Significance
All known terrestrial protein enzymes fold robustly and explore the complex energy landscapes that are required for activity. The catalytic residues must be held together in precise positions that may conflict with the correct folding dynamics of the domain itself. We quantified these energetic conflicts and found that these are effectively present in extant enzymes, regardless of the protein oligomeric state, overall topology, and enzymatic class. The conservation of these energetic signatures is higher than the conservation quantified in the primary structures.
Keywords: local frustration, protein enzymes, catalytic sites, evolution, bioinformatics
Abstract
Conflicting biological goals often meet in the specification of protein sequences for structure and function. Overall, strong energetic conflicts are minimized in folded native states according to the principle of minimal frustration, so that a sequence can spontaneously fold, but local violations of this principle open up the possibility to encode the complex energy landscapes that are required for active biological functions. We survey the local energetic frustration patterns of all protein enzymes with known structures and experimentally annotated catalytic residues. In agreement with previous hypotheses, the catalytic sites themselves are often highly frustrated regardless of the protein oligomeric state, overall topology, and enzymatic class. At the same time a secondary shell of more weakly frustrated interactions surrounds the catalytic site itself. We evaluate the conservation of these energetic signatures in various family members of major enzyme classes, showing that local frustration is evolutionarily more conserved than the primary structure itself.
Along with heredity, the evolution of specific catalytic activity is the physicochemical core of life. By controlling the dissipation of chemical energy, modern enzymes allow organisms to coordinate how the energy flows with astounding efficiency and robustness (1). The availability of both the sequences and inferred structures of many enzymes makes possible new modes of exploration of how specific biochemical activities have come in to existence and evolved. To achieve catalytic activity, folding of the polypeptide chains of an enzyme must bring residues together in space with some precision. Because of this functional necessity, the residues that are directly involved in the catalytic act must be supported by the folding of the rest of the structure, but may themselves be in conflict with that overall fold (2). Many studies have shown that mutations at catalytic sites often make protein folds more stable, while destabilizing mutations elsewhere are the norm (3, 4); however, this is not always the case (5). Both catalytic and folding studies in the laboratory are time consuming so examining the connections between folding and catalysis has been carried out only on an anecdotal basis. Here we use energy landscape ideas along with the now available rich sequence and structural data to explore these questions more thoroughly.
To locate the conflicts encoded in protein sequence and structure one needs a reliable way to measure how well a sequence fits into a structure. A simple heuristic method based on the energy landscape theory of protein folding has been shown to be very useful for analyzing how the stabilization energy in protein domains is distributed (6). Briefly, the contribution of a particular interaction in a natural protein is compared with the free energy of alternative sequence–structure pairings. A local frustration index can then be defined as the Z score of the free energy of those energetic contributions to the native structure with respect to the distribution of the energy of decoys with rearranged sequences or structures. If a native pair of interacting residues has an energy that lies in the most favorable end of the distribution among alternative decoys, the interaction is labeled as minimally frustrated, as most changes in that location will destabilize the overall structure. Conversely, the regions in which most local sequence or structural changes would lower the free energy of the system are labeled as highly frustrated and must be held there not only over the functional time but also over generations of evolutionary time at the expense of other interactions (7). In this paper, we analyze the local frustration patterns for all protein enzymes with currently known structures and experimentally characterized catalytic residues from the Catalytic Site Atlas (8). We compare the resulting frustration patterns of the proteins according to their oligomeric state, overall protein topology, and enzymatic activity. We further evaluate the energetic signatures across the family members of two popular enzymatic classes and find that local frustration is evolutionarily conserved close to the catalytic and other functionally related residues.
Results
Catalytic Sites of Enzymes Are Spatially Surrounded by Highly Frustrated Interactions.
