Abstract
Free fatty acids (FFAs) are an energy source, and induce activation of signal transduction pathways that mediate several biological processes. In breast cancer cells, oleic acid (OA) induces proliferation, matrix metalloproteinase-9 (MMP-9) secretion, migration and invasion. However, the signal transduction pathways that mediate migration and invasion induced by OA in breast cancer cells have not been studied in detail. We demonstrate here that FFAR1 and FFAR4 mediate migration induced by OA in MDA-MB-231 and MCF-7 breast cancer cells. Moreover, OA induces migration, invasion, AKT1 and AKT2 activation, 12-LOX secretion and an increase of NFκB-DNA binding activity in breast cancer cells. Cell migration requires FFAR1, FFAR4, EGFR, AKT and PI3K activity, whereas invasion is mediated though a PI3K/Akt-dependent pathway. Furthermore, OA promotes relocalization of paxillin to focal contacts and it requires PI3K and EGFR activity, whereas NFκB-DNA binding activity requires PI3K and AKT activity.
Keywords: breast cancer, oleic acid (OA), migration, invasion, 12-LOX
Introduction
Epidemiological studies strongly suggest an association between a higher intake of dietary fat and obesity with an increased risk of developing breast cancer. Moreover, obesity is characterized for an elevation of circulating free fatty acids (FFAs) (1, 2, 3, 4). FFAs are an energy source, and induce activation of signal transduction pathways in breast cancer cells (5, 6, 7, 8). Particularly, oleic acid (OA) induces proliferation, migration and invasion of breast cancer cells (5, 6, 8, 9, 10).
Free fatty acid receptor 1 (FFAR1) and FFAR4 are G protein-coupled receptors activated by medium- to long-chain fatty acids, such as OA; but they are not activated by short chain FFAs (11, 12). FFAR1 is expressed in pancreatic β-cells and enhances insulin secretion mediated by glucose; whereas FFAR4 is expressed in adipose tissue, macrophages and taste buds and is involved in anti-inflammatory response, taste perception and insulin signaling (13, 14, 15, 16). Moreover, FFAR1 and FFAR4 are expressed in MDA-MB-231 and MCF-7 breast cancer cells and mammary non-tumorigenic epithelial cells MCF10A (9, 11, 17).
The PI3K/AKT/mTOR pathway has been involved in growth, proliferation, survival, metabolism and motility (18). In breast cancer, mutations in the PI3K/AKT pathway are frequently found, and approximately 60% of tumors have mutations that hyperactive this pathway (19).The family of PI3K lipid kinases generates phosphoinositol lipids that act as second messengers in a number of signaling pathways, including the activation of PDK1 and AKT. The AKT family of serine-threonine kinases consists of three members, namely AKT1, AKT2 and AKT3. The AKT maximum activation requires phosphorylation at threonine (Thr)-308, Thr-309 and Thr-305, and the phosphorylation at serine (Ser)-473, Ser-474 and Ser-472 in AKT1, AKT2 and AKT3, respectively (20, 21). Particularly, PI3K catalyzes the synthesis of the membrane phospholipid PI (3, 4, 5), P3 from PI (3, 4) P2, which recruit AKT to the plasma membrane and then AKT is activated by its phosphorylation. Activation of AKT promotes its translocation to several subcellular locations, and phosphorylation of targets (22, 23).
Arachidonic acid (AA) is an essential fatty acid, which is obtained from dietary sources, and can be derived from linoleic acid. In mammalian cells, AA is esterified into membrane phospholipids and is released mainly by the action of cytosolic phospholipase A2α (cPLA2α). However, AA is also released by other enzymes including PLC and PLD (24, 25). Free AA is enzymatically metabolized by three major pathways, namely lipoxygenases (LOXs), cyclooxygenases (COXs) and cytochrome P450 epoxygenases. The LOX pathway generates hydroeicosatetraenoic acids (HETEs), which are the precursors of leukotrienes (LKs), lipoxins (LOs) and hepoxillins (HOs) (26). The COX pathway is mediated by two members, COX-1 and COX-2, which produce prostaglandins (PGs), prostacyclin D2 (PGD2), prostacyclin E2 (PGE2), prostacyclin F2α (PGF2α), prostacyclin I2 (PGI2) and thromboxane A2 (TXA2) (27). In general, AA metabolites (eicosanoids) mediate a variety of cell processes including chemotaxis, inflammation, angiogenesis and apoptosis (28). In MDA-MB-231 breast cancer cells, OA induces FAK activation and migration through a PLC- and LOXs-dependent pathway (8).
In the present study, we studied the signal transduction pathways that mediate migration and invasion induced by OA in breast cancer cells.
Materials and methods
Materials
OA sodium salt and A6730 were from Sigma. AG1478 was from Calbiochem. LY294002, Akt2 siRNAs, antibodies (Abs) against Akt1 (5C10), Akt2 (F-7), and FAK (C-20) were from Santa Cruz Biotechnology. Paxillin Ab was from Invitrogen. FFAR4 Ab was from OriGene (Rockville, MD, USA). Phosphospecific Ab to Thr-308/Thr-309 of Akt1/Akt2 (anti-p-Akt-Thr) (244F9), and Ser-473/Ser-474 of Akt1/Akt2 (anti-p-Akt-Ser) (9271S) were from Cell Signaling. Phosphospecific Ab to tyrosine (Tyr)-397 of FAK (anti-p-FAK) was from Invitrogen. Actin monoclonal Ab was kindly provided by PhD Manuel Hernandez (Cinvestav-IPN). DC260126 and AH7614 were from Tocris (Minneapolis, MN, USA). Basement membrane matrix (BD Matrigel) was from BD Biosciences (Bedford, MA, USA). (γ-32P) ATP was from Perkin-Elmer.
Cell culture
The human MDA-MB-231 and MCF-7 breast cancer cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 3.7 g/L sodium bicarbonate, 5% fetal bovine serum (FBS) and antibiotics in a humidified atmosphere containing 5% CO2 and 95% air at 37°C. For experimental purposes, cultures were serum-starved for 24 h before treatment.
Preparation of OA solution
OA solution was prepared with bovine serum albumin (BSA) as described previously (6, 29). Briefly, OA bound to BSA (BSA-OA) was prepared by stirring OA sodium salt at 37°C with 5% fatty acid-free BSA. Solution was adjusted to pH 7.4 and filtered through 0.22 μm filter and the fatty acid concentration was measured using a fatty acid assay kit (Bio Vision). When BSA-OA was added to serum-free medium, final concentration of BSA was adjusted to 0.005%.
Cell stimulation
Confluent cultures were washed twice with DMEM without FBS, equilibrated in the same medium at 37°C for 30 min, and treated with inhibitors and/or BSA-OA. The stimulation was terminated by aspirating the medium and cells were solubilized with 0.5 mL of ice-cold radioimmunoprecipitation assay (RIPA) buffer containing 50 mM HEPES pH 7.4, 150 mM NaCl, 1 mM EGTA, 1 mM sodium orthovanadate, 100 mM NaF, 10 mM sodium pyrophosphate, 10% glycerol, 1% Triton X-100, 1% sodium deoxycholate, 1.5 mM MgCl2, 0.1% SDS and 1 mM PMSF. Lysates were clarified by centrifugation at 13,539 g for 10 min at 4°C. Supernatants were transferred to fresh tubes and the protein level of each sample was determined by the micro Bradford protein assay (Bio-Rad).
Western blotting
Equal amounts of protein were separated by SDS-PAGE using 10% separating gels and transferred to nitrocellulose membranes. Next, membranes were blocked using 5% non-fat dried milk in phosphate buffered saline (PBS) pH 7.2/0.1% Tween 20 (wash buffer), and incubated overnight at 4°C with primary Ab. The membranes were washed three times with wash buffer and incubated with secondary Ab (horseradish peroxidase-conjugated Abs to rabbit) (1:5000) for 2 h at 22°C. After washing, immunoreactive bands were visualized using ECL detection reagent. Autoradiograms were scanned and the labeled bands were quantified using the ImageJ software (https://imagej.nih.gov/ij/).
Immunoprecipitation
Lysates were clarified by centrifugation at 13,539 g for 10 min. Supernatants were transferred to fresh tubes, and proteins were immunoprecipitated overnight at 4°C with protein A-agarose linked to a specific Ab against the target protein. Immunoprecipitates were washed three times with RIPA buffer.
Scratch-wound assay
Cells were grown to confluence in 35 mm culture dishes, starved for 24 h in DMEM and treated for 2 h with 12 μM mitomycin C to inhibit proliferation. Next, cultures were scratch-wounded using a sterile 200 µL pipette tip, washed twice with DMEM and re-fed with DMEM without or with inhibitors and/or BSA-OA. Progress of cell migration into the wound was photographed at 48 h using an inverted microscope coupled to camera. Each experiment was repeated three times.
