indicate the percentage of the niche space that overlaps with either tissue as the Portion of niche overlap.