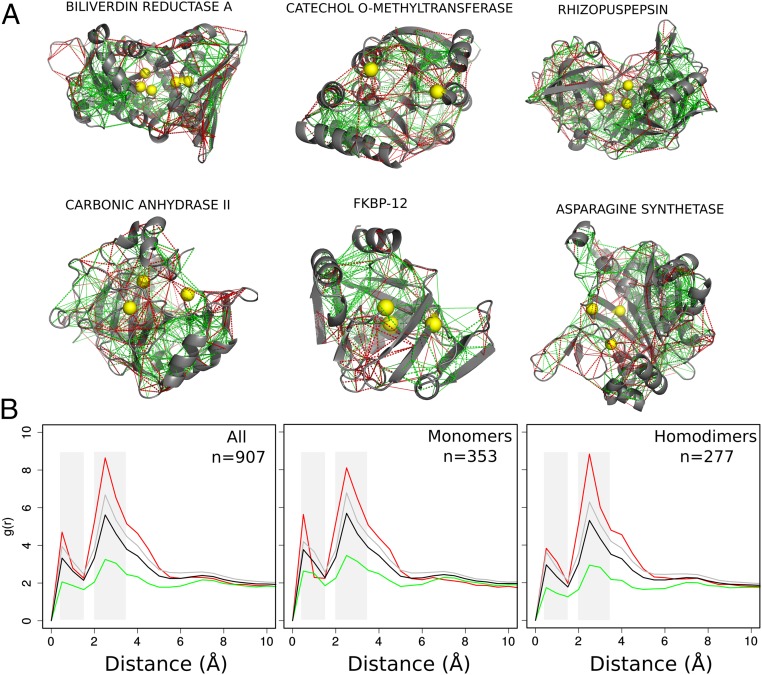
To analyze the local frustration distribution in protein enzymes, we collected all entries in the Catalytic Site Atlas (CSA) (8) for which one can find at least one high-resolution structure and for which catalytic residues have been experimentally assigned (907 nonredundant entries). We then calculated the local frustration patterns using the Frustratometer server (9, 10). Fig. 1A shows examples of the local frustration patterns in enzymes. It is apparent that the macromolecular frameworks are strongly interconnected by minimally frustrated interactions and that, in contrast, highly frustrated interactions typically form clusters. Many of these can be found at the surface of the globules, perhaps reflecting binding or allosteric sites, in line with the observations of general local frustration patterns in globular proteins as described earlier (7, 11, 12). Catalytic residues are also found close in space and again appear to be depleted in minimally frustrated contacts. To quantify the local frustration patterns we calculated the pair distribution functions for the various classes of contacts as measured by the frustration index as a function of distance from the C of the catalytic residues to the center of mass of the interactions (Methods). The for each frustration class is then compared with the of all contacts, which accounts for the geometry of the residue–residue interaction network defined by the protein topology. Fig. 1B shows the function for the mutational frustration index for all of the entries in the dataset. The distribution of interactions around the catalytic sites displays two characteristic peaks [black lines in Fig. 1B: one located around 1 Å, corresponding to those interactions of the catalytic residues themselves (first shell), and a second peak between 2 Å and 3.5 Å, which comprises interactions between residues that coordinate the catalytic residues (second shell)]. The density of minimally frustrated contacts is depleted in the first and second shells while both neutral and highly frustrated interactions are enriched in these shells compared with the overall contacts distribution (Fig. 1B, Left). Since catalytic sites in oligomers are frequently located near the interface between chains and protein–protein interaction regions that are typically enriched in highly frustrated interactions (7); therefore we divided the dataset according to the oligomeric state of the protein (SI Appendix, Table S1). Fig. 1B shows that the overall patterns of finding depletion of the minimally frustrated interactions around catalytic sites still hold in these sets. Both monomers and homodimers show an enrichment of highly frustrated interactions in both shells (Fig. 1B). To investigate whether these patterns are specific for catalytic sites, we performed controls of the for randomly chosen noncatalytic residues that show no enrichment of any type of interactions (SI Appendix, Fig. S1). Additionally, we observed that the solvent accessibility is not a determinant for the signal, since both buried and exposed catalytic residues show an enrichment of highly frustrated interactions (SI Appendix, Fig. S2). A complementary analysis based on the location of annotated metallic cofactors shows that these are also spatially enriched with highly frustrated interactions (SI Appendix, Fig. S3).
Fig. 1.
Local frustration patterns in enzymes. (A) Examples of frustration patterns in various enzymes. The backbones of the proteins are shown as gray cartoons, minimally frustrated contacts are depicted with green lines, and highly frustrated interactions with red lines. Neutral interactions were omitted to help visualization. The alpha carbons (C) of catalytic sites are marked with yellow spheres. (B) Pair distribution functions, , between the C of the annotated catalytic residues and the center of mass of the contacts, divided by local frustration class, for the monomeric enzymes, homodimers, and all of the dataset. Green, minimally frustrated contacts; red, highly frustrated contacts; gray, neutral contacts; black, all contacts. g(r) plots were adjusted in their axis ranges to enhance visualizations; however, in all cases g(r) values were normalized such that g(20) = 1.
There are two complementary ways of defining local frustration (7). The mutational frustration index (discussed above) measures how favorable native residues are relative to other residues that could have been in the same location. The configurational frustration index indicates how favorable the native interactions between the two residues are with respect to other interactions they can form in other compact structures. Interestingly, the two types of local frustration patterns do not overlap precisely in the enzymes sets. SI Appendix, Fig. S4 shows the results for the function for the configurational frustration index for all of the entries as well as for the monomers and homodimers. There is a slight enrichment of highly frustrated and neutral interactions in the second shell only and most highly frustrated interactions are depleted around catalytic sites. The distributions of all other types of interactions are not visibly enriched in proximity to the catalytic residues when the configurational index is used. Thus, the principal factor that accounts for the energetic conflicts around catalytic sites is the identity of the residues that compose the surroundings of the catalytic sites and not their conformational localization. The energetic conflict comes from having those specific identities in those specific locations, as expected for the strong functional requirement of keeping chemically active residues close in space.
Local Frustration Enrichment Is Independent of Catalytic Mechanism.