Invasion assay
Invasion assays were performed by the modified Boyden chamber method in 24-well plates containing 12 cell-culture inserts with 8 µm pore size (Costar, Corning, Inc). An amount of 50 µL BD Matrigel was added into culture inserts and kept overnight at 37°C to form a semisolid matrix. Cells were plated at 1 × 105 cells per insert in serum-free DMEM on the top chamber. The lower chamber contained 600 µL DMEM without or with BSA-OA. Chambers were incubated for 48 h at 37°C in a 5% CO2 atmosphere, and then cells and Matrigel on the upper surface of membrane were removed with cotton swabs, and cells on the lower surface of membrane were washed and fixed with methanol for 5 min. Number of invaded cells was estimated by staining with 0.1% crystal violet in PBS. Dye was eluted with 300 µL 10% acetic acid, and absorbance at 600 nm was measured. Background value was obtained from wells without cells.
Determination of 12(S)-HETE
MDA-MB-231 cells were treated with 100 μM OA or 15 μM AA for 30 min, and supernatants were collected. The concentration of 12(S)-HETE was determined by using the 12(S)-HETE ELISA kit (Enzo Life Sciences, Farmingdale, NY, USA), according to the manufacturer´s guidelines.
RNA interference
AKT2 expression was silenced in breast cancer cells by using the Silencer siRNA kit from Santa Cruz Biotechnology, according the manufacturer’s guidelines. One control of scramble siRNAs was included according to the manufacturer’s guidelines.
Silencing of FFAR4 with shRNA
Lentiviral shRNA vectors from Santa Cruz Biotechnology targeting human FFAR4 were utilized for generation of stable knockdown in MDA-MB-231 cells, according the manufacturer’s guidelines. Transfected cells were selected by their resistance to puromycin (5 µg/mL).
Immunofluorescence confocal microscopy
Cells grown on coverslips were stimulated with OA for various times. After stimulation, cells were fixed with 4% paraformaldehyde in PBS for 20 min, permeabilized with 0.1% Triton X-100 in PBS for 20 min, and blocked for 1 h with 3% BSA. Cells were stained with TRITC-conjugated phalloidin to reveal F-actin and with anti-paxillin Ab for 12 h to reveal focal adhesions, followed by incubation with FITC-labeled anti-mouse secondary Ab for 2 h at room temperature. Cells were viewed using a Leica confocal microscope (Model TCS SP2; Leica Microsystems). Serial optical sections of 0.8−0.9 µm thick were taken in both xyz and xzy. To prevent interference from the fluorescent probes, images of the same optical section were taken as separate channels, and they were analyzed by using ImageJ software.
Preparation of nuclear extracts
Briefly, 1.5 × 106 cells were lysed with 0.1% nonionic detergent Nonidet P40 in Buffer A (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 6 mM MgCl2, 10 mM NaF, 1 mM Na3VO4, 1 mM DTT, 1 mM PMSF). Lysates were pelleted at 636 g for 15 min and resuspended in Buffer B (20 mM HEPES, pH 7.9, 420 mM NaCl, 20% glycerol 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM Na3VO4, 10 mM NaF, 1 mM DTT, 0.2 mM PMSF). Nuclear extracts were recovered by centrifugation at 17,136 g for 15 min at 4°C and the protein level of each sample was determined by the micro Bradford protein assay.
Electrophoretic mobility shift assay (EMSA)
Double-stranded oligonucleotide containing specific binding sites for NFκB, 5′ AGCTAAGGGACTTTCCGCTGGGGACTTTCCAGG 3′, was used as probe. A total amount of 20 pmol of annealed NFκB oligonucleotide was labeled with (γ-32P) ATP using T4 polynucleotide kinase. The 32P-labeled oligonucleotide probe (~1 ng) was incubated with 5 µg of nuclear extract in a reaction mixture containing 3 µg of poly (dI-dC), 0.25 M HEPES pH 7.5, 0.6 M KCl, 50 mM MgCl2, 1 mM EDTA, 7.5 mM DTT and 9% glycerol for 20 min at 4°C. One hundred-fold excess of unlabeled NFκB probe and an irrelevant oligonucleotide were used as specific and nonspecific competitors. The samples were fractionated on a 6% polyacrylamide gel in 0.5X Tris borate-EDTA buffer. Gels were dried and analyzed by autoradiography.
Statistical analysis
Results are expressed as mean ± s.d. of at least three independent experiments. Data were statistically analyzed using one-way ANOVA and the pairwise comparison was performed using Newman–Keuls multiple comparison test. Control comparison was performed using Dunnett’s test. Statistical probability of P < 0.05 was considered significant.
Results
FFAR1 and FFAR4 mediate migration induced by OA in breast cancer cells
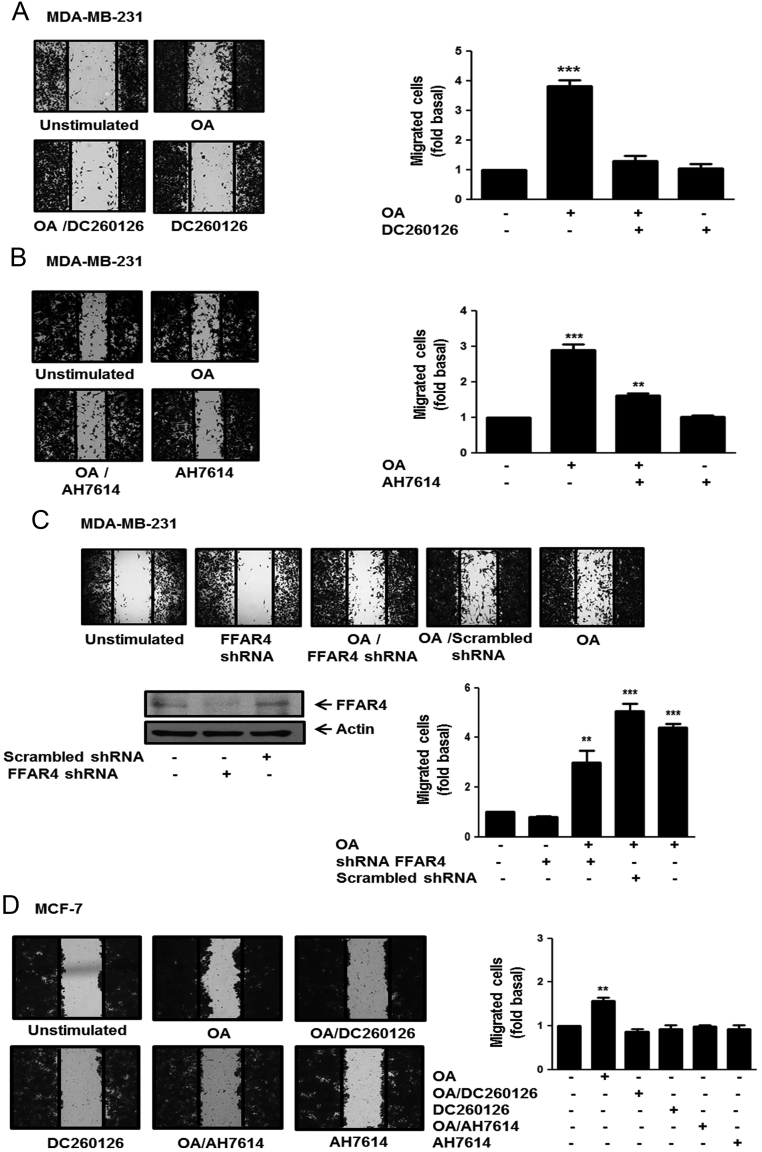
OA induces migration of MDA-MB-231 breast cancer cells (8, 30). We determined the role of FFAR1 and FFAR4 on migration induced by OA in breast cancer cells. Cultures of MDA-MB-231 and MCF-7 breast cancer cells were treated for 1 h with 3 μM DC260126 or 10 µM AH7614, which are selective inhibitors of FFAR1 and FFAR4 respectively (31, 32, 33), and then they were scratch-wounded and stimulated with 100 µM OA for 48 h. As illustrated in Fig. 1A and B, treatment with DC260126 completely inhibited migration, whereas treatment with AH7614 partly inhibited migration induced by OA in MDA-MB-231 cells.
Figure 1.
FFAR1 and FFAR4 mediate migration induced by OA in breast cancer cells. Panel (A), (B) and (D) Cultures of MDA-MB-231 and MCF-7 cells were treated with DC260126 or AH7614, scratch-wounded and stimulated with 100 µM OA for 48 h. Panel (C) MDA-MB-231 cells were transfected with FFAR4 shRNA or scramble shRNA and cell lysates were analyzed by Western blotting with anti-FFAR4 Ab. Cultures of MDA-MB-231 cells transfected with FFAR4 shRNA or scramble shRNA were scratch-wounded and stimulated with 100 μM OA for 48 h. Graphs represent the mean ± s.d. of at least three independent experiments and are expressed as fold of migrated cells above unstimulated cells. **P < 0.01, ***P < 0.001.
To further substantiate the role of FFAR4 in cell migration of MDA-MB-231 cells. Cultures of MDA-MB-231 cells, in which FFAR4 expression was knocked down by using shRNA against FFAR4, were scratch-wounded and treated with 100 μM OA for 48 h. In agreement with our previous results, migration induced by OA is partly dependent of FFAR4 expression in MDA-MB-231 cells (Fig. 1C).
