Significance
Photosystems catalyze photosynthetic light absorption and energy conversion, and yet require photoprotection mechanisms for avoiding oxidative damage from excess light absorption. The major short-term mechanism is thermal dissipation of the excitation energy absorbed in excess, called nonphotochemical quenching (NPQ). Although NPQ of photosystem II (PSII) excitons has been extensively investigated, we focus here on NPQ of PSI in the model organism for green algae: Chlamydomonas reinhardtii. By analyzing several NPQ and state transition mutants, we show that NPQ occurs within antenna proteins excitonically connected to PSI and is catalyzed by LHCSR (Light Harvesting Complex Stress Related) subunits, with a major role observed for LHCSR3. State transitions have a limited effect in quenching by redistributing Light Harvesting Complexes II, the major target of quenching by LHCSR, between photosystems.
Keywords: photosynthesis, photoprotection, NPQ, LHCSR, microalgae
Abstract
Photosynthetic organisms prevent oxidative stress from light energy absorbed in excess through several photoprotective mechanisms. A major component is thermal dissipation of chlorophyll singlet excited states and is called nonphotochemical quenching (NPQ). NPQ is catalyzed in green algae by protein subunits called LHCSRs (Light Harvesting Complex Stress Related), homologous to the Light Harvesting Complexes (LHC), constituting the antenna system of both photosystem I (PSI) and PSII. We investigated the role of LHCSR1 and LHCSR3 in NPQ activation to verify whether these proteins are involved in thermal dissipation of PSI excitation energy, in addition to their well-known effect on PSII. To this aim, we measured the fluorescence emitted at 77 K by whole cells in a quenched or unquenched state, using green fluorescence protein as the internal standard. We show that NPQ activation by high light treatment in Chlamydomonas reinhardtii leads to energy quenching in both PSI and PSII antenna systems. By analyzing quenching properties of mutants affected on the expression of LHCSR1 or LHCSR3 gene products and/or state 1–state 2 transitions or zeaxanthin accumulation, namely, npq4, stt7, stt7 npq4, npq4 lhcsr1, lhcsr3-complemented npq4 lhcsr1 and npq1, we showed that PSI undergoes NPQ through quenching of the associated LHCII antenna. This quenching event is fast-reversible on switching the light off, is mainly related to LHCSR3 activity, and is dependent on thylakoid luminal pH. Moreover, PSI quenching could also be observed in the absence of zeaxanthin or STT7 kinase activity.
Photosystem II (PSII) and I (PSI) are pigment–protein complexes, located in the thylakoid membranes, composed of a core complex, hosting photochemical reactions, and a peripheral antenna system formed by Light Harvesting Complexes (LHC) (1, 2). PSI and PSII fuel a light-dependent electron transport chain from water to NADPH coupled with proton transport to the lumen driving ATP synthesis. ATP and NADPH are then used by the Calvin-Benson-Bassham cycle to reduce CO2 into sugars. In excess light, the rate of Calvin-Benson-Bassham cycle reactions is saturated and ATP and NADPH are produced in excess compared with their metabolic demand, leading to ATPase limitation from lack of ADP substrate, which reduces the return of H+ to the stroma compartment and causes lumen acidification. Lack of electron acceptors causes charge recombination in PSII with triplet chlorophyll (Chl) excited state formation and reaction with oxygen, forming toxic reactive oxygen species (3). A major photoprotective mechanism, nonphotochemical quenching (NPQ), is activated when luminal pH drops, safely dissipating up to 80% of the excitation energy absorbed into heat (4). In Chlamydomonas reinhardtii, NPQ activity requires LHCSR1 and LHCSR3 proteins, which are triggered to a quenching state on sensing low luminal pH (5). Both LHCSR1 and LHCSR3 subunits are overexpressed on prolonged high-light (HL) treatment, whereas LHCSR1 expression depends on high CO2 (5, 6). LHCSRs expression has been reported to be triggered by blue light, involving phototropins as photoreceptors that activate a signal transduction pathway leading to LHCSR3 accumulation (7). LHCSR1 has been reported to be triggered by UV light through the activity of the UVR8 photoreceptor. LHCSR3 is accumulated to a far higher level than LHCSR1, making the former the major player in NPQ activity (8, 9). The npq4 mutant lacks LHCSR3 and retains a low NPQ, which is abolished in npq4 lhcsr1 also lacking LHCSR1 (5, 9). In HL, violaxanthin is converted to zeaxanthin, a strong NPQ enhancer in plants (10, 11) but not in C. reinhardtii (npq1) (12). Photoprotection is also favored by reversible phosphorylation of PSII antenna subunits LHCII and CP29, on which they are released from PSII and connect to PSI, enhancing its cross-section and balancing PSI vs. PSII electron transport rates. This process, called state 1–state 2 transition, depends on the STT7 kinase (13, 14), which, in turn, is activated by interaction with Cytochrome b6f complex on reduction of the PQ pool (15–17). Despite STT7 activity decreases under HL (18), a photoprotective effect of state transitions was reported based on enhanced photoinhibition observed in the stt7 npq4 double mutant compared to npq4 single mutant under HL (19). Evidence for interaction between NPQ and state transitions relies on LHCSR3 being phosphorylated by STT7 (12, 20) and interacting with the mobile LHCII fraction (19, 21). However, STT7-independent phosphorylation sites have also been reported in LHCSR3 and LHCSR1 (20). LHCSR3 was reported to interact with both PSI and PSII complexes (20, 22, 23), with phosphorylation negatively affecting LHCSR3 binding to PSI (20). Phosphorylation of LHCSR3 and LHCII was reported not to affect NPQ (12). Although both LHCSR1 and LHCR3 have been reported to be quenchers for LHCII and PSII complexes (8, 21, 24), their involvement in PSI photoprotection is still under debate. In the moss Physcomitrella patens, LHCSR1 was found to be localized in stroma membranes and to be a quencher of both photosystems (25). Recently, LHCSR1 was reported to be involved in PSI quenching via excitation energy transfer from LHCII in C. reinhardtii (26). In this work, we investigated the quenching properties of LHCSR proteins toward LHCII, PSII and PSI-LHCII complexes in C. reinhardtii.
Results
NPQ at Room Temperature.