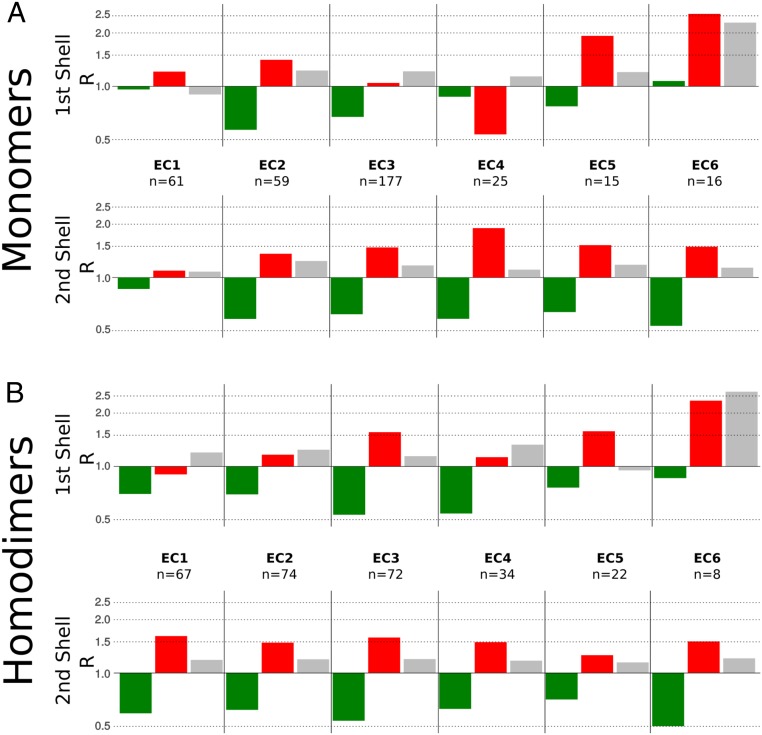
We observe a general enrichment of highly frustrated interactions around catalytic sites. To analyze whether this a common feature of all enzymes or is attributable only to some subgroups, we constructed datasets according to the first level of the Enzyme Commission classification (EC number) which hierarchically segregates enzymes as oxidoreductases (EC 1), transferases (EC 2), hydrolases (EC 3), lyases (EC 4), isomerases (EC 5), and ligases (EC 6) (SI Appendix, Table S2). A new class EC 7, translocases, was added to the EC list in October 2018; however, we did not include it in this analysis. Fig. 2 shows the ratio between the value of the for each contact type ()) and that for all of the contacts ()), for both coordinations shells and every enzyme type: . SI Appendix, Fig. S5 shows the results for these functions. All of the enzyme classes show depletion of the minimally frustrated interactions in both shells with the exception of EC 1 and EC 6 which do not show this behavior in the first shell. All EC classes show various degrees of enrichment of frustrated interactions in the second shell, with monomers of EC 1 showing the lowest enrichment. The signal at the first shell shows more variability with monomeric EC 3 and dimeric EC 2 and EC 4. Similarly, monomeric EC 4 and dimeric EC 1 display an exclusion of highly frustrated interactions. The enrichment of highly frustrated interactions around active sites holds for all enzyme classes, with distinct patterns. EC classes group together enzymes that catalyze very diverse chemical reactions by different mechanisms. To further explore the frustration distributions we analyzed the set of monomers corresponding to the hydrolases (EC 3), which corresponds to the most populous class, and went down one level in the EC number classification (SI Appendix, Table S3) to inspect whether there were differences among the subclasses that have more than five structures reported in the CSA (SI Appendix, Fig. S6). Although again one finds slight differences in the frustration distributions, all of the subclasses show an enrichment of highly frustrated interactions around their catalytic sites.
Fig. 2.
(A and B) Local frustration patterns of EC classes for (A) monomers and (B) homodimers. The ratio of the value for each type of contact (green, minimally frustrated; red, highly frustrated; gray, neutral) and the total involving all interaction functions is shown. The first shell is defined from 0.5 Å to 1.5 Å with respect to the catalytic residue positions, and the second shell is defined from 2 Å to 3.5 Å.
Local Frustration Enrichment Is Independent of Fold Topology.