We also studied the role of FFAR1 and FFAR4 in migration induced by OA in another breast cancer cell (MCF-7). Our findings showed that treatment with DC260126 and AH7614 completely inhibited migration induced by OA in MCF-7 cells (Fig. 1D).
OA induces migration through a PI3K/AKT2-dependent pathway
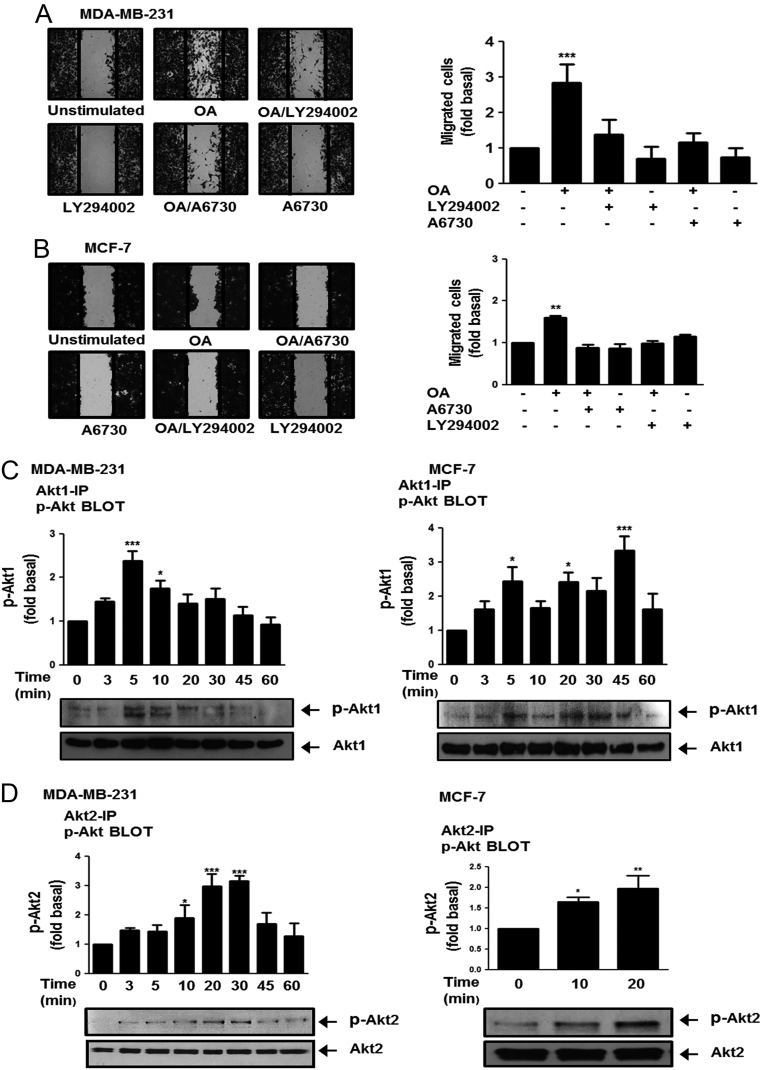
In order to determine the role of PI3K and AKT1/2 on cell migration, we used selective inhibitors of PI3K and Akt1/2, namely LY294002 and A6730 respectively (34, 35). Cultures of MDA-MB-231 and MCF-7 cells were treated for 1 h with 10 μM LY294002 or 2 µM A6730, and then they were scratch-wounded and stimulated with 100 µM OA for 48 h. Our results showed that inhibition of PI3K and AKT1/2 activity inhibited migration induced by OA in MDA-MB-231 and MCF-7 cells (Fig. 2A and B).
Figure 2.
OA induces migration through a PI3K/AKT-dependent pathway. Panel (A) and (B) Cultures of MDA-MB-231 and MCF-7 cells were treated with LY294002 or A6730, scratch-wounded and treated with OA. Panel (C) and (D) Lysates from MDA-MB-231 and MCF-7 cells treated with OA for various times were analyzed by immunoprecipitation (IP) with Akt1 Ab or Akt2 Ab followed by Western blotting with anti-p-Akt-Thr Ab and anti-p-Akt-Ser Ab, respectively. Membranes were analyzed further by Western blotting with anti-Akt1 or Akt2 Ab. Graphs represent the mean ± s.d. of at least three independent experiments and are expressed as fold of migrated cells or p-Akt-1 or -2 above unstimulated cells. *P < 0.05, **P < 0.01 ***P < 0.001.
Since migration induced by OA required AKT1/2 activity; we determined whether OA induces AKT1/2 activation and the role of AKT2 on cell migration. First, AKT1/2 activation was studied by using the phosphorylation of Akt1 at Thr-308 and phosphorylation of Akt2 at Ser-474 (22). MDA-MB-231 and MCF-7 cells were stimulated with 100 μM OA for various times and lysed. Lysates were analyzed by immunoprecipitation (IP) with anti-Akt1 Ab or anti-Akt2 Ab, followed by Western blotting with anti-p-Akt-Thr Ab and anti-p-Akt-Ser Ab, respectively. As illustrated in Fig. 2C and D, OA induced AKT1 and AKT2 phosphorylation at Thr-308/309 and Ser-473/474, respectively, in MDA-MB-231 and MCF-7 cells.
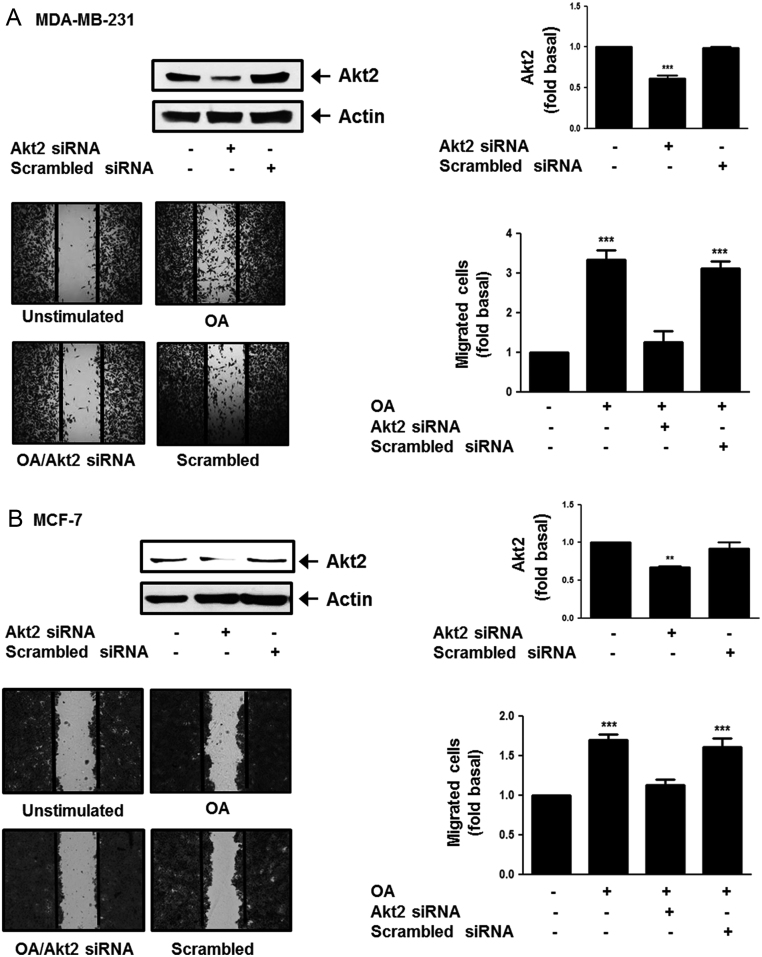
Next, we inhibited Akt2 expression in MDA-MB-231 and MCF-7 cells by using an Akt2 siRNA. Our findings showed a clear inhibition of Akt2 expression. Moreover, cultures of MDA-MB-231 and MCF-7 cells transfected with Akt2 siRNA and scramble siRNA were scratch-wounded and treated with 100 μM OA for 48 h. Our results showed that migration induced by OA required Akt2 expression in MDA-MB-231 and MCF-7 cells (Fig. 3A and B).
Figure 3.
OA induces migration through an AKT2-dependent pathway. Panel (A) and (B) MDA-MB-231 and MCF-7 cells were transfected with Akt2 siRNA or scrambled siRNA and cell lysates were analyzed by Western blotting with anti-Akt2 Ab. Cultures of MDA-MB-231 and MCF-7 cells transfected with Akt2 siRNA or scrambled siRNA were scratch-wounded and stimulated with OA. Graphs represent the mean ± s.d. of at least three independent experiments and are expressed as fold of Akt2 or migrated cells above unstimulated cells. **P < 0.01 ***P < 0.001.