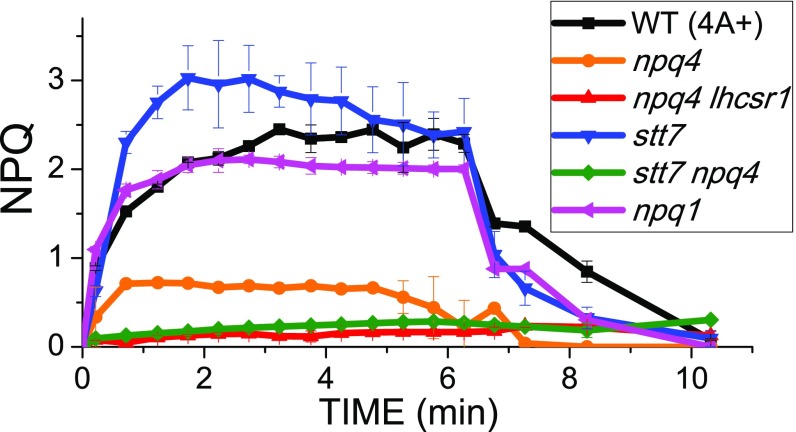
Although room temperature fluorescence analysis is effective as a probe for NPQ of PSII, the low quantum yield of PSI makes its fluorescence a poor signal. As reported in Fig. 1, the npq4 mutant showed low residual NPQ activity from LHCSR1 when the effect of excess light was measured at room temperature; that is, in conditions specific for detection of fluorescence from PSII. NPQ activity was further reduced in the double-mutant npq4 lhcsr1. NPQ induction of the stt7 mutant was faster than in WT, and its amplitude was enhanced, suggesting STT7 kinase is not essential for NPQ. Interestingly, stt7 npq4 mutant exhibited lower NPQ compared with npq4, suggesting that the LHCSR1-dependent quenching might depend on STT7 activity, despite the fact that LHCSR1 was not a STT7 substrate. Analysis of WT vs. npq1 mutant (27) showed NPQ was independent from zeaxanthin (Fig. 1), in agreement with previous reports (12). Because the amplitude of NPQ in C. reinhardtii is modulated by the amount of LHCSR subunits, their accumulation was quantified in the genotypes investigated by immunoblotting. LHCSR3 content per PSI or PSII was similar in WT, npq1, and stt7 mutants. In WT and npq1 strains, LHCSR3 appeared as a double band, related to the presence of the phosphorylated form, increasing its apparent molecular weight. The LHCSR3 phosphorylated form was lost in the absence of the STT7 kinase. In stt7 mutant, LHCSR1 accumulation was rather increased compared with WT, as in the case of npq4 and stt7 npq4 (SI Appendix, Fig. S1).
Fig. 1.
NPQ induction kinetics measured at room temperature. Pulse-amplitude fluorometric time course at room temperature of WT and npq1, npq4, npq4 lhcsr1, stt7, and stt7 npq4 mutants. SDs are reported as error bars (n = 5).
Light-Dependent Quenching of PSII and PSI in C. reinhardtii Measured by 77 K Fluorescence Emission Spectra.
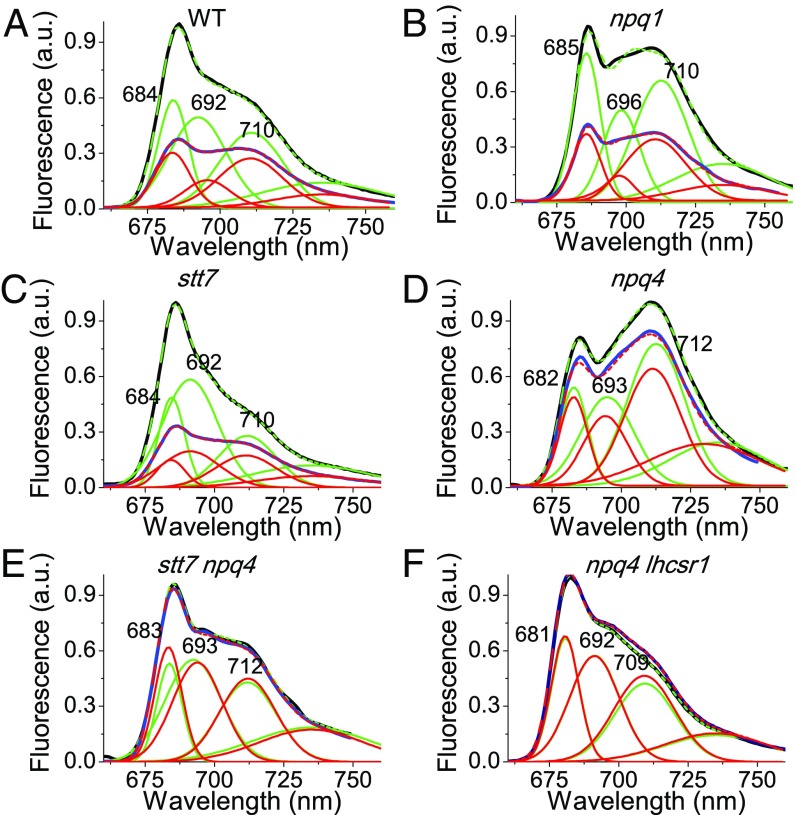
PSI fluorescence contribution to the overall fluorescence emitted by C. reinhardtii can be investigated at 77 K, where the PSI photochemistry is essentially blocked and the fluorescence quantum yield is significantly increased (28). Fluorescence emission spectra at the low temperature of the WT show two peaks, at 687 nm and 710 nm, which can be mainly attributed to PSII and PSI contribution, respectively. Spectral deconvolution with Gaussian forms allowed for extrapolating the contributions of the different emitting components (SI Appendix, Fig. S2): two Gaussians peaking at 684 and 694 nm can be associated with PSII-LHCII complexes, whereas the Gaussian form peaking at 712 nm can be associated with PSI contribution. The Gaussian component peaking at 735 nm was instrumental in compensating for deviation of the Chl fluorescence spectral form from Gaussian shape. These attributions were then confirmed by deconvolution analysis on 77 K fluorescence emission spectra obtained from mutants with a reduced amount of PSI (psaB mutant) (29) or depleted of PSII (psbD mutant) (30) or LHCI and LHCII complexes (cbs3) (31).