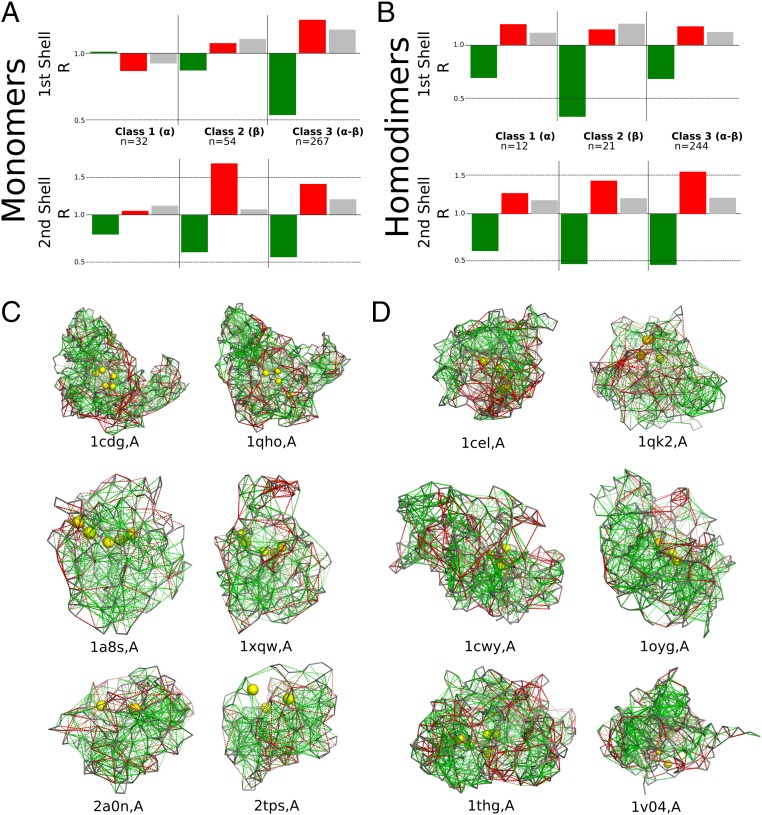
Enzymes show local energetic conflicts around their catalytic sites. Sometimes the highly frustrated interactions directly involve the catalytic residues themselves but sometimes they involve residues in close proximity to the catalytic sites. One question that arises is whether the frustration patterns around the catalytic sites are related to the protein topology. We note that proteins with the same EC classification are not necessarily similar in structure (13). We therefore also classified the monomer and homodimeric sets according to their class, architecture, topology, and homologous superfamily (CATH) class (14), which describes the overall secondary-structure composition of the domain, hierarchically classifying proteins into mainly (class 1), mainly (class 2), and - (class 3). Fig. 3 shows the for the three different CATH classes, for both monomers (Fig. 3A) and homodimers (Fig. 3B). SI Appendix, Fig. S7 shows the results for these functions. For most proteins, we see there are highly frustrated interactions near their catalytic sites, regardless of their CATH classification. In all cases we observe an enrichment of highly frustrated interactions at the second shell, but the first shell is enriched in this type of interactions only for CATH classes 2 and 3 which have structures. Although CATH classes are a useful first approximation toward classifying topology, the relation between CATH number and structural identity is not definitive. Therefore, we aligned all of the pairs of monomer structures using Topmatch (15) and analyzed the frustration patterns of protein pairs that either have a high structural identity but a different EC number or catalyze the same reaction but are structurally very dissimilar (SI Appendix, Table S4). It is interesting to note that for enzymes having similar structures that catalyze different reactions, the active sites are located in the same region even when their residue composition differs. Some examples of the structures compared are shown in Fig. 3C. The cyclomaltodextrin glucanotransferases [Protein Data Bank (PDB) ID code 1cdg] and glucan 1,4--maltohydrolases (PDB ID code 1qho) are both TIM barrels; they also share the same acidic catalytic triad, D-D-E, and catalytic histidine, but the transferase has an additional catalytic arginine. Likewise, the chloride peroxidases (PDB ID code 1a8s) and prolyl aminopeptidases (PDB ID code 1xqw) both have Rossman folds and share an acid–base–nucleophile catalytic triad, where the nucleophile is a serine for the peroxidase and an alanine for the peptidase. Moreover, these enzymes share a similar mechanism in which an additional pair of aromatic and neutral amino acids help form the oxoanion hole which stabilizes the intermediate. Although the imidazole glycerol phosphate synthase subunit hisF (PDB ID code 2a0n) and the thiamine–phosphate diphosphorylase (PDB ID code 2tps) share the same fold and their catalytic sites are localized in the same area of the protein, their catalytic residues are different. On the other hand, we also can find that for proteins with a low structural similarity but a shared function, the location of the catalytic site and its residue composition may vary. Exoglucanases I (PDB ID code 1cel) and II (PDB ID code 1qk2) catalyze the exact same reaction but their structures and topologies differ, as the first class has a -fold structure and the second class consists of TIM barrels (Fig. 3D). Both proteins have at least two acidic residues at their catalytic site, but otherwise their compositions differ. The 4--glucanotransferases (PDB ID code 1cwy) and levansucrases (PDB ID code 1oyg) are both hexosyltransferases. Although the reactions catalyzed by the two classes are similar and both display an acidic catalytic triad, the glucanotransferases are TIM barrels, while the levansucrases are propellers. Furthermore, triacylglycerol lipases ((PDB ID code 1thg) and arylesterases ((PDB ID code 1v04) are both carboxylic ester hydrolases; however, their folds and catalytic residues differ. While the lipases have a Rossmann fold, the arylesterases are propellers. It is worth noting that we did not find any isomerases (EC 5) or ligases (EC 6) that display structural similarity to proteins that catalyze other reactions (i.e., have a different EC class). Moreover, for some combinations of EC classes, such as oxidoreductases (EC 1) and transferases (EC 2), there are no highly similar structures either. This suggests that certain topologies make enzymes switch more easily between types of reactions, which has been reported already for Rossmann folds and TIM barrels (16, 17). On the other hand, looking at proteins that share an activity but have very different topology, most belong to EC class 3.2.1.-, which is the glycosidases, i.e., enzymes hydrolyzing O- and S-glycosyl compounds, suggesting that this activity may be easier to acquire. None of these observations is a side effect of the abundance of these proteins, as the EC number distribution is balanced in these sets.