Baicalein inhibits migration and activation of AKT2 and FAK induced by OA
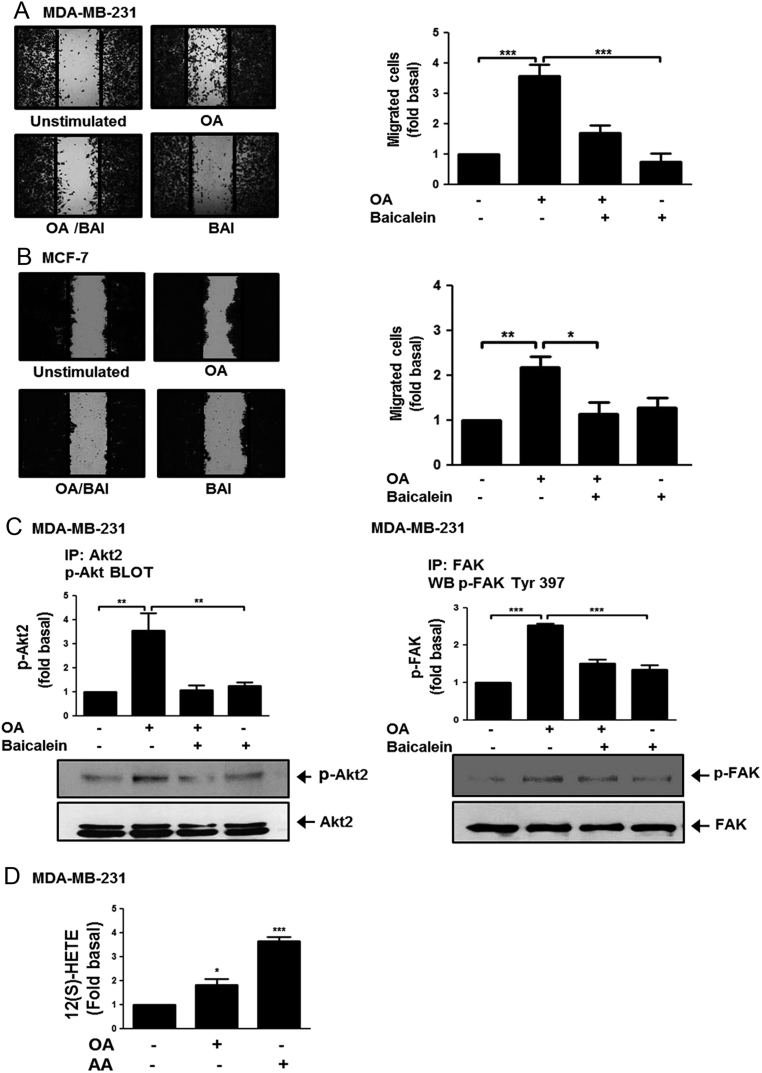
We studied the effect of baicalein in migration induced by OA in MDA-MB-231 and MCF-7 cells, because it is an inhibitor of 12-LOX and 15-LOX (36, 37). Migration assays were performed by using MDA-MB-231 and MCF-7 cells treated with 20 µM baicalein for 24 h and stimulated with 100 µM OA for 48 h. Our results showed that treatment with baicalein inhibited migration induced by OA in MDA-MB-231 and MCF-7 cells (Fig. 4A and B).
Figure 4.
Baicalein inhibits migration and activation of AKT2 and FAK induced by OA. Panel (A) and (B) Cultures of MDA-MB-231 and MCF-7 cells were treated with baicalein (BAI), scratch-wounded and treated with OA. Panel (C) Lysates from MDA-MB-231 cells treated with BAI and stimulated with OA were analyzed by IP with Akt2 Ab or FAK Ab, followed by Western blotting with anti-p-Akt-Thr Ab or p-FAK Ab. Membranes were analyzed further by Western blotting with anti-Akt2 or FAK Ab. Panel (D) Concentration of 12(S)-HETE was analyzed by ELISA in supernatants from MDA-MB-231 cells treated with OA or AA. Graphs represent the mean ± s.d. of at least three independent experiments and are expressed as fold of migrated cells, p-Akt2, p-FAK or 12(S)-HETE above unstimulated cells. *P < 0.05, **P < 0.01, ***P < 0.001.
Next, we studied the effect of baicalein in FAK and AKT2 activation. MDA-MB-231 cells were treated with 20 µM baicalein for 24 h and stimulated with 100 µM OA for 10 min (FAK) and 20 min (Akt2). AKT2 and FAK activation were analyzed by IP with anti-Akt2 Ab and anti-FAK Ab followed of Western blotting with anti-p-Akt-Thr Ab and anti-p-FAK Ab. As illustrated in Fig. 4C and D, treatment with baicalein inhibited AKT2 and FAK activation induced by OA in MDA-MB-231 cells.
We also determined whether OA induced 12(S)-HETE secretion. Conditioned medium from MDA-MB-231 cells treated with 100 µM OA for 15 min was analyzed to determine the concentration of 12(S)-HETE. Our findings demonstrated that OA induced the secretion of 12(S)-HETE in MDA-MB-231 cells (Fig. 4E). One control of 12(S)-HETE secretion was included (AA).
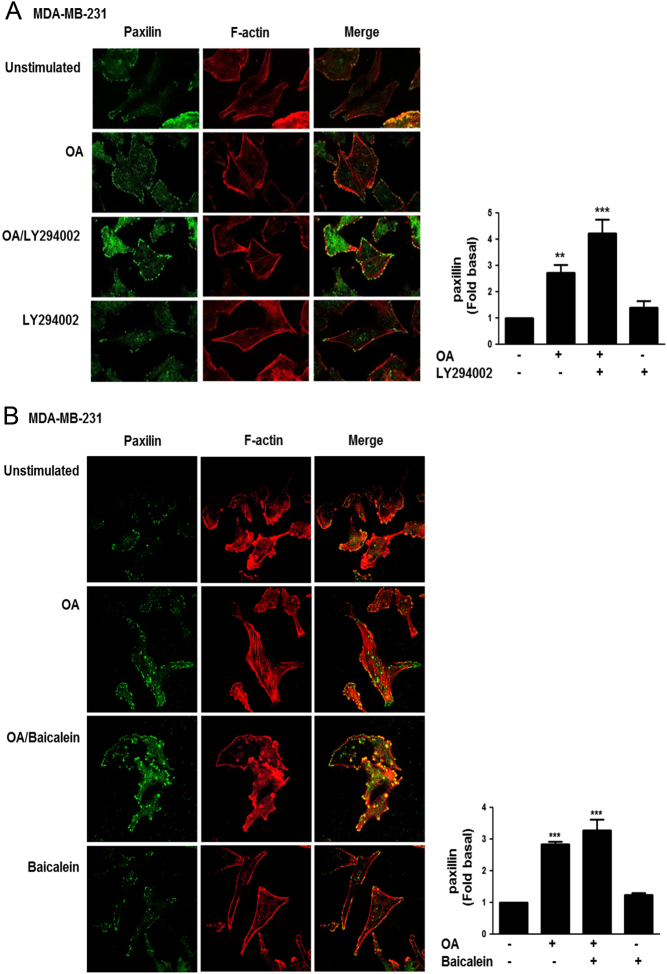
Focal contacts formation induced by OA requires PI3K activity
We studied whether OA induced focal contacts formation and the effect of a PI3K inhibitor and baicalein. MDA-MB-231 cells were cultured on coverslips and treated for 1 h with 10 μM LY294002 or 20 µM baicalein for 24 h, and then cells were stimulated with 100 μM OA for 15 min. The presence of focal contacts was analyzed by immunofluorescence analysis of paxillin, because it is a protein localized in focal contacts (38). Our findings showed that OA induced an increase in the number of focal contacts in MDA-MB-231 cells. Interestingly, treatment with a PI3K inhibitor and baicalein increased the number and size of focal contacts induced by OA in MDA-MB-231 cells (Fig. 5A and B).
Figure 5.
Focal contacts formation induced by OA requires PI3K activity. MDA-MB-231 cells cultured on coverslips were treated with LY294002 (Panel A) or baicalein (Panel B) and stimulated with OA. Cells were fixed and focal contacts were analyzed by staining with anti-paxillin Ab conjugated to FITC. F-actin was stained with TRITC-conjugated phalloidin. F-actin structures are shown in red, and focal adhesions are shown in green. Graphs represent the mean ± s.d. of at least three independent experiments and are expressed as mean fluorescent intensities of paxillin above unstimulated cells. **P < 0.01, ***P < 0.001.