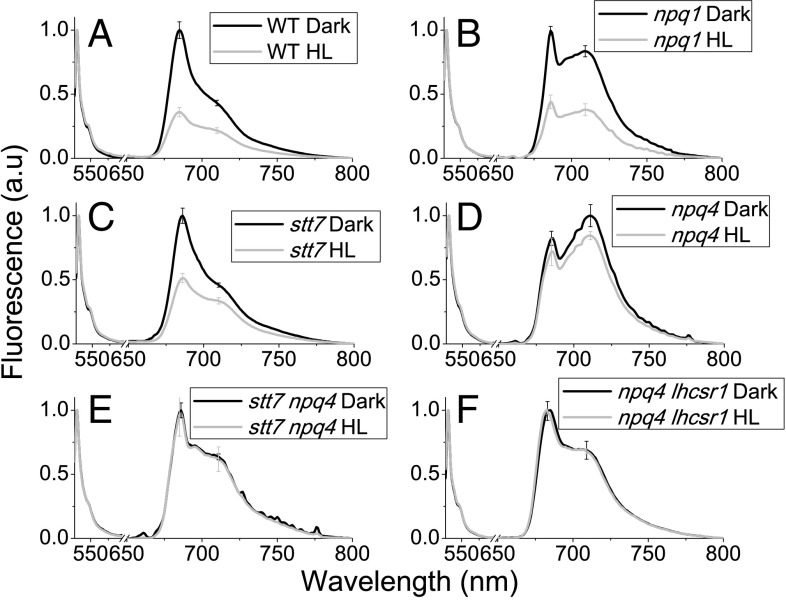
To investigate the role of LHCSR proteins in the quenching of PSI and PSII, C. reinhardtii cells from WT, npq4, npq4 lhcsr1, stt7, npq1, and stt7 npq4 mutants were acclimated to HL (400 μE m−2⋅s−1) for at least 10 generations to induce LHCSR1 and LHCSR3 expression in the genotypes in which the genes were expressed. Dark-adapted, HL-acclimated cells were added with green fluorescent protein (GFP) as the internal fluorescence intensity standard, and split into aliquots for different treatments on which samples were rapidly frozen in liquid nitrogen and stored in the dark at 77 K until fluorescence measurements were performed. As reported in SI Appendix, Fig. S3, HL treatment for 6 min with strong light (1,500 μE) did not change the GFP fluorescence emission spectrum or amplitude, enabling its use as internal standard, as previously reported (25). Because a light-independent transthylakoidal ΔpH was previously reported to form in green algae, especially in the presence of high reducing power in the mitochondria (32), and to exclude any potential quenching on PSII or PSI in dark-adapted cells, 77 K fluorescence emission spectra were measured in the presence of the uncoupler nigericin, obtaining no significant effect on fluorescence emission spectra (SI Appendix, Fig. S4). HL treatment gave a similar reduction of both PSI and PSII peaks in WT and npq1 (Fig. 2 A and B). The quenching on PSI and PSII observed was confirmed by deconvolution of WT and npq1 77 K fluorescence emission spectra into Gaussian components, as described in SI Appendix, Fig. S2, resulting into a reduced amplitude for both PSI and PSII spectral components (Fig. 3 A and B). In the case of stt7, the overall fluorescence emission of PSI was reduced in stt7 mutant, likely as a result of the depletion of phosphorylated LHCII contributing to PSI emission in this mutant (19), where HL treatment caused a more evident quenching of the main peak (PSII), rather than on the 709-nm shoulder from PSI (Fig. 2C). Gaussian deconvolution analysis, however, allowed for detection of decreased emission from PSI (Fig. 3C), implying the onset of STT7-independent quenching on PSI. In the case of npq4, a minor effect was detected on PSII components on HL treatment (Figs. 2D and 3D), whereas a more evident reduction of PSI contribution at 709 nm was observed (Fig. 2D). These results suggest that both PSII and PSI quenching were still active in npq4, where LHCSR1 is the only LHCSR subunit, even if to a reduced extent compared with WT. It is worth noting that the highest PSI/PSII fluorescence ratio was detected in npq4, consistent with previous findings (9, 19). This residual quenching on PSI was, however, almost absent in the double-mutant stt7 npq4 (Figs. 2E and 3E). Both PSI and PSII quenching activities were absent in npq4 lhcsr1 (Figs. 2F and 3F). To further investigate the possible role of LHCSR1 in PSI and PSII quenching, a genotype with only a LHCSR3 subunit was generated by complementation of the npq4 lhcsr1 mutant with the lhcsr3.1 gene under the control of its endogenous promoter, as previously described (33). Complemented lines, here called C-lhcsr3-4 and C-lhcsr3-24, were characterized by a similar level of LHCSR3 compared with WT, but no LHCSR1 (SI Appendix, Fig. S1). The resulting NPQ at room temperature was similar to the WT with even a faster rise upon illumination and faster recovery under low far-red light (SI Appendix, Fig. S5), as previously reported (33). In addition, 77 K fluorescence emission spectra demonstrated that both PSI and PSII contributions were quenched on HL treatment, even in absence of LHCSR1 in C-lhcsr3-4 and C-lhcsr3-24 lines, obtaining similar results compared with WT (SI Appendix, Fig. S5).
Fig. 2.
Here, 77 K fluorescence emission spectra of C. reinhardtii WT (A) and npq1 (B), stt7 (C), npq4 (D), stt7 npq4 (E), and npq4 lhcsr1 (F) mutant cells normalized to GFP before and after HL exposure. Fluorescence emission spectra of C. reinhardtii were recorded for whole cells that were dark adapted (black) or HL treated (1,500 μE m−2⋅s−1) for 6 min (gray). GFP was added as internal standard for normalization. SDs are reported as error bars (n = 4).
Fig. 3.
Fluorescence emission spectra of dark-adapted (black) or HL-treated (blue) WT (A) and npq1 (B), stt7 (C), npq4 (D), stt7 npq4 (E), and npq4 lhcsr1 (F) mutant cells were reconstructed by spectral deconvolution with Gaussians: cumulative fit results are reported in dashed green (dark-adapted samples) or dashed red (HL-treated samples), whereas the different Gaussians used are reported in green (dark-adapted samples) or red (HL-treated samples). GFP was added as internal standard for normalization.
To characterize the kinetics of quenching, 77 K fluorescence emission spectra were followed on HL treatment for 2, 4, and 6 min and after 2, 5, or 10 min of dark recovery in the presence of far-red light. As reported in SI Appendix, Figs. S6–S8, HL treatment of WT, stt7, npq1, and complemented lines C-lhcsr3-4 and C-lhcsr3-24 induced a progressive decrease of fluorescence emissions from both the main PSII peak (685 nm) and the PSI peak (709 nm). After 6 min of HL treatment cells were further incubated in the dark using a dim far-red light to maintain plastoquinone pool oxidized: their fluorescence emission almost recovered the amplitude of the dark-adapted cells (SI Appendix, Fig. S6). Differently, npq4 and stt7 npq4 mutants only underwent a transient decrease of both 686-nm and 710/711-nm emission peaks during the first 4 min HL, whereas a slight reduction of the 711-nm peak was observed in the case of the npq4 mutant only, with poor, if any, recovery in the dark (SI Appendix, Fig. S7). No significant quenching could be observed in the case of npq4 lhcsr1 mutant, neither on 682-nm nor on 709-nm peaks (SI Appendix, Fig. S6 D and E). Rather, a minor reduction of a 682-nm peak was detected during dark recovery in the presence of far-red light, possibly related to activation of PSII repair system.