Fig. 3.
Local frustration and topology. (A and B) Local frustration patterns of CATH classes for (A) monomers and (B) homodimers. The ratio of the value for each type of contact (green, minimally frustrated contacts; red, highly frustrated contacts; gray, neutral contacts) and all interactions is shown. The first shell is defined from 0.5 Å to 1.5 Å with respect to the catalytic residue positions, and the second shell is defined from 2 Å to 3.5 Å. (C) Frustration patterns in enzyme pairs that share a high structural similarity but catalyze different reactions. (D) Frustration patterns in enzyme pairs that catalyze similar reactions but are structurally different. The backbones of the proteins are shown as gray ribbons, minimally frustrated contacts are depicted with green lines, and highly frustrated interactions with red lines. Neutral interactions were omitted to help visualization. The Cs of the catalytic residues are marked with yellow spheres.
Conservation of Energetic Conflicts at Catalytic Sites.
Since the number of structures with experimentally annotated catalytic residues is still small for some specific classifications in the EC number schema, we decided to analyze frustration patterns in catalytic residues at the level of the protein family. By analyzing various members of a common family, it is possible to study the conservation of frustration patterns in homologous positions across all members and compare it to the sequence conservation. To evaluate the conservation of energetic local frustration at specific positions of a multiple-sequence alignment (MSA) of a given protein family, we measured the information content of frustration states (FrstIC) by comparing the single-residue frustration index for all of the structures of members of a family. Schneider’s approach was used to compute the information content for both sequence (SeqIC) and the FrstIC, as developed in ref. 18.
-Lactamases Class A Family.
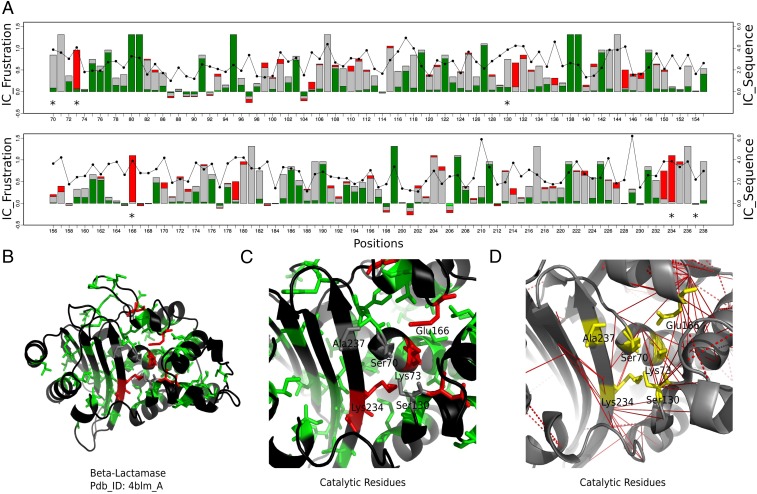
Class A -lactamases have six catalytic residues: Ser70, Lys73, Ser130, Glu166, Lys234, and Ala237. Of these residues Lys73, Glu166, and Lys234 are always found to be highly frustrated in the entire family (Fig. 4). In contrast, Ser70 and Ser130 are neutral in most cases and the frustration state of Ala237 is not conserved (FrstIC 0.5). Many of these catalytic sites display high FrstIC values based on the single-residue frustration index, while at the same time, many interactions with high FrstIC values based on the pairwise frustration index are found around them as well (Fig. 4D).
Fig. 4.
Conservation of local frustration and sequence identity for -lactamases class A family. (A) FrstIC based on the single-residue level frustration index. Green, minimally frustrated contacts; red, highly frustrated contacts; gray, neutral contacts. Black circles show the values of the SeqIC. (B) -Lactamase structure: in green and red, residues with FrstIC values greater than 0.5, minimally frustrated and highly frustrated, respectively. (C) Catalytic residues at the active site. (D) Mutational frustration at the catalytic site. Red lines correspond to highly frustrated interactions of the catalytic residues (in yellow) among themselves and with nearby residues in the structure.
Five noncatalytic residues display high FrstIC values and are highly frustrated. Asp131 when mutated negatively affects fitness in TEM-1 -lactamase (19). Asp233 belongs to a group of residues that are essential for wild-type levels of activity (20). Both Asp131 and Asp233 are located right next to the catalytic residues. Pro145 is one of two candidate prolines whose isomerization is relevant to folding (21). Tyr105 is a conserved residue involved in substrate recognition and binding (22). Mutations of Asp179 compromise the level of resistance of -lactamases to different antibiotics (23).