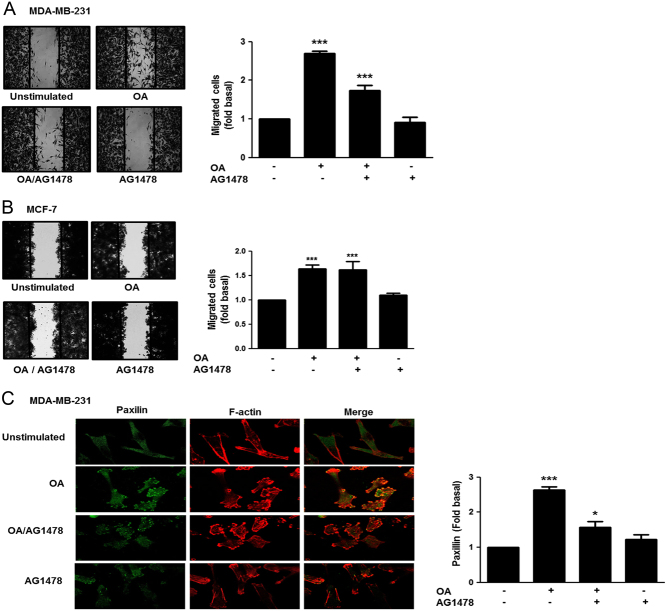
Role of EGFR on migration and focal contacts formation induced by OA
We studied the role of EGFR on cell migration and focal contacts formation. Migration assays were performed by using MDA-MB-231 and MCF-7 cells treated for 30 min with 500 nM AG1478, a selective inhibitor of EGFR (39), and stimulated with 100 μM OA for 48 h. As illustrated in Fig. 6A, treatment with AG1478 partly inhibited the migration induced by OA in MDA-MB-231 cells. In contrast, treatment with AG1478 did not inhibit migration induced by OA in MCF-7 cells (Fig. 6B). In addition, treatment with AG1478 inhibited the increased number and size of focal contacts induced by OA in MDA-MB-231 cells (Fig. 6C).
Figure 6.
Role of EGFR in migration and focal contacts formation induced by OA. Panel (A) and (B) Cultures of MDA-MB-231 and MCF-7 cells were treated with AG1478, scratch-wounded and treated with OA. Panel (C) MDA-MB-231 cells cultured on coverslips were treated with AG1478 and stimulated with OA. Cells were fixed and focal contacts were analyzed by staining with anti-paxillin Ab conjugated to FITC. F-actin was stained with TRITC-conjugated phalloidin. F-actin structures are shown in red, and focal adhesions are shown in green. Graphs represent the mean ± s.d. of at least three independent experiments and are expressed as fold of migrated cells or mean fluorescent intensities of paxillin above unstimulated cells. *P < 0.05, ***P < 0.001.
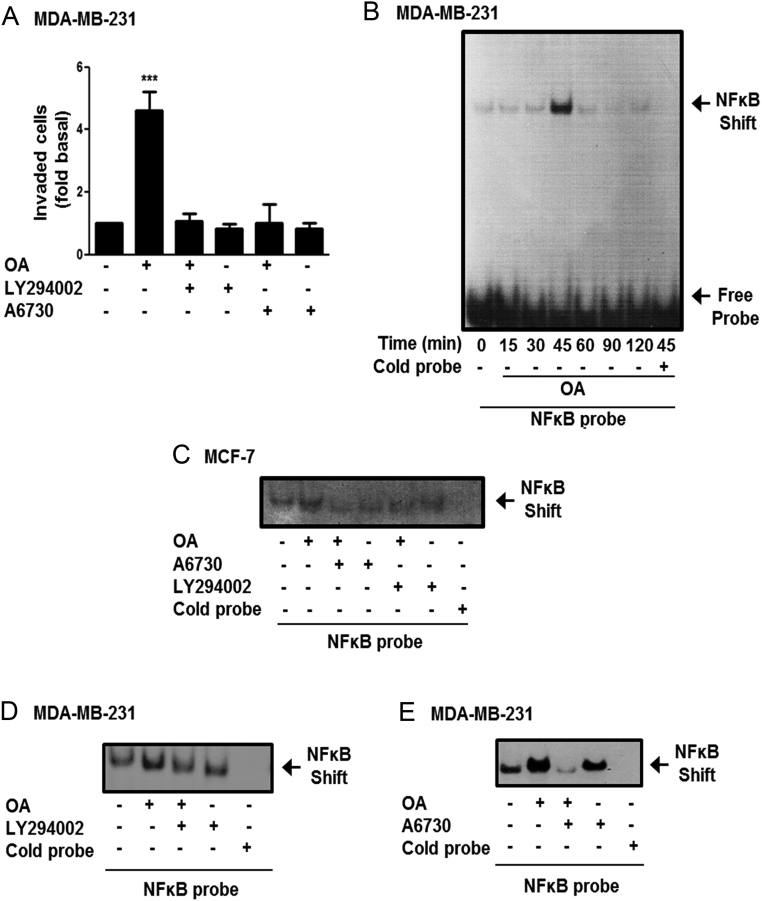
OA induces invasion and NFκB-DNA binding activity
Since, OA induces invasion in MDA-MB-231 cells (10), we studied the role of PI3K and AKT1/2 in the invasion process. Invasion assays were performed by using the Boyden chamber method and MDA-MB-231 cells treated for 1 h with 10 μM LY294002 or 2 μM A6730 and stimulated with 100 μM OA for 48 h. Our findings demonstrated that invasion induced by OA in MDA-MB-231 cells is dependent on PI3K and AKT1/2 activity (Fig. 7A).
Figure 7.
OA induces invasion and NFkB-DNA binding activity. Panel (A) Invasion assays were performed by using the Boyden chamber method and MDA-MB-231 cells treated with LY294002 or A6730 and stimulated with OA. Graph represents the mean ± s.d. of three independent experiments and is expressed as fold of invasion above unstimulated cells. Panel (B) MDA-MB-231 cells were treated with OA for various times and nuclear extracts were obtained. Panel (C, D and E) MCF-7 and MDA-MB-231 cells were treated with LY294002 or A6730, stimulated with OA and nuclear extracts were obtained. NFκB-DNA binding activity was analyzed by EMSA. Control included EMSA reactions with 100-fold excess of cold NFκB competitor (Cold probe). Results shown are representative of three independent experiments. ***P < 0.001.
The NFκB transcription factor mediates expression of genes implicated in invasion process. We determined whether OA induced an increase of NFκB-DNA binding activity in MDA-MB-231 cells. Nuclear extracts from MDA-MB-231 cells stimulated with 100 μM OA for various times were analyzed by EMSA using a radiolabeled probe representing a canonical NFκB binding site. Our findings demonstrated that OA induced an increase of NFκB-DNA binding activity at 45 min of stimulation in MDA-MB-231 cells (Fig. 7B). Next, we studied whether OA also induced an increase of NFκB-DNA binding activity in MCF-7 cells and the role of PI3K and AKT1/2. MCF-7 and MDA-MB-231 cells were treated with 10 μM LY294002 or 2 μM A6730 and stimulated with 100 μM OA for 45 min, nuclear extracts were obtained and analyzed by EMSA. As illustrated in Fig. 7C, D and E, OA induced an increase of NFκB binding activity through a PI3K- and AKT1/2-dependent pathway in MCF-7 and MDA-MB-231 cells.
Discussion
Epidemiological studies in women strongly suggest that obesity and dietary factors including a high intake of fat and meat enhance the risk of breast cancer (4, 40, 41). We previously demonstrated that OA induces proliferation, invasion, MMP-9 secretion, activation of ERK1/2, FAK and Src, and an increase of AP1-DNA binding activity in breast cancer cells (8, 9, 10). However, the signal transduction pathways involved in migration and invasion induced by OA in breast cancer cells remains to be studied.
FFAR1 and FFAR4 have been involved in cancer progression (42). Particularly, OA induces an increase of intracellular Ca2+ levels via FFAR1 in MCF-7 breast cancer cells, whereas it promotes proliferation through a FFAR1-dependent pathway in MDA-MB-231 breast cancer cells (6, 43). We demonstrate here that OA induces migration through a FFAR1- and FFAR4-dependent pathway in MCF-7 cells. However, OA induces migration through a FFAR1-dependent pathway and it is partly dependent on FFAR4 activity in MDA-MB-231 cells. In contrast to our findings, n − 3 fatty acids inhibit proliferation via FFAR1 and FFAR4 in MDA-MB-231 and MCF-7 cells (44). Moreover, inhibition of FFAR1 expression promotes migration but loss of FFAR4 expression inhibits migration in pancreatic cancer cells PANC-1 (45). In addition, FFAR1 is a negative regulator of migration and invasion in HT1080 fibrosarcoma cells (46). We propose that natural ligands of FFARs, including OA, mediate specific responses in breast cancer cells through activation of FFAR1 and/or FFAR4. Therefore, the role of FFARs in the cell responses mediated by different free fatty acids should be studied in different cell types.
Mouse models and cell cultures suggest that AKT family members do not have redundant functions, because each AKT member fulfills unique functions in normal and tumor cells (47, 48). Particularly, AKT1 activation promotes tumorigenesis and suppresses invasion in a mouse model of mammary tumorigenesis induced by ErbB-2 expression; whereas overexpression of Akt2 induces upregulation of integrin β1 and an increase of invasion and metastasis in breast and ovarian cancer cells (49, 50). We demonstrate here that OA induces migration through a PI3K- and AKT-dependent pathway in MDA-MB-231 and MCF-7 cells. In addition, OA induces invasion via PI3K and AKT activity, whereas migration requires AKT2 activity in MDA-MB-231 cells. Since, OA induces activation of AKT1 and AKT2 in MDA-MB-231 and MCF-7 cells, it remains to be investigated, the role of AKT1 in migration and the role of AKT1 in the invasion process. We propose that OA induces FFAR1/FFAR4 activation, and then activation of PI3K/AKT pathway, which mediate migration and invasion in breast cancer cells. Supporting our proposal, an agonist of FFAR4 (TUG891) induces phosphorylation of AKT at Ser-473 (51).