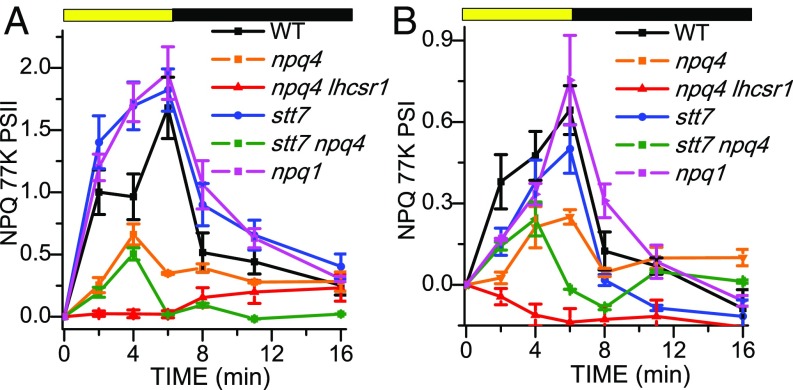
To reconstruct the kinetics of PSI and PSII quenching from the fluorescence emission spectra, spectral deconvolution into Gaussians components was performed, as described in Fig. 3, for the 77 K fluorescence emission spectra obtained at different times of illumination or dark recovery. Fitting analysis on WT, C-lhcsr3-4 and C-lhcsr3-24, stt7, and npq1 curves showed similar quenching kinetics for all the components retrieved, with the 692–696-nm component (PSII) showing the highest quenching amplitude on light treatment (SI Appendix, Figs. S9–S13). In npq4 and stt7 npq4 mutants, small and virtually irreversible quenching effects were detected for the different components (SI Appendix, Figs. S14 and S15). In npq4 lhcsr1, instead, no quenching was observed in any component on HL treatment (SI Appendix, Fig. S16). According to the amplitude of the PSII and PSI Gaussians, the quenching on PSII vs. PSI was estimated (Fig. 4). The amplitude at time X (AX) of the different Gaussians components was then used to calculate the quenching on PSI or PSII according to the formula (ADark − AX)/AX. PSII quenching estimated from Gaussian deconvolution was faster in stt7, npq1, and C-lhcsr3-4 and C-lhcsr3-24 lines compared with WT, while being strongly reduced in npq4 and stt7 npq4 mutants and negligible in npq4 lhcsr1 (Fig. 4A and SI Appendix, Fig. S17), consistent with the NPQ kinetics at room temperature (Fig. 1 and SI Appendix, Fig. S5). PSI quenching was observed in WT, stt7, C-lhcsr3-4 and C-lhcsr3-24 lines and npq1 strains, with a faster induction kinetic in the case of WT and C-lhcsr3-4 and C-lhcsr3-24 lines. PSI quenching was partially detectable in npq4 and in stt7 npq4, even if strongly reduced, but absent in the npq4 lhcsr1 mutant. These results imply that mainly LHCSR3 is involved in quenching of both PSI and PSII, whereas STT7 activity and zeaxanthin have a minor effect. In the absence of LHCSR3a, a small amount of LHCSR1-dependent quenching could be also measured.
Fig. 4.
Calculated NPQ induction kinetics at 77 K. The NPQ curves were calculated for PSII (A) and PSI (B) from the area of the sum of the Gaussians used for the fitting according to the formula (ADark − AX)/AX, where AX and ADark are, respectively, the amplitude at time X (AX) or at time 0 (ADark, dark-adapted samples) of the different Gaussians attributable to PSII or PSI. SDs are reported as error bars (n = 4).
77 K Fluorescence Excitation Spectra: PSI Quenching Is Specifically Located on Antenna Complexes.
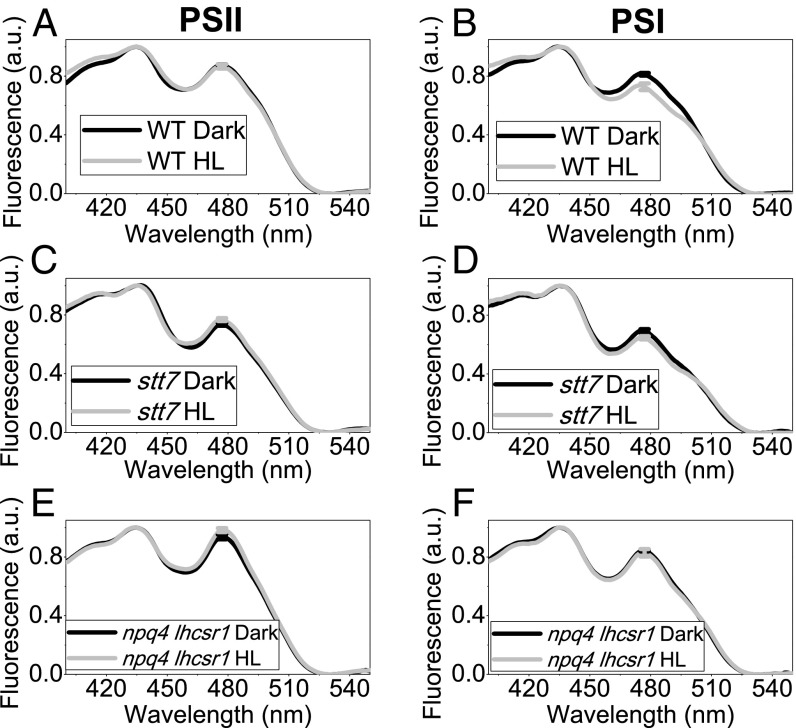
LHCSRs-dependent quenching can be active specifically on LHC antenna proteins, acting as an alternative trap for excitation energy, or on the whole PSI or PSII supercomplexes. To discriminate between these two possibilities, the fluorescence excitation spectra were measured for PSI (emission at 710 nm) or PSII (emission at 685 nm) in dark-adapted or HL-treated cells. In addition, 77 K excitation spectra were characterized by two main peaks: 435 nm (Chl a) and 480 nm (Chl b). Because LHC antenna proteins bind both Chl a and Chl b, whereas core complexes bind Chl a only, a preferential quenching of LHC antenna in HL is expected to yield excitation spectra with a decreased Chl b contribution. In WT, NPQ induction did not change the PSII excitation spectrum compared with a dark-adapted sample, but reduced the Chl b peak in the PSI excitation spectrum (Fig. 5 A and B). These results indicate that in the case of PSII, NPQ activation quenches the overall PSII-LHCII supercomplex, whereas in the case of PSI, the HL treatment specifically reduces the contribution of LHC complexes to the fluorescence emission of PSI. This observation was even more striking when the 480 nm/440 nm ratio observed in 77 K fluorescence excitation spectra was corrected for the partial overlapping of PSII and PSI emission at 710 nm, according to the Gaussian deconvolution of 77 K fluorescence emission spectra (SI Appendix, Table S1). Similar results were then obtained in the case of C-lhcsr3-4 and C-lhcsr3-24 or npq1 (SI Appendix, Figs. S18 and S19). In the case of stt7 mutant, HL treatment caused a decrease of Chl b contribution in PSI excitation spectrum, although the decrease was smaller compared with that for WT (Fig. 5 C and D). No decrease in Chl b contribution was observed in npq4 lhcsr1 (Fig. 5 E and F), npq4, and stt7 npq4 (SI Appendix, Fig. S19), consistent with LHCSR being involved in specifically quenching LHC proteins connected to PSI while homogeneously quenching the LHCII-PSII core pigment bed.