When calculating the single-residue frustration, a residue that establishes both highly and minimally frustrated interactions simultaneously will be assigned a neutral state that masks the existence of specific pair–residue conflictive interactions. Therefore, we calculated the mutational FrstIC contact map, i.e., for each pairwise interaction among residues (Fig. 4D). Although Ser70 and Ser130 have a neutral FrstIC at the single-residue level, they are found to participate in several conserved and highly frustrated interactions. Interestingly, most of these involve the other catalytic residues (SI Appendix, Table S5). It is worth noting that sequence conservation (black circles and lines on top of the FrstIC logo, Fig. 4A) does not correlate with the local frustration state conservation. For some positions that display high sequence variability, the local frustration signature is retained. For some other positions, the frustration state is sensitive to changes in sequence.
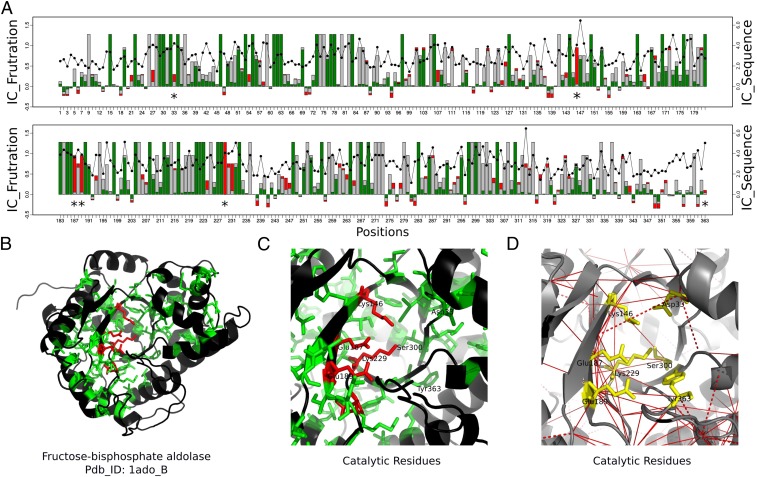
Fructose-Bisphosphate Aldolase Family.
Fructose-bisphosphate aldolase (EC 4.1.2.13), often just called aldolase, belongs to a superfamily of enzymes that catalyze a reversible reaction that splits the aldol, fructose 1,6-bisphosphate, into the triose phosphates dihydroxyacetone phosphate (DHAP) and glyceraldehyde 3-phosphate (G3P). The aldolases class I superfamily (CATH 3.20.20.70) has been classified into different functional families (24), and here we use the 1aldA00 functional family for which 17 nonredundant structures are known.
This family has seven catalytic residues annotated in the CSA database: Asp33, Lys146, Glu187, Glu189, Lys229, Ser300, and Tyr363. Lys146, Glu187, Glu189, and Lys229 have high FrstIC values with the highly frustrated state being the one that contributes the most to the conservation of local frustration (Fig. 5). Ser300 has a highly conserved FrstIC value in the neutral state. The catalytic residues at positions Asp33 and Tyr363 do not display high FrstIC values, showing that the energy distribution of their interactions is not conserved across the family but varies for each protein. As in the case of the -lactamase family, catalytic residues with neutral single-residue FrstIC values participate in a network of conserved highly frustrated interactions as shown in the FrstIC contact map (Fig. 5D and SI Appendix, Table S6). Asp33 has conserved highly frustrated interactions with Lys146. Also, Ser300 interacts in a highly frustrated way with Lys146, Glu187, and Lys229. Arg148, a residue in the active site, is known to be important to orient the substrate during the cleavage reaction and is involved in highly frustrated interactions with Glu189 and Lys229 (SI Appendix, Fig. S8).
Fig. 5.
Conservation of local frustration and sequence for the aldolases family. (A) FrstIC based on the single-residue level frustration index. Green, minimally frustrated contacts; red, highly frustrated contacts; gray, neutral contacts. Black circles show the values of the SeqIC. (B) Aldolase structure: in green and red, residues with FrstIC values greater than 0.5, minimally frustrated and highly frustrated, respectively. (C) Catalytic residues at the active site. (D) Frustration index at the contact level. Red lines correspond to highly frustrated interactions of the catalytic residues (in yellow) among themselves and with nearby residues in the structure.
Discussion
Local frustration has been linked to many functional aspects of proteins (6). Enzymes are particularly good test cases for inquiring about the relations between folding, dynamics, and biological function (25), as in addition to robust foldability one of their functions is clear: catalytic power which must persist over evolutionary times. Moreover, the chemical activities enzymes perform need also to be regulated by presumably optional interactions, coupling biological functions in metabolic space (26). The exploration of the excited states of the energy landscapes dictates the local rearrangements that occur in response to the environment (27). By analyzing the local energetic satisfaction or conflict in extant sequence–structure pairs, we can analyze the impact of these external constraints on the coding of biological information in linear strings of amino acids.