Cell adhesion is the attachment among cells and surrounding environment, including extracellular matrix (ECM). Cellular motility is an essential process that mediates metastasis and involves the adhesion and detachment of ECM. Moreover, adhesion induces activation of signal transduction pathways that mediate proliferation and survival (52, 53). Focal contacts are structures wherein integrin receptors mediate interaction between actin cytoskeleton and ECM, which are composed of diverse cell components including scaffolding proteins, adaptor proteins, GTPases, kinases and phosphatases (54, 55).
We demonstrate here that stimulation of MDA-MB-231 cells with OA induces formation of focal contacts and treatment with a PI3K inhibitor and OA increased the number and size of focal contacts and inhibition of migration. We propose that PI3K/AKT activation induced by OA mediates the disassembly of focal contacts during migration in MDA-MB-231 cells. In agreement with our findings, migration of MCF-7 cells requires Rac1-dependent actin organization through activation of PI3K, whereas, migration and invasion are regulated by Rho-A/PI3K activity in BLM melanoma cells (56, 57, 58).
LOXs pathway is composed of a dioxygenases family including 5-, 12- and 15-LOX-1/-2, and their main products are 5(S)-, 12(S)- and 15(S)-HETE, respectively (26). Particularly, 12-LOX is expressed in a variety of tumors, including breast cancer, whereas, 12(S)-HETE has been involved in the invasion and metastasis of tumor cells (59, 60). We propose that 12-LOX and/or 15-LOX mediate migration and focal contact formation through AKT2 and FAK activation in MDA-MB-231 and MCF-7 cells. Supporting our proposal, we demonstrate here that OA induces secretion of 12(S)-HETE in MDA-MB-231 cells, whereas 12-LOX activates PI3K/AKT and MMP-9 expression through NFκB in prostate cancer cells PC-3 (61).
Transactivation of EGFR induced by GPCRs occurs via MMPs activation and release of EGF-like ligands (HB-EGF) from precursors in the plasmatic membrane (62). We previously demonstrated that OA induces ERK1/2 activation, an increase of AP-1 DNA binding activity and invasion through an EGFR transactivation-dependent pathway in breast cancer cells (9, 10). We demonstrate here that OA induces migration through EGFR in MDA-MB-231 and MCF-7 cells, whereas it partly mediates focal contacts formation in MDA-MB-231 cells. We propose that OA induces EGFR transactivation and then activated EGFR mediates AKT activation, migration and gene expression in breast cancer cells. Supporting our proposal, migration and proliferation of non-small cell lung cancer cells require EGFR activity and phospho-ERK, whereas knockdown of glucose transporter decreases proliferation, migration and invasion via modulation of EGFR/MAPK signaling pathway in MDA-MB-231 cells (63, 64).
The transcription factor NFκB has been associated broadly with the development and progression of several cancers, because it mediates expression of genes implicated in angiogenesis and metastasis, including TNF, IL-1, iNOS, MMP-9 and urokinase-type plasminogen activator (uPA) (65, 66). In PTEN-null/inactive prostate cancer cells, AKT mediates NFκB activity via interaction and stimulation of IKK, whereas a model of prostate cancer progression in TRAMP mice show a differential protein expression of PI3K, AKT phosphorylated at Ser-473, IKK kinase activity and NFκB DNA binding activity (67, 68). We demonstrate here that OA induces an increase of NFκB-DNA binding activity and it requires the activity of PI3K and AKT in MDA-MB-231 and MCF-7 cells. We propose that OA induces NFκB activation and expression of genes implicated in migration and invasion in breast cancer cells.
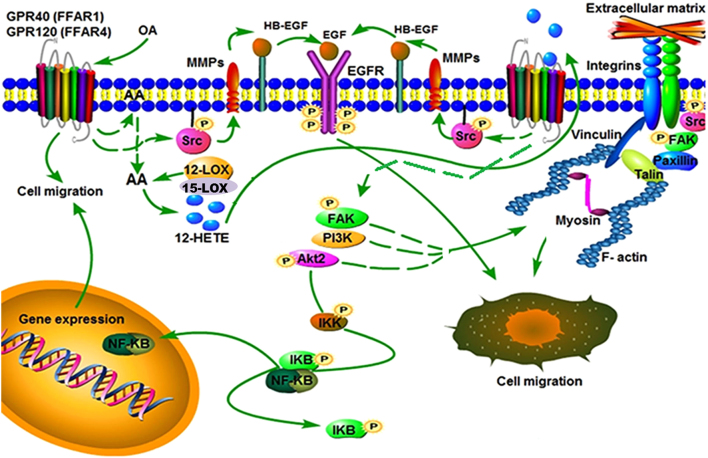
In conclusion, our findings demonstrate FFAR1/FFAR4, AKT, PI3K, EGFR and NFκB activation play a pivotal role in the migration and invasion processes in breast cancer cells (Fig. 8).
Figure 8.
Model of signal transduction pathways that mediate migration induced by OA in breast cancer cells.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This work was supported by a grant from CONACYT-Mexico (255429). C M-M and C G-R are supported by a CONACYT predoctoral training grant.
Author contribution statement
C M-M and E P S designed and analyzed the experiments, and wrote the manuscript. C M-M and P C-R performed the experiments. C G-R participated in critical revision of manuscript.
Acknowledgements
We are grateful to Nora Ruiz for her technical assistance.
References
- 1.Rossini A, Zanobbio L, Sfondrini L, Cavalleri A, Secreto G, Morelli D, Palazzo M, Sommariva M, Tagliabue E, Rumio C, et al Influence of fatty acid-free diet on mammary tumor development and growth rate in HER-2/Neu transgenic mice. Journal of Cellular Physiology 2013. 228 242–249. ( 10.1002/jcp.24130) [DOI] [PubMed] [Google Scholar]
- 2.Carmichael AR. Obesity and prognosis of breast cancer. Obesity Reviews 2006. 7 333–340. ( 10.1111/j.1467-789X.2006.00261.x) [DOI] [PubMed] [Google Scholar]
- 3.Xue F, Michels KB. Diabetes, metabolic syndrome, and breast cancer: a review of the current evidence. American Journal of Clinical Nutrition 2007. 86 s823–s835. ( 10.1093/ajcn/86.3.823S) [DOI] [PubMed] [Google Scholar]
- 4.Calle EE, Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nature Reviews Cancer 2004. 4 579–591. ( 10.1038/nrc1408) [DOI] [PubMed] [Google Scholar]
- 5.Byon CH, Hardy RW, Ren C, Ponnazhagan S, Welch DR, McDonald JM, Chen Y. Free fatty acids enhance breast cancer cell migration through plasminogen activator inhibitor-1 and SMAD4. Laboratory Investigation 2009. 89 1221–1228. ( 10.1038/labinvest.2009.97) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hardy S, St-Onge GG, Joly E, Langelier Y, Prentki M. Oleate promotes the proliferation of breast cancer cells via the G protein-coupled receptor GPR40. Journal of Biological Chemistry 2005. 280 13285–13291. ( 10.1074/jbc.M410922200) [DOI] [PubMed] [Google Scholar]
- 7.McArthur MJ, Atshaves BP, Frolov A, Foxworth WD, Kier AB, Schroeder F. Cellular uptake and intracellular trafficking of long chain fatty acids. Journal of Lipid Research 1999. 40 1371–1383. [PubMed] [Google Scholar]
- 8.Navarro-Tito N, Soto-Guzman A, Castro-Sanchez L, Martinez-Orozco R, Salazar EP. Oleic acid promotes migration on MDA-MB-231 breast cancer cells through an arachidonic acid-dependent pathway. International Journal of Biochemistry and Cell Biology 2010. 42 306–317. ( 10.1016/j.biocel.2009.11.010) [DOI] [PubMed] [Google Scholar]
- 9.Soto-Guzman A, Robledo T, Lopez-Perez M, Salazar EP. Oleic acid induces ERK1/2 activation and AP-1 DNA binding activity through a mechanism involving Src kinase and EGFR transactivation in breast cancer cells. Molecular and Cellular Endocrinology 2008. 294 81–91. ( 10.1016/j.mce.2008.08.003) [DOI] [PubMed] [Google Scholar]
- 10.Soto-Guzman A, Navarro-Tito N, Castro-Sanchez L, Martinez-Orozco R, Salazar EP. Oleic acid promotes MMP-9 secretion and invasion in breast cancer cells. Clinical and Experimental Metastasis 2010. 27 505–515. ( 10.1007/s10585-010-9340-1) [DOI] [PubMed] [Google Scholar]
- 11.Hirasawa A, Tsumaya K, Awaji T, Katsuma S, Adachi T, Yamada M, Sugimoto Y, Miyazaki S, Tsujimoto G. Free fatty acids regulate gut incretin glucagon-like peptide-1 secretion through GPR120. Nature Medicine 2005. 11 90–94. ( 10.1038/nm1168) [DOI] [PubMed] [Google Scholar]