Fig. 5.
Here, 77 K fluorescence excitation spectra were recorded measuring fluorescence emission at 685 nm (A, C, E), where mainly PSII emits, and 710 nm (B, D, F), where mainly PSII emits. Fluorescence excitation spectra were measured in the case of dark-adapted (black) and HL treated (gray) WT (A, B) and stt7 (C, D) and npq4 lhcsr1 (E, F) mutant cells and normalized to the Chl a contribution at 436 nm. SDs are reported as error bars (n = 4).
77 K Time-Resolved Analysis.
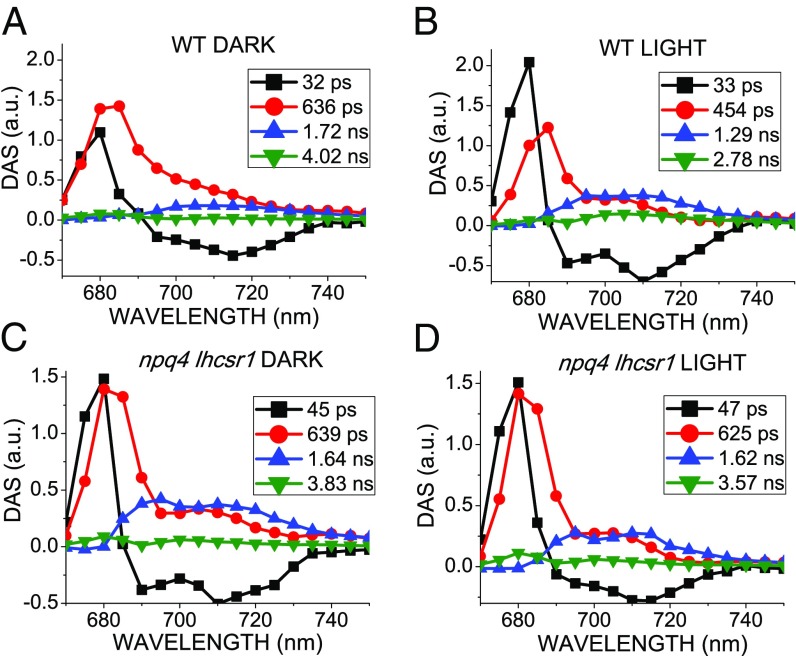
Time-resolved fluorescence analysis at 77 K was performed on WT and npq4 lhcsr1 strain in the dark-adapted state or on activating quenching by HL treatment. Fluorescence decay traces were then submitted to global analysis, as previously described (34), to identify the different associated spectral components and relative decay time constants. Four DAS (decay-associated spectral components) were required for best fit of the fluorescence decay maps (Fig. 6). The first component (DAS 1) was characterized by a positive/negative DAS and a time constant of 32–47 ps: this component reflects the excitation energy transfer from LHC proteins to PSII and PSI core complex, which is not significantly affected by activation of NPQ mechanisms (35). The second component (DAS 2) peaked in the 680–690-nm region, with a shoulder at 710 nm and a decay constant of 630 ps in dark-adapted WT and in npq4 lhcsr1. Differently, in the case of HL-treated WT, DAS 2 showed a shorter time constant of 454 ps. DAS 2 is principally related to PSII (680–690-nm emission), with a small contribution from PSI (emission at 710 nm), in agreement with reports of subnanosecond component at 77 K from both PSII and PSI (36). The third DAS (DAS 3) showed a wide emission in the 690–740-nm, best-fitting PSI spectrum. The decay constant was 1.72 ns, reduced to 1.29 ns in HL. In the npq4 lhcsr1 mutant, the decay constant of this DAS 3 was not significantly affected by light treatment. These results agree with a LHCSR-dependent activation of quenching mechanisms for both PSI and PSII on HL treatment. Finally, a small component (DAS 4) with a decay constant of 2.7–4 ns was observed in all samples: this component has been previously attributed to loosely bound LHCII and to the longest decay contribution from PSI at 77 K (36). In WT, DAS 4 was shortened on HL exposure, from 4.01 to 2.70 ns. It should be noticed that the npq4 lhcsr1 mutant also underwent a limited reduction in DAS 4 lifetime from 3.8 to 3.6 ns. DAS 4, however, was specifically decreased at 685 nm in WT compared to npq4 lhcsr1 mutant, suggesting a preferential LHCSR-dependent quenching of loosely bound LHCII. Time-resolved fluorescence analysis was also performed on stt7, npq1, npq4, stt7 npq4, and C-lhcsr3-4 and C-lhcsr3-24 strains (SI Appendix, Fig. S20). In the case of dark-adapted samples, global analysis yielded similar results compared with WT and npq4 lhcsr1 genotypes. On HL treatment, only stt7, npq1, and C-lhcsr3-4 and C-lhcsr3-24 strains exhibited a shortening of the decay constants of DAS 2, DAS 3, and DAS 4, as in the case of WT. A minor shortening of decay constant of DAS 3 from 1.77 ns to 1.49 nm was also observed for npq4, in agreement with a minor quenching activity on PSI from residual LHCSR1.
Fig. 6.
Global analysis of time-resolved fluorescence kinetics at 77 K. Fluorescence decay kinetics of dark-adapted (A, C) or HL-treated (B, D) WT (A, C) and npq4 lhcsr1 mutant (B, D) were acquired at 77 K in the 670–750 nm range with 5 nm step and globally fitted with four exponentials. The DAS obtained are reported as normalized to the same total area for each sample, whereas the associated time constants are indicated in the legend. SD associated to time constants is less than 5% for each component.