Here we have computed the local frustration patterns of all protein enzyme structures for which catalytic residues have been experimentally assigned in the CSA database. We found that regardless of chemical activity (Figs. 1 and 2) or topological classifications (Fig. 3), the enrichment of highly frustrated interactions around catalytic residues is a general feature of proteins. Frustration is manifested directly in the interactions within the catalytic residues (first shell) but also appears at residues that these contact (second shell). This is also a feature found in the vicinity of active sites that require metallic cofactors (SI Appendix, Fig. S3) and is not found at random (SI Appendix, Fig. S1). Thus, the multitude of biological functions that are coded in enzymatic polypeptides generally require there being local frustration in their folding energy landscapes.
When analyzing the frustration patterns of similar structures that catalyze different reactions, we noted that the catalytic sites are located in analogous frustrated regions, even when residue composition at those sites differs. This hints that the active sites emerge most likely in restricted places among the structures, constrained by the folded topology. On the other hand, we observe that proteins with a low structural similarity which catalyze the same reaction nevertheless retain highly frustrated interactions in their catalytic site, even when the enzymes are clearly nonhomologous. The fact that all types of catalytic sites in many different topologies show similar signs of local energetic frustration precludes a simple characterization of EC based on the local frustration pattern.
We evaluated the conservation of the local frustration patterns in many distinct members of two protein families. Both -lactamases (Fig. 4) and aldolases (Fig. 5) show that catalytic residues are consistently found to be highly frustrated. The general pattern of conserved minimally frustrated regions corresponds with the folding cores and some positions are found consistently neutral. It is notable that the degree of conservation of the local frustration state is stronger than the variations given by the sequences. Even for positions that display high sequence variability, the local frustration signature is retained, suggesting that the sequences can be tuned in multiple ways as long as local frustration is conserved in these sites. For some other positions, the frustration state is sensitive to changes in sequence, marking sites where sequence changes may modulate folding stability.
Methods
Dataset of Enzyme Protein Structures and Local Frustration.
We obtained the PDB ID codes of all of the structures whose catalytic residues are annotated with experimental evidence at the CSA database (CSA version 2.0) (8). The protein structures were downloaded from the PDB (https://www.rcsb.org/) and the frustration patterns were calculated using the protein Frustratometer software (9, 10) (www.frustratometer.tk/). The enzymes were classified in different sets according to their oligomeric state, EC number, and CATH classifications. The calculation of the degree of conservation of local frustration patterns for a family of proteins is described in ref. 18. Structures were aligned using Topmatch (15).
Pair Distribution Function to Quantify Local Frustration Patterns.
Since mutational frustration is assigned to the contact interaction between two residues, to quantify the density of contacts of each frustration type around a catalytic residue, or any residue in general, we first need to create virtual particles (VPs). These are points in the space that lies in the middle of the interaction between two residues, that is, the center of mass of the interaction. For each protein structure, we obtained the list of contacts for each frustration class and hence calculated a set of VPs coordinates. Subsequently, distances from the C from selected residues, catalytic or control, or cofactor molecules were calculated with respect to the VPs coordinates. g(r) plots were adjusted in their axis ranges to enhance visualizations; however, in all cases g(r) values were normalized such that g(20) = 1.
Supplementary Material
Acknowledgments
We acknowledge Prof. Christine Orengo and Dr. Sayone Das for providing data regarding the aldolase family. This work was supported by the Consejo de Investigaciones Cientificas y Tecnicas (CONICET), the Agencia Nacional de Promocion Cientifica y Tecnologica (Grant PICT2016/1467 to D.U.F.), ECOS Sud, Secretaría de Ciencia y Tecnología-A14E04, and NASA Astrobiology Institute-Enigma. D.U.F. is a CONICET researcher and A.B.G. holds a CONICET fellowship. This work was supported by the National Institute of General Medical Sciences (Grant R01GM44557 to P.G.W.). Additional support was provided by the D. R. Bullard-Welch Chair at Rice University (Grant C-0016 to P.G.W.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819859116/-/DCSupplemental.