- 12.Briscoe CP, Tadayyon M, Andrews JL, Benson WG, Chambers JK, Eilert MM, Ellis C, Elshourbagy NA, Goetz AS, Minnick DT, et al The orphan G protein-coupled receptor GPR40 is activated by medium and long chain fatty acids. Journal of Biological Chemistry 2003. 278 11303–11311. ( 10.1074/jbc.M211495200) [DOI] [PubMed] [Google Scholar]
- 13.Briscoe CP, Peat AJ, McKeown SC, Corbett DF, Goetz AS, Littleton TR, McCoy DC, Kenakin TP, Andrews JL, Ammala C, et al Pharmacological regulation of insulin secretion in min6 cells through the fatty acid receptor GPR40: identification of agonist and antagonist small molecules. British Journal of Pharmacology 2006. 148 619–628. ( 10.1038/sj.bjp.0706770) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cartoni C, Yasumatsu K, Ohkuri T, Shigemura N, Yoshida R, Godinot N, le Coutre J, Ninomiya Y, Damak S. Taste preference for fatty acids is mediated by GPR40 and GPR120. Journal of Neuroscience 2010. 30 8376–8382. ( 10.1523/JNEUROSCI.0496-10.2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozdener MH, Subramaniam S, Sundaresan S, Sery O, Hashimoto T, Asakawa Y, Besnard P, Abumrad NA, Khan NA. CD36- and GPR120-mediated Ca(2)(+) signaling in human taste bud cells mediates differential responses to fatty acids and is altered in obese mice. Gastroenterology 2014. 146 995–1005. ( 10.1053/j.gastro.2014.01.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oh DY, Walenta E, Akiyama TE, Lagakos WS, Lackey D, Pessentheiner AR, Sasik R, Hah N, Chi TJ, Cox JM, et al A Gpr120-selective agonist improves insulin resistance and chronic inflammation in obese mice. Nature Medicine 2014. 20 942–947. ( 10.1038/nm.3614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navarro-Tito N, Robledo T, Salazar EP. Arachidonic acid promotes FAK activation and migration in MDA-MB-231 breast cancer cells. Experimental Cell Research 2008. 314 3340–3355. ( 10.1016/j.yexcr.2008.08.018) [DOI] [PubMed] [Google Scholar]
- 18.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nature Reviews Genetics 2006. 7 606–619. ( 10.1038/nrg1879) [DOI] [PubMed] [Google Scholar]
- 19.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nature Reviews Cancer 2009. 9 550–562. ( 10.1038/nrc2664) [DOI] [PubMed] [Google Scholar]
- 20.Fresno Vara JA, Casado E, de Castro J, Cejas P, Belda-Iniesta C, Gonzalez-Baron M. PI3K/Akt signalling pathway and cancer. Cancer Treatment Reviews 2004. 30 193–204. ( 10.1016/j.ctrv.2003.07.007) [DOI] [PubMed] [Google Scholar]
- 21.Dillon RL, White DE, Muller WJ. The phosphatidyl inositol 3-kinase signaling network: implications for human breast cancer. Oncogene 2007. 26 1338–1345. ( 10.1038/sj.onc.1210202) [DOI] [PubMed] [Google Scholar]
- 22.Dillon RL, Muller WJ. Distinct biological roles for the Akt family in mammary tumor progression. Cancer Research 2010. 70 4260–4264. ( 10.1158/0008-5472.CAN-10-0266) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irie HY, Pearline RV, Grueneberg D, Hsia M, Ravichandran P, Kothari N, Natesan S, Brugge JS. Distinct roles of Akt1 and Akt2 in regulating cell migration and epithelial-mesenchymal transition. Journal of Cell Biology 2005. 171 1023–1034. ( 10.1083/jcb.200505087) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maloberti P, Cornejo Maciel F, Castillo AF, Castilla R, Duarte A, Toledo MF, Meuli F, Mele P, Paz C, Podesta EJ. Enzymes involved in arachidonic acid release in adrenal and Leydig cells. Molecular and Cellular Endocrinology 2007. 265–266 113–120. ( 10.1016/j.mce.2006.12.026) [DOI] [PubMed] [Google Scholar]
- 25.Dessen A. Structure and mechanism of human cytosolic phospholipase A(2). Biochimica and Biophysica Acta 2000. 1488 40–47. ( 10.1016/S1388-1981(00)00108-6) [DOI] [PubMed] [Google Scholar]
- 26.Brash AR. Lipoxygenases: occurrence, functions, catalysis, and acquisition of substrate. Journal of Biological Chemistry 1999. 274 23679–23682. ( 10.1074/jbc.274.34.23679) [DOI] [PubMed] [Google Scholar]
- 27.Urade Y, Watanabe K, Hayaishi O. Prostaglandin D, E, and F synthases. Journal of Lipid Mediators and Cell Signalling 1995. 12 257–273. ( 10.1016/0929-7855(95)00032-L) [DOI] [PubMed] [Google Scholar]
- 28.Harizi H, Corcuff JB, Gualde N. Arachidonic-acid-derived eicosanoids: roles in biology and immunopathology. Trends in Molecular Medicine 2008. 14 461–469. ( 10.1016/j.molmed.2008.08.005) [DOI] [PubMed] [Google Scholar]
- 29.Soto-Guzman A, Villegas-Comonfort S, Cortes-Reynosa P, Perez Salazar E. Role of arachidonic acid metabolism in Stat5 activation induced by oleic acid in MDA-MB-231 breast cancer cells. Prostaglandins, Leukotrienes, and Essential Fatty Acids 2013. 88 243–249. ( 10.1016/j.plefa.2012.12.003) [DOI] [PubMed] [Google Scholar]
- 30.Li S, Zhou T, Li C, Dai Z, Che D, Yao Y, Li L, Ma J, Yang X, Gao G. High metastaticgastric and breast cancer cells consume oleic acid in an AMPK dependent manner. PLoS One 2014. 9 e97330 ( 10.1371/journal.pone.0097330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sparks SM, Chen G, Collins JL, Danger D, Dock ST, Jayawickreme C, Jenkinson S, Laudeman C, Leesnitzer MA, Liang X, et al Identification of diarylsulfonamides as agonists of the free fatty acid receptor 4 (FFA4/GPR120). Bioorganic and Medicinal Chemistry Letters 2014. 24 3100–3103. ( 10.1016/j.bmcl.2014.05.012) [DOI] [PubMed] [Google Scholar]
- 32.Sun P, Wang T, Zhou Y, Liu H, Jiang H, Zhu W, Wang H. DC260126: a small-molecule antagonist of GPR40 that protects against pancreatic beta-Cells dysfunction in db/db mice. PLoS One 2013. 8 e66744 ( 10.1371/journal.pone.0066744) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu H, He LY, Gong Z, Li N, Lu YN, Zhai QW, Liu H, Jiang HL, Zhu WL, Wang HY. A novel class of antagonists for the FFAs receptor GPR40. Biochemical and Biophysical Research Communications 2009. 390 557–563. ( 10.1016/j.bbrc.2009.10.004) [DOI] [PubMed] [Google Scholar]
- 34.Hu C, Huang L, Gest C, Xi X, Janin A, Soria C, Li H, Lu H. Opposite regulation by PI3K/Akt and MAPK/ERK pathways of tissue factor expression, cell-associated procoagulant activity and invasiveness in MDA-MB-231 cells. Journal of Hematology and Oncology 2012. 5 16 ( 10.1186/1756-8722-5-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). Journal of Biological Chemistry 1994. 269 5241–5248. [PubMed] [Google Scholar]
- 36.Xu J, Zhang Y, Xiao Y, Ma S, Liu Q, Dang S, Jin M, Shi Y, Wan B, Zhang Y. Inhibition of 12/15-lipoxygenase by baicalein induces microglia PPARbeta/delta: a potential therapeutic role for CNS autoimmune disease. Cell Death and Disease 2013. 4 e569 ( 10.1038/cddis.2013.86) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deschamps JD, Kenyon VA, Holman TR. Baicalein is a potent in vitro inhibitor against both reticulocyte 15-human and platelet 12-human lipoxygenases. Bioorganic and Medicinal Chemistry 2006. 14 4295–4301. ( 10.1016/j.bmc.2006.01.057) [DOI] [PubMed] [Google Scholar]
- 38.Schaller MD. Paxillin: a focal adhesion-associated adaptor protein. Oncogene 2001. 20 6459–6472. ( 10.1038/sj.onc.1204786) [DOI] [PubMed] [Google Scholar]
- 39.Ward WH, Cook PN, Slater AM, Davies DH, Holdgate GA, Green LR. Epidermal growth factor receptor tyrosine kinase. Investigation of catalytic mechanism, structure-based searching and discovery of a potent inhibitor. Biochemical Pharmacology 1994. 48 659–666. ( 10.1016/0006-2952(94)90042-6) [DOI] [PubMed] [Google Scholar]