Discussion
LHCSR3 and LHCSR1 are pigment-binding proteins involved in NPQ activation in C. reinhardtii (5, 8, 9, 26). NPQ measurements have been mainly based on room temperature fluorescence measurements monitoring changes of PSII fluorescence, whereas the low-fluorescence quantum yield of PSI prevents analysis in the presence of strong emissions by PSII. At 77 K, fluorescence quantum yield is high for both PSI and PSII, allowing for proper quantification of both emissions. Moreover, freezing samples in liquid nitrogen preserves the conformations previously induced by actinic light, thus allowing for spectral characterization of quenched states (25, 37). The direct involvement of LHCSR in activating quenching is evident from Figs. 2 and 3, showing that HL was effective in reducing the amplitude of both PSII and PSI emissions in the presence of LHCSR subunits. Deconvolution of fluorescence emission spectra into Gaussian components allowed us to isolate the contributions of PSI from PSII and showed that both were quenched on HL treatment (Fig. 3). Indeed, time-resolved fluorescence analysis, also at 77 K, on dark-adapted vs. HL-treated cells showed a strong reduction of time constants of both PSI and PSII in the presence of LHCSR subunits only (Fig. 6). The activity of LHCSR subunits as quenchers for LHCII trimers of either components of PSII or PSI supercomplexes or loosely bound, is consistent with previous reports. LHCSR1 was found active vs. LHCII, either free or bound to PSI (8, 26), whereas the LHCSR3 subunit was found to bind to both PSI and PSII in C. reinhardtii (20, 22, 23) and to be active in quenching purified PSII–LHCII supercomplexes (23). Interestingly, the absence of LHCSR1 in C-lhcsr3-4 and C-lhcsr3-24 lines did not affect PSI or PSII quenching (SI Appendix, Fig. S17), whereas only a partial quenching was observed on LHCSR1 up-regulation in npq4 or stt7 npq4 mutant (Fig. 4). These results indicate that LHCSR3 is the major actor in PSI and PSII quenching, whereas only a minor role, if any, can be attributed to LHCSR1. Nevertheless, the residual quenching observed in npq4 and stt7 npq4 indicates that LHCSR1 might be a quencher for both photosystems, even if with a much lower efficiency compared with LHCSR3. The increased quenching activity of LHCSR3 compared with LHCSR1 might be related simply to a dose effect and/or to some specific interactions with potential partners and/or to a specific intrinsic quenching activity. Quenching activity toward PSI or PSII by LHCSR3 differs: Although the fluorescence excitation spectra of PSII-LHCII complex were similar in dark-adapted or HL-treated samples, suggesting homogeneous quenching in HL, a specific reduction of Chl b contribution to PSI emission was observed in the case of LHCSR3-dependent quenching of PSI (Fig. 5 and SI Appendix, Table S1). This result suggests that in the case of PSI, LHCSR3 subunits preferentially quench antenna proteins, rather than the PSI core complex. Both LHCSR1 and LHCSR3 have been previously reported to interact with the “mobile” fraction of LHCII preferentially involved in state transitions: LHCSR3 was suggested to modulate coupling/decoupling of this LHCII population to PSII (21) and LHCSR1 to modulate the excitation energy transfer from LHCII to PSI (26). The results reported here are consistent with the above reports, highlighting a specific role of LHCSR subunits, and in particular LHCSR3, in quenching “mobile” LHCII trimers and thus reducing the fraction of LHC subunits involved in efficient energy transfer to PSI (Fig. 6). At variance with results reported by Kosuge et al. (26), 77 K fluorescence excitation spectra and time-resolved fluorescence analysis suggest that the LHCSR-dependent quenching on LHCII trimers does not correlate with increased excitation energy transfer to PSI. This interpretation is supported by several pieces of evidence: the Chl b contribution to PSI fluorescence emission was reduced, whereas an increased excitation energy transfer to PSI would be expected to increase the contribution of antenna proteins to PSI emission; the amplitude of the shortest DAS (DAS 1, ∼30 ps), obtained by time-resolved fluorescence analysis from excitation energy transfer to PSI, did not increase on HL treatment, as expected from increasing the antenna size; time constants of DAS attributable to both PSII and PSI in WT dark-adapted cells were reduced on HL treatment, consistent with the onset of a quenching mechanism, rather than as a result of excitation energy transfer to the PSI reaction center; and PSI fluorescence was observed at 77 K, that is, with photochemical activity, if any, strongly reduced. This finding implies that the possible LHCSR-dependent quenching mechanism at room temperature should be extremely quick to compete with PSI photochemistry: a quenching conformation decaying in 80 ps was reported in vitro in the case of LHCSR1 from the moss Physcomitrella patens (38). This time scale is consistent with competition with PSI photochemical traps, even if further investigation is required to properly evaluate this point. Nevertheless, we cannot fully rule out the possibility that LHCSR-dependent quenching might in part involve energy transfer to PSI reaction centers, as suggested by Kosuge et al. (26), or detachment of antenna proteins from the PSI core, which could account for the preferential loss of Chl b contribution to PSI excitation spectra (Fig. 5).
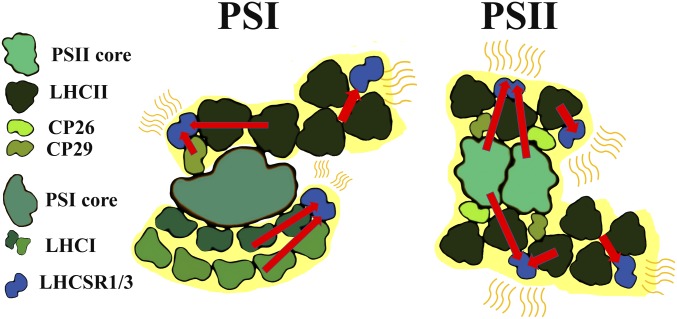
Kinetics of NPQ on PSII and PSI were reconstructed according to Gaussian deconvolution at different illumination times (Fig. 4). Zeaxanthin accumulation on HL treatment had little effect on PSII quenching, whereas PSI quenching was only slightly affected in terms of decay kinetics (Fig. 4). These results are consistent with a minor role, if any, of zeaxanthin in increasing the kinetic for activation of PSI quenching in C. reinhardtii (12). In the case of stt7 mutant, the absence of an active STT7 kinase did not impair NPQ induction at the level of either PSI or PSII (Fig. 4). Although LHCII phosphorylation is dependent on STT7 activity and related to state transitions (14), LHCSR3 was also reported to harbor phosphorylation sites that are not substrates for STT7, and the binding of LHCSR3 to PSI was negatively affected by its phosphorylation (20). The results presented here, together with previous findings, suggest that LHCSR3-dependent quenching of PSI occurs on LHC proteins bound to PSI-complex, which could be either identified as phosphorylated LHCII trimers or LHCI proteins (Fig. 7), thus explaining the different Chl b contribution observed in fluorescence excitation spectra in stt7 mutant on HL treatment.
Fig. 7.