References
- 1.Bar-Even A, Milo R, Noor E, Tawfik DS. The moderately efficient enzyme: Futile encounters and enzyme floppiness. Biochemistry. 2015;54:4969–4977. doi: 10.1021/acs.biochem.5b00621. [DOI] [PubMed] [Google Scholar]
- 2.Ferreiro DU, Komives EA, Wolynes PG. Frustration in biomolecules. Q Rev Biophys. 2014;47:285–363. doi: 10.1017/S0033583514000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shoichet BK, Baase WA, Kuroki R, Matthews BW. A relationship between protein stability and protein function. Proc Natl Acad Sci USA. 1995;92:452–456. doi: 10.1073/pnas.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meiering EM, Serrano L, Fersht AR. Effect of active site residues in barnase on activity and stability. J Mol Biol. 1992;225:585–589. doi: 10.1016/0022-2836(92)90387-y. [DOI] [PubMed] [Google Scholar]
- 5.Sanchez I, Tejero J, Gomez-Moreno C, Medina M, Serrano L. Point mutations in protein globular domains: Contributions from function, stability and misfolding. J Mol Biol. 2006;363:422–432. doi: 10.1016/j.jmb.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 6.Ferreiro DU, Komives EA, Wolynes PG. Frustration, function and folding. Curr Opin Struct Biol. 2018;48:68–73. doi: 10.1016/j.sbi.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreiro DU, Hegler JA, Komives EA, Wolynes PG. Localizing frustration in native proteins and protein assemblies. Proc Natl Acad Sci USA. 2007;104:19819–19824. doi: 10.1073/pnas.0709915104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furnham N, et al. The Catalytic Site Atlas 2.0: Cataloging catalytic sites and residues identified in enzymes. Nucleic Acids Res. 2014;42:D485–D489. doi: 10.1093/nar/gkt1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parra RG, et al. Protein Frustratometer 2: A tool to localize energetic frustration in protein molecules, now with electrostatics. Nucleic Acids Res. 2016;44:W356–W360. doi: 10.1093/nar/gkw304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jenik M, et al. Protein Frustratometer: A tool to localize energetic frustration in protein molecules. Nucleic Acids Res. 2012;40:W348–W351. doi: 10.1093/nar/gks447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreiro DU, Hegler JA, Komives EA, Wolynes PG. On the role of frustration in the energy landscapes of allosteric proteins. Proc Natl Acad Sci USA. 2011;108:3499–3503. doi: 10.1073/pnas.1018980108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, Wolynes PG, Takada S. Frustration, specific sequence dependence, and nonlinearity in large-amplitude fluctuations of allosteric proteins. Proc Natl Acad Sci USA. 2011;108:3504–3509. doi: 10.1073/pnas.1018983108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newton MS, Arcus VL, Gerth ML, Patrick WM. Enzyme evolution: Innovation is easy, optimization is complicated. Curr Opin Struct Biol. 2018;48:110–116. doi: 10.1016/j.sbi.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Orengo CA, et al. CATH–A hierarchic classification of protein domain structures. Structure. 1997;5:1093–1109. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 15.Sippl MJ, Wiederstein M. A note on difficult structure alignment problems. Bioinformatics. 2008;24:426–427. doi: 10.1093/bioinformatics/btm622. [DOI] [PubMed] [Google Scholar]
- 16.Nagano N, Orengo CA, Thornton JM. One fold with many functions: The evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J Mol Biol. 2002;321:741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- 17.Dellus-Gur E, Toth-Petroczy A, Elias M, Tawfik DS. What makes a protein fold amenable to functional innovation? Fold polarity and stability trade-offs. J Mol Biol. 2013;425:2609–2621. doi: 10.1016/j.jmb.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 18.Parra RG, Espada R, Verstraete N, Ferreiro DU. Structural and energetic characterization of the ankyrin repeat protein family. PLoS Comput Biol. 2015;11:e1004659. doi: 10.1371/journal.pcbi.1004659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stiffler MA, Hekstra DR, Ranganathan R. Evolvability as a function of purifying selection in TEM-1 -lactamase. Cell. 2015;160:882–892. doi: 10.1016/j.cell.2015.01.035. [DOI] [PubMed] [Google Scholar]
- 20.Palzkill T, Botstein D. Probing -lactamase structure and function using random replacement mutagenesis. Proteins Struct Funct Bioinform. 1992;14:29–44. doi: 10.1002/prot.340140106. [DOI] [PubMed] [Google Scholar]
- 21.Vanhove M, Lejeune A, Pain R. .-lactamases as models for protein-folding studies. Cell Mol Life Sci. 1998;54:372–377. doi: 10.1007/s000180050166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langan PS, et al. The structure of toho1 -lactamase in complex with penicillin reveals the role of tyr105 in substrate recognition. FEBS Open Bio. 2016;6:1170–1177. doi: 10.1002/2211-5463.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vakulenko SB, Tóth M, Taibi P, Mobashery S, Lerner SA. Effects of Asp-179 mutations in TEM pUC19 beta-lactamase on susceptibility to beta-lactams. Antimicrob Agents Chemother. 1995;39:1878–1880. doi: 10.1128/aac.39.8.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das A, Plotkin SS. SOD1 exhibits allosteric frustration to facilitate metal binding affinity. Proc Natl Acad Sci USA. 2013;110:3871–3876. doi: 10.1073/pnas.1216597110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 26.Monod J. Chance and Necessity: An Essay on the Natural Philosophy of Modern Biology. Vintage Books; New York: 1973. [Google Scholar]
- 27.Pabis A, Risso VA, Sanchez-Ruiz JM, Kamerlin SC. Cooperativity and flexibility in enzyme evolution. Curr Opin Struct Biol. 2018;48:83–92. doi: 10.1016/j.sbi.2017.10.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.