- 40.Boyd NF, Stone J, Vogt KN, Connelly BS, Martin LJ, Minkin S. Dietary fat and breast cancer risk revisited: a meta-analysis of the published literature. British Journal of Cancer 2003. 89 1672–1685. ( 10.1038/sj.bjc.6601314) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lahmann PH, Hoffmann K, Allen N, van Gils CH, Khaw KT, Tehard B, Berrino F, Tjonneland A, Bigaard J, Olsen A, et al Body size and breast cancer risk: findings from the European Prospective Investigation into Cancer and Nutrition (EPIC). International Journal of Cancer 2004. 111 762–771. ( 10.1002/ijc.20315) [DOI] [PubMed] [Google Scholar]
- 42.Houthuijzen JM. For better or worse: FFAR1 and FFAR4 signaling in cancer and diabetes. Molecular Pharmacology 2016. 90 738–743. ( 10.1124/mol.116.105932) [DOI] [PubMed] [Google Scholar]
- 43.Yonezawa T, Katoh K, Obara Y. Existence of GPR40 functioning in a human breast cancer cell line, MCF-7. Biochemical and Biophysical Research Communications 2004. 314 805–809. ( 10.1016/j.bbrc.2003.12.175) [DOI] [PubMed] [Google Scholar]
- 44.Hopkins MM, Zhang Z, Liu Z, Meier KE. Eicosopentaneoic acid and other free fatty acid receptor agonists inhibit lysophosphatidic acid- and epidermal growth factor-induced proliferation of human breast cancer cells. Journal of Clinical Medicine 2016. 5 16. ( 10.3390/jcm5020016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fukushima K, Yamasaki E, Ishii S, Tomimatsu A, Takahashi K, Hirane M, Fukushima N, Honoki K, Tsujiuchi T. Different roles of GPR120 and GPR40 in the acquisition of malignant properties in pancreatic cancer cells. Biochemical and Biophysical Research Communications 2015. 465 512–515. ( 10.1016/j.bbrc.2015.08.050) [DOI] [PubMed] [Google Scholar]
- 46.Ishii S, Kitamura Y, Hirane M, Tomimatsu A, Fukushima K, Takahashi K, Fukushima N, Honoki K, Tsujiuchi T. Negative effects of G-protein-coupled free fatty acid receptor GPR40 on cell migration and invasion in fibrosarcoma HT1080 cells. Molecular Carcinogenesis 2016. 55 1553–1559. ( 10.1002/mc.22408) [DOI] [PubMed] [Google Scholar]
- 47.Clark AR, Toker A. Signalling specificity in the Akt pathway in breast cancer. Biochemical Society Transactions 2014. 42 1349–1355. ( 10.1042/BST20140160) [DOI] [PubMed] [Google Scholar]
- 48.Chin YR, Toker A. Akt isoform-specific signaling in breast cancer: uncovering an anti-migratory role for palladin. Cell Adhesion and Migration 2011. 5 211–214. ( 10.4161/cam.5.3.15790) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hutchinson JN, Jin J, Cardiff RD, Woodgett JR, Muller WJ. Activation of Akt-1 (PKB-alpha) can accelerate ErbB-2-mediated mammary tumorigenesis but suppresses tumor invasion. Cancer Research 2004. 64 3171–3178. ( 10.1158/0008-5472.CAN-03-3465) [DOI] [PubMed] [Google Scholar]
- 50.Arboleda MJ, Lyons JF, Kabbinavar FF, Bray MR, Snow BE, Ayala R, Danino M, Karlan BY, Slamon DJ. Overexpression of AKT2/protein kinase Bbeta leads to up-regulation of beta1 integrins, increased invasion, and metastasis of human breast and ovarian cancer cells. Cancer Research 2003. 63 196–206. [PubMed] [Google Scholar]
- 51.Prihandoko R, Alvarez-Curto E, Hudson BD, Butcher AJ, Ulven T, Miller AM, Tobin AB, Milligan G. Distinct phosphorylation clusters determine the signaling outcome of free fatty acid receptor 4/G protein-coupled Receptor 120. Molecular Pharmacology 2016. 89 505–520. ( 10.1124/mol.115.101949) [DOI] [PubMed] [Google Scholar]
- 52.Wells A, Grahovac J, Wheeler S, Ma B, Lauffenburger D. Targeting tumor cell motility as a strategy against invasion and metastasis. Trends in Pharmacological Sciences 2013. 34 283–289. ( 10.1016/j.tips.2013.03.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alizadeh AM, Shiri S, Farsinejad S. Metastasis review: from bench to bedside. Tumour Biology 2014. 35 8483–8523. ( 10.1007/s13277-014-2421-z) [DOI] [PubMed] [Google Scholar]
- 54.Guo W, Giancotti FG. Integrin signalling during tumour progression. Nature Reviews. Molecular Cell Biology 2004. 5 816–826. ( 10.1038/nrm1490) [DOI] [PubMed] [Google Scholar]
- 55.Wozniak MA, Modzelewska K, Kwong L, Keely PJ. Focal adhesion regulation of cell behavior. Biochimica and Biophysica Acta 2004. 1692 103–119. ( 10.1016/j.bbamcr.2004.04.007) [DOI] [PubMed] [Google Scholar]
- 56.Kallergi G, Agelaki S, Markomanolaki H, Georgoulias V, Stournaras C. Activation of FAK/PI3K/Rac1 signaling controls actin reorganization and inhibits cell motility in human cancer cells. Cellular Physiology and Biochemistry 2007. 20 977–986. ( 10.1159/000110458) [DOI] [PubMed] [Google Scholar]
- 57.Monterrubio M, Mellado M, Carrera AC, Rodriguez-Frade JM. PI3Kgamma activation by CXCL12 regulates tumor cell adhesion and invasion. Biochemical and Biophysical Research Communications 2009. 388 199–204. ( 10.1016/j.bbrc.2009.07.153) [DOI] [PubMed] [Google Scholar]
- 58.Reiske HR, Kao SC, Cary LA, Guan JL, Lai JF, Chen HC. Requirement of phosphatidylinositol 3-kinase in focal adhesion kinase-promoted cell migration. Journal of Biological Chemistry 1999. 274 12361–12366. ( 10.1074/jbc.274.18.12361) [DOI] [PubMed] [Google Scholar]
- 59.Honn KV, Tang DG, Gao X, Butovich IA, Liu B, Timar J, Hagmann W. 12-lipoxygenases and 12(S)-HETE: role in cancer metastasis. Cancer Metastasis Reviews 1994. 13 365–396. ( 10.1007/BF00666105) [DOI] [PubMed] [Google Scholar]
- 60.Mashima R, Okuyama T. The role of lipoxygenases in pathophysiology; new insights and future perspectives. Redox Biology 2015. 6 297–310. ( 10.1016/j.redox.2015.08.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dilly AK, Ekambaram P, Guo Y, Cai Y, Tucker SC, Fridman R, Kandouz M, Honn KV. Platelet-type 12-lipoxygenase induces MMP9 expression and cellular invasion via activation of PI3K/Akt/NF-kappaB. International Journal of Cancer 2013. 133 1784–1791. ( 10.1002/ijc.28165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fischer OM, Hart S, Gschwind A, Ullrich A. EGFR signal transactivation in cancer cells. Biochemical Society Transactions 2003. 31 1203–1208. ( 10.1042/BST0311203) [DOI] [PubMed] [Google Scholar]
- 63.Oh S, Kim H, Nam K, Shin I. Glut1 promotes cell proliferation, migration and invasion by regulating epidermal growth factor receptor and integrin signaling in triple-negative breast cancer cells. BMB Reports 2017. 50 132–137. ( 10.5483/BMBRep.2017.50.3.189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Han T, Guo M, Zhang T, Gan M, Xie C, Wang JB. A novel glutaminase inhibitor-968 inhibits the migration and proliferation of non-small cell lung cancer cells by targeting EGFR/ERK signaling pathway. Oncotarget 2017. 8 28063–28073. ( 10.18632/oncotarget.14188) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prasad S, Ravindran J, Aggarwal BB. NF-kappaB and cancer: how intimate is this relationship. Molecular and Cellular Biochemistry 2010. 336 25–37. ( 10.1007/s11010-009-0267-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature 2006. 441 431–436. ( 10.1038/nature04870) [DOI] [PubMed] [Google Scholar]
- 67.Shukla S, Maclennan GT, Marengo SR, Resnick MI, Gupta S. Constitutive activation of P I3 K-Akt and NF-kappaB during prostate cancer progression in autochthonous transgenic mouse model. Prostate 2005. 64 224–239. ( 10.1002/pros.20217) [DOI] [PubMed] [Google Scholar]
- 68.Dan HC, Cooper MJ, Cogswell PC, Duncan JA, Ting JP, Baldwin AS. Akt-dependent regulation of NF-{kappa}B is controlled by mTOR and Raptor in association with IKK. Genes and Development 2008. 22 1490–1500. ( 10.1101/gad.1662308) [DOI] [PMC free article] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a