Model for LHCSRs quenching on PSI and PSII. LHCSR1 and LHCSR3 interaction with PSI and PSII supercomplexes is schematically reported as a model for their quenching activity. LHCSR3 has the major role in quenching, whereas a residual LHCSR1 dependent quenching activity can be observed in the absence of LHCSR3. Red arrows indicate excitation energy transfer. Peripheral LHCII trimers loosely connected to PSI or PSII are also reported. In the model, the PSI and PSII subunits quenched by LHCR proteins are highlighted in yellow.
We conclude that in C. reinhardtii, LHCSR3 is involved in quenching both PSI and PSII by directly interacting both with PSII supercomplexes and with PSI-bound LHCII (Fig. 7). Because in C. reinhardtii, photosystem II subunit S protein is only transiently expressed (39, 40) and not detected in the conditions applied here (SI Appendix, Fig. S1), LHCSR3 proteins appear to account for most or all NPQ activity, as shown by lack of quenching in npq4 lhcsr1. Quenching activity occurs at different sites: PSII complexes, where LHCSR3 was found docking to LHCII trimers or CP26 (24); LHC complexes bound to PSI complexes; and a LHCII “mobile” pool loosely connected to photosystems (Fig. 7). In the WT, on HL stress, PSII quenching rapidly occurred, whereas PSI quenching was slower (Fig. 4), possibly as a result of a time lag for LHCSR-dependent detachment of LHCII proteins from PSI. The photoprotective relevance for the observed fluorescence quenching on PSI was confirmed by the strong PSI photoinhibition observed when PSI quenching was completely abolished in the stt7 npq4 mutant (19).
Methods
Strains and Culture Conditions.
C. reinhardtii cells were grown at 25 °C in flask with white light (70 μE m−2⋅s−1; 16 h light/8 h dark photoperiod) in TAP medium. HL acclimation was induced by growing cells at 400 μE m−2⋅s−1 in HS medium. npq4 lhcsr1 complementation was performed as described in ref. 33.
NPQ Measurements at Room Temperature.
NPQ measurements were performed with a PAM-101 (Waltz) with actinic and saturating light of 1,500 μE m−2⋅s−1 and 4,000 μE m−2⋅s−1, respectively. The far-red LED was kept on during dark recovery. During dark adaptation, cells were shaken in HS medium.
Quenching Measurements at Low Temperature.
In addition, 77 K fluorescence emission and excitation spectra were recorded using a Fluoromax3 (Horiba scientific) on whole C. reinhardtii cells dark-adapted or HL-treated (1,500 μE m−2⋅s−1) as described in the text. GFP protein was added to the sample as internal standard for normalization of fluorescence emission spectra. Additional details on fluorescence spectra acquisition and analysis are reported on SI Appendix.
Time-Resolved Fluorescence.
Time-resolved fluorescence measurements were performed at 77 K, using a Chronos BH ISS Photon Counting instrument with picosecond laser excitation at 447 nm operating at 50 MHz. Laser power was kept below 0.1 μW. Fluorescence decay maps were then globally fitted with exponential functions, as previously reported (34), using Glotaran v.1.5.1 software (41).
SDS/PAGE and Immunoblotting.
SDS/PAGE and immunoblotting were performed as described in ref. 12. LHCSR1- and LHCSR3-specific antibodies (AS142819 and AS142766, respectively) were acquired from Agrisera company.
Supplementary Material
Acknowledgments
The research was supported by the Marie Curie Actions Initial Training Networks “Environmental Acclimation of Photosynthesis” (AccliPhot) (Grant PITN-GA-2012-316427 to R.B.) and “Solar Energy to Biomass - Optimisation of light energy conversion in plants and microalgae” (SE2B) (Grant 675006 SE2B to R.B.) and by the European Research Council Starting Grant “Improving photosynthetic solar energy conversion in microalgal cultures for the production of biofuels and high value products” (SOLENALGAE) (679814 to M.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809812116/-/DCSupplemental.
References
- 1.Mazor Y, Borovikova A, Caspy I, Nelson N. Structure of the plant photosystem I supercomplex at 2.6 Å resolution. Nat Plants. 2017;3:17014. doi: 10.1038/nplants.2017.14. [DOI] [PubMed] [Google Scholar]
- 2.Wei X, et al. Structure of spinach photosystem II-LHCII supercomplex at 3.2 Å resolution. Nature. 2016;534:69–74. doi: 10.1038/nature18020. [DOI] [PubMed] [Google Scholar]
- 3.Niyogi KK. photoprotection revisited: Genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:333–359. doi: 10.1146/annurev.arplant.50.1.333. [DOI] [PubMed] [Google Scholar]
- 4.Rees D, et al. pH dependent chlorophyll fluorescence quenching in spinach thylakoids from light treated or dark adapted leaves. Photosynth Res. 1992;31:11–19. doi: 10.1007/BF00049532. [DOI] [PubMed] [Google Scholar]
- 5.Peers G, et al. An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature. 2009;462:518–521. doi: 10.1038/nature08587. [DOI] [PubMed] [Google Scholar]
- 6.Maruyama S, Tokutsu R, Minagawa J. Transcriptional regulation of the stress-responsive light harvesting complex genes in Chlamydomonas reinhardtii. Plant Cell Physiol. 2014;55:1304–1310. doi: 10.1093/pcp/pcu068. [DOI] [PubMed] [Google Scholar]
- 7.Petroutsos D, et al. A blue-light photoreceptor mediates the feedback regulation of photosynthesis. Nature. 2016;537:563–566. doi: 10.1038/nature19358. [DOI] [PubMed] [Google Scholar]
- 8.Dinc E, et al. LHCSR1 induces a fast and reversible pH-dependent fluorescence quenching in LHCII in Chlamydomonas reinhardtii cells. Proc Natl Acad Sci USA. 2016;113:7673–7678. doi: 10.1073/pnas.1605380113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berteotti S, Ballottari M, Bassi R. Increased biomass productivity in green algae by tuning non-photochemical quenching. Sci Rep. 2016;6:21339. doi: 10.1038/srep21339. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Ware MA, Belgio E, Ruban AV. Comparison of the protective effectiveness of NPQ in Arabidopsis plants deficient in PsbS protein and zeaxanthin. J Exp Bot. 2015;66:1259–1270. doi: 10.1093/jxb/eru477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niyogi KK, Grossman AR, Björkman O. Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell. 1998;10:1121–1134. doi: 10.1105/tpc.10.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonente G, et al. Analysis of LhcSR3, a protein essential for feedback de-excitation in the green alga Chlamydomonas reinhardtii. PLoS Biol. 2011;9:e1000577. doi: 10.1371/journal.pbio.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bellafiore S, Barneche F, Peltier G, Rochaix JD. State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature. 2005;433:892–895. doi: 10.1038/nature03286. [DOI] [PubMed] [Google Scholar]
- 14.Depège N, Bellafiore S, Rochaix JD. Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science. 2003;299:1572–1575. doi: 10.1126/science.1081397. [DOI] [PubMed] [Google Scholar]
- 15.Lemeille S, Turkina MV, Vener AV, Rochaix JD. Stt7-dependent phosphorylation during state transitions in the green alga Chlamydomonas reinhardtii. Mol Cell Proteomics. 2010;9:1281–1295. doi: 10.1074/mcp.M000020-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dumas L, et al. A stromal region of cytochrome b6f subunit IV is involved in the activation of the Stt7 kinase in Chlamydomonas. Proc Natl Acad Sci USA. 2017;114:12063–12068. doi: 10.1073/pnas.1713343114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shapiguzov A, et al. Activation of the Stt7/STN7 kinase through dynamic interactions with the cytochrome b6f complex. Plant Physiol. 2016;171:82–92. doi: 10.1104/pp.15.01893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemeille S, et al. Analysis of the chloroplast protein kinase Stt7 during state transitions. PLoS Biol. 2009;7:e45. doi: 10.1371/journal.pbio.1000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allorent G, et al. A dual strategy to cope with high light in Chlamydomonas reinhardtii. Plant Cell. 2013;25:545–557. doi: 10.1105/tpc.112.108274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bergner SV, et al. STATE TRANSITION7-dependent phosphorylation is modulated by changing environmental conditions, and its absence triggers remodeling of photosynthetic protein complexes. Plant Physiol. 2015;168:615–634. doi: 10.1104/pp.15.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roach T, Na CS. LHCSR3 affects de-coupling and re-coupling of LHCII to PSII during state transitions in Chlamydomonas reinhardtii. Sci Rep. 2017;7:43145. doi: 10.1038/srep43145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue H, et al. PHOTOSYSTEM II SUBUNIT R is required for efficient binding of LIGHT-HARVESTING COMPLEX STRESS-RELATED PROTEIN3 to photosystem II-light-harvesting supercomplexes in Chlamydomonas reinhardtii. Plant Physiol. 2015;167:1566–1578. doi: 10.1104/pp.15.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tokutsu R, Minagawa J. Energy-dissipative supercomplex of photosystem II associated with LHCSR3 in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2013;110:10016–10021. doi: 10.1073/pnas.1222606110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Semchonok DA, et al. Interaction between the photoprotective protein LHCSR3 and C2S2 photosystem II supercomplex in Chlamydomonas reinhardtii. Biochim Biophys Acta Bioenerg. 2017;1858:379–385. doi: 10.1016/j.bbabio.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 25.Pinnola A, et al. Light-harvesting complex stress-related proteins catalyze excess energy dissipation in both photosystems of Physcomitrella patens. Plant Cell. 2015;27:3213–3227. doi: 10.1105/tpc.15.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kosuge K, et al. LHCSR1-dependent fluorescence quenching is mediated by excitation energy transfer from LHCII to photosystem I in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2018;115:3722–3727. doi: 10.1073/pnas.1720574115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niyogi KK, Bjorkman O, Grossman AR. Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell. 1997;9:1369–1380. doi: 10.1105/tpc.9.8.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho F, Govindjee Low-temperature (4-77 degrees K) spectroscopy of Chlorella: Temperature dependence of energy transfer efficiency. Biochim Biophys Acta. 1970;216:139–150. doi: 10.1016/0005-2728(70)90166-0. [DOI] [PubMed] [Google Scholar]
- 29.Lee H, Bingham SE, Webber AN. Function of 3′ non-coding sequences and stop codon usage in expression of the chloroplast psaB gene in Chlamydomonas reinhardtii. Plant Mol Biol. 1996;31:337–354. doi: 10.1007/BF00021794. [DOI] [PubMed] [Google Scholar]
- 30.Erickson JM, et al. Lack of the D2 protein in a Chlamydomonas reinhardtii psbD mutant affects photosystem II stability and D1 expression. EMBO J. 1986;5:1745–1754. doi: 10.1002/j.1460-2075.1986.tb04422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka A, et al. Chlorophyll a oxygenase (CAO) is involved in chlorophyll b formation from chlorophyll a. Proc Natl Acad Sci USA. 1998;95:12719–12723. doi: 10.1073/pnas.95.21.12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finazzi G, Rappaport F. In vivo characterization of the electrochemical proton gradient generated in darkness in green algae and its kinetic effects on cytochrome b6f turnover. Biochemistry. 1998;37:9999–10005. doi: 10.1021/bi980320j. [DOI] [PubMed] [Google Scholar]
- 33.Ballottari M, et al. Identification of pH-sensing sites in the light harvesting complex stress-related 3 protein essential for triggering non-photochemical quenching in Chlamydomonas reinhardtii. J Biol Chem. 2016;291:7334–7346. doi: 10.1074/jbc.M115.704601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Stokkum IH, Larsen DS, van Grondelle R. Global and target analysis of time-resolved spectra. Biochim Biophys Acta. 2004;1657:82–104. doi: 10.1016/j.bbabio.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 35.Chukhutsina VU, Büchel C, van Amerongen H. Disentangling two non-photochemical quenching processes in Cyclotella meneghiniana by spectrally-resolved picosecond fluorescence at 77K. Biochim Biophys Acta. 2014;1837:899–907. doi: 10.1016/j.bbabio.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 36.Wlodarczyk LM, Dinc E, Croce R, Dekker JP. Excitation energy transfer in Chlamydomonas reinhardtii deficient in the PSI core or the PSII core under conditions mimicking state transitions. Biochim Biophys Acta. 2016;1857:625–633. doi: 10.1016/j.bbabio.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 37.Wlodarczyk LM, Snellenburg JJ, Dekker JP, Stokkum IHM. Development of fluorescence quenching in Chlamydomonas reinhardtii upon prolonged illumination at 77 K. Photosynth Res. 2018;137:503–513. doi: 10.1007/s11120-018-0534-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinnola A, et al. Functional modulation of LHCSR1 protein from Physcomitrella patens by zeaxanthin binding and low pH. Sci Rep. 2017;7:11158. doi: 10.1038/s41598-017-11101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Correa-Galvis V, et al. Photosystem II subunit PsbS is involved in the induction of LHCSR protein-dependent energy dissipation in Chlamydomonas reinhardtii. J Biol Chem. 2016;291:17478–17487. doi: 10.1074/jbc.M116.737312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tibiletti T, Auroy P, Peltier G, Caffarri S. Chlamydomonas reinhardtii PsbS protein is functional and accumulates rapidly and transiently under high light. Plant Physiol. 2016;171:2717–2730. doi: 10.1104/pp.16.00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snellenburg J, Laptenok S, Seger R, Mullen K, van Stokkum I. Glotaran: A Java-based graphical user interface for the R package TIMP. J Stat Softw. 2012;49:1–22